Abstract
Posttranscriptional RNA silencing of many endogenous transcripts, viruses, and transgenes involves the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 (RDR6/DCL4)-dependent short interfering RNA (siRNA) biogenesis pathway. Arabidopsis thaliana contains several families of trans-acting siRNAs (tasiRNAs) that form in 21-nucleotide phased arrays through the RDR6/DCL4-dependent pathway and that negatively regulate target transcripts. Using deep sequencing technology and computational approaches, the phasing patterns of known tasiRNAs and tasiRNA-like loci from across the Arabidopsis genome were analyzed in wild-type plants and silencing-defective mutants. Several gene transcripts were found to be routed through the RDR6/DCL4-dependent pathway after initial targeting by one or multiple miRNAs or tasiRNAs, the most conspicuous example of which was an expanding clade of genes encoding pentatricopeptide repeat (PPR) proteins. Interestingly, phylogenetic analysis using Populus trichocarpa revealed evidence for small RNA–mediated regulatory mechanisms within a similarly expanded group of PPR genes. We suggest that posttranscriptional silencing mechanisms operate on an evolutionary scale to buffer the effects of rapidly expanding gene families.
INTRODUCTION
RNA-based silencing in eukaryotes is involved in inactivation or repression of a variety of endogenous and exogenous genetic elements, including genes, transposons and retroelements, viruses, and transgenes (Baulcombe, 2004; Mello and Conte, 2004; Martienssen et al., 2005). The core silencing mechanism involves a double-stranded RNA (dsRNA) trigger formed by any of several mechanisms, including bidirectional transcription of DNA, self-complementary RNA foldbacks, or RNA-dependent RNA transcription. In canonical RNA silencing, small RNAs of ∼21 to 24 nucleotides in either of two biogenesis classes—short interfering RNA (siRNA) from perfectly complementary dsRNA or microRNA (miRNA) from imperfect foldbacks—are formed, although recently discovered classes of small RNAs (PIWI-interacting RNAs and 21U-RNAs) with novel characteristics and biogenesis mechanisms have been identified (Aravin et al., 2006; Girard et al., 2006; Grivna et al., 2006a, 2006b; Kim, 2006; Lau et al., 2006; Ruby et al., 2006). siRNAs and miRNAs arise by the activities of RNase III–type activities of DICER or DICER-LIKE (DCL) enzymes in complexes that catalyze formation of small RNA duplexes (Henderson et al., 2006; Vazquez, 2006). Generally, one strand of the duplex is incorporated into RNase H–like ARGONAUTE (AGO)-containing effector complex, conferring sequence-specific guide functions to the AGO protein (Tomari and Zamore, 2005; Tolia and Joshua-Tor, 2007).
Plants evolved several classes of siRNAs that form by distinct mechanisms and function in separate effector pathways (Brodersen and Voinnet, 2006; Vaucheret, 2006). The most abundant siRNA class (24 nucleotides) arises by RNA-DEPENDENT RNA POLYMERASE2 (RDR2)/DCL3/PolIV activities and functions through AGO4 to initiate or maintain transcriptional silencing through DNA methylation and histone modifications (Zilberman et al., 2003, 2004; Chan et al., 2004; Xie et al., 2004; Herr et al., 2005; Kanno et al., 2005; Onodera et al., 2005; Pontier et al., 2005; Tran et al., 2005). This pathway is associated with transposons, retroelements, and other repeat classes (Xie et al., 2004; Lu et al., 2006; Rajagopalan et al., 2006; Kasschau et al., 2007). Other siRNA classes function at the posttranscriptional level. Silencing of viral RNA and transgene transcripts involves RDR6/DCL4-dependent siRNAs (21 nucleotides) and, for at least some viruses, DCL2-dependent siRNAs (22 nucleotides) (Waterhouse et al., 2001; Xie et al., 2004; Bouche et al., 2006; Deleris et al., 2006; Fusaro et al., 2006; Waterhouse and Fusaro, 2006; Mourrain et al., 2007). Dual siRNA biogenesis pathways may be an important adaptation during antiviral defense due to the ability of viruses to suppress individual pathways (Deleris et al., 2006). The RDR6/DCL4 pathway also catalyzes formation of trans-acting siRNAs (tasiRNAs), which are endogenous regulators of several mRNAs, as discussed below (Peragine et al., 2004; Vazquez et al., 2004b; Allen et al., 2005; Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005). In addition, natural antisense transcript-derived siRNAs are formed through induced, bidirectionally oriented transcript pairs, but through biogenesis mechanisms that are not yet clear (Borsani et al., 2005; Wang et al., 2005; Katiyar-Agarwal et al., 2006; Munroe and Zhu, 2006).
The tasiRNAs form through a mechanism that yields 21-nucleotide, phased siRNAs from TAS transcripts that are initially processed by miRNA-guided cleavage (reviewed in Vaucheret, 2005; Willmann and Poethig, 2005). Specifically, miR173 (TAS1 and TAS2), miR390 (TAS3), and miR828 (TAS4) function as processing guides on primary transcripts to yield defined 5′ or 3′ ends on precursor RNAs, respectively (Allen et al., 2005; Yoshikawa et al., 2005; Rajagopalan et al., 2006). SUPPRESSOR OF GENE SILENCING3 (SGS3) may stabilize the precursor after miRNA-guided cleavage and facilitate dsRNA formation catalyzed by RDR6 (Mourrain et al., 2000; Yoshikawa et al., 2005). The dsRNA precursor is then processed in an end-dependent manner by DCL4, which yields duplexes containing 21-nucleotide siRNAs with 2-nucleotide overhangs at each 3′ end (Dunoyer et al., 2005; Gasciolli et al., 2005; Xie et al., 2005). Functional tasiRNAs with complementarity to target transcripts arise from multiple processing cycle positions, depending on the locus (Vazquez et al., 2004b; Allen et al., 2005; Yoshikawa et al., 2005).
Biological functions have been assigned to the TAS3 family tasiRNAs, which downregulate mRNAs encoding several AUXIN RESPONSE FACTORS, including ARF3 (ETTIN) and ARF4 (Allen et al., 2005; Williams et al., 2005). TAS3-mediated regulation of ARF3 and ARF4 mRNAs is required for proper timing of vegetative shoot development and establishment of leaf polarity (Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Hunter et al., 2006). Formation, stability, or activity of TAS3 tasiRNAs requires AGO7 (ZIPPY), which was originally identified genetically due to its role in vegetative phase change (Hunter et al., 2003; Adenot et al., 2006; Fahlgren et al., 2006). miR390-guided TAS3-like loci evolved in ancient land plants as revealed by comparative phylogenetic analyses (Allen et al., 2005; Axtell et al., 2006; Talmor-Neiman et al., 2006). Biological functions for the nonconserved TAS1 and TAS2 tasiRNAs are not yet clear. These tasiRNAs target mRNAs encoding several members of the pentatricopeptide repeat protein (PPR) family as well as mRNAs encoding proteins of unknown function (Peragine et al., 2004; Vazquez et al., 2004b; Allen et al., 2005; Yoshikawa et al., 2005).
In this article, the phasing patterns of tasiRNAs identified from sequenced libraries were analyzed. These libraries were generated by highly parallel sequencing methods using wild-type plants and RNA silencing-defective mutants (Fahlgren et al., 2006; Kasschau et al., 2007; http://asrp.cgrb.oregonstate.edu/db/). Genome-wide scans for small RNAs resembling tasiRNAs revealed several new RDR6-dependent loci that yield phased siRNAs. Many of these loci correspond to protein-coding genes, including many members of the large PPR family, with transcripts targeted by miRNAs and other tasiRNAs.
RESULTS
Analysis of TAS1, TAS2, and TAS3 Families
A detailed analysis of small RNAs from known TAS loci was done using populations of sequenced small RNAs from wild-type (Col-0), dcl mutant (dcl1-7, dcl2-1, dcl3-1, and dcl4-2), and rdr mutant (rdr1-1, rdr2-1, and rdr6-15) plants (Fahlgren et al., 2007; Kasschau et al., 2007; http://asrp.cgrb.oregonstate.edu/db/). tasiRNAs from TAS1, TAS2, and TAS3 families were represented by at least 836 reads (Table 1), and all were RDR6 dependent (Table 2). Only two reads were obtained for the TAS4 locus. Small RNAs from TAS loci were predominantly 21 nucleotides in length, and this size class was greatly reduced in the dcl4-2 mutant (all values normalized for sequence population size; Table 2). Additionally and as shown previously, tasiRNAs in these sequenced populations were dependent on DCL1 (see Supplemental Table 1 online; Kasschau et al., 2007).
Table 1.
Arabidopsis TAS Families
| TAS Familya | No. Loci/Family | Conservedb | No. Readsc | Target Family | Target Function | Target Validationd |
|---|---|---|---|---|---|---|
| TAS1 | 3 | N | 1739 | Unclassified | Unknown | Y |
| PPR | RNA binding | Y | ||||
| TAS2 | 1 | N | 3570 | PPR | RNA binding | Y |
| TAS3 | 3 | Y | 836 | ARF | Transcription factor | Y |
| TAS4 | 1 | NC | 2 | MYB | Transcription factor | Y |
Peragine et al. (2004), Vazquez et al. (2004b), Allen et al. (2005), and Rajagopalan et al. (2006).
Conserved between Arabidopsis and Populus. NC, not clear (Rajagopalan et al., 2006).
Total reads from each TAS locus from all libraries in the ASRP database (http://asrp.cgrb.oregonstate.edu/db/).
Reviewed by Vaucheret (2006).
Table 2.
