Abstract
Historical and genetic evidences suggest that the recently founded population of Antioquia (Colombia) is potentially useful for the genetic mapping of complex traits. This population was established in the 16th–17th centuries through the admixture of Amerinds, Europeans, and Africans and grew in relative isolation until the late 19th century. To examine the origin of the founders of Antioquia, we typed 11 markers on the nonrecombining portion of the Y chromosome and four markers on mtDNA in a sample of individuals with confirmed Antioquian ancestry. The polymorphisms on the Y chromosome (five biallelic markers and six microsatellites) allow an approximation to the origin of founder men, and those on mtDNA identify the four major founder Native American lineages. These data indicate that ∼94% of the Y chromosomes are European, 5% are African, and 1% are Amerind. Y-chromosome data are consistent with an origin of founders predominantly in southern Spain but also suggest that a fraction came from northern Iberia and that some possibly had a Sephardic origin. In stark contrast with the Y-chromosome, ∼90% of the mtDNA gene pool of Antioquia is Amerind, with the frequency of the four Amerind founder lineages being closest to Native Americans currently living in the area. These results indicate a highly asymmetric pattern of mating in early Antioquia, involving mostly immigrant men and local native women. The discordance of our data with blood-group estimates of admixture suggests that the number of founder men was larger than that of women.
Introduction
Genetic data are being used extensively in human evolutionary studies and for following the dispersal of humans throughout the globe (Cavalli-Sforza et al. 1994). Evaluation of the genetic history of human populations is also relevant for the optimal design of studies aimed at the identification of genetic determinants of complex diseases. For such a task, approaches that use the association between genetic variants and disease in population samples have been proposed as a more powerful alternative to linkage studies (Risch and Merikangas 1996). The strength of such association is going to be strongly influenced by evolutionary factors such as drift and admixture, and it has been proposed that the linkage disequilibrium (LD) generated by them could be used for genetic mapping (Terwilliger and Weiss 1998). We are interested in evaluating the potential for complex trait mapping of the population living in the province of Antioquia, in northwest Colombia, because demographic history data indicate that this population could combine two sources of LD: recent founding by a small number of individuals and population admixture. Furthermore, founder effects have been documented in Antioquia for monogenic diseases such as early-onset Alzheimer’s (Lendon et al. 1997; Lopera et al. 1997) and juvenile Parkinson’s (A.R.-L., unpublished data) diseases.
Historical records indicate that immigrants from the Iberian peninsula founded the first non-Amerind settlements in northwest South America (in present-day Colombia) in the early 16th century. Soon after that, the Atlantic slave trade introduced African individuals, who originated mostly from western Africa (Curtin 1969). The arrival of immigrants led to the establishment of a rapidly growing admixed criollo population and a concomitant decline in the Amerind population. The Andes divides into three major mountain ranges (“cordilleras”) in southern Colombia, which cover most of the western side of the country. The ruggedness of the terrain has been a formidable obstacle to communications for centuries, and the demographic growth of various populations in the region occurred in relative isolation until the late 19th century. During that time, a distinct regional identity (termed paisa) developed around the fertile highland valleys of Aburrá and Rionegro, in the province of Antioquia, where some of the most successful settlements had been established in the mid-17th century (Alvarez 1996). From these two valleys, the Antioquian population expanded southward, following the central cordillera (Parsons 1968). This expansion was given considerable impetus in the 19th century with the introduction of the coffee crop (Juan Valdez, the logo used by the Colombian Coffee Federation, is an iconic paisa). Blood-group data indicate that the genetic background of the population of Antioquia is ∼70% white, 15% Amerindian, and 15% African (Sandoval et al. 1993).
In order to evaluate the origin of male and female founders of Antioquia, we examined mtDNA and Y-chromosome markers in a sample of descendants of the Aburrá and Rionegro expansion. Our results reveal the imprint of an early admixture involving mostly native women and immigrant men. Interestingly, the female contribution still reflects the genetic makeup of South Amerinds inhabiting the region. As expected, the founder male contribution seems to be predominantly from southern Spain, although a northern Spanish (possibly Basque) component is detectable. Finally, Y-chromosome data suggest that some of the Antioquian founders could have a Sephardic ancestry.
Subjects and Methods
Study Population
Samples were taken from 80 individuals in Medellín from among students and staff of the Universidad de Antioquia Medical School and San Vicente de Paul University Hospital (fig. 1). To select individuals of Antioquian ancestry, we applied a genealogical interview that recorded the names, dates, and places of birth of ancestors up to the great-grandparents. None of the selected individuals shared any of the ancestors recorded in the interview. Each individual provided informed consent for this research (this project was approved by the Bioethics committee of Universidad de Antioquia). Whole blood was collected, and genomic DNA was extracted using the Nucleon DNA extraction kit, following the manufacturer’s instructions.
