Abstract
It has been proposed that defects in the assembly of spliceosomal uridine-rich small nuclear ribonucleoprotein (U snRNP) complexes could account for the death of motor neurons in spinal muscular atrophy (SMA). We discovered that infection of cultured cells with poliovirus results in the specific cleavage of the host factor Gemin3 by a virus-encoded proteinase, 2Apro. Gemin3 is a component of the macromolecular SMN complex that mediates assembly of U snRNP complexes by aiding the heptameric oligomerization of Sm proteins onto U snRNAs. Using in vitro Sm core assembly assays, we found that lowering the intracellular amounts of Gemin3 by either poliovirus infection or small interfering RNA (siRNA)-mediated knockdown of Gemin3 resulted in reduced assembly of U snRNPs. Immunofluorescence analyses revealed a specific redistribution of Sm proteins from the nucleoplasm to the cytoplasmic periphery of the nucleus in poliovirus-infected cells. We propose that defects in U snRNP assembly may be shared features of SMA and poliomyelitis.
Keywords: snRNP assembly, picornaviral proteinases, Gemins
Neuromuscular disorders define a wide array of diseases resulting in the progressive degeneration of motor neurons with symptoms ranging from muscle weakness and atrophy to paralysis and death in severe cases. While factors leading to these disorders have been identified in many cases, the molecular mechanisms that cause the specificity of neuronal cell death remain unknown.
Poliovirus, a member of the Enterovirus genus in the Picornaviridae family, is best recognized as the causative agent of poliomyelitis (for review, see Mueller et al. 2005). Following oral entry, it replicates in the gastrointestinal tract as is characteristic of enteroviruses. In rare cases, poliovirus can spread to the CNS, where its replication leads to the degeneration of motor neurons, ultimately resulting in poliomyelitis. While poliovirus is one of the most well-characterized viruses, its pathogenesis and propensity to specifically destroy motor neurons in infected individuals are not understood.
Spinal muscular atrophy (SMA) (for review, see Frugier et al. 2002) is an autosomal recessive disorder and one of the leading genetic causes of infant death. It is characterized by reduced levels of the 38-kDa “survival of motor neurons” (SMN) protein due to deletion or mutation of the SMN1 gene (Lefebvre et al. 1995; Coovert et al. 1997). A second copy of the gene, SMN2, has arisen from a duplication event; however, a mutation in exon 7 of SMN2 results in frequent skipping of this exon and low levels of functional protein (Lorson et al. 1999). Curiously, SMN is expressed in a wide range of tissues (Coovert et al. 1997), and the question of how deficiency in an ubiquitously expressed protein results in the specific destruction of motor neurons remains a major question in the field (Monani 2005).
Clues to the molecular mechanisms underlying SMA have come from studying the functions of the SMN complex—a large and dynamic macromolecular complex composed of SMN and a class of proteins called Gemins. The core SMN complex consists of SMN and Gemins2–8, and localizes to both the cytoplasm and nucleoplasm (Fischer et al. 1997; Charroux et al. 1999, 2000; Baccon et al. 2002; Gubitz et al. 2002; Pellizzoni et al. 2002; Carissimi et al. 2006). Recent work by Otter et al. (2007) revealed that SMN, Gemin7, and Gemin8 form a central complex to which the other Gemins peripherally associate via multiple interactions. Furthermore, this study supported previous findings that Gemin3 is responsible for the recruitment of Gemin4 into the SMN complex (Otter et al. 2007). The most well-characterized function of the SMN complex is its role in the biogenesis of spliceosomal complexes. Specifically, “uridine-rich small nuclear ribonucleoprotein” (U snRNP) complexes have been shown to be assembled by the SMN complex in vitro (Meister et al. 2001). The major U snRNAs (U1, U2, U4, and U5) are transcribed in the nucleus and transported to the cytoplasm, where direct interaction with the SMN complex has been demonstrated (Yong et al. 2004; Golembe et al. 2005). All of the SMN core complex components, with the exception of Gemin2, have been shown to associate with various subsets of so-called Sm proteins (SmB/B′, SmD1–3, SmE, SmF, and SmG) that can form a heptameric ring-like structure, known as the Sm core, in the presence of target U snRNAs (Fischer et al. 1997; Charroux et al. 1999, 2000; Baccon et al. 2002; Gubitz et al. 2002; Pellizzoni et al. 2002; Carissimi et al. 2006). Sm core formation depends on the presence of a 6- to 8-nucleotide (nt) “Sm site” on the U snRNAs (Raker et al. 1999). Following assembly of the Sm core, the U snRNPs move into the nucleus and localize to discrete foci named Cajal bodies, characterized by the presence of the 80-kDa protein coilin. In the Cajal bodies, U snRNPs undergo further maturation prior to spliceosome assembly (for review, see Kiss 2004). SMN has also been found to localize to Cajal bodies through a direct interaction with coilin (Hebert et al. 2001).
A correlation between reduced SMN protein levels and motor neuron degeneration in individuals with SMA has been established (Buhler et al. 1999). Importantly, SMN levels have been shown to be proportional to snRNP formation, and reduced Sm core assembly has been observed in cell extracts derived from SMA patients (Wan et al. 2005). Additionally, in the absence of endogenous SMN, ectopic expression of SMN protein containing SMA-causing mutations was shown to result in Sm core assembly defects (Shpargel and Matera 2005). Some of the most convincing evidence linking motor neuron defects to reduced levels of snRNPs was reported by Winkler et al. (2005), who observed axon outgrowth defects in zebrafish embryos following silencing of SMN expression. This motor neuron abnormality could be rescued by coinjection of purified snRNPs, suggesting that the axonal defects were a consequence of reduced snRNP levels induced by loss of the SMN protein (Winkler et al. 2005).
Although the causative agents of SMA and poliomyelitis are diverse, the specific effect each has on motor neurons led us to question whether U snRNP assembly was affected by poliovirus infection. We used an in vitro assembly assay to examine the effects of poliovirus infection on Sm core formation and found that Sm core assembly activity was reduced in poliovirus-infected cells. Analyses of SMN complex component levels revealed that Gemin3 is a novel target of the poliovirus-encoded proteinase 2Apro. Proteolysis of Gemin3 in poliovirus-infected cells correlated with reduced Sm core assembly activity, which was also observed upon RNA interference (RNAi)-mediated knockdown of Gemin3. Additionally, Sm proteins were found to be redistributed in poliovirus-infected cells, resulting in reduced colocalization of the core proteins with coilin in nuclear foci. Our findings suggest that decreased levels of U snRNPs may exacerbate the specific motor neuron degeneration observed in poliomyelitis.
Results
Poliovirus infection reduces Sm core assembly
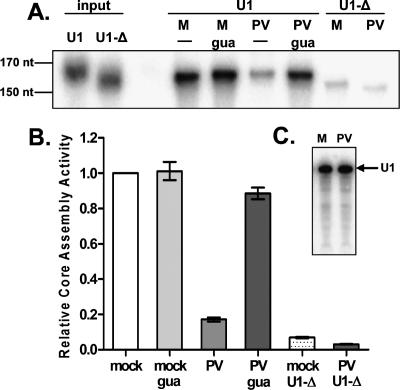
To assess whether U snRNP biogenesis was affected during poliovirus infection, an in vitro Sm core assembly assay was used to examine the effect of poliovirus on snRNP formation. Radiolabeled U1 snRNAs were added to assembly reactions containing extracts prepared from mock- or poliovirus-infected cells. Guanidine-HCl, a compound that inhibits poliovirus RNA replication, was used to distinguish whether observed effects could be induced by viral entry alone or required subsequent RNA replication and amplification. To control for the specificity of Sm core formation, a U1 snRNA containing a 9-nt deletion of the Sm site (U1-Δ) was also tested. Assembled U1 cores were isolated by immunoprecipitation with the monoclonal antibody Y12, directed against epitopes on Sm proteins B′/B and D (Hirakata et al. 1993), and the amount of associated U1 snRNA was used as a measure of assembly activity.
Sm core assembly activity was found to be reduced more than fivefold in extracts prepared from poliovirus-infected cells compared with mock-treated extracts (Fig. 1A,B). Treatment with guanidine-HCl in the absence of viral infection had no effect on Sm core assembly (Fig. 1B, mock gua), suggesting that this compound does not interfere with the core assembly reaction. Infection in the presence of guanidine-HCl did not result in a significant decrease in core assembly (Fig. 1B, PV gua), indicating that the observed defect required active viral RNA replication or increased amounts of viral proteins. Core formation was dependent on the Sm site sequence, as the amount of immunoprecipitated U1-Δ snRNA, which migrates faster due to deletion of the Sm site, was 10-fold less than that observed for the wild-type U1 snRNA (Fig. 1A,B). To ensure that the reduction in Sm core assembly observed in extracts prepared from poliovirus-infected cells was not due to degradation of the input U1 snRNA, assembly reactions were performed, and the integrity of the extracted RNA was inspected. As shown in Figure 1C, the U1 snRNA remained intact during Sm core assembly reactions in both mock- and poliovirus-infected extracts. In addition, steady-state amounts of endogenous U1 snRNAs were found to be unaltered during poliovirus infection (data not shown). Together, these data demonstrate that Sm core formation on U1 snRNAs is reduced during poliovirus infection.
