Abstract
Gammaherpesviruses are associated with a number of diseases including lymphomas and other malignancies. Murine gammaherpesvirus 68 (MHV-68) constitutes the most amenable animal model for this family of pathogens. However experimental characterization of gammaherpesvirus gene expression, at either the protein or RNA level, lags behind that of other, better-studied alpha- and beta-herpesviruses. We have developed a cDNA array to globally characterize MHV-68 gene expression profiles, thus providing an experimental supplement to a genome that is chiefly annotated by homology. Viral genes started to be transcribed as early as 3 h postinfection (p.i.), and this was followed by a rapid escalation of gene expression that could be seen at 5 h p.i. Individual genes showed their own transcription profiles, and most genes were still being expressed at 18 h p.i. Open reading frames (ORFs) M3 (chemokine-binding protein), 52, and M9 (capsid protein) were particularly noticeable due to their very high levels of expression. Hierarchical cluster analysis of transcription profiles revealed four main groups of genes and allowed functional predictions to be made by comparing expression profiles of uncharacterized genes to those of genes of known function. Each gene was also categorized according to kinetic class by blocking de novo protein synthesis and viral DNA replication in vitro. One gene, ORF 73, was found to be expressed with α-kinetics, 30 genes were found to be expressed with β-kinetics, and 42 genes were found to be expressed with γ-kinetics. This fundamental characterization furthers the development of this model and provides an experimental basis for continued investigation of gammaherpesvirus pathology.
The gammaherpesviruses are an important group of pathogens that cause serious disease in humans and animals. The two human gammaherpesviruses, Epstein-Barr virus (EBV) and human herpesvirus 8 (HHV-8), are associated with a number of lymphomas and other cancers. EBV has been linked to Burkitt's lymphoma as well as nasopharyngeal carcinomas (9), and HHV-8 has been associated with Kaposi's sarcoma, body cavity-based lymphomas, and multicentric Castleman's disease (6). In vitro work on these human gammaherpesviruses is restricted to a limited number of latently infected cell lines (5, 10, 18, 27), as these viruses do not readily infect cells in tissue culture. This has left the natural progression of disease relatively uncharted, with primary lytic infection being particularly unclear. Therefore the lytic life cycle has only been studied by artificially reactivating these viruses (9a), raising problems such as synchronicity and physiological differences between primary infection and reactivation.
Murine gammaherpesvirus 68 (MHV-68) has rapidly gained recognition as a small-animal model for studying gammaherpesvirus biology, partly because it is a natural pathogen of murid rodents (34). Infection of mice in laboratory conditions shows an acute phase that is cleared approximately 2 weeks postinfection (p.i.) and a latent phase in B lymphocytes and other cell types that persists (4). MHV-68 has also been shown to enhance atherosclerosis in apolipoprotein E-deficient mice (2), as well as being able to trigger lymphoproliferative disease (39). The virus also has the property of being easily manipulated in tissue culture systems, where it causes a fully productive infection in a wide range of cell lines (23). Recently a bacterial artificial chromosome mutagenesis system has been developed; this system allows rapid generation of mutant strains (1) which, combined with the existing breeds of transgenic mice, allow experimental manipulation of both the pathogen and its environment.
MHV-68 has been fully sequenced (42) and has been shown to be closely related to all gammaherpesviruses at a genetic level, in particular to the gamma-2 subfamily which includes HHV-8 and herpesvirus saimiri. In a genome of around 80 genes, 63 are homologous with those of other gamma-2 herpesviruses, while the others are unique to the virus and are designated M1 to M14. In common with the human gammaherpesviruses, MHV-68 encodes a number of host protein homologues, including a viral D-type cyclin, a G protein-coupled receptor similar to the interleukin-8 receptor, a Bcl-2 homologue, and a complement activation regulator. There is also a protein with homology to poxvirus serpins. While much of MHV-68's genome has been annotated and functions have been assigned to its genes, most of this information has been attributed via homology to other better-studied herpesviruses. Experimental confirmation is necessary to verify these predicted functions and develop this model further. Gene expression profiles are a useful initial characterization, as the origin of a gene's function lies in its expression and a fundamental characterization of the virus life cycle in vitro would greatly enhance the elucidation of MHV-68 pathogenesis.
One method of examining gene expression is by using the recently developed array technology (32). This technique allows a global analysis of transcription for an entire viral genome in a single step. To date arrays have been successfully used to examine the transcriptional program of HHV-8 (16, 26), herpes simplex virus type 1 (37), and human cytomegalovirus (8), as well as the host's transcriptional response to these viral infections (36, 45).
This study set out to develop a cDNA array for MHV-68 and apply this technique to characterize the expression profiles of the virus throughout an infection in vitro. The transcription of each gene was monitored during the lytic life cycle and also categorized into a kinetic class by its dependence on protein synthesis or viral DNA replication. The identification of gene expression patterns for MHV-68 provides a valuable insight into primary lytic gammaherpesvirus infections and, together with the kinetic classification, furthers the development of the MHV-68 model system.
MATERIALS AND METHODS
Cells and virus.
BHK-21 cells (ECACC CB2857) were maintained in Glasgow modified Eagle's medium (Sigma) supplemented with 10% newborn calf serum, 5% tryptose phosphate broth (Sigma), penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM l-glutamine. NIH 3T3 cells (ATCC CCL1658) were cultured in Dulbecco's modified Eagle medium (Life Technologies) containing 10% newborn calf serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM l-glutamine. The original stock of MHV-68 (strain g2.4) was kindly provided by S. Efstathiou (Cambridge University). Working stocks of virus were prepared as previously described (19). Briefly, BHK-21 cells were infected at a multiplicity of infection (MOI) of 0.001 and harvested as soon as cytopathic effect was complete. Cellular debris was removed from the viral suspension by centrifugation at 1,600 × g for 30 min. The extracellular virus was pelleted by centrifugation at 16,000 × g for 90 min. The resulting pellet was resuspended in phosphate-buffered saline (Life Technologies) and stored in aliquots at −80°C.
Infectious virus was assayed by plaque titration (19). Briefly, 10-fold dilutions of virus were used to infect NIH 3T3 monolayers, and the cells were then overlaid with 0.8% agarose-containing medium. Cells were stained after 4 to 5 days with 0.1% crystal violet solution to determine the number of plaques.
To monitor changing gene expression levels through lytic infection, the following time course was set up. Virus was adsorbed onto NIH 3T3 monolayers at an MOI of 10 and incubated at 37°C for 1 h. Unattached virus was then removed by washing cells with medium containing 2% newborn calf serum (MM). Infected cells were then overlaid with further MM and 4 × 106 cells were harvested at 0, 1, 3, 5, 8, 12, and 18 h p.i. for RNA isolation.
Protein synthesis was inhibited with cycloheximide (CX; Calbiochem). To establish the efficiency of inhibition, the drug was titrated and its effect was determined by measuring the incorporation of radiolabeled methionine as follows. Seventy-five percent confluent monolayers of cells were incubated in medium containing 1 μCi of [35S]methionine/105 cells and concentrations of CX between 100 μg/ml and 100 pg/ml. Monolayers were incubated at 37°C for 8 h before being washed with MM and lysed with NP-40 lysis buffer (150 mM NaCl, 1.0% NP-40, 50 mM Tris, pH 8.0) on ice for 30 min. Cellular debris was removed by centrifugation at 10,000 × g for 10 min. Aliquots of the supernatant were spotted onto glass fiber filters (Wallac), and proteins were precipitated with ice-cold 5% trichloroacetic acid (TCA) for 30 min. Filters were washed twice with 5% TCA for 5 min and once with 95% ethanol for 5 min before being dried at room temperature. Incorporation of labeled methionine was then quantified with a Beckman scintillation counter.
To block de novo protein synthesis, monolayers of NIH 3T3 cells were pretreated with CX for 30 min prior to and throughout infection. Cells (4 × 106) were harvested at 5 and 8 h p.i. Negative (mock-infected) and positive (infections without CX) controls at both time points were also included.
Viral DNA replication was inhibited by 2′-deoxy-5-ethyl-β-4′-thiouridine (4′-S-EtdU) at a concentration of 200 ng/ml (3). Again, monolayers were pretreated for 30 min prior to and throughout infection. Samples of 4 × 106 cells were taken at 5 and 18 h p.i. Negative (mock-infected) and positive (infections without 4′-S-EtdU) controls at both time points were included.
Array design and manufacture.
