Abstract
Cowpea mosaic virus (CPMV) replicates in close association with small membranous vesicles that are formed by rearrangements of intracellular membranes. To determine which of the viral proteins are responsible for the rearrangements of membranes and the attachment of the replication complex, we have expressed individual CPMV proteins encoded by RNA1 in cowpea protoplasts by transient expression and in Nicotiana benthamiana plants by using the tobacco rattle virus (TRV) expression vector. The 32-kDa protein (32K) and 60K, when expressed individually, accumulate in only low amounts but are found associated with membranes mainly derived from the endoplasmic reticulum (ER). 24K and 110K are freely soluble and accumulate to high levels. With the TRV vector, expression of 32K and 60K results in rearrangement of ER membranes. Besides, expression of 32K and 60K results in necrosis of the inoculated N. benthamiana leaves, suggesting that 32K and 60K are cytotoxic proteins. On the other hand, during CPMV infection 32K and 60K accumulate to high levels without causing necrosis.
It has become increasingly clear that replication of positive-strand RNA viruses takes place in association with intracellular membranes (for a review see reference 6). Often these membranes are induced upon infection by vesiculation or rearrangement of membranes from different organelles including the early and late endomembrane system (24, 27, 36). The importance of membranes for viral replication is evident from various studies. In vitro synthesis of positive-strand RNAs of picornaviruses and nodaviruses depends on membranes (25, 49). Poliovirus, Semliki Forest virus, and cowpea mosaic virus (CPMV) RNA replication is inhibited in cells treated with cerulenin, an inhibitor of lipid biosynthesis (7, 17, 28). Brome mosaic virus RNA replication is severely inhibited in yeast cells that carry a mutation in the host Δ9 fatty acid desaturase, which affects the lipid composition of the membranes (21). Despite the central role of membranes in viral replication, it remains poorly understood how viral replication complexes are targeted to and assembled on specific membrane sites.
Upon infection of cowpea (Vigna unguiculata L.) or Nicotiana benthamiana cells with CPMV, typical cytopathological structures are formed which consist of an amorphous matrix of electron-dense material traversed by rays of small membranous vesicles (7, 10, 20). The membranous vesicles are closely associated with CPMV RNA replication, as was revealed by autoradiography in conjunction with electron microscopy on sections of CPMV-infected leaves treated with [3H]uridine (10). Fractionation of homogenates from infected leaves further demonstrated that the viral replicase activity is present in the crude membrane fraction, which corroborated the notion that viral replication complexes are physically associated with membranes (13). Based on the observation that the endoplasmic reticulum (ER) undergoes a drastic proliferation upon CPMV infection and that the rearranged ER membranes colocalize with the cytopathological structure, we have proposed that the small membranous vesicles originate from the ER (7).
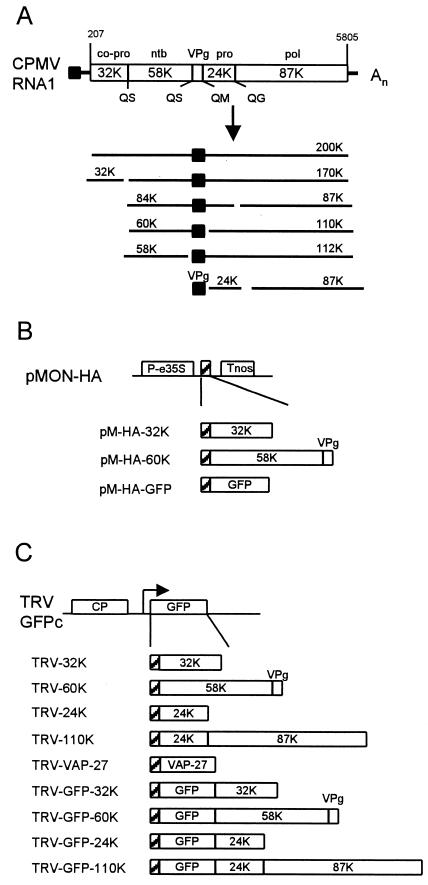
CPMV is the type member of the comoviruses and bears strong resemblance to animal picornaviruses both in gene organization and in the amino acid sequence of replication proteins. Both RNA1 and RNA2 express large polyproteins, which are proteolytically cleaved into different proteins by the 24-kDa proteinase (24K) (Fig. 1A ). The proteins encoded by RNA1 are necessary and sufficient for virus replication, whereas RNA2 codes for the capsid proteins and the movement protein. Based on amino acid sequence comparison with other viruses and different experimental data, functions have been attributed to the different RNA1-encoded proteins. The 32-kDa protein (32K) is a hydrophobic protein which contains no motifs common to other positive-strand RNA viruses outside the Comoviridae. It is involved in regulation of the RNA1 polyprotein processing and is required as a cofactor in the cleavage of the RNA2 polyprotein (32). 60K (58K + VPg) is able to bind ATP via a conserved Walker nucleotide-binding motif and has been proposed elsewhere to be a viral helicase (31). 87K has a domain specific to RNA-dependent RNA polymerases (RdRp); however, 110K (87K plus 24K) is the only viral protein present in highly purified RdRp capable of elongating nascent viral RNA chains (13), suggesting that fusion to 24K is required for replicase activity.
FIG. 1.
Schematic diagram of CPMV RNA1 and the TRV RNA2-based expression vector used to express the CPMV nonstructural proteins. (A) Genetic organization and translational expression of CPMV RNA1. Open reading frames in the RNA molecules (open bars), VPg (black square), cleavage sites (Q/G, Q/M, and Q/S), and nucleotide positions of start and stop codons are indicated. Abbreviations: pro, proteinase; co-pro, cofactor for proteinase; ntb, nucleotide binding protein; pol, core RdRp. (B) Organization of the constructs in the plant expression vector pMON-HA used to transiently express CPMV RNA1-encoded proteins in cowpea protoplasts. Expression is driven by an enhanced CaMV 35S promoter (P-e35S), and transcription is terminated at a nopaline synthase terminator sequence (Tnos). An HA epitope tag was added to facilitate immunodetection of the expressed proteins (hatched box). 60K is composed of the 58K nucleotide-binding protein and VPg. (C) Organization of the expression vector TRV-GFPc and the TRV constructs used to express CPMV RNA1-encoded proteins in N. benthamiana leaves. The duplicated promoter regionwith downstream cloning sites (arrow) permits transcription of a subgenomic RNA for expression of foreign gene inserts. The 110K polymerase is composed of the 24K proteinase and the 87K core polymerase as indicated. Additional constructs were made that contain a fusion with GFP. VAP27 is an A. thaliana protein that is inserted in the ER membrane. The position of the HA epitope tag is indicated with a hatched box.
