Abstract
Small RNA (sRNA) molecules have gained much interest lately, as recent genome-wide studies have shown that they are widespread in a variety of organisms. The relatively small family of 10 known sRNA-encoding genes in Escherichia coli has been significantly expanded during the past two years with the discovery of 45 novel genes. Most of these genes are still uncharacterized and their cellular roles are unknown. In this survey we examined the sequence and genomic features of the 55 currently known sRNA-encoding genes in E.coli, attempting to identify their common characteristics. Such characterization is important for both expanding our understanding of this unique gene family and for improving the methods to predict and identify sRNA-encoding genes based on genomic information.
INTRODUCTION
Traditionally, most RNA molecules were thought to function as the mediators that carry the information from the gene to the translational machinery. Exceptions to these were the transfer RNAs and ribosomal RNAs that had long been known to carry out functions of their own, associated also with translation. However, it is now widely acknowledged that other types of untranslated RNA molecules exist that are involved in a diverse range of functions, from structural through regulatory to catalytic. Untranslated RNA molecules are present in many different organisms, ranging from bacteria to mammals, affecting a large variety of processes including plasmid replication, phage development, bacterial virulence, developmental control and more (1–7). These unique RNA molecules have recently drawn much attention, as many novel RNA-encoding genes were discovered during the past two years (for review see 8–11).
Small RNA (sRNA)-encoding genes have been hard to detect both experimentally and computationally. Their small size complicates their detection by biochemical and genetic methods. Furthermore, sRNA genes are not translated into proteins and are thus immune to frameshift and nonsense mutations. For the same reason these genes are impossible to predict by the conventional computer algorithms that rely on the presence of protein coding sequences. Indeed, until the year 2000 and for the last three decades only 10 sRNA-encoding genes were known in Escherichia coli (6), most of which were discovered fortuitously.
During the last two years six studies were published that described systematic searches for novel sRNA genes in E.coli (12–17). One study provided 562 predictions of novel non-translated RNA genes using a machine learning approach (15). A second study used a whole genome array approach to examine the transcription of E.coli coding regions as well as intergenic regions (17), and identified 317 novel transcripts with unknown functions. Out of these, nine transcripts were suggested by the authors to be good candidates for sRNA molecules, based on their size, expression pattern or conservation. The remaining four studies used a combination of computational and experimental approaches (12–14,16). The study by Argaman et al. (12) relied on transcription signals and sequence conservation in empty intergenic regions of the genome, and resulted in the prediction of 24 candidate sRNA-encoding genes. Experimental testing of these candidates showed that 14 of them were expressed, and their 5′ and 3′ ends were determined (12). Two of these sRNA genes, gcvB and rprA, were independently discovered in two separate studies (18,19). The study by Wassarman et al. (13) relied on sequence conservation and DNA array data, and led to the prediction of 60 candidate sRNA genes. These candidates were examined by northern blots and 17 of them were found to be sRNA genes (13). Rivas et al. (14) relied on the premise that functional RNAs should have a conserved structure in the different organisms in which they exist. Based on an algorithm that detects such structural conservation, 275 candidates were predicted. Experimental testing of 49 candidates detected the expression of 11 genes (14). The algorithm by Chen et al. relied on transcription signals alone, and resulted in the prediction of 227 candidate sRNA-encoding genes (16). Eight of the predictions were tested experimentally, out of which seven were found to be expressed. Since several genes were identified in more than one study, all in all 45 novel sRNA genes were discovered. Thus, there are currently 55 known sRNA-encoding genes in the E.coli genome (Table 1). These probably comprise only a portion of the population of sRNA-encoding genes in E.coli, as many predicted genes still await their detailed experimental testing. In the current survey we focus on the genomic characterization of the sRNAs encoded in the genome of E.coli.
