Abstract
The plant immune response known as systemic acquired resistance (SAR) is a general defense mechanism that confers long-lasting resistance against a broad spectrum of pathogens. SAR triggers many molecular changes including accumulation of antimicrobial pathogenesis-related (PR) proteins. Transcription of PR genes in Arabidopsis is regulated by the coactivator NPR1 and the repressor SNI1. Pathogen infection also triggers an increase in somatic DNA recombination, which results in transmission of changes to the offspring of infected plants. However, it is not known how the induction of homologous recombination during SAR is controlled. Here, we show that SNI1 and RAD51D regulate both gene expression and DNA recombination. In a genetic screen for suppressors of sni1, we discovered that RAD51D is required for NPR1-independent PR gene expression. As a result, the rad51d mutant has enhanced disease susceptibility. Besides altered PR gene expression, rad51d plants are hypersensitive to DNA-damaging agents and are impaired in homologous recombination. The dual role of RAD51D and SNI1 in PR gene transcription and DNA recombination suggests a mechanistic link between the short-term defense response and a long-term survival strategy.
Keywords: NPR1, pathogenesis-related gene expression, systemic acquired resistance
Combating infection is a never-ending struggle for all organisms. Different immune strategies have evolved to protect individual organisms and ensure long-term survival of the species. Plant genomes have clusters of resistance (R) genes encoding R proteins that recognize specific pathogenic signals and trigger a hypersensitive response to confer resistance (1) (see summary in Fig. 1). Concomitant with the hypersensitive response at the site of infection is the induction of systemic acquired resistance (SAR), which restricts the spread of the pathogen and prevents infection of systemic tissues (2). Pathogen infection can also lead to a systemic increase in somatic DNA recombination, which leads to heritable changes in the progeny of infected plants (3, 4). However, it is unknown whether and how such a strategy, which may create new adaptive traits for long-term survival (5), is associated with the regulatory mechanism of SAR.
Fig. 1.
A simplified model for plant defense signaling. Plant immunity is determined by the hundreds of resistance (R1, R2, R3 …) genes that are present in clusters in the genome (hollow and filled tandem arrows) (27). When an R protein recognizes a specific signal produced by the pathogen, it can trigger a series of rapid physiological responses to restrict pathogen growth. These local resistance responses also result in an increase in SA levels through an unknown systemic signal(s) (2). SA is necessary and sufficient for the induction of defense-related genes and systemic resistance to a broad-spectrum of pathogens. NPR1 is required for SA signaling (7). In the npr1 mutant, SA-induced gene expression and resistance is completely abolished. SA controls the translocation of NPR1 to the nucleus where it serves as a cofactor of TGA transcription factors (TFTGAs). NPR1 is also proposed to inactivate the transcriptional repressor SNI1, as the sni1 mutation can suppress the npr1 phenotype. In the sni1 single mutant, NPR1-dependent genes are specifically derepressed and induced-state chromatin modification (red dots) was observed at a defense gene (PR1) promoter (9). Because in the sni1 and sni1 npr1 mutants, SA is still required for full induction, there must be signaling components, including RAD51D, whose activities depend on SA but not NPR1. RAD51D activity makes the chromosome more accessible for transcription and homologous recombination. Defense-associated homologous DNA recombination may result in generation of new R genes (R4) and new pathogen recognition capability.
The hallmark of SAR is the coordinated induction of pathogenesis-related (PR) genes, which encode vacuole-targeted or secreted proteins with antimicrobial activities (6). In Arabidopsis, PR gene induction requires the signal molecule salicylic acid (SA) and the transcriptional coactivator NPR1 (nonexpresser of PR genes 1, also known as NIM1) (2, 7) (Fig. 1). In the npr1 mutant, SA-induced PR gene expression and SAR are completely abolished. The phenotype of npr1 can be suppressed by a recessive mutation in a transcriptional repressor SNI1 (suppressor of NPR1 inducible 1) (8). In the sni1 npr1 double mutant, PR gene induction and SAR are restored. In the absence of SAR induction, SNI1 prevents PR gene expression. Loss of SNI1 function causes preferential derepression of NPR1-dependent genes (9). However, the sni1 and sni1 npr1 mutants still require SA or its analogues INA (2,6-dichloroisonicotinic acid) or BTH (benzothiadiazole S-methyl ester) to fully induce PR genes and SAR, albeit at much lower concentrations than WT (8) (Fig. 2 B–D). This finding indicates that, besides NPR1 and SNI1, there are additional SA-dependent regulatory components controlling PR gene expression, whose functions are revealed only in the sni1 and sni1 npr1 mutants. To identify these components, we carried out a genetic screen for suppressors of sni1. Here, we describe the ssn1 (suppressor of sni1 1) mutant, which was found to have a recessive mutation in the Arabidopsis RAD51D gene; therefore, we refer to it as rad51d. The rad51d mutation abolished not only the enhanced PR gene expression but also the high homologous DNA recombination observed in sni1, suggesting that these processes are coregulated by SNI1 and RAD51D.
