Abstract
Fifty years after Briggs and King first succeeded in obtaining normal tadpoles from transplanted embryo nuclei in vertebrates, two general principles have emerged from work in amphibia and mammals. One is the conservation of the genome during cell differentiation. A small percentage of adult or differentiated cells have totipotent nuclei, and a much higher percentage of cells committed to one pathway of cell differentiation have multipotent nuclei. The other is the remarkable reprogramming capacity of cell, and especially egg, cytoplasm. The eventual identification of reprogramming molecules and mechanisms could facilitate a route toward cell replacement therapy in humans.
It has now been 50 years since Briggs and King (1) (Fig. 1) published their paper showing that normal hatched tadpoles can be obtained by transplanting the nucleus of a blastula cell to the enucleated eggs of Rana pipiens. This finding provided an initial answer to the long-standing question of whether the process of development and cell differentiation requires a loss or stable change in the genetic constitution of cells. This question had occupied the minds of developmental biologists since the time of Weissmann (2), who proposed that, as cells progress along their various pathways of differentiation, genes no longer required for other divergent lineages are cast off or permanently inactivated. The developmental equivalence of nuclei at the eight-cell stage of a newt embryo was established by a temporary ligation experiment of Spemann (3), and the technical ability to transplant a living cell nucleus had been achieved long before in the single-celled organisms Amoeba (4) and Acetabularia (5). Nevertheless, Briggs and King were the first to open the way to a direct test of the genetic equivalence of somatic cell nuclei in development and cell differentiation.
Fig. 1.
Robert Briggs (A; 1911–1983) and Thomas J. King (B; 1921–2000). Photographs were kindly supplied by Marie A. DiBerardino through the courtesy of the Institute for Cancer Research of the Fox Chase Cancer Center, Philadelphia.
Amphibia
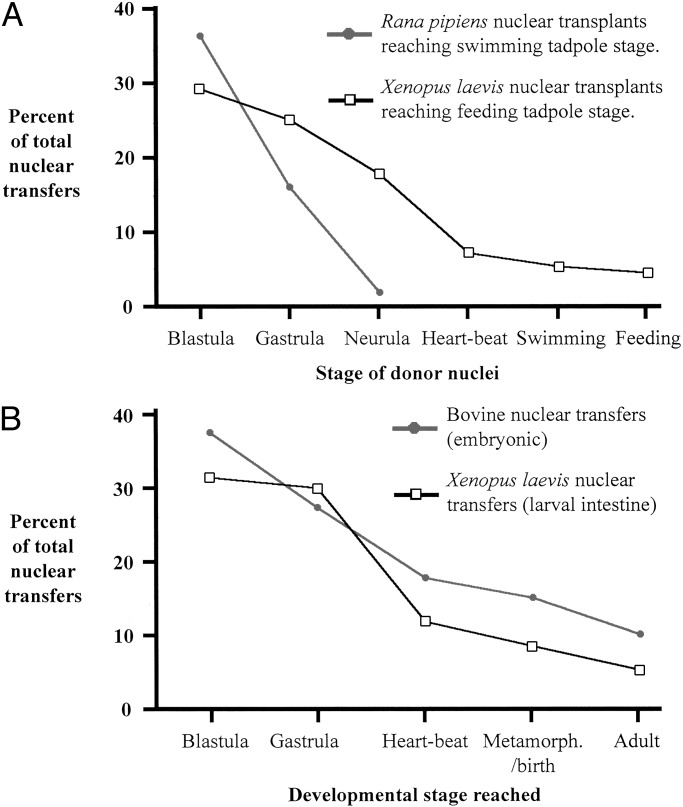
Briggs and King's immediate pursuit of their 1952 breakthrough gave the somewhat surprising result that, whereas blastula nuclei supported normal tadpole development in up to 40% of all tests, gastrula nuclei were markedly less successful. By the tail-bud stage, nuclei of the endoderm (and in later work nuclei of other germ layers) gave only abnormal embryo development (Fig. 2A) (6), even though the nuclei of tail-bud germ cells gave a high proportion of tadpole development (7). This observation implied that, already in early development, nuclei undergo some change restricting their ability to substitute for a zygote nucleus. They furthermore found that this loss of developmental capacity was heritable as judged by the results of serial nuclear transplantation (8). For this last procedure, a blastula resulting from the original transplantation of a somatic cell nucleus serves as a source of nuclei for a further set of nuclear transfers to enucleated eggs, creating a “serial transfer clone.” The development of embryos in any one such clone differed dramatically from those in other clones from the same original germ layer. A good explanation for this was the finding, primarily of DiBerardino (9), that nuclear transplantation from gastrulae and later stages often resulted in chromosome damage, whereas nuclei from blastula cells were damaged a great deal less. This, in turn, can be attributed to the slowing cell cycle as cells differentiate and to other changes undergone as cells progress toward a specialized state. These characteristics seem to make most nuclei unable to switch, within 2 h, to the rapid cell cycle of an activated amphibian egg. We should explain that, once activated, as happens at fertilization or in some species by penetration with a pipette, an egg will always divide at its own rapid rate, and will not wait for a transplanted nucleus to complete its chromosome replication. Incomplete replication of transplanted nuclei seems to us to be the best explanation, at present, for the developmental abnormalities so commonly obtained after transplanting nuclei from differentiating or differentiated cells of Amphibia.