Figure 1.
Sm core assembly in extracts from uninfected and poliovirus-infected cells. (A) Representative phosphorimage of U1 and U1-Δ snRNA core assembly reactions. Cells were infected with poliovirus (PV) or mock-infected (M), and assembly extracts were prepared 5 h post-infection. Addition of 2 mM guanidine-HCl (gua) was used to inhibit viral replication. Following assembly reactions, Sm cores were immunoprecipitated with an anti-Y12 antibody directed against epitopes of Sm proteins B′/B and D, and the associated snRNAs were resolved on 5% TBE-urea gels. The U1-Δ snRNA, containing a 9-nt deletion of the Sm core-binding site, served as a negative control. The input lanes on the left were loaded with 500 cpm of the indicated snRNAs. (B) Quantitation of three independent Sm core assembly experiments performed as described in A. Data were graphed as fractions of the U1 snRNA signal observed in mock-infected extracts. Error bars indicate the standard error. (C) Stability of U1 snRNAs during Sm core assembly reactions. Core assembly reactions were treated with proteinase K to disrupt RNA–protein interactions, and snRNAs were purified by phenol-chloroform extraction. Twenty percent of the material obtained from reactions containing mock-infected (M) or poliovirus-infected (PV) extracts was separated on 5% TBE-urea gels. A phosphorimage of the gel is shown. The position of the 164-nt U1 snRNA is indicated by an arrow.
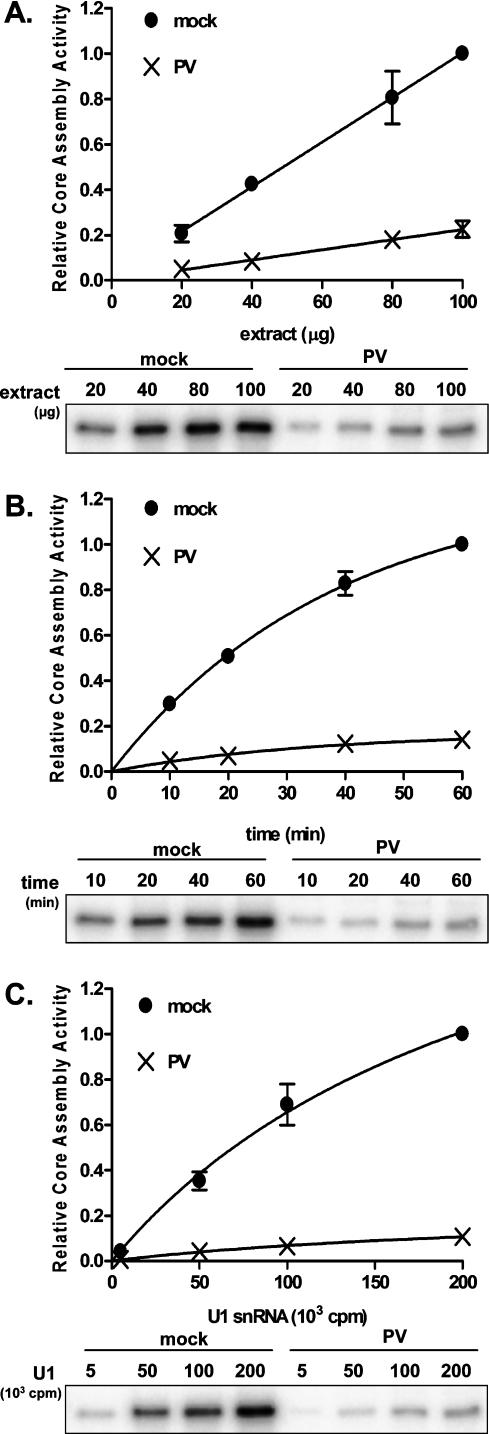
To characterize the core assembly defect further, Sm core assembly assays were performed using increasing amounts of extracts prepared from mock- or poliovirus-infected cells. As shown in Figure 2A, the amount of core assembly increased linearly with increasing extract concentration under both conditions. We observed that the slope of the curve obtained from mock-infected extracts was more than fourfold greater than that calculated from experiments performed in poliovirus-infected extracts. Therefore, the capacity for Sm core assembly, measured as assembly activity per microgram of extract, is reduced by >75% in extracts prepared from infected cells. Next, a time course was performed in which standard assembly reactions were allowed to proceed from 10 to 60 min prior to immunoprecipitation of the Sm cores. Figure 2B shows that Sm core formation increased over time in both mock- and poliovirus-infected extracts. Comparison of the curve plateaus, obtained by fitting each data set to a one-phase exponential association equation, indicated that the maximal amount of Sm core formation was reduced more than fivefold in infected extracts compared with extracts prepared from mock-infected cells. However, comparative analysis of the curve fits indicated that the rates at which the respective maximal assembly activities were reached were similar. Thus, the decreased core assembly activity observed in infected extracts likely reflects a reduction in the concentration of some vital component of the U snRNP assembly machinery during poliovirus infection. Finally, we monitored Sm core assembly by varying the amount of input U1 snRNA in the assays. After fitting the data to a one-site-binding hyperbola (Fig. 2C), comparison of the extrapolated plateaus indicated that the maximum possible amount of snRNA that could be bound by Sm cores in poliovirus-infected extracts was significantly less than that in mock-infected extracts. Comparison of the curve fits suggested that the nature of the association between the U1 snRNA and the protein core did not differ significantly in mock- or poliovirus-infected extracts. Together, these analyses show that poliovirus infection causes a reduction in U snRNP assembly, likely by decreasing the capacity of infected extracts to form U1 snRNPs rather than by altering the stability of the U1 snRNA or the rate of the assembly process.
Figure 2.
Concentration dependence and kinetics of snRNP assembly in extracts from uninfected and infected cells. (A) U1 Sm core assembly as a function of extract concentration. (Top) Reactions were performed for 20 min at 30°C, and the amount of core-associated snRNA was plotted as a fraction of the core assembly observed in 100 μg of mock-infected extract. Data were fit to a first-order polynomial using GraphPad Prism to obtain a slope for each curve. Bottom panel shows a phosphorimage of a representative experiment. (B) Kinetics of U1 Sm core assembly. Standard Sm core assembly reactions were incubated for the indicated lengths of times and processed as in A. Data were plotted as the fraction of U1 snRNA signal observed at 60 min in mock-infected extracts. A one-phase exponential association equation (GraphPad Prism) was used to fit the data. Bottom panel shows a representative phosphorimage. (C) Sm core assembly as a function of U1 snRNA concentration. Increasing amounts of U1 snRNAs were used in standard core assembly reactions. Reactions were processed as in A. Top panel shows results plotted as the fraction of core assembly observed with 200,000 cpm U1 snRNA in mock-infected extracts. Data were fit to a one-site-binding hyperbola (GraphPad Prism). A representative phosphorimage is shown in the bottom panel. For each graph in A–C, error bars represent the standard error of three independent experiments. Error bars for some data points are too small to be visible.
Gemin3 is proteolyzed in poliovirus-infected cells
Poliovirus infection induces a myriad of antiviral defenses that are avoided by the virus. For example, the virus inhibits host transcription through proteolysis of factors that aid in transcription by RNA polymerases I, II, and III, such as the TATA-box-binding and CRE-binding proteins (for review, see Weidman et al. 2001). Similarly, several translation initiation factors are proteolyzed by virus-encoded proteinases, resulting in the inhibition of cap-dependent host mRNA translation while allowing the cap-independent translation of the viral polypeptide mediated by an internal ribosome entry sequence (for review, see Lloyd 2006). Given these precedents for destruction of specific host proteins during viral infection, we hypothesized that the reduction in Sm core assembly observed in poliovirus-infected extracts could be due to destruction of a specific component of the assembly machinery. To test this notion, the steady-state amounts of SMN complex components and Sm proteins were compared in infected and uninfected cells.