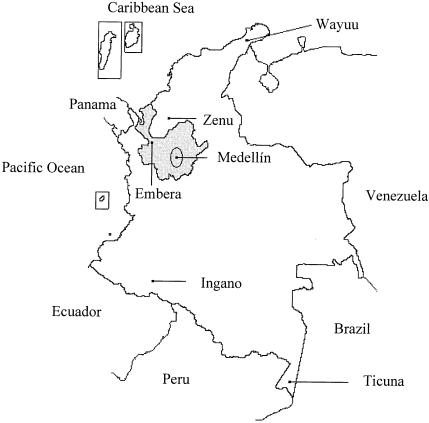
Figure 1 .
Map of Colombia, in northwest South America. The province of Antioquia is highlighted in gray. Also shown is the location of the five Amerind populations discussed in the text. The Antioquian population was sampled in Medellín, the provincial capital, centered in the Aburrá Valley (indicated by an ellipse).
Y-Chromosome Polymorphisms
The following biallelic markers were typed as reported: DYS271 (Hammer and Horai 1995), DYS287 (Seielstad et al. 1994), 92R7, SRY-2627 (Hurles et al. 1999), and DYS199 (Hurles et al. 1998). An additional six microsatellites were examined in a subset of the samples. Markers DYS19, DYS388, DYS390, DYS391, DYS392, and DYS393 were typed fluorescently, using the experimental conditions reported by Thomas et al. (1999), on an ABI310 genetic analyzer. Here we refer to the lineages defined by the biallelic markers as “haplogroups” and reserve the term “haplotypes” for those defined by microsatellites.
mtDNA Polymorphisms
Three variable restriction sites and the polymorphic 9-bp COII/tRNALys intergenic deletion were examined, to identify the four major founder Native American lineages (denoted “A–D”). The enzymes used and the location of the restriction sites examined (following the Cambridge Reference Sequence coordinates) were HaeIII (663 bp), HincII (3259 bp), and AluI (5176 bp). PCR primers and amplification conditions were as described by Bailliet et al. (1994).
Data Analysis
Allele and haplotype frequencies were estimated by counting. The program ARLEQUIN (Schneider et al. 2000) was used to calculate pairwise Fst genetic distances, using the estimator proposed by Reynolds et al. (1983), with nonzero significance evaluated by a randomization test. ARLEQUIN was also used to estimate the gene diversity (Nei 1978) on the basis of mtDNA haplotypes and the average gene diversity across Y-chromosome microsatellites. Admixture was estimated by the least-squares approximation of Long (1991), as implemented in the ADMIX program (kindly provided by J. C. Long).
Results
Y-Chromosome Biallelic Polymorphisms
Four of the markers examined allow the identification of five haplogroups, some of which have marked differences in frequency among the putative parental populations of Antioquia: Europeans, western Africans, and southern Amerinds (table 1). In this comparison, haplogroup B is restricted to South America and haplogroup E to western Africa (although both extend into other geographic areas that are unlikely to have contributed to Antioquia). Among the 80 Y chromosomes examined in Antioquia, three carry the G allele at locus DYS271 and are most likely African in origin. One individual carries the T allele at locus DYS199, thus identifying this chromosome as Amerind. Overall, the haplogroup frequencies in Antioquia are closest to those seen in Europe (table 1). Genetic distances, based on haplogroup frequencies, among these four groups are shown in table 2. The smallest distance is seen between Antioquia and Europe, and the value obtained (.008) is not significantly different from zero. Also, Antioquia is seen to be slightly closer to western Africa (.41) than to South America (.45), as suggested by the higher frequency in Antioquia of haplogroup E relative to haplogroup B (table 1). Admixture estimation, based on the haplogroup frequencies of table 1, indicates that the Antioquian Y chromosomes are ∼94% European, 5% African, and 1% Amerind, in agreement with the genetic distance analysis.
Table 1.
Biallelic Y-Chromosome Haplogroups as Defined by the Alleles Present at Four Markers and Their Frequency in Antioquia and in Putative Parental Populations
| A | B | C | D | E | |
| Marker:a | Haplogroup |
||||
| 92R7 | T | T | C | C | C |
| DYS287 (Yap) | (−) | (−) | (−) | (+) | (+) |
| DYS271 | A | A | A | A | G |
| DYS199 | C |
T |
C |
C |
C |
| Population:b | % Frequency |
||||
| Europeans (n = 337) | 50 | 0 | 35 | 15 | 0 |
| Western Africans (n = 56) | 0 | 0 | 12 | 25 | 63 |
| Southern Amerinds (n = 144) | 19 | 74 | 7 | 0 | 0 |
| Antioquians (n = 80) | 58 | 1 | 33 | 4 | 4 |
For marker DYS287, (−) and (+) refer to the absence and presence of an Alu insertion (denoted Yap), respectively. For the other markers, the nucleotide present at the polymorphic site is given.
Table 2.
Pairwise Fst Distances between Antioquians and Putative Parental Populations, Based on Biallelic Y-Chromosome Haplogroup Frequencies
| Europeans | WesternAfricans | SouthernAmerinds | |
| Western Africans | .365 | ||
| Southern Amerinds | .406 | .536 | |
| Antioquians | .008a | .406 | .450 |
P>.05.
