Abstract
The GPCRDB is a molecular class-specific information system that collects, combines, validates and disseminates heterogeneous data on G protein-coupled receptors (GPCRs). The database stores data on sequences, ligand binding constants and mutations. The system also provides computationally derived data such as sequence alignments, homology models, and a series of query and visualization tools. The GPCRDB is updated automatically once every 4–5 months and is freely accessible at http://www.gpcr.org/7tm/.
INTRODUCTION
G protein-coupled receptors (GPCRs) are a major target for the pharmaceutical industry as is reflected by the fact that more than 50% of all medicines available today act on a GPCR (1). GPCRs consist of seven transmembrane helices that are connected by loops. The N-terminal extremity is always located extra-cellularly, while the C-terminus extends into the cytoplasm. GPCRs are found in a diverse range of species where they are involved in signalling from outside the cell to inside the cell. Most GPCRs detect a ligand, that upon binding, elicits a response that is felt by the heterotrimeric G protein at the cytosolic side. This G protein in turn activates a second messenger system by modulating the activity of enzymes such as adenylyl cyclase or phospholipase C. Ligands for GPCRs are heterogeneous molecules and include ions, hormones, neurotransmitters, peptides and proteins. Stimuli such as light, taste or odor can activate sensory GPCRs [see (2) for review].
In a related application, we have used the molecular class-specific information system (MCSIS) technology to maintain the NucleaRDB nuclear receptor information system (8). In this current report, the authors explain how the MCSIS technology is used to maintain the GPCRDB.
The GPCRDB is presently the only regularly updated GPCR information system available on the World Wide Web [WWW; see (3) for a list of GPCR-related websites]. Since its introduction in late 1994, the GPCRDB has received increasing attention from the scientific community.
DATA CONTENT
The GPCRDB contains sequences, mutation and ligand binding data that are regularly imported from the original sources [SWISS-PROT (4), EMBL (5), TinyGRAP (6)]. Recently, a significant amount of ligand binding data was incorporated from Organon (7). Table 1 summarises statistics about the data content of the GPCRDB.
Table 1. Statistics about the September 2002 release of the GPCRDB.
| GPCR sequences | 4609 |
| Including fragments | 1654 |
| cDNA/protein pairwise alignments | 3978 |
| Families, sub families, etc. | 283 |
| Multiple sequences alignments (HSSP format) | 280 |
| Phylogenetic trees | 352 |
| Ligand bindind data (from P. Seeman) for | 28 receptor sub-families |
| Ligand bindind data (from Organon) for | 54 receptors and 300 ligands |
| Point mutations extracted from the literature | 2577 (517 articles) |
| 3D Models (11 depositors) | 1898 |
| PDB files (links to) | 78 |
PRIMARY DATA
Sequences are updated as described previously (8).
Structure data is only available for bovine rhodopsin (9). For years, there was no high-resolution structure of a GPCR and the structure of the bacteriorhodopsin was used as a reference. Thus, many individuals within and outside industry have built 3D models of a variety of GPCRs. Some of these models were deposited in the GPCRDB, in addition to the 3D models we automatically built based on three different templates.
Mutation data is obtained from the manually curated TinyGRAP (6) and is fully integrated throughout the GPCRDB. We also provide point mutation data extracted from online literature using an automated procedure (Horn and Cohen, in preparation).
Ligand-binding data has been obtained from a collection by P. Seeman (10) and from Organon (7). Information on agonists and antagonists binding is very difficult to collect because of the complexity of the nomenclature (antagonists to activators and agonists to repressors tend to show similar effects in assays, which leads to confusion when extracting information from literature). We encourage academic and industrial researchers to submit their ligand binding data to the GPCRDB, in order to make this information searchable and more accessible to the scientific community.
COMPUTATIONALLY-DERIVED DATA
Multiple sequence alignments are performed with WHAT IF (11) using an iterative sequence-based profile (12). The multiple sequence alignments are provided in different formats (HSSP, MSF). In all the alignments, the seven transmembrane helices are annotated and numbered using a general numbering scheme suggested by Oliveira et al. (1993).
cDNA-protein alignments and phylogenetic trees are generated as described previously (8).
Correlated Mutation Analysis (CMA) is used to identify pairs of residues that remained conserved or mutated in tandem during evolution. The rationale behind this analysis is that when a mutation occurs at a functionally important site, the protein either becomes non-functional or may acquire its original or a different function due to a compensatory mutation at another position. Residues detected by the CMA method are often involved in intermolecular interactions [e.g. between ligands and receptors; see (13–15) for an explanation of the methods and examples of CMA application]. Residues detected by the CMA method are indicated in multiple sequence alignments and in snake-like diagrams.