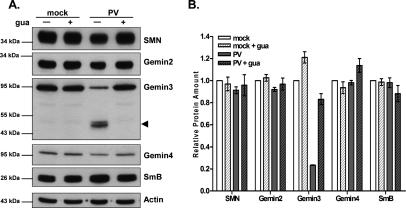
The immunoblot in Figure 3A displays the amounts of SMN, Gemin2, Gemin3, Gemin4, and SmB/B′ in extracts prepared for core assembly assays. Most proteins were equally abundant in infected and uninfected cells. However, a striking decrease in the amount of full-length Gemin3 protein and the concomitant appearance of an ∼45-kDa cleavage product was observed in poliovirus-infected extracts. Due to the specificity of the antibody used, this fragment must originate from the C terminus of Gemin3. The amounts of full-length Gemin3 were reduced by >75% in infected extracts (Fig. 3B). Furthermore, proteolysis of Gemin3 was dependent on viral RNA amplification, as addition of guanidine-HCl to infected cells abolished Gemin3 cleavage. Quantitation of the bands in the immunoblots (Fig. 3B) confirmed the observations that the relative abundance of SMN, Gemin2, Gemin4, and SmB/B′ were not significantly affected by viral infection or after guanidine-HCl treatment and that, of the proteins tested, only Gemin3 was specifically cleaved during poliovirus infection. Furthermore, immunoprecipitation analyses revealed that association of uncleaved Gemin3 with SMN is reduced in poliovirus-infected cells (Supplementary Fig. S1). Together, these results show that the reduction in Sm core assembly observed in infected extracts is not due to limiting pools of SmB/B′ proteins and suggest a role for Gemin3 in snRNP assembly.
Figure 3.
Abundance and integrity of SMN complex and Sm core components in uninfected and infected cells. (A) Relative amounts of SMN complex components and Sm proteins in mock- and poliovirus-infected cells. Infection and treatment with guanidine-HCl (gua) were carried out as described in the legend for Figure 1A, and immunoblot analysis was performed using the indicated antibodies. Each lane contains 20 μg of total protein. The Gemin3 cleavage product is marked by an arrowhead. Molecular weight markers are shown on the left. (B) Quantitation of immunoblot analyses from experiments performed as described in A. For each antibody, the signal intensity was normalized to actin as a loading control, and the ratio obtained for mock-infected extracts was set equal to 1. Error bars represent the standard error of three independent experiments.
Reduction in Gemin3 levels results in decreased Sm core assembly
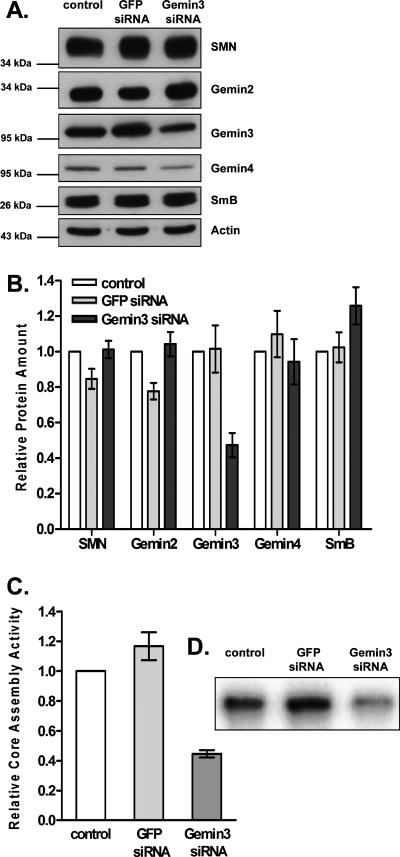
To test the hypothesis that Gemin3 is required for U snRNP formation, the intracellular abundance of Gemin3 protein was reduced by the addition of small interfering RNAs (siRNAs) directed against Gemin3 mRNA in the absence of viral infection. Figure 4, A and B, shows that Gemin3 protein abundance was decreased approximately twofold in cells treated with Gemin3 siRNAs, but remained unaffected in cells treated with control siRNAs targeting the green fluorescent protein (GFP). The amounts of SMN, Gemin2, and Gemin4 were only slightly reduced, and SmB/B′ abundance was not affected in the presence of Gemin3 or GFP siRNAs.
Figure 4.
Effect of RNAi-mediated reduction of Gemin3 on Sm core assembly. (A) Immunoblot analysis of extracts prepared 48 h after transfection of siRNAs directed against GFP or Gemin3. Control extracts were obtained from cells treated with transfection reagent alone. Twenty micrograms of total protein from each sample were resolved by SDS-PAGE and blotted with the indicated antibodies. Molecular weight markers are shown on the left. (B) Quantitative analysis of the immunoblot analyses shown in A. For each antibody, the signal intensity was normalized to actin as a loading control, and the ratio obtained for control-treated extracts was set equal to 1. Error bars represent the standard error of three independent experiments. (C) Sm core assembly activity. Sm core assembly assays were performed using U1 snRNAs in extracts prepared from cells treated with transfection reagent alone (control), GFP siRNAs, or Gemin3 siRNAs. Assembly activity was plotted as the fraction of immunoprecipitated U1 snRNA observed in control extracts. Error bars represent the standard error of four independent experiments. (D) A representative phosphorimage of the Sm core assembly assays performed in C.
To examine the potential correlation between Gemin3 protein amounts and U snRNP assembly, the same extracts were then tested in Sm core assembly assays. As shown in Figure 4, C and D, RNAi-mediated reduction of Gemin3 led to a greater than twofold decrease in core formation, while treatment with GFP siRNAs did not significantly affect assembly activity.
Our findings indicate that reduced abundance of Gemin3 leads to a decrease in U snRNP formation that roughly corresponds to the RNAi-mediated decrease in Gemin3 protein levels (Fig. 4B,C). Comparison of Gemin3 levels and Sm core formation in poliovirus-infected extracts also reveals a rough correlation between the relative amount of protein and core assembly activity as both are reduced by >75% (Figs. 3B, 1B). Interestingly, ectopic expression of an N-terminally truncated Gemin3 protein did not affect Sm core formation (Supplementary Fig. S2). This observation argues that the C-terminal fragment of Gemin3 does not act as a dominant-negative inhibitor of core formation. Overall, our data suggest that the Sm core assembly defect observed in infected extracts could be a direct result of the reduction in full-length Gemin3 by specific cleavage of the protein during poliovirus infection.
Poliovirus infection induces the redistribution of Sm proteins
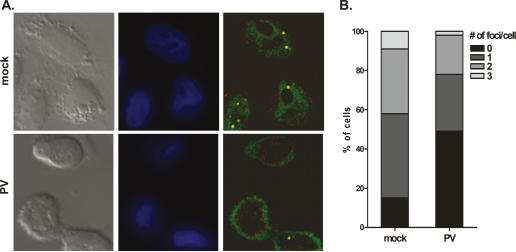
SnRNPs are imported into the nucleus and localize to Cajal bodies for further maturation, which involves the addition of other snRNP-specific proteins and snRNA modifications (Sleeman and Lamond 1999; Kiss 2004). To assess whether reduced Sm core formation in poliovirus infection has effects on snRNP biogenesis in intact cells, indirect immunofluorescence was used to analyze the localization of Sm proteins and the Cajal body-specific marker protein coilin. Cells were infected for 5 h and costained with anti-Sm and anti-coilin antibodies. In mock-treated cells, Sm proteins (green) exhibited characteristic diffuse nucleoplasmic staining and were also observed to colocalize with coilin (red) in discrete nuclear foci (yellow) (Fig. 5A, top). In contrast, poliovirus infection resulted in a redistribution of Sm proteins to the cytoplasmic periphery of the nucleus (Fig. 5A, bottom). This phenotype was observed in >95% of infected cells and was not seen during mock infections. Colocalization of Sm proteins and coilin in nuclear foci was still observed, suggesting that Sm proteins are not entirely excluded from the nucleus during infection (Fig. 5A, bottom right). The percentage of cells lacking colocalized foci increased from ∼15% in mock-infected cells to 50% in infected cells (Fig. 5B). Additionally, a statistically significant reduction in the number of cells containing one, two, or three foci was observed upon poliovirus infection (p < 0.0001). Sm protein colocalization with coilin was also assessed following siRNA-mediated reduction of Gemin3. In this case, redistribution of Sm proteins was not observed, and colocalization of Sm proteins and coilin was only slightly affected (Supplementary Fig. S3). These data are in agreement with findings reported by Shpargel and Matera, who noted that Sm protein distribution was unaltered by reduced amounts of Gemin3 and Gemin4 (Shpargel and Matera 2005). In addition, we observed that colocalization of SMN and coilin was not affected by infection with poliovirus or transfection of Gemin3 siRNAs (data not shown).
Figure 5.
Effects of poliovirus infection on the colocalization of Sm proteins with coilin in nuclear foci. (A) Indirect immunofluorescence analysis of endogenous Sm and coilin proteins. HeLa cells plated on glass coverslips were fixed at 5 h post-mock or poliovirus infection and costained with anti-Y12 and anti-coilin antibodies. (Left panels) Differential interference contrast (DIC) images. (Middle panels) Hoechst-stained nuclei (blue). Representative maximum-intensity projections of deconvolved serial sections show the localization of Sm proteins (green) and coilin (red). (B) The number of colocalized foci observed in uninfected or poliovirus-infected cells were quantified. The graph represents the analysis of 100 cells for each condition. The p-value from χ2 analyses was <0.0001.