Fig. 2.
(A) In Amphibia, nuclear transfer success declines rapidly as cells differentiate. Details are from Briggs and King (6) and Gurdon (20). Feeding tadpoles are more advanced than swimming tadpoles. (B) Nuclear transfer survival in mammals compared with Amphibia. Abnormalities and death occur progressively during development. Original results are from refs. 13, 70, and 71.
Soon after Briggs and King's first report of successful nuclear transplantation in Rana pipiens, similar success began to be achieved with other species. Largely as a result of the adventures of a peripatetic scientist, Laurence Hogben, the south African frog Xenopus was becoming, in the 1950s, a favored species for embryological research in Europe, eventually replacing, in this regard, the European newts of Triturus species that had reigned supreme since the 1890s (10). Xenopus laevis offered the enormous advantages of a short life cycle and of responding to commercial mammalian hormone preparations by laying eggs at any time of year, in contrast to the limited breeding season and requirement for pituitary extracts to induce egg laying in newts, salamanders, and most frogs and toads. Even more importantly, Fischberg, working in Oxford, had isolated, in Xenopus, the anucleolate mutation that, in heterozygous form, provided a living cell nuclear marker with which to distinguish cells of solely donor origin from those that could have resulted from failed recipient egg enucleation (11).
Following the first report of successful nuclear transfer in Xenopus (12), a series of publications described the developmental capacity of nuclei transplanted from the endoderm lineage using donors from blastulae up to the intestinal epithelium of feeding larvae (13). These results confirmed the conclusion of Briggs and King that the ability of transplanted nuclei to promote normal development declines as development progresses (Fig. 2A). However the main conclusion was entirely different, in that some normal development was obtained from nuclei of even the most differentiated cells. This phase of work culminated in the description of “fertile intestine nuclei”; fertile adult male and female frogs, genetically marked as of solely donor origin, were obtained from the transplantation of nuclei from intestinal epithelial cells of feeding larvae (14). This result established the general principle that the process of cell differentiation does not necessarily require any stable change to the genetic constitution of a cell. Thus, cell differentiation depends on changes in the expression not content of the genome.
Nuclear transfer from adult amphibian cells has given limited success. In no case was an adult animal obtained by nuclear transplantation from the cell of an adult frog. However, the multipotent properties of nuclei from many different adult organs, including lung, heart, and liver, was demonstrated by finding that 1–2% of transplanted nuclei from all these adult sources gave nuclear transplant embryos that reached feeding larval stages (15). In most cases the successful donor cells were not defined (fibroblasts, stem cells, etc.), but in the case of adult skin and adult erythrocytes, donor cells contained keratin (16) or haemoglobin (17), respectively.