Fig. 2.
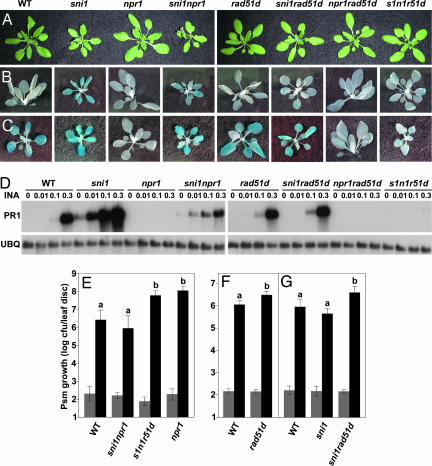
The rad51d mutant is a suppressor of sni1 and is impaired in disease resistance. (A) Morphological phenotypes of 4-week-old, soil-grown WT and mutant plants. (B and C) Expression of BGL2:GUS in untreated plants (B) or plants sprayed with 0.3 mM BTH 2 days previously (C). (D) PR1 gene expression in WT and mutant plants. (E–G) Growth of Psm ES4326 in WT and mutant plants. (E) Leaf discs were collected from six plants for day 0 (gray bars) and 10–12 plants for day 3. Error bars represent 95% confidence intervals of log-transformed data. The data were analyzed by Student's t test. (F and G) Leaf discs were collected from 4–16 plants for day 0 (gray bars) and 8–16 plants for day 3 (black bars). The data were analyzed by two-way ANOVA. Results show means for five pooled experiments (n = 60 plants per genotype; ANOVA, genotype: P < 0.0001; experiment: P < 0.0001; genotype × experiment: P = 0.5255) (F) and three pooled experiments (n = 28 plants per genotype; ANOVA, genotype: P < 0.0001; experiment: P < 0.0001; genotype × experiment: P = 0.3645) (G). Letters above bars indicate statistically significant differences between genotypes (Bonferroni correction, P < 0.01).
Results
Mutation of RAD51D Suppresses the sni1 and sni1 npr1 Mutant Phenotypes.
Derepression of genes, predominantly NPR1-dependent genes, in the sni1 and sni1 npr1 mutants results in growth retardation (8, 9). The mutant plants are smaller than WT and have narrower leaves (Fig. 2A). We used this phenotype to screen for suppressors of sni1. Full details of the genetic screen are given in supporting information (SI) Text. As shown in Fig. 2A, rad51d restored WT morphology to both sni1 and sni1 npr1. To determine whether rad51d affects PR gene transcription, we examined the expression of an SA-responsive reporter gene BGL2:GUS, a fusion of the BGL2 (PR2) promoter with the Escherichia coli uidA gene encoding the β-glucuronidase (GUS) enzyme. GUS activity was assayed by using histochemical staining, resulting in a blue color. The low constitutive GUS activity detected in sni1 and sni1 npr1 was eliminated in sni1 rad51d and sni1 npr1 rad51d (Fig. 2B). We hypothesized that the NPR1-independent PR gene induction in sni1 npr1 would also be abolished in sni1 npr1 rad51d. Indeed, in the presence of BTH, BGL2:GUS was induced to WT levels in the sni1 rad51d double mutant, whereas in the sni1 npr1 rad51d triple mutant, the reporter gene was not responsive, resembling npr1 plants (Fig. 2C). We then examined the effect of rad51d on another PR gene, PR1, over a range of INA concentrations (Fig. 2D). As observed previously, PR1 expression in sni1 and sni1 npr1 was induced at a 10-fold lower concentration of INA than in WT. In the sni1 rad51d double mutant, a WT pattern of induction was restored. On the other hand, the induction pattern in sni1 npr1 rad51d reverted to that of npr1. The sni1 npr1 rad51d mutant also exhibited other phenotypes characteristic of npr1, including enhanced susceptibility to the bacterial pathogen Pseudomonas syringae pv maculicola ES4326 (Psm ES4326) (Fig. 2E) and a loss of protection against Psm ES4326 after BTH treatment (data not shown). These data clearly demonstrate that rad51d is a true suppressor of sni1. In other words, both the background and the SA-induced NPR1-independent PR gene expression and resistance observed in sni1 and sni1 npr1 require the function of RAD51D.