Arabidopsis Phased Small RNA–Generating Loci
| Gene | Gene/Gene Family | 21-nt Small RNA Readsa
|
21-nt/Non-21-nt Small RNAa
|
miRNA Phase Initiatorb | miRNA-Initiated Phasing (Peak P Score)c | tasiRNA Phase Initiatorb | tasiRNA-Initiated Phasing (Peak P Score)c | ||
|---|---|---|---|---|---|---|---|---|---|
| Col-0 and rdr2 | rdr6 | Col-0 | dcl4 | ||||||
| tasiRNA loci | |||||||||
| At2g27400 | TAS1a | 384 | 0 | 347/60 | 1/7 | miR173 | Y (33.1) | ||
| At1g50055 | TAS1b | 217 | 0 | 176/17 | 6/18 | miR173 | Y (25.4) | ||
| At2g39675 | TAS1c | 377 | 0 | 333/99 | 2/23 | miR173 | Y (22.2) | ||
| At2g39681 | TAS2 | 855 | 0 | 698/192 | 18/98 | miR173 | Y (31.8) | ||
| At3g17185 | TAS3a | 380 | 0 | 283/78 | 31/23 | 5′ miR390 | N | TAS3a 3′D2(−) | Y (3.25) |
| 3′ miR390 | Y (7.7) | ||||||||
| At5g49615 | TAS3b | 36 | 0 | 14/4 | 1/8 | 5′ miR390 | N | ||
| 3′ miR390 | Y (2.2) | ||||||||
| Other phased small RNA–generating loci | |||||||||
| At1g62910 | PPR | 290 | 0 | 243/65 | 0/1 | 5′ miR161.1 | Y (7.4) | 5′ TAS2 3′D6(−) | Y (18.2) |
| 5′ miR161.2 | Y (2.2) | 5′ TAS2 3′D11(−) | N | ||||||
| 5′ miR400 | N | ||||||||
| 3′ miR161.1 | Y (2.3) | 3′ TAS2 3′D6(−) | Y (5.3) | ||||||
| 3′ miR161.2 | Y (1.8) | 3′ TAS2 3′D11(−) | N | ||||||
| 3′ miR400 | N | ||||||||
| At1g63130 | PPR | 231 | 0 | 201/45 | 0/1 | miR161.1 | Y (7.7) | TAS2 3′D6(−) | Y (17.7) |
| miR161.2 | Y (4.8) | TAS2 3′D9(−) | N | ||||||
| miR400 | N | TAS2 3′D11(−) | N | ||||||
| At1g62930 | PPR | 153 | 0 | 125/40 | 0/1 | miR161.1 | Y (2.0) | TAS1a 3′D9(−) | N |
| miR161.2 | Y (5.5) | TAS2 3′D11(−) | N | ||||||
| miR400 | N | ||||||||
| At1g63080 | PPR | 138 | 0 | 125/30 | 0/0 | miR161.1 | Y (2.3) | TAS2 3′D6(−) | Y (9.9) |
| miR161.2 | Y (2.2) | TAS2 3′D9(−) | N | ||||||
| miR400 | N | TAS2 3′D11(−) | N | ||||||
| At1g63400 | PPR | 87 | 0 | 74/26 | 0/1 | miR161.1 | Y (1.9) | TAS2 3′D6(−) | N |
| miR161.2 | N | TAS2 3′D11(−) | N | ||||||
| miR400 | N | ||||||||
| At1g63150 | PPR | 96 | 0 | 76/15 | 0/0 | miR161.1 | N | TAS2 3′D6(−) | Y (17.2) |
| miR161.2 | Y (5.8) | TAS2 3′D11(−) | N | ||||||
| miR400 | N | ||||||||
| At1g63070 | PPR | 68 | 0 | 55/12 | 0/0 | miR161.2 | N | TAS2 3′D6(−) | Y (9.9) |
| miR400 | N | TAS2 3′D9(−) | N | ||||||
| TAS2 3′D11(−) | N | ||||||||
| At1g63330 | PPR | 37 | 0 | 31/16 | 0/0 | miR161.2 | N | TAS2 3′D6(−) | N |
| miR400 | N | TAS1a 3′D9(−) | N | ||||||
| TAS2 3′D11(−) | N | ||||||||
| At1g62590 | PPR | 33 | 0 | 26/10 | 0/0 | miR161.2 | N | TAS2 3′D6(−) | Y (2.6) |
| miR400 | N | TAS1a 3′D9(−) | N | ||||||
| TAS2 3′D11(−) | N | ||||||||
| At5g38850 | TIR-NBS-LRR | 81 | 0 | 75/11 | 0/1 | NId | N (4.4) | ||
| At1g12820 | AFB3 | 10 | 0 | 7/0 | 0/1 | miR393 | Y (1.4)e | ||
| At5g41610 | ATCHX18 | 23 | 0 | 6/4 | 1/3 | miR856 | Y (1.6) | ||
| miR780 | N | ||||||||
Raw small RNA reads were not normalized for repeats or library size. nt, nucleotide.
miRNAs or tasiRNAs that were experimentally validated to guide cleavage of the target are indicated in bold.
Y indicates that at least three small RNAs within eight 21-nucleotide cycles are in phase with the target site. The peak score is based on 20- to 25-nucleotide total small RNA reads from Col-0 and rdr2-1 libraries.
NI, not identified.
Phasing is one nucleotide offset from the miR393-guided cleavage site.
The fact that validated tasiRNAs, or tasiRNAs with high-quality predicted targets, arise from the TAS locus positions residing up to 12 processing cycles away from the initiation cleavage sites (Figure 1) points to the importance of accurate phasing to generate small RNAs with sufficient complementarity to target sequences. Therefore, the phasing patterns of small RNAs from the miR173-guided cleavage site (TAS1 and TAS2 loci) and 3′ miR390-guided cleavage site (TAS3 loci) were analyzed. Small RNA sequence data from wild-type and rdr2-1 mutant (no effect on tasiRNA biogenesis) plants were used to develop an algorithm that converted phase position and amplitude (small RNA abundance) data into phase signals. First, a sense-antisense small RNA consolidation step was developed to unify sequence data from both strands. Small RNA pairs from sense-antisense positions that were offset by two nucleotides, with the overhang at the 3′ ends, were combined. This accounted for the possibility that either sense or antisense small RNA from a given small RNA duplex might accumulate. Second, a phase score, P, was calculated using the following formula:
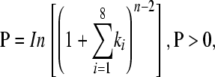 |
in which n = number of phase cycle positions occupied by at least one small RNA read within an eight-cycle window, and k = the total number of reads for all small RNAs with consolidated start coordinates in a given phase within an eight-cycle window. Phase cycle length was set at 21 nucleotides. A positive phase signal, therefore, was limited by the power function (n − 2) to those loci with small RNAs occupying at least three cycle positions in a phased, 21-nucleotide register. Requiring occupation of three positions in an eight-cycle window limited the possibility of a positive phase score by chance. It should also be noted that the phase score amplitude is influenced by both small RNA abundance and number of positions occupied. This calculation was applied in a scrolling window format with single nucleotide advances, and scores were assigned to the fourth cycle position in phasing plots (Figure 1).
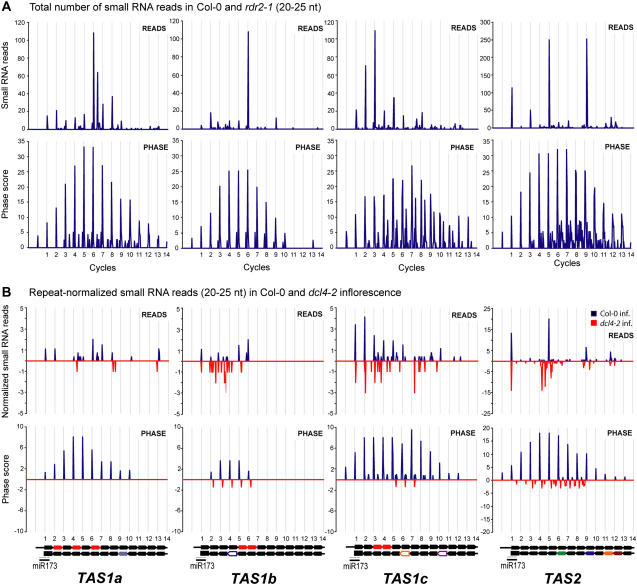
Figure 1.
Distribution and Abundance of 21-Nucleotide Small RNAs in TAS1a, TAS1b, TAS1c, and TAS2.
(A) Abundance of Col-0 and rdr2-1 small RNA reads in TAS1a, TAS1b, TAS1c, and TAS2 loci. The top graphs represent the total number of small RNA reads, whereas the bottom graphs shows phase signals from Col-0 and rdr2-1 small RNA reads. Gridlines correspond to 21-nucleotide cycles of small RNAs from the miR173-guided cleavage site. nt, nucleotides.
(B) Comparison of Col-0 and dcl4-2 inflorescence small RNAs. The top graphs represent 20- to 25-nucleotide small RNAs in Col-0 inflorescence versus 20- to 25-nucleotide small RNAs in dcl4-2 inflorescence for TAS1a, TAS1b, TAS1c, and TAS2 loci. The bottom graphs show phase signals from Col-0 inflorescence small RNA reads versus dcl4-2 inflorescence small RNA reads. Small RNA counts were normalized to the size of the dcl4-2 library and also repeat normalized to account for redundant small RNAs in each locus. The schematics below the phase plots represent miR173-guided cleavage and 21-nucleotide small RNA positions in TAS1a, TAS1b, TAS1c, and TAS2 transcripts. The position of the miR173 target site is shown as a thin gray line. Thick black arrows represent 21-nucleotide phase positions from the miR173-guided cleavage site. Colored arrows indicate small RNAs previously validated to target various transcripts. White arrows with colored outlines indicate small RNAs with a predicted target. Arrows with identical colors (solid and outlined) indicate small RNAs within the same family. Red arrows indicate small RNA positions for the ASRP255-like family.
Small RNAs were detected unidirectionally downstream of the miR173-guided cleavage site from each TAS1 and TAS2 locus (Figure 1A; http://asrp.cgrb.oregonstate.edu/db/). Phase plots clearly revealed the dominant phasing event to be coincident with the miR173 target site for each locus, although both the raw read and phase transform plots revealed out-of-phase noise at each locus. Phase drift was detected at TAS1c, with small RNAs accumulating in the phase position one nucleotide forward from the initial miR173-directed phase, starting at cycle five (Figure 1A). Between cycles 1 and 5, the phase-forward:initial phase ratio of small RNA reads at TAS1c was 1:5; however, between cycles 6 and 10, the phase-forward:initial phase ratio at TAS1c was 22:1. For TAS3a, small RNAs were detected exclusively upstream of the 3′ miR390 target site (Figure 2A). The miR390-initiated phase signal, however, was slightly lower than an out-of-phase signal, which likely originated from a secondary targeting event guided by an siRNA from the 5′D2(−) position (Figure 2B; Allen et al., 2005; Axtell et al., 2006). Accurately phased, miR390-initiated RNAs were detected close to the 3′ target site at TAS3a, but phase-forward drift was detected after several cycles (Figure 2). Between cycles 1 and 6, the phase-forward:initial phase ratio of small RNA reads at TAS3a was 1:3. However, the phase-forward:initial phase ratio was 32:1 at cycles 7 and 8. This may be an adapted feature of TAS3a, as the functional tasiRNAs occur in the forward phase at the 5′D7(+) and 5′D8(+) positions (positional nomenclature from Allen et al. [2005]).