Table 1. Summary of the 55 sRNA genes in E.coli.
| sRNA gene | Minutes | Adjacent genes | Stranda | Length | 3′ End position | %GC | Reference |
|---|---|---|---|---|---|---|---|
| tpke11 | 0.3 | dnaK/dnaJ | → → → | 370 | (14) | ||
| tp2 | 2.6 | pdhR/aceE | → ← → | 120 | (14) | ||
| t44 | 4.1 | map/rpsB | ← → → | 135 | 189847 | 50.7 | (14) |
| C0067 | 5.1 | yafT/yafU | ← → ← | 124 | 238586 | 29.6 | (17) |
| sraA / t15 | 9.9 | clpX/lon | → ← → | 120 | (12) | ||
| ffs | 10.3 | ybaZ/ybaA | ← → → | 138 | 475785 | 61.6 | (34) |
| rybA | 18.4 | ybiP/ybiQ | → ← → | 205 | (13) | ||
| rybB / p25 | 19.1 | ybjK/ybjL | → ← ← | 80 | (13) | ||
| sraB / pke20 | 24.7 | yceF/yceD | ← → → | 168 | 1145980 | 38.5 | (12) |
| C0293 | 25.8 | icd/ymfD | → → ← | 72 | 1196009 | 58.9 | (17) |
| C0299 | 26.5 | hlyE/umuD | ← → → | 78 | 1229930 | 43 | (17) |
| IS061 | 30.2 | abgR/ydaL | → ← → | 158 | 1403676b | 45.3 | (16) |
| C0343 | 30.3 | ydaN/dbpA | → → → | 74 | 1407461 | 58.7 | (17) |
| IS063 / tke8 | 30.9 | ompN/ydbK | ← → ← | 117 | 1435259b | 39.0 | (16) |
| C0362 | 33.4 | fdnI/yddM | → → ← | 385 | 1550410 | 45.6 | (17) |
| dicF | 35.5 | rzpQ/dicB | → → → | 53 | 1647458 | 54.7 | (35) |
| rydB / tpe7 / IS082 | 38.0 | ydiC/ydiH | ← ← ← | 67 | 1762726 | 57.4 | (13) |
| rprA / IS083 | 38.1 | ydiK/ydiL | → → → | 105 | 1768500 | 39.0 | (12,19) |
| ryeB / tpke79 | 41.4 | pphA/yebY | ← ← ← | 100 | (13) | ||
| sraC / ryeA / tpke79 / IS091 | 41.4 | pphA/yebY | ← → ← | 249 | 1921338 | 45.8 | (12,13) |
| C0465 | 42.5 | tar/cheW | ← → ← | 77 | 1970840 | 46.2 | (17) |
| IS092 | 42.8 | yecJ/yecR | ← ← → | 165 | 1985862b | 43.4 | (16) |
| dsrA | 43.6 | dsrB/yedP | ← ← → | 85 | 2023250 | 44.7 | (36) |
| IS102 | 44.6 | yeeP/flu | → → → | 203 | 2069540b | 42.6 | (16) |
| ryeC / tp11 | 46.4 | yegL/yegM | ← → → | 143 | (13) | ||
| ryeD / tpe60 | 46.4 | yegL/yegM | ← → → | 137 | (13) | ||
| ryeE | 46.7 | yegQ/ogrK | → → ← | 47 | (13) | ||
| micF | 49.8 | ompC/yojN | ← → → | 93 | 2311196 | 35.5 | (37) |
| tpke70 | 53.8 | ddg/yfdZ | → ← ← | 40 | (14) | ||
| C0614 | 57.2 | sseA/sseB | → ← ← | 86 | 2651472 | 36.8 | (17) |
| IS128 | 57.2 | sseA/sseB | → → ← | 208 | 2651743b | 35.3 | (16) |
| ryfA / tp1 | 57.2 | sseA/sseB | → → ← | 302 | 2652177 | 52.8 | (13,14) |
| tke1 | 58.0 | yfhK/purL | ← ← ← | 158 | 2689212 | 51.0 | (14) |
| ssrA | 59.4 | smpB/intA | → → → | 363 | 2753974 | 52.9 | (38) |
| sraD | 60.6 | ygaG/gshA | ← → ← | 70 | 2812897 | 43.4 | (12) |
| C0664 | 61.1 | ygbD/hypF | → → ← | 57.5 | 2833189 | 57.5 | (17) |
| csrB | 63.0 | yqcC/syd | ← ← ← | 360 | 2922178 | 51.1 | (39) |
| gcvB / IS145 | 63.4 | gcvA/ygdI | ← → ← | 205 | 2940922 | 42.9 | (12,18) |
| sraE / rygA / t59 | 64.1 | aas/galR | ← ← → | 88 | 2974124 | 50.0 | (12,13) |
| rygB / t59 | 64.1 | aas/galR | ← ← → | 83 | 2974332b | 51.2 | (13) |
| ssrS | 65.8 | ygfE/ygfA | → → → | 183 | 3054185 | 55.2 | (40) |
| rygC / t27 | 65.8 | ygfA/serA | → → ← | 139 | (13) | ||
| C0719 | 67.2 | yghK/glcB | ← → ← | 221 | 3119595 | 43.7 | (17) |
| tp8 / C0730 | 68.8 | yqiK/rfaE | → ← ← | 144 | 3192737 | 48.3 | (14,17) |
| sraF / tpk1 / IS160 | 69.8 | ygjR/ygjT | → → → | 189 | 3236203 | 49.7 | (12,14) |
| rnpB | 70.4 | yhaC/yhaD | → ← ← | 377 | 3267857 | 61.8 | (41) |
| sraG / p3 | 71.3 | pnp/rpsO | ← → ← | 174 | 3309039 | 49.4 | (12) |
| sraH / ryhA | 72.2 | elbB/arcB | ← → ← | 108 | 3348325 | 53.7 | (12,13) |
| sraI / ryhB / IS176 | 77.1 | yhhX/yhhY | ← ← → | 94 | 3578554 | 51.1 | (12,13,42) |
| IS183 | 78.9 | yhiW/yhiX | ← → ← | 113 | 3662604b | 48.2 | (16) |
| sraJ / ryiA / k19 | 85.9 | aslA/hemY | ← → ← | 172 | 3984216 | 47.7 | (12,13) |
| spf | 87.2 | polA/yihA | → → ← | 109 | 4047587 | 42.2 | (43) |
| sraK / ryiB / tpk2 / csrC | 87.3 | yihA/yihI | ← → → | 245 | 4048860 | 48.6 | (12,13) |
| oxyS | 89.6 | argH/oxyR | → ← → | 110 | 4155864 | 49.1 | (44) |
| sraL / ryjA | 92.2 | soxR/yjcD | → ← → | 140 | 4275506 | 52.1 | (12,13) |
aThe middle arrow represents the sRNA gene while the flanking arrows indicate the orientation of the adjacent genes. Genes present on the strand given in the E.coli genome database (Watson strand) are indicated by (→), and genes present on the complementary strand (Crick strand) are indicated by (←).
bPredicted 3′ ends.