The rad51d Mutant Has Increased Susceptibility to Pathogen Infection.
A direct role for RAD51D in plant defense was established by infection experiments performed on rad51d. The rad51d single mutant showed a moderate, but significant, increase in susceptibility to Psm ES4326 (Fig. 2F). This effect was augmented in the sni1 and sni1 npr1 backgrounds, where greater differences in Psm ES4326 growth (10- to 70-fold) were observed (Fig. 2 E and G). Using the sensitized sni1 and sni1 npr1 backgrounds revealed a positive role for RAD51D in PR gene expression and defense, which we would not have discovered otherwise. Indeed, except for the moderate disease susceptibility phenotype, rad51d plants are indistinguishable from WT in morphology and expression of the BGL2:GUS and PR1 genes (Fig. 2 A–D).
Map-Based Cloning of RAD51D.
The rad51d mutation was mapped to the RAD51D locus (At1g07745) (SI Fig. 6), which encodes a member of the RecA/Rad51 family of recombination and repair proteins (10, 11). A 7-bp deletion was discovered in exon 4 of this gene (base pair 974–980, relative to the ATG), which would cause a frameshift resulting in truncation of the protein. The identity of the mutation was confirmed by genetic complementation. Two cosmids (cosmids 24 and 84 in SI Fig. 6) containing RAD51D complemented the rad51d mutation, and transgenic plants homozygous for constructs containing the genomic RAD51D sequence or the RAD51D cDNA (in the sni1 rad51d background) regained sni1 leaf morphology and expression of BGL2:GUS (Fig. 3 A and B). Full details of the mapping and cloning are given in SI Text.
Fig. 3.
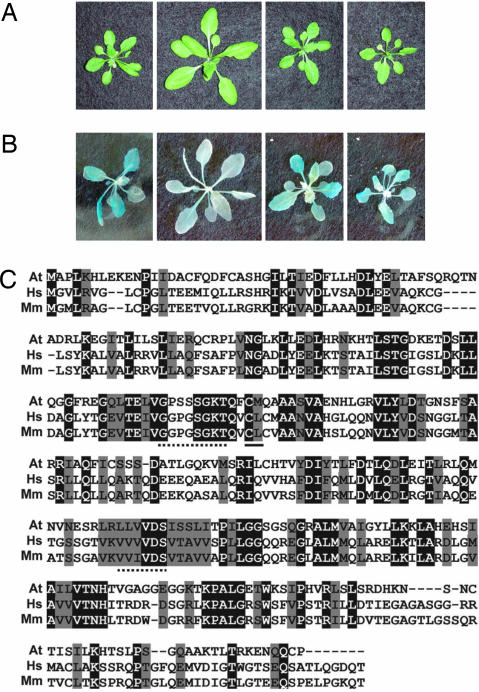
SSN1 corresponds to At1g07745, which encodes RAD51D. (A and B) Complementation of the ssn1 mutation by RAD51D. The RAD51D gene driven by its own promoter and the RAD51D cDNA driven by the constitutive 35S promoter were transformed into sni1 rad51d plants. Complementation restored sni1 morphology (A) and BGL2:GUS expression (B). From left to right are photographs of sni1, sni1rads51d, the RAD51D gene in the sni1rad51d background, and the RAD51D cDNA in the sni1rad51d background. (C) Sequence alignment of Arabidopsis (At), human (Hs), and mouse (Mm) RAD51D proteins. Identical and conserved amino acids are highlighted in black and gray, respectively. Dashes indicate gaps in the sequence to optimize the alignment. The conserved Walker A and B motifs and the 7-bp deletion in Arabidopsis rad51d are indicated by dashed, dotted, and solid lines, respectively.