DATABASE CROSS-LINKING
For each GPCR, a table of cross-references lists all the available pointers to local and remote information. This is done automatically by reading the SWISS-PROT entries and querying other remote resources and the GPCRDB itself. In addition, we are currently trying to map the GPCR classifications in the GPCRDB to Gene Ontology (16) identifiers. The Gene Ontology consortium provides a controlled vocabulary for the description of molecular function, biological process and cellular components of gene products. As a first step into this direction, each table now contains a link to the Gene Ontology database via the QuickGO system (http://golgi.ebi.ac.uk/ego/index.html). In addition, we will integrate the IUPHAR receptor codes, which will be publicly available in December 2002.
DISSEMINATION FACILITIES
MCSIS provide fast and easy access to all information related to an underlying molecular class. For this purpose we have implemented and will continue to develop the four basic information system tools: browsing, retrieval, query and inferring. Inference engine facilities are as described previously (8).
Browsing
The data organisation is based on the pharmacological classification of GPCRs and the main way to access the data is via a hierarchical list of known families in agreement with this classification. For a specific family, users can access individual sequences, multiple sequence alignments, the profiles used to perform the latter, snake-like diagrams and phylogenetic trees. Each type of data is displayed in a WWW page with hyperlinks to other data, where appropriate. Another way to access the data is to browse lists that display one type of data (e.g. all chromosomal locations, all mutations, etc). In addition, a dynamic page lists all SWISS-PROT and TrEMBL entries present in the database. Two-dimensional snake-like diagrams are used to represent and combine GPCR sequence, 2D structure and mutation information. These diagrams are automatically generated using the Viseur program (17). There are three types of snake-like diagrams: the first one displays mutation data for each receptor, the second superimposes all the mutation data available for one receptor family onto the corresponding consensus sequence and the last one shows the residues detected by the CMA method for each receptor family. In the first two types of diagrams, the ‘mutated’ residues are hyperlinked to the TinyGRAP database (6). In the CMA snake-like diagram, residues with a predicted important functional role are hyperlinked to the multiple sequence alignments and to details of the CMA results.
Retrieval
Most data can be retrieved in its native form using the ‘save as’ option of the browser or via anonymous FTP from ftp://www.gpcr.org.7tm/. A complete copy of the GPCRDB can be made available to academic and industrial researchers upon request (G.V.).
Query
A basic query system is also available to retrieve GPCR entries using keywords and SWISS-PROT identifiers. In addition, users can also run a BLAST search against the GPCRDB sequence data via the CMBI server.
Inferences
CMA is a powerful tool for the computational discovery of novel inter-residue relationships, and thus represent a useful inference engine. With little computational effort, the potential functionally or structurally relevant residues are selected from among the thousands of residues in each alignment. We make these residue positions available for browsing purposes by mapping them onto multiple sequence alignments.
DISCUSSION
The GPCRDB started in 1993 as an Email-based system and was converted in 1994 to the WWW. It has been in continuous service now for almost 10 years and is probably the only non-government sponsored database/information system with such longevity. It is routinely used from fifty to a hundred thousand times per month by researchers in industry and academia from more than 120 countries (see the usage statistics at http://www.gpcr.org/7tm/htmls/analog.html).
Although users are not obliged to inform us why they use the GPCRDB, we have reason to believe that the main usage is simple to retrieve information that would otherwise have to be collected from multiple sites. Furthermore, the system is often used to obtain knowledge about orphan receptors. Correlated mutations, the GPCR specific BLAST, multiple sequence alignments and phylogenetic trees are used for this purpose.
The existence of the GPCRDB as a recognizable site for GPCR-related data also leads to the increased dissemination of data. As an example, we received two large batches of ligand binding data (7,10).
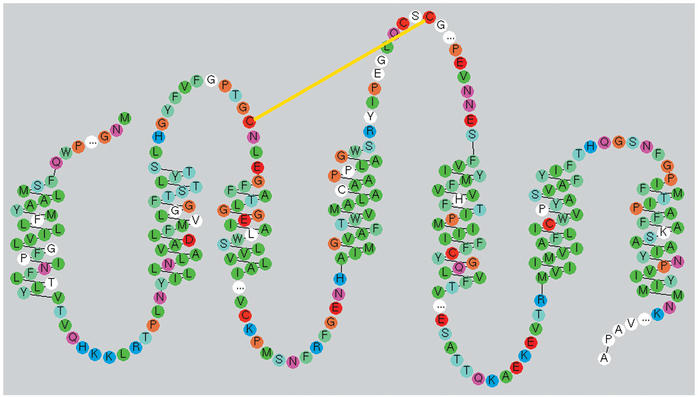
The so-called snake-like diagrams are a popular way to access mutation data (Fig. 1). Some of the snake-like diagrams have been used more than ten thousand times for this purpose.
Figure 1.
Snake-like diagram of the human Rhodopsin receptor. This plot was generated with the RbDe software (18), which will be used for the next update of the snake-like diagrams. Residues are colored based on their chemical properties. White residues indicate that mutation data is available. The yellow line represents a disulfide bridge and ‘…’ indicates hidden residues.
