Abstract
Standard methods for the detection of enteroviruses in environmental samples involve the use of cell culture, which is expensive and time-consuming. The polymerase chain reaction (PCR) is an attractive method for the detection of enteroviruses in water because primary cell culture is not needed and the increased sensitivity of PCR allows detection of the low numbers of target DNAs and RNAs usually found in environmental samples. However, environmental samples often contain substances that inhibit PCR amplification of target DNA and RNA. Procedures that remove substances that interfere with the amplification process need to be developed if PCR is to be successfully applied to environmental samples. An RNA-PCR assay for the detection of enteroviruses in water was developed and used to test a variety of groundwater concentrates and humic acid solutions seeded with poliovirus type 1. The groundwater samples and humic acid solutions were treated with Sephadex G-50, Sephadex G-100, Sephadex G-200, Chelex-100 resin, and a mixed bed resin to remove PCR-inhibitory material from the samples. Sephadex G-100 in combination with Chelex-100 was found to be very effective in removing inhibitory factors for the detection of enteroviruses in groundwater concentrates by PCR. Viruses were detected in two of the groundwater concentrates by the RNA-PCR assay after treatment with Sephadex G-100 plus Chelex-100. This was confirmed by tissue culture, suggesting that the treatment protocol and, subsequently, the RNA-PCR assay are applicable for the detection of enteroviruses in environmental samples.
Full text
PDF


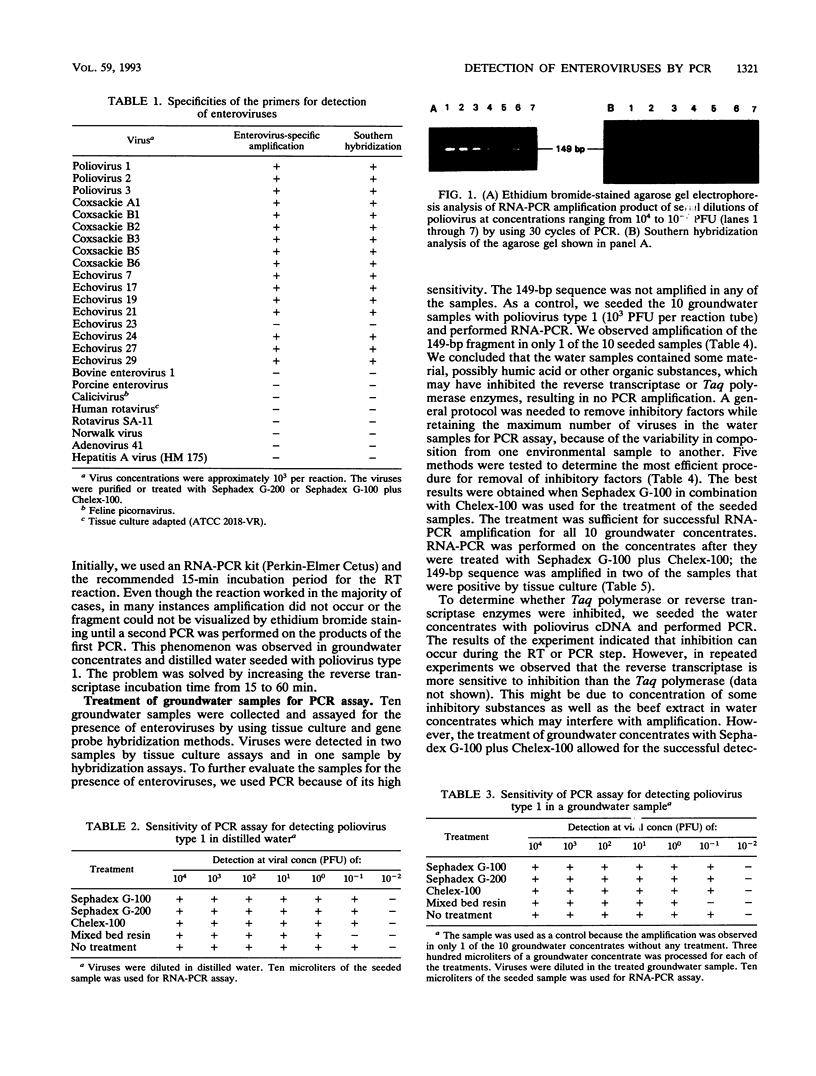


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Guttman-Bass N., Catalano-Sherman J. Effects of humic materials on virus recovery from water. Appl Environ Microbiol. 1985 May;49(5):1260–1264. doi: 10.1128/aem.49.5.1260-1264.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman-Bass N., Catalano-Sherman J. Humic acid interference with virus recovery by electropositive microporous filters. Appl Environ Microbiol. 1986 Sep;52(3):556–561. doi: 10.1128/aem.52.3.556-561.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hyypiä T., Auvinen P., Maaronen M. Polymerase chain reaction for human picornaviruses. J Gen Virol. 1989 Dec;70(Pt 12):3261–3268. doi: 10.1099/0022-1317-70-12-3261. [DOI] [PubMed] [Google Scholar]
- Hyypiä T., Maaronen M., Auvinen P., Stålhandske P., Pettersson U., Stanway G., Hughes P., Ryan M., Almond J., Stenvik M. Nucleic acid sequence relationships between enterovirus serotypes. Mol Cell Probes. 1987 Jun;1(2):169–176. doi: 10.1016/0890-8508(87)90025-9. [DOI] [PubMed] [Google Scholar]