RESULTS AND DISCUSSION
Distribution of sRNA-encoding genes along the E.coli genome
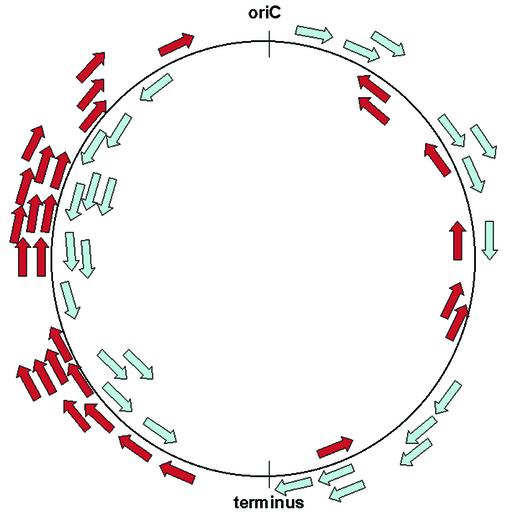
All known E.coli sRNA-encoding genes are located in ‘empty’ intergenic regions, where no other gene resides on either of the two strands (based on the Colibri and Ecogene database annotations of E.coli K12 MG1655 genes [http://genolist. pasteur.fr/Colibri, (20)]. In order to find out whether these genes are located in certain genomic regions, their distribution along the genome with regard to the origin of replication was examined (Fig. 1). The DNA replication of E.coli starts from a single origin and is bidirectional, creating two replicores: the left replicore and the right one (21). While 51% of the open reading frames (ORFs) are located on the left replicore and 49% on the right replicore, the sRNA genes show different proportions, as 64% of them (35/55) are located on the left replicore (p < 0.05 by a binomial test). There are no significant differences in the number of ‘empty’ intergenic regions and in their length between the two replicores. Therefore, there is no obvious reason for the difference in the number of sRNA-encoding genes between the two replicores. We also examined the distribution of sRNA genes between the lagging and leading strands and found that they are distributed about equally.
Figure 1.
Distribution of the sRNA-encoding genes along the E.coli genome. The origin and terminus of replication are indicated. sRNA genes on the leading and lagging strands are colored blue and red, respectively.
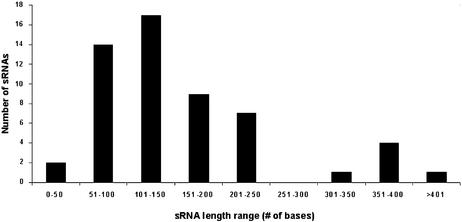
In general, the genome of E.coli is quite compact. There are only 33 ‘empty’ intergenic regions that are >900 nt, many of which contain repetitive sequences (21). Most intergenic regions in the E.coli genome are relatively short: 1155 are <50 nt, 1889 range between 50 and 300 nt, and 478 are 300–900 nt long. No sRNA gene resides in an intergenic region that is <50 nt. Of the sRNAs, 71% (39 genes) reside in larger intergenic regions of lengths between 300 and 900 nt, which make up only 20% of the total intergenic regions that are >50 nt and 39% of the nucleotides of these regions. Twenty-seven percent of the sRNA genes reside in relatively short intergenic regions (50–300 nt), which make up 79% of intergenic regions >50 nt and 52% of the total nucleotides of these regions. Only one sRNA gene (rnpB) resides in an empty intergenic region >900 nt. Analysis of the size distribution of the sRNAs themselves shows that most of them (47 genes) are between 50 and 250 nt long (Fig. 2). A weak correlation was found between the size of an sRNA and the size of the intergenic region in which it resides (r = 0.26, p ≤ 0.05). In addition, we examined possible clustering of the sRNA genes in the intergenic regions. It seems that the sRNA-encoding genes do not tend to be clustered, as only in three cases two genes are located in a single intergenic region (ryeC and ryeD, sraE and rygB, sraC and ryeB), and only in one case three genes are located in the same intergenic region (ryfA, IS128 and C0614). In each of the pairs of genes, ryeC and ryeD, sraE and rygB, and ryfA and IS128, there is some sequence similarity and they are transcribed in the same direction. C0614 and sraC are opposites of ryfA/IS128 and reyB, respectively.
Figure 2.
Length distribution of the 55 known sRNA genes.
Base composition of sRNAs
In hyperthermophiles it was shown that sRNAs differ in their base composition from the rest of the genome (22,23). In fact, novel sRNA-encoding genes were identified in the AT-rich genomes of hyperthermophiles such as Methanococcus jannachii and Pyrococcus furiosus by searching for GC-rich regions (22,23). We examined whether the sRNAs in E.coli also have some distinct base composition. For comparison, we examined the base composition of the ORFs, tRNA genes, rRNA genes and the intergenic sequences. Base composition was analyzed for sRNAs for which either the 5′ or the 3′ end was experimentally determined or the 3′ end was predicted (a total of 44 sRNAs). As a group, the sRNA molecules seem to be only slightly richer in guanines and cytosines (48.2%) in comparison with intergenic regions where sRNA-encoding genes have not been identified (42.4%) (Table 2). This analysis also revealed that the sRNAs’ GC content is lower than that of the tRNAs (59%) and rRNAs (54%).
Table 2. Composition of sRNAs and other sequences in the E.coli genomea.
| Type of sequence | %GC |
|---|---|
| All sRNAs | 48.2 |
| Known housekeeping sRNAsb | 57.6 |
| Intergenic sequences | 42.4 |
| Open reading frames | 51.9 |
| rRNAs | 54.0 |
| tRNAs | 59.0 |
aThe percent of total GC out of total nucleotides in each group of sequences was computed.
bThe known housekeeping sRNAs are ssrA, rnpB, ssrS and ffs.
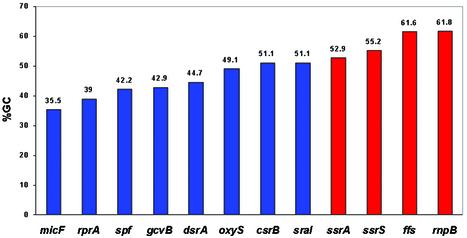
The sRNA-encoding genes can be divided into two subgroups based on their functions: the regulatory sRNAs that act as regulators of gene expression, and the housekeeping sRNAs that affect different aspects of cellular metabolism. Analysis of the GC content of 12 sRNAs with known function showed that the base composition of the housekeeping sRNAs (ssrA 53%, rnpB 62%, ffs 62% and ssrS 55%) is closer to the composition of the tRNAs and rRNAs, while the regulatory sRNAs show a lower GC content (Fig. 3 and Table 2). dicF, which is not a housekeeping sRNA, is also GC rich (55%); however, dicF was not included since it is known to originate from a phage, and is therefore expected to have a composition different to that of the E.coli genome. It is conceivable that, in order to meet functional requirements that are based on specific structures, tRNAs and perhaps also rRNAs have a high GC content which is associated with a more rigid structure. It is intriguing, however, that the housekeeping genes, as opposed to the regulatory genes, also have a high GC content which could indicate the need for a rigid versus a more flexible structure. Unlike the housekeeping sRNA genes, the regulatory sRNAs such as oxyS or dsrA often regulate the expression of a number of different genes. Thus, it is possible that they might need a more flexible structure. It would be interesting to find out whether novel sRNAs with a relatively high GC content such as sraH or rydB would be classified as housekeeping.