Polymorphism at the fifth biallelic marker examined (SRY-2627) seems to have originated in the Iberian peninsula, with the derived T allele being relatively common in northern Spanish populations, particularly in Basque and Catalan, where it reaches frequencies of 11% and 22%, respectively (Hurles et al. 1999). The T allele was detected at a frequency of 5% in Antioquia, in agreement with an Iberian origin for the European migrants to the province and suggesting that some of these founders came from northern Spain.
Y-Chromosome Microsatellites
To refine the assessment of the origin of Antioquian founders, we compared microsatellite allele frequencies at Y-linked loci in Antioquia with those available for the Iberian peninsula and for northern Africa (since a considerable part of Spain was under Arab rule between the 8th and 15th centuries). From the initial Antioquian sample, 55 individuals were randomly selected for the microsatellite analyses. The genetic distance estimates obtained between these populations on the basis of Y-microsatellite data are shown in table 3. A clear separation is seen between Spanish and northern African populations, with most genetic distances between populations within those two regions being not significantly different from zero. Antioquia is seen to be much closer to the Spanish than to northern African populations. Interestingly, there is a gradient in genetic distance between Spanish, Antioquian, and northern African populations (table 3), with the greatest distance being between the Basque and the Saharawis (.381) and the smallest distance across regions between Antioquians and Arabs (.149). Because southern Spain is known to have contributed the largest proportion of migrants to the American colonies (Boyd-Bowman 1976), this gradient could reflect the more intense occupation of southern Spain by the Arabs.
Table 3.
Pairwise Fst Distances between Antioquia, Spanish, and Northern African Populations, Based on Y-Chromosome Microsatellite Allele Frequencies[Note]
|
Pairwise Fst Distances |
||||||
| Basque | Catalan | Antioquia | Arab | Tachelhit | Berber | |
| Catalan | .020a | |||||
| Antioquia | .049 | .037 | ||||
| Arab | .259 | .167 | .149 | |||
| Tachelhit | .329 | .242 | .219 | .001a | ||
| Berber | .372 | .293 | .252 | .032a | −.014a | |
| Saharawi | .381 | .305 | .264 | .081 | .045 | .034a |
Note.— Data for populations from Spain (Basque and Catalan) and northern Africa (Arab, Tachelhit, Berber, and Saharawi) are from Pérez-Lezaun et al. (1997) and Bosch et al. (1999), respectively. The Arab, Tachelhit, and Berber samples are from Morocco, and those of the Saharawi are from the western Sahara (Bosch et al. 1999, 2000).
P>.05.
Table 4 shows the 36 Y-chromosome microsatellite haplotypes detected in the Antioquian sample and their frequency in Spanish and northern African samples. Of the 36 Antioquian haplotypes, 19 were seen only once. Singletons include chromosomes of African (haplogroup E) and Amerind (haplogroup B) origin. Two instances of convergent microsatellite haplotypes across biallelic haplogroups were detected. One of the five chromosomes carrying haplotype 2 in Antioquia occurred in haplogroup C, whereas the other four and those detected in Iberia and northern Africa occurred in haplogroup A. Haplotype 8 occurred in Antioquia in haplogroup A, whereas this haplotype has been detected in four other populations in haplogroup D.
Table 4.
Y-Chromosome Microsatellite Haplotypes Detected in Antioquia and Their Frequency in Iberian and Northern African Populations[Note]
|
Haplotype |
Frequency in |
|||||||||||||
| HaplotypeDesignation | Haplogroup | DYS19 | DYS388 | DYS390 | DYS391 | DYS392 | DYS393 | Basque(n = 51) | Catalan(n = 27) | Arab(n = 44) | Tachelhit(n = 44) | Berber(n= 12) | Saharawi(n = 29) | Antioquia(n = 55) |
| 1 | A | 14 | 12 | 24 | 11 | 13 | 13 | .294 | .148 | .023 | 0 | 0 | 0 | .200 |
| 2 | A,C | 14 | 12 | 24 | 10 | 13 | 13 | .196 | .074 | .023 | 0 | 0 | 0 | .091 |
| 3 | C | 13 | 12 | 23 | 10 | 13 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | .036 |
| 4 | C | 14 | 16 | 23 | 10 | 11 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | .036 |
| 5 | C | 14 | 15 | 22 | 10 | 11 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | .036 |
| 6 | A | 14 | 12 | 23 | 10 | 13 | 13 | .020 | .037 | 0 | 0 | 0 | 0 | .036 |
| 7 | A | 14 | 12 | 25 | 11 | 13 | 13 | 0 | .037 | 0 | 0 | 0 | 0 | .036 |
| 8 | A,D | 13 | 12 | 24 | 10 | 11 | 13 | 0 | .037 | .091 | .046 | .083 | 0 | .018 |
| 9 | D | 13 | 12 | 23 | 10 | 11 | 13 | 0 | 0 | .046 | 0 | 0 | 0 | .018 |
| 10 | A | 16 | 12 | 24 | 11 | 13 | 13 | 0 | 0 | .023 | 0 | 0 | 0 | .018 |
| 11 | D | 13 | 12 | 25 | 10 | 11 | 13 | 0 | 0 | 0 | .023 | 0 | 0 | .018 |
| 12 | C | 14 | 15 | 23 | 10 | 11 | 12 | 0 | 0 | 0 | .023 | 0 | 0 | .018 |
| 13 | A | 15 | 12 | 23 | 11 | 13 | 13 | .020 | 0 | 0 | 0 | 0 | 0 | .018 |