The most important reason for the popularity of the GPCRDB is probably its long term presence as a regularly updated one-stop-resource for GPCR data.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jacob de Vlieg, Robert Bywater and the MCSIS team at the CMBI for stimulating discussions. The MCSIS project is financially supported by Organon and Unilever. F.H. and F.E.C. acknowledge the NIH for support.
REFERENCES
- 1.Gudermann T., Nurnberg,B. and Schultz,G. (1995) Receptors and G proteins as primary components of transmembrane signal transduction. Part 1. G-protein-coupled receptors: structure and function. J. Mol. Med., 73, 51–63. [DOI] [PubMed] [Google Scholar]
- 2.Watson S. and Arkinstall,S. (eds) (1994) The G Protein Linked Receptor Facts Book. Academic Press, London, UK.
- 3.Rana B.K. and Insel,P.A. (2002) G-protein-coupled receptor websites. Trends Pharmacol. Sci., 23, 535–536. [DOI] [PubMed] [Google Scholar]
- 4.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoesser G., Baker,W., van den Broek,A., Camon,E., Garcia-Pastor,M., Kanz,C., Kulikova,T., Leinonen,R., Lin,Q., Lombard,V., Lopez,R., Redaschi,N., Stoehr,P., Tuli,M.A., Tzouvara,K. and Vaughan,R. (2002) The EMBL Nucleotide Sequence Database. Nucleic Acids Res., 30, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beukers M.B., Kristiansen,K., IJzerman,A.P. and Edvardsen,O. (1999) TinyGRAP database: a bioinformatics tool to mine G-protein-coupled receptor mutant data. Trends Pharmacol. Sci., 20, 475–477. [DOI] [PubMed] [Google Scholar]
- 7.Cutler D., Barbier,A. and Pestell,K. (2002) In brief. Trends Pharmacol. Sci., 23, 258–259. [PubMed] [Google Scholar]
- 8.Horn F., Vriend,G. and Cohen,F.E. (2001) Collecting and harvesting biological data: the GPCRDB and NucleaRDB information systems. Nucleic Acids Res., 29, 346–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palczewski K., Kumasaka,T., Hori,T., Behnke,C.A., Motoshima,H., Fox, B.A., Le Trong,I., Teller,D.C., Okada,T., Stenkamp,R.E., Yamamoto,M. and Miyano,M. (2000) Crystal structure of rhodopsin: a G protein-coupled receptor. Science, 289, 739–745. [DOI] [PubMed] [Google Scholar]
- 10.Seeman P. (1993) Receptor Tables Vol. 2: Drug dissociation constants for neuroreceptors and transporters, SZ Research, Toronto.
- 11.Vriend G. (1990) WHAT IF: a molecular modeling and drug design program. J. Mol. Graph., 8, 29, 52–56. [DOI] [PubMed] [Google Scholar]
- 12.Oliveira L., Paiva,A.C. and Vriend,G. (1993) A common motif in G protein-coupled seven transmembrane helix receptors. J. Comp. Aided Mol. Des., 7, 649–658. [Google Scholar]
- 13.Oliveira L., Paiva,A.C.M. and Vriend,G. (1995) In Kaumaya,P.T.P. and Hodges,R.S. (eds), Peptides: Chemistry, Structure and Biology. Mayflower Scientific Ltd., Kingswinford, UK, pp. 408–409.
- 14.Kuipers W., Oliveira,L., Paiva,A.C.M., Rippman,F., Sander,C., Vriend,G. and IJzerman,A.P. (1996) In Findlay,J.B.C. (ed.), Membrane Protein Models. BIOS Scientific Publishers Ltd, Oxford, UK, pp. 27–45.
- 15.Horn F., Bywater,R., Krause,G., Kuipers,W., Oliveira,L., Paiva,A.C., Sander,C. and Vriend,G. (1998) The interaction of class B G protein-coupled receptors with their hormones. Receptors Channels, 5, 305–314. [PubMed] [Google Scholar]
- 16.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T., Harris,M.A., Hill,D.P., Issel-Tarver,L., Kasarskis,A., Lewis,S., Matese,J.C., Richardson,J.E., Ringwald,M., Rubin,G.M. and Sherlock,G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campagne F., Jestin,R., Reversat,J.L., Bernassau,J.M. and Maigret,B. (1999) Visualisation and integration of G protein-coupled receptor related information help the modelling: description and applications of the Viseur program. J. Comput. Aided Mol. Des., 13, 625–643. [DOI] [PubMed] [Google Scholar]
- 18.Konvicka K., Campagne,F. and Weinstein,H. (2000) Interactive construction of residue-based diagrams of proteins: the RbDe web service. Protein Eng., 13, 395–396. [DOI] [PubMed] [Google Scholar]