During infection, the bulk of the nonstructural proteins encoded by RNA1 is present in electron-dense structures adjacent to the small membranous vesicles (48). It was proposed elsewhere that the replication complexes are deposited onto these electron-dense structures after taking part in RNA replication located on the membranes (43). Aggregation of the virus nonstructural proteins in this structure complicates the analysis of the membrane binding properties of the individual proteins during normal CPMV infection. Expression of RNA1-encoded proteins in insect cells, by using the baculovirus expression system, showed that 60K but not 110K is able to induce and associate with small membranous vesicles in the cytoplasm in these cells (41).
In this study, the nature of the interaction of different RNA1-encoded proteins and membranes was further explored in plant cells. By using a virus vector-based expression system, selected RNA1 proteins were expressed alone or in a fusion with a reporter protein, which allowed us to identify and visualize specific membranes interacting with the viral proteins in live plant cells. In addition, expression of certain viral proteins led to drastic rearrangements of intracellular membranes and ultimately cell death.
MATERIALS AND METHODS
Construction of the transient expression vectors.
Plasmid pMON999 is a plant expression vector that contains the enhanced cauliflower mosaic virus (CaMV) 35S promoter, followed by a multiple cloning site and the terminator of the nopaline synthase gene (43). pMON-HA is derived from pMON999 and contains the coding region of the hemagglutinin (HA) epitope tag (YPYDVPDYA) between the promoter and terminator (44). The individual CPMV RNA1 proteins were PCR amplified with gene-specific primers (Table 1) and cloned in these vectors. Because the individual proteins are expressed in a polyprotein, the primers were designed to add a start codon at the 5′ end and a stop codon at the 3′ end of each gene when appropriate. Furthermore, the primers contained restriction sites to facilitate cloning. pM-HA-32K was constructed by amplification of the 32K coding region from pTB1G (14) with primers EcoRI_32K_F and BamHI_32K_R and insertion in pMON-HA. pM-HA-60K was constructed by PCR amplification of the 60K coding region from pTB1G with EcoRI_60K_F and BamHI_60K_R, and pM-HA-GFP was constructed by amplification of the green fluorescent protein (GFP) coding region from pM19GFP10 (16) with primers EcoRI_GFP_F and BamHI_GFP_R. pM-60K was constructed by amplification of the 60K coding region from pTB1G (14) with primers XbaI_60K_F and BamHI_60K_R and insertion in pMON999. pM-32K was constructed by insertion of an XbaI-SstI fragment of pTB32*, which contains the 32K coding sequence followed by a stop codon and the SstI restriction site (47), into pMB200K. pMB200K contains the entire RNA1-encoded 200K polyprotein followed by the SstI restriction site in pMON999 (43). The XbaI restriction site is present internally in the coding sequence of 32K.
TABLE 1.
Oligonucleotides used in this study to insert coding sequences of CPMV replication proteins and GFP in different expression vectors
| Primer | Sequence |
|---|---|
| EcoRI_32K_F | GAGAATTCGGTCTCCCAGAATATGAGG |
| BamHI_32K_R | GGGGATCCTCACTGTGCATTGTTCTTTTCAC |
| EcoRI_60K_F | CGGAATTCAGTAGTCCTGTTATCCTCTTAG |
| BamHI_60K_R | CCGGATCCTTATTGTGCGTCTGCCCAAA |
| EcoRI_GFP_F | CGGAATTCATGAGTAAAGGAGAAGAACTTTTCACT |
| BamHI_GFP_R | CCGGATCCTTATTTGTATAGTTCATCCATGC |
| NcoI_HA_F | GGGACCATGGCTTATCCATACGATGTTCCA |
| NcoI_24K_F | GGGGCCATGGCTTCTTTGGATCAGAGTAGTGTT |
| SstI_24K_R | CCCGAGCTCTTATTGCGCTTGTGCTATTGG |
| NcoI_32K_F | GGGGCCATGGGTCTCCCAGAATAT |
| EcoRI_TAA_32K_R | CCCGAATTCTTACTGTGCATTGTCCTTTTCACC |
| NcoI_60K_F | GGGGCCATGGCTAGTAGTCCTGTTATCCTCTTA |
| EcoRI_TAA_60K_R | CCCGAATTCTTATTGTGCGTCTGCCCAAACTC |
| SstI_TAA_110K_R | CCCGAGCTCCTAAACATCAGAAAAAGCGAAATTGA |
| SalI_GFP5_F | GGGGGGTCGACGATGAGTAAAGGAGAAGAACTTTTC |
| SalI_GFP5_R | CCCCCGTCGACTCTTTGTATAGTTCATCCATGC |
Construction of TRV expression vectors.
Tobacco rattle virus (TRV) vectors carrying CPMV RNA1-encoded proteins were constructed with TRV-GFPc. This plasmid contains cDNA from TRV RNA2 and an engineered subgenomic promoter followed by the GFP gene (23). First this plasmid was modified to contain a start codon followed by the coding sequence of the HA epitope tag and a multiple cloning site. This was achieved by replacement of the NcoI fragment released from TRV-GFPc with a PCR fragment amplified from pM-HA-32K with primers NcoI_HA_F and SstI_TAA_32K_R digested with the same enzyme. This intermediate plasmid was designated TRV-HA-Δ32K and contains the SalI, PstI, and EcoRI sites directly following the coding sequence for the HA epitope. These restriction sites and the EcoRI, SstI, or KpnI present at the 3′ end can be used to insert a gene of choice as an N-terminal fusion with the HA tag.
TRV-HA-24K was constructed by PCR amplification of the 24K coding sequence from pTB1G with primers NcoI_24K_F and SstI_TAA_24K_R. This PCR product was digested with NcoI (filled in with Klenow polymerase) and SstI and cloned in TRV-HA-Δ32K digested with EcoRI (filled in with Klenow polymerase) and SstI. TRV-HA-32K was constructed by PCR amplification of the 32K coding sequence from pTB1G with primers NcoI_32K_F and SstI_TAA_32K_R. This PCR product was digested with HindIII and SstI and was cloned in TRV-HA-Δ32K digested with HindIII and SstI. TRV-HA-60K was constructed by PCR amplification of the 60K coding sequence from pMON-HA-60K with primers NcoI_60K_F and EcoRI_TAA_60K_R. This PCR product was digested with EcoRI and cloned in TRV-HA-Δ32K digested with EcoRI. To construct TRV-HA-110K, the 110K coding sequence was PCR amplified from pTB1G with primers NcoI_110K_F and SstI_TAA_110K_R, digested with NcoI and SstI, and cloned in TRV-GFPc digested with the same enzymes, resulting in clone TRV-110K. Subsequently TRV-HA-24K was digested with KpnI, and the released fragment was replaced with the KpnI fragment of TRV-110K to construct TRV-HA-110K. The fusion constructs of the different viral proteins with GFP were made by using the SalI restriction site directly downstream of the HA epitope sequence. The GFP coding sequence was PCR amplified from pM19GFP10 (16) with primers SalI_GFP5_F and SalI_GFP5_R. For the construction of TRV-HA-VAP27, the complete coding sequence of the 27-kDa VAMP-associated protein (VAP27) was released as an EcoRI fragment from pGAD10-VAP27, a clone isolated from an Arabidopsis thaliana cDNA library. This fragment was ligated in TRV-HA-Δ32K digested with the same enzyme.
