Abstract
Parkinson's disease (PD) is the most frequent neurodegenerative movement disorder. Mutations in the PINK1 gene are linked to the autosomal recessive early onset familial form of PD. The physiological function of PINK1 and pathological abnormality of PD-associated PINK1 mutants are largely unknown. We here show that inactivation of Drosophila PINK1 (dPINK1) using RNAi results in progressive loss of dopaminergic neurons and in ommatidial degeneration of the compound eye, which is rescued by expression of human PINK1 (hPINK1). Expression of human SOD1 suppresses neurodegeneration induced by dPINK1 inactivation. Moreover, treatment of dPINK1 RNAi flies with the antioxidants SOD and vitamin E significantly inhibits ommatidial degeneration. Thus, dPINK1 plays an essential role in maintaining neuronal survival by preventing neurons from undergoing oxidative stress, thereby suggesting a potential mechanism by which a reduction in PINK1 function leads to PD-associated neurodegeneration.
Keywords: neurodegeneration, oxidative stress, Parkinson's disease, SOD1
Parkinson's disease (PD) is a progressive neurodegenerative movement disorder characterized by age-dependent resting tremor, muscular rigidity, and akinesia. The disease affects ≈1–2% of the population over 65 years of age (1, 2). The pathological hallmarks of PD patients include progressive loss of dopaminergic (DA) neurons in the substantia nigra pars compacta as well as the presence of ubiquitin-positive Lewy neurites and Lewy bodies in the remaining neurons. No treatment is currently available to prevent disease progression and neurodegeneration, although administration of l-dopa temporarily relieves parkinsonism. Understanding the molecular basis of PD is likely to facilitate development of effective therapies of the disease.
The molecular pathways that result in the PD-specific pathological changes and concomitant motor deficits are largely unknown. Nevertheless, significant progress on molecular genetics of PD using early onset familial cases has been made during the last several years. Majority of the PD cases appear to be sporadic. However, specific genetic defects are linked to familial form of PD that resemble idiopathic PD. Mutations in at least six genes are individually linked to familial forms of PD, including autosomal dominant mutations in α-synuclein, uchL1, and LRRK2 and autosomal recessive mutations in parkin, PINK1, and DJ-1 (3–6). Characterization of these genes has provided important insights into the pathogenesis of PD. For example, α-synuclein is a major structural component of Lewy bodies in PD (7). PD-associated α-synuclein mutant proteins show an increased propensity to self-aggregate to form oligomeric structures and Lewy body-like fibrils comparing to wild-type α-synuclein, thereby directly linking the disease-associated α-synuclein mutant proteins to PD pathology (8, 9).
Among the three genes linked to the autosomal recessive early onset familial form of PD, mutations in PINK1 appear to be the second-most-common genetic cause in autosomal recessive PD (after parkin), found in 8–15% early onset PD cases (4, 10, 11). Heterozygous mutations of PINK1 were also detected in sporadic PD cases (11, 12). The PINK1 gene encodes a putative kinase that acts on yet unidentified substrates and contains an N-terminal mitochondrial targeting motif (4). PINK1 localization to mitochondria has indeed been reported in transfected cells and human brain neurons (4, 13, 14), but its biological function remains unclear. Functional studies suggest that wild-type PINK1 may have a neuroprotective role (4) that is abrogated by pathogenic mutations in the PINK1 gene. Consistent with the notion, we have recently shown that PINK1 and DJ-1 physically associate and collaborate to protect cells against oxidative stress (15). These results suggest a potential role of PINK1 in maintaining mitochondrial homeostasis and in defending against oxidative stress.
In the present study, we have examined the function of dPINK1 in neuronal survival and in protection against oxidative stress by transgenic RNAi-mediated inactivation of PINK1 in Drosophila. Our results suggest that dPINK1 plays an essential role in maintaining neuronal survival by protecting (DA) neurons from undergoing oxidative stress, thus indicating a potential cause for PD-associated neurodegeneration triggered by loss-of-PINK1-function.
Results and Discussion
PINK1 gene encodes a 581-aa putative serine/threonine kinase with unknown substrates and function. Sequence analysis revealed a single Drosophila homolog (CG4523) of hPINK1with 52% similarity at the amino acid level that was designated as Drosophila PINK1 (dPINK1) (Fig. 5, which is published as supporting information on the PNAS web site). Quantitative real-time PCR showed that dPINK1 is expressed throughout the lifespan of Drosophila, with the highest levels in pupal stages and slightly reduced levels in adults (Fig. 6A, which is published as supporting information on the PNAS web site). In adult flies, dPINK1 mRNA is highly abundant in brain including the retina of the eye (Fig. 6B), suggesting a role for dPINK1 in the developing or adult nervous system of Drosophila.