The array was designed as previously described (16) with the following modifications. Primers were designed (Table 1) to amplify by PCR approximately 300-bp fragments of DNA from the 5′ end of predicted MHV-68 open reading frames (ORFs) (42). In addition to the predicted ORFs, regions of MHV-68's genome between ORFs of greater than 100 bp were also included. If the intergene regions exceeded 300 bp (e.g., ∼840 bp between ORFs K3 and M5), then two cDNAs were amplified to correspond to the 5′ end of a potential transcript that could be transcribed in either direction. The following were chosen as positive and negative hybridization controls: genes encoding glyceraldehyde 3-phosphate dehydrogenase (NM008084), myosin 1 (L00923), murine ornithine decarboxylase (M10624), β-actin (X03672), calcium binding protein Cab45 (U45977), ribosomal protein S29 (NM009093), ubiquitin (AF285162), phospholipase A2 (D78647), and hypoxanthine phosphoribosyltransferase (J00423); pBluescript II SK(+) plasmid (Stratagene); and the gene encoding the tobacco mosaic virus 180-kDa protein (D78608). Water was also used as a negative control. Luciferase (E15166) was included as an internal control for normalization of arrays.
TABLE 1.
Primer sequences used for amplification of DNA elements for the MHV-68 array
| ORF | Sequence (5′-3′) of:
|
|
|---|---|---|
| Forward primer | Reverse primer | |
| M1 | GCCAAAGCATAGCTCACTGG | ACACCTTGATGACCCTTCC |
| M2 | CAAGGAAAGATTCCCAATCC | CAGGACTTGGTACAGGACTCG |
| M3 | GCTCATTAAATGCTGCATCC | TCTCTTGACCCAGCTCTTCC |
| M4 | CACCCAGCCTAGATTTTTAG | TACGCAGACATAATAGCTGC |
| ORF4 | GGGATTGTGGGTGTAAATGG | GGGATCACAGTAAAACACTGC |
| ORF6 | CTCCCAGGCTCCTCTTGG | CCACGTCCATGAAAAATTGG |
| ORF7 | GATTATGCATGGACCTCAGC | CACCATAGAAGCAGGCAAGG |
| ORF8 | ATGAGAGTCGCCCACCTAAC | TGAAGATGTAGGGCACGATG |
| ORF9 | CAGCCTTGCCAAGAAAGAGC | CTGAACGGAATGTCCTCGC |
| ORF10 | TTAATCACTGGTGCGTGACC | CGGGAGATTAGTTTTTATAGCC |
| ORF11 | ACGAAGGTCATCTACATTTGC | TGTGAGGACAACCTGGAGG |
| K3 | AGGAGAGTTCTGTTGGATCTGC | TCCCCATCACTATCATCAGC |
| M5 | AGCATACAGCGTGGGAATGG | AGGGGATTTCCAGGTAGAGG |
| M6 | GGACCCTCCATTCTATAAACC | CCAGCTCGGGAGGGGGC |
| ORF17 | CGTGGGAGGATATGTGGAC | ATGTGTGCAACATCTGCAGC |
| ORF18 | AGGTGCATCAGCCATGTTGG | TGAGGTCCTTCGTTGTCAGG |
| ORF19 | TTTGTTTACCTGAACCCAAACC | ACTCCACCCGACTGGAAGG |
| ORF20 | GAATGAATTAGGAGCCAAGC | CTTCTCTGGTGGTACTGTGC |
| ORF21 | AACAACCCTGGATTCCAACC | GCCCATATGCCCCTATATCC |
| ORF22 | GTTGGTGCATGGGGTAGG | GCTGGTAATGGTGGGATAGC |
| ORF23 | AAAGACTGTGCCTTAACAGG | GTCATGGTCAGGATTTTTGC |
| ORF24 | TGTGAATATGATGAGAATAGCC | ATATCACTGGAACATAGGTGC |
| ORF25 | TTTGCCTATGTCAGGACTGG | ACTTGGTCTTCTTCCATCACC |
| ORF26 | CTCTAACATCTAGATTGTATGC | CTAAATCATCATGCTCATCCC |
| ORF27 | TGGTGAGGTTGTGCTAAATGG | GGCAGGATAATAGGGATGTTTG |
| ORF29b | TGTTACAGAAGGATGCCAAG | AGACTTCCGTGTGATTGC |
| ORF30 | GGATGCTCTGTGAATAAATCAAAG | TGTCTTGGCCGCGCTTGC |
| ORF31 | CCAAGACAAGTGTATGATGC | ACAATGTCACCAAACAGTGC |
| ORF32 | TTCCTTTACAACAAGATGAGC | TCTAAAGAAGCCCTCATCCC |
| ORF33 | GACATTTCTGAATAAAGAGTGTATATGG | GTACTGGAATGGCTGACTTCC |
| ORF34 | TTTTCCCCGAAGCAGATCC | TATGCTGTGCTCACTGATGG |
| ORF35 | AATTGGGAGTAGTTTAAGGGC | TTAACAGTGTCTTCCAACTCC |
| ORF36 | TGGATTACCGACAGTTACCG | TCTGATGCCATGGTAGAAACC |
| ORF37 | GGAAGGGTCGATTATTCTGG | ATACACGGCAGACACCTCAC |
| ORF38 | TCGGATGTTGCAAGAAAACC | CGTGTTTGCCTCGTTCATATTC |
| ORF39 | AAAAGTTGGAGCCGTCTAGG | CAGACGGTACACTGTTACTGC |
| ORF40 | CAATGGCACGATGTTCAGG | TCTCCTTGCCAGAAGTACCC |
| ORF42 | CTTCCCAGGCTGATGTTGG | CCACCATGGAAATCAGTGC |
| ORF43 | TCTGTCAGAGAGTGCCATGC | AAGTGTTGCTACCCAAGAAGC |
| ORF44 | ACCTCAGATGCCAAAATTCG | TGTGGGACAATATGACTTGAGC |
| ORF45 | ATGGACCCCTTTAAGAAACC | CTTGTACTTGACTCGCTGACC |
| ORF46 | TGGACACTTGGCTAAAAACG | TGAACACTAAAGGCCAAACC |
| ORF47 | TGGTCCTTTTGTGTATTAATGC | TGGGCCACATTGATATTCCC |
| ORF48 | ACCTTGAAACCCGTGAAGG | GAGCTGGCACACAAAGAAGG |
| ORF49 | CCTTCTCTGGAAAGCGTGG | AATTGACAGTGCCTATGGCC |
| ORF50 | CAAAGTCCATAACAGGCATCC | AAATGCCTCAACTTCTCTGG |
| M7 | TGTGGCGTTAAATCCCTAGC | TGGGGAAGTTGGTTCTGAGG |
| ORF52 | TGGTCAAGGAAGTAGAAAGG | CTGGCACCCACAGTAGTTTC |
| ORF53 | ATCACCCAAGAAACCACACC | GCGTAGATCAAAAAGACACCAC |
| ORF54 | ATACTCCTTTGTGCCCAAGC | ATTGGCCCGGTTGAATCTCC |
| ORF55 | TGTACCTTACAAGAGGCTCG | GCCTCATCTACACTTATTGC |
| ORF56 | ATGGCCAGATACCACAGC | AAAACTGTTTGGCATCATCG |
| M8 | ACCAGTTGAGGAGCCAACG | TTTCTGGTTCCATCTGTTTGG |
| ORF57 | CTTGCCTGAAACACGGTAGG | AGAGGGTCCCTGATTACACG |
| ORF58 | ATTGTGGGAGGAATGTCTGC | GGAACCCATGTGGAAAGC |
| ORF59 | CTTCCAGCTTGACACTGAGC | AAGACAGGGAGGCAGATTCC |
| ORF60 | CTGGGTTCCTTGAACTGACC | CTTCAGCTTCATCATGTCACC |
| ORF61 | ACGTGGAGCCTGTTCAGACC | GCTGCCAGATAACCATTTTCC |
| ORF62 | TCTTCAGTAGTCACATCAGC | AGGCTCTTGTTGTGTTTTAGG |
| ORF63 | CCATCAGTGAGCGATAGTGG | TGCAGCACAAAGAAGACTGG |
| ORF64 | TTCGCATCGAAGGTACAGC | GCCAAACATTTCAGCAGAGG |
| M9 | CCCAGAGCTCCATAACAAGC | AAATGCTCCAGAAGAGGAAGG |
| ORF66 | CAGTGGATGAGTTTAGAAGC | AGGGCACAGTGAGTATTTTGG |
| ORF67 | CTGATAGACGAGCTCTGTGG | GCCATATTGACCCTGTTGC |
| ORF68 | TCCTTCCTCTCAAATACACTGG | ATGCAGCACGTAGAAGCAGAGG |
| ORF69 | GCGCTCAACAGGCTCTGC | ACCCAGATTGTCCGTGTGG |
| M10C | AAGCAGGAGCAGCACAGC | TGGTATGTCAACCCCTGACC/PICK> |
| ORF72 | TCCAAGGATTTCTTGACAGC | ATGTAGGCCCTGACCTTTCC |
| M11 | TGGGCAACCCTGATTACAGC | ATGATCCTCCGTCCAACTGC |
| ORF73 | ACACAACCTCAGGCAAACC | CCTTCAACATCAACATCTGG |
| ORF74 | CTTAGAAAACTCATCATTGTCC | ACACAATAGCATAGATCCTGG |
| ORF75C | AGACAGAAAAAGAACTCATCG | GAATCAGATCGTGAAGATAGG |
| ORF75B | GGATGAGGACGTCTGGGC | ATAAAATCTAGCCGCTGGGC |
| M12 | GGGGAAAATATGCGTGATACC | CCAGTGGTCTGTTCTGATGG |
| M13 | TAGTAGGGGGCCTCCTGC | CAAAGTTTAAAGTGAAAGTAAGC |
| M14 | GCTACCGCCCGGGCCTGAGG | AGCAGGGCCCGAGCCCCTCTT |
| M1-M2 | CTGGGGACCAGATGTAAAGC | TCCATGGGTGCATATTTGG |
| M2-M1 | TAACAGTGAAGGTGCTAACG | CAGGTTCTCGGTTCAAGTCC |
| M2-M3 | AAACCCCCTCCAGTAAAAGG | CAAGGCCCCAGAGAAAGC |
| M3-M2 | TGATACTCAAGTAACTTAAGAGG | GCCCTGAATCAGTTTTGGC |
| M3-M2 | TTAAAAAAGATACGAGTCAGGTGG | CGGTCGAGAAGACATATCCC |
| M3-M4 | AAATATGCTCCATGGTTTGG | TCCCCAAAAGATACATCAAGC |
| M4-M3 | TCATCTGACTTGCTGCATACG | CTTGTGAGCCTCAAGAGTGG |
| K3-M5 | TTTTGGCCAGGAGAATATAAGC | CGAGACAGGTTGTGGAAAGC |
| M5-K3 | CAGGAGACATGGCCTATCG | CCCCTGAAGCATAACTCTCG |
| ORF27-29B | TCTGGATTTTCATTTCATGTG | CACGTGACAAGAGTGCTTGG |