| 14 | C | 14 | 17 | 23 | 10 | 11 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 15 | A | 12 | 12 | 23 | 10 | 13 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 16 | C | 16 | 12 | 22 | 10 | 11 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 17 | B | 13 | 12 | 22 | 10 | 14 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 18 | A | 16 | 12 | 23 | 10 | 12 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 19 | C | 13 | 16 | 24 | 10 | 11 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 20 | A | 14 | 14 | 24 | 10 | 13 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 21 | A | 14 | 12 | 24 | 12 | 11 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 22 | C | 16 | 13 | 23 | 10 | 11 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 23 | A | 14 | 12 | 24 | 11 | 9 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 24 | E | 12 | 12 | 24 | 12 | 13 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 25 | E | 14 | 12 | 21 | 10 | 11 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 26 | A | 14 | 12 | 24 | 11 | 13 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 27 | C | 14 | 13 | 22 | 10 | 11 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 28 | C | 13 | 14 | 22 | 10 | 11 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 29 | C | 13 | 16 | 23 | 10 | 11 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 30 | C | 14 | 12 | 25 | 10 | 13 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 31 | A | 17 | 12 | 23 | 10 | 13 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 32 | A | 14 | 12 | 24 | 9 | 13 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 33 | A | 14 | 14 | 24 | 10 | 11 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 34 | C | 16 | 16 | 23 | 9 | 11 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 35 | A | 14 | 12 | 23 | 11 | 14 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| 36 | A | 13 | 12 | 23 | 11 | 13 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | .018 |
| Sum | .529 | .333 | .205 | .091 | .083 | 0 | 1.001 | |||||||
| h | .360 | .423 | .458 | .400 | .351 | .408 | .531 | |||||||
Note.— Haplogroup designation refers to the nomenclature in table 1. Numbers at microsatellite loci indicate repeats at each locus, based on the reference provided by Kayser et al. (1997). Haplotypes shared among populations are underlined. The two bottom lines indicate, for each population, the aggregate (Sum) frequency of the haplotypes found in Antioquia and the average gene diversity (h).
In agreement with the genetic distances shown in table 3, most of the common haplotypes detected in Antioquia (1, 2, 6, and 7) are also found in Spain but are rare in northern Africa. Three of these haplotypes (1, 2, and 7) differ by one mutational step, with an additional two haplotypes being one mutational step away from them (haplotypes 26 and 32). Apart from the occurrence of one haplotype 2 in haplogroup C, this set of closely related haplotypes occurs in haplogroup A and represents 38% of the chromosomes seen in Antioquia. The two most common Antioquian haplotypes are also the most frequent among the Basque. The most frequent haplotypes present in the Catalan and in northern African populations, which reach frequencies >20% in those populations (data not shown), were not detected in Antioquia. The average gene diversity for the Y microsatellites is higher in Antioquia than in the Spanish and northern African populations. The Basque show a relatively lower diversity, which is also reflected in the fact that the Antioquian sample includes ∼53% of the haplotypes seen in the Basque, the highest proportion among the populations examined (table 3).
A number of the Antioquian Y-microsatellite haplotypes shown in table 4 carry large alleles at locus DYS388 (alleles with >14 repeats). These alleles are absent or have low frequencies in European and African populations but reach high frequencies in Middle Eastern populations (Kayser et al. 1997; Thomas et al. 2000). Large alleles were detected in the Basque and Catalan populations, at frequencies of 3.9% and 3.7%, respectively, and, in Antioquia, at a frequency of 16.2%. Among the Arabs, Berbers, Saharawis, and Tachelhits, such alleles were found at frequencies of 8.9%, 0%, 10%, and 11%, respectively. This suggests some Semitic ancestry for Antioquia and is consistent with the genetic distance analysis of table 3. Interestingly, haplotype 4, which carries a DYS388 allele with 16 repeats, corresponds to the Cohen modal haplotype (CMH) of Thomas et al. (1998). This haplotype has frequencies >10% among Jewish populations but seems to be rare in Arab populations and has been proposed as an indicator of Jewish ancestry (Thomas et al. 2000). Two other haplotypes (12 and 29) are one mutational step away from the CMH. Haplotypes 3 and 5 also match haplotypes detected among Jewish populations; they correspond to haplotypes 2 and 27 in Thomas et al. (2000). In that survey, Antioquian haplotype 3 was observed only among Sephardic Jews. These matches occur in haplogroup C and, on aggregate, imply that ∼14% of the Antioquian haplotypes could have a Jewish ancestry.