Transfection of cowpea protoplasts and infection of plants.
Cowpea mesophyll protoplasts were prepared and transfected by polyethylene glycol-mediated transformation of plasmid DNA as described previously (43). Five-week-old wild-type N. benthamiana plants or N. benthamiana plants carrying the mGFP5-ER transgene (34) were dusted with carborundum and infected with TRV RNA1 together with transcripts of the TRV RNA2 recombinants as described previously (23).
Subcellular fractionation and immunoblotting.
Protoplasts transfected with the transient expression vectors were collected 16 h posttransfection and centrifuged at 30,000 × g, and proteins from the supernatant and pellet fraction were electrophoresed in 10% polyacrylamide gels and immunoblotted as described previously (5) with the mouse monoclonal antibody anti-HA (clone 12CA5; Boehringer) recognizing the HA epitope tag.
For immunoblot analysis of the proteins produced by the TRV expression vector, infected leaves were collected 1 day postinfection and ground with a mortar and pestle with 2 ml of extraction buffer (50 mM Tris-acetate [pH 7.4], 10 mM potassium acetate, 1 mM EDTA, 5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride per g of fresh weight). After extraction, the homogenates were centrifuged, electrophoresed, and immunoblotted as described earlier (5) with anti-HA or anti-GFP (Clontech). In some cases, homogenates were not centrifuged, but total protein was extracted with sample buffer (10% glycerol, 5% β-mercaptoethanol, 2% sodium dodecyl sulfate, 0.01% bromophenol blue, 75 mM Tris-HCl, pH 6.8), boiled for 3 min, and loaded on the gel.
Fluorescence microscopy.
A Zeiss LSM 510 confocal microscope was used to obtain images. GFP fluorescence was observed with standard settings (excitation wavelength, 488 nm; emission band-pass filter, 505 to 550 nm). Red chlorophyll fluorescence was detected with an excitation wavelength of 543 nm and an emission long-pass filter of 560 nm. To observe GFP fluorescence in living epidermal cells, large samples (2 by 2 cm) were cut from N. benthamiana leaves infected with the TRV recombinants 1 day postinfection. The samples were placed on a glass slide, and after a drop of water was applied on top of the samples, they were covered with a coverslip. The samples were directly analyzed by confocal laser scanning microscopy.
RESULTS
Expression of individual CPMV RNA1-encoded proteins in plant cells.
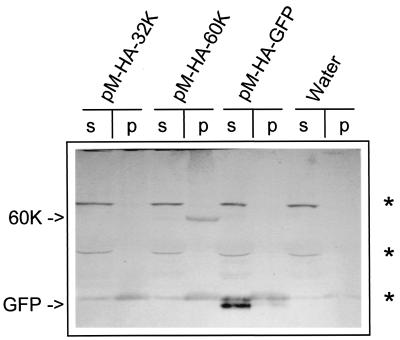
In previous studies several individual CPMV proteins were transiently expressed in cowpea protoplasts by using the plant expression vector pMON999, which carries the enhanced CaMV 35S promoter (30, 43). Those studies concerned the viral RdRp and therefore were focused on the C-terminal 110K region of the 200K polyprotein. In the present study the proteins contained in the N-terminal region were included also. For this purpose, the coding sequences for the 32K proteinase cofactor and the 60K nucleoside triphosphate (NTP) binding protein were inserted into the multiple cloning site of pMON999 to produce pM-32K and pM-60K. The constructs were used to transfect protoplasts, which were then harvested 16 h posttransfection. Homogenates of the protoplasts were subjected to centrifugation at 30,000 × g, crudely separating soluble proteins from membrane-bound proteins. It was shown previously by immunoblotting procedures that 110K and 87K accumulate to high levels in protoplasts and are found mainly in the soluble fraction (S30) (43). 32K and 60K were not detectable in either S30 or the pellet fraction (P30) with anti-32K and anti-VPg sera (data not shown). The failure to detect 32K and 60K might be due to low expression levels of these proteins and/or to low titers of the antisera. To avoid the latter possibility, constructs were made with the HA epitope tag fused to the N terminus of 32K and 60K (Fig. 1B). As a positive control, the HA epitope was also fused to GFP. Western blot analysis of extracts from transfected protoplasts with mouse monoclonal antibodies raised against the HA epitope revealed that HA-GFP accumulated to readily detectable levels and was present mainly in the S30 fraction (Fig. 2). HA-60K accumulated to much lower levels and was present mainly in the P30 fraction (Fig. 2), whereas 32K was not detectable in either S30 or P30.
FIG. 2.
Immunodetection of CPMV RNA1-encoded proteins in different subcellular fractions of cowpea protoplasts transfected with transient expression vectors. Protoplasts were harvested 16 h posttransfection, and homogenates were separated in the supernatant (s) and pellet fraction (p) by centrifugation at 30,000 × g. Detection of the proteins was done with a mouse monoclonal anti-HA antibody. 60K and GFP are indicated by arrows. Cowpea proteins that cross-react with this antibody are indicated by asterisks. “Water” indicates lanes with no DNA added.
