Abstract
Proteins destined for the mitochondrial matrix are imported by the translocase of the outer membrane—the TOM complex—and the presequence translocase of the inner membrane—the TIM23 complex. At present, there is no structural information on components of the presequence translocase. Tim21, a subunit of the presequence translocase consisting of a membrane anchor and a carboxy-terminal domain exposed to the intermembrane space, directly connects the TOM and TIM23 complexes by binding to the intermembrane space domain of the Tom22 receptor. We crystallized the binding domain of Tim21 of Saccharomyces cerevisiae and determined its structure at 1.6 Å resolution. The Tim21 structure represents a new α/β-mixed protein fold with two α-helices flanked by an extended eight-stranded β-sheet. We also identified a core sequence of Tom22 that binds to Tim21. Furthermore, negatively charged amino-acid residues of Tom22 are important for binding to Tim21. Here we suggest a mechanism for the TOM–TIM interaction.
Keywords: contact sites, mitochondria, presequence translocase, Tim21, TOM
Introduction
The mitochondria import about 1,000 different precursor proteins from the cytosol, and the translocase of the outer membrane—the TOM complex—functions as the general import gate of the outer membrane. From here, the precursor proteins are distributed to four different translocases and sorting machineries (Neupert, 1997; Jensen & Johnson, 2001; Koehler, 2004; Rehling et al, 2004)—(i) the precursors of β-barrel proteins are transferred to the sorting and assembly machinery (SAM complex) that mediates integration into the outer membrane; (ii) small proteins of the intermembrane space (IMS) use the mitochondrial IMS import and assembly machinery; (iii) hydrophobic carrier proteins interact with the Tim9–Tim10 complex of the IMS and are inserted into the inner membrane by the twin-pore translocase; and (iv) the main mitochondrial protein import pathway involves cleavable targeting signals (presequences) at the amino termini of preproteins. The TIM23 complex mediates the translocation of the preproteins into, or across, the inner membrane. The presequence translocase-associated motor complex (PAM) drives the completion of translocation into the matrix, in which the presequences are proteolytically removed.
Presequence-carrying preproteins are transferred from the TOM complex to the TIM23 complex at translocation contact sites. Although their first description by Schleyer and Neupert (1985) raised significant interest, the molecular nature of translocation contact sites has remained unknown. It was shown that a preprotein arrested during transport across both mitochondrial membranes stably connected the TOM and TIM23 complexes (Rassow et al, 1989; Dekker et al, 1997; Chacinska et al, 2003). Tim50, an essential subunit of the TIM23 complex, interacts with preproteins on their emergence on the IMS side of the outer membrane; however, Tim50 does not form a direct contact with the TOM complex (Koehler, 2004; Rehling et al, 2004). The identification of Tim21 provided the first evidence for a direct contact between TOM and TIM23 complexes (Chacinska et al, 2005).
Tim21 consists of a single transmembrane segment in the inner membrane, and a large IMS domain that has been shown to bind to the IMS domain of the outer membrane of the Tom22 receptor (Chacinska et al, 2005; Mokranjac et al, 2005). This interaction is required for efficient cell growth under conditions that require high mitochondrial activity (Chacinska et al, 2005). Presequence peptides inhibit the Tim21–Tom22 interaction (Chacinska et al, 2005), suggesting a regulatory role for Tim21 in protein import. However, the nature and mechanism of the TOM–TIM interaction remain unknown owing to the current lack of detailed structural information of most components of the mitochondrial protein import machinery, which has strongly limited a mechanistic understanding of the import process. So far, only five high-resolution structures have been reported: the cytosolic domains of the receptors Tom20 and Tom70 (Abe et al, 2000; Perry et al, 2006; Wu & Sha, 2006); the Tim9–Tim10 chaperone complex (Webb et al, 2006); the carboxy-terminal domain of Tim44 of the motor PAM (Josyula et al, 2006); and the matrix processing peptidase (Taylor et al, 2001). Here, we report the structure of the IMS domain of Tim21 at 1.6 Å resolution and propose a mechanism for the TOM–TIM interaction.