To investigate the mechanism of PINK1 mutants in PD pathogenesis using Drosophila as a model, we reduced dPINK1 function with a transgenic dPINK1-RNAi by using the GAL4/UAS system (16). Knockdown of dPINK1 in multiple transgenic lines was verified by RT-PCR (Fig. 6C). Inactivation of dPINK1 driven by the ubiquitous driver daughterless (da)-GAL4 resulted in embryonic lethality of second-instar larvae. To determine RNAi specificity for dPINK1, we generated transgenic Drosophila lines expressing hPINK1. Expression of hPINK1 variants in flies driven by da-GAL4 was verified by immunoprecipitation followed by immunoblotting (Fig. 6D). Expression of hPINK1, but not lacZ or GFP, fully rescued the lethality caused by dPINK1 RNAi (Table 1, which is published as supporting information on the PNAS web site; data not shown). In contrast, expression of a mutant form of PINK1, hPINK1G309D, which was identified in patients with PD (4), failed to rescue (data not shown). The observed rescue is likely to be specific to hPINK1 activity, because little dPINK1 was detected in flies expressing both dPINK1 RNAi and hPINK1 cDNA (Fig. 6E). These results suggest that dPINK1 apparently is the functional homolog of hPINK1.
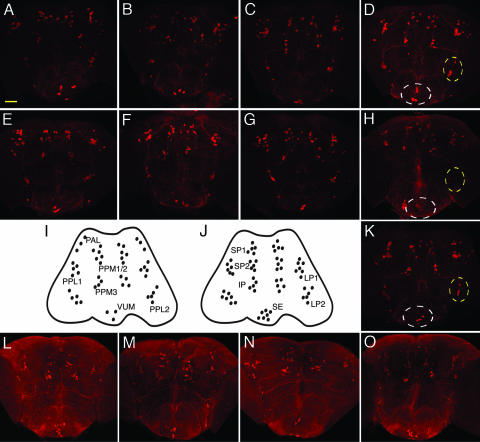
We next examined the effect of dPINK1 inactivation on DA neurons. There are 13 well defined DA neuronal clusters normally present in Drosophila adult brain, including six paired clusters in each brain hemisphere and an unpaired ventral medial cluster (VUM) (Fig. 1I). dPINK1 inactivation in DA neurons or all neurons was achieved by using Ddc-GAL4 or elav-GAL4 drivers, respectively. DA neurons were detected by immunofluorescent staining of whole-mount fly brains at 1 and 10 days of age by using an anti-tyrosine hydroxylase (TH) antibody (17). When dPINK1 was inactivated by using the elav-GAL4 driver, flies at 1 day of age showed little difference in the total number and distribution of DA neurons in brain compared to control flies expressing the elav-GAL4 driver alone, elav-GAL4 driven lacZ, or elav-GAL4 driven hPINK1 (Fig. 1 A–D). In contrast, a dramatic reduction in the number of DA neurons was seen in fly brains with dPINK1 inactivation at 10 days of age compared to age-matched controls (Fig. 1 E–H). Significant neuronal loss was observed in most DA neuron clusters including in PAL, PPM1/2, PPM3, PPL2, and less so in PPL1 (Fig. 1; see Fig. 3E). In contrast, the number of DA neurons in the VUM regions of the brain were not significantly affected, although the intensity of TH staining in this and other clusters seem to be reduced. These results agree with previously reported selective loss of DA neurons seen as early as 10 days of age in transgenic flies overexpressing human α-synuclein (18, 19). A similar loss of DA neurons was observed in flies with Ddc-GAL4 driven dPINK1 RNAi (data not shown). Loss of DA neurons in flies expressing dPINK1 RNAi in both cases is likely specific to the loss of dPINK1 function because it was rescued by coexpression of hPINK1 (Fig. 1K; see Fig. 3E). Staining of serotonergic neurons showed little change between flies expressing elav-GAL4 alone and elav-GAL4 driven dPINK1 RNAi (Fig. 1 L–O). Thus, inactivation of dPINK1 results in progressive and apparently selective degeneration of DA neurons in the Drosophila brain.
Fig. 1.