| M10C-ORF72 | TCATGGCAACAGTCAAAAGG | AAACGTGCTAGCCAATCAGG |
| ORF72-M10C | CTGTGCGAGATTTGCGTATG | ATGTATCGATCCCCGTCCTC |
All PCR products were cloned into pGEM T-easy vectors (Promega), and sequences were verified on a Beckman automated sequencer. DNA species were then reamplified from the relevant plasmids and purified (QIAquick PCR purification kit; Qiagen). Fifty nanograms of each was spotted in duplicate onto charged nylon membranes (Hybond N+; Amersham) with a 384-pin multiblotter (V & P Scientific). The DNA arrays were then denatured (0.66 M NaCl, 0.5 M NaOH) and washed (double-distilled water and then 40 mM phosphate buffer, pH 7.3).
RNA preparation.
Total RNA was isolated with TRIzol reagent (Life Technologies). Residual DNA contamination was eliminated by incubation with 20 U of DNase I (RNase free; Roche) in DNase buffer (40 mM Tris-HCl [pH 7.5], 10 mM NaCl, 6 mM MgCl2) for 30 min at 37°C before termination with 10 mM EDTA. RNA was reisolated by phenol-chloroform extraction and precipitated with ethanol. Quality was assessed on a denaturing agarose gel. Only samples with 260/280-nm absorbance ratios of greater than 1.9 were used for subsequent experiments.
Luciferase RNA was transcribed in vitro from a sequence-confirmed luciferase plasmid with a RiboMAX kit (Promega).
Array hybridizations.
All arrays were prehybridized for 1 h in ExpressHyb (Clontech) with sheared salmon sperm DNA (1 mg/ml) and murine C0T-1 DNA (1 μg/ml). C0T-1 DNA was used to suppress cross-hybridization to repetitive DNA. Ten micrograms of RNA, spiked with 10 ng of luciferase RNA, was primed with a specific primer mixture (0.2 μM) consisting of the 3′ primers used to amplify the DNA species for the array membranes. RNA and primers were annealed at 70°C for 5 min before RNA was reverse transcribed with Superscript II (Life Technologies) and labeled with [α-33P]dATP at 50°C for 90 min. The labeled cDNA was purified with Nucleospin columns (Clontech), denatured (95°C for 5 min), and hybridized in ExpressHyb (Clontech) to arrays at 60°C overnight. Following hybridization, arrays were washed three times for 15 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-1% sodium dodecyl sulfate (SDS) and three times for 15 min in 0.1× SSC-0.5% SDS.
Northern blot analysis.
RNA was extracted and size separated by electrophoresis on 1% agarose gels containing 2.2 M formaldehyde. The RNA was then transferred onto nylon membranes (Hybond N+; Amersham) and cross-linked (Stratagene). Plasmids used to create the array probes were used as the template for a PCR producing the Northern blot probes. These probes were then labeled with [α-32P]dATP by the High Prime labeling method (Roche) according to the manufacturer's protocol. Probes were purified with Nucleospin columns (Clontech) before hybridization to prehybridized membranes in Expresshyb (Clontech) for 1 h at 68°C. Membranes were finally washed three times for 15 min in 2× SSC-1% SDS and three times for 15 min in 0.1× SSC-0.5% SDS. Hybridization signals were quantified with a phosphorimager (Packard).
Data analysis.
Signals on arrays were quantified by OptiQuant phosphorimager analysis software (Packard). Means of duplicate spots were calculated before the background (mean signal for negative controls) was subtracted. To allow comparison of arrays with each other, they were normalized by expressing all signals as ratios of the internal luciferase control. In this way, variation in the specific activity of radioisotopes, labeling, and hybridization efficiencies could be accounted for. The log2 values of these ratios were calculated to allow greater resolution of the large linear range of signals produced on the arrays.
Normalized log2 ratios were converted into percentages of each gene's respective maximal expression and imported into Cluster analysis software (11). This conversion of expression profiles onto a relative scale allowed grouping of genes whose levels of expression show similar patterns without taking the absolute levels of expression into account. Hierarchical clustering was performed using the average-linkage algorithm and an uncentered correlation matrix. This method compares levels of gene expression pairwise and groups them according to similarity. These results were visualized with Treeview software (11).
RESULTS
MHV-68 array.
A DNA-based array was designed to characterize the life cycle of MHV-68 on a transcriptional basis. Specific sets of primers (Table 1) were used to amplify by PCR the predicted ORFs of MHV-68 as well as intergene regions. ORFs M10a, M10b, and M13 could not be amplified by PCR as they overlap with repeat regions of the genome. In addition, the cloned DNA fragments of ORF 75a and ORF M14 could not be verified by DNA sequencing. Thus these five genes were not represented on the array. All other DNA fragments were checked for potential cross-hybridization against each other and cellular genes by BLAST. None of the array probes showed any significant potential for such cross-hybridization.
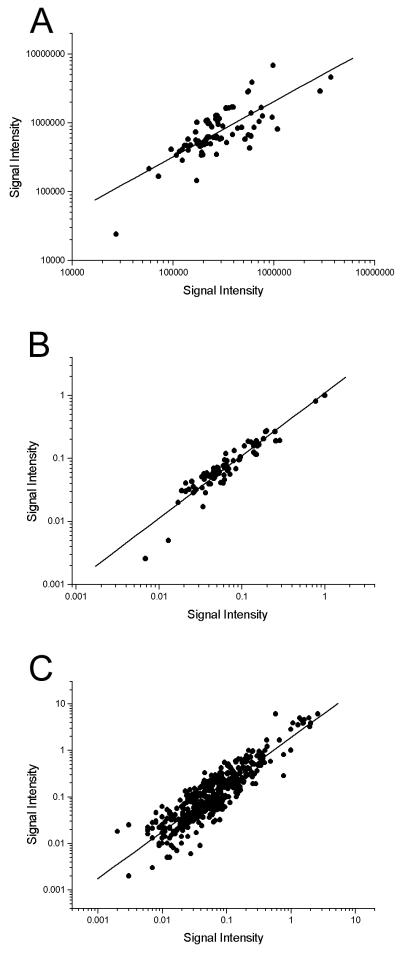
Initial experiments indicated that the nine cellular housekeeping genes included on the array were down-regulated to various degrees p.i., relative to mock-infected controls (data not shown). As these genes could therefore not be used to normalize data and compare separate arrays, an internal luciferase control was incorporated into the array for subsequent experiments. To confirm that normalizing data by this method was effective, arrays from repeat experiments were analyzed on scatter plots before and after normalization (Fig. 1). Linear regression analysis of the scatter plots showed gradients of 0.80 (correlation coefficient [r] = 0.76) (Fig. 1A) before normalization and 0.99 (r = 0.94) after normalization (Fig. 1B). This shows the internal control to be an effective method for normalizing data. Furthermore, to assess the reproducibility of arrays, the data from four repeat arrays were plotted against each other in all combinations and the same regression analysis was performed. A gradient of 1.01 (r = 0.91) confirmed the reproducibility of the arrays (Fig. 1C).