As noted above, the success of nuclear transfers decreases as cells differentiate. Fully differentiated intestinal epithelium cells of feeding larvae yielded fertile adults in ≈1% of total nuclear transfers. In Amphibia, ≈70% of nuclei transplanted from differentiated cells fail to elicit any normal recipient egg cleavage, but ≈25% result in partial cleavage. The latter is thought to result from incomplete replication of donor cell chromosomes, such that the transplanted nucleus does not divide at the first recipient egg mitosis, but enters one of the first two blastomeres, where it has a second chance to complete replication (13). In accord with this idea, nuclei from these partial blastulae give good nuclear transfer results after serial nuclear transfer or after grafting to host embryos (18). When assessing these results, it is important to keep in mind the efficiency with which fully committed cells are reprogrammed by nuclear transfers (19). For example, tail-bud endoderm cells are specified and determined, i.e., they cannot form any nonendodermal cell when explanted or transplanted. Yet their nuclei can be reprogrammed for functional muscle and nerve development in 25% of first transfers (20), and in ≈50% of cases if the results of serial transfers from partial blastulae are included (13).
Mammals
Historically, a primary difficulty in performing somatic cell nuclear transfer in mammals has been the small size of the mammalian egg. The mammalian egg (in second meiotic metaphase) is <0.1% the volume of an amphibian egg. Hence, before nuclear transfer could succeed in mammals, micromanipulation techniques were required that could handle, enucleate, and fuse a very small mammalian egg with a single somatic cell. These techniques were principally developed in the late 1960s and early 1970s (for example, see refs. 21–23).
The first report of development to the morula stage following mammalian nuclear transfer was by Bromhall (24), who used both microinjection and Sendai virus induced fusion to transfer labeled rabbit morula cell nuclei into enucleated rabbit eggs. These experiments produced embryos that arrested during cleavage, with a low percentage reaching the morula stage. The first claim to have created a cloned adult mammal by using somatic cell nuclei was in 1981. Illmensee and Hoppe (25) reported that they had obtained three cloned mice by transferring inner cell mass (ICM) nuclei into enucleated zygotes. However, these results were not repeatable (26, 27). In 1983, McGrath and Solter (28) obtained live mice when they transferred a zygote donor nucleus into an enucleated zygote. However, they were unable to obtain any successful development when they used donor cell nuclei from later developmental stages (26). In retrospect, the primary problem affecting these early murine nuclear transfer experiments was that they transferred donor nuclei into enucleated zygotes rather than into unfertilized eggs. Although all amphibian experiments had used unfertilized eggs as recipients, it was thought that zygote cytoplasm would support development better than unfertilized egg cytoplasm. McGrath and Solter's (28) zygotic nuclear transfer experiment was successful because both donor and recipient were in the same developmental stage. A particularly interesting outcome of these early mammalian nuclear transfer experiments was the discovery of imprinting; different genes are stably repressed during oogenesis and spermatogenesis in mammals, and normal development requires a contribution from both the male and female pronuclei (29, 30).
In 1986, Willadsen used electrofusion or Sendai virus to fuse cells of 8 or 16 cell embryos into enucleated eggs of sheep, and obtained two healthy cloned animals (31). Nuclear transfer using embryonic donor cell nuclei was subsequently performed successfully in rabbits (32), pigs (33), mice (34), cows (35), and monkeys (36). In species where nuclear transfer is difficult, nuclei are transferred first to egg cytoplasm and then, the next day, to zygote cytoplasm (33).
In 1996, with the practical applications of cloning technology in mind, Campbell and Wilmut performed nuclear transfer with the nuclei of an established cell line, originating from a day-9 embryo, that had differentiated in vitro. Campbell induced these cells to enter a quiescent state before electrofusing them into enucleated sheep eggs. These nuclear transfers resulted in two healthy cloned sheep (37). The next year they used the same technique with nuclei of cultured adult mammary gland cells and succeeded in producing a single cloned sheep “Dolly” (38). Since the creation of Dolly, many other mammals have been successfully cloned from adult donor cell nuclei. These include mice (39), cows (40), goats (41), pigs (42), rabbits (43), and a cat (44). In the mouse, it has even been possible to derive adult mice from the nuclei of adult lymphocytes, by growing embryonic stem cells from nuclear transplant blastocysts, and by using tetraploid complementation to bypass the need for lymphocyte-derived placental tissue. These mice had the lymphocyte type of rearranged immunoglobin genes in all their cell-types (45).
The efficiency of mammalian nuclear transfer experiments is very similar to that obtained in amphibia (Fig. 2B). Less than 1% of all nuclear transfers from adult or differentiated cells result in apparently normal offspring, and developmental and physiological abnormalities have been observed in a significant proportion of the fetuses obtained (46), especially in their placentas (47). Because many of these abnormalities are not inherited, it is thought that they are not caused by deficiencies in chromosome replication, but rather by a failure to reprogram epigenetic characteristics of somatic cells, especially imprinted genes (48).