Knockdown of dPINK1 results in age-dependent loss of DA neurons, but not serotonergic neurons, in Drosophila. Brains dissected from flies expressing elav-GAL4 driver alone (A, E, and L), elav-GAL4/UAS-lac Z (B, F, and M), elav-GAL4/UAS-hPINK1 (C, G, and N), elav-GAL4/UAS-dPINK1 RNAi (D, H, and O), and elav-GAL4/UAS-dPINK1 RNAi/UAS-hPINK1 (K) aged 1 day (A–D) or 10 days (E–G, K, and L–O) were immunostained with either anti-Drosophila TH antibody or anti-5HT antibody followed by an Alexa Fluor 594-labeled secondary antibody to identify DA (A–H and K) or serotonergic (L–O) neurons, respectively. Representative pictures shown were collected by using confocal microscopy. Localization of Drosophila DA and serotonergic neurons are illustrated in I and J, respectively. Note that changes of DA neurons in VUM (white circle) and PPL2 (yellow circle) regions in 1-day (D) and 10-day (H) fly brains with dPINK1 knockdown as well as 10-day fly brains (K) with dPINK1 knockdown rescued by hPINK1 are indicated. Serotonergic neurons remain similar among all groups (L–O). (Scale bar, 50 μm.)
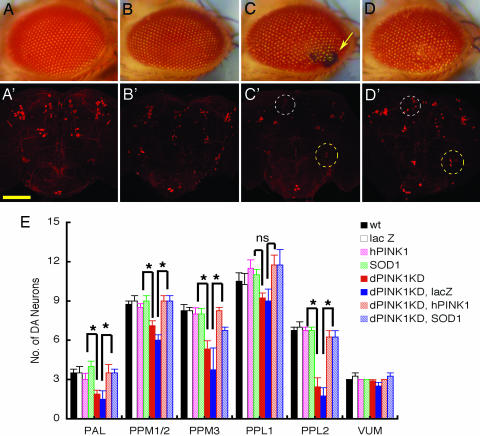
Fig. 3.
Expression of hSOD1 suppresses degeneration of ommatidia and DA neurons. Representative images of external eye phenotypes of 3-day-old flies expressing GMR-GAL4 driver alone (A), GMR-GAL4/UAS-lac Z (B), GMR-GAL4/UAS-dPINK1 RNAi (C), and GMR-GAL4/UAS-dPINK1 RNAi/UAS-hSOD1 (D), as well as images of DA neurons identified by anti-Drosophila TH staining of 10-day-old flies expressing elav-GAL4 driver alone (A′), elav-GAL4/UAS-lac Z (B′), elav-GAL4/UAS-dPINK1 RNAi (C′), and elav-GAL4/UAS-dPINK1 RNAi/UAS-hSOD1 (D′). Note that ommatidial degeneration (C, yellow arrow) and loss (C′) and rescue (D′) of DA neurons in PPM1/2 (white circle) and PPL2 (yellow circle) are indicated. (Scale bar, 50 μm.) (E) Quantification of DA neurons is shown for 10-day-old flies expressing elav-GAL4 driver alone (WT), elav-GAL4/UAS-lac Z (lac Z), elav-GAL4/UAS-hPINK1 (hPINK1), elav-GAL4/UAS-hSOD1 (SOD1), elav-GAL4/UAS-dPINK1 RNAi (dPINK1KD), elav-GAL4/UAS-dPINK1 RNAi/UAS-lac Z (dPINK1KD, lacZ), elav-GAL4/UAS-dPINK1 RNAi/UAS-hPINK1 (dPINK1KD, hPINK1), and elav-GAL4/UAS-dPINK1 RNAi/UAS-hSOD1 (dPINK1KD, SOD1). DA neurons in six brain regions, including PAL, PPM1/2, PPM3, PPL1, PPL2, and VUM, were quantitated, and differences were statistically analyzed. ∗, P < 0.05. ns, no statistical significance.