FIG. 1.
Scatter plots of array data. (A) Background-subtracted data from two repeated arrays (time point, 5 h p.i.) were plotted against each other. Linear regression analysis gives a line of gradient 0.80 (r = 0.76). (B) The data from the array in panel A were subjected to normalization via the internal control, and these normalized data were plotted against each other. Linear regression analysis gives a line of gradient 0.99 (r = 0.94). (C) Reproducibility of arrays was assessed by plotting four repeated arrays (time point, 5 h p.i.) against each other in all combinations. Linear regression analysis gives a line of gradient 1.01 (r = 0.91).
Transcriptional profiling.
To establish a standard transcriptional program for MHV-68, a primary lytic infection in a fully permissive tissue culture system was monitored. NIH 3T3 cell monolayers were infected at an MOI of 10, and cells were harvested at 0, 1, 3, 5, 8, 12, and 18 h p.i. Total RNA was extracted at each time point and reverse transcribed, and the resulting radiolabeled cDNAs were hybridized to arrays.
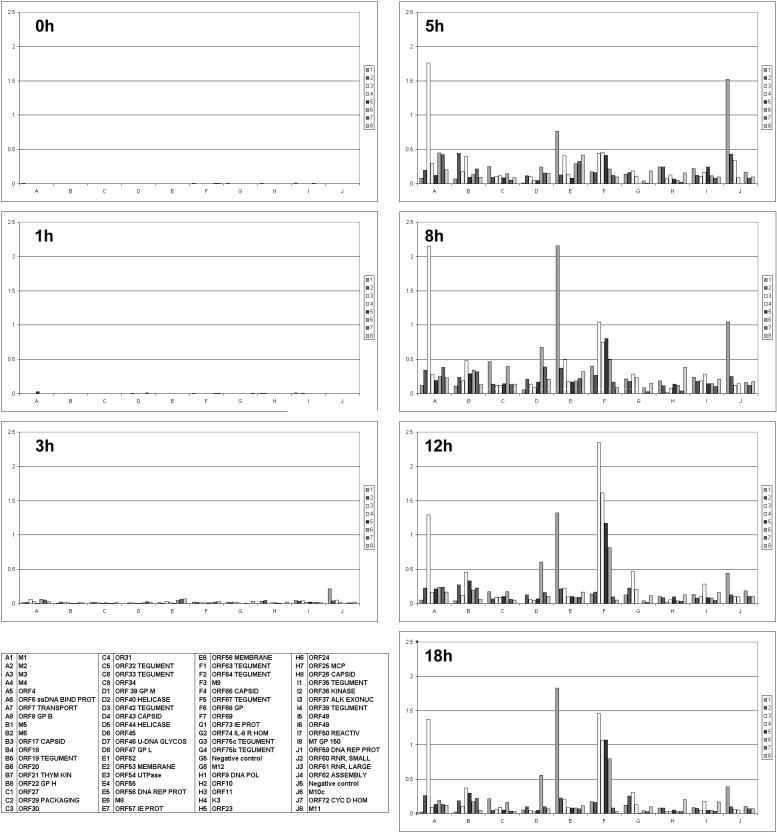
The normalized ratios for the time course are shown plotted on sequential bar charts (Fig. 2). Little transcription was detected at 1 h p.i., but at 3 h p.i. ORF 59 (DNA replication protein) was detected at noticeably higher levels than other viral transcripts, suggesting that DNA replication may start early in the virus life cycle. By 5 h p.i., signals for the majority of genes could be seen, indicating a rapid escalation of viral gene expression. These results show that MHV-68's global transcriptional profile changes as it progresses through the life cycle and that individual genes have their individual expression kinetics.
FIG. 2.
Array analysis of MHV-68 gene expression. NIH 3T3 cells were infected and harvested for isolation of RNA at various time points p.i. RNA was reverse transcribed to produce radiolabeled cDNAs that were hybridized to arrays. Bar charts represent the global profile of gene expression at various times p.i. as indicated. Each chart is based on mean values of independent experiments (duplicates; n = 2 to 6), normalized to internal controls. The identity of each bar is given in the key. Abbreviations are as follows: ssDNA BIND PROT, single-stranded DNA binding protein; GP B, glycoprotein B; THYM KIN, thymidine kinase; U-DNA GLYCOS, uracil DNA glycosylase; DNA REP PROT, DNA replication protein; IE PROT, immediate-early protein; IL-8 R HOM, interleukin-8 receptor homologue; DNA POL, DNA polymerase; MCP, major capsid protein; ALK EXONUC, alkaline exonuclease; REACTIV, transcriptional activator; GP150, glycoprotein 150; RNR, ribonucleotide reductase; CYC D HOM, cyclin D homologue.
A number of genes stand out as being highly expressed, including ORFs M3 (chemokine-binding protein) (25), M9, and 52. The array data showed that M3 was highly expressed by 5 h p.i. and then continued to be expressed at high levels up to 18 h p.i. M9 was also detected by 5 h p.i. but then showed a different profile, as its expression increased steadily up to 12 h p.i. before falling again. This suggests that M3 and M9 encode proteins that play roles at different times in the life cycle. ORF 52 was also heavily expressed up to 18 h p.i., which suggests a significant role for the gene product. As there has been no function assigned to ORF 52, this gene would be an interesting target for further study.
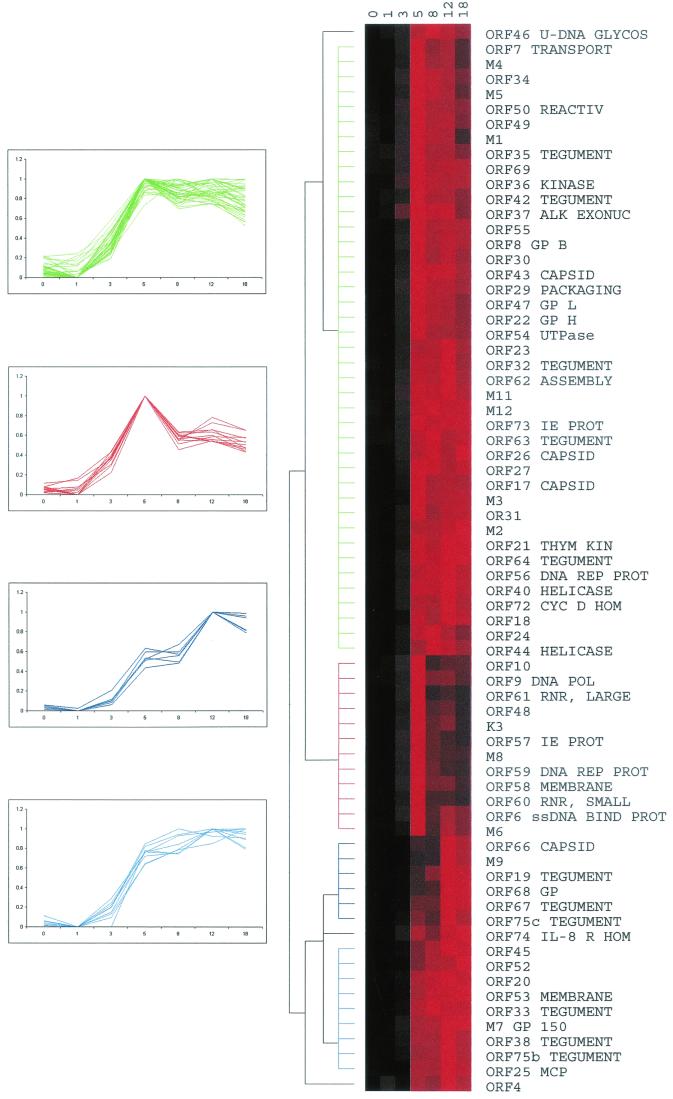
For other genes, like ORF 52, with no designated role, it is possible to make functional predictions by comparing their expression profiles to those of better-characterized genes. Therefore the time course data were subjected to cluster analysis (11) (Fig. 3). Overall, the majority of genes started to be expressed after 3 h p.i., and their profiles clustered into groups representing four general profiles of expression, as shown in the line graphs of Fig. 3. ORFs 6 (single-stranded DNA binding protein), 9 (DNA polymerase), 60 (ribonucleotide reductase, small subunit), and 61 (ribonucleotide reductase, large subunit) (42) cluster together (red line graphs) with a correlation coefficient of 0.992, suggesting similar functional roles, which is clearly the case here. These DNA replication-related genes also cluster with ORFs 57 (immediate-early protein) and K3 (bovine herpesvirus 4 immediate-early protein 1 homologue), suggesting that DNA replication may be an early event in the life cycle of MHV-68, and indeed the line graphs show that the transcription profiles of all these genes have a very defined and early peak in expression. Interestingly M6, which has no homologues or a predicted function, also clustered within this group.