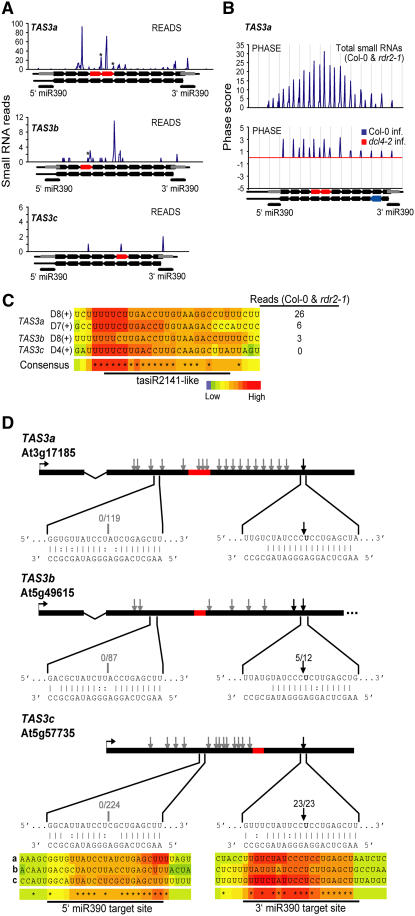
Figure 2.
Identification of New Loci and miR390 Target Sites in the TAS3 Family.
(A) The total number of small RNA reads in Col-0 and rdr2-1 libraries for TAS3a and TAS3b and all libraries for TAS3c are shown. The 21-nucleotide cycle positions from the 3′ miR390-guided cleavage site for each locus are indicated below each graph. Predicted and validated miR390 target sequences are indicated by gray boxes, and ASRP2141-like sequences are indicated by red arrows. Asterisks indicate ASRP2141-like sequences.
(B) Phase plots for TAS3a tasiRNAs from Col-0 and rdr2-1 (top) and from normalized reads from Col-0 (bottom, above the x axis) or dcl4-2 (bottom, below the x axis). Vertical gridlines indicate 21-nucleotide cycles.
(C) T-Coffee alignment of ASRP2141-like sequences from TAS3a, TAS3b, and TAS3c. Identical positions are indicated by asterisks in the consensus plot. The numbers of ASRP2141-like tasiRNA reads in Col-0 and rdr2-1 libraries are listed.
(D) Diagrammatic representation of primary TAS3 transcripts and validation of 3′ miR390 cleavage sites by 5′ RACE. ASRP2141-like regions are highlighted in red. The miR390 and target mRNA duplexes are shown in the expanded regions. Bases in bold indicate predicted cleavage sites. Arrowheads indicate positions corresponding to the 5′ end of at least one cloned 5′ RACE PCR product. The 5′ RACE products sequenced for each miR390 target site are indicated in gray and black, respectively. The number of specific 5′ RACE products/total products sequenced at each miR390 target site is indicated. The 3′ miR390 target site for TAS3a was reported by Allen et al. (2005). T-Coffee alignment of 5′ and 3′ miR390 target sites in TAS3a (a), TAS3b (b), and TAS3c (c). Identical positions are indicated by asterisks in the consensus plot.
The basis for phase-forward drift at TAS3a and TAS1c could be due, at least in part, to non-21-nucleotide siRNAs resulting from misprocessing by DCL4 or alternate DCLs. Alternate-size small RNAs were detected at relatively low levels for most TAS loci, although an abundant 22-nucleotide RNA from TAS1c was detected at the fifth processing cycle from the miR173 target site (http://asrp.cgrb.oregonstate.edu/db/). Loss of DCL4 in the dcl4-2 mutant resulted in off-sized RNAs at each TAS locus (Table 2, Figure 1B). Although the abundance of total small RNAs arising from TAS1, TAS2, and TAS3 loci was only modestly affected, the ratio of 21-nucleotide to non-21-nucleotide RNAs was considerably lower in the mutant (Table 2). Importantly, this was accompanied by loss of miRNA-guided phasing (Figures 1B and 2B), and loss of functional tasiRNAs, at each locus. Therefore, although DCL4 is not required to process dsRNA formed from each TAS locus, it is clearly necessary for biogenesis of active, properly phased tasiRNAs.
A search for previously unrecognized members of TAS1, TAS2, and TAS3 families revealed two additional TAS3 family members (TAS3b and TAS3c), which contained sequences similar to ARF3- and ARF4-targeting tasiR2141 from the originally characterized TAS3a locus (Figure 2C). TAS3b was previously identified as a tasiRNA-like region by Lu et al. (2006). Unlike TAS3a, both TAS3b and TAS3c contain only a single tasiR2141-like sequence. RDR6-dependent small RNAs of predominantly 21 nucleotides were detected from both TAS3b and TAS3c (Table 2; http://asrp.cgrb.oregonstate.edu/db/), although TAS3c generated relatively few RNAs. Importantly, nucleotide sequence alignments and target validation assays revealed a functional miR390-guided cleavage site on the 3′ side of both TAS3b and TAS3c tasiRNA-generating regions (Figure 2D). The tasiR2141-like sequence was detected in the miR390-initiated phase for TAS3c, but in the +4 forward phase for TAS3b. Transcripts for each TAS3 locus were detected, and 5′ transcription start sites were mapped (see Supplemental Figure 1 online). Therefore, TAS3 tasiRNAs arise from a small multigene family, all members of which have conserved miR390-guided target sites.
Interestingly, in addition to the conserved 3′ miR390 target sites and tasiR2141-related sequences, conserved miR390 target sites on the 5′ side of TAS3a, TAS3b, and TAS3c tasiRNA-generating sequences were detected (Figure 2D). In each case, however, the site deviates from canonical plant miRNA target sites in that mispairs and G:U pairs occur at positions 9 to 11 (relative to the 5′ end of miR390; Figure 2D). Based on mutational and informatic analyses of naturally occurring target sites, mispairs and G:U pairs at these positions were predicted to have restricted miR390-guided cleavage (Jones-Rhoades and Bartel, 2004; Mallory et al., 2004; Allen et al., 2005; Schwab et al., 2005). Indeed, 5′ rapid amplification of cDNA ends (RACE) analysis failed to detect any cleavage products corresponding to an active 5′ miR390 target site, although 5′ RACE products were detected at low frequencies at several sites flanking the noncanonical target site at each locus (Figure 2D). Given the high degree of conservation, we conclude that the site is likely important, but functional in a mode that does not depend on target cleavage. Axtell et al. (2006) also detected the noncanonical 5′ miR390 site in TAS3a and showed that it functions in a noncleavage mode in vivo for TAS3-derived tasiRNA biogenesis and function.
Genome-Wide Scans for tasiRNA-Like Loci
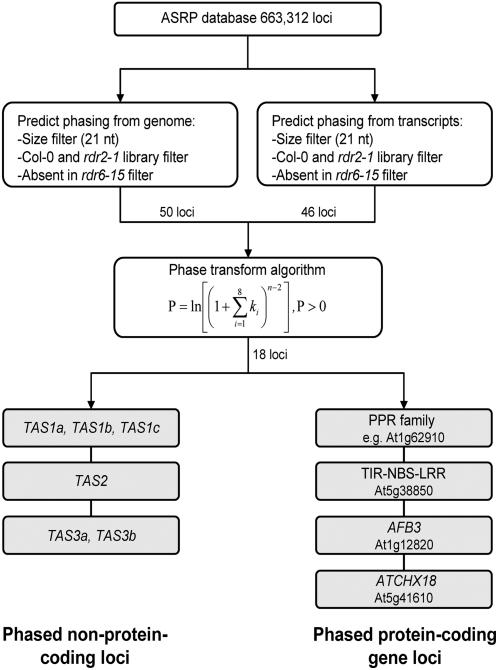
To identify loci with tasiRNA-like features or that spawn RDR6/DCL4-dependent siRNAs, a genome-wide scan using Columbia (Col-0) and mutant small RNA sequence libraries was done. These loci were predicted to yield RDR6-dependent, 21-nucleotide RNAs in phased arrays. A set of filters to identify clusters of 21-nucleotide small RNAs that were present in either Col-0 and/or rdr2-1 mutant libraries, and absent in rdr6-15 mutant libraries, yielded overlapping sets of 50 and 46 loci in scans of genomic and transcript sequences, respectively (Figure 3; see Supplemental Table 1 online). Thirty-six of these loci were known to be targeted by at least one miRNA or tasiRNA (see Supplemental Table 1 online). These included overlapping sets of 30 miRNA targets (including seven known TAS loci) and 18 tasiRNA targets. Twenty of the 36 loci contained multiple miRNA or tasiRNA target sites. Therefore, among the 50 RDR6-dependent small RNA clusters identified, 72% arose from miRNA/tasiRNA-targeted transcripts and 40% from multiply targeted transcripts. Small RNAs (21 nucleotides) from these loci were generally lost, or accumulated to low levels, in both the dcl1-7 and dcl4-2 mutants (see Supplemental Table 1 online), indicating dependence on both miRNA-based and tasiRNA-like mechanisms.
Figure 3.
Flowchart for Identification of RDR6-Dependent, Phased, Small RNA–Generating Loci.
Eighteen loci, including 17 that were targeted by miRNAs/tasiRNAs, contained at least three in-phase (21-nucleotide cycle) positions occupied by at least one small RNA within an eight-cycle window, yielding peak phase scores ranging from 1.4 to 33.1 (Table 2). Except for the known TAS1, TAS2, and TAS3 loci that came through the screen, no other non-protein-coding loci were identified. However, three noncoding loci yielded RDR6-dependent, 21-nucleotide RNAs that were phased using a more permissive algorithm in which signal required occupation of only two out of 12 cycle positions (see Supplemental Table 1 online). Two of these loci (from chromosome 4, between coordinate positions 1472767 and 1476524) were embedded in a region with duplications of ubiquitin-like genes or gene sequences (such as At4g03350; see Supplemental Table 1 online). In fact, both small RNA-generating loci contained extensive, continuous similarity to these genes and appear to have resulted from recent duplication events. The other noncoding locus was identified between opposingly oriented genes At5g27650 and At5g27660 but was represented by too few small RNAs in Col-0 plants to allow a meaningful analysis (see Supplemental Table 1 online). Lu et al. (2006) recently identified two TAS-like loci from noncoding regions (Chr1:25282658-25283382 and Chr4:13295428-13296124). These loci were not identified here because small RNAs from both loci were RDR6 independent (http://asrp.cgrb.oregonstate.edu/db/). The recently identified TAS4 locus (Rajagopalan et al., 2006) was not identified due to a lack of sequenced small RNAs.