Our data indicate that poliovirus infection induces a redistribution of Sm proteins and results in reduced nuclear colocalization of Sm proteins with coilin. In contrast, siRNA-mediated reduction of Gemin3 did not alter the colocalization of coilin with Sm proteins and did not cause the relocalization of Sm proteins to the cytoplasmic periphery of the nucleus. Therefore, it appears that poliovirus infection has a specific effect on the nuclear localization of Sm proteins. The relationship between these observations and cleavage of Gemin3 is not yet known.
Gemin3 is cleaved by poliovirus 2Apro
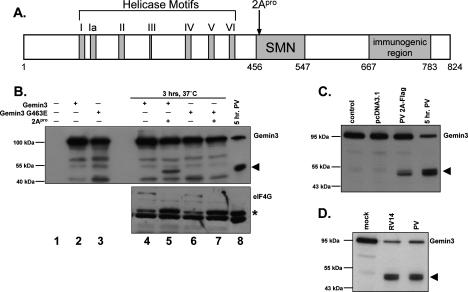
Having established a correlation between decreased levels of Gemin3 protein and reductions in Sm core assembly activity, we wished to identify the proteinase responsible for mediating Gemin3 cleavage during infection. As shown in Figure 3, Gemin3 cleavage requires amplification of viral RNA, viral proteins, or both. Infections performed in the presence of cycloheximide or actinomycin D revealed that Gemin3 cleavage required ongoing translation, but was independent of ongoing host transcription (Supplementary Fig. S4). Poliovirus encodes two proteinases, 2Apro and 3Cpro, known to specifically target certain host proteins (for review, see Lloyd 2006). Studies have defined specific consensus cleavage motifs for the 2A and 3C proteinases (Sommergruber et al. 1992, 1994; Lamphear et al. 1993; Tong 2002). Examination of the Gemin3 amino acid sequence revealed the presence of a potential 2Apro consensus cleavage site, VHTYG, where cleavage would be predicted to occur between amino acids Tyr462 and Gly463 (Fig. 6A, 2Apro). Cleavage at this position would result in a C-terminal fragment with a predicted size similar to that of the cleavage product observed during poliovirus infection (Fig. 3A). No obvious 3C proteinase consensus sites were found. We therefore explored the possibility that Gemin3 is targeted by 2Apro during poliovirus infection, and that cleavage occurs between Tyr462 and Gly463.
Figure 6.
Cleavage activity of picornaviral protease 2Apro in vitro and in vivo. (A) Schematic representation of the Gemin3 protein sequence. The seven conserved helicase motifs (I–VI), the SMN interaction domain, and the region recognized by the Gemin3 antibody used in this study are indicated. The predicted 2A proteinase cleavage site is designated by an arrow. Amino acid residues are indicated below. (B) Immunoblot analysis of 2A proteinase cleavage reactions using antibodies against Gemin3 (top panel) or eIF4G (bottom panel). Lanes 1–3 contain samples from translation reactions in rabbit reticulocyte lysate with no RNA, wild-type Gemin3 RNA, or mutant Gemin3 RNA (Gemin3-G463E), respectively. Purified coxsackievirus B3 2A protease (2Apro) was added to the indicated samples, and cleavage reactions were incubated for 3 h at 37°C. For comparison, Gemin3 levels in total extracts prepared from cells that were infected for 5 h with poliovirus are shown in lane 8. An arrowhead marks the Gemin3 cleavage product. As a control, the blot was also incubated with antibodies directed against eIF4G to demonstrate that the recombinant protease was active as predicted. Due to the large molecular weight of eIF4G, the full-length protein transfers poorly and can be seen as a faint band marked as eIF4G. An asterisk shows the position of the cleaved eIF4G protein. (C) Cleavage of Gemin3 in cultured cells by transiently expressed polioviral 2A proteinase. Extracts were prepared from cells treated with transfection reagent alone (control), an expression vector lacking 2A coding sequences (pcDNA3.1), or a vector expressing the poliovirus 2A protease gene (2A-Flag) at 24 h post-transfection. Total protein extracts (20 μg) were separated using SDS-PAGE and blotted with an antibody directed against Gemin3. For comparison, 20 μg of total protein obtained from cells infected for 5 h with poliovirus was included. The Gemin3 cleavage product is indicated by an arrowhead. (D) Effects of poliovirus and rhinovirus 14 infection on Gemin3 cleavage. Extracts were prepared from mock-, rhinovirus 14-, and poliovirus-infected HeLa cells. Each lane contains 20 μg of total protein, and the blot was probed with the Gemin3 antibody. An arrowhead indicates the cleavage product.
Previous analysis of translation factor eIF4G proteolysis by poliovirus 2Apro showed that mutation of the Gly486 cleavage site residue to glutamic acid reduced cleavage in vitro by ∼100-fold (Lamphear and Rhoads 1996). Thus, we introduced a single glycine-to-glutamic acid mutation into a human Gemin3 cDNA at the predicted position to generate a G463E mutant. Wild-type and mutated Gemin3 mRNAs were synthesized in vitro and added to rabbit reticulocyte translation lysates. Purified 2Apro from coxsackievirus B3, a picornavirus that is very closely related to poliovirus, was then added, and Gemin3 protein synthesis and susceptibility to cleavage monitored by immunoblot analysis (Fig. 6B). Addition of 2Apro to the translation reaction containing wild-type Gemin3 resulted in the appearance of a C-terminal cleavage product similar in size to that seen in poliovirus infection (Fig. 6B, lanes 5,8). In contrast, Gemin3 protein containing the Glu463 mutation was resistant to 2Apro cleavage (Fig. 6B, lane 7). Neither wild-type nor mutant Gemin3 protein was affected by incubation in the absence of protease (Fig. 6B, lanes 4,6). Control experiments in which the proteolysis of eIF4G by 2Apro was examined confirmed that the recombinant 2Apro was active in the cleavage reactions (Fig. 6B, bottom panel). These data indicate that Gemin3 can be cleaved by the 2A proteinase in vitro, most likely between Tyr462 and Gly463.
To test whether the polioviral 2A proteinase is able to cleave Gemin3 in tissue culture, cells were transfected with a plasmid that directs the expression of polioviral 2Apro. As shown in Figure 6C, expression of 2Apro in cultured cells led to the appearance of a cleavage product similar in size to that observed during poliovirus infection. Unfortunately, we were unable to express either wild-type or mutant G463E Gemin3 proteins in cultured cells to test whether this mutation would render the protein resistant to cleavage during infection. While Gemin3 cleavage was not as efficient upon expression of polioviral 2Apro compared with infection, these data support our hypothesis that Gemin3 is cleaved by the 2A proteinase.
Finally, we infected cells with rhinovirus, a related member of the Enterovirus genus. Rhinovirus encodes a 2Apro protein that has been shown to cleave eIF4G similarly to poliovirus 2Apro (Haghighat et al. 1996). Figure 6D shows that Gemin3 is cleaved during rhinovirus infection to produce a C-terminal cleavage product similar in size to that observed in poliovirus infection. Taken together with the in vitro cleavage assays and the expression of 2Apro in cells, these data argue that Gemin3 cleavage is mediated by the polioviral 2A proteinase during infection.
Discussion
Poliovirus is the causative agent of poliomyelitis, a neuromuscular disease resulting in muscle weakness and paralysis. Despite extensive characterization of the virus, the molecular mechanisms leading to its pathogenesis are unclear. In studies of SMA, an autosomal recessive disorder also resulting in neuromuscular degeneration, correlations have been found between disease severity and defects in spliceosomal snRNP biogenesis. Here we have shown that poliovirus infection results in reduced Sm core formation. Examination of SMN complex components and Sm proteins revealed the specific cleavage of Gemin3 during poliovirus infection, leading to a >75% decrease in the intracellular abundance of Gemin3 in infected cells. The core assembly defect observed in poliovirus-infected extracts appears to reflect a reduced capacity for core formation rather than alterations in the assembly or stability of snRNPs. A decrease in Sm core assembly was also observed upon siRNA-mediated knockdown of Gemin3. Comparison of the data obtained from virus infection and siRNA transfection suggests a correlation between Gemin3 protein levels and Sm core assembly activity. While no alteration in the colocalization of SMN and coilin was observed in poliovirus-infected cells, suggesting that the virus does not disrupt Cajal bodies, infection resulted in the relocalization of Sm proteins and reduced nuclear colocalization with coilin. These findings raise questions as to the specific role of Gemin3 in snRNP biogenesis and the consequences of its cleavage by poliovirus.