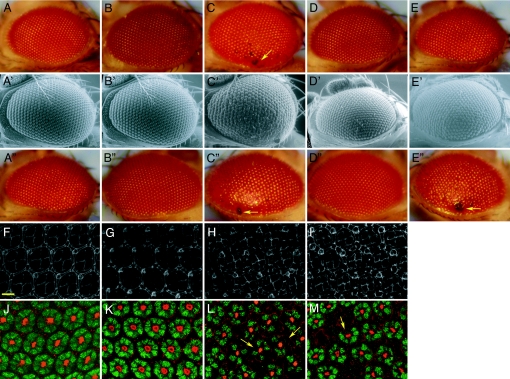
However, dPINK1 inactivation-mediated neurodegeneration is not restricted to DA neurons of the brain. Expression of dPINK1 RNAi in Drosophila eyes using the GMR-GAL4 driver resulted in age-dependent progressive ommatidial degeneration as manifested in black lesions in the external eyes that is rescued by coexpression of wild-type hPINK1 (Fig. 2A–E and Fig. 7, which is published as supporting information on the PNAS web site). Scanning electron microscopic imaging also revealed a “rough eye” phenotype, disorganized interommatidial bristles, and degeneration of ommatidia in flies with dPINK1 knockdown (Fig. 2 A′–D′). However, ommatidial degeneration induced by dPINK1 RNAi could not be rescued by coexpression of a PD-associated PINK1G309D mutant (Figs. 2 A″–E″ and 6D). The results suggest that mutant forms of PINK1 that are associated with PD in humans, unlike the wild-type counterpart, are no longer able to protect ommatidia with dPINK1-RNAi from degeneration. To elucidate the cellular manifestation of dPINK1-RNAi-induced ommatidial degeneration, we examined the arrangement and integrity of ommatidial cells and pupal retinal neurons 44 h after pupae formation (APF). Immunostaining for Discs Large (Dlg), a Drosophila membrane-associated guanylate kinase protein (MAGUK) (20), revealed morphologically disrupted ommatidia, increased and misoriented mechanosensory bristle groups, and disorganized and enlarged pigment cells after dPINK1 inactivation compared with controls (Fig. 2 F–I). Colabeling with phalloidin (revealing actin organization) and an anti-elav antibody (marking neuronal nuclei) shows a significant loss of photoreceptor neurons 44 h APF in pupal retinas with dPINK1-RNAi (Fig. 2 J–M). These results reveal that dPINK1 inactivation in the Drosophila eye induces degeneration of photoreceptors. This rough-eye and ommatidial degeneration phenotype appears to be autonomous to photoreceptors because dPINK1-RNAi driven by pan-neuronal elav-GAL4 was indistinguishable to that of GMR-GAL4 driven dPINK1-RNAi (Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 2.
Knockdown of dPINK1 induces degeneration of ommatidia and retinal neurons in Drosophila. External eye phenotypes of flies aged 1 day expressing GMR-GAL4 driver alone (A, A′, and A″), GMR-GAL4/UAS-lac Z (B, B′, and B″), GMR-GAL4/UAS-dPINK1 RNAi (C, C′, C″), GMR-GAL4/UAS-hPINK1 (D and D′), GMR-GAL4/UAS-hPINK1G309D (D″), GMR-GAL4/UAS-dPINK1 RNAi/UAS-hPINK1 (E and E′), and GMR-GAL4/UAS-dPINK1 RNAi/UAS-hPINK1 G309D (E″) under light microscopy (A–E and A″–E″) and electronic microscopy (A′–E′). Magnification of light microscopic images and electronic microscopic images is ×25 and ×400, respectively. Arrows indicate lesions of ommatidial degeneration (C, C′, C″, and E″). Microscopic images of DLG staining (F–I) as well as phalloidin and elav double staining of 44 h AFP retina dissected from flies expressing GMR-GAL4 driver alone (F and J), GMR-GAL4/UAS-lac Z (G and K), and GMR-GAL4/UAS-dPINK1 RNAi (H, I, L, and M). Images from two independent lines expressing GMR-GAL4/UAS-dPINK1 RNAi are shown (H and L are from one line, whereas I and M are from another independent line). Note that dPINK1 knockdown induces cellular disorganization (H and I) and loss of neurons (L and M; yellow arrows). (Scale bar, 10 μm.)
Rough eyes and the loss of ommatidia induced by dPINK1-RNAi is unlikely to occur via apoptosis, because TUNEL assay detected little signal in the pupal retina of both RNAi flies and their controls 44 h after pupae formation (data not shown). Moreover, expression of dIAP1, a Drosophila inhibitor of apoptosis protein (21), did not inhibit ommatidial degeneration induced by dPINK1-RNAi (Fig. 9, which is published as supporting information on the PNAS web site). Interestingly, expression of PD-associated PINK1 mutants potentiates oxidative stress-induced death of transfected SH-SY5Y cells in vitro (4). To investigate possible molecular mechanisms underlying neuronal loss induced by dPINK1 inactivation, we determined whether expression of SOD1 could prevent degeneration of DA neurons and ommatidia in flies with dPINK1 inactivation. SOD1 is an antioxidant enzyme found in the cytosol, nucleus, peroxisomes, and mitochondrial intermembrane space of eukaryotic cells (22). Expression of human SOD1, but not lacZ, in flies with dPINK1 knockdown under the control of the GMR-Gal4 driver markedly suppressed ommatidial degeneration (Fig. 3 A–D). Consistent with this finding, expression of human SOD1 driven by elav-GAL4 remarkably inhibited dPINK1 inactivation-induced degeneration of DA neurons (Fig. 3 A′–D′ and E). These results suggest that dPINK1 inactivation is likely to induce neuronal death via an oxidative stress pathway.