Figure 3.
Base composition of the sRNAs for which a function has been assigned. dicF is not included as it originated from a phage and is expected to differ in its composition. sRNAs that were assigned a housekeeping function are colored red, while those with a regulatory function are colored blue.
Conservation of the sRNA-encoding genes and their adjacent genes in other bacteria
The sequences of nine of the previously known sRNAs were shown to be conserved in closely related bacteria. Based on this and on the premise that functional sequences are expected to be conserved in related organisms, three of the genome-wide studies used sequence conservation as a criterion for prediction (12–14). However, the three studies used different stringency for conservation analysis: Rivas et al. (14) allowed a lower level of sequence conservation, and relied on structural conservation, while Argaman et al. (12) and Wassarman et al. (13) requested a higher level of sequence conservation. We therefore attempted to conduct a consistent conservation analysis for all 55 sRNA-encoding genes.
For the conservation analysis, the sequences of the 55 sRNAs in E.coli were used as queries and compared by BLAST (24,25) to the sequences of 102 complete bacterial genomes, downloaded from the NCBI ftp server (ftp.ncbi.nlm.nih.gov) and listed in the Supplementary Material. An sRNA-encoding gene was considered as conserved in another organism if the alignment had an E-value lower than 0.001 (Table 3); however, in the Supplementary Material we list alignments up to an E-value of 1.0. Since the majority of the sRNAs did not show any significant sequence similarities beyond Yersinia pestis, only these comparisons are summarized in Table 3. Out of the 55 sRNAs examined, two genes were found uniquely in E.coli K12. Fifty sRNA sequences were conserved in the two E.coli O157:H7 genomes and in E.coli CFT073. All but three of the sRNAs were conserved in Shigella flexneri. In Salmonella typhimurium and in Salmonella typhi 42 of the sRNAs were conserved. Sixteen sRNAs were conserved in Y.pestis, with an E-value below 0.001. We also compared the sRNA sequences with all available archeal sequences and did not find any significant similarities.
Table 3. Conservation of the sRNA genes and their adjacent genesa.
| Left adjacent gene | sRNA-encoding gene | Right adjacent gene | Escherichia coli O157:H7 | Escherichia coli O157:H7 EDL933 | Escherichia coli CFT073 | Shigella flexneri 2a | Salmonella typhimurium LT2 | Salmonella typhi | Yersinia pestis CO92 | Yersinia pestis KIM |
|---|---|---|---|---|---|---|---|---|---|---|
| dnaK | tpke11b | dnaJ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □□□ | □□□ |
| pdhR | tp2b | aceE | ▪▪▪ | ▪▪▪ | □▪□ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □□□ | □□□ |
| map | t44c | rpsB | ▪▪▪ | ▪▪▪ | □▪□ | □▪□ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □▪□ |
| yafT | C0067 | yafU | □□□ | □□□ | □□□ | □□□ | □□□ | □□□ | □□□ | □□□ |
| clpX | sraAb,c | lon | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □▪□ |
| ybaZ | ffsd | ybaA | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪□ | ▪▪□ | ▪▪□ | ▪▪□ |
| ybiP | rybAb | ybiQ | ▪▪▪ | □▪□ | □▪□ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □□□ | □□□ |
| ybjK | rybBb | ybjL | ▪▪▪ | ▪▪▪ | □▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □▪▪ | □▪▪ |
| yceF | sraB | yceD | ▪▪▪ | ▪▪▪ | □▪□ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □□□ | □□□ |
| icdA | C0293 | ymfD | □□□ | □□□ | □□□ | □□□ | □□□ | □□□ | □□□ | □□□ |
| hlyE | C0299 | umuD | ▪▪▪ | ▪▪▪ | ▪▪▪ | □▪▪ | □□□ | □□□ | □□□ | □□□ |
| abgR | IS061 | ydaL | ▪▪▪ | ▪▪▪ | □▪□ | □▪□ | □□□ | □□□ | □□□ | □□□ |
| ydaN | C0343 | dbpA | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □□□ | □□□ | □□□ | □□□ |
| ompN | IS063 | ydbK | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □▪▪ | □▪□ | □□□ | □□□ |
| fdnI | C0362 | yddM | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □▪□ | □▪□ | □□□ | □□□ |
| rzpQ | dicF | dicB | □▪▪ | ▪▪▪ | □▪□ | □□□ | □□□ | □□□ | □□□ | □□□ |
| ydiC | rydB | ydiH | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □▪▪ | □▪▪ | □□□ | □□□ |
| ydiK | rprAc | ydiL | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪□ | ▪▪□ | ▪▪□ |