Results And Discussion
Crystallization of the Tim21 binding domain
We expressed the full-length IMS domain of Saccharomyces cerevisiae Tim21 (residues 103–239) with an N-terminal His tag. This domain binds to the IMS domain of Tom22 (Chacinska et al, 2005). A concentrated solution of the purified Tim21 domain was subjected to various crystallization screens, but no crystals were obtained. As flexible termini often prevent proteins from crystallizing, we used subtilisin to tentatively remove terminal amino-acid residues and obtained a protease-resistant domain. Analysis using mass spectrometry identified two fragments—residues 103–227 and 103–225. The shorter fragment, termed Tim21IMS, was expressed with an N-terminal His tag (Fig 1A). We used two assays to analyse whether this domain of Tim21 still contained the TOM interaction site. First, a glutathione S-transferase (GST) fusion protein containing Tom22IMS was incubated with Tim21IMS and the mixture was subjected to affinity purification using Ni-NTA chromatography. GST–Tom22IMS was co-eluted with Tim21IMS (Fig 1A). We then analysed the interaction of the untagged domains using 1H NMR spectroscopy. The addition of increasing amounts of Tim21IMS led to significant changes in intensity in several Tom22IMS peaks (Fig 1B), indicating an interaction of both domains. We conclude that Tim21IMS is competent to interact with Tom22IMS.
Figure 1.
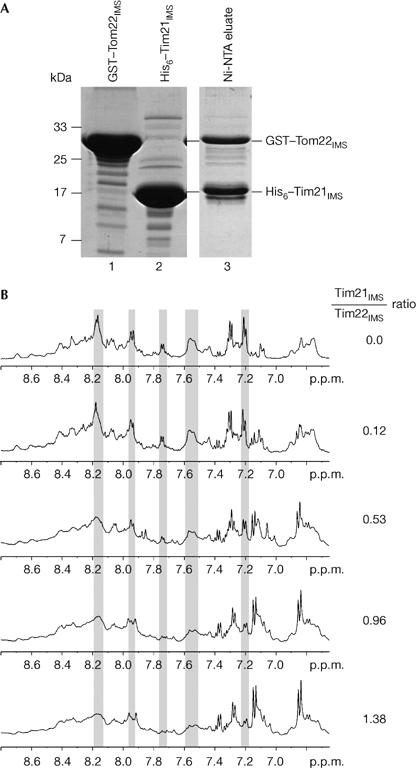
Tim21IMS interaction with Tom22IMS. (A) Purified GST–Tom22IMS and His6–Tim21IMS were mixed and applied to Ni-NTA-agarose. After washing, bound proteins were eluted with imidazole and analysed by SDS–polyacrylamide gel electrophoresis and stained with Coomassie R-250. (B) 1H NMR spectroscopic detection of complex formation between Tim21IMS and Tom22IMS. Spectra of Tom22IMS were recorded in the absence or presence of Tim21IMS. Grey panels indicate changes in peak intensity. GST, glutathione S-transferase; Tim21IMS, translocase of the inner membrane; Tom22IMS, translocase of the outer membrane.
Initial crystallization trials with untagged Tim21IMS did not lead to any crystalline material. However, we noticed that an unusually high number of crystallization drops remained clear, indicating that the protein concentration, although 25–30 mg/ml, was still too low. On concentrating Tim21IMS to approximately 90 mg/ml, cubical-shaped crystals were obtained after 2–4 weeks.
Structure of the Tim21 binding domain
Tim21IMS crystallized in the orthorhombic space group P212121 with one monomer of the protein in the asymmetric unit (Table 1). We recorded a native data set of the Tim21IMS crystals to a resolution limit of 1.6 Å. The structure was solved by a multi-wavelength anomalous dispersion (MAD) experiment carried out on a seleno-methionine (SeMet)-labelled protein. A large part of the model was built automatically, and subsequent model building enabled us to fit all amino acids of the Tim21IMS chain into the electron density map except for the side chains of three lysine residues (Lys 118, Lys 120 and Lys 193), which could not be localized and were thus modelled as alanine residues. The structure was refined to a crystallographic R-factor of 19.7% (Rfree=24.8%). The final model consisted of residues 103–225 and 167 ordered solvent molecules. In the crystal lattice, the molecules are densely packed, resulting in a low solvent content of 39%.