FIG. 3.
Hierarchical cluster analysis of MHV-68 expression profiles. Data in the form of log2 normalized means (duplicates; n = 2 to 6) were converted into a percentage of each gene's maximum. These percentages were imported into Cluster software, and hierarchical clustering was performed using the average-linkage algorithm and an uncentered correlation similarity matrix. The data are shown as a color matrix with columns representing time points p.i. and rows representing each gene's expression profile. Black boxes, no expression; brighter shades of red, increasing expression. The dendrogram shows related expression profiles on the same branch, with branch lengths representing the degree of similarity between individual profiles. Line graphs of clustered genes are shown to the left, with the color of each line graph corresponding to the dendrogram branch of the same color. Abbreviations are defined in the legend for Fig. 2.
The largest cluster of genes (green line graphs) shows a peak in expression at 5 h p.i., followed by a more gradual fall, in comparison to the cluster shown in red. This group contains many functional classes of genes including ORFs 50 (transcriptional activator), 37 (alkaline exonuclease), and 47 (glycoprotein L) (42). Therefore further studies of these genes and their expression may be required to yield more functionally defined groups.
ORFs 25 (major capsid protein), 33 (tegument), 38 (membrane), M7 (glycoprotein 150), and 53 (membrane) all cluster together (light blue line graphs) as genes that reach and maintain a peak of expression at around 8 h p.i. This group of late structural genes (by homology to other herpesviruses) also contains ORFs 20, 45, and 52, which have no predicted function. The cluster represented by the dark blue line graphs tends to peak around 12 h p.i. and are the last set of genes to reach high levels of expression. This cluster consists of another group of structural genes, e.g., ORFs 19 (tegument), 66 (capsid), and 68 (glycoprotein), as well as M9, which, although not designated a function originally (42), has now been found to have homology to a capsid protein-encoding gene of other gammaherpesviruses (24).
MHV-68 transcription in the absence of de novo protein synthesis.
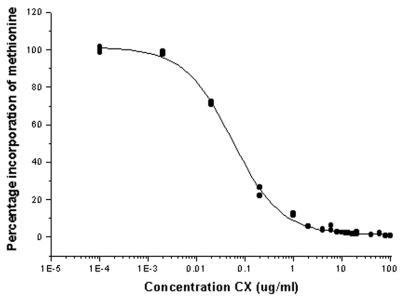
To further dissect the transcriptional behavior of MHV-68, viral genes were categorized into the following three groups: immediate-early (α), early (β), and late (γ). The α genes were characterized by blocking de novo protein synthesis with CX. Initial studies indicated that 100- and 50-μg/ml concentrations of CX triggered apoptosis in NIH 3T3 cells (data not shown), as has been shown in a variety of other cell types (22). As apoptotic cells are unlikely to present a normal pattern of infection, the metabolism of radiolabeled methionine by NIH 3T3 cells was measured in the presence of various amounts of CX. The resulting dose-response curve shows that, in NIH 3T3 cells, 95% inhibition of protein synthesis is obtained with only 2 μg of CX/ml (Fig. 4).
FIG. 4.
Incorporation of radiolabeled methionine into protein by NIH 3T3 cells in the presence of CX. NIH 3T3 cell monolayers were pretreated with various concentrations of CX before introduction of radiolabeled methionine into the medium. Cells were harvested after 8 h, and the level of methionine incorporation was measured. y axis, percent incorporation of labeled methionine relative to that for uninhibited controls; x axis, concentration of CX present in cell medium.
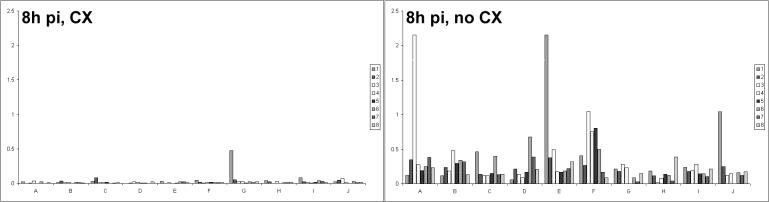
NIH 3T3 monolayers were pretreated and infected in the presence of CX. Negative controls (uninfected cells with CX) and positive controls (cells infected without CX) were also included. Cells were harvested at 5 and 8 h p.i., and RNA was isolated for subsequent array hybridizations. At 5 h p.i. in the presence of a 95% protein synthesis block, a weak signal for ORF 73 was detected (data not shown), and by 8 h p.i. (n = 3), it had accumulated as shown in Fig. 5. In addition, the level of ORF 73 detected was higher than that detected in positive-control infections without the protein synthesis block. This suggests that ORF 73 was transcribed without a requirement for de novo viral proteins and continued to be transcribed in the absence of a switch to β-class gene expression. No other genes with α-class transcriptional profiles were detected on the array.
FIG. 5.
MHV-68 gene expression in the absence of de novo protein synthesis. NIH 3T3 cells were infected with or without CX inhibition. RNA was isolated from cells harvested 8 h p.i. and used to produce radiolabeled cDNAs, which were hybridized to arrays. (Left) Array analysis of gene expression with a 95% protein synthesis block. (Right) Gene expression in a control, uninhibited infection. The identity of each bar is as given in the key of Fig. 2.
MHV-68 transcription in the absence of viral DNA replication.
The γ class of genes are defined as those requiring viral DNA replication for their transcription, and 4′-S-EtdU has been shown to be a potent inhibitor of MHV-68 DNA replication (3). Thus NIH 3T3 cells were pretreated and infected in the presence of 200 ng of 4′-S-EtdU/ml. Negative (uninfected with 4′-S-EtdU) and positive (infected without inhibition) controls were also set up, and cells were harvested at 5 and 18 h p.i.
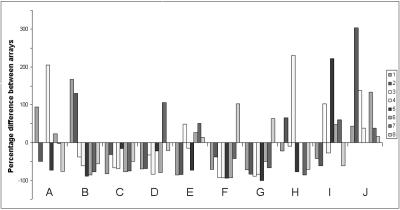
The results of these experiments are summarized in Fig. 6 (n = 4), where negative values represent a transcript that is less abundant when viral DNA replication is inhibited. Only genes whose expression was down-regulated by more than 2 measures of standard deviation were considered significantly inhibited in the presence of 4′-S-EtdU. Forty-two transcripts showed reduced abundance, and thus these can be said to show γ-class kinetics. They include ORFs 4 (complement regulatory protein), 8 (glycoprotein B), 17 (capsid), 19 (tegument), and 22 (glycoprotein H) (42), as well as ORFs 20, 45, 52, and M9, which were already highlighted as potential γ genes from the cluster analysis of their transcription profiles (Fig. 3).
FIG. 6.
Relative expression of MHV-68 genes in the absence of viral DNA replication. NIH 3T3 cells were infected with or without inhibition of viral DNA replication. RNA was isolated from cells harvested 18 h p.i. and used to produce radiolabeled cDNAs that were hybridized to arrays. Array data from an uninhibited infection were subtracted from data for a viral DNA replication-inhibited infection (n = 4). y axis, percent difference in expression between inhibited and uninhibited infections. Negative values represent percent reduction in signals following inhibition and vice versa. Genes that were down-regulated by more than 2 measures of standard deviation were classified as γ genes. The identity of each bar is as given in the key of Fig. 2.
Interestingly, many γ genes showed reduced expression as early as 5 h p.i. (data not shown). For example, ORFs M9, 66 (capsid), and 67 (tegument) all showed a reduction in expression relative to uninhibited positive controls. Although these data are relative and therefore small differences can be exaggerated, they indicate that viral DNA replication occurs as early as 5 h p.i.
Northern blot analysis.