Nuclear Reprogramming
Although complete nuclear reprogramming takes place in only a small percentage of nuclear transfers from differentiated cells, it is remarkable that it takes place at all. Whether brought about by nuclear transfer or by cell fusion (49), the causative agent is a change of cytoplasm. This opens up the attractive possibility of understanding mechanisms of reprogramming and of identifying molecules that possess reprogramming activity (50). As a result, it might be possible to use this information to improve the efficiency of reprogramming by egg cytoplasm. Eventually, it may be possible to use molecules derived from eggs to convert adult somatic cells directly into multipotent embryonic cells for the purpose of cell replacement.
The magnitude and rapidity of reprogramming is revealed most impressively by the morphological changes undergone by the same kind of somatic nuclei injected into amphibian eggs or oocytes. The term oocyte is best applied to the growing egg, a cell in the diplotene phase of meiotic prophase with lampbrush chromosomes intensely active in transcription. When fully grown, oocytes undergo hormone-induced maturation, passing through meiotic divisions with highly condensed chromosomes. The unfertilized egg in second meiotic metaphase can be fertilized, and chromosome replication in the egg and sperm pronuclei starts ≈20 min later in frogs. The same kind of somatic nuclei injected into growing oocytes, into meiotic oocytes in division, or into eggs, undergo completely different changes within a few hours to conform to the characteristics of the host cells (Fig. 3).
Fig. 3.
Adult frog brain nuclei rapidly assume the morphology and synthetic activity of recipient oocytes or eggs a few hours after injection. (A) Brain nuclei of an adult frog. (B) Brain nuclei active in transcription. (C) Condensed chromosomes. (D) Brain nuclei active in replication. Further details can be obtained from Graham et al. (72) and Gurdon (73).
Morphological changes are accompanied by changes in nuclear activity: these include the rapid induction of DNA replication in nondividing somatic nuclei transplanted to eggs, and a massive enhancement of RNA synthesis in somatic nuclei in growing oocytes. In the case of nuclei injected into oocytes, reprogramming includes a qualitative change in gene expression. This was demonstrated by transferring nuclei from Xenopus cultured kidney cells into oocytes of the Urodele Pleurodeles (51). Some Xenopus proteins normally expressed in oocytes, but not those specific to kidney cells, were activated in the Pleurodeles oocytes containing Xenopus nuclei, and these proteins were distinguishable from the equivalent Pleurodeles-specific proteins by 2D electrophoresis. In nuclear transfers to oocytes, the transplanted nuclei do not replicate or divide, and the induced changes in gene expression take place on the original somatic cell DNA; this is in contrast to nuclei injected into eggs, because these nuclei undergo several rounds of cell division, diluting out the original somatic cell DNA before new transcription starts. Reprogramming without replication is also observed in mammalian heterokaryons (49).
Somatic cell nuclei transplanted to enucleated eggs undergo rapid morphological changes, but alterations in gene expression have not been seen before the 5,000-cell blastula stage in Amphibia (5 h) or before the 4-cell stage in mice (36 h), when new zygotic gene expression starts. The major classes of RNA made by Amphibian nuclear transplant embryos are the same as in embryos grown from fertilized eggs (52), though quantitative abnormalities are seen for some early zygotic genes (18). In mammals, a microarray study showed that 96% of 10,000 genes were transcribed correctly (53), although abnormalities in the expression of some early zygotic genes were observed more commonly in somatic cell nuclear transplant embryos than in embryos obtained by in vitro fertilization or sperm injection. A prevailing view is that many of the abnormalities of mouse nuclear transplant embryo development can be accounted for by a deficiency or abnormality of early zygotic gene expression, especially of Oct4 (54).