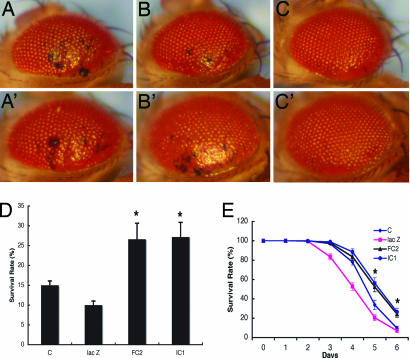
To further explore the involvement of oxidative stress in dPINK1 inactivation-dependent neurodegeneration, we treated GMR-GAL4 driven dPINK1-RNAi flies with compounds that exhibit antioxidant activity, including recombinant SOD1 protein (as in refs. 38 and 39) and vitamin E, by adding the antioxidants to the flies' diet. Treatment with SOD or vitamin E inhibited ommatidial degeneration in a dose-dependent manner (Fig. 4 A–C and A′–C′ and Fig. 10, which is published as supporting information on the PNAS web site). In GMR-GAL4 driven dPINK1 RNAi flies, SOD treatment also inhibited degeneration of DA neuron, especially of the PPL3 cluster (Fig. 11, which is published as supporting information on the PNAS web site). These results suggest that inactivation of dPINK1 results in oxidative stress in Drosophila and that specific antioxidants can suppress dPINK1 inactivation-induced neurodegeneration. Consistent with this notion, overexpression of hPINK1 results in flies with reduced sensitivity to treatment with paraquat, an environmental toxin linked to sporadic PD (23), and of H2O2 (Fig. 4 D and E), known inducers of oxidative stress. In contrast, expression of hPINK1 did not protect flies from damage induced by protein unfolding-promoting 2-mecaptoethanol (not shown). Together, these results indicate that PINK1 plays an important role in preventing oxidative stress-induced neuronal injury and death.
Fig. 4.
Inhibition of ommatidial degeneration by the antioxidants SOD1 and vitamin E. Flies (7 days old) with dPINK1 knockdown driven by GMR-GAL4 (GMR-GAL4/UAS-dPINK1 RNAi) were treated without (A) or with 100 units/ml (B) and 1,000 units/ml (C) SOD, or without (A′) or with 20 μg/ml (B′) and 200 μg/ml (C′) vitamin E. Representative images of external eye phenotype of each group are shown. (Magnification, ×25.) Flies overexpressing hPINK1 were treated with either 20 mM paraquat for 24 h (D) or 1% H2O2 over 6 days (E) and were quantitated for survival rates. Results from lines expressing Da-GAL4 driver alone (Da-GAL4) or Da-GAL4/UAS-lac Z (lac Z) and two independent lines expressing Da-GAL4/UAS-hPINK1 (hPINK1FC2 and hPINK1TC1) are shown. ∗∗, P < 0.001.