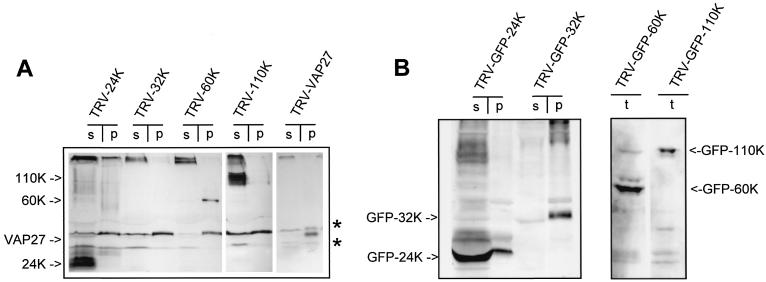
To confirm the results obtained in the protoplast system, the individual CPMV RNA1-encoded proteins were expressed in plants by using the TRV expression system (23). TRV does not replicate and spread in cowpea plants, which are the natural host of CPMV. However, in N. benthamiana, which is also a systemic host for CPMV, the TRV-based expression system was shown previously to give high levels of foreign protein expression (23). Since TRV does not cause visible morphological changes to the host endomembrane system as observed by electron microscopy (9) and is not related to CPMV, a TRV infection probably does not interfere with proper targeting of the CPMV proteins. Vector TRV-GFPc contains the coding sequence of GFP in a cDNA clone of TRV RNA2 in which GFP can be replaced with other genes of choice (Fig. 1C) (23). To facilitate immunodetection of the CPMV proteins, TRV-GFPc was modified to encode the HA epitope followed by a multiple cloning site. Subsequently, the coding sequences for the 24K proteinase, the 32K proteinase cofactor, the 60K NTP binding protein, and the 110K polymerase were each inserted into the TRV vector immediately downstream of the HA tag, thereby replacing the GFP gene (Fig. 1C). In vitro transcripts of these constructs were combined with TRV RNA1 and used as inoculum on N. benthamiana. Homogenates of infected leaves were separated into P30 and S30 fractions. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed on samples of these fractions followed by Western blot analysis with mouse monoclonal antibodies recognizing the HA epitope present at the N terminus of the proteins. 24K and 110K were present predominantly in the soluble fraction, whereas 60K was present in the crude membrane fraction (Fig. 3A). We were unable to detect 32K in either fraction (Fig. 3A). To confirm that membrane-associated proteins are localized to the pellet during fractionation, cDNA of an A. thaliana integral membrane protein, VAP27, was cloned into TRV as a fusion with the HA epitope tag. VAP27 is a SNARE-like protein that is inserted posttranslationally into the ER membrane via a C-terminal transmembrane domain (37). Homogenates of TRV-VAP27-infected leaves were fractionated, and like 60K, VAP27 was detected predominantly in the P30 fraction (Fig. 3A).
FIG. 3.
Immunodetection of CPMV RNA1-encoded proteins, either tagged with HA (A) or fused to GFP (B), in different subcellular fractions of N. benthamiana leaves infected with TRV expression vectors. Infected leaves were collected 1 dpi, and total homogenates (t) were prepared that were either loaded directly for TRV-GFP-60K and TRV-GFP-110K or centrifuged at 30,000 × g to separate proteins from the supernatant (s) and pellet fraction (p) for the other constructs. Detection of the proteins was done with mouse monoclonal anti-HA antibody (A) or with a rabbit polyclonal anti-GFP serum (B). The bands corresponding to the CPMV proteins are indicated by arrows, while N. benthamiana proteins that cross-react with anti-HA are indicated by asterisks.
These data suggest that 60K associates with membranes when expressed in the absence of other CPMV-encoded proteins whereas 24K and 110K behave as cytoplasmic, soluble proteins. We were unable to detect the 32K protein by immunoblotting either expressed in N. benthamiana leaves with the TRV expression system or transiently expressed in cowpea protoplasts.
Intracellular localization of the replication proteins.
To visualize and characterize the protein-membrane interaction in live cells, TRV expression vectors encoding fusions of the CPMV proteins with GFP were constructed. First it was established by immunoblotting that the constructs were properly expressed. Homogenates of infected leaves were separated by centrifugation in the P30 and S30 fractions. By using anti-GFP serum, GFP-24K was found present mainly in the S30 fraction (Fig. 3B). In contrast to native 32K, GFP-32K was detectable by immunoblotting and was found to be present mainly in the P30 fraction (Fig. 3B). Expression of GFP-60K and GFP-110K was detected in the leaf homogenates (Fig. 3B); however, after fractionation in the P30 and S30 fractions no specific bands could be visualized for unknown reasons (data not shown).
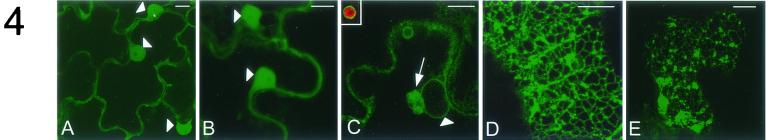
The intracellular localization of the fusion proteins was examined in infected leaf epidermal cells 1 day postinoculation (dpi) by confocal laser scanning microscopy. Cells containing GFP-24K (data not shown) or GFP-110K (Fig. 4A) displayed a pattern of fluorescence similar to that of nonfused GFP (Fig. 4B), and fluorescence was present diffusely in the cytoplasm and in the nucleus. Since GFP is excluded from the vacuole that takes up more than 90% of the cell volume in epidermal cells, cytoplasmic GFP is confined to a small layer between the vacuole and the cell wall (the cortex) and to a small region near the nucleus (Fig. 4B). The occurrence of GFP in the nucleus is in line with earlier reports that indicate that GFP alone or fused to a soluble protein accumulates partly in the nucleus, probably by means of passive diffusion through the nuclear pore (46). The fluorescence pattern of GFP-60K differed from that of nonfused GFP. GFP-60K fluorescence was most obvious in a ring surrounding the nucleus, in small rings surrounding spherical organelles in the cytoplasm, and in one or several spherical aggregates often near the nucleus, while no fluorescence was present in the nucleus (Fig. 4C). The fluorescent ring around the nucleus was reminiscent of what we have observed in transgenic N. benthamiana plants expressing ER-GFP (7), a GFP fusion protein that contains an N-terminal cleavable signal peptide and a C-terminal HDEL retention signal and therefore is targeted to the lumen of the ER (18) (compare Fig. 4C with Fig. 5B). It thus appears that GFP-60K localizes to the ER membranes that are contiguous with the nuclear envelope. Alternatively, 60K is present in the nuclear envelope itself. Red chlorophyll autofluorescence was observed in the small spherical organelles (Fig. 4C, inset), suggesting that GFP-60K is also present in the outer membrane of plastids. The nature of the spherical aggregates near the nucleus (Fig. 4C, arrow) is unknown, but it might be a structure derived from ER membranes, as pointed out below. Also the fluorescence observed with GFP-32K differed markedly from that for nonfused GFP. The fluorescence was most obvious at the cortex of the infected cells and was present in a clear reticulate pattern (Fig. 4D) closely resembling the pattern observed with ER-GFP in transgenic plants (compare Fig. 4D with Fig. 5A). It thus appears that GFP-32K localizes to the cortical ER network. In some cells the reticulate pattern was less bright and GFP-32K was additionally present in large highly fluorescent aggregates at the cortex, which may be derived from ER membranes (Fig. 4E). No fluorescence of GFP-32K was observed in the nuclear envelope (data not shown), which is in contrast to the fluorescence observed in transgenic plants that express ER-GFP or in TRV-GFP-60K-infected plants.
FIG.4.