mtDNA Haplotypes
Table 5 shows the mtDNA haplotype frequencies in Antioquia and in five Colombian Native American populations (Mesa et al. 2000). These populations have a wide geographic distribution within Colombia (fig. 1). The Embera live in Antioquia, the Zenu and Wayuu live to the north of the province (with part of the Zenu territory overlapping with Antioquia), and the Ingano and the Ticuna are located in southern Colombia (the Ticuna live in a remote Amazonian location). Approximately 90% of the Antioquian samples carry variants characteristic of the major Amerind mtDNA founder lineages, with a predominance of haplogroups A (45%) and B (37%). These same haplogroups predominate in the Embera, which have a more extreme frequency of haplogroup A (73%). The other populations have high frequencies of haplogroups B and C, the first one predominating among the Zenu and Ingano and the second one among the Wayuu and the Ticuna. The Ticuna also show a relatively high frequency of haplogroup D. Approximately 10% of the haplotypes detected in Antioquia, and, at most, 5% of those seen in the Amerinds examined lack diagnostic changes of the major Amerind founder lineages. These could represent reversions at diagnostic sites or additional founder Amerind lineages or they could correspond to non-Amerind haplotypes. Preliminary sequence data indicate that these haplotypes in Antioquia are nearly equally distributed among European and African lineages. On the basis of haplogroup frequencies, Antioquia shows an mtDNA diversity in the lower range of that seen in the Amerind populations, except the Embera, which show a lower diversity. A low genetic diversity of the Embera relative to the other four Amerind populations is consistently seen with autosomal and Y-chromosome markers (Mesa et al. 2000), suggesting a recent demographic constriction of the Embera settlement sampled.
Table 5.
Haplogroup Frequency of mtDNA in Antioquians and in Five Amerind Populations from Colombia[Note]
|
Haplogroup |
||||||
| Population (n) | A | B | C | D | Other | Gene Diversity |
| Ticuna (54) | .13 | .15 | .39 | .33 | 0 | .721 |
| Ingano (27) | .15 | .44 | .37 | 0 | .04 | .667 |
| Wayuu (40) | .25 | .35 | .38 | 0 | .03 | .692 |
| Zenu (37) | .19 | .41 | .30 | .05 | .05 | .725 |
| Embera (22) | .73 | .23 | 0 | 0 | .05 | .437 |
| Antioquia (80) | .45 | .37 | .06 | .01 | .10 | .646 |
Note.— Amerind data are from Mesa et al. (2000).
Genetic distances between Antioquia and the five Amerind populations tested are shown in table 6. The distance between Antioquia and the Embera is not significantly different from zero, as is also the case for the distances between Wayuu, Zenu, and Ingano. The Embera have the largest interpopulation distances, a finding consistent with stronger drift in this population, as is suggested by its lower genetic diversity. The mean distance between Antioquia and the three Amerind populations of western Colombia (other than the Embera) is similar to that seen between these and the Amazonian Ticuna (0.91 vs. 0.92, respectively).
Table 6.
Pairwise Fst Distances between Antioquians and Five Colombian Amerind Populations, Based on mtDNA Haplogroup Frequencies
|
Pairwise Fst Distance |
|||||
| Ticuna | Ingano | Wayuu | Zenu | Embera | |
| Ingano | .104 | ||||
| Wayuu | .092 | −.014a | |||
| Zenu | .081 | −.024a | −.012a | ||
| Embera | .317 | .296 | .216 | .229 | |
| Antioquia | .212 | .114 | .085 | .074 | .054a |
P>.05.
Discussion
Our results indicate a marked difference in the population origin of the female and male founders of Antioquia. Although the great majority of female founders were Amerind, >90% of the male founders were European in origin. An important contribution of Amerinds to the mtDNA gene pool of Latin Americans has been documented for Mexican and Brazilian populations, but no equivalent study has yet been performed with Y-chromosome markers (Merriwether et al. 1997; Alves-Silva et al. 2000; Green et al. 2000). The high frequency of Amerind mtDNA founder haplotypes found in Antioquia indicates that the number of nonnative women who immigrated into the region was quite small. Although no estimate is available for Colombia, this result agrees with Spanish records showing that, during the colonial period (late 15th to the early 19th centuries), particularly during its initial stages, a relatively small number of women embarked for the New World. Of the 15,000 names recorded in the “Catálogo de Pasajeros a Indias,” a list of passengers embarking for the New World between 1509 and 1559, only 10% were women (Sanchez-Albornoz 1977). Furthermore, after achieving independence in the early 19th century, Colombia has not received the important currents of immigration that have moved into many other Latin American countries (Sanchez-Albornoz 1977; Bushnell 1993). For example, in the Brazilian study by Alves-Silva et al. (2000), ∼40% of the mtDNA lineages were found to be European, with the highest frequency (66%) being observed in regions of important recent immigration, such as the southern Brazilian states.