Our results provide strong in vivo evidence that PINK1 plays an important role in the survival of DA neurons. PD-associated PINK1 mutants are likely to fail in protecting neurons from stress and potentiate susceptibility to neuronal death. These results agree with previously reported inhibition of basal and staurosporine-induced death of SH-SY5Y cells by overexpressing wild-type PINK1 but not PD-associated PINK1 mutants in vitro (24). Our findings that SOD1 and antioxidant treatments suppress Drosophila neuronal death due to dPINK1 knockdown indicate that PINK1 in vivo acts to protect neurons that seem to be particularly susceptible to oxidative stress. The mechanism for this remains to be determined, but a potential clue may be the mitochondrial localization of PINK1 (4, 13, 25). Mitochondria are the main source of reactive oxygen and nitrogen species, and are the cellular compartments critical for integration of intrinsic death pathways. PD-associated PINK1 mutants may disrupt homeostasis of mitochondria, resulting in oxidative stress and eventual neuronal death. Alternatively, PINK1 could functionally collaborate with parkin and DJ-1, two other recessive, early-onset PD-linked genes, to regulate neuronal sensitivity to oxidative stress. In vitro, parkin and DJ-1 have been shown to protect cells from oxidative stress (26–31). Moreover, PINK1 interacts with DJ-1 and digenic mutations of PINK1 and DJ-1 are associated in early onset familial form of PD (15). Consistent with our findings that antioxidants can ameliorate the dPINK1-dependent PD pathology in Drosophila are two recent reports that show that deletion mutants of dPINK1 result in abnormally functioning mitochondria, sensitization to oxidative stress, and mild degeneration of DA neurons (32, 33). The phenotype of the dPINK1 deletion mutants in these studies appears to be milder, with no or less DA neuron loss than we observe. Possible reasons include: (i) maternal dPINK1present in zygotic mutants may be less in our “systemic” RNAi flies and thus cause earlier (larval) lethality, and importantly, (ii) the genetic background perhaps compounded by slight environmental differences may be sufficiently different to render our knock-down flies more sensitive to a reduced PINK1 function.
This study also introduces a fly model for Parkinson's disease that complements and extends previous models based on the expression of human α-synuclein or rotenone treatment (18, 34). Flies undergoing dPINK1 inactivation show degeneration of both DA neurons and ommatidia. The obvious ommatidial degeneration seen after dPINK1 inactivation should enable efficient screening of compounds preventing PD-related neurodegeneration and greatly facilitate identification of novel factors involved in PD pathogenesis.
Materials and Methods
Plasmids and Reagents.
cDNAs encoding the entire coding sequence of human PINK1 and parkin were amplified from a human brain cDNA library and cloned into the GAL4-responsive pUAST expression vector. Nucleotides encoding the flag-tag were designed on the reverse primer to add a C-terminal tag to PINK1. Anti-Drosophila TH antibody (1:500) was described (17). Anti-5HT antibody (1:500) was from Sigma (St. Louis, MO). Antibodies for Drosophila DLG and elav were obtained from American Type Culture Collection (Manassas, VA). Phalloidin and all secondary antibodies were from Invitrogen (San Diego, CA).
Drosophila Stocks.
UAS-hSOD1 transgenic flies were kindly provided by J. P. Philipps (35). UAS-dIAP1 was from S.O. (Burnham Institute for Medical Research). Flies expressing Ddc-GAL4, elav-GAL4, daughterless-GAL4, GMR-GAL4, and UAS-LacZ were obtained from the Bloomington Drosophila stock center. Drosophila were grown on standard cornmeal medium at 25°C.
Transgenic Drosophila.
Transgenic strains were created by embryo injection. cDNAs encoding human wild-type PINK1 and PINK1G309D mutant with a C-terminal flag tag were cloned into the pUAST expression vector. To make the dPINK1 RNAi construct, a cDNA fragment corresponding to base pair 1410 to base pair 1727 of dPINK1 (CG4523) was PCR-amplified (forward primer, 5′-GCTCTAGATCTGCGGCCAGTGATTTC-3′; reverse primer, 5′-GCTCTAGACTCGAGCAAACGTTCCCACTCATC-3′). Two copies of the PCR fragments were cloned in opposite orientations into the pWIZ expression vector (36). Expression of human PINK1 and knockdown of dPINK1 were verified by immunoblotting and RT-PCR, respectively.
RT-PCR.
Total RNA samples (1 μg) were reverse-transcribed by using a Quantitect reverse transcription kit (Qiagen, Valencia, CA). To verify dPINK1 knockdown, semiquantitative RT-PCR was carried out by using standard protocols. Quantitative real-time PCR was performed by using a LightCycler FastStart DNA Masterplus SYBR Green I kit (Roche, Basel, Switzerland). Primers for semiquantitative RT-PCR of dPINK1 were 5′-GCTCTAGACGCCAACATTTTGGACCAG-3′ (forward) and 5′-GCTCTAGACTCGAGTCGTGATTCCAGGCGTTCT-3′ (reverse). Primers for quantitative real-time PCRs included dPINK1 primers 5′-GCTCAGCAAGGAGGATGAAC-3′ and 5′-AAATCCGCTGCATAGACGAC-3′ (reverse) and Drosophila actin primers 5′-ACTTCTGCTGGAAGGTGGAC-3′ (forward) and 5′-ATCCGCAAGGATCTGTATGC-3′ (reverse).