Subcellular distribution of CPMV proteins fused to GFP in N. benthamiana epidermal cells infected with TRV expression vectors. Fluorescent signals were visualized by confocal microscopy 1 dpi. GFP-110K (A) is present in the nucleus (arrowheads) and in a diffuse pattern in the cytoplasm, as is nonfused GFP (B). GFP-60K (C) localizes in a ring surrounding the nucleus (arrowhead), in one or several bodies (arrow) often near the nucleus, and in a ring surrounding plastids that autofluoresce in red (inset). GFP-32K localizes in a cortical reticulated network (D) and in cortical fluorescent aggregates (E). The images shown in panels A to C correspond to single optical sections of 1 μm, while panels D and E correspond to projections of serial optical sections. Bars, 10 μm.
FIG.5.
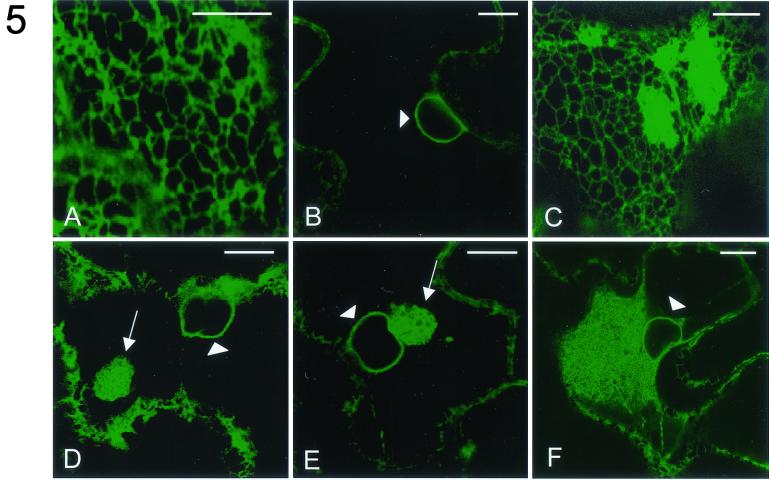
Effect of expression of different CPMV proteins on the morphology of the ER. N. benthamiana plants transgenic with GFP targeted to the lumen of the ER (ER-GFP) were infected with TRV expression vectors. Fluorescent signals were visualized by confocal microscopy 1 dpi. In epidermal cells infected with TRV the pattern of ER-GFP fluorescence is not distinguishable from that in mock-infected cells and is present in a cortical reticulate pattern (A) and in a ring around the nucleus (arrowhead; B). In cells infected with TRV-32K ER-GFP fluorescence is additionally present in large aggregates connected to the cortical network (C) and in one or several spherical aggregates (arrow; D) often near the nucleus (arrowhead). In TRV-60K-infected cells the latter structure is also present (arrow; E). Extensive ER proliferation near the nucleus (arrowhead) of a CPMV-infected cell is shown (F). The images shown in panels B, D, E, and F correspond to single optical sections of 1 μm, while panels A and C correspond to projections of serial optical sections. Bars, 10 μm.
These data corroborate the cell fractionation data and indicate that individually expressed 24K and 110K are cytosolic, soluble proteins and that 32K and 60K associate with membranes mainly derived from the ER.
Expression of 32K and 60K modifies ER morphology.
CPMV infection induces an extensive rearrangement of ER membranes, a process independent of RNA2-encoded proteins (7). To investigate whether a specific RNA1-encoded protein is responsible for these membrane modifications, the TRV constructs each carrying an individual RNA1-encoded protein (not fused to GFP) were used to infect N. benthamiana plants transgenic for GFP targeted to the lumen of the ER (18, 34). At 1 dpi the morphology of the ER membranes in infected epidermal cells was observed and compared to that of mock-infected cells. TRV infection itself had no influence on ER membrane morphology, since, in plants infected with TRV, ER-GFP fluorescence in epidermal cells did not differ from that in mock-infected cells and fluorescence was detected in the typical cortical ER network (Fig. 5A) and the nuclear envelope (Fig. 5B). In TRV-24K- and TRV-110K-infected cells the fluorescence pattern of ER-GFP was also indistinguishable from that of mock-infected cells (data not shown). In contrast, the ER in cells infected with TRV-32K or TRV-60K differed radically from the ER in mock-infected cells. In TRV-32K-infected cells, ER-GFP fluorescence was present in large amorphous aggregates that were connected with the cortical ER network (Fig. 5C) and in spherical aggregates (Fig. 5D) which were often near the nucleus. While in TRV-60K-infected cells these spherical aggregates were also found near the nucleus (Fig. 5E), cortical ER aggregates were not observed (data not shown). Both types of aggregates resembled the regions of proliferative ER membranes induced in CPMV-infected cells, although the extent of ER proliferation in CPMV-infected cells was generally larger (Fig. 5F). The ER modifications seen with TRV-32K and with TRV-60K may be artifacts that occur upon expression of any ER membrane-associated protein with the TRV-based expression system. This was tested by expressing the integral ER membrane protein VAP27 with the TRV vector. When TRV-VAP27 was used to infect ER-GFP transgenic plants, no changes were found in the ER-GFP fluorescence pattern from that of mock-infected cells (data not shown).
From these data we conclude that expression of 32K and 60K induced drastic morphological changes in the ER, resembling CPMV-induced ER membrane proliferation, whereas 24K and 110K expression did not influence ER morphology.
Expression of 32K and 60K induces necrosis.