Table 1.
Statistics of data collection and refinement on Tim21IMS
| Data collection | SeMet crystal | Native crystal | ||
|---|---|---|---|---|
| Space group | P212121 | P212121 | ||
| Unit cell constants | ||||
| a (Å) | 32.1 | 32.3 | ||
| b (Å) | 59.2 | 58.8 | ||
| c (Å) | 62.6 | 62.6 | ||
| Inflection | Peak | Remote | ||
| Wavelength (Å) | 0.9794 | 0.9792 | 0.9763 | 1.05 |
| Resolution (Å) | 1.9 | 1.9 | 1.9 | 1.58 |
| (2.02–1.9) | (2.02–1.9) | (2.02–1.9) | (1.68–1.58) | |
| Unique reflections | 9,503 | 9,785 | 9,541 | 16,515 |
| Completeness (%) | 96.2 | 99.4 | 96.4 | 97.5 |
| (85.7) | (97.2) | (88.3) | (94.0) | |
| Redundancy | 3.4 | 6.8 | 3.4 | 3.33 |
| Anomalous diffractors | 3 | |||
| Rmerge (%) | 4.9 (51) | 7.1 (62.3) | 5.4 (55.6) | 3.8 (26.4) |
| FOM* (SOLVE) | 0.42 | |||
| FOM* (RESOLVE) | 0.54 | |||
| I/σ(I) | 14.48 (2.07) | 15.41 (2.42) | 13.05 (1.9) | 18.0 (3.7) |
| Refinement statistics | ||||
| Resolution range (Å) | 20.0–1.6 (1.7–1.6) | |||
| No. of reflections used | 16,047 | |||
| No. of reflections in working set | 15,244 | |||
| No. of reflections in test set | 803 | |||
| No. of non-hydrogen protein atoms | 1,003 | |||
| No. of solvent atoms | 167 | |||
| Rcryst (%)/Rfree (%) † | 19.7/24.8 (22.2/28.1) | |||
| R.m.s.d. of bond lengths (Å) | 0.006 | |||
| R.m.s.d. of bond angles (deg) | 1.3 | |||
| Mean β factor (Å2) | 22.9 | |||
| Ramachandran plot statistics‡ | ||||
| Most favoured | 111 (91.7%) | |||
| Additionally allowed | 8 (6.6%) | |||
| Generously allowed | 2 (1.6%) | |||
| Disallowed | 0 | |||
| Values in parentheses refer to the highest resolution shell. | ||||
| *FOM, figure of merit. | ||||
| †Rfree was calculated using 5% randomly selected reflections. | ||||
| ‡As defined by PROCHECK. | ||||
Tim21IMS belongs to the class of α/β mixed proteins (Fig 2A,B). In total, the secondary structure is composed of two α-helices (α1 and α2) and eight β-strands (β1–β8) connected by loops, some of which are extended but nonetheless well defined. About 50% of the residues are part of well-defined secondary structure elements (Fig 2B,C). At the N terminus, the chain folds into a four-repeat long α-helix followed by a short loop that connects α1 helix to the tilted α2 helix. The α1 helix is flanked by the antiparallel β-strands β4–β8, which together form an extended β-sheet. Three hydrogen bonds exist between the helices and the β-sheet (Fig 2D; supplementary Table 1 online). Helix α1 is flanked by two loop structures—the extended loop connects β-strands 6 and 7 to the right side, and parts of the loop connect the α2 helix and β1 to the left side. Residues 219–225 are stabilized in the crystal lattice by interactions with neighbouring molecules of the crystal packing. A DALI database search for structures related to Tim21IMS did not find a protein with a similar fold; thus, we conclude that the Tim21IMS structure represents a new protein fold.