Four gene families were represented among the 12 protein-coding loci that emerged from the phase scan (Table 2). These included a Toll/Interleukin1 receptor–nucleotide binding site–leucine-rich repeat (TIR-NBS-LRR) disease resistance gene (At5g38850; Meyers et al., 2003), the TIR1-like AFB3 gene encoding an F-box protein involved in auxin signaling (At1g12820; Dharmasiri et al., 2005), a cation/hydrogen antiporter gene ATCHX18 (At5g41610; Sze et al., 2004), and nine genes encoding closely related PPR proteins (discussed below). Transcripts from AFB3 and ATCHX18 were targeted and phase-initiated by miR393 and miR856, respectively. Interestingly, two miRNAs (miR856 and miR780) were shown to target the ATCHX18 transcript (Fahlgren et al., 2007), but phasing is initiated only at the miR856 site. Although the TIR-NBS-LRR gene generated abundant, highly phased siRNAs, we did not identify a miRNA or tasiRNA that initiates phasing. In addition to the miRNA target transcripts that yielded phased siRNAs, targets for miR168 (AGO1), miR472 (several CC-NBS-LRR genes), miR393 (AFB2), miR773 (MET2-like), and miR172 (AP2) yielded RDR6-dependent siRNAs that failed to generate a positive phase score (see Supplemental Table 1 online). siRNAs from many of these genes were also identified in other analyses (Axtell et al., 2006; Lu et al., 2006; Rajagopalan et al., 2006; Ronemus et al., 2006; see Supplemental Table 1 online).
Targeting of PPR-P Gene Transcripts and Formation of Phased siRNAs
By far, the most abundant phased, 21-nucleotide siRNA populations were identified from two clusters of PPR genes on chromosome 1 (Table 2). The Arabidopsis thaliana genome contains at least 448 PPR-related genes, which encode putative RNA binding proteins with repeats of a 35–amino acid motif (Lurin et al., 2004). Some PPR proteins are known to regulate RNA processing, stability, editing, or translation in chloroplasts or mitochondria (Lurin et al., 2004; Cushing et al., 2005; Kotera et al., 2005; Okuda et al., 2006; Shikanai, 2006). The PPR proteins are classified into subfamilies containing only tandem arrays of PPR motifs (PPR-P) or containing diversified PPR motifs interspersed with non-PPR domains (Aubourg et al., 2000; Lurin et al., 2004; Rivals et al., 2006). A few PPR-P genes from Raphanus sativus (Rfo), Zea mays (Rf-1), and Petunia hybrida (Rf) are known restorers of fertility in lines with cytoplasmic male sterility conferred by certain mitochondrial ATPase alleles (Bentolila et al., 2002; Desloire et al., 2003; Kazama and Toriyama, 2003; Wang et al., 2006; Chase, 2007).
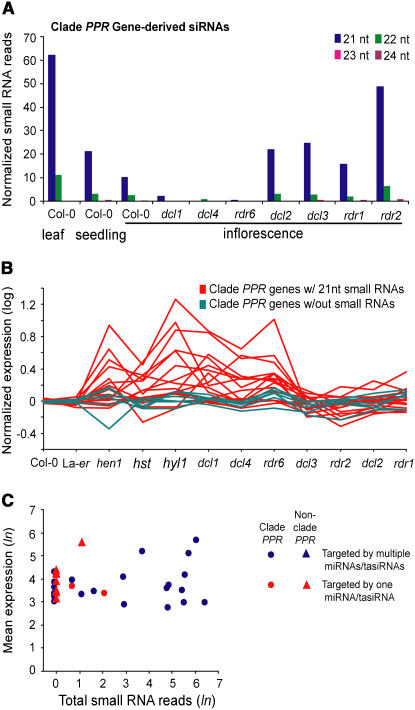
Two regions on chromosome 1 contain 21 PPR-P genes and pseudogenes, of which 17 (81%) spawned primarily 21-nucleotide siRNAs (Figure 4A). These were dependent on RDR6, DCL4, and DCL1 but independent of RDR1, RDR2, DCL2, and DCL3 (Figure 4A, Table 2; see Supplemental Table 1 online). Phylogenetic analysis of the entire PPR-P subfamily revealed that the siRNA-generating loci belonged to a clade of 28 PPR-P genes (hereafter referred to as the PPR-P clade) that has undergone relatively recent expansion (see below). Using expression profiling arrays, the steady state levels of 23 of the 28 PPR-P clade members in inflorescence tissue was measured in each of the dcl and rdr mutants, in hen1, hst, and hyl1 mutants, and in Col-0 and Landsberg erecta (Ler) plants. The levels of transcripts for the siRNA-generating PPR-P genes were generally elevated in the mutants with miRNA or tasiRNA defects (dcl1, dcl4, rdr6, hen1, hst, and hyl1) but not in mutants with defects in other small RNA classes (dcl2, dcl3, rdr1, and rdr2) (Figure 4B). Fifty-three percent of siRNA-generated clade genes were significantly upregulated in at least one mutant involved in miRNA and tasiRNA biogenesis (23-gene set, analysis of variance, false discovery rate = 0.05). These data indicate that a high proportion of genes within the clade not only spawn siRNAs with genetic requirements similar to those of tasiRNAs, but that the PPR-P transcripts are repressed by posttranscriptional silencing mechanisms.
Figure 4.
Analysis of PPR-P Clade Genes and siRNAs.
(A) Abundance of small RNA size classes from clade PPR-P transcripts in various tissues and genotypes.
(B) Expression profile of 23 of the 28 PPR-P clade genes in Col-0, Ler, and 10 mutants with defects in silencing factors. Transcripts that yield at least one 21-nucleotide small RNA (red) or no small RNAs (teal) are color-coded. The list of PPR genes in the expression profile is as follows: At4g26800, At1g63070, At1g62670, At1g63130, At1g62930, At1g63080, At1g63320, At1g63150, At1g62590, At1g62910, At1g63400, At5g41170, At3g16710, At5g16640, At1g62720, At1g62680, At1g64580, At1g06580, At1g12700, At1g12620, At1g64100, At1g62860, and At1g63630.
(C) Comparison of small RNA reads and expression level of PPR-P genes that are targeted by miR161, miR400, and/or tasiRNAs from TAS1 or TAS2 loci.
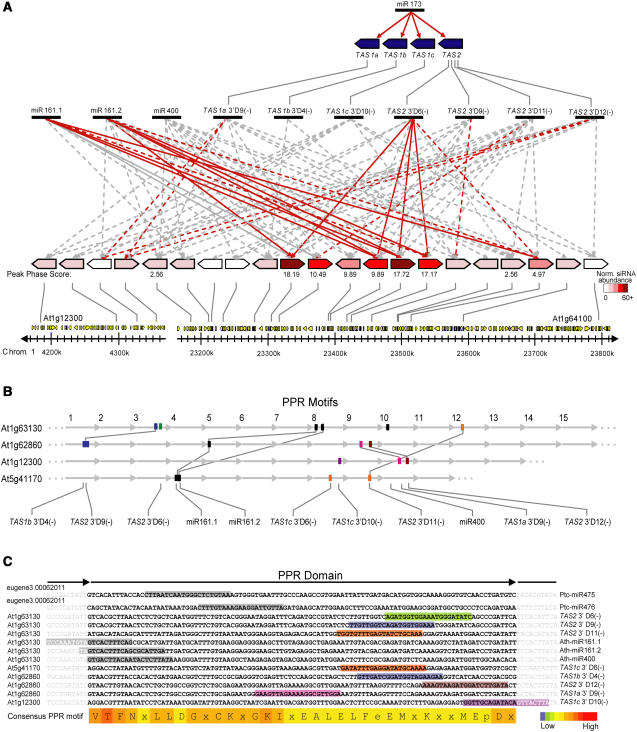
Computational analysis and experimental validation assays revealed 11 Arabidopsis miRNAs and tasiRNAs with potential or validated function to guide cleavage of PPR-P mRNAs (Rhoades et al., 2002; Allen et al., 2004; Sunkar and Zhu, 2004; Yoshikawa et al., 2005). These include both forms of miR161 (miR161.1 and miR161.2), miR400, four tasiRNAs from the TAS1 family, and four tasiRNAs from the TAS2 locus (Table 2; see Supplemental Figure 2 online). Remarkably, all of these small RNAs were predicted or validated to target PPR-P transcripts predominantly from the clade, with most clade members targeted by multiple small RNAs (Figure 5A, Table 2; see Supplemental Table 1 online). Five of these genes were validated here by 5′ RACE as both miR161 and TAS2 3′D6(−) targets (Table 2; see Supplemental Figure 2 online). Combined with previously validated transcript targets (Allen et al., 2004; Yoshikawa et al., 2005), eight PPR-P transcripts were validated experimentally as targets for at least one small RNA. As a group, these miRNAs and tasiRNAs were predicted to target a total of 40 PPR-P transcripts, including all 28 PPR genes in the clade. Although siRNA generation and targeting of clade PPR-P transcripts by miRNAs and tasiRNAs were correlated (see below), it was also possible that the high siRNA levels were a reflection of high accumulation levels of the target transcripts. To determine if PPR gene–derived siRNA levels were correlated with transcript abundance, the mean expression value in wild-type plants (Col-0 inflorescence tissues) and repeat-normalized siRNA reads were compared. There was no significant correlation between siRNA abundance and transcript level (Pearson correlation coefficient = 0.077; Figure 4C), suggesting that siRNA abundance was related to miRNA and/or tasiRNA targeting rather than to transcript level.
Figure 5.
Targeting of the PPR Gene Family by miRNAs and tasiRNAs.
miR161.1, miR161.2, miR400, and tasiRNAs from TAS1 and TAS2 are predicted to target several PPR-P clade transcripts from two gene clusters in chromosome 1.
(A) A schematic representation of PPR targeting by miR161.1, miR161.2, miR400, TAS1a 3′D9(−) (also named TAS1a siR9 by Yoshikawa et al. [2005]), TAS1b 3′D4(−) (siR4), TAS1c 3′D10(−) (siR10), and TAS2 3′D6(−), 3′D9(−), 3′D11(−), and 3′D12(−) (siR6, siR9, siR11, and siR12). Red and gray arrows indicate experimentally validated and predicted-only targets, respectively. Solid arrows indicate targeting events that direct phasing of small RNAs from the PPR-P transcript. The peak P score is listed under each gene with phased small RNAs. PPR genes are color-coded based on repeat normalized abundance of small RNAs from Col-0 and rdr2-1 libraries. The PPR gene clusters are shown relative to genomic position on chromosome 1. From left to right, the PPR-P genes are as follows: At1g12300, At1g12620, At1g12700, At1g12775, At1g62590, At1g62670, At1g62680, At1g62720, At1g62860, At1g62910, At1g62930, At1g63070, At1g63080, At1g63130, At1g63150, At1g63230, At1g63320, At1g63330, At1g63400, At1g63630, and At1g64100.