Analysis of RAD51D cDNAs showed that, similar to the mammalian orthologs (12), Arabidopsis RAD51D mRNA undergoes alternative splicing (SI Table 1, SI Fig. 7, and SI Text). The most abundant splice variant (≈50% of the transcripts analyzed) encodes the full-length protein, as determined by alignment with mammalian RAD51D (Fig. 3C). The functionality of this transcript was confirmed by genetic complementation when the cDNA was expressed from the constitutive 35S promoter (Fig. 3 A and B).
Yeast Rad51 (ScRad51) is a eukaryotic homologue of the E. coli RecA protein, which plays a central role in DNA recombination and repair (12). In higher eukaryotes such as mammals and Arabidopsis, in addition to RAD51 and the meiosis-specific DMC1, there are five RAD51 paralogs: RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3 (10–16). The RAD51 paralogs show limited sequence similarity (≈15–20% identity) to ScRad51, and their biological functions are poorly understood (13). Arabidopsis RAD51D has only 14% identity to ScRad51; however, phylogenetic analysis indicates that Arabidopsis RAD51D is the ortholog of human RAD51D (27% identity) (10) (SI Fig. 8).
Expression studies of RAD51D using a fusion of the RAD51D promoter to GUS showed detectable levels of GUS staining only in pollen grains (SI Fig. 9 A and B). Expression in leaf tissues is low, but detectable by RT-PCR (data not shown). Despite the high expression of RAD51D in pollen grains, the rad51d mutant has viable pollen (SI Fig. 9D) and is fertile. Furthermore, rad51d partially suppresses the reduced fertility phenotype of sni1 (data not shown).
The rad51d Mutant Is Hypersensitive to DNA Damaging Agents.
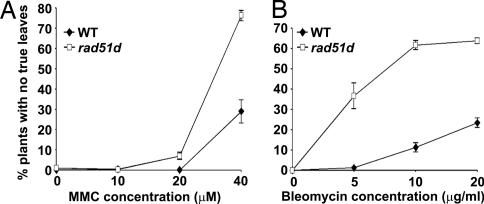
Mutants lacking proteins involved in DNA repair often show increased sensitivity to treatments that induce DNA damage (17). Mutations in all of the other Arabidopsis RAD51 paralogs result in increased sensitivity to the DNA-cross-linking agent mitomycin C (MMC) (10, 14). We therefore tested the sensitivity of rad51d to MMC and the γ-ray mimetic bleomycin, which causes double-strand breaks. The rad51d mutant was hypersensitive to both MMC and bleomycin (Fig. 4). DNA cross-linking and double-strand breaks can be repaired through homologous recombination (17), therefore it is likely that RAD51D plays an essential role in this homologous recombination repair pathway.
Fig. 4.
RAD51D plays a role in DNA repair. The rad51d mutant is hypersensitive to the DNA-damaging agents MMC (A) and bleomycin (B). For each data point values represent three replicate Petri plates with each replicate containing ≈100 plants. Error bars represent standard error.
RAD51D Is Required for Somatic Homologous Recombination.
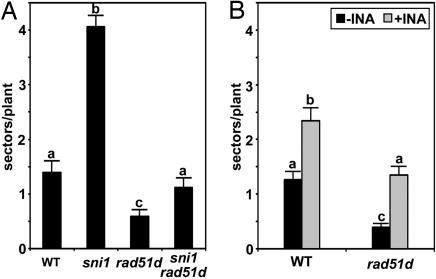
In plants, both abiotic and biotic stresses can cause an increase in homologous recombination (5). Treatment of Arabidopsis with BTH, INA, or pathogen challenge increases the rate of somatic homologous recombination (18). The cim3 (constitutive immunity 3) mutant, which shows constitutive activation of SAR, also has elevated levels of somatic recombination. The involvement of RAD51D in defense-related gene expression suggests a possible link between induced gene expression and DNA recombination during plant defense. To test this hypothesis, we used plants containing reporter transgenes consisting of two overlapping segments of the GUS gene in direct or inverted orientations, lines 1418 and 1445, respectively (18). Homologous recombination between the overlapping regions results in a functional GUS gene and groups of cells expressing GUS appear as blue sectors after staining. We crossed each reporter line with sni1 npr1 rad51d to generate lines homozygous for the recombination reporter transgene in a homozygous sni1, rad51d, or sni1 rad51d background. We observed that, in the 1445 background, the sni1 mutant had constitutively elevated levels of recombination compared with WT, as indicated by an increased frequency of recombination sectors (Fig. 5A). This increase was not observed in the sni1 rad51d double mutant, indicating that RAD51D is required for the somatic recombination observed in sni1. Similar results were observed for the 1418 reporter (data not shown). Moreover, the rad51d single mutant has fewer sectors than WT under both uninduced and INA-induced conditions (Fig. 5), indicating that the rad51d mutation alone results in a defect in homologous recombination.