Recently, Shpargel and Matera (2005) reported that siRNA-mediated reduction in levels of SMN, Gemin2, Gemin3, and Gemin4 were each found to decrease Sm core assembly. Reduction in Gemin3 protein levels, in this case by 90%, resulted in the inhibition of Sm core assembly activity by ∼40% (Shpargel and Matera 2005). In a similar study, Feng et al. (2005) achieved an ∼30% siRNA-mediated reduction in Gemin3 protein levels, which had no significant effect on the efficiency of Sm core assembly. Thus, it is possible that Gemin3 levels must fall below a critical threshold, such as that observed during polioviral infection, to have a measurable effect on Sm core formation.
Treatment of cells with siRNAs directed against Gemin3 was previously observed to induce a reduction in Gemin4 protein levels (Feng et al. 2005; Shpargel and Matera 2005). We also noted a slight decrease in Gemin4 (Fig. 4) upon Gemin3 siRNA transfection. These observations suggest that interaction with Gemin3 is important for Gemin4 stability. However, no significant changes in Gemin4 levels were observed in poliovirus-infected cells at a time when most Gemin3 protein was proteolyzed (Fig. 3). This difference could be explained by a model in which proteolyzed Gemin3 fragments can still interact with Gemin4 independently or as part of the SMN complex. In the first case, association with either the N- or C-terminal Gemin3 cleavage products could stabilize Gemin4. Alternatively, the C-terminal fragment of Gemin3, which contains the SMN-binding site (Fig. 6A), could interact with Gemin4 and anchor it to the SMN complex (Gubitz et al. 2004; Carissimi et al. 2006), resulting in enhanced intracellular stability. Indeed, preliminary studies have shown that ectopically expressed, epitope-tagged C-terminal fragments of Gemin3 associate with SMN, Gemin2, and Gemin4 in cultured cells. Polioviral 2Apro-generated C-terminal fragments of Gemin3 were also observed to cosediment with SMN in fractions containing molecules larger than 20S (data not shown). These results argue that SMN complexes containing N-terminally truncated Gemin3 molecules accumulate in virus-infected cells, and that these complexes could be defective in Sm core assembly. This scenario appears unlikely because ectopic expression of the C-terminal Gemin3 proteolysis product did not inhibit core assembly activity (Supplementary Fig. S2). Overall, these findings point to important roles for the N-terminal ATPase, RNA helicase, and RNA interaction domains of Gemin3 (Fig. 6A) in Sm core assembly.
It is intriguing that an RNA virus whose replication cycle is entirely cytoplasmic encodes a proteinase that targets the spliceosomal Sm core assembly machinery. It is highly unlikely that picornaviruses need access to the splicing machinery in the infected host cell. However, it is possible that truncated Gemin3 molecules or altered SMN complexes have unsuspected regulatory functions in the viral replication cycle that are unrelated to the known roles of the SMN complex. To test this possibility, we measured viral yields in cells that ectopically expressed the C terminus of Gemin3. In comparison with control cells, no differences in viral yield were observed in this experiment (data not shown). Thus, the C terminus of Gemin3 does not appear to regulate viral gene expression in this experimental system. In addition, siRNA-mediated reduction of Gemin3 had no effect on poliovirus mRNA translation or viral yield (data not shown).
Our analyses suggest that cleavage of Gemin3 is also mediated by 2A proteases encoded by two other picornaviruses, coxsackievirus and rhinovirus, which do not cause specific motor neuron degeneration. It is possible that the tissue tropism of poliovirus could cause an snRNP assembly defect in motor neurons. Of course, Gemin3 proteolysis could also affect other cellular functions of Gemin3. For example, Gemin3 is known to interact with a variety of cellular transcription factors such as steriodogenic factor 1 (SF-1), early growth response proteins 1–4 (Ergs 1–4), and forkhead transcription factor FOXL2 (Ou et al. 2001; Gillian and Svaren 2004; Lee et al. 2005). Cleavage of Gemin3 may therefore alter the rate of formation or intracellular amounts of Gemin3–transcription factor complexes, and specifically affect transcription of particular host or viral genes in neuronal and nonneuronal cells. Additionally, Gemin3 was reported to be associated with Argonaute2 in a miRNP complex (Mourelatos et al. 2002). It is unlikely that Gemin3 cleavage by poliovirus affects its suggested role in miRNP function, because we found that endogenous let7a and miR122 microRNAs could still regulate reporter gene expression in infected cells at a time when Gemin3 was cleaved (data not shown). It cannot be excluded, however, that Gemin3 proteolysis could alter miRNP activity in a manner not detected by our analyses.
Poliovirus infection also resulted in altered distribution of Sm proteins and in reduced colocalization of Sm proteins to Cajal bodies (Fig. 5). Cytoplasmic accumulation of Sm proteins would not be surprising given the drastic reduction in Sm core assembly in infected cells. However, siRNA-mediated reduction of Gemin3, while also decreasing core formation, did not affect Sm protein distribution (Supplementary Fig. S3). Our data are in agreement with those of Shpargel and Matera (2005), who reported that reduced levels of Gemin3 and Gemin4 had no effect on Sm protein localization. Interestingly, Shpargel and Matera (2005) found that reduction of SMN protein levels, which led to the disruption of Cajal bodies, altered the localization of an overexpressed GFP-tagged SmB protein although endogenous Sm proteins were unaffected. They hypothesized that the differential effects were due to nuclear import of endogenous Sm proteins in the absence of core formation and the potential saturation of this alternative import pathway by GFP-SmB (Shpargel and Matera 2005). Poliovirus is known to inhibit multiple nuclear import pathways including the importin-α-mediated classical pathway (for review, see Gustin 2003). One hypothesis for the specific effects of poliovirus on the localization of Sm proteins is that the importin-β-mediated nuclear import of snRNPs occurs during infection, while the import of individual Sm proteins is blocked. The cytoplasmic tails of several Sm proteins have been found to contain classical nuclear import signals (Girard et al. 2004), suggesting that import in the absence of core formation could be mediated by the classical pathway inhibited in poliovirus infection.
Defects in snRNP biogenesis could lead to downstream effects on pre-mRNA splicing, which would be an efficient way for the virus to down-regulate the expression of genes that encode proteins involved in innate immune responses. While an overall inhibition of translation and transcription occurs after infection of cultured cells, it is clear that the turnover of SMN, Gemin2, Gemin4, and SmB/B′ is negligible in infected cells. Proteolysis of Gemin3 would therefore provide a mechanism to inhibit SMN complex-mediated snRNP formation. We found, however, that a transiently expressed reporter construct was efficiently spliced following siRNA-mediated reduction of Gemin3 (Supplemental Material; Supplementary Fig. S5) and upon expression of polioviral 2A proteinase (data not shown). The reporter precursor RNA was designed to be efficiently spliced, and thus might be insensitive to slight effects on spliceosomal activity. Somewhat curiously, splicing defects have not been observed in SMA patients or in a mouse model of the disease (Jablonka et al. 2000). Multiple roles in both neuron-specific and general cellular functions have been proposed for SMN (for review, see Briese et al. 2005). To support the hypothesis that neuronal degeneration is linked to reduced snRNP formation, Eggert et al. (2006) cite evidence that motor neurons have a high turnover rate of mRNAs encoding tissue-specific proteins, and therefore a subset of mRNAs might be uniquely susceptible to slight effects on the splicing machinery. A recent report, however, suggested that the motor neuron defects observed in SMA are not linked to the role of SMN in U snRNP biogenesis, because motor axon defects induced by reduction of endogenous SMN could be rescued by mutant SMN proteins defective in functions critical for Sm core formation (Carrel et al. 2006).
It is possible that analyses of U snRNP assembly defects induced by poliovirus could help to elucidate the role of reduced Sm core formation in motor neuron degeneration. Gemin3 cleavage also has the potential to alter host activities unconnected to its effects on snRNP assembly. Further studies to identify other cellular functions affected by Gemin3 cleavage will point to additional roles of this DEAD-box helicase protein in cellular pathways.
Materials and methods
Cell culture and viral infection
HeLa cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; GIBCO) supplemented with 10% fetal bovine serum (Omega Scientific), 100 U/mL penicillin-streptomycin (GIBCO), and 2 mM L-glutamine (GIBCO). For poliovirus infections, viral stocks were diluted in phosphate-buffered saline supplemented with 0.1 mg/mL CaCl2 plus 0.1 mg/mL MgCl2 (CPBS). Cells were washed once with CPBS and infected at a multiplicity of infection (MOI) of 50. Following incubation for 30 min at 37°C, cells were washed with CPBS and media was replaced. To inhibit viral replication, 2 mM guanidine-HCl (gua) was added with the media. Infections were allowed to proceed for 5 h at 37°C. Rhinovirus 14 infections were carried out as above with the following exceptions. Cells were infected at an MOI of <0.1, incubated at 32°C, and harvested at 26 h post-infection. Mock infections were carried out in parallel with CPBS alone.