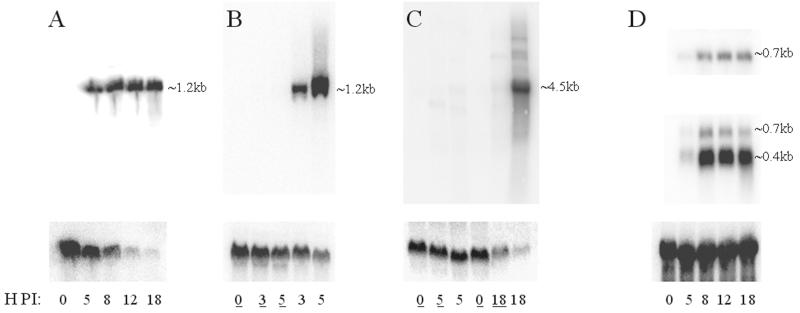
The array data presented here are reflected in previous work using alternate methods of examining transcription (14, 21, 29, 35, 44). Northern blot analysis was also employed here to confirm the data presented. A radiolabeled probe specific for ORF M3 (chemokine-binding protein) was hybridized to a blot of RNA isolated at various times p.i. (Fig. 7A). A single transcript of ∼1.2 kb was detected, showing strong expression of ORF M3 from 5 to 18 h p.i., as shown by the array (Fig. 2). ORF M3 transcripts could not be detected in the presence of CX (Fig. 7B), again confirming the array data (Fig. 5).
FIG. 7.
Northern blot analysis of MHV-68 transcripts. NIH 3T3 cells were infected and harvested for isolation of RNA at various time points p.i. The RNA was run out on a denaturing gel and blotted onto nylon membranes. Radiolabeled probes specific for various transcripts (see Materials and Methods) were hybridized to the membranes. The time p.i. when cells were taken for RNA isolation is indicated at the bottom of each lane. An underlined number indicates the presence of a metabolic inhibitor. Approximate sizes of transcripts are indicated to the right of each panel. Control hybridizations are shown below the viral transcript hybridizations. (A) ORF M3-specific probe (top). β-Actin (bottom) is reduced following infection. (B) ORF M3-specific probe (top). Underlined time points indicate presence of CX. Again β-actin (bottom) shows reduced expression following infection. (C) ORF 67-specific probe (top). Underlined time points indicate presence of 4′-S-EtdU. Bottom, β-actin. (D) ORF 53-specific (top), ORF 52-specific (middle), and luciferase-specific (bottom) probes. Note that the same blot was used for panels A and D.
An ORF 67 (tegument)-specific probe was hybridized to a blot of RNA isolated at various times p.i. with and without the presence of 4′-S-EtdU (Fig. 7C). One predominant transcript of around 4.5 kb was detected at the later time point of 18 h p.i. No such band was seen in lanes corresponding to infections in the presence of 4′-S-EtdU. This confirms the array data which categorized ORF 67 as a γ gene (Fig. 6). Interestingly, faint bands could be seen at 5 h p.i., indicating that γ genes start to be expressed early in the life cycle of MHV-68, which was also observed with the array (Fig. 2).
Probes corresponding to ORFs 52 and 53 (Fig. 7D) were also used. ORF 52 is a gene of unknown function and was shown by the array to be highly expressed. ORF 53 is another protein of no designated function which was found to be expressed with a pattern similar to that for ORF 52 but to a far lesser degree. The Northern analysis showed that indeed ORF 52 was highly expressed by 8 h p.i. and continued to be highly expressed up to 18 h p.i. and that ORF 53 was weakly expressed from 8 to 18 h p.i., confirming the array data (Fig. 2). The probe for ORF 52 also detected a second band, which was likely to be ORF 53, as these two ORFs are predicted to share a polyadenylation signal (24).
It should be noted that the control hybridizations for β-actin showed a transcript that was reduced in abundance following infection (Fig. 7A). This confirms the initial observations made with the array, which led to the housekeeping genes being considered redundant in terms of normalizing expression data from cells infected in vitro. Furthermore, β-actin transcript levels appear less affected when the infection was inhibited by CX (Fig. 7B), suggesting that host protein synthesis shutoff is only partly dependent on MHV-68 α-gene expression. The relative abundance of β-actin in the uninfected lanes suggests that any toxicity to the cells caused by CX was minimal.
To test the efficacy of the luciferase internal control used to normalize the arrays, 10 ng of luciferase RNA was spiked into 10 μg of total cell RNA isolated at various time points p.i. before the RNA was separated on a denaturing gel. Hybridization of the resulting blot with a luciferase-specific probe shows equal signal intensities in each lane, indicating the same quantity of luciferase in each RNA sample and therefore allowing confidence in the array internal control.
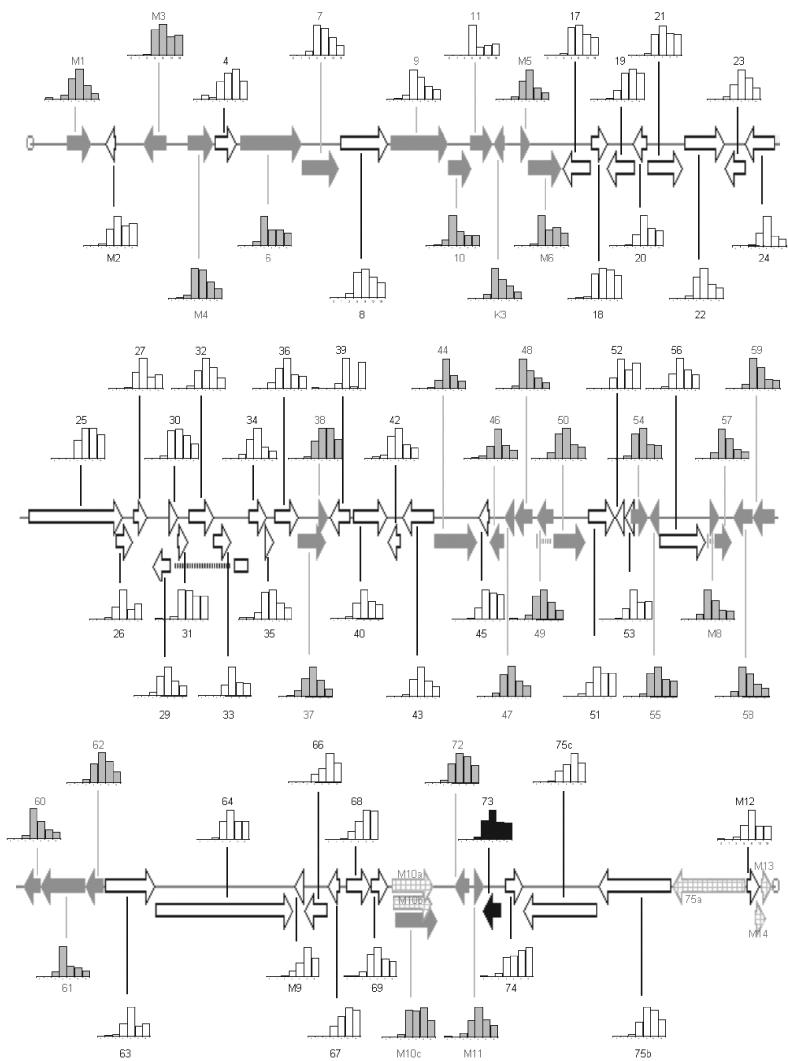
Kinetics of transcription.
The data from the experiments presented here can be summarized as a map of MHV-68 transcriptional profiles (Fig. 8). Black arrows correspond to α genes, defined as genes that are transcribed when de novo protein synthesis is blocked. White arrows show γ genes, defined as genes whose expression is reduced if viral DNA replication is inhibited. The remaining genes are shown with grey arrows and most likely show β-class kinetics, although this is as yet not a defined group. It should be noted that the signals generated for ORFs 11 and 39 (Fig. 2) are very low and most likely at the threshold of sensitivity for the array. Therefore further studies will be required to fully characterize the transcriptional profiles of these genes. Interestingly, with the exception of these two, it seems unlikely that any strictly latent genes are represented on the array, as all the other genes show expression during a lytic infection.
FIG. 8.
Map of MHV-68 transcriptional profiles as produced by array analysis. The expression profiles for MHV-68 were placed onto a physical map of the virus. Genes are color coded according to their expression kinetics: black arrows, α-class genes; white arrows, γ-class genes; grey arrows, remaining set of genes; checked arrows, genes not included on the array. Transcriptional profiles normalized to maximal expression for each gene are shown as bar charts.
It is clear that genes with similar kinetics tend to cluster on the genome. The expression of γ genes tends to follows one of two profiles. These are a relatively late initiation of transcription followed by sustained or reduced expression. However, there is a continuous range of profiles between these two extremes, suggesting that there is a range of regulatory mechanisms controlling the expression of these genes.