The high frequency of Amerind mtDNA lineages in Antioquia conflicts with blood-group data indicating that ∼70% of the genetic background of this population is white (Sandoval et al. 1993). This discordance suggests that the number of male founders was larger than that of women. This could result from the incorporation of male founders into the Antioquian gene pool over a longer time period than that of women. This is consistent with the very rapid decline of the native population after the first years of contact. It has been estimated that the overall decimation of the native population of the American continents during the first century of contact was ∼90% (from an initial population anywhere between 10 and 100 million [Sanchez-Albornoz 1977]). In the case of Antioquia, chronicles of the first forays of conquistadors into the Aburrá valley (in the mid-16th century) mention the existence of ∼3,000 natives (Alvarez 1996). About a century later, the census drawn at the founding of Medellín reports 29 native families among a total of 221 living in the area (Alvarez 1996). In addition, colonial administration and society imposed many pressures, favoring the establishment of families with mestizo rather than with native women (Tirado Mejía 1989), further reducing the likelihood of incorporating additional female Amerind founders into the expanding Antioquian population.
Interestingly, the genetic distance between Antioquia and the neighboring Embera population is not statistically significant. Furthermore, the distance between Antioquia and three other native populations from western Colombia is similar to that seen between them and the isolated Ticuna population. This suggests that the Amerind female founders of Antioquia belonged to a population or populations closely related to the Embera. Thus, there seems to be a genetic continuity (at the mtDNA level) of pre- and post-Columbian populations living in this area. The existence of such genetic continuity suggests that the migration rate of mestizo women within Colombia, since the founding of Antioquia, has been relatively small and not sufficient to erase the pre-Columbian Amerind population structure. This is consistent with the reported historical isolation of the province (Parsons 1968; Alvarez 1996). A more thorough sampling of local native populations and of other admixed Colombian populations is required, to evaluate the extent of pre- and post-Columbian genetic continuity of the populations living in this part of South America and to assess the extent of female migration between them during the last centuries.
Data from Y-chromosome polymorphisms are consistent with the notion that a large proportion of the male founders of Antioquia originated in southern Spain, but they also suggest a northern Spanish and possibly a Sephardic contribution. The frequency of allele T at marker SRY-2627 has been assessed in several Iberian and European populations (Hurles et al. 1999). This allele is absent or has low frequencies in the populations surveyed, except for the Basque and Catalan, and its presence at a frequency of 5% in Antioquia suggests a northern Spanish origin for some Antioquian founders. The absence in Antioquia of the most common Catalan microsatellite haplotype and the relatively high frequency in Antioquia of the two most common Basque haplotypes suggests a direct contribution from the latter population. The relatively high frequency of large alleles at locus DYS388 and the genetic distances shown in table 3 are consistent with a Semitic contribution to the gene pool of Antioquia. This is in agreement with an origin of a considerable fraction of the Antioquian founders in southern Spain, since this area was under Arab rule for ∼8 centuries. Furthermore, recent autosomal microsatellite data from Andalucía confirm that there has been some genetic exchange between northern Africa and southern Spain (Bosch et al. 2000). Haplotype analysis suggests that the Semitic contribution to Antioquia might include a Sephardic component, since ∼14% of the Antioquian Y-chromosome haplotypes are shared with Jewish populations, including the CMH and other closely related haplotypes. Greater confidence in the possibility that these haplotypes reflect a Jewish ancestry in Antioquia requires a more extensive evaluation of their frequency in Mediterranean populations, particularly in northern Africa and southern Spain. Other genetic evidence indicative of a Sephardic ancestry for some of the initial Iberian migrants to the New World has recently been reported on the basis of the observation, in non-Jewish Latin American pedigrees, of disease-causing mutations mostly restricted to Jewish populations (Ellis et al. 1998; Lee et al. 1999).
Our attempt to refine the assessment of the origin of Antioquian male founders is complicated by the limited number of populations that have been examined in Iberia and northern Africa and by the possible distortion of allele/haplotype frequencies because of drift. In addition, microsatellites are prone to recurrent mutation—consequently, the haplotype analysis in particular should be regarded with caution. Notwithstanding such limitations, it is noticeable that our findings agree with historical accounts about the origin of Antioquian founders. A substantial migration from southern Spain to the New World is well documented (Boyd-Bowman 1976), and a relatively important Basque and Sephardic contribution to Antioquia has been proposed by several historians (Mesa Bernal 1996). A relatively common Basque origin for Antioquian founders is supported by an estimated 20% frequency of Basque surnames in the province (Twinam 1980). Proposals about a Sephardic contribution rely on less-suggestive evidence. The Jewish ancestry has been documented for only a few Antioquian founders, and this hypothesis has been the subject of intense debate among historians (Mesa Bernal 1996). Persecuted by the Crown, Spanish Jews were expulsed or forced into conversion in the 15th century. The colonial administration officially prohibited the travel of converts to America, and the Spanish Inquisition established itself in Cartagena (in present-day Colombia) to oversee issues of faith, although it was reputedly less severe than it was in Spain (Bushnell 1993). Consequently, even if converts circumvented these circumstances and established themselves in the New World, it is not surprising that very little documentary evidence exists. If the Sephardic origin of the haplotypes detected in Antioquia is confirmed, it will be interesting to evaluate whether this is a common phenomenon in Latin American populations or, as some historians claim, whether it might be more pronounced in Colombia and particularly in Antioquia (Mesa Bernal 1996).