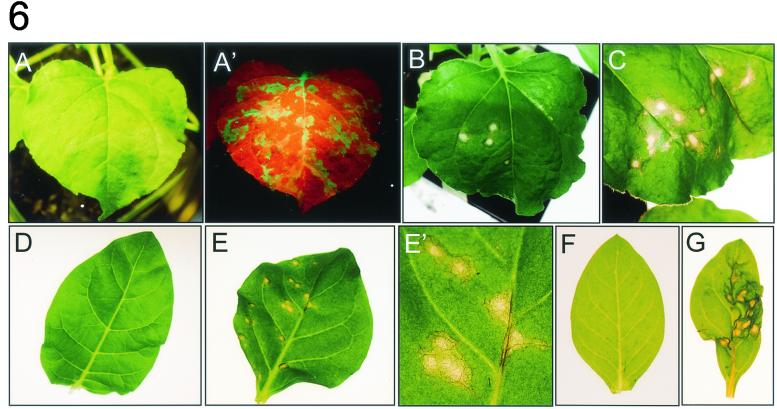
During the course of the experiments a severe effect of prolonged expression of 32K and 60K on cell viability was observed. TRV infection caused no visible symptoms on inoculated leaves of N. benthamiana, but a slight curling of the systemically infected leaves was apparent 4 dpi. Infection of leaves with TRV-GFP allowed us to monitor the spread of TRV from the initial infection sites to the systemic leaves by using a handheld UV lamp. One day after infection GFP fluorescence was visible as green spots on inoculated leaves. With time these spots grew larger, and 3 dpi fluorescence was observed in large patches and in the major veins of the inoculated leaves as well as in the veins of the systemically infected leaves (Fig. 6A ′). In contrast to TRV and TRV-GFP (Fig. 6A), infection with TRV-32K or TRV-60K triggered formation of necrotic spots on the inoculated leaves. The first sign of lesion formation occurred from approximately 2 dpi onward when infected tissue developed a glassy appearance. At 3 dpi the developing lesions were more distinct from the surrounding tissue, appearing more clearly necrotic and collapsed (Fig. 6B and C). The necrotic tissue was typically confined to the inoculated leaves, but occasionally some necrosis was observed in the upper, noninoculated leaves at 4 to 6 dpi (data not shown). This suggests that movement of the recombinant TRV RNA2 is severely inhibited. Also the TRV construct carrying the GFP-32K fusion produced formation of necrotic spots, but TRV-GFP-60K did not trigger necrosis (data not shown). Infection with TRV-24K, TRV-110K, TRV-GFP-24K, TRV-GFP-110K, and TRV-VAP27 did not result in the formation of necrotic lesions, although a slight yellowing of infection sites was observed for TRV-110K at 6 dpi (data not shown). These results suggest that prolonged expression of 32K or 60K or the fusion of 32K with GFP from the TRV expression vector leads to cell death. This was rather surprising, since CPMV does not cause necrosis on N. benthamiana plants and moves systemically to the upper leaves and high amounts of 32K and 60K accumulate during the infection. To determine whether 32K and 60K expression also produced necrosis in other host plants, Nicotiana clevelandii, Nicotiana tabacum, and Nicotiana glutinosa plants were infected with TRV-32K, TRV-60K, and TRV-GFP as control. In all these Nicotiana species, necrosis was observed at 2 to 3 dpi on inoculated leaves infected with TRV-32K and TRV-60K, whereas leaves infected with TRV-GFP were symptomless (Fig. 6D to G and data not shown).
FIG.6.
Necrosis of leaves from different host plants infected with TRV-32K and TRV-60K. Leaves were infected with the TRV constructs and photographed 3 dpi. (A and A′) N. benthamiana leaf infected with TRV-GFP photographed either under white light (A) or under UV light (A′), where GFP fluorescence appears green. The red fluorescence in A′ is due to chlorophyll. (B to G) Leaves were photographed under white light. (B) N. benthamiana leaf infected with TRV-32K. (C) N. benthamiana leaf infected with TRV-60K. (D) N. tabacum leaf infected with TRV-GFP. (E) N. tabacum leaf infected with TRV-32K. (E′) Close-up of panel E. (F) N. clevelandii infected with TRV-GFP. (G) N. clevelandii infected with TRV-60K.
DISCUSSION
In this study, the CPMV 32K and 60K replication proteins were shown to associate with membranes mainly derived from the ER, when expressed in isolation in plant cells. Moreover, expression of the 32K protein and to a lesser extent 60K effected proliferation of the ER resembling what occurs upon CPMV infection. Other RNA1-encoded proteins, the 110K polymerase and the N-terminal cleavage product 24K proteinase, behaved as freely soluble proteins when expressed in isolation. We propose that localization signals in 32K and 60K target the replication complexes to ER membranes and that this physical association causes an ER membrane rearrangement that may result in formation of the small membranous vesicles that are the site of CPMV RNA synthesis.
During CPMV infection the replication proteins accumulate in both electron-dense structures and the adjacent small membranous vesicles where CPMV replication occurs (10, 48). To analyze the properties of the individual RNA1-encoded nonstructural proteins, they were expressed separately with a transient expression system in cowpea protoplasts and with the TRV expression vector in N. benthamiana. In contrast to the 110K (87K plus 24K) polymerase or the 24K proteinase, which accumulated to high levels in both systems, the 60K nucleotide-binding protein accumulated poorly, while the 32K cofactor for the protease was not detectable by immunoblot analysis, which may suggest that 32K and 60K have a high turnover rate. This is in line with previous findings in which expression of 32K and 60K in insect cells or in Escherichia coli cells yielded considerably lower protein levels than did expression of 110K (29, 33, 41, 42). The high levels of 32K and 60K accumulating during CPMV infection may be the result of aggregation in the electron-dense material. The role of the electron-dense structures in the viral life cycle and how the replication proteins end up in this structure remain unclear. Alternative explanations for the low expression levels of 32K and 60K may be the cytotoxicity of the proteins or the possibility that sequences in the RNA either inhibit mRNA translation or stimulate mRNA degradation. Fusion of GFP to 32K improved the expression levels, and GFP-32K, like 60K, was found to be present mainly in the crude membrane fraction. Comparison of the GFP-32K fluorescence with that of GFP-60K revealed that 32K was present mainly in the cortical ER whereas GFP-60K was found mainly in the nuclear envelope, the plastidial membrane, and aggregates presumably derived from the ER. This suggests that 32K is more specifically targeted to and/or retained in the cortical ER, in contrast to 60K, which displays affinity for various intracellular membranes. It should be noted, however, that fusion of 60K to GFP abolished the ability of 60K to induce necrosis, which may suggest that 60K could not engage in the proper interactions when fused to GFP. Therefore, the localization pattern observed with GFP-60K might not represent the actual localization of the nonfused 60K.
Expression of either 32K or 60K was shown to influence ER morphology, with 32K inducing proliferation of the cortical ER and formation of one or several small bodies of proliferated ER near the nucleus, whereas 60K induced only the latter structure. The alterations of the ER morphology resembled the proliferations that occur in CPMV-infected cells, although the regions of proliferated ER are generally larger in the latter case. Maybe the combined action of 32K and 60K is necessary to induce proliferations of the ER that mimic those observed for CPMV-infected cells. The observation that CPMV nonstructural proteins, when expressed in isolation, target ER membranes and cause ER membrane rearrangements is in agreement with the previously proposed notion that the small membranous vesicles that are involved in virus replication originate from the ER (7).
Previous studies showed that 60K, when expressed in insect cells by the baculovirus expression system, induced formation of small membranous vesicles (41). Here we make the first report that 32K is also involved in the process of membrane attachment and rearrangement. Unlike the other replication proteins of CPMV, the 326-amino-acid hydrophobic 32K contains no conserved domains such as an NTP-binding motif (present in 60K), a proteinase domain (24K), or a polymerase domain (110K). In vitro translation studies pointed to a role for 32K as a cofactor required for processing of the RNA2-encoded polyprotein (32). Deletion of 32K from the RNA1-encoded 200K polyprotein resulted in more rapid cleavage of the 170K processing intermediate both in vivo and in vitro, suggesting that 32K inhibits processing of 200K (32). It is likely that targeting of and insertion in the membrane of replicase proteins, polyprotein cleavage, and the initiation of viral RNA synthesis occur in a coordinated fashion during CPMV replication. Since it was shown previously that 32K remains associated with 170K after cleavage from the 200K polyprotein (15), 32K might first target the 32K/170K complex to the membranous site of replication where, after further processing, initiation of replication occurs.