X-chromosome inactivation is efficiently reversed and randomized in embryo, though not in trophectoderm, cells of mouse nuclear transplants (55). Likewise, telomeres are efficiently extended when nuclei of low telomere length are transplanted in cows (56). In these respects, somatic cell nuclei are efficiently reprogrammed by egg cytoplasm. In the case of DNA methylation and imprinting, the situation is less certain. In some cases the expression of imprinted genes is, and in other cases is not, changed by nuclear transplantation (57, 58). Conclusions regarding imprinting are complicated by a high degree of variation in the expression of imprinted genes, such as H19 and Igf2, in embryonic stem cell lines (59), and these cells are commonly used as nuclear transplant donors. The erratic expression of imprinted genes in nuclear transplant embryos may be responsible for the large size of many mammalian nuclear transplant fetuses and their placentas (large offspring syndrome). However, to a remarkable extent the observed variation in gene expression does not seem to prevent the generation of morphologically normal mammals (60, 61).
Attempts to understand the mechanisms of nuclear reprogramming have so far been limited to a description of events that (i) rapidly follow the transplantation of nuclei to the cytoplasm of living eggs or oocytes, and (ii) take place in nuclei or permeabilized cells incubated in cell extracts in vitro. A massive enlargement of up to 100× volume, dispersal of chromatin, and extensive exchange of nuclear proteins from cytoplasm to nucleus and nucleus to cytoplasm are seen in somatic nuclei transplanted to amphibian eggs or oocytes (52). Somatic histone H1 is rapidly lost in transplanted bovine nuclei (62). Much protein exchange takes place in vitro in permeabilized cells (63), though it is not known whether these exchanged proteins are causally connected with reprogramming. Reversibly permeabilized cells may allow the reprogramming effect of imported or exported molecules to be assessed. Thus, Håkelien (64) exposed permeabilized fibroblasts to neural stem cell protein extracts, and saw polarized cell outgrowths, suggesting a neuronal reprogramming.
The Future of Nuclear Transplantation
The two principles to emerge from the first half-century of nuclear transplantation are the conservation of the genome during cell differentiation, and the ability of cell cytoplasm to reprogram gene activity and hence to redirect cell differentiation. Although certainly helping us to understand the processes of development and cell differentiation, the original purpose of nuclear transfer in multicellular animals, these two principles constitute essential requirements for reproductive and therapeutic cloning; if either condition did not prevail, cloning would not be possible.
Reproductive cloning, the production of adult animals by the transplantation of somatic cell nuclei to eggs, is of potential value for animal husbandry, for the preservation of rare genetic stocks, and perhaps for the production of genetically identical stocks for research. As a means of alleviating human infertility, scientists and many others argue that human reproductive cloning should be made illegal on account of the many defects observed postnatally in cloned mammals (65, 66).
Therapeutic cloning, on the other hand, that is the production by nuclear transfer of cells for replacement, could have many potential benefits if applied to humans. It would provide donor cells of the same genetic constitution as the recipient. This would avoid the need for immunosuppression that is required for most cases when donor and recipient are not genetically matched. There would be no genetic alteration of the product of a natural fertilization because the donated somatic cells would not persist beyond the life of the recipient. Therapeutic cloning would be expected to follow the route of deriving embryonic stem cells from nuclear transplant embryos (67) and the supply of such cells to a recipient in need of replacement cells (68).
What are the practical objections or limitations to cell replacement cloning in humans? The chief ethical objection is that the combination of a transplanted somatic nucleus (e.g., skin) and an unfertilized egg constitute a potential human being and should not be used as a source of spare parts. However, in the absence of implantation, a reconstituted embryo has no possibility of becoming a human being. Furthermore, it has been shown (18) that seriously defective nuclear transplant embryos that cannot survive can nevertheless provide a useful source of replacement cells.
There are many practical constraints with current methodology. A sufficient supply of human recipient eggs might be a limitation; the use of nonhuman eggs is unlikely to be a viable alternative because nucleo-cytoplasmic combinations between species do not develop beyond the blastocyst stage. We believe that the remarkable reprogramming activity of egg and oocyte cytoplasm (69) will eventually be understood in terms of identified molecules, and it may well be possible to apply the equivalent human molecules to reprogram somatic cells, which would have to be proliferated in vitro as are embryonic stem cells. It is possible that cells produced in this way would constitute a cancer risk. However, the disadvantage of a potential cancer risk might be preferable to the immediate suffering that would otherwise afflict those in urgent need of cell replacement. A second half-century of nuclear transplantation should identify the molecules and mechanisms that achieve nuclear reprogramming, and will almost certainly continue to help us understand normal mechanisms of development and cell differentiation.
Acknowledgments
We thank Davor Solter for discussion, and the Wellcome Trust, the Biotechnology and Biological Sciences Research Council, and the Manifold Trust for support of our own work.