Whole-Mount Immunostaining.
Whole-mount immunostaining of fly brain was essentially done as described (37). Briefly, fly heads were fixed with 4% paraformaldehyde containing 0.2% Triton X-100 overnight and washed with PBT (PBS containing 0.2% Triton X-100) three times. Brains were dissected in blocking buffer (PBS, 5% heat-inactivated normal goat serum, 0.2% Triton X-100), followed by blocking at room temperature for 1 h. Brains were immunostained with corresponding primary antibodies at 4°C overnight followed by respective secondary antibodies at room temperature for 3 h. DA neurons were quantitated from confocal images and analyzed statistically by using InStat 3 (GraphPad, San Diego, CA).
For eye disk staining, whole fly brains containing eye discs were dissected from white pupae aged for 44 h under the microscope, fixed in 4% paraformaldehyde in PBS for 30 min, rinsed with PBT three times, and blocked in blocking buffer at room temperature for 1 h. Eye discs were incubated in primary antibodies at 4°C overnight, followed by the respective secondary antibodies. After several washes, eye discs were dissected from brain and mounted onto poly(l-lysine)-coated coverslips. All samples were examined by confocal microscopy.
Antioxidant Treatment.
Antioxidants were dissolved in solvents suggested by suppliers and mixed with instant baby food. Solvents used were also individually mixed with instant baby food as controls. For treatment, flies were crossed in vials with controls and antioxidant reagents. After the new generation was produced, offspring flies were transferred to new vials with the same antioxidant reagents and aged for another 7 days. Eye phenotypes were scored under microscopy. Antioxidative activity of endogenous SOD1 has been shown in both cultured cells in vitro and dog in vivo (38, 39).
Supplementary Material
Acknowledgments
We thank Dr. Andrew Zelhof for teaching us brain dissection and Dr. J. P. Philipps (University of Guelph, Guelph, ON, Canada) for UAS-SOD1 transgenic flies. This work was supported by grants from the National Institutes of Health (to Z.Z. and R.B.), the American Parkinson's Disease Association, and the Alzheimer's Disease Association (to Z.Z.).
Abbreviations
- DA
dopaminergic
- PD
Parkinson's disease
- VUM
unpaired ventral medial cluster
- TH
tyrosine hydroxylase
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
See Commentary on page 13269.
References
- 1.Lang A. E., Lozano A. M. N. Engl. J. Med. 1998;339:1130–1143. doi: 10.1056/NEJM199810153391607. [DOI] [PubMed] [Google Scholar]
- 2.Lang A. E., Lozano A. M. N. Engl. J. Med. 1998;339:1044–1053. doi: 10.1056/NEJM199810083391506. [DOI] [PubMed] [Google Scholar]
- 3.Dawson T. M., Dawson V. L. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 4.Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G., et al. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 5.Paisan-Ruiz C., Jain S., Evans E. W., Gilks W. P., Simon J., van der Brug M., Lopez de Munain A., Aparicio S., Gil A. M., Khan N., et al. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 6.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R. J., Calne D. B., et al. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Spillantini M. G., Schmidt M. L., Lee V. M., Trojanowski J. Q., Jakes R., Goedert M. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 8.Conway K. A., Lee S. J., Rochet J. C., Ding T. T., Williamson R. E., Lansbury P. T., Jr. Proc. Natl. Acad. Sci. USA. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conway K. A., Harper J. D., Lansbury P. T. Nat. Med. 1998;4:1318–1320. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- 10.Li Y., Tomiyama H., Sato K., Hatano Y., Yoshino H., Atsumi M., Kitaguchi M., Sasaki S., Kawaguchi S., Miyajima H., Toda T., et al. Neurology. 2005;64:1955–1957. doi: 10.1212/01.WNL.0000164009.36740.4E. [DOI] [PubMed] [Google Scholar]
- 11.Hatano Y., Li Y., Sato K., Asakawa S., Yamamura Y., Tomiyama H., Yoshino H., Asahina M., Kobayashi S., Hassin-Baer S., et al. Ann. Neurol. 2004;56:424–427. doi: 10.1002/ana.20251. [DOI] [PubMed] [Google Scholar]