Cell extract preparation
To prepare HeLa cell extracts, cells were harvested and washed once with cold PBS. Three pellet volumes of lysis buffer containing 10 mM Tris-HCl (pH 8), 60 mM KCl, 1 mM DTT, 0.3% NP-40, 1 mM EDTA, and a cocktail of Complete Protease Inhibitors (Roche) were added. Cells were incubated on ice for 5 min and sedimented at 2500 rpm for 5 min. The lysates were cleared by an additional 5 min sedimentation at 14,000 rpm. Total protein concentration was determined using the standard Bradford protein assay (Bio-Rad) according to the manufacturer’s instructions.
In vitro transcription of U snRNAs and Sm core assembly assays
32P-labeled U1 and U1-Δ snRNAs were transcribed in vitro using T7 polymerase (Promega) from vectors generously provided by Greg Matera (Case Western Reserve University, Cleveland, OH). Reactions containing 500 μM rATP, 500 μM rCTP, 100 μM rGTP, 100 μM rUTP, 50 μCi of [α-32P]UTP (Perkin-Elmer), 2 mM m7G cap analog (New England Biolabs), and RNasin (Promega) were incubated for 4 h at 37°C. Following DNase (Promega) treatment, transcripts were purified using RNeasy Mini Kits (Qiagen).
In vitro Sm core assembly (SCA) reaction mixtures included 20 mM HEPES-KOH (pH 7.9), 50 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 2.5 mM rATP, 0.01% NP-40, 0.5 mg/mL yeast tRNA, and 20 U of RNasin (Promega). Unless otherwise indicated, 40 μg of total protein and 100,000 counts of U snRNA were incubated in reaction mixtures for 20 min at 30°C. Protein extracts were diluted with lysis buffer such that equal volumes were added to each reaction. Following incubation, 1 mL of cold SCA Buffer (20 mM Tris-HCl at pH 7.5, 600 mM NaCl, 2.5 mM MgCl2, 0.01% NP-40) was added, and reactions were precleared by addition of Protein-G beads (Pierce) for 30 min at 4°C. Sm cores were immunoprecipitated with the monoclonal anti-Y12 antibody (Labvision) that detects epitopes in the B′/B and D species of Sm proteins (Hirakata et al. 1993). Beads were washed four times with SCA Buffer prior to boiling in FLB (80% formamide, 10 mM EDTA). A mix of bromophenol blue and xylene cyanol dye was added, and samples were separated on 5% TBE-urea gels. snRNA signals were quantitated using a Storm PhosphorImager (Molecular Dynamics). Data were analyzed and plotted using GraphPad Prism, version 4.00. To test the stability of the U1 snRNA, SCA reactions were performed as described above. Following immunoprecipitation, proteinase K was added at a final concentration of 0.2 mg/mL, and samples were incubated for 10 min at 50°C. RNA was phenol-chloroform-extracted from the supernatant and resolved on a 5% TBE-urea gel.
RNA and DNA transfections
Cells were plated in antibiotic-free media 24 h prior to transfection. All transfections were carried out using Lipofectamine-2000 (Invitrogen) according to the manufacturer’s instructions. For siRNA transfections, cells were treated with 500 nM Gemin3 siGENOME SMARTpool siRNAs (Dharmacon) or 500 nM GFP Duplex I siRNAs (Dharmacon), and the media was changed at 5 and 24 h post-transfection. Protein extracts were prepared 48 h post-transfection. For 2A protease expression, cells were transfected with an expression vector encoding the poliovirus 2A protease with a C-terminal Flag-tag (PAK12-2A; a generous gift from Kurt Gustin, University of Idaho, Moscow, ID). The media was changed after 5 h, and cells were harvested 24 h post-transfection.
Immunoblot analyses and antibodies
The following antibodies were used for immunoblot analysis: anti-SMN (BD Biosciences), anti-Gemin2 (Immuquest), anti-Gemin4 (Santa Cruz Biotechnology), anti-Y12 (Labvision), anti-actin (Santa Cruz Biotechnology), anti-eIF4G (BD Biosciences), and anti-DP103/Gemin3 (BD Biosciences), which recognizes the C-terminal fragment of Gemin3. Quantitative immunoblots were developed with an ECF Detection Kit (GE Healthcare Life Sciences), and images were quantitated using a Storm PhosphorImager (Molecular Dynamics). Results were graphed using GraphPad Prism, version 4.00. The figure images in Figures 3A, 4A, and 6 were obtained using an ECL Detection Kit (GE Healthcare Life Sciences).
Indirect immunofluorescence assays
For immunofluorescence analysis, HeLa cells were grown on glass coverslips coated with poly-L-lysine (Sigma-Aldrich). Poliovirus infections and siRNA transfections were carried out as described above. At 5 h post-infection or 48 h post-transfection, cells were fixed for 15 min with 4% paraformaldehyde (w/v) in PBS, washed three times in PBS, and permeabilized for 15 min using 0.5% Triton X-100 in PBS containing 1% fish gelatin (PBS/FG; Sigma-Aldrich). Following three washes in PBS/FG, primary antibodies diluted in PBS/FG were added to coverslips and allowed to incubate for 1 h. Cells were washed three times with PBS/FG, and secondary antibodies diluted in PBS/FG were applied. After 1 h of incubation, three PBS/FG washes were performed, and Hoechst dye was included in the second wash to stain the nuclei. Coverslips were mounted on glass slides using Fluormount-G (SouthernBiotech) and sealed with nail polish. All steps were carried out at room temperature. The primary antibodies used were monoclonal anti-SMN (BD Biosciences; dilution 1:1000), monoclonal anti-Y12 (Labvision; dilution 1:1000), and polyclonal anti-coilin (generous gift from Greg Matera; dilution 1:200). The secondary antibodies used were Alexa-Fluor 488 donkey anti-mouse and Alexa-Fluor 647 donkey anti-rabbit (Molecular Probes; dilution 1:200). Fluorescence microscopy was carried out using a Zeiss Axiovert 200M inverted microscope (100× oil objective; Zeiss). Images were collected with a Hamamatsu ORCA-ER digital camera (Hamamatsu Photonics) using OpenLab software (Improvision). Serial z-sections were obtained in 0.2-μm intervals following acquisition of differential phase contrast (DIC) and nuclear staining images. Vertical stacks were deconvolved with Volocity software (Improvision) to remove out-of-focus light, and four images were used to obtain a 3D reconstruction. Images were further processed using Adobe Photoshop.
In vitro cleavage assays
A plasmid containing the human Gemin3 sequence with a C-terminal HA-tag (pcDNA-Gemin3) was generated from total HeLa cell RNA using an Advantage 2 PCR Kit (Clontech) with the following primers: Gemin3-5′ KpnI (GACGGTAC CATGGCGGCGGCATTTGAAG) and Gemin3-3′ HA-XhoI (GACCTCGCGTTATCAAGCGTAATCTGGAACATCGTATG GGTACATCTGGTTACTATGCATTTGT). PCR products were cloned into TOPO-pCR2.1 (Invitrogen) and sequenced. The Gemin3-HA sequence was then cloned into pcDNA3.1 Zeo+ (Invitrogen) using the KpnI and XhoI restriction sites. A mutant Gemin3 plasmid, containing a glycine-to-glutamic acid change at position 463 (pcDNA-G463E), was generated from pcDNA-Gemin3 using Gemin3 G463E Forward primers (GCT GCTGTGCATACATATGAAATAGCAAGTGTACCTAACC) and Gemin3 G463E Reverse primers (GGTTAGGTACACTT GCTATTTCATATGTATGCACAGCAGC) with the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene).
Capped transcripts were produced in vitro from DraIII-linearized pcDNA-Gemin3 and pcDNA-G463E plasmids using RiboMax T7 (Promega) according to the manufacturer’s protocol. Reactions were incubated for 4 h at 37°C. Following treatment with DNase (Promega), transcripts were purified using RNeasy Mini Kits (Qiagen). In vitro translation reactions were carried out in nuclease-treated rabbit reticulocyte lysate (Promega) according to the manufacturer’s protocol. Reactions were incubated for 90 min at 30°C. One microliter of coxsackievirus B3 2A proteinase (0.5 mg/mL; Richard Lloyd, Baylor College of Medicine, Houston, TX) was added to 8 μL of the in vitro translation reaction and incubated for 3 h at 37°C. Gemin3 cleavage was monitored by immunoblot analysis.