Fig. 5.
Somatic recombination is affected in the sni1 and rad51d mutants. Frequency of recombination sectors per plant in WT or mutant lines containing the reporter transgene 1445 without (A) and with 50 mM INA induction (B). Error bars represent standard error; letters above bars indicate statistically significant differences (Bonferroni correction, P < 0.01; for Col n = 71, sni1 n = 107, rad51d n = 220, sni1 rad51d n = 214).
Discussion
Through a screen for suppressors of the sni1 mutant, we have discovered that SNI1 and RAD51D play dual roles in the regulation of both defense gene transcription and homologous recombination, suggesting an intriguing mechanistic link between the two processes. Both of these processes require access to DNA, which is achieved through ATP-dependent chromatin remodeling machines (19). SNI1 has structural similarity to Armadillo repeat proteins (9) and may serve as a scaffold for formation of a complex involved in both chromatin remodeling and homologous recombination.
A connection between chromatin remodeling proteins and DNA recombination machinery has been made before. Recent studies showed that the chromatin remodeling activities of yeast and Drosophila RAD54 proteins are strongly enhanced by the addition of RAD51 (20, 21). RAD51 is also involved in activation of the HIV-1 LTR through interaction with the C/EBP family of transcription factors (22). A dual role in transcription and recombination or DNA repair was also recently described for INO80, a member of the SWI/SNF family of ATPases, in both yeast and Arabidopsis (23–25). However, our results provide a biological context to this connection that has not been described before to our knowledge. Our study demonstrates that in Arabidopsis SNI1 and RAD51D are involved in both defense-related transcription and homologous DNA recombination.
As summarized in Fig. 1, in the sni1 mutant, the chromatin at PR gene promoters is in a more accessible conformation and therefore sni1 plants have elevated gene expression (9) and recombination (Fig. 5A). We hypothesize that components of the recombination machinery, including RAD51D, contribute to this open chromatin structure. Therefore, in the sni1 rad51d double mutant, the DNA is less accessible, preventing the PR gene expression and homologous recombination observed in sni1. From these data, we propose that in WT plants SNI1 is an important negative regulator preventing PR gene expression and recombination in the absence of induction. Upon activation of SAR, SNI1 repression is relieved, allowing PR gene induction and facilitating homologous DNA recombination. RAD51D, and possibly additional components of the recombination machinery such as other RAD51 paralogs or RAD54, are important for both processes and may form a complex with SNI1.
It has been proposed that pathogen-induced somatic recombination may be an adaptive mechanism to generate new resistance specificities (5, 26). Plant development is indeterminate and plant reproductive tissues appear late in the lifecycle, therefore changes to the genome in somatic cells may not only give rise to altered tissues and organs, but also be transmitted to the offspring (3–5). Recent data from Hohn and colleagues (4) support this idea. In plants treated with flagellin, a peptide elicitor of plant defense, rates of homologous recombination were elevated. Furthermore, in the untreated progeny of these plants, levels of somatic recombination were constitutively elevated. These changes were not observed when plants were treated with an inactive peptide. Possible targets of such recombination include R gene loci, which commonly consist of tandem duplications. New resistance specificities can be generated by recombination between or within such R gene clusters (27). The evolutionary dynamics of 27 R genes was recently studied by sequencing the leucine-rich repeat region in a sample of 96 accessions (28). R genes were shown to have higher rates of recombination compared with an empirical sample of randomly distributed genomic fragments. Furthermore, nearly all of the R genes were highly polymorphic for protein variants, suggesting selection pressure to generate new alleles. Therefore, it was suggested that the high level of recombination among R genes reflects an adaptation to generate new resistance specificities. A future challenge will be to identify other members of the SNI1–RAD51D complex and examine whether this complex affects R gene recombination.
Materials and Methods
Mutant Screen.