| pphA | ryeBb | yebY | ▪▪▪ | ▪▪▪ | ▪▪□ | □▪▪ | □▪□ | □▪□ | □▪▪ | □▪▪ |
| pphA | sraC | yebY | ▪▪▪ | ▪▪▪ | ▪▪□ | □▪▪ | □▪□ | □▪□ | □▪▪ | □▪▪ |
| tar | C0465 | cheW | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □□□ | □□□ | □□□ | □□□ |
| yecJ | IS092 | yecR | ▪▪▪ | ▪▪▪ | □▪□ | ▪▪▪ | □□□ | □□□ | □□□ | □□□ |
| dsrB | dsrA | yedP | □▪▪ | □▪▪ | □▪▪ | □▪□ | □▪▪ | □▪▪ | □□□ | □□□ |
| yeeP | IS102 | flu | ▪▪▪ | □▪□ | ▪▪▪ | ▪▪▪ | □□□ | □□□ | □□□ | □□□ |
| yegL | ryeCb | yegM | ▪▪▪ | ▪▪□ | ▪▪▪ | ▪▪□ | □▪▪ | □▪▪ | □□□ | □□□ |
| yegL | ryeDb | yegM | ▪▪▪ | ▪▪□ | ▪▪▪ | ▪▪□ | □▪▪ | □▪▪ | □□□ | □□□ |
| yegQ | ryeEb | ogrK | ▪▪□ | ▪▪□ | □▪□ | ▪▪□ | ▪▪□ | ▪▪□ | □□□ | □□□ |
| ompC | micF | yojN | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □□□ | □□□ |
| ddg | tpke70b | yfdZ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪□ | □□□ | □□□ |
| sseA | C0614 | sseB | □□□ | □□□ | □▪□ | □▪▪ | □□□ | □□□ | □□□ | □□□ |
| sseA | IS128 | sseB | □□□ | □□□ | □▪□ | □▪▪ | □□□ | □□□ | □□□ | □□□ |
| sseA | ryfA/tp1 | sseB | ▪▪▪ | ▪▪▪ | □▪□ | □▪▪ | □▪▪ | □▪▪ | □□□ | □□□ |
| yfhK | tke1 | purL | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □□□ | □□□ |
| smpBd | ssrAd | intA | ▪▪□ | ▪▪□ | ▪▪□ | ▪▪▪ | □▪□ | □▪□ | ▪▪□ | ▪▪□ |
| ygaG | sraD | gshA | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ |
| ygbD | C0664 | hypF | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □▪□ | □▪□ | □□□ | □□□ |
| yqcC | csrB | syd | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □▪▪ | □▪▪ |
| gcvA | gcvB | ygdI | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪□ | ▪▪▪ |
| aas | sraE | galR | ▪▪▪ | ▪▪▪ | ▪▪□ | ▪▪▪ | ▪▪▪ | □▪▪ | □□□ | □□□ |
| aas | rygBc | galR | ▪▪▪ | ▪▪▪ | □▪□ | ▪▪▪ | ▪▪▪ | □▪▪ | ▪▪□ | ▪▪□ |
| ygfE | ssrS | ygfA | ▪▪▪ | ▪▪▪ | □▪□ | □▪□ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ |
| ygfA | rygCb | serA | □▪▪ | □▪▪ | ▪▪▪ | ▪▪▪ | □▪□ | □▪□ | □□□ | □□□ |
| yghK | C0719 | glcB | □□□ | □□□ | ▪▪▪ | ▪▪▪ | □□□ | □□□ | □□□ | □□□ |
| yqiK | tp8 | rfaE | ▪▪□ | ▪▪▪ | ▪▪▪ | □▪□ | ▪▪▪ | ▪▪▪ | □□□ | □□□ |
| ygjR | sraF | ygjT | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □□□ | □□□ |
| yhaC | rnpBd | yhaD | ▪▪▪ | □▪▪ | □▪▪ | □▪▪ | □▪▪ | □▪▪ | □▪□ | □▪□ |
| pnp | sraG | rpsO | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪□ | ▪▪□ |
| elbB | sraH | arcB | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □▪□ | ▪▪▪ |
| yhhX | sraI | yhhY | ▪▪▪ | ▪▪▪ | □▪□ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □▪□ | □▪□ |
| yhiW | IS183 | yhiX | ▪▪▪ | ▪▪▪ | ▪▪▪ | ▪▪▪ | □□□ | □□□ | □□□ | □□□ |
| aslA | sraJ | hemY | ▪▪▪ | ▪▪▪ | □▪▪ | □▪▪ | □▪▪ | □▪▪ | □□□ | □□□ |
| polA | spfc,d | yihAd | ▪▪▪ | ▪▪▪ | □▪□ | ▪▪▪ | □▪□ | ▪▪▪ | ▪▪□ | ▪▪□ |
| yihA | sraKc | yihI | ▪▪▪ | □▪▪ | □▪□ | ▪▪▪ | □▪□ | □▪□ | ▪▪▪ | ▪▪▪ |
| argH | oxyS | oxyR | ▪▪▪ | ▪▪▪ | □▪▪ | □▪▪ | ▪▪▪ | ▪▪▪ | □□□ | □□□ |
| soxR | sraL | yjcD | ▪▪▪ | ▪▪▪ | ▪▪□ | ▪▪▪ | ▪▪□ | ▪▪□ | □□□ | □□□ |
aThe middle box represents the sRNA gene itself while the right and left boxes represent the right and left adjacent genes, respectively. A black box represents conservation (E-value < 0.001) while a white box represents lack of conservation. Only the sequences of complete genomes were compared.
bNo data was available for the exact location of the sRNA. Conservation of the entire intergenic region in which the sRNA resides was examined.
cThe conservation of this sRNA in Y.pestis was not very significant (E-values ranging between 0.07 and 10E-04). Conservation of the adjacent gene increased the likelihood that this sRNA is in fact conserved in Y.pestis.
dConserved in additional genomes. For details see Supplementary Material.
Out of the 13 sRNAs with known functions, five regulatory and all four housekeeping sRNAs were conserved in Y.pestis. Remarkably, three out of the four housekeeping sRNAs (ssrA, ffs and rnpB) are conserved beyond Y.pestis, each in several additional organisms (marked in Table 3 as such and detailed in the Supplementary Material). The only regulatory sRNA sequence that is conserved beyond Y.pestis is spf, showing a significant similarity to a sequence in the Shewanella oneidensis genome. None of the new sRNAs with unknown functions were conserved beyond Y.pestis.