Figure 2.
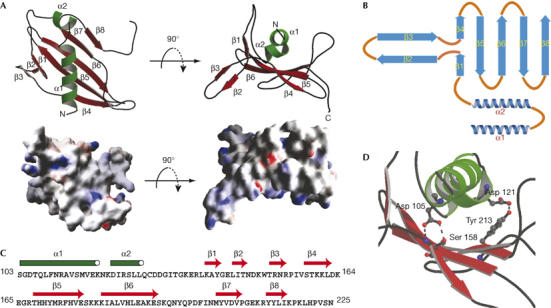
Crystal structure of Tim21IMS. (A) Views of the molecular structure of Tim21IMS (top) with labelled α-helices (green) and β-strands (red). Images were generated using Molscript (Kraulis, 1991). Surface presentations of the Tim21IMS molecule are shown (bottom). Positive and negative potentials are coloured blue and red, respectively. The images were generated using Grasp (Nicholls et al, 1991). (B) Schematic representation of the Tim21IMS fold. (C) Sequence of Tim21IMS and distribution of secondary structure elements. (D) Hydrogen bonds (dashed) between the helical part and the antiparallel β-sheet. Tim21IMS, translocase of the inner membrane.
Negative charges in Tom22–Tim21 interaction
Surface-exposed residues of Tim21IMS, which were conserved between different species, were grouped into four classes—positively charged, negatively charged, amphipathic residues, and hydrophobic residues. Patches of positively charged residues represented the most abundant form of the exposed conserved residues (Fig 3). By contrast, Tom22IMS, consisting of the C-terminal 33 residues, has a net charge of −5 (Court et al, 1996; Moczko et al, 1997; Komiya et al, 1998). The different charge distribution between Tim21IMS (with a net charge of +8) and Tom22IMS, and the reported inhibition of the Tim21–Tom22 interaction by positively charged presequences (Chacinska et al, 2005), raised the possibility that electrostatic interactions are involved in the binding of Tim21 to the TOM complex.
Figure 3.
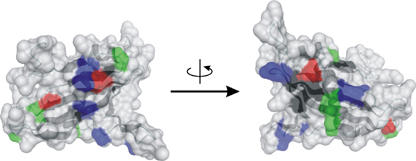
Surface presentations of Tim21IMS. Conserved positively charged (blue), negatively charged (red), amphipathic (green) and hydrophobic (black) residues on the surface are shown. Ten unicellular organisms were compared (Saccharomyces cerevisiae, Aspergillus fumigatus, Neurospora crassa, Candida albicans, Kluyveromyces lactis, Aphis gossypii, Candida glabrata, Desulfomusa hansenii, Yarrowia lipolytica, Schizosaccharomyces pombe). Images were generated using DINO (http://www.dino3d.org/). Tim21IMS, translocase of the inner membrane.
To analyse the interaction between the IMS domain of Tim21 and the TOM complex, the purified domain was attached to an affinity column and incubated with solubilized mitochondria (Chacinska et al, 2005). Although the TOM complex efficiently bound to Tim21 at 80 mM NaCl, the interaction was strongly decreased on washing with higher salt concentrations (Fig 4A). This salt sensitivity indicates an involvement of electrostatic interactions in the binding of Tim21 to the TOM complex.
Figure 4.