In conclusion, our data are consistent with historical accounts about the origin of Antioquia and its relative isolation. However, the extent of sex bias among native and European founders is surprising. It remains to be explored whether this founding admixture resulted in levels of LD that could currently facilitate complex trait mapping in Antioquia. Although mtDNA data are consistent with some degree of isolation of this population, the level of founder diversity seems substantial and does not support a strong bottleneck at the founding of Antioquia. However, our sample covers a wide geographic area within the Aburrá/Rionegro expansion, and it is possible that internal subisolates exist within the province.
Acknowledgments
We are very grateful to the individuals who contributed samples for this study. We thank Cátira Bortolini for assistance with the admixture calculation and Jaume Bertranpetit, Elena Bosch, and Anna Pérez-Lezaun for facilitating data files. We also thank Mark Thomas, Neil Bradman, and David Goldstein for providing CMH reference DNA and for stimulating discussions. This work was funded by the Royal Society (grant 20779), the Wellcome Trust (grant 056081), Colciencias (grant 1115-04-414-98), and Universidad de Antioquia (CODI grants CPT-9811 and CPT-9812).
References
- Alvarez VM (1996) Poblamiento y población en el valle de Aburrá y Medellín, 1541-1951. In: Melo JO (ed) Historia de Medellín. Suramericana, Medellín [Google Scholar]
- Alves-Silva J, da Silva SM, Guimaraes PE, Ferreira AC, Bandelt HJ, Pena SD, Prado VF (2000) The ancestry of Brazilian mtDNA lineages. Am J Hum Genet 67:444–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailliet G, Rothhammer F, Carnese FR, Bravi CM, Bianchi NO (1994) Founder mitochondrial haplotypes in Amerindian populations. Am J Hum Genet 55:27–33 [PMC free article] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Perez-Lezaun A, Clarimon J, Comas D, Mateu E, Martinez-Arias R, Morera B, Brakez Z, Akhayat O, Sefiani A, Hariti G, Cambon-Thomsen A, Bertranpetit J (2000) Genetic structure of north-west Africa revealed by STR analysis. Eur J Hum Genet 8:360–366 [DOI] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Santos FR, Perez-Lezaun A, Comas D, Benchemsi N, Tyler-Smith C, Bertranpetit J (1999) Variation in short tandem repeats is deeply structured by genetic background on the human Y chromosome. Am J Hum Genet 65:1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd-Bowman P (1976) Patterns of Spanish emigration to the Indies until 1600. Hispanic Am Hist Rev 66:580–604 [Google Scholar]
- Bushnell D (1993) The making of modern Colombia: a nation in spite of itself. University of California Press, Berkeley [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton [Google Scholar]
- Curtin PD (1969) The Atlantic slave trade: a census. University of Wisconsin Press, Madison [Google Scholar]
- Ellis NA, Ciocci S, Proytcheva M, Lennon D, Groden J, German J (1998) The Ashkenazic Jewish Bloom syndrome mutation blmAsh is present in non-Jewish Americans of Spanish ancestry. Am J Hum Genet 63:1685–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LD, Derr JN, Knight A (2000) mtDNA affinities of the peoples of North-Central Mexico. Am J Hum Genet 66:989–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Horai S (1995) Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 56:951–962 [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Redd AJ, Wood ET, Bonner MR, Jarjanazi H, Karafet T, Santachiara-Benerecetti S, Oppenheim A, Jobling MA, Jenkins T, Ostrer H, Bonne-Tamir B (2000) Jewish and middle eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc Natl Acad Sci USA 97:6769–6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Spurdle AB, Karafet T, Bonner MR, Wood ET, Novelletto A, Malaspina P, Mitchell RJ, Horai S, Jenkins T, Zegura SL (1997) The geographic distribution of human Y chromosome variation. Genetics 145:787–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Irven C, Nicholson J, Taylor PG, Santos FR, Loughlin J, Jobling MA, Sykes BC (1998) European Y-chromosomal lineages in Polynesians: a contrast to the population structure revealed by mtDNA. Am J Hum Genet 63:1793–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Veitia R, Arroyo E, Armenteros M, Bertranpetit J, Perez-Lezaun A, Bosch E, Shlumukova M, Cambon-Thomsen A, McElreavey K, Lopez DM, Rohl A, Wilson IJ, Singh L, Pandya A, Santos FR, Tyler-Smith C, Jobling MA (1999) Recent male-mediated gene flow over a linguistic barrier in Iberia, suggested by analysis of a Y-chromosomal DNA polymorphism. Am J Hum Genet 65:1437–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, Zegura SL, Posukh O, Osipova L, Bergen A, Long J, Goldman D, Klitz W, Harihara S, de Knijff P, Wiebe V, Griffiths RC, Templeton AR, Hammer MF (1999) Ancestral Asian source(s) of new world Y-chromosome founder haplotypes. Am J Hum Genet 64:817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Caglià A, Corach D, Fretwell N, Gehrig C, Graziosi G, Heidorn F et al (1997) Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med 110:125–133 [DOI] [PubMed] [Google Scholar]