Read more about this Classic PNAS article online at www.pnas.org/misc/classics.shtml.
References
- 1.Briggs, R. & King, T. J. (1952) Proc. Natl. Acad. Sci. USA 38 455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weismann, A. (1893) The Germ-Plasm: A Theory of Heredity (Walter Scott Ltd., London).
- 3.Spemann, H. (1938) Embryonic Development and Induction (Yale Univ. Press, New Haven, CT).
- 4.Comandon, J. & de Fonbrune, P. (1939) C. R. Seanc. Soc. Biol. 130 740-748. [Google Scholar]
- 5.Hammerling, J. (1934) Arch EntwMech. Org. 132 424-462. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, R. & King, T. J. (1957) J. Morphol. 100 269-312. [Google Scholar]
- 7.Smith, L. D. (1965) Proc. Natl. Acad. Sci. USA 54 101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King, T. J. & Briggs, R. (1956) Cold Spring Harbor Symp. Quant. Biol. 21 271-290. [DOI] [PubMed] [Google Scholar]
- 9.DiBerardino, M. A. & King, T. J. (1967) Dev. Biol. 15 102-128. [DOI] [PubMed] [Google Scholar]
- 10.Gurdon, J. B. & Hopwood, N. (2000) Int. J. Dev. Biol. 44 43-50. [PubMed] [Google Scholar]
- 11.Elsdale, T. R., Fischberg, M. & Smith, S. (1958) Exp. Cell Res. 14 642-643. [DOI] [PubMed] [Google Scholar]
- 12.Fischberg, M., Gurdon, J. B. & Elsdale, T. R. (1958) Nature 181 424. [DOI] [PubMed] [Google Scholar]
- 13.Gurdon, J. B. (1962) J. Embryol. Exp. Morphol. 10 622-640. [PubMed] [Google Scholar]
- 14.Gurdon, J. B. & Uehlinger, V. (1966) Nature 210 1240-1241. [DOI] [PubMed] [Google Scholar]
- 15.Laskey, R. A. & Gurdon, J. B. (1970) Nature 228 1332-1334. [DOI] [PubMed] [Google Scholar]
- 16.Gurdon, J. B., Laskey, R. A. & Reeves, O. R. (1975) J. Embryol. Exp. Morphol. 34 93-112. [PubMed] [Google Scholar]
- 17.DiBerardino, M. A. & Hoffner, N. J. (1983) Science 219 862-864. [DOI] [PubMed] [Google Scholar]
- 18.Byrne, J. A., Simonsson, S. & Gurdon, J. B. (2002) Proc. Natl. Acad. Sci. USA 99 6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurdon, J. B., Byrne, J. A. & Simonsson, S. (2003) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 20.Gurdon, J. B. (1960) J. Embryol. Exp. Morphol. 8 327-340. [PubMed] [Google Scholar]
- 21.Graham, C. F. (1969) Wistar Inst. Symp. Monogr. 9 19-35. [PubMed] [Google Scholar]
- 22.Baranska, W. & Koprowski, H. (1970) J. Exp. Zool. 174 1-14. [DOI] [PubMed] [Google Scholar]
- 23.Lin, T. P. (1971) in Methods in Mammalian Embryology (Freeman, San Francisco).