It is important to note that in this study we examined conservation only through sequence similarity. In many cases the function of an sRNA may depend on the structure of the molecule rather than on a specific sequence. In addition, there are instances in which the function of the sRNA depends on sequence complementarity to a target gene. In such cases, complementary mutations in the sRNA and its target gene may cause a change in the sequence of the sRNA. Indeed, there are E.coli sRNA-encoding genes (ssrA, rnpB and micF) that are known to have homologs in other organisms. Yet, these homologs do not share enough sequence similarity with the E.coli sequences to allow a BLAST search to find them when the E.coli sequence is used as a query. For example, ssrA is known to exist in organisms as distant as archea. Indeed, it has been shown that additional ssrA homologs could be identified when a representative sequence from an organism distant from E.coli was used as a query (26). It is therefore quite likely that some of the sRNAs surveyed have homologs in additional genomes, yet these were not detected by our analysis due to sequence drift. As the algorithms for the optimal alignment of a sequence to an RNA secondary structure (27) improve, it may become possible to find these more distant orthologs.
It has previously been observed that conservation of gene adjacency in different organisms is associated with functional relationships between the conserved genes. Moreover, it was shown that in many cases proteins encoded by pairs of adjacent genes whose adjacency is conserved, work in concert, by physical interaction (28–30). This property, in turn, was used to infer relationships between conserved adjacent genes (30). The same rationale may hold for sRNA-encoding genes and genes to which they are functionally related. We therefore examined the conservation in other genomes of the genes flanking the sRNA-encoding genes. When comparing the E.coli K12 sRNA sequences to the sequences of the 102 complete bacterial genomes we noted the positions in which the sRNAs were found in the other genomes. Based on the GenBank annotation of the relevant genomes, the genes flanking the conserved sRNAs were extracted and a database was created. For this analysis an sRNA was considered conserved if it had an E-value lower than 1. The adjacent genes of the E.coli K12 sRNA genes were translated and used as queries for a TBLASTN search against the database of the flanking genes described above. We found that conservation of adjacency was very widespread in the bacteria closer to E.coli K12. For 44 out of the 50 sRNAs conserved in E.coli O157:H7 both flanking genes were conserved (Table 3). For the remaining six sRNAs only one flanking gene was conserved. This high rate of conservation may be due to the close relatedness between the two E.coli strains. For 23 of the 42 conserved sRNAs in S.typhimurium LT2, we observed a conservation of both adjacent genes. For 11 additional sRNAs one of the adjacent genes was conserved. In Y.pestis CO92, out of the sRNAs for which BLAST found similar sequences with an E-value below 1, only in five cases were both adjacent genes conserved. In 11 additional cases one of the adjacent genes in Y.pestis was conserved. In some cases conservation of adjacency was used also to support the prediction of an sRNA as conserved in Y.pestis. BLAST hits with an E-value above 0.001 were normally ignored; however, if an sRNA was found in Y.pestis with an E-value between 0.001 and 1, and one or more of its adjacent genes was conserved, this strengthened our certainty in the conservation of this sRNA in Y.pestis. In this manner three additional sRNA-encoding genes (sraA, sraK and rygB) were found to be conserved in Y.pestis, thus raising the number of sRNAs conserved in Y.pestis to 19 (Table 3). For two sRNA genes (gcvB and ssrA) the conserved adjacent genes (gcvA and smpB, respectively) are known to be functionally associated with the sRNAs: GcvA is known to activate the transcription of the adjacent sRNA, gcvB (18); SmpB is an RNA-binding protein, which is necessary for the activity of ssrA (31). These observations could suggest that in the other cases of conservation observed in Y.pestis (Table 3), the conserved adjacent genes may be functionally associated with the sRNAs, and provide some hint as to the function of these sRNAs.
Sequence comparison of the sRNAs
In order to reveal whether there is sequence conservation among some of the sRNA molecules, an all against all comparison of the sRNA sequences was conducted using the GCG program bestfit that implements the Smith–Waterman algorithm (32). For sRNAs with defined 5′ and 3′ ends, the precise sequences were used; otherwise, the entire sequence of the intergenic region that contains the sRNA was used. For most sRNA-encoding genes the sequence comparison did not reveal any similarity with any of the other sRNAs. sraE and rygB, which reside in the same intergenic region between aas and galR, show significant sequence similarity with 77% identity over 84 nt. These two sRNAs overlap a previously identified intergenic repeat called PAIR2 (33). ryeC and ryeD, which reside in the same intergenic region between yegL and yegM, and two additional sRNA genes named tp8 and rygC, which reside in different intergenic regions, also seem to share sequence similarity (with percent identities ranging between 68 and 83%, covering substantial regions of the sRNAs). These sRNA genes overlap repeats that have been previously identified and named QUAD repeats (33). Also ryfA and IS128, which reside in the same intergenic region, show some sequence similarity (94% identity over 18 nt). A pair of intergenic repeats called PAIR3 was identified in this region; however, IS128, as well as ryfA, correspond only partially to the locations of the previously identified repeats (33). This could be partially due to the fact that the exact 5′ and 3′ ends of IS128 were not experimentally identified. C0614 and IS128 are opposite and therefore complementary. C0362 and C0664 are complementary along 31 bases due to the inclusion of REP elements (21) within their sequences.
Predicted sRNAs
In addition to the 45 novel sRNA-encoding genes that were discovered, a large number of additional candidate genes were predicted. In the studies of Argaman et al. (12) and Wassarman et al. (13), a relatively small number of sRNAs were predicted and all of the candidates were tested experimentally. In the studies of Chen et al. (16), Rivas et al. (14) and Carter et al. (15) a large number of sRNAs were predicted but only a small portion of these were experimentally examined. Tjaden et al. (17) used a whole genome array to detect transcription from the intergenic regions of the E.coli genome. These transcripts were further characterized and classified as either 5′ or 3′ untranslated regions, operon elements, sRNAs or as transcripts with unknown function.