The properties of 32K and 60K in ER membrane attachment and modification resemble those of replication proteins encoded by different viruses of which poliovirus is the best-studied example. Membrane association of the poliovirus proteins encoded by the 2BC3AB region was shown previously to be mediated by a stretch of hydrophobic amino acids (3AB) or by an amphipathic helix (2BC and 2C) (4, 26, 40). 32K contains three stretches of hydrophobic amino acids at the C-terminal end (unpublished observations), and 60K contains an amphipathic helix at the N terminus (amino acids 45 to 61 [32]) and a 22-amino-acid stretch of hydrophobic amino acids (amino acids 544 to 565 [unpublished observations]) at the C terminus immediately upstream of VPg. These domains are conserved among the comoviruses. Further experiments will be necessary to verify that these regions are involved in membrane binding, and the TRV expression system might provide a suitable experimental system for that purpose. Apart from binding to intracellular membranes, expression of poliovirus proteins in different cell types has revealed that proteins from the 2BC3AB region could extensively modify the endomembrane system. 2BC and 2C induced formation of small membranous vesicles probably derived from the ER that were morphologically similar to poliovirus-induced vesicles (2, 3, 8). The 3A protein, when expressed in isolation, associated with ER membranes, causing swelling of these membranes, but no small membranous vesicles were formed (11, 12). Recently, it was reported that coexpression of 3A and 2BC resulted in formation of vesicles that were similar, both in ultrastructure and in biochemical properties, to the vesicles induced in a poliovirus infection (39). Also, for the equine arteritis virus it was shown elsewhere that coexpression of the membrane proteins nsp2 and nsp3 induces the formation of double membrane vesicles probably derived from the ER (38). The tobacco etch virus 6-kDa protein has been shown to localize specifically to ER membranes when expressed as a fusion with GFP or glucuronidase and was proposed to cause the ER modifications that are observed during tobacco etch virus infection (35). Such membrane-associated proteins may play a dual role. Firstly, the induction and accumulation of vesicles could compartmentalize viral RNA synthesis. Secondly, these proteins could promote the interaction of viral RdRp with the proliferating membranes.
Infection with TRV-32K and TRV-60K led to formation of necrotic tissue in inoculated leaves from N. benthamiana and other Nicotiana species, suggesting that 32K and 60K are cytotoxic proteins that cause cell death. Fusion of 60K with GFP abolished its ability to induce necrosis for unknown reasons. The cytotoxic properties of 60K have been reported before for insect cells expressing 60K with the baculovirus system (42). These cells showed abnormal cytopathic effects and rapid cell lysis usually within 48 h postinfection. In the same system expression of 32K did not lead to clear effects on cell viability (41). It is remarkable that 32K and 60K induce necrosis only when expressed separately from other RNA1-encoded proteins, since CPMV infection of N. benthamiana leads to high accumulation of 32K and 60K but no visible necrosis. The aggregation of 32K and 60K in electron-dense structures during CPMV infection may somehow prevent induction of cell death by these proteins. The significance of the cytotoxic properties of 32K and 60K for the viral life cycle is unclear. Unlike animal viruses, plant viruses do not rely on lysis of host cells for their spread through the plant. It has been reported that expression in mammalian cells of poliovirus 2B and 2BC or 2B from the related coxsackievirus resulted in an increase in membrane permeability ultimately leading to cell lysis, suggesting that these proteins play an important role in the release of viral progeny (1, 45).
Recently it was reported that tobacco mosaic virus modified to express the 2B protein of tomato aspermy virus triggered the induction of a hypersensitive response (HR) resulting in tissue necrosis, despite the fact that infection of this plant species with either wild-type tobacco mosaic virus or wild-type tomato aspermy virus did not lead to necrosis (22). The HR is a plant defense mechanism that occurs upon attack by certain pathogens and involves rapid death of plant cells in association with the restriction of pathogen spread (for a review see reference 19). At the moment we cannot exclude the possibility that 32K and 60K are recognized by the plant and elicit an HR rather than causing cell death directly by increasing membrane permeability.
Acknowledgments
We gratefully acknowledge Jan Verver and Jeroen Pouwels for technical assistance and comments and Janneke Saaijer and Aart van Ommeren for growing the N. benthamiana plants.
J.E.C. was supported by The Netherlands Foundation for Chemical Research (CW) with financial aid from The Netherlands Foundation for Scientific Research (NWO).
REFERENCES
- 1.Aldabe, R., A. Barco, and L. Carrasco. 1996. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 271:23134-23137. [DOI] [PubMed] [Google Scholar]
- 2.Aldabe, R., and L. Carrasco. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64-76. [DOI] [PubMed] [Google Scholar]
- 3.Barco, A., and L. Carrasco. 1995. A human virus protein, poliovirus protein 2BC, induces membrane proliferation and blocks the exocytic pathway in the yeast Saccharomyces cerevisiae. EMBO J. 14:3349-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barco, A., and L. Carrasco. 1998. Identification of regions of poliovirus 2BC protein that are involved in cytotoxicity. J. Virol. 72:3560-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertens, P., J. Wellink, R. Goldbach, and A. van Kammen. 2000. Mutational analysis of the cowpea mosaic virus movement protein. Virology 267:199-208. [DOI] [PubMed] [Google Scholar]
- 6.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carette, J. E., M. Stuiver, J. van Lent, J. Wellink, and A. van Kammen. 2000. Cowpea mosaic virus infection induces a massive proliferation of endoplasmic reticulum but not Golgi membranes and is dependent on de novo membrane synthesis. J. Virol. 74:6556-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 9.De Zoeten, G. A. 1966. California tobacco rattle virus, its intracellular appearance, and the cytology of the infected cell. Phytopathology 56:744-754. [Google Scholar]
- 10.De Zoeten, G. A., A. M. Assink, and A. Van Kammen. 1974. Association of cowpea mosaic virus-induced double-stranded RNA with a cytopathological structure in infected cells. Virology 59:341-355. [DOI] [PubMed] [Google Scholar]