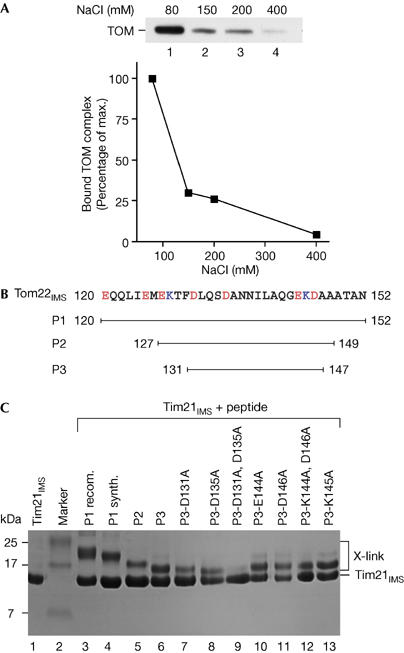
The Tim21IMS–Tom22IMS association involves electrostatic interaction. (A) The immobilized IMS domain of Tim21 was incubated with solubilized yeast mitochondria. After washing in the presence of NaCl, bound proteins were eluted and analysed by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and western blotting with Tom40 antibodies. (B) Sequence of Tom22IMS and synthetic peptides. (C) Isolated Tim21IMS was incubated with purified Tom22IMS (P1 recom.) or the indicated peptides and crosslinked with glutaraldehyde. Samples were analysed by SDS–PAGE and Coomassie staining. Tim21IMS, translocase of the inner membrane; Tom22IMS, translocase of the outer membrane.
We examined which regions of Tom22 are required for interaction with Tim21. A Tom22 peptide library, comprising the region from the membrane span to the C terminus, was probed with the purified IMS domain of Tim21, followed by quantitative analysis. Binding was maximal in a region between residues 123 and 147 (supplementary Fig 1A online). We then analysed three peptides for an interaction with Tim21IMS in solution: the entire 33-residue Tom22IMS (P1); a 23-residue fragment (P2); and a 17-residue fragment (P3; Fig 4B). All three peptides were crosslinked to Tim21IMS (Fig 4C). The shortest peptide P3, corresponding to residues 131–147—that is, the peak and plateau area of binding to the peptide library—was linked to Tim21IMS with an efficiency similar to that with full-length Tom22IMS. We thus studied whether this short peptide affected the interaction of Tim21IMS with Tom22 in the context of the in vivo/in organello situation, that is, when full-length Tom22 was present in the TOM complex and the complete set of mitochondrial proteins was present. The peptide indeed impaired the interaction of Tim21IMS with the TOM complex in solubilized mitochondria (supplementary Fig 1B online). Taken together, we conclude that the segment of Tom22 represented by P3 contains sufficient information for binding to Tim21.
We systematically replaced the five charged residues of P3 by alanines and analysed the interaction of the modified peptides with Tim21IMS by crosslinking (Fig 4C, lanes 7–13). A replacement of both Asp 131 and Asp 135 blocked the interaction of P3 with Tim21IMS, whereas the interaction took place when the two other negatively charged residues (Glu 144 and Asp 146) or the positively charged Lys 145 were replaced. We studied whether the replacement of Asp 131 and Asp 135 affected the influence of P3 on the interaction of Tim21IMS with the TOM complex under in organello conditions. Indeed, the modified peptide P3-D131A,D135A did not compete for binding of full-length Tom22 to Tim21IMS, in contrast with the wild-type form of P3 (supplementary Fig 1C online). The crucial role of two aspartate residues thus supports the model of an electrostatic interaction between Tom22 and Tim21.
Conclusions
We solved the structure of the binding domain of Tim21 that contains a new mixed α/β-protein fold. The surface of Tim21 shows patches of conserved, positively charged residues and interacts with the negatively charged IMS domain of Tom22. Binding of Tim21 to Tom22 involves electrostatic interactions, with a 17-residue segment of Tom22IMS being sufficient for binding to Tim21IMS. Two negatively charged residues in this core segment of Tom22IMS are involved in the association with Tim21IMS. An electrostatic interaction between Tim21 and Tom22 agrees with the observation that the positively charged presequences compete with Tim21 for binding to Tom22 (Chacinska et al, 2005). Tim21 represents the first structure of a component of the mitochondrial presequence translocase, as well as of a component that connects protein complexes located in the outer and inner membranes of the mitochondria.