The remaining set of genes all show early peaks followed by reduced expression, which is characteristic of β genes. However it has been shown here that this is also a feature of α genes. Although the time course data show genes to have distinct transcriptional profiles, there is an overlap between genes of different kinetic classes. This demonstrates the need to block β- or γ-gene transcription to elucidate the kinetic organization of the genome.
DISCUSSION
In this study, we have developed a membrane-based cDNA array for MHV-68 and performed a rapid global analysis of changing gene expression through time. The transcriptional program of MHV-68 was monitored through a lytic life cycle in vitro by taking RNA samples at various times p.i. for subsequent array analysis. The resulting data impart a transcriptional profile to each gene during such an infection. These patterns of expression were then categorized into α-, β-, and γ-kinetic classes by inhibiting de novo protein synthesis and viral DNA replication. For the majority of MHV-68's ORFs, these data are the first experimental evidence for the homology-based functions assigned to them.
Evaluation of the MHV-68 array.
We have realized the array technology for MHV-68 by applying a proven design strategy that has been developed successfully to study the transcriptional behavior of HHV-8 (16) and herpes simplex virus type 1 (HSV-1) (33). Our results indicate that the MHV-68 array is a reproducible system as shown by scatter plot analyses. It can also detect RNA species that are present in both large and small amounts in a semiquantitative manner and so possesses a large range of detection. Most of the results from the array analyses are reflected in the expression kinetics of the small subset of MHV-68 genes which have been studied with various Northern blots, including the ones in this study, and RNase protection assay experiments (14, 21, 29, 35, 44). Many of the remaining genes have homologues in other herpesviruses which have been better characterized and which allow useful comparison with the data presented here. For example HSV-1 genes UL24, UL37, and UL49a have been shown to be γ genes (31), and their predicted homologues in MHV-68, ORFs 20, 53, and 63, have been shown here to have γ kinetics as well. Similarly, HSV-1 genes UL2, UL12, and UL50 are β genes, and in MHV-68 ORFs 46, 37, and 54 have been shown here to be β genes as well. In conclusion the array has been shown to be a reliable global tool to rapidly analyze transcription levels of the virus under a variety of conditions during infection.
It should be pointed out that, while the array technique is a powerful method to examine global gene expression, it does possess limitations. The complex organization of viral genomes in particular brings to light these limitations. Nested sets of coterminal genes sharing a polyadenylation site are a relatively common feature of herpesviruses (30), and preliminary sequence analysis has identified such occurrences in the MHV-68 genome (24). For example, ORFs 52 and 53 are predicted to be coterminal transcripts encoded on the right strand of the genome, i.e., the ORF 53 transcript also contains the sequence for ORF 52. Resolving individual transcription profiles from such gene structures can be problematic for methods of transcription analysis. On the array, the ORF 52 probe detects both the ORF 52 and 53 transcripts and therefore is not specific for ORF 52 alone. In fact three labeled cDNA species result from reverse transcription of these two mRNAs, one deriving from the ORF 52 transcript and two deriving from the ORF 53 transcript. Of the latter two cDNAs (primed by the ORF 52- and 53-specific primers, respectively), one is an artifact that binds to both the ORF 52 and 53 probes. It is also longer than the ∼300 bp of specific cDNAs and therefore incorporates more label as well. These factors can combine to make the signals observed on the array for such gene structures unreliable indicators of transcript abundance. In nested sets of many genes, these artifact cDNAs add unequal background signals to the array probes, and therefore an alternate method of transcription analysis must be used to elucidate the levels of transcription occurring in such sets of genes.
In fact, as there are only two members in this nested set, the transcription levels for ORF 52 and 53 on the array were still representative of actual transcription, albeit with a higher background, and this was confirmed by Northern analysis in this study. Northern analysis, which resolves transcription by both abundance and size, allowed differentiation between the transcripts for ORF 52 and 53; although Northern analysis suffers from the same redundancy in probe specificity, multiple analyses with individual probes can resolve this issue. The occurrence of nested sets of coterminal transcripts highlights the need to be aware of the organization of transcription, and therefore transcript maps are a very useful reference when both designing and interpreting array data.
Gene expression data.
The time course experiments have allowed us to monitor the expression of each gene through a primary lytic infection in vitro. It is clear that different genes have different expression profiles and that genes of similar functions generally have similar profiles. These results have been combined with those from experiments where de novo protein synthesis and viral DNA replication have been inhibited to provide a global picture of transcription and kinetics.
The transcription profiles of ORFs M3, 52, and M9 were highlighted due to their high levels of expression. ORF M3 has been shown to be an abundantly expressed chemokine-binding protein (7, 25, 40, 41), and similarly M9 is also known to be an abundantly expressed gene (43). As their expression patterns differ, it seems likely that the two play separate roles: M3 is required to exert its function from early on and throughout a lytic infection, whereas M9 is required in large amounts but principally at later stages of the lytic life cycle. ORF 52 is a well-conserved protein of herpesviruses that has yet to be characterized functionally. It shares transcriptional characteristics with viral structural proteins, and the abundance of its RNA message indicates a major role in that area.
Cluster analysis can sometimes seem to be of limited use when gene expression profiles are very similar to each other, as is the case for some of the data presented here. However, it is still a valuable analytical tool and indeed has proven itself a useful method for interpreting array data. For example clustering the time course data suggested that ORFs 20, 45, 52, and M9 were late genes and subsequent characterization of γ genes by inhibiting DNA synthesis has shown this to be so. Genes involved in DNA replication were found to cluster with α genes, suggesting that DNA replication is an early event in the life cycle of MHV-68, and this has also been corroborated in subsequent experiments. For example, γ genes were found to be expressed as early 5 h p.i. and their expression was inhibited when DNA replication was inhibited at that time point. This suggests that MHV-68 replicates its DNA as early as 5 h p.i. In addition, the first gene to be detected on the array was ORF 59, which encodes a predicted DNA replication protein. Furthermore, even when the transcription patterns of many genes are very similar, as seen in the green cluster of Fig. 3, the cluster algorithms manage to place ORFs 47 (glycoprotein L) and 22 (glycoprotein H) adjacent to each other. These two genes have homologues in HSV-1, which have been shown to form a heterocomplex and which are therefore expressed together (15).
By placing a protein synthesis block, we saw clear accumulation of one RNA species, the ORF 73 species, to higher levels than in positive-control infections without inhibition, a facet that has not been shown previously. While other studies of MHV-68 α-gene expression have suggested that ORFs 57/M8, 50, and K3 are also expressed in the absence of protein synthesis (20, 29), these studies used techniques and experimental conditions different from those employed here. However, given the detection of only a single α transcript, reverse transcription-PCR (RT-PCR) was also employed to examine α-gene expression (data not shown). ORF 73 was readily detected by RT-PCR in the presence of CX, which confirmed the results of the array. However faint bands corresponding to ORFs 50, 57, and K3 could also be detected, indicating that these too could be α genes. Therefore it may be beyond the threshold of sensitivity of the array method to detect low abundance α genes, and a more sensitive, dedicated study to elucidate α-gene transcription may be required to characterize the gene expression events that occur immediately following infection by this virus. Interestingly, there have been similar problems elucidating α-gene transcription in HHV-8 (28, 38), with a lack of agreement concerning the α status of various genes.
It should also be noted, however, that the initial categorization of viral genes into α, β, and γ kinetic groups was based on observations made at a protein level (13, 17). It remains unclear how well these categories relate back to the RNA level, especially considering that the relationship between RNA abundance and protein abundance is not understood fully (12). Nonetheless, the kinetic classification of MHV-68's genes is in good accordance with previous studies on this and other herpesviruses.
Following the design of this array, the MHV-68 virus was sequenced a second time and reanalyzed with respect to its ORFs (24). It is useful to compare the differences between the two sequences and see how they are resolved on the array. Following reannotation, the previously undesignated region between ORFs 27 and 29 has now been designated a gene, ORF 28. Due to the strategy of including inter-ORF regions on the array, ORF 28 was preemptively represented on the array. Indeed, the time course shows an γ expression profile for the inter-27/29 region, with highest levels detected at 8 h p.i. None of the other intergene regions produced patterns of expression through a time course, supporting the analysis based on sequence data alone.