Acknowledgments
We are grateful to Karla Kirkegaard and Cara Pager for critical reading of the manuscript. We are especially thankful to Greg Matera and Richard Lloyd for reagents. This work was supported by National Institutes of Health Grants T32 GM07276 (to L.L.A.) and R01 069007 (to P.S.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1535607
References
- Baccon J., Pellizzoni L., Rappsilber J., Mann M., Dreyfuss G., Pellizzoni L., Rappsilber J., Mann M., Dreyfuss G., Rappsilber J., Mann M., Dreyfuss G., Mann M., Dreyfuss G., Dreyfuss G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J. Biol. Chem. 2002;277:31957–31962. doi: 10.1074/jbc.M203478200. [DOI] [PubMed] [Google Scholar]
- Briese M., Esmaeili B., Sattelle D.B., Esmaeili B., Sattelle D.B., Sattelle D.B. Is spinal muscular atrophy the result of defects in motor neuron processes? Bioessays. 2005;27:946–957. doi: 10.1002/bies.20283. [DOI] [PubMed] [Google Scholar]
- Buhler D., Raker V., Luhrmann R., Fischer U., Raker V., Luhrmann R., Fischer U., Luhrmann R., Fischer U., Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: Implications for spinal muscular atrophy. Hum. Mol. Genet. 1999;8:2351–2357. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- Carissimi C., Saieva L., Baccon J., Chiarella P., Maiolica A., Sawyer A., Rappsilber J., Pellizzoni L., Saieva L., Baccon J., Chiarella P., Maiolica A., Sawyer A., Rappsilber J., Pellizzoni L., Baccon J., Chiarella P., Maiolica A., Sawyer A., Rappsilber J., Pellizzoni L., Chiarella P., Maiolica A., Sawyer A., Rappsilber J., Pellizzoni L., Maiolica A., Sawyer A., Rappsilber J., Pellizzoni L., Sawyer A., Rappsilber J., Pellizzoni L., Rappsilber J., Pellizzoni L., Pellizzoni L. Gemin8 is a novel component of the survival motor neuron complex and functions in small nuclear ribonucleoprotein assembly. J. Biol. Chem. 2006;281:8126–8134. doi: 10.1074/jbc.M512243200. [DOI] [PubMed] [Google Scholar]
- Carrel T.L., McWhorter M.L., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E., McWhorter M.L., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E., Bassell G.J., Burghes A.H., Beattie C.E., Burghes A.H., Beattie C.E., Beattie C.E. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J. Neurosci. 2006;26:11014–11022. doi: 10.1523/JNEUROSCI.1637-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B., Pellizzoni L., Perkinson R.A., Shevchenko A., Mann M., Dreyfuss G., Pellizzoni L., Perkinson R.A., Shevchenko A., Mann M., Dreyfuss G., Perkinson R.A., Shevchenko A., Mann M., Dreyfuss G., Shevchenko A., Mann M., Dreyfuss G., Mann M., Dreyfuss G., Dreyfuss G. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B., Pellizzoni L., Perkinson R.A., Yong J., Shevchenko A., Mann M., Dreyfuss G., Pellizzoni L., Perkinson R.A., Yong J., Shevchenko A., Mann M., Dreyfuss G., Perkinson R.A., Yong J., Shevchenko A., Mann M., Dreyfuss G., Yong J., Shevchenko A., Mann M., Dreyfuss G., Shevchenko A., Mann M., Dreyfuss G., Mann M., Dreyfuss G., Dreyfuss G. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H., Androphy E.J., Prior T.W., Burghes A.H., Prior T.W., Burghes A.H., Burghes A.H. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- Eggert C., Chari A., Laggerbauer B., Fischer U., Chari A., Laggerbauer B., Fischer U., Laggerbauer B., Fischer U., Fischer U. Spinal muscular atrophy: The RNP connection. Trends Mol. Med. 2006;12:113–121. doi: 10.1016/j.molmed.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Feng W., Gubitz A.K., Wan L., Battle D.J., Dostie J., Golembe T.J., Dreyfuss G., Gubitz A.K., Wan L., Battle D.J., Dostie J., Golembe T.J., Dreyfuss G., Wan L., Battle D.J., Dostie J., Golembe T.J., Dreyfuss G., Battle D.J., Dostie J., Golembe T.J., Dreyfuss G., Dostie J., Golembe T.J., Dreyfuss G., Golembe T.J., Dreyfuss G., Dreyfuss G. Gemins modulate the expression and activity of the SMN complex. Hum. Mol. Genet. 2005;14:1605–1611. doi: 10.1093/hmg/ddi168. [DOI] [PubMed] [Google Scholar]
- Fischer U., Liu Q., Dreyfuss G., Liu Q., Dreyfuss G., Dreyfuss G. The SMN–SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- Frugier T., Nicole S., Cifuentes-Diaz C., Melki J., Nicole S., Cifuentes-Diaz C., Melki J., Cifuentes-Diaz C., Melki J., Melki J. The molecular bases of spinal muscular atrophy. Curr. Opin. Genet. Dev. 2002;12:294–298. doi: 10.1016/s0959-437x(02)00301-5. [DOI] [PubMed] [Google Scholar]
- Gillian A.L., Svaren J., Svaren J. The Ddx20/DP103 dead box protein represses transcriptional activation by Egr2/Krox-20. J. Biol. Chem. 2004;279:9056–9063. doi: 10.1074/jbc.M309308200. [DOI] [PubMed] [Google Scholar]
- Girard C., Mouaikel J., Neel H., Bertrand E., Bordonne R., Mouaikel J., Neel H., Bertrand E., Bordonne R., Neel H., Bertrand E., Bordonne R., Bertrand E., Bordonne R., Bordonne R. Nuclear localization properties of a conserved protuberance in the Sm core complex. Exp. Cell Res. 2004;299:199–208. doi: 10.1016/j.yexcr.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Golembe T.J., Yong J., Dreyfuss G., Yong J., Dreyfuss G., Dreyfuss G. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol. Cell. Biol. 2005;25:10989–11004. doi: 10.1128/MCB.25.24.10989-11004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubitz A.K., Mourelatos Z., Abel L., Rappsilber J., Mann M., Dreyfuss G., Mourelatos Z., Abel L., Rappsilber J., Mann M., Dreyfuss G., Abel L., Rappsilber J., Mann M., Dreyfuss G., Rappsilber J., Mann M., Dreyfuss G., Mann M., Dreyfuss G., Dreyfuss G. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 2002;277:5631–5636. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- Gubitz A.K., Feng W., Dreyfuss G., Feng W., Dreyfuss G., Dreyfuss G. The SMN complex. Exp. Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Gustin K.E. Inhibition of nucleo-cytoplasmic trafficking by RNA viruses: Targeting the nuclear pore complex. Virus Res. 2003;95:35–44. doi: 10.1016/S0168-1702(03)00165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat A., Svitkin Y., Novoa I., Kuechler E., Skern T., Sonenberg N., Svitkin Y., Novoa I., Kuechler E., Skern T., Sonenberg N., Novoa I., Kuechler E., Skern T., Sonenberg N., Kuechler E., Skern T., Sonenberg N., Skern T., Sonenberg N., Sonenberg N. The eIF4G–eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol. 1996;70:8444–8450. doi: 10.1128/jvi.70.12.8444-8450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert M.D., Szymczyk P.W., Shpargel K.B., Matera A.G., Szymczyk P.W., Shpargel K.B., Matera A.G., Shpargel K.B., Matera A.G., Matera A.G. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes & Dev. 2001;15:2720–2729. doi: 10.1101/gad.908401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakata M., Craft J., Hardin J.A., Craft J., Hardin J.A., Hardin J.A. Autoantigenic epitopes of the B and D polypeptides of the U1 snRNP. Analysis of domains recognized by the Y12 monoclonal anti-Sm antibody and by patient sera. J. Immunol. 1993;150:3592–3601. [PubMed] [Google Scholar]
- Jablonka S., Schrank B., Kralewski M., Rossoll W., Sendtner M., Schrank B., Kralewski M., Rossoll W., Sendtner M., Kralewski M., Rossoll W., Sendtner M., Rossoll W., Sendtner M., Sendtner M. Reduced survival motor neuron (Smn) gene dose in mice leads to motor neuron degeneration: An animal model for spinal muscular atrophy type III. Hum. Mol. Genet. 2000;9:341–346. doi: 10.1093/hmg/9.3.341. [DOI] [PubMed] [Google Scholar]
- Kiss T. Biogenesis of small nuclear RNPs. J. Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- Lamphear B.J., Rhoads R.E., Rhoads R.E. A single amino acid change in protein synthesis initiation factor 4G renders cap-dependent translation resistant to picornaviral 2A proteases. Biochemistry. 1996;35:15726–15733. doi: 10.1021/bi961864t. [DOI] [PubMed] [Google Scholar]