We compiled a list of the candidates that were not yet verified experimentally (Table 4 and Supplementary Material). In this compilation the number of non-redundant candidates is 1001. Out of these, 906 candidates were predicted by one study alone, 85 candidates were predicted by two studies, and 10 candidates were predicted by three independent studies. As seen in Table 4, only very few of the candidate sRNAs are located within 5′ and 3′ UTRs or operons, as determined by Tjaden et al. (17).
Table 4. Summary of candidate sRNA-encoding genes with no experimental verification.
| Source | Number of candidates | Number of unique candidatesa | Overlapping a 5′ UTRb | Overlapping a 3′ UTRb | Located within an operonb |
|---|---|---|---|---|---|
| Argaman et al. (12) | 10 | 5 | 0 | 0 | 1 |
| Rivas et al. (14) | 255 | 222 | 21 | 4 | 43 |
| Chen et al. (16) | 137c | 104 | 10 | 1 | 1 |
| Tjaden et al. (17) | 278 | 220 | 0 | 0 | 0 |
| Carter et al. (15) | 423c | 355 | 27 | 14 | 25 |
aUnique candidates are candidates that were predicted only in one study.
bBased on the whole genome array analysis performed by Tjaden et al. (17).
cOverlapping predictions were united into a single candidate sRNA.
CONCLUSIONS
The 55 sRNA-encoding genes in E.coli seem to be a much more varied class of genes than tRNA genes and rRNA genes. It is hard to determine a genomic or sequence feature that is shared by all of them. Still, common characteristics could be identified.
By examining the distribution of the sRNA-encoding genes on the genome we found a preference for the left replicore and no preference for the leading or lagging strand. We also found that the sRNA-encoding genes are not clustered in certain intergenic regions, as usually no more than one sRNA gene exists per intergenic region. These genes usually reside in intergenic regions ranging in size from 300 to 900 nt. They very rarely reside in intergenic regions >900 nt, which in E.coli usually contain repetitive sequences.
Our analysis of base composition revealed that all genes are richer in GC in comparison with intergenic sequences. sRNAs are, however, less GC-rich than the other types of genes. There does exist a sub-group of sRNAs, ‘housekeeping sRNAs’, that are richer in GC compared with the other sRNAs. These sRNAs seem to have a base composition more similar to that of structural RNA genes, such as tRNAs and rRNAs. The difference in the GC content could point to the different structural requirements associated with the function of the sRNAs; regulatory sRNAs versus housekeeping sRNAs.
Since three out of the five studies that led to the discovery of the 45 novel sRNAs relied on sequence conservation, it is not surprising that most of the known sRNAs are conserved in closely related bacteria. The conservation is strongest in the other E.coli strains and in S.flexneri, while only 19 of the sRNAs are conserved also in Y.pestis. Only four of the sRNAs were conserved beyond Y.pestis. Three of these sRNAs carry out housekeeping functions. No sRNA homologs were found in archea. It is important to note that since we examined conservation through sequence similarity alone, it is possible that some of the sRNA homologs that maintain only structural conservation may have been missed.
Conservation analysis of the sRNA-encoding genes along with their flanking genes revealed stronger conservation in the other strains of E.coli, S.flexneri and the two Salmonella strains, than in Y.pestis. In two of the cases in which gene order was conserved in Y.pestis, the adjacent gene and the sRNA were known to be associated. This may point to a functional association for the other sRNA-encoding genes and their adjacent genes in cases where such a conservation was observed.
Most sRNAs do not show sequence similarity with other sRNAs. We found that in most cases where sRNAs were similar in sequence they were also located in neighboring genomic locations, suggesting that they may have resulted from duplication events. Indeed, most of these were previously identified as intergenic repeats (33). It could therefore be interesting to check other intergenic repeats, to see whether they too encode for sRNA molecules.
We compiled the candidate sRNA genes predicted in the different studies and compared them with the annotation of 5′ UTRs, 3′ UTRs and operons reported by the study of Tjaden et al. (17). After uniting overlapping predictions into single candidates there remain 906 candidates that are unique to a single study, 85 candidates that were predicted by two studies and 10 candidates that were predicted by three studies. We find that most candidates are not located within annotated operons, 5′ UTRs or 3′ UTRs.
No function is known at present for 42 out of the 55 discovered sRNAs. Our survey provides pointers that may aid in associating function to some of these molecules. Also, the various characteristics we have identified may be used for the development of a refined algorithm for predicting additional sRNA-encoding genes in E.coli, as well as in closely related organisms.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Elena Rivas for sending us unpublished data. This study was supported by a grant from the Human Frontier Science Program granted to H.M. and S.A.