- Katzenelson E., Fattal B., Hostovesky T. Organic flocculation: an efficient second-step concentration method for the detection of viruses in tap water. Appl Environ Microbiol. 1976 Oct;32(4):638–639. doi: 10.1128/aem.32.4.638-639.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keswick B. H., Gerba C. P., DuPont H. L., Rose J. B. Detection of enteric viruses in treated drinking water. Appl Environ Microbiol. 1984 Jun;47(6):1290–1294. doi: 10.1128/aem.47.6.1290-1294.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payment P. Isolation of viruses from drinking water at the Point-Viau water treatment plant. Can J Microbiol. 1981 Apr;27(4):417–420. doi: 10.1139/m81-063. [DOI] [PubMed] [Google Scholar]
- Pillai S. D., Josephson K. L., Bailey R. L., Gerba C. P., Pepper I. L. Rapid method for processing soil samples for polymerase chain reaction amplification of specific gene sequences. Appl Environ Microbiol. 1991 Aug;57(8):2283–2286. doi: 10.1128/aem.57.8.2283-2286.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rivera V. M., Welsh J. D., Maizel J. V., Jr Comparative sequence analysis of the 5' noncoding region of the enteroviruses and rhinoviruses. Virology. 1988 Jul;165(1):42–50. doi: 10.1016/0042-6822(88)90656-3. [DOI] [PubMed] [Google Scholar]
- Rotbart H. A. Enzymatic RNA amplification of the enteroviruses. J Clin Microbiol. 1990 Mar;28(3):438–442. doi: 10.1128/jcm.28.3.438-442.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Schmidt N. J., Ho H. H., Riggs J. L., Lennette E. H. Comparative sensitivity of various cell culture systems for isolation of viruses from wastewater and fecal samples. Appl Environ Microbiol. 1978 Sep;36(3):480–486. doi: 10.1128/aem.36.3.480-486.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobsey M. D., Hickey A. R. Effects of humic and fulvic acids on poliovirus concentration from water by microporous filtration. Appl Environ Microbiol. 1985 Feb;49(2):259–264. doi: 10.1128/aem.49.2.259-264.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy S. A comparison of genomic homologies among the coxsackievirus B group: use of fragments of the cloned coxsackievirus B3 genome as probes. J Gen Virol. 1984 Dec;65(Pt 12):2167–2172. doi: 10.1099/0022-1317-65-12-2167. [DOI] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl Environ Microbiol. 1992 Feb;58(2):754–757. doi: 10.1128/aem.58.2.754-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn J. M., Landry E. F., Baranosky L. J., Beckwith C. A., Dahl M. C., Delihas N. C. Survey of human virus occurrence in wastewater-recharged groundwater on Long Island. Appl Environ Microbiol. 1978 Jul;36(1):47–51. doi: 10.1128/aem.36.1.47-51.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]