- 24.Bromhall, J. D. (1975) Nature 258 719-722. [DOI] [PubMed] [Google Scholar]
- 25.Illmensee, K. & Hoppe, P. C. (1981) Cell 23 9-18. [DOI] [PubMed] [Google Scholar]
- 26.McGrath, J. & Solter, D. (1984) Science 226 1317-1319. [DOI] [PubMed] [Google Scholar]
- 27.McLaren, A. (1984) Nature 309 671-672. [DOI] [PubMed] [Google Scholar]
- 28.McGrath, J. & Solter, D. (1983) Science 220 1300-1302. [DOI] [PubMed] [Google Scholar]
- 29.McGrath, J. & Solter, D. (1984) Cell 37 179-183. [DOI] [PubMed] [Google Scholar]
- 30.Surani, M. A., Barton, S. C. & Norris, M. L. (1984) Nature 308 548-550. [DOI] [PubMed] [Google Scholar]
- 31.Willadsen, S. M. (1986) Nature 320 63-65. [DOI] [PubMed] [Google Scholar]
- 32.Stice, S. L. & Robl, J. M. (1988) Biol. Reprod. 39 657-664. [DOI] [PubMed] [Google Scholar]
- 33.Prather, R. S., Sims, M. M. & First, N. L. (1989) Biol. Reprod. 41 414-418. [DOI] [PubMed] [Google Scholar]
- 34.Cheong, H. T., Takahashi, Y. & Kanagawa, H. (1993) Biol. Reprod. 48 958-963. [DOI] [PubMed] [Google Scholar]
- 35.Sims, M. & First, N. L. (1994) Proc. Natl. Acad. Sci. USA 91 6143-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng, L., Ely, J. J., Stouffer, R. L. & Wolf, D. P. (1997) Biol. Reprod. 57 454-459. [DOI] [PubMed] [Google Scholar]
- 37.Campbell, K. H., McWhir, J., Ritchie, W. A. & Wilmut, I. (1996) Nature 380 64-66. [DOI] [PubMed] [Google Scholar]
- 38.Wilmut, I., Schnieke, A. E., McWhir, J., Kind, A. J. & Campbell, K. H. (1997) Nature 385 810-813. [DOI] [PubMed] [Google Scholar]
- 39.Wakayama, T., Perry, A. C., Zuccotti, M., Johnson, K. R. & Yanagimachi, R. (1998) Nature 394 369-374. [DOI] [PubMed] [Google Scholar]
- 40.Kato, Y., Tani, T., Sotomaru, Y., Kurokawa, K., Kato, J., Doguchi, H., Yasue, H. & Tsunoda, Y. (1998) Science 282 2095-2098. [DOI] [PubMed] [Google Scholar]
- 41.Baguisi, A., Behboodi, E., Melican, D. T., Pollock, J. S., Destrempes, M. M., Cammuso, C., Williams, J. L., Nims, S. D., Porter, C. A., Midura, P., et al. (1999) Nat. Biotechnol. 17 456-461. [DOI] [PubMed] [Google Scholar]
- 42.Polejaeva, I. A., Chen, S. H., Vaught, T. D., Page, R. L., Mullins, J., Ball, S., Dai, Y., Boone, J., Walker, S., Ayares, D. L., et al. (2000) Nature 407 86-90. [DOI] [PubMed] [Google Scholar]
- 43.Chesne, P., Adenot, P. G., Viglietta, C., Baratte, M., Boulanger, L. & Renard, J. P. (2002) Nat. Biotechnol. 20 366-369. [DOI] [PubMed] [Google Scholar]
- 44.Shin, T., Kraemer, D., Pryor, J., Liu, L., Rugila, J., Howe, J., Buck, S., Murphy, K., Lyons, L. & Westhusin, M. (2002) Nature 415 859-860. [DOI] [PubMed] [Google Scholar]
- 45.Hochedlinger, K. & Jaenisch, R. (2002) Nature 415 1035-1038. [DOI] [PubMed] [Google Scholar]
- 46.Tsunoda, Y. & Kato, Y. (2002) Differentiation 69 158-161. [DOI] [PubMed] [Google Scholar]
- 47.Hill, J. R., Burghardt, R. C., Jones, K., Long, C. R., Looney, C. R., Shin, T., Spencer, T. E., Thompson, J. A., Winger, Q. A. & Westhusin, M. E. (2000) Biol. Reprod. 63 1787-1794. [DOI] [PubMed] [Google Scholar]
- 48.Eggan, K. & Jaenisch, R. (2002) in Principles of Cloning, eds. Cibelli, J. B., Lanza, R. P., Campbell, K. H. & West, M. D. (Academic, San Diego), pp. 85-98.