- 12.Valente E. M., Salvi S., Ialongo T., Marongiu R., Elia A. E., Caputo V., Romito L., Albanese A., Dallapiccola B., Bentivoglio A. R. Ann. Neurol. 2004;56:336–341. doi: 10.1002/ana.20256. [DOI] [PubMed] [Google Scholar]
- 13.Silvestri L., Caputo V., Bellacchio E., Atorino L., Dallapiccola B., Valente E. M., Casari G. Hum. Mol. Genet. 2005;14:3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 14.Gandhi S., Muqit M. M., Stanyer L., Healy D. G., Abou-Sleiman P. M., Hargreaves I., Heales S., Ganguly M., Parsons L., Lees A. J., et al. Brain. 2006;12:1720–1731. doi: 10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- 15.Tang B., Xiong H., Sun P., Zhang Y., Wang D., Hu Z., Zhu Z., Ma H., Pan Q., Xia J. H., et al. Hum. Mol. Genet. 2006;15:1816–1825. doi: 10.1093/hmg/ddl104. [DOI] [PubMed] [Google Scholar]
- 16.Kennerdell J. R., Carthew R. W. Nat. Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 17.Neckameyer W., O'Donnell J., Huang Z., Stark W. J. Neurobiol. 2001;47:280–294. doi: 10.1002/neu.1035. [DOI] [PubMed] [Google Scholar]
- 18.Feany M. B., Bender W. W. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 19.Wittmann C. W., Wszolek M. F., Shulman J. M., Salvaterra P. M., Lewis J., Hutton M., Feany M. B. Science. 2001;293:711–714. doi: 10.1126/science.1062382. [DOI] [PubMed] [Google Scholar]
- 20.Woods D. F., Bryant P. J. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 21.Hay B. A., Wassarman D. A., Rubin G. M. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 22.Selverstone Valentine J., Doucette P. A., Zittin Potter S. Annu. Rev. Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 23.Uversky V. N. Cell Tissue Res. 2004;318:225–241. doi: 10.1007/s00441-004-0937-z. [DOI] [PubMed] [Google Scholar]
- 24.Petit A., Kawarai T., Paitel E., Sanjo N., Maj M., Scheid M., Chen F., Gu Y., Hasegawa H., Salehi-Rad S., et al. J. Biol. Chem. 2005;280:34025–34032. doi: 10.1074/jbc.M505143200. [DOI] [PubMed] [Google Scholar]
- 25.Beilina A., Van Der Brug M., Ahmad R., Kesavapany S., Miller D. W., Petsko G. A., Cookson M. R. Proc. Natl. Acad. Sci. USA. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonifati V., Rizzu P., van Baren M. J., Schaap O., Breedveld G. J., Krieger E., Dekker M. C., Squitieri F., Ibanez P., Joosse M., et al. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 27.Taira T., Saito Y., Niki T., Iguchi-Ariga S. M., Takahashi K., Ariga H. EMBO Rep. 2004;5:213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menzies F. M., Yenisetti S. C., Min K. T. Curr. Biol. 2005;15:1578–1582. doi: 10.1016/j.cub.2005.07.036. [DOI] [PubMed] [Google Scholar]
- 29.Meulener M., Whitworth A. J., Armstrong-Gold C. E., Rizzu P., Heutink P., Wes P. D., Pallanck L. J., Bonini N. M. Curr. Biol. 2005;15:1572–1577. doi: 10.1016/j.cub.2005.07.064. [DOI] [PubMed] [Google Scholar]
- 30.Goldberg M. S., Fleming S. M., Palacino J. J., Cepeda C., Lam H. A., Bhatnagar A., Meloni E. G., Wu N., Ackerson L. C., Klapstein G. J., et al. J. Biol. Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- 31.Palacino J. J., Sagi D., Goldberg M. S., Krauss S., Motz C., Wacker M., Klose J., Shen J. J. Biol. Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 32.Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 33.Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., Chung J. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 34.Coulom H., Birman S. J. Neurosci. 2004;24:10993–10998. doi: 10.1523/JNEUROSCI.2993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkes T. L., Elia A. J., Dickinson D., Hilliker A. J., Phillips J. P., Boulianne G. L. Nat. Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y. S., Carthew R. W. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 37.Whitworth A. J., Theodore D. A., Greene J. C., Benes H., Wes P. D., Pallanck L. J. Proc. Natl. Acad. Sci. USA. 2005;102:8024–8029. doi: 10.1073/pnas.0501078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tilly J. L., Tilly K. I. Endocrinology. 1995;136:242–252. doi: 10.1210/endo.136.1.7828537. [DOI] [PubMed] [Google Scholar]
- 39.Skorohod N., Yeates D. B. J. Appl. Physiol. 2005;98:1478–1486. doi: 10.1152/japplphysiol.00910.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.