(B) Schematic representation of PPR motifs within four PPR-P genes and the location of small RNA target sites relative to the four genes illustrated that they collectively contain targets for all 11 PPR-targeting miRNAs and tasiRNAs. Numbers above the diagram indicate the PPR motif. Identical colors for miRNA or tasiRNA target sites represent members of the same family.
(C) PPR domain conservation within four Arabidopsis PPR genes and one Populus PPR gene. PPR domains listed are those that collectively contain target sites for all 11 miRNA or tasiRNA predicted targets from Arabidopsis and two known miRNA target sites from Populus. Each PPR domain consists of 105 nucleotides. Colored boxes indicate the position of the target sites within each PPR gene and correspond to colors in (B). A consensus PPR motif, with conservation highlighted by a heat map, is shown.
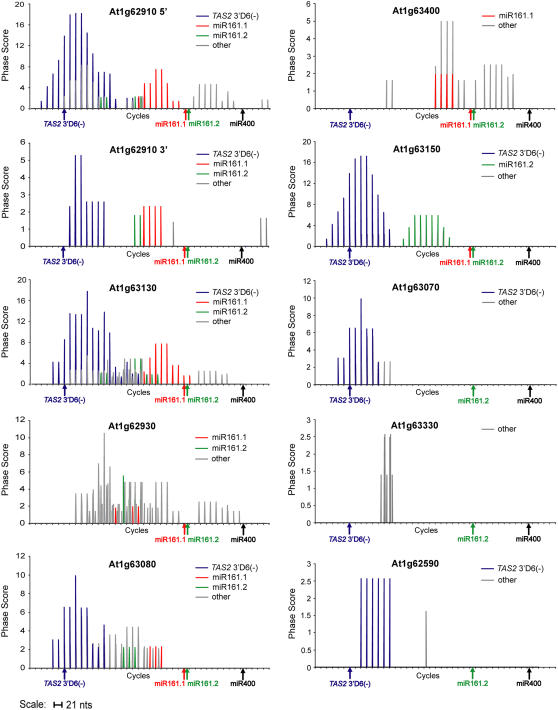
Eight of the nine PPR-P genes identified in the genome-wide phase scan contained phase initiation sites corresponding to miR161.1, miR161.2, or TAS2 3′D6(−) (Table 2, Figure 6). In fact, loci spawning the highest numbers of siRNAs appeared to correspond to genes with transcripts that were dual-targeted by both miR161 forms and TAS2 3′D6(−) (Table 2). In the case of At1g62910, which contains a duplicated coding region resulting in nearly twice as many PPR domains as most other clade proteins, two sets of miR161 and TAS2 3′D6(−) target sites and phased siRNAs were identified (Figure 6). None of the other phase patterns from the PPR-P sequences were coincident with any other predicted or validated small RNA-guided target sites, although target sites downstream of miR400 were not tested in 5′ RACE assays. Curiously, although miR400 was predicted to interact with all but one PPR transcript yielding phased siRNAs, no phased RNAs were detected in the vicinity of the miR400 target sites. In fact, each of five PPR sequences tested were negative in miR400 target validation assays, even though each miR161 and TAS2 3′D6(−) target site tested from the same genes was positive (see Supplemental Figure 2 online). miR400 target validation assays were only done using Col-0 inflorescence tissue, and it is possible that miR400-guided cleavage occurs in other tissues or under different conditions than those tested here.
Figure 6.
Phase Transform Plots for Nine Arabidopsis PPR Genes.
Phased cycles from miR161.1, miR161.2, and TAS2 3′D6(−) target sites are indicated by color coding. Arrows below each plot indicate the miRNA and tasiRNA target sites within each PPR transcript.
The phasing patterns from miR161 and TAS2 3′D6(−) target sites were distinctive. Phased siRNAs from the TAS2 3′D6(−) target site generally occurred at the 3′ side (Figure 6); indeed, nearly all small RNAs (phased and nonphased) from the targeted PPR-P genes occurred at the 3′ side of the TAS2 3′D6(−) target site, much like tasiRNAs that originate from the 3′ side of the miR173-guided cleavage site in TAS1 and TAS2 transcripts (Figure 1). By contrast, nearly all of the phased siRNAs initiated at the miR161.1 and miR161.2 target sites occurred on the 5′ side of the initiation sites (Figure 6), which is reminiscent of tasiRNAs originating from the 5′ side of the miR390-guided cleavage site at TAS3 loci (Figure 2).
The distribution of predicted or validated target sites for the 11 miRNAs or tasiRNAs within the Arabidopsis PPR-P clade sequences was analyzed. The miR161.1/miR161.2 and miR400 target sites were separated by several hundred nucleotides in their common target transcripts (Figure 5B). However, in alignments of individual PPR domain–coding regions, it was clear that miR161 and miR400 target sequences span domain-homologous sequences (Figure 5C). In alignments of miR161 and miR400 foldback sequences, a modest similarity was detected (ClustalW score = 67), raising the possibility that their respective MIRNA genes share a common origin. The TAS1 and TAS2 tasiRNAs targeted sequences within PPR domain 3′ coding sequences, which were generally more diverse than the domain 5′ regions (Figure 5C). Although some target sequences overlap domain-homologous regions, the tasiRNA target sites represent positions across 65 nucleotides of domain sequence space (Figure 5C). This comparison suggests that targeting of PPR transcripts by miRNAs and tasiRNAs likely originated independently on multiple occasions. Interestingly, two Populus miRNAs (miR475 and miR476) also have target sites within PPR protein–coding transcripts (Lu et al., 2005). These miRNAs target PPR domain–coding sequences that are spatially distinct from the Arabidopsis small RNA target sites, again suggesting independent origination of PPR transcript targeting by distinct small RNAs.
Secondary Targeting of PPR-P Gene Transcripts
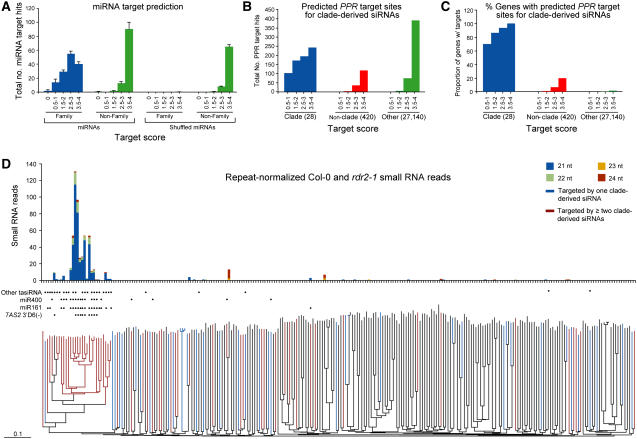
Prolific generation and amplification of RDR6/DCL4-dependent siRNAs after initial targeting by miRNAs and tasiRNAs could potentially reinforce silencing of PPR-P clade transcripts. They could also potentially trigger a cascade of reactions in which silencing spreads through the PPR gene family. To assess the potential of secondary targeting by siRNAs from the 28-member PPR-P clade, genome-wide targets were computationally predicted for antisense 21-nucleotide small RNAs arising from clade sequences. Targets were predicted based on the same methods used to predict miRNA targets (Figure 7A; Allen et al., 2005; Fahlgren et al., 2007). Secondary siRNA targets were grouped by target score and by their identity as members of the clade, nonclade PPR-P genes, or non-PPR genes. Hits resulting from perfect complementarity between siRNA and PPR gene transcripts (target score = 0) were eliminated, as these could not be assessed in terms of potential targeting in trans. Clade PPR-derived siRNAs were more likely to target members of the clade, regardless of their predicted target score (Figures 7B and 7C). The number of nonclade PPR and non-PPR hits by clade-derived siRNAs increased at higher target score levels (2.5 to 4.0; Figure 7B), as expected for increasing the allowance for mispairs and G:U pairs. This was similar to that observed for miRNA target predictions, where off-target hits were detected at higher scores using both authentic miRNAs and sequence-shuffled miRNAs (Figure 7A). The percentage of non-clade PPR hits, however, was much greater than the percentage of non-PPR hits (Figure 7C), suggesting a greater likelihood of clade-derived siRNAs interacting with other PPR transcripts rather than non-PPR transcripts. Whereas all PPR-P clade members, except for one, were potential targets of multiple clade-derived siRNAs, only 72 of the 255 PPR-P subfamily members were predicted as targets of at least one siRNA. Thirty-three nonclade PPR-P transcripts were predicted targets for two or more clade-derived siRNAs (Figure 7D). Most of these had predicted target scores of 3.5 to 4.0 (Figures 7B and 7C).
Figure 7.
Primary and Secondary Targeting of PPR-P Transcripts.
(A) miRNA target prediction for randomly selected miRNA family members (one/family). Target predictions were done with 10 sets of either miRNAs or sequence-shuffled RNAs, and the mean (±sd) target score distribution was plotted. miRNA target hits for a validated gene family (blue) or a nonvalidated gene family (green) are grouped according to target score. miRNA families used for target prediction were miR156/157, miR158, miR159/319, miR160, miR161, miR162, miR163, miR164, miR166/165, miR167, miR168, miR169, miR171/170, miR172, miR173, miR390/391, miR393, miR394, miR395, miR396, miR397, miR398, miR399, miR400, miR402, miR403, miR408, miR447, miR773, miR775, miR824, miR827, miR842, miR844, miR856, miR857, and miR858.
(B) Secondary siRNA target prediction for all antisense 21-nucleotide small RNAs arising from all clade PPR-P transcripts. Clade-derived siRNAs predicted to target the clade PPR-Ps (blue), nonclade PPRs (red), or non-PPRs (green) were plotted by target score. The total number of genes in each group is listed in parenthesis.
(C) The percentage of clade PPR-P (blue), nonclade PPR (red), and non-PPR (green) genes that have transcripts targeted by clade-derived siRNAs. Target hits in (A) to (C) were grouped according to target score.