- Lee HS, Sambuughin N, Cervenakova L, Chapman J, Pocchiari M, Litvak S, Qi HY, Budka H, del Ser T, Furukawa H, Brown P, Gajdusek DC, Long JC, Korczyn AD, Goldfarb LG (1999) Ancestral origins and worldwide distribution of the PRNP 200K mutation causing familial Creutzfeldt-Jakob disease. Am J Hum Genet 64:1063–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendon CL, Martinez A, Behrens IM, Kosik KS, Madrigal L, Norton J, Neuman R, Myers A, Busfield F, Wragg M, Arcos M, Arango Viana JC, Ossa J, Ruiz A, Goate AM, Lopera F (1997) E280A PS-1 mutation causes Alzheimer's disease but age of onset is not modified by ApoE alleles. Hum Mutat 10:186–195 [DOI] [PubMed] [Google Scholar]
- Long JC (1991) The genetic structure of admixed populations. Genetics 127:417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopera F, Ardilla A, Martinez A, Madrigal L, Arango-Viana JC, Lemere CA, Arango-Lasprilla JC, Hincapie L, Arcos-Burgos M, Ossa JE, Behrens IM, Norton J, Lendon C, Goate AM, Ruiz-Linares A, Rosselli M, Kosik KS (1997) Clinical features of early-onset Alzheimer disease in a large kindred with an E280A presenilin-1 mutation. JAMA 277:793–799 [PubMed] [Google Scholar]
- Merriwether DA, Huston S, Iyengar S, Hamman R, Norris JM, Shetterly SM, Kamboh MI, Ferrell RE (1997) Mitochondrial versus nuclear admixture estimates demonstrate a past history of directional mating. Am J Phys Anthropol 102:153–159 [DOI] [PubMed] [Google Scholar]
- Mesa NR, Mondragón MC, Soto ID, Parra MV, Duque C, Ortíz-Barrientos D, García LF, Velez ID, Bravo ML, Múnera JG, Bedoya G, Bortolini M-C, Ruiz-Linares A (2000) Autosomal, mtDNA, and Y-chromosome diversity in Amerinds: pre- and post-Columbian patterns of gene flow in South America. Am J Hum Genet 67:1277–1286 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa Bernal D (1996) De los Judíos en la Historia de Colombia. Planeta, Bogotá [Google Scholar]
- Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JJ (1968) Antioqueño colonization in western Colombia. Vol. 32, 2d ed. University of California Press, Berkeley [Google Scholar]
- Pérez-Lezaun A, Calafell F, Seielstad M, Mateu E, Comas D, Bosch E, Bertranpetit J (1997) Populations genetics of Y-chromosome short tandem repeats in humans. J Mol Evol 45:265–270 [DOI] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC (1983) Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105:767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Sánchez-Albornoz N (1977) La población de America Latina: desde los tiempos precolombinos al año 2000. 2d ed. Alianza Editorial, Madrid [Google Scholar]
- Sandoval C, de la Hoz A, Yunis E (1993) Estructura Genética de la Población Colombiana. Rev Fac Med Univ Nac Colombia 41:3–14 [Google Scholar]
- Schneider S, Roessli D, Excoffier L (2000) ARLEQUIN v. 2000. A software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva [Google Scholar]
- Seielstad MT, Hebert JM, Lin AA, Underhill PA, Ibrahim M, Vollrath D, Cavalli-Sforza LL (1994) Construction of human Y-chromosomal haplotypes using a new polymorphic A to G transition. Hum Mol Genet 3:2159–2161 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Weiss KM (1998) Linkage disequilibrium mapping of complex disease: fantasy or reality? Curr Opin Biotechnol 9:578–594 [DOI] [PubMed] [Google Scholar]
- Thomas MG, Bradman N, Flinn HM (1999) High throughput analysis of 10 microsatellite and 11 diallelic polymorphisms on the human Y-chromosome. Hum Genet 105:577–581 [DOI] [PubMed] [Google Scholar]
- Thomas MG, Parfitt T, Weiss DA, Skorecki K, Wilson JF, le Roux M, Bradman N, Goldstein DB (2000) Y chromosomes traveling south: the Cohen modal haplotype and the origins of the Lemba—the “Black Jews of Southern Africa.” Am J Hum Genet 66:674–686 [DOI] [PMC free article] [PubMed]
- Thomas MG, Skorecki K, Ben Ami H, Parfitt T, Bradman N, Goldstein DB (1998) Origins of Old Testament priests. Nature 394:138–140 [DOI] [PubMed] [Google Scholar]
- Tirado Mejía A (ed) (1989) Nueva historia de Colombia. Planeta, Bogotá [Google Scholar]
- Twinam A (1980) From Jew to Basque: ethnic myths and Antioqueño entrepreneurship. J Interamerican Stud World Affairs 22:81–107 [Google Scholar]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, Vullo CM, Wallace DC (1993) Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet 53:563–590 [PMC free article] [PubMed] [Google Scholar]