- 49.Blau, H. M., Chiu, C. P. & Webster, C. (1983) Cell 32 1171-1180. [DOI] [PubMed] [Google Scholar]
- 50.Rossant, J. (2002) Nature 415 967-969. [DOI] [PubMed] [Google Scholar]
- 51.De Robertis, E. M. & Gurdon, J. B. (1977) Proc. Natl. Acad. Sci. USA 74 2470-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gurdon, J. B. (1986) J. Cell Sci. Suppl. 4 287-318. [DOI] [PubMed] [Google Scholar]
- 53.Humpherys, D., Eggan, K., Akutsu, H., Friedman, A., Hochedlinger, K., Yanagimachi, R., Lander, E. S., Golub, T. R. & Jaenisch, R. (2002) Proc. Natl. Acad. Sci. USA 99 12889-12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bortvin, A., Eggan, K., Skaletsky, H., Akutsu, H., Berry, D. L., Yanagimachi, R., Page, D. C. & Jaenisch, R. (2003) Development (Cambridge, U.K.) 130 1673-1680. [DOI] [PubMed] [Google Scholar]
- 55.Eggan, K., Akutsu, H., Hochedlinger, K., Rideout, W., III, Yanagimachi, R. & Jaenisch, R. (2000) Science 290 1578-1581. [DOI] [PubMed] [Google Scholar]
- 56.Lanza, R. P., Cibelli, J. B., Blackwell, C., Cristofalo, V. J., Francis, M. K., Baerlocher, G. M., Mak, J., Schertzer, M., Chavez, E. A., Sawyer, N., et al. (2000) Science 288 665-669. [DOI] [PubMed] [Google Scholar]
- 57.Daniels, R., Hall, V. & Trounson, A. O. (2000) Biol. Reprod. 63 1034-1040. [DOI] [PubMed] [Google Scholar]
- 58.Inoue, K., Kohda, T., Lee, J., Ogonuki, N., Mochida, K., Noguchi, Y., Tanemura, K., Kaneko-Ishino, T., Ishino, F. & Ogura, A. (2002) Science 295 297. [DOI] [PubMed] [Google Scholar]
- 59.Dean, W., Santos, F., Stojkovic, M., Zakhartchenko, V., Walter, J., Wolf, E. & Reik, W. (2001) Proc. Natl. Acad. Sci. USA 98 13734-13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Humpherys, D., Eggan, K., Akutsu, H., Hochedlinger, K., Rideout, W. M., III, Biniszkiewicz, D., Yanagimachi, R. & Jaenisch, R. (2001) Science 293 95-97. [DOI] [PubMed] [Google Scholar]
- 61.Reik, W. & Walter, J. (2001) Nat. Rev. Genet. 2 21-32. [DOI] [PubMed] [Google Scholar]
- 62.Bordignon, V., Clarke, H. J. & Smith, L. C. (1999) Biol. Reprod. 61 22-30. [DOI] [PubMed] [Google Scholar]
- 63.Kikyo, N., Wade, P. A., Guschin, D., Ge, H. & Wolffe, A. P. (2000) Science 289 2360-2362. [DOI] [PubMed] [Google Scholar]
- 64.Hakelien, A. M., Landsverk, H. B., Robl, J. M., Skalhegg, B. S. & Collas, P. (2002) Nat. Biotechnol. 20 460-466. [DOI] [PubMed] [Google Scholar]
- 65.Jaenisch, R. & Wilmut, I. (2001) Science 291 2552. [DOI] [PubMed] [Google Scholar]
- 66.McLaren, A. (2000) Science 288 1775-1780. [DOI] [PubMed] [Google Scholar]
- 67.Munsie, M. J., Michalska, A. E., O'Brien, C. M., Trounson, A. O., Pera, M. F. & Mountford, P. S. (2000) Curr. Biol. 10 989-992. [DOI] [PubMed] [Google Scholar]
- 68.Rideout, W. M., III, Hochedlinger, K., Kyba, M., Daley, G. Q. & Jaenisch, R. (2002) Cell 109 17-27. [DOI] [PubMed] [Google Scholar]
- 69.Bolani, M., Eckardt, S., Schöler, H. R. & McLaughlin, K. J. (2002) Genes Dev. 16 1209-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pennisi, E. & Vogel, G. (2000) Science 288 1722-1727. [DOI] [PubMed] [Google Scholar]
- 71.Hill, J. R. & Chavatte-Palmer, P. (2002) in Principles of Cloning, eds. Cibelli, J. B., Lanza, R. P., Campbell, K. H. & West, M. D. (Academic, San Diego).
- 72.Graham, C. F., Arms, K. & Gurdon, J. B. (1966) Dev. Biol. 14 349-381. [Google Scholar]
- 73.Gurdon, J. B. (1968) J. Embryol. Exp. Morphol. 20 401-414. [PubMed] [Google Scholar]