(D) Repeat normalized abundance of small RNAs with identity to each PPR-P transcript. Dots indicate PPR-P transcripts with predicted target sites for miR161, miR400, TAS2 3′D6(−), or other tasiRNAs. Transcripts that are singly (blue) or multiply (red) targeted by one or more clade-derived siRNAs are color-coded within the phylogenetic tree. PPR amino acid sequences were aligned using ClustalW with the BLOSUM matrix series. A tree that only differed in the placement of PPR proteins outside of the clade was obtained using the GONNET matrix series (data not shown).
Does targeting of siRNAs from clade transcripts lead to amplification of secondary siRNAs from nonclade members? In total, 25 nonclade PPR-P loci (11%) yielded at least one siRNA. This is actually below the genome-wide level of protein-coding genes with identity to at least one small RNA (18.2%; Kasschau et al., 2007). Among the 72 and 33 nonclade PPR-P transcripts that were plausible predicted targets of at least one or two clade-derived siRNAs, respectively, only nine (12.5%) and six (18.2%) yielded siRNAs. Neither of these percentages were significantly different (P > 0.27) from the genome-wide percentage. Additionally, four of these nonclade PPR-P small RNA-generating loci (At1g43980, At4g17910, At3g60960, and At5g27300) overlapped with transposons or retroelements and produced predominantly 23- to 24-nucleotide small RNAs (Figure 7D), a feature associated with RDR2/DCL3-dependent siRNAs. Furthermore, there was no correlation between siRNA generation from nonclade PPR-P loci and predicted targeting by any of miR161, miR400, or TAS1 or TAS2 tasiRNAs (Figure 7D; data not shown). These results argue against secondary targeting by clade-derived siRNAs on nonclade target transcripts.
DISCUSSION
TAS Loci and siRNA Phasing
The tasiRNA biogenesis mechanism employs a novel use for miRNAs as site-specific RNA processing factors. Functional tasiRNAs must be in (or very close to) a 21-nucleotide register with the miRNA-guided cleavage site, otherwise the resulting siRNAs would have limited targeting potential (Vazquez et al., 2004b; Allen et al., 2005; Yoshikawa et al., 2005). Importantly, correct phasing also depends on DCL4. Although small RNAs form in the absence of DCL4, due to alternate activities provided by DCL2 and DCL3 (Dunoyer et al., 2005; Gasciolli et al., 2005; Xie et al., 2005), phasing is lost in the dcl4-2 mutant. Given the developmental phenotypes and derepression characteristics of tasiRNA target transcripts in dcl4 mutants, we infer that the off-sized, nonphased small RNAs that originate from TAS loci in dcl4-2 are largely nonfunctional. The structure of DICER from a lower eukaryote indicates that size characteristics of siRNAs are controlled by the spacing between the binding pocket of the PAZ domain, which grips the end of a substrate dsRNA with a 2-nucleotide 3′ overhang and the catalytic features at the dual active sites (Macrae et al., 2006). The 21-nucleotide siRNA-generating structure of DCL4, therefore, is a critical property of the enzyme and has been incorporated as a molecular ruler with physiologic importance. An open question concerns phase drift, which occurs after several DCL4 processing cycles of several tasiRNA precursors. Phase drift occurs at discrete points, such as processing cycle 5 for the TAS1c precursor (Figure 1), and may relate to unusual properties of the dsRNA interval interacting with DCL4 at a given cycle.
Two additional TAS3 loci (TAS3b and TAS3c) were identified (Figure 2; see Supplemental Table 1 and Supplemental Figure 1 online). These were assigned to the TAS3 family for two reasons. First, they contained sequenced or predicted tasiRNAs that were similar to the ARF3/ARF4 transcript-interacting tasiRNAs from TAS3a (Figure 2C; Allen et al., 2005; Williams et al., 2005; Hunter et al., 2006). Second, they contained conserved miR390 target sites on both the 5′ and 3′ sides of the tasiRNA sequences. Whether or not the TAS3b and TAS3c tasiRNAs are functional remains to be determined. A mutant carrying a T-DNA insertion in TAS3a displayed accelerated phase-change phenotypes similar to those seen in rdr6 and dcl4 mutants (Adenot et al., 2006), suggesting that TAS3b and TAS3c cannot substitute for TAS3a function. For each TAS3 family member, the 3′ miR390 target site, but not the 5′ target site, was cleaved (Figure 2D). The base pair interactions between miR390 and 5′ target sites contain key mispairs that prevent cleavage, and this is a highly conserved and functional feature of the TAS3 family (Axtell et al., 2006). The consequences of dual targeting of transcripts by miRNAs are discussed in greater detail below.
Genome-Wide Populations of RDR6-Dependent siRNAs
Analysis of the TAS1, TAS2, and TAS3 tasiRNA biogenesis and phasing patterns allowed development of an algorithm to detect RDR6-dependent small RNA phase signals from across the Arabidopsis genome (Figure 3). Three categories of RDR6-dependent siRNAs were detected (see Supplemental Table 1 online). In the first, tasiRNAs from six TAS loci were identified. Neither TAS3c, nor the recently recognized TAS4 (Rajagopalan et al., 2006), loci emerged from the phase scan due to low tasiRNA abundance. Second, three RDR6-dependent loci that were annotated as protein noncoding were identified, although none of these yielded a strong phase signal. Third, 38 protein-coding gene transcripts were identified as RDR6-dependent, as well as DCL1- and DCL4-dependent, generators of 21-nucleotide siRNAs. Nearly all of these are targets of at least one miRNA and/or tasiRNA, and nearly half were targeted by multiple small RNAs. Twelve of these loci yielded strong phase signals that were initiated by one or more miRNA/tasiRNA-guided cleavage events. The absence of a phase signal from the remaining 26 protein-coding genes may have been due, in part, to relatively low siRNA abundance, with more sequencing reads likely to yield more phase signals. Among the loci with a phase signal, only one (At5g38850, encoding a TIR-NBS-LRR protein) lacked a known miRNA or tasiRNA phase initiator (Table 2). These data reveal the strong linkage between endogenous deployment of the RDR6/DCL4 siRNA biogenesis system and small RNA-guided initiation.
The high proportion of dual-targeted transcripts that yield phased siRNAs through the RDR6/DCL4-dependent pathway strongly supports the two-hit trigger hypothesis proposed by Axtell et al. (2006). How efficiently a transcript gets routed through the RDR6/DCL4 siRNA biogenesis pathway may depend on how many small RNA target sites exist and on at least one of these target sites being cleaved. Small RNA-guided effector complexes may interact with factors, such as SGS3, that stabilize one of the cleavage products (Mourrain et al., 2000; Peragine et al., 2004; Yoshikawa et al., 2005). This would then lead to recruitment of RDR6 to the stabilized RNA fragment for dsRNA synthesis and subsequent DCL4-mediated processing in an end-dependent manner. The features governing which small RNA-guided cleavage fragment is stabilized and converted to dsRNA is not clear. Perhaps a cleavage product that is associated with another small RNA-AGO effector complex, such as that which associates with the 5′ fragment from miR390-guided TAS3 transcript cleavage products, marks a preferred template. The abundance of small RNA target sites, some of which do not guide cleavage, may be what directs the PPR transcripts so effectively through the pathway.
The miRNA and RDR6/DCL4-dependent siRNA systems may have coevolved as a posttranscriptional silencing mechanism to limit the activity of the RDR6/DCL4 system on most endogenous transcripts and RNAs. The need for one, and preference for multiple, small RNA–mediated initiation events may disqualify most cellular RNAs as targets. The preference for multiple targeting events also explains why most single-site miRNA target transcripts, which are the vast majority of plant miRNA targets, do not spawn siRNAs (Axtell et al., 2006). Furthermore, it may explain why viral RNAs and other targets of complex populations of siRNAs effectively amplify silencing signals (Brodersen and Voinnet, 2006).
Targeting of a Rapidly Expanding PPR-P Gene Clade
Except for TAS1 and TAS2 loci, the rapidly evolving PPR-P clade yields the most easily recognizable set of RDR6/DCL4-dependent siRNAs in Arabidopsis. Most of the PPR-P clade transcripts are targeted by multiple miRNAs and/or tasiRNAs, which efficiently drive the cleavage fragments through the RDR6/DCL4 pathway. This very likely reinforces silencing that might be directed via targeting by miR161, miR400, and TAS1/TAS2 tasiRNAs. Given the abundance of the resulting clade-derived siRNAs, it is puzzling why siRNA biogenesis does not spread outside of the clade. Despite 33 nonclade PPR-P transcripts possessing multiple, plausible target sites for sequenced, clade-derived siRNAs, the RDR6/DCL4 system was not directed to this group of transcripts. It is possible that the target prediction algorithm, which we have used successfully with miRNAs and tasiRNAs (Allen et al., 2005; Fahlgren et al., 2007), did not accurately identify prospective targets for this analysis. It is also possible that clade-derived siRNAs had the potential to trigger silencing of nonclade PPR-P transcripts, but they were in cellular concentrations that were too low to function. Functional siRNAs may have also been compartmentalized away from nonclade PPR-P target transcripts.
What functions might targeting of the PPR-P clade by the RDR6/DCL4-dependent silencing system serve? This is not immediately obvious from genetic analysis, as most of the mutant phenotypes associated with loss of RDR6 and DCL4 appear to relate specifically to loss of TAS3 tasiRNAs (Xie et al., 2005; Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Hunter et al., 2006). Of course, physiologic functions for silencing of the clade may lay beyond those that are revealed in short-term growth and development assays. The PPR gene family is widespread and has undergone a dramatic evolutionary expansion in plants (Small and Peeters, 2000; Rivals et al., 2006). An important observation is the close phylogenetic relationship between the radish (R. sativus) fertility restorer genes (PPR-A and PPR-B) and members of the targeted PPR-P clade in Arabidopsis (see Supplemental Figure 3 online; Desloire et al., 2003). The fertility restorer genes compensate for defects in mitochondrion-encoded ATPase alleles in other species (Kazama and Toriyama, 2003; Wen et al., 2003; Akagi et al., 2004; Wang et al., 2006), although in Arabidopsis there are no known mitochondrion-based cytoplasmic male sterility systems and, therefore, no corresponding fertility restorer functions identified. A group of Populus PPR-P genes also shows relatively close phylogenetic relationship to fertility restorer genes based on amino acid sequence similarity (data not shown). Interestingly, these genes have expanded in a clade, similar to what has occurred in Arabidopsis (see Supplemental Figure 3 online). Expansion of these PPR-P clades in several plant species may reflect mechanisms to diversify alleles for interactions with multiple mitochondrial gene alleles. As the functions of the PPR-P genes are not known in Arabidopsis or Populus, the putative allele interactions may not necessarily relate to fertility or be mitochondrion specific. Remarkably, the Populus PPR-P clade members are singly or dually targeted by two miRNAs, miR475 and miR476, indicating that this clade is under posttranscriptional regulation (Lu et al., 2005). While primary targeting of PPR transcripts in Arabidopsis and Populus has occurred independently, it is unclear if Populus PPR transcripts also spawn secondary siRNAs. Regardless, silencing of the expanding clades in multiple species may serve a dosage compensation function, especially if gene expansion is providing a reservoir of PPR-P alleles to balance infrequently dispersed mitochondrial alleles. Posttranscriptional silencing, as we proposed previously (Allen et al., 2004; Fahlgren et al., 2007), may enable rapid expansion of gene families while minimizing detrimental dosage effects.