Methods
Protein interaction analysis. Tim21IMS (190 μM) was incubated with a twofold molar excess of Tom22 peptides in 10 mM HEPES, 50 mM NaCl, 50 mM KCl and 3 mM MgCl2. Glutaraldehyde was added at 0.02% and the mixture was incubated at 30°C for 15 min, followed by SDS–polyacrylamide gel electrophoresis. Salt-dependent interaction of the IMS domain of Tim21 with the TOM complex was analysed as described previously (Chacinska et al, 2005), except that the wash buffer contained the indicated amounts of NaCl. 1H NMR spectra were recorded on a DRX-500 spectrometer (Bruker BioSpin, Billerica, MA, USA) at 27°C. Tim21IMS (5.3 mM in 10 mM HEPES and 50 mM NaCl (pH 7.5)) was added stepwise to Tom22IMS (∼2.1 mM in PBS, pH 6.9; H2O:D2O=9:1). Water suppression was achieved using the WATERGATE sequence (Sklenar et al, 1993) and data processing was carried out using XWINNMR-v3 (Bruker). Protein expression and purification are described in the supplementary information online.
Protein crystallization and data collection. Crystallization was carried out by the hanging-drop set-up by mixing 1 μl of protein solution and reservoir solution using the Crystal Screens I and II and the Index Screen (Hampton, Aliso Viejo, CA, USA). Initial Tim21IMS crystals were obtained from numbers 9 and 40 of the Crystal Screen I and from numbers 67 and 75 of the Index Screen after 2–4 weeks. Crystallization could be reproduced and improved by seeding. SeMet protein crystals were obtained from variations of number 75 of the Index Screen by seeding with crystals of the native protein. Best results were achieved with 20% (w/v) polyethylene glycol 3350, 0.2 M Li2SO4 and 0.1 M Bis–Tris (pH 6.5). Before measurement, crystals were stabilized in crystallization buffer supplemented with 15% (v/v) glycerol or 20% (w/v) xylitol and flash frozen in liquid nitrogen. A native data set with a resolution limit of 1.6 Å was obtained at beamline BW6 (DESY, Hamburg, Germany). For structure solution, a MAD experiment was carried out on an SeMet crystal at beamline ID29 (ESRF, Grenoble, France). Data were recorded at Se peak, inflection and remote energies to a resolution limit of 1.9 Å (Table 1).
Structure determination, model building and structure refinement. Images were indexed, integrated and scaled using the XDS program package (Kabsch, 1993). Three of the four Se atoms could be located with the Patterson routine implemented in the program SOLVE, and density modification was carried out in RESOLVE (Terwilliger, 2000). A partial model consisting of amino acids 104–161 and 171–216 was automatically generated with the program ARP/wARP (Lamzin et al, 2001). Inspection of the model, refinements, improvements of density maps and manual completion of the molecule were carried out using repetitive combinatorial cycles of the programs ‘O' (Jones et al, 1991) and CNS (Brunger et al, 1998). Water molecules were picked automatically using CNS and their positions included in the refinements. During the refinement rounds, 5% of the reflections were used for cross-validation. The cloning and TEV cleavage-derived additional amino acids could not be seen in the electron density map. The stereochemical quality of the model was validated with PROCHECK (Laskowski et al, 1993). The coordinates and diffraction amplitudes were deposited in the RCSB Protein Data Bank with accession code 2CIU.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank S. Uebel and co-workers, and the staffs of beamlines BW6 (Deutsches Elektronen-Synchrotron) and ID29 (European Synchrotron Radiation Facility) for their support and R. Wagner and J. Schneider-Mergener for discussion. We are grateful to I. Perschil, A. Schulze-Specking and E. Weyher-Stingl for excellent technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft, the Sonderforschungsbereich 388, Gottfried Wilhelm Leibniz Program, Max Planck Research Award, Alexander von Humboldt Foundation, Bundesministerium für Bildung und Forschung, and the Fonds der Chemischen Industrie.