To screen for suppressors of sni1 we took advantage of the sni1 morphological phenotype (smaller and narrower leaves, shorter roots, and lower fertility than WT) by looking for mutants that are similar in size to WT plants, in a homozygous sni1 background. Approximately 40,000 homozygous sni1 seed were mutagenized by using fast neutron bombardment, and M2 plants were examined for suppression of the sni1 morphology. M3 seeds from each putative mutant were grown on MS media and tested for loss of expression of the BGL2:GUS reporter gene and the PR1 gene. Full details of the mutant screen are given in SI Text.
Double and Triple Mutant Generation.
To generate the sni1 npr1 rad51d triple mutant, sni1 rad51d was crossed to sni1 npr1–1. The rad51d single mutant and the npr1 rad51d double mutant were generated by outcrossing sni1 rad51d to Col BGL2:GUS and npr1–1, respectively. Cleaved Amplified Polymorphic Sequence analysis was used to identify the sni1, npr1 (8), and rad51d mutations (see SI Text for details of sni1 and rad51d markers).
Map-Based Cloning of RAD51D.
Map-based cloning was carried out using standard techniques; full details are given in SI Text.
Pseudomonas syringae Infection.
Whole leaves of 4-week-old soil-grown plants were infiltrated with a Psm ES4326 suspension (OD600 = 0.0001) in 10 mM MgSO4. The infected tissue was harvested immediately after infection and after 3 days. Two 6-mm leaf discs were collected per plant from 4–16 plants. The leaf discs were placed in 500 μl of MgSO4 in a 96-well plate and ground by using a metal bead in a Geno/Grinder 2000 (Spex CertiPrep; Spex Industries, Metuchen, NJ) at 1,000 rpm for 30 s followed by 1,500 rpm for 10 s. Serial 10-fold dilutions were made in 10 mM MgSO4 solution. Aliquots (10 μl) were plated in rows onto King's B medium containing 100 μg/ml streptomycin by using an eight-channel multipipettor. The plates were incubated for 2–3 days at room temperature before counting colonies. Statistical analyses were performed using Student's t test of the differences between two means of log-transformed data (29) or by two-way ANOVA using the STATLETS program (StatPoint LLC, Herndon, VA).
MMC and Bleomycin Assays.
Plants were grown on MS media containing MMC (0, 10, 20, or 40 μM) or bleomycin (0, 5, 10, or 20 μg/ml) (Sigma, St. Louis, MO) for 14 days. In the absence of the genotoxic agent, all plants were able to grow true leaves. The percentage of plants with no true leaves was therefore used as an indication of sensitivity.
Somatic Recombination Assays.
The recombination reporter lines 1418 Col and 1445 Col were crossed to the sni1 npr1 rad51d triple mutant containing the BGL2:GUS transgene. F2 plants containing the recombination reporters were selected by growth on MS media containing 10 μg/ml hygromycin. The presence of the sni1, npr1, and rad51d mutations was determined by CAPS analysis as described in SI Text. The absence of the BGL2:GUS transgene was determined by PCR using the primers NptIIIF (CTTGGGTGGAGAGGCTATTC) and NptIIIR (CTTGAGCCTGGCGAACAGTT) and confirmed in subsequent generations by growth on MS media containing 50 μg/ml kanamycin. The following lines were identified: sni1 1445, rad51d 1445, and sni1 rad51d 1445. The same mutants were generated with the 1418 reporter (data not shown). Only lines homozygous for the recombination reporter transgene and the mutations of interest and lacking the BGL2:GUS transgene were used for recombination assays. For analysis of recombination WT, sni1, and sni1 rad51d plants were grown on MS media with or without 50 mM INA for 2 weeks. Recombination frequencies were determined by histochemical staining for GUS activity, as described (18). The data were analyzed by Student's t test. The experiments were repeated at least three times with similar results.
Supplementary Material
Acknowledgments
We thank Neal Goldenburg and Edward Nam for technical assistance; Barbara Hohn (Friedrich Miescher Institute, Basel, Switzerland) for the recombination reporter lines; and Frederick Ausubel, Christian Danna, Barbara Hohn, Hong Ma, and Thomas Petes for helpful discussions and critiques of the manuscript. This work was supported by a postdoctoral fellowship from the International Human Frontier Science Program Organization (to W.E.D.) and National Science Foundation Grant MCB-0445621 (to X.D.).
Abbreviations
- SAR
systemic acquired resistance
- PR
pathogenesis-related
- SA
salicylic acid
- INA
2,6-dichloroisonicotinic acid
- BTH
benzothiadiazole S-methyl ester
- GUS
β-glucuronidase
- MMC
mitomycin C.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ436466–DQ436473).