Finally, while there is compelling evidence that many MIRNA genes had evolutionary origins in duplicated segments of protein-coding genes (Allen et al., 2004; Axtell et al., 2006; Fahlgren et al., 2007), the origins of TAS gene loci are not clear. However, in view of targeting of the PPR-P clade by miRNA and RDR6/DCL4 systems, and the resulting phased siRNA patterns, we speculate that TAS loci may originate from miRNA-targeted protein-coding genes. In fact, the abundant siRNA-generating PPR-P loci look very similar to known TAS loci, except that most still appear to encode functional proteins. However, one can easily imagine loss of protein-coding potential through genetic drift and/or recombination events, but maintenance of one or more functional siRNA modules in phase with initiator small RNA cleavage sites. A nascent TAS locus would be retained if suppression of gene-of-origin family members provided an advantage.
METHODS
Small RNA Libraries
Small RNA libraries were described by Kasschau et al. (2007). Small RNA sequencing for wild-type (Col-0), dcl1-7, dcl2-1, dcl3-1, dcl4-2, rdr1-1, rdr2-1, and rdr6-15 inflorescence, Col-0 and rdr6-15 seedlings, and Col-0 leaf tissues was done by picoliter-scale pyrosequencing (454 Life Sciences; Margulies et al., 2005). All small RNA sequences are available for download at the Gene Expression Omnibus (GEO) (Edgar et al., 2002; Barrett et al., 2005; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE6682 and the Arabidopsis Small RNA Project Database (http://asrp.cgrb.oregonstate.edu/db/).
Arabidopsis thaliana Mutants and Target Validation
Mutant lines for dcl1-7, dcl2-1, dcl3-1, dcl4-2, rdr1-1, rdr2-1, rdr6-15, hen1-1, hst-15, and hyl1-2 were described previously (Park et al., 2002; Reinhart et al., 2002; Peragine et al., 2004; Vazquez et al., 2004a; Xie et al., 2004, 2005; Allen et al., 2005). Targets were validated using a 5′ RACE assay with the GeneRacer kit (Invitrogen) as described previously (Llave et al., 2002; Kasschau et al., 2003; Allen et al., 2004, 2005). Total RNA was isolated from inflorescence tissue of 28-d-old, stage 1 to 12 flowers of both Col-0 and hen1-1 plants. RNA was subjected to batch and spin column purification for poly(A)+ enrichment with an Oligotex mRNA Midi kit (Qiagen). mRNA was subsequently ligated to the GeneRacer RNA oligo adaptor without further modification. The GeneRacer oligo(dT) primer was used to prime cDNA synthesis via reverse transcription. Two rounds of 5′ RACE reactions were performed with two gene-specific primers designed ∼200 to 500 nucleotides from the 3′ end of the predicted target sites using PCR parameters defined by the GeneRacer kit and an annealing temperature of 65°C. The 5′ RACE products were gel purified, TOPO TA cloned (Invitrogen), and sequenced.
Alignments, Phase Transformations, and Expression Profiling
Alignments between TAS3a, TAS3b, and TAS3c were performed using T-Coffee (Notredame et al., 2000). Phase transform data were produced using the following equation:
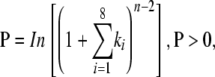 |
where n = number of phase cycle positions occupied by at least one small RNA read within an eight-cycle window, and k = the total number of reads for all small RNAs with consolidated start coordinates in a given phase within an eight-cycle window.
Phase cycle length was set at 21 nucleotides (see Figure 3). All microarray data were generated using Affymetrix ATH1 arrays and are available at GEO (Edgar et al., 2002; Barrett et al., 2005; http://www.ncbi.nlm.nih.gov/geo/).
Phylogenetic Analysis
PPR amino acid sequences were aligned using ClustalW (Higgins et al., 1996) with either the BLOSUM or the GONNET matrix series and with a gap opening penalty of 10 for both the pairwise and multiple alignments and a gap extension penalty of 0.1 and 0.2 for pairwise and multiple alignments, respectively. Gaps in the multiple alignments, defined as regions with at least five consecutive gap characters in all but one or two sequences, were removed. In addition, the first and last 80 positions of the multiple alignments were deleted manually to restrict the analysis to the most conserved region. Trees were produced with ClustalW using the neighbor-joining method (Saitou and Nei, 1987) and 1000 bootstrap replicates. Trees were viewed and rooted using TREEVIEW (Page, 1996).
Accession Numbers
The cDNA sequence for TAS3b was deposited in The Arabidopsis Information Resource database under accession number At5g49615. Sequence data from this article can be found in the EMBL/GenBank data libraries under accession number GSE6682 (GEO small RNA data). Col-0 (control for dcl1-7, rdr6-15, and dcl4-2), dcl1-7, rdr6-15, and dcl4-2 were from experiments described by Xie et al. (2005) (GEO accession number GSE3011, samples GSM65929, GSM65930, GSM65931, GSM65932, GSM65933, GSM65934, GSM65935, GSM65936, GSM65937, GSM65938, GSM65939, GSM65940, GSM65941, GSM65942, and GSM65943). Col-0 (control for hst-15, hyl1-2, dcl2-1, dcl3-1, rdr1-1, and rdr2-1), Ler (control for hen1-1), hst-15, hyl1-2, dcl2-1, dcl3-1, rdr1-1, and rdr2-1 were from experiments described previously (Allen et al., 2005) (GEO accession number GSE2473, samples GSM47011, GSM47012, GSM47013, GSM47014, GSM47015, GSM47016, GSM47020, GSM47021, GSM47022, GSM47028, GSM47029, GSM47030, GSM47031, GSM47032, GSM47033, GSM47034, GSM47035, GSM47036, GSM47037, GSM47038, GSM47039, GSM47040, GSM47041, GSM47042, GSM47043, GSM47044, GSM47045, GSM47046, GSM47047, and GSM47048).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mapping of Transcription Start Sites of TAS3 Family Genes by RNA Ligase-Mediated 5′ RACE.
Supplemental Figure 2. Mapping of miR161- and TAS2 3′D6(−)-Mediated Cleavage Sites in PPR Gene Transcripts by 5′ RACE.
Supplemental Figure 3. Phylogenetic Tree of PPR-P Clade Proteins from Arabidopsis, PPR-P–Related Proteins from Populus and R. sativus, and Outgroup PPR proteins CRP1 (Z. mays), PET309 (Saccharomyces cerevisiae), PTCD1 (Homo sapiens), and RF1A and RF1B (Oryza sativa).
Supplemental Table 1. RDR6-Dependent 21-Nucleotide RNA-Generating Loci.