REFERENCES
- 1.Hildebrandt M. and Nellen,W. (1992) Differential antisense transcription from the Dictyostelium EB4 gene locus: implications on antisense-mediated regulation of mRNA stability. Cell, 69, 197–204. [DOI] [PubMed] [Google Scholar]
- 2.Lankenau S., Corces,V.G. and Lankenau,D.H. (1994) The Drosophila micropia retrotransposon encodes a testis-specific antisense RNA complementary to reverse transcriptase. Mol. Cell. Biol., 14, 1764–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morfeldt E., Taylor,D., von Gabain,A. and Arvidson,S. (1995) Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J., 14, 4569–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharp T.V., Schwemmle,M., Jeffrey,I., Laing,K., Mellor,H., Proud,C.G., Hilse,K. and Clemens,M.J. (1993) Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res., 21, 4483–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner E.G. and Simons,R.W. (1994) Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol., 48, 713–742. [DOI] [PubMed] [Google Scholar]
- 6.Wassarman K.M., Zhang,A. and Storz,G. (1999) Small RNAs in Escherichia coli. Trends Microbiol., 7, 37–45. [DOI] [PubMed] [Google Scholar]
- 7.Wightman B., Ha,I. and Ruvkun,G. (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C.elegans. Cell, 75, 855–862. [DOI] [PubMed] [Google Scholar]
- 8.Eddy S.R. (2001) Non-coding RNA genes and the modern RNA world. Nature Rev. Genet., 2, 919–929. [DOI] [PubMed] [Google Scholar]
- 9.Wassarman K.M. (2002) Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell, 109, 141–144. [DOI] [PubMed] [Google Scholar]
- 10.Storz G. (2002) An expanding universe of noncoding RNAs. Science, 296, 1260–1263. [DOI] [PubMed] [Google Scholar]
- 11.Eddy S.R. (2002) Computational genomics of noncoding RNA genes. Cell, 109, 137–140. [DOI] [PubMed] [Google Scholar]
- 12.Argaman L., Hershberg,R., Vogel,J., Bejerano,G., Wagner,E.G., Margalit,H. and Altuvia,S. (2001) Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol., 11, 941–950. [DOI] [PubMed] [Google Scholar]
- 13.Wassarman K.M., Repoila,F., Rosenow,C., Storz,G. and Gottesman,S. (2001) Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev., 15, 1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivas E., Klein,R.J., Jones,T.A. and Eddy,S.R. (2001) Computational identification of noncoding RNAs in E.coli by comparative genomics. Curr. Biol., 11, 1369–1373. [DOI] [PubMed] [Google Scholar]
- 15.Carter R.J., Dubchak,I. and Holbrook,S.R. (2001) A computational approach to identify genes for functional RNAs in genomic sequences. Nucleic Acids Res., 29, 3928–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S., Lesnik,E.A., Hall,T.A., Sampath,R., Griffey,R.H., Ecker,D.J. and Blyn,L.B. (2002) A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems, 65, 157–177. [DOI] [PubMed] [Google Scholar]
- 17.Tjaden B., Saxena,R.M., Stolyar,S., Haynor,D.R., Kolker,E. and Rosenow,C. (2002) Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res., 30, 3732–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urbanowski M.L., Stauffer,L.T. and Stauffer,G.V. (2000) The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol., 37, 856–868. [DOI] [PubMed] [Google Scholar]
- 19.Majdalani N., Chen,S., Murrow,J., St John,K. and Gottesman,S. (2001) Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol., 39, 1382–1394. [DOI] [PubMed] [Google Scholar]
- 20.Rudd K.E. (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res., 28, 60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F., Gregor,J., Davis,N.W., Kirkpatrick,H.A., Goeden,M.A., Rose,D.J., Mau,B. and Shao,Y. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 22.Schattner P. (2002) Searching for RNA genes using base-composition statistics. Nucleic Acids Res., 30, 2076–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein R.J., Misulovin,Z. and Eddy,S.R. (2002) Noncoding RNA genes identified in AT-rich hyperthermophiles. Proc. Natl Acad. Sci. USA, 99, 7542–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 25.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zwieb C., Gorodkin,J., Knudsen,B., Burks,J. and Wower,J. (2003) tmRDB (tmRNA database). Nucleic Acids Res., 31, 446–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eddy S.R. (2002) A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMCBioinformatics, 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huynen M., Snel,B., Lathe,W.,III and Bork,P. (2000) Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res., 10, 1204–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dandekar T., Snel,B., Huynen,M. and Bork,P. (1998) Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci., 23, 324–328. [DOI] [PubMed] [Google Scholar]
- 30.Yanai I., Mellor,J.C. and DeLisi,C. (2002) Identifying functional links between genes using conserved chromosomal proximity. Trends Genet., 18, 176–179. [DOI] [PubMed] [Google Scholar]
- 31.Karzai A.W., Susskind,M.M. and Sauer,R.T. (1999) SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J., 18, 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 33.Rudd K.E. (1999) Novel intergenic repeats of Escherichia coli K-12. Res. Microbiol., 150, 653–664. [DOI] [PubMed] [Google Scholar]
- 34.Brown S. and Fournier,M.J. (1984) The 4.5 S RNA gene of Escherichia coli is essential for cell growth. J. Mol. Biol., 178, 533–550. [DOI] [PubMed] [Google Scholar]
- 35.Tetart F. and Bouche,J.P. (1992) Regulation of the expression of the cell-cycle gene ftsZ by DicF antisense RNA. Division does not require a fixed number of FtsZ molecules. Mol. Microbiol., 6, 615–620. [DOI] [PubMed] [Google Scholar]
- 36.Sledjeski D. and Gottesman,S. (1995) A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl Acad. Sci. USA, 92, 2003–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizuno T., Chou,M.Y. and Inouye,M. (1984) A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc. Natl Acad. Sci. USA, 81, 1966–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keiler K.C., Waller,P.R. and Sauer,R.T. (1996) Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science, 271, 990–993. [DOI] [PubMed] [Google Scholar]
- 39.Liu M.Y., Gui,G., Wei,B., Preston,J.F.,III, Oakford,L., Yuksel,U., Giedroc,D.P. and Romeo,T. (1997) The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem., 272, 17502–17510. [DOI] [PubMed] [Google Scholar]
- 40.Brownlee G.G. (1971) Sequence of 6S RNA of E. coli. Nature New Biol., 229, 147–149. [DOI] [PubMed] [Google Scholar]
- 41.Jain S.K., Gurevitz,M. and Apirion,D. (1982) A small RNA that complements mutants in the RNA processing enzyme ribonuclease P. J. Mol. Biol., 162, 515–533. [DOI] [PubMed] [Google Scholar]
- 42.Masse E. and Gottesman,S. (2002) A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA, 99, 4620–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polayes D.A., Rice,P.W. and Dahlberg,J.E. (1988) DNA polymerase I activity in Escherichia coli is influenced by spot 42 RNA. J. Bacteriol., 170, 2083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Argaman L. and Altuvia,S. (2000) fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol., 300, 1101–1112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.