References
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nisnikawa S-I, Endo T, Kohda D (2000) Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100: 551–560 [DOI] [PubMed] [Google Scholar]
- Brunger AT et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chacinska A, Rehling P, Guiard B, Frazier AE, Schulze-Specking A, Pfanner N, Voos W, Meisinger C (2003) Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM–TIM–preprotein supercomplex. EMBO J 22: 5370–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A et al. (2005) Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120: 817–829 [DOI] [PubMed] [Google Scholar]
- Court DA, Nargang FE, Steiner H, Hodges RS, Neupert W, Lill R (1996) Role of the intermembrane-space domain of the preprotein receptor Tom22 in protein import into mitochondria. Mol Cell Biol 16: 4035–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker PJT, Martin F, Maarse AC, Bömer U, Müller H, Guiard B, Meijer M, Rassow J, Pfanner N (1997) The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70–Tim44. EMBO J 16: 5408–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RE, Johnson AE (2001) Opening the door to mitochondrial protein import. Nat Struct Biol 8: 1008–1010 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Josyula R, Jin Z, Fu Z, Sha B (2006) Crystal structure of yeast mitochondrial peripheral membrane protein Tim44p C-terminal domain. J Mol Biol 359: 798–804 [DOI] [PubMed] [Google Scholar]
- Kabsch WJ (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. Appl Crystallogr 26: 795–800 [Google Scholar]
- Koehler CM (2004) New developments in mitochondrial assembly. Annu Rev Cell Dev Biol 20: 309–335 [DOI] [PubMed] [Google Scholar]
- Komiya T, Rospert S, Koehler M, Looser R, Schatz G, Mihara K (1998) Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: evidence for the ‘acid chain' hypothesis. EMBO J 17: 3886–3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Lamzin VS, Perrakis A, Wilson KS (2001) The ARP/WARP suite for automated construction and refinement of protein models. In Rossmann MG, Arnold E (eds) International Tables for Crystallography. Vol. F: Crystallography of Biological Macromolecules, pp 720–772. Dordrecht, The Netherlands: Kluwer Academic [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Moczko M, Bömer U, Kübrich M, Zufall N, Hönlinger A, Pfanner N (1997) The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol Cell Biol 17: 6574–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Popov-Celeketic D, Hell K, Neupert W (2005) Role of Tim21 in mitochondrial translocation contact sites. J Biol Chem 280: 23437–23440 [DOI] [PubMed] [Google Scholar]
- Neupert W (1997) Protein import into mitochondria. Annu Rev Biochem 66: 863–917 [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct Funct Genet 11: 281–296 [DOI] [PubMed] [Google Scholar]
- Perry AJ, Hulett JM, Likic VA, Lithgow T, Gooley PR (2006) Convergent evolution of receptors for protein import into mitochondria. Curr Biol 16: 221–229 [DOI] [PubMed] [Google Scholar]
- Rassow J, Guiard B, Wienhues U, Herzog V, Hartl FU, Neupert W (1989) Translocation arrest by reversible folding of a precursor protein imported into mitochondria: a means to quantitate translocation contact sites. J Cell Biol 109: 1421–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P, Brandner K, Pfanner N (2004) Mitochondrial import and the twin-pore translocase. Nat Rev Mol Cell Biol 5: 519–530 [DOI] [PubMed] [Google Scholar]
- Schleyer M, Neupert W (1985) Transport of proteins into mitochondria: translocational intermediates spanning contact sites between outer and inner membranes. Cell 43: 339–350 [DOI] [PubMed] [Google Scholar]
- Sklenar V, Piotto M, Leppik R, Saudek V (1993) Gradient-tailored water suppression for 1H–15N HSQC experiments optimized to retain full sensitivity. J Magn Reson 102: 241–245 [Google Scholar]
- Taylor AB, Smith BS, Kitada S, Kojima K, Miyaura H, Otwinowski Z, Ito A, Deisenhofer J (2001) Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure 9: 615–625 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2000) Maximum-likelihood density modification. Acta Crystallogr D 56: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CT, Gorman MA, Lazarou M, Ryan MT, Gulbis JM (2006) Crystal structure of the mitochondrial chaperone TIM9-10 reveals a six-bladed α-propeller. Mol Cell 21: 123–133 [DOI] [PubMed] [Google Scholar]
- Wu Y, Sha B (2006) Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat Struct Mol Biol 13: 589–593 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