A number of genes have been downgraded to the status of being unlikely to encode proteins following this second annotation: ORFs M5, M6, 29a (now part of 29), M8 (now part of 57), M10a, M10b, M10c, M12, M13, and M14. While some of these predicted ORFs lie in repeat regions and hence did not make good templates for PCR amplification, the rest were included on the array. As mentioned above, ORF M8 is now reclassified as part of ORF 57, and the array analysis confirms that both have the same transcriptional profile. Furthermore, the cluster analysis algorithm placed them next to each other on the dendrogram (correlation coefficient = 0.999), again indicating that the array data strongly support this analysis. ORFs M5 and M12 show very-low-level signals throughout, from 0 to 18 h p.i., and this adds support for the downgrading of these genes. However ORFs M6 and M10c show higher signals, suggesting that these could encode proteins and indicating that more detailed analysis of these ORFs is required.
In summary, a novel cDNA array for MHV-68 has been developed and used to provide an initial analysis of global gene expression by monitoring transcriptional profiles through a lytic infection in vitro. Furthermore we have used the array to classify each gene into α, β, or γ kinetic classes, providing further context in which to view individual expression profiles. We have demonstrated the power of this tool, which has many potential applications, including the ability to analyze mutant strains of the virus, to test the efficacy and mode of action of antiviral agents, and also, crucially, to examine the pathogenesis and replication of the virus in its natural setting in vivo. By comparison to profiles achieved in fully permissive tissue culture systems, arrays provide a rapid global screen to assess points of restriction imposed on the virus during a natural infection. They can then also be used to investigate the transcriptional circuitry affected by such restrictions. These studies should impart a greater understanding of the mechanisms involved in viral replication and pathogenesis and therefore allow the synthesis of informed strategies to combat disease.
Acknowledgments
This work was funded by the Biotechnology and Biological Sciences Research Council (J.W.A.), Arrow Theraputics (J.W.A.), and the British Heart Foundation (D.G.A.).
We thank Stephen Henderson for helpful advice on scatter plot analyses and Richard Jenner for helpful discussion concerning the HHV-8 array.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alber, D. G., K. L. Powell, P. Vallance, D. A. Goodwin, and C. Grahame-Clarke. 2000. Herpesvirus infection accelerates atherosclerosis in the apolipoprotein E-deficient mouse. Circulation 102:779-785. [DOI] [PubMed] [Google Scholar]
- 3.Barnes, A., H. Dyson, N. P. Sunil-Chandra, P. Collins, and A. A. Nash. 1999. 2′-Deoxy-5-ethyl-beta-4′-thiouridine inhibits replication of murine gammaherpesvirus and delays the onset of virus latency. Antivir. Chem. Chemother. 10:321-326. [DOI] [PubMed] [Google Scholar]
- 4.Belz, G. T., and P. C. Doherty. 2001. Virus-specific and bystander CD8+ T-cell proliferation in the acute and persistent phases of a gammaherpesvirus infection. J. Virol. 75:4435-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornkamm, G. W., and W. Hammerschmidt. 2001. Molecular virology of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:437-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boshoff, C., and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus: a new DNA tumor virus. Annu. Rev. Med. 52:453-470. [DOI] [PubMed] [Google Scholar]
- 7.Bridgeman, A., P. G. Stevenson, J. P. Simas, and S. Efstathiou. 2001. A secreted chemokine binding protein encoded by murine gammaherpesvirus-68 is necessary for the establishment of a normal latent load. J. Exp. Med. 194:301-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford, D. H. 2001. Biology and disease associations of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:461-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Doherty, P. C., J. P. Christensen, G. T. Belz, P. G. Stevenson, and M. Y. Sangster. 2001. Dissecting the host response to a gammaherpesvirus. Philos. Trans. Soc. Lond. B Biol. Sci.. 356:581-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donofrio, G., and V. L. van Santen. 2001. A bovine macrophage cell line supports bovine herpesvirus-4 persistent infection. J. Gen. Virol. 82:1181-1185. [DOI] [PubMed] [Google Scholar]
- 11.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gygi, S. P., Y. Rochon, B. R. Franza, and R. Aebersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Husain, S. M., E. J. Usherwood, H. Dyson, C. Coleclough, M. A. Coppola, D. L. Woodland, M. A. Blackman, J. P. Stewart, and J. T. Sample. 1999. Murine gammaherpesvirus M2 gene is latency-associated and its protein a target for CD8+ T lymphocytes. Proc. Natl. Acad. Sci. USA 96:7508-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, P. C., and B. Roizman. 1979. Regulation of herpesvirus macromolecular synthesis. VIII. The transcription program consists of three phases during which both extent of transcription and accumulation of RNA in the cytoplasm are regulated. J. Virol. 31:299-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung, J. U., J. K. Choi, A. Ensser, and B. Biesinger. 1999. Herpesvirus saimiri as a model for gammaherpesvirus oncogenesis. Semin. Cancer Biol. 9:231-239. [DOI] [PubMed] [Google Scholar]
- 19.Killington, R. A., and K. L. Powell. 1985. Growth, assay and purification of herpesviruses, p. 207-236. In B. W. J. Mahy (ed.), Virology—a practical approach. IRL Press, Oxford, United Kingdom.
- 20.Liu, S., I. V. Pavlova, H. W. Virgin IV, and S. H. Speck. 2000. Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol. 74:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackett, M., J. P. Stewart, S. D. V. Pepper, M. Chee, S. Efstathiou, A. A. Nash, and J. R. Arrand. 1997. Genetic content and preliminary transcriptional analysis of a representative region of murine gammaherpesvirus 68. J. Gen. Virol. 78:1425-1433. [DOI] [PubMed] [Google Scholar]
- 22.Martin, S. J., C. P. Reutelingsperger, A. J. McGahon, J. A. Rader, R. C. van Schie, D. M. LaFace, and D. R. Green. 1995. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 182:1545-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mistrikova, J., H. Raslova, M. Mrmusova, and M. Kudelova. 2000. A murine gammaherpesvirus. Acta Virol. 44:211-226. [PubMed] [Google Scholar]
- 24.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. 2001. Natural history of murine gamma-herpesvirus infection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parry, B. C., J. P. Simas, V. P. Smith, C. A. Stewart, A. C. Minson, S. Efstathiou, and A. Alcami. 2000. A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J. Exp. Med. 191:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renne, R., D. Blackbourn, D. Whitby, J. Levy, and D. Ganem. 1998. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 72:5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rimessi, P., A. Bonaccorsi, M. Sturzl, M. Fabris, E. Brocca-Cofano, A. Caputo, G. Melucci-Vigo, M. Falchi, A. Cafaro, E. Cassai, B. Ensoli, and P. Monini. 2001. Transcription pattern of human herpesvirus 8 open reading frame K3 in primary effusion lymphoma and Kaposi's sarcoma. J. Virol. 75:7161-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rochford, R., M. L. Lutzke, R. S. Alfinito, A. Clavo, and R. D. Cardin. 2001. Kinetics of murine gammaherpesvirus 68 gene expression following infection of murine cells in culture and in mice. J. Virol. 75:4955-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roizman, B. 1996. Herpesviridae, p. 2221-2230. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 31.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 2231-2278. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 32.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 33.Sciortino, M. T., M. Suzuki, B. Taddeo, and B. Roizman. 2001. RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions. J. Virol. 75:8105-8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simas, J. P., and S. Efstathiou. 1998. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 6:276-282. [DOI] [PubMed] [Google Scholar]
- 35.Simas, J. P., D. Swann, R. Bowden, and S. Efstathiou. 1999. Analysis of murine gammaherpesvirus-68 transcription during lytic and latent infection. J. Gen. Virol. 80:75-82. [DOI] [PubMed] [Google Scholar]
- 36.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. USA 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stingley, S. W., J. J. Ramirez, S. A. Aguilar, K. Simmen, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2000. Global analysis of herpes simplex virus type 1 transcription using an oligonucleotide-based DNA microarray. J. Virol. 74:9916-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sunil-Chandra, N. P., J. Arno, J. Fazakerley, and A. A. Nash. 1994. Lymphoproliferative disease in mice infected with murine gammaherpesvirus 68. Am. J. Pathol. 145:818-826. [PMC free article] [PubMed] [Google Scholar]
- 40.van Berkel, V., J. Barrett, H. L. Tiffany, D. H. Fremont, P. M. Murphy, G. McFadden, S. H. Speck, and H. I. Virgin. 2000. Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J. Virol. 74:6741-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Berkel, V., K. Preiter, H. W. Virgin IV, and S. H. Speck. 1999. Identification and initial characterization of the murine gammaherpesvirus 68 gene M3, encoding an abundantly secreted protein. J. Virol. 73:4524-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Virgin, H. W., IV, R. M. Presti, X. Y. Li, C. Liu, and S. H. Speck. 1999. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J. Virol. 73:2321-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, T. T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]