- 11.Doedens, J. R., T. H. Giddings, Jr., and K. Kirkegaard. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J. Virol. 71:9054-9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eggen, R., A. Kaan, R. Goldbach, and A. Van Kammen. 1988. Cowpea mosaic virus RNA replication in crude membrane fractions from infected cowpea and Chenopodium armaranticolor. J. Gen. Virol. 69:2711-2720. [Google Scholar]
- 14.Eggen, R., J. Verver, J. Wellink, A. De Jong, R. Goldbach, and A. van Kammen. 1989. Improvements of the infectivity of in vitro transcripts from cloned cowpea mosaic virus cDNA: impact of terminal nucleotide sequences. Virology 173:447-455. [DOI] [PubMed] [Google Scholar]
- 15.Franssen, H., M. Moerman, G. Rezelman, and R. Goldbach. 1984. Evidence that the 32,000-dalton protein encoded by bottom-component RNA of cowpea mosaic virus is a proteolytic processing enzyme. J. Virol. 50:183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gopinath, K., J. Wellink, C. Porta, K. M. Taylor, G. P. Lomonossoff, and A. van Kammen. 2000. Engineering cowpea mosaic virus RNA-2 into a vector to express heterologous proteins in plants. Virology 267:159-173. [DOI] [PubMed] [Google Scholar]
- 17.Guinea, R., and L. Carrasco. 1990. Phospholipid biosynthesis and poliovirus genome replication, two coupled phenomena. EMBO J. 9:2011-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haseloff, J., K. R. Siemering, D. C. Prasher, and S. Hodge. 1997. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heath, M. C. 2000. Hypersensitive response-related death. Plant Mol. Biol. 44:321-334. [DOI] [PubMed] [Google Scholar]
- 20.Hibi, T., G. Rezelman, and A. Van Kammen. 1975. Infection of cowpea mesophyll protoplasts with cowpea mosaic virus. Virology 64:308-318. [DOI] [PubMed] [Google Scholar]
- 21.Lee, W. M., M. Ishikawa, and P. Ahlquist. 2001. Mutation of host Δ9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 75:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, H. W., A. P. Lucy, H. S. Guo, W. X. Li, L. H. Ji, S. M. Wong, and S. W. Ding. 1999. Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 18:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacFarlane, S. A., and A. H. Popovich. 2000. Efficient expression of foreign proteins in roots from tobravirus vectors. Virology 267:29-35. [DOI] [PubMed] [Google Scholar]
- 24.Mackenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molla, A., A. V. Paul, and E. Wimmer. 1993. Effects of temperature and lipophilic agents on poliovirus formation and RNA synthesis in a cell-free system. J. Virol. 67:5932-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul, A. V., A. Molla, and E. Wimmer. 1994. Studies of a putative amphipathic helix in the N-terminus of poliovirus protein 2C. Virology 199:188-199. [DOI] [PubMed] [Google Scholar]
- 27.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez, L., R. Guinea, and L. Carrasco. 1991. Synthesis of Semliki Forest virus RNA requires continuous lipid synthesis. Virology 183:74-82. [DOI] [PubMed] [Google Scholar]
- 29.Peters, S. A. 1994. Thesis. Wageningen University, Wageningen, The Netherlands.
- 30.Peters, S. A., J. M. Mesnard, I. M. Kooter, J. Verver, J. Wellink, and A. van Kammen. 1995. The cowpea mosaic virus RNA 1-encoded 112 kDa protein may function as a VPg precursor in vivo. J. Gen. Virol. 76:1807-1813. [DOI] [PubMed] [Google Scholar]
- 31.Peters, S. A., J. Verver, E. A. Nollen, J. W. van Lent, J. Wellink, and A. van Kammen. 1994. The NTP-binding motif in cowpea mosaic virus B polyprotein is essential for viral replication. J. Gen. Virol. 75:3167-3176. [DOI] [PubMed] [Google Scholar]
- 32.Peters, S. A., W. G. Voorhorst, J. Wery, J. Wellink, and A. van Kammen. 1992. A regulatory role for the 32K protein in proteolytic processing of cowpea mosaic virus polyproteins. Virology 191:81-89. [DOI] [PubMed] [Google Scholar]
- 33.Richards, O. C., R. Eggen, R. Goldbach, and A. van Kammen. 1989. High-level synthesis of cowpea mosaic virus RNA polymerase and protease in Escherichia coli. Gene 78:135-146. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz, M. T., O. Voinnet, and D. C. Baulcombe. 1998. Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skehel, P. A., R. Fabian-Fine, and E. R. Kandel. 2000. Mouse VAP33 is associated with the endoplasmic reticulum and microtubules. Proc. Natl. Acad. Sci. USA 97:1101-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82:985-994. [DOI] [PubMed] [Google Scholar]
- 39.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 41.van Bokhoven, H., J. W. van Lent, R. Custers, J. M. Vlak, J. Wellink, and A. van Kammen. 1992. Synthesis of the complete 200K polyprotein encoded by cowpea mosaic virus B-RNA in insect cells. J. Gen. Virol. 73:2775-2784. [DOI] [PubMed] [Google Scholar]
- 42.van Bokhoven, H., J. Wellink, M. Usmany, J. M. Vlak, R. Goldbach, and A. van Kammen. 1990. Expression of plant virus genes in animal cells: high-level synthesis of cowpea mosaic virus B-RNA-encoded proteins with baculovirus expression vectors. J. Gen. Virol. 71:2509-2517. [DOI] [PubMed] [Google Scholar]
- 43.van Bokhoven, H., J. Verver, J. Wellink, and A. van Kammen. 1993. Protoplasts transiently expressing the 200K coding sequence of cowpea mosaic virus B-RNA support replication of M-RNA. J. Gen. Virol. 74:2233-2241. [DOI] [PubMed] [Google Scholar]
- 44.Van Der Heijden, M. W., J. E. Carette, P. J. Reinhoud, A. Haegi, and J. F. Bol. 2001. Alfalfa mosaic virus replicase proteins P1 and P2 interact and colocalize at the vacuolar membrane. J. Virol. 75:1879-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Kuppeveld, F. J., J. G. Hoenderop, R. L. Smeets, P. H. Willems, H. B. Dijkman, J. M. Galama, and W. J. Melchers. 1997. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 16:3519-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Arnim, A. G., X. W. Deng, and M. G. Stacey. 1998. Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221:35-43. [DOI] [PubMed] [Google Scholar]
- 47.Vos, P., J. Verver, M. Jaegle, J. Wellink, A. van Kammen, and R. Goldbach. 1988. Two viral proteins involved in the proteolytic processing of the cowpea mosaic virus polyproteins. Nucleic Acids Res. 16:1967-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wellink, J., J. Van Lent, and R. Goldbach. 1988. Detection of viral proteins in cytopathic structures in cowpea protoplasts infected with cowpea mosaic virus. J. Gen. Virol. 69:751-755. [Google Scholar]
- 49.Wu, S. X., P. Ahlquist, and P. Kaesberg. 1992. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc. Natl. Acad. Sci. USA 89:11136-11140. [DOI] [PMC free article] [PubMed] [Google Scholar]