This article contains supporting information online at www.pnas.org/cgi/content/full/0609357104/DC1.
References
- 1.Hammond-Kosack KE, Jones JDG. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durrant WE, Dong X. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 3.Kovalchuk I, Kovalchuk O, Kalck V, Boyko V, Filkowski J, Heinlein M, Hohn B. Nature. 2003;423:760–762. doi: 10.1038/nature01683. [DOI] [PubMed] [Google Scholar]
- 4.Molinier J, Ries G, Zipfel C, Hohn B. Nature. 2006;442:1046–1049. doi: 10.1038/nature05022. [DOI] [PubMed] [Google Scholar]
- 5.Schuermann D, Molinier J, Fritsch O, Hohn B. Trends Genet. 2005;21:172–181. doi: 10.1016/j.tig.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Van Loon LC, Van Strien EA. Physiol Mol Plant Pathol. 1999;55:85–97. [Google Scholar]
- 7.Dong X. Curr Opin Plant Biol. 2004;7:547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Zhang Y, Clarke JD, Li Y, Dong X. Cell. 1999;98:329–339. doi: 10.1016/s0092-8674(00)81962-5. [DOI] [PubMed] [Google Scholar]
- 9.Mosher RA, Durrant WE, Wang D, Song J, Dong X. Plant Cell. 2006;18:1750–1765. doi: 10.1105/tpc.105.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bleuyard JY, Gallego ME, Savigny F, White CI. Plant J. 2005;41:533–545. doi: 10.1111/j.1365-313X.2004.02318.x. [DOI] [PubMed] [Google Scholar]
- 11.Osakabe K, Abe K, Yamanouchi H, Takyuu T, Yoshioka T, Ito Y, Kato T, Tabata S, Kurei S, Yoshioka Y, et al. Plant Mol Biol. 2005;57:819–833. doi: 10.1007/s11103-005-2187-1. [DOI] [PubMed] [Google Scholar]
- 12.Kawabata M, Kawabata T, Nishibori M. Acta Med Okayama. 2005;59:1–9. doi: 10.18926/AMO/31987. [DOI] [PubMed] [Google Scholar]
- 13.Thacker J. Cancer Lett. 2005;219:125–135. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 14.Bleuyard JY, White CI. EMBO J. 2004;23:439–449. doi: 10.1038/sj.emboj.7600055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abe K, Osakabe K, Nakayama S, Endo M, Tagiri A, Todoriki S, Ichikawa H, Toki S. Plant Physiol. 2005;139:896–908. doi: 10.1104/pp.105.065243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Yang X, Lin Z, Timofejeva L, Xiao R, Makaroff CA, Ma H. Plant Physiol. 2005;138:965–976. doi: 10.1104/pp.104.058347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bray CM, West CE. New Phytol. 2005;168:511–528. doi: 10.1111/j.1469-8137.2005.01548.x. [DOI] [PubMed] [Google Scholar]
- 18.Lucht JM, Mauch-Mani B, Steiner HY, Metraux JP, Ryals J, Hohn B. Nat Genet. 2002;30:311–314. doi: 10.1038/ng846. [DOI] [PubMed] [Google Scholar]
- 19.Lusser A, Kadonaga JT. BioEssays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- 20.Alexeev A, Mazin A, Kowalczykowski SC. Nat Struct Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 21.Alexiadis V, Kadonaga JT. Genes Dev. 2002;16:2767–2771. doi: 10.1101/gad.1032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chipitsyna G, Sawaya BE, Khalili K, Amini S. J Cell Physiol. 2006;207:605–613. doi: 10.1002/jcp.20612. [DOI] [PubMed] [Google Scholar]
- 23.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 24.van Attikum H, Fritsch O, Hohn B, Gasser SM. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 25.Fritsch O, Benvenuto G, Bowler C, Molinier J, Hohn B. Mol Cell. 2004;16:479–485. doi: 10.1016/j.molcel.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 26.Dong X. Trends Plants Sci. 2004;9:60–61. doi: 10.1016/j.tplants.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Leister D. Trends Genet. 2004;20:116–122. doi: 10.1016/j.tig.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Bakker EG, Toomajian C, Kreitman M, Bergelson J. Plant Cell. 2006;18:1803–1818. doi: 10.1105/tpc.106.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokal RR, Rohlf FJ. Biometry. New York: Freeman; 1981. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.