- Lamphear B.J., Yan R., Yang F., Waters D., Liebig H.D., Klump H., Kuechler E., Skern T., Rhoads R.E., Yan R., Yang F., Waters D., Liebig H.D., Klump H., Kuechler E., Skern T., Rhoads R.E., Yang F., Waters D., Liebig H.D., Klump H., Kuechler E., Skern T., Rhoads R.E., Waters D., Liebig H.D., Klump H., Kuechler E., Skern T., Rhoads R.E., Liebig H.D., Klump H., Kuechler E., Skern T., Rhoads R.E., Klump H., Kuechler E., Skern T., Rhoads R.E., Kuechler E., Skern T., Rhoads R.E., Skern T., Rhoads R.E., Rhoads R.E. Mapping the cleavage site in protein synthesis initiation factor eIF-4 γ of the 2A proteases from human Coxsackievirus and rhinovirus. J. Biol. Chem. 1993;268:19200–19203. [PubMed] [Google Scholar]
- Lee K., Pisarska M.D., Ko J.J., Kang Y., Yoon S., Ryou S.M., Cha K.Y., Bae J., Pisarska M.D., Ko J.J., Kang Y., Yoon S., Ryou S.M., Cha K.Y., Bae J., Ko J.J., Kang Y., Yoon S., Ryou S.M., Cha K.Y., Bae J., Kang Y., Yoon S., Ryou S.M., Cha K.Y., Bae J., Yoon S., Ryou S.M., Cha K.Y., Bae J., Ryou S.M., Cha K.Y., Bae J., Cha K.Y., Bae J., Bae J. Transcriptional factor FOXL2 interacts with DP103 and induces apoptosis. Biochem. Biophys. Res. Commun. 2005;336:876–881. doi: 10.1016/j.bbrc.2005.08.184. [DOI] [PubMed] [Google Scholar]
- Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., Benichou B., Cruaud C., Millasseau P., Zeviani M., Cruaud C., Millasseau P., Zeviani M., Millasseau P., Zeviani M., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lloyd R.E. Translational control by viral proteinases. Virus Res. 2006;119:76–88. doi: 10.1016/j.virusres.2005.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson C.L., Hahnen E., Androphy E.J., Wirth B., Hahnen E., Androphy E.J., Wirth B., Androphy E.J., Wirth B., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Buhler D., Pillai R., Lottspeich F., Fischer U., Buhler D., Pillai R., Lottspeich F., Fischer U., Pillai R., Lottspeich F., Fischer U., Lottspeich F., Fischer U., Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- Monani U.R. Spinal muscular atrophy: A deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48:885–896. doi: 10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z., Dostie J., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G., Dostie J., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G., Abel L., Rappsilber J., Mann M., Dreyfuss G., Rappsilber J., Mann M., Dreyfuss G., Mann M., Dreyfuss G., Dreyfuss G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S., Wimmer E., Cello J., Wimmer E., Cello J., Cello J. Poliovirus and poliomyelitis: A tale of guts, brains, and an accidental event. Virus Res. 2005;111:175–193. doi: 10.1016/j.virusres.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Otter S., Grimmler M., Neuenkirchen N., Chari A., Sickmann A., Fischer U., Grimmler M., Neuenkirchen N., Chari A., Sickmann A., Fischer U., Neuenkirchen N., Chari A., Sickmann A., Fischer U., Chari A., Sickmann A., Fischer U., Sickmann A., Fischer U., Fischer U. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J. Biol. Chem. 2007;282:5825–5833. doi: 10.1074/jbc.M608528200. [DOI] [PubMed] [Google Scholar]
- Ou Q., Mouillet J.F., Yan X., Dorn C., Crawford P.A., Sadovsky Y., Mouillet J.F., Yan X., Dorn C., Crawford P.A., Sadovsky Y., Yan X., Dorn C., Crawford P.A., Sadovsky Y., Dorn C., Crawford P.A., Sadovsky Y., Crawford P.A., Sadovsky Y., Sadovsky Y. The DEAD box protein DP103 is a regulator of steroidogenic factor-1. Mol. Endocrinol. 2001;15:69–79. doi: 10.1210/mend.15.1.0580. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Baccon J., Rappsilber J., Mann M., Dreyfuss G., Baccon J., Rappsilber J., Mann M., Dreyfuss G., Rappsilber J., Mann M., Dreyfuss G., Mann M., Dreyfuss G., Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 2002;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- Raker V.A., Hartmuth K., Kastner B., Luhrmann R., Hartmuth K., Kastner B., Luhrmann R., Kastner B., Luhrmann R., Luhrmann R. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol. 1999;19:6554–6565. doi: 10.1128/mcb.19.10.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel K.B., Matera A.G., Matera A.G. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl. Acad. Sci. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J.E., Lamond A.I., Lamond A.I. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 1999;9:1065–1074. doi: 10.1016/s0960-9822(99)80475-8. [DOI] [PubMed] [Google Scholar]
- Sommergruber W., Ahorn H., Zophel A., Maurer-Fogy I., Fessl F., Schnorrenberg G., Liebig H.D., Blaas D., Kuechler E., Skern T., Ahorn H., Zophel A., Maurer-Fogy I., Fessl F., Schnorrenberg G., Liebig H.D., Blaas D., Kuechler E., Skern T., Zophel A., Maurer-Fogy I., Fessl F., Schnorrenberg G., Liebig H.D., Blaas D., Kuechler E., Skern T., Maurer-Fogy I., Fessl F., Schnorrenberg G., Liebig H.D., Blaas D., Kuechler E., Skern T., Fessl F., Schnorrenberg G., Liebig H.D., Blaas D., Kuechler E., Skern T., Schnorrenberg G., Liebig H.D., Blaas D., Kuechler E., Skern T., Liebig H.D., Blaas D., Kuechler E., Skern T., Blaas D., Kuechler E., Skern T., Kuechler E., Skern T., Skern T. Cleavage specificity on synthetic peptide substrates of human rhinovirus 2 proteinase 2A. J. Biol. Chem. 1992;267:22639–22644. [PubMed] [Google Scholar]
- Sommergruber W., Ahorn H., Klump H., Seipelt J., Zoephel A., Fessl F., Krystek E., Blaas D., Kuechler E., Liebig H.D., Ahorn H., Klump H., Seipelt J., Zoephel A., Fessl F., Krystek E., Blaas D., Kuechler E., Liebig H.D., Klump H., Seipelt J., Zoephel A., Fessl F., Krystek E., Blaas D., Kuechler E., Liebig H.D., Seipelt J., Zoephel A., Fessl F., Krystek E., Blaas D., Kuechler E., Liebig H.D., Zoephel A., Fessl F., Krystek E., Blaas D., Kuechler E., Liebig H.D., Fessl F., Krystek E., Blaas D., Kuechler E., Liebig H.D., Krystek E., Blaas D., Kuechler E., Liebig H.D., Blaas D., Kuechler E., Liebig H.D., Kuechler E., Liebig H.D., Liebig H.D., et al. 2A proteinases of coxsackie- and rhinovirus cleave peptides derived from eIF-4 γ via a common recognition motif. Virology. 1994;198:741–745. doi: 10.1006/viro.1994.1089. [DOI] [PubMed] [Google Scholar]
- Tong L. Viral proteases. Chem. Rev. 2002;102:4609–4626. doi: 10.1021/cr010184f. [DOI] [PubMed] [Google Scholar]
- Wan L., Battle D.J., Yong J., Gubitz A.K., Kolb S.J., Wang J., Dreyfuss G., Battle D.J., Yong J., Gubitz A.K., Kolb S.J., Wang J., Dreyfuss G., Yong J., Gubitz A.K., Kolb S.J., Wang J., Dreyfuss G., Gubitz A.K., Kolb S.J., Wang J., Dreyfuss G., Kolb S.J., Wang J., Dreyfuss G., Wang J., Dreyfuss G., Dreyfuss G. The survival of motor neurons protein determines the capacity for snRNP assembly: Biochemical deficiency in spinal muscular atrophy. Mol. Cell. Biol. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidman M.K., Yalamanchili P., Ng B., Tsai W., Dasgupta A., Yalamanchili P., Ng B., Tsai W., Dasgupta A., Ng B., Tsai W., Dasgupta A., Tsai W., Dasgupta A., Dasgupta A. Poliovirus 3C protease-mediated degradation of transcriptional activator p53 requires a cellular activity. Virology. 2001;291:260–271. doi: 10.1006/viro.2001.1215. [DOI] [PubMed] [Google Scholar]
- Winkler C., Eggert C., Gradl D., Meister G., Giegerich M., Wedlich D., Laggerbauer B., Fischer U., Eggert C., Gradl D., Meister G., Giegerich M., Wedlich D., Laggerbauer B., Fischer U., Gradl D., Meister G., Giegerich M., Wedlich D., Laggerbauer B., Fischer U., Meister G., Giegerich M., Wedlich D., Laggerbauer B., Fischer U., Giegerich M., Wedlich D., Laggerbauer B., Fischer U., Wedlich D., Laggerbauer B., Fischer U., Laggerbauer B., Fischer U., Fischer U. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes & Dev. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J., Golembe T.J., Battle D.J., Pellizzoni L., Dreyfuss G., Golembe T.J., Battle D.J., Pellizzoni L., Dreyfuss G., Battle D.J., Pellizzoni L., Dreyfuss G., Pellizzoni L., Dreyfuss G., Dreyfuss G. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol. Cell. Biol. 2004;24:2747–2756. doi: 10.1128/MCB.24.7.2747-2756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]