Acknowledgments
We thank Heather Sweet and Amy Shatswell for excellent technical assistance, Christine Gustin for assistance with 5′ RACE analysis, and Erin McWhorter for media preparation. We thank Mark Dasenko for sequence analysis. We also thank Robert Burton for advice on the phase transform equation. This work was supported by grants from the National Science Foundation 2010 Project (MCB-0618433), the National Institutes of Health (AI43288), and the USDA (2006-35301-17420).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: James C. Carrington (carrington@cgrb.oregonstate.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouche, N., Gasciolli, V., and Vaucheret, H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16 927–932. [DOI] [PubMed] [Google Scholar]
- Akagi, H., Nakamura, A., Yokozeki-Misono, Y., Inagaki, A., Takahashi, H., Mori, K., and Fujimura, T. (2004). Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor. Appl. Genet. 108 1449–1457. [DOI] [PubMed] [Google Scholar]
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. [DOI] [PubMed] [Google Scholar]
- Allen, E., Xie, Z., Gustafson, A.M., Sung, G.H., Spatafora, J.W., and Carrington, J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36 1282–1290. [DOI] [PubMed] [Google Scholar]
- Aravin, A., et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442 203–207. [DOI] [PubMed] [Google Scholar]
- Aubourg, S., Boudet, N., Kreis, M., and Lecharny, A. (2000). In Arabidopsis thaliana, 1% of the genome codes for a novel protein family unique to plants. Plant Mol. Biol. 42 603–613. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J., Jan, C., Rajagopalan, R., and Bartel, D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127 565–577. [DOI] [PubMed] [Google Scholar]
- Barrett, T., Suzek, T.O., Troup, D.B., Wilhite, S.E., Ngau, W.C., Ledoux, P., Rudnev, D., Lash, A.E., Fujibuchi, W., and Edgar, R. (2005). NCBI GEO: Mining millions of expression profiles–database and tools. Nucleic Acids Res. 33 D562–D566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431 356–363. [DOI] [PubMed] [Google Scholar]
- Bentolila, S., Alfonso, A.A., and Hanson, M.R. (2002). A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. USA 99 10887–10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani, O., Zhu, J., Verslues, P.E., Sunkar, R., and Zhu, J.K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche, N., Lauressergues, D., Gasciolli, V., and Vaucheret, H. (2006). An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P., and Voinnet, O. (2006). The diversity of RNA silencing pathways in plants. Trends Genet. 22 268–280. [DOI] [PubMed] [Google Scholar]
- Chan, S.W., Zilberman, D., Xie, Z., Johansen, L.K., Carrington, J.C., and Jacobsen, S.E. (2004). RNA silencing genes control de novo DNA methylation. Science 303 1336. [DOI] [PubMed] [Google Scholar]
- Chase, C.D. (2007). Cytoplasmic male sterility: A window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 23 81–90. [DOI] [PubMed] [Google Scholar]
- Cushing, D.A., Forsthoefel, N.R., Gestaut, D.R., and Vernon, D.M. (2005). Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta 221 424–436. [DOI] [PubMed] [Google Scholar]
- Deleris, A., Gallego-Bartolome, J., Bao, J., Kasschau, K.D., Carrington, J.C., and Voinnet, O. (2006). Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313 68–71. [DOI] [PubMed] [Google Scholar]
- Desloire, S., et al. (2003). Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 4 588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., Weijers, D., Lechner, E., Yamada, M., Hobbie, L., Ehrismann, J.S., Jurgens, G., and Estelle, M. (2005). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9 109–119. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P., Himber, C., and Voinnet, O. (2005). DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 37 1356–1360. [DOI] [PubMed] [Google Scholar]
- Edgar, R., Domrachev, M., and Lash, A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N., Howell, M.D., Kasschau, K.D., Chapman, E.J., Sullivan, C.M., Cumbie, J.S., Givan, S.A., Law, T.F., Grant, S.R., Dangl, J.L., and Carrington, J.C. (2007). High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE 2 e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N., Montgomery, T.A., Howell, M.D., Allen, E., Dvorak, S.K., Alexander, A.L., and Carrington, J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16 939–944. [DOI] [PubMed] [Google Scholar]
- Fusaro, A.F., Matthew, L., Smith, N.A., Curtin, S.J., Dedic-Hagan, J., Ellacott, G.A., Watson, J.M., Wang, M.B., Brosnan, C., Carroll, B.J., and Waterhouse, P.M. (2006). RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 7 1168–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D., Collier, S.A., Byrne, M.E., and Martienssen, R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16 933–938. [DOI] [PubMed] [Google Scholar]
- Gasciolli, V., Mallory, A.C., Bartel, D.P., and Vaucheret, H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15 1494–1500. [DOI] [PubMed] [Google Scholar]
- Girard, A., Sachidanandam, R., Hannon, G.J., and Carmell, M.A. (2006). A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442 199–202. [DOI] [PubMed] [Google Scholar]
- Grivna, S.T., Beyret, E., Wang, Z., and Lin, H. (2006. b). A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20 1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna, S.T., Pyhtila, B., and Lin, H. (2006. a). MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc. Natl. Acad. Sci. USA 103 13415–13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, I.R., Zhang, X., Lu, C., Johnson, L., Meyers, B.C., Green, P.J., and Jacobsen, S.E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38 721–725. [DOI] [PubMed] [Google Scholar]
- Herr, A.J., Jensen, M.B., Dalmay, T., and Baulcombe, D.C. (2005). RNA polymerase IV directs silencing of endogenous DNA. Science 308 118–120. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G., Thompson, J.D., and Gibson, T.J. (1996). Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266 383–402. [DOI] [PubMed] [Google Scholar]
- Hunter, C., Sun, H., and Poethig, R.S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13 1734–1739. [DOI] [PubMed] [Google Scholar]
- Hunter, C., Willmann, M.R., Wu, G., Yoshikawa, M., de la Luz Gutierrez-Nava, M., and Poethig, S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133 2973–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades, M.W., and Bartel, D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14 787–799. [DOI] [PubMed] [Google Scholar]
- Kanno, T., Huettel, B., Mette, M.F., Aufsatz, W., Jaligot, E., Daxinger, L., Kreil, D.P., Matzke, M., and Matzke, A.J. (2005). Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 37 761–765. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Fahlgren, N., Chapman, E.J., Sullivan, C.M., Cumbie, J.S., Givan, S.A., and Carrington, J.C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5 e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4 205–217. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal, S., Morgan, R., Dahlbeck, D., Borsani, O., Villegas, A., Jr., Zhu, J.K., Staskawicz, B.J., and Jin, H. (2006). A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 103 18002–18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama, T., and Toriyama, K. (2003). A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett. 544 99–102. [DOI] [PubMed] [Google Scholar]
- Kim, V.N. (2006). Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 20 1993–1997. [DOI] [PubMed] [Google Scholar]
- Kotera, E., Tasaka, M., and Shikanai, T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433 326–330. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Seto, A.G., Kim, J., Kuramochi-Miyagawa, S., Nakano, T., Bartel, D.P., and Kingston, R.E. (2006). Characterization of the piRNA complex from rat testes. Science 313 363–367. [DOI] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297 2053–2056. [DOI] [PubMed] [Google Scholar]
- Lu, C., Kulkarni, K., Souret, F.F., Muthuvalliappan, R., Tej, S.S., Poethig, R.S., Henderson, I.R., Jacobsen, S.E., Wang, W., Green, P.J., and Meyers, B.C. (2006). MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 16 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S., Sun, Y.H., Shi, R., Clark, C., Li, L., and Chiang, V.L. (2005). Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17 2186–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae, I.J., Zhou, K., Li, F., Repic, A., Brooks, A.N., Cande, W.Z., Adams, P.D., and Doudna, J.A. (2006). Structural basis for double-stranded RNA processing by Dicer. Science 311 195–198. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Reinhart, B.J., Jones-Rhoades, M.W., Tang, G., Zamore, P.D., Barton, M.K., and Bartel, D.P. (2004). MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 23 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies, M., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R.A., Zaratiegui, M., and Goto, D.B. (2005). RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 21 450–456. [DOI] [PubMed] [Google Scholar]
- Mello, C.C., and Conte, D., Jr. (2004). Revealing the world of RNA interference. Nature 431 338–342. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101 533–542. [DOI] [PubMed] [Google Scholar]
- Mourrain, P., van Blokland, R., Kooter, J.M., and Vaucheret, H. (2007). A single transgene locus triggers both transcriptional and post-transcriptional silencing through double-stranded RNA production. Planta 225 365–379. [DOI] [PubMed] [Google Scholar]
- Munroe, S.H., and Zhu, J. (2006). Overlapping transcripts, double-stranded RNA and antisense regulation: a genomic perspective. Cell. Mol. Life Sci. 63 2102–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D.G., and Heringa, J. (2000). T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302 205–217. [DOI] [PubMed] [Google Scholar]
- Okuda, K., Nakamura, T., Sugita, M., Shimizu, T., and Shikanai, T. (2006). A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J. Biol. Chem. 281 37661–37667. [DOI] [PubMed] [Google Scholar]
- Onodera, Y., Haag, J.R., Ream, T., Nunes, P.C., Pontes, O., and Pikaard, C.S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120 613–622. [DOI] [PubMed] [Google Scholar]
- Page, R.D. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. [DOI] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier, D., Yahubyan, G., Vega, D., Bulski, A., Saez-Vasquez, J., Hakimi, M.A., Lerbs-Mache, S., Colot, V., and Lagrange, T. (2005). Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19 2030–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110 513–520. [DOI] [PubMed] [Google Scholar]
- Rivals, E., Bruyere, C., Toffano-Nioche, C., and Lecharny, A. (2006). Formation of the Arabidopsis pentatricopeptide repeat family. Plant Physiol. 141 825–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus, M., Vaughn, M.W., and Martienssen, R.A. (2006). MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell 18 1559–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby, J.G., Jan, C., Player, C., Axtell, M.J., Lee, W., Nusbaum, C., Ge, H., and Bartel, D.P. (2006). Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127 1193–1207. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Schwab, R., Palatnik, J.F., Riester, M., Schommer, C., Schmid, M., and Weigel, D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8 517–527. [DOI] [PubMed] [Google Scholar]
- Shikanai, T. (2006). RNA editing in plant organelles: Machinery, physiological function and evolution. Cell. Mol. Life Sci. 63 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, I.D., and Peeters, N. (2000). The PPR motif - A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25 46–47. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., and Zhu, J.K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze, H., Padmanaban, S., Cellier, F., Honys, D., Cheng, N.H., Bock, K.W., Conejero, G., Li, X., Twell, D., Ward, J.M., and Hirschi, K.D. (2004). Expression patterns of a novel AtCHX gene family highlight potential roles in osmotic adjustment and K+ homeostasis in pollen development. Plant Physiol. 136 2532–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmor-Neiman, M., Stav, R., Klipcan, L., Buxdorf, K., Baulcombe, D.C., and Arazi, T. (2006). Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J. 48 511–521. [DOI] [PubMed] [Google Scholar]
- Tolia, N.H., and Joshua-Tor, L. (2007). Slicer and the Argonautes. Nat. Chem. Biol. 3 36–43. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., and Zamore, P.D. (2005). Perspective: Machines for RNAi. Genes Dev. 19 517–529. [DOI] [PubMed] [Google Scholar]
- Tran, R.K., Zilberman, D., de Bustos, C., Ditt, R.F., Henikoff, J.G., Lindroth, A.M., Delrow, J., Boyle, T., Kwong, S., Bryson, T.D., Jacobsen, S.E., and Henikoff, S. (2005). Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis. Genome Biol. 6 R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H. (2005). MicroRNA-dependent trans-acting siRNA production. Sci. STKE 2005 pe43. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H. (2006). Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes Dev. 20 759–771. [DOI] [PubMed] [Google Scholar]
- Vazquez, F. (2006). Arabidopsis endogenous small RNAs: Highways and byways. Trends Plant Sci. 11 460–468. [DOI] [PubMed] [Google Scholar]
- Vazquez, F., Gasciolli, V., Crete, P., and Vaucheret, H. (2004. a). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14 346–351. [DOI] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A.C., Hilbert, J.L., Bartel, D.P., and Crete, P. (2004. b). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16 69–79. [DOI] [PubMed] [Google Scholar]
- Wang, X.J., Gaasterland, T., and Chua, N.H. (2005). Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 6 R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., et al. (2006). Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, P.M., and Fusaro, A.F. (2006). Plant science. Viruses face a double defense by plant small RNAs. Science 313 54–55. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Wang, M.B., and Lough, T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411 834–842. [DOI] [PubMed] [Google Scholar]
- Wen, L., Ruesch, K.L., Ortega, V.M., Kamps, T.L., Gabay-Laughnan, S., and Chase, C.D. (2003). A nuclear restorer-of-fertility mutation disrupts accumulation of mitochondrial ATP synthase subunit alpha in developing pollen of S male-sterile maize. Genetics 165 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, L., Carles, C.C., Osmont, K.S., and Fletcher, J.C. (2005). A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. USA 102 9703–9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann, M.R., and Poethig, R.S. (2005). Time to grow up: The temporal role of smallRNAs in plants. Curr. Opin. Plant Biol. 8 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Allen, E., Wilken, A., and Carrington, J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2 E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa, M., Peragine, A., Park, M.Y., and Poethig, R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299 716–719. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., Johansen, L.K., Xie, Z., Carrington, J.C., and Jacobsen, S.E. (2004). Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 14 1214–1220. [DOI] [PubMed] [Google Scholar]