Abstract
The Dnmt3a DNA methyltransferase is essential for mammalian development and is responsible for the generation of genomic methylation patterns, which lead to transcriptional silencing. Here, we show that Dnmt3a associates with RP58, a DNA-binding transcriptional repressor protein found at transcriptionally silent heterochromatin. Dnmt3a acts as a co-repressor for RP58 in a manner that does not require its de novo methyltransferase activity. Like other characterized co-repressors, Dnmt3a associates with the histone deacetylase HDAC1 using its ATRX-homology domain. This domain of Dnmt3a represents an independent transcriptional repressor domain whose silencing functions require HDAC activity. These results identify Dnmt3a as a co-repressor protein carrying deacetylase activity and show that Dnmt3a can be targeted to specific regulatory foci via its association with DNA-binding transcription factors.
Keywords: Dnmt3a/histone deacetylation/methylation/transcriptional repression
Introduction
CpG methylation is the major epigenetic modification of mammalian genomes and is essential for normal development, most likely due to its important roles in genomic imprinting, X chromosome inactivation, silencing of parasitic elements and tissue-specific gene expression (Jaenisch, 1997; Surani, 1998; Bestor, 2000). The role played by CpG methylation in these diverse biological processes may be related to its transcriptional gene silencing functions (Razin, 1998; Ng and Bird, 1999).
DNA methylation occurs in a non-random manner within the genome, generating a pattern of methylation that is gene and tissue specific. The generation of methylation patterns is a dynamic process that requires de novo methylation during embryogenesis (Monk et al., 1987; Sanford et al., 1987; Kafri et al., 1992). Despite the known role played by de novo methylation to establish the methylation patterns, the mechanisms underlying this process remain largely undefined. Understanding these mechanisms will require a precise knowledge of the enzymes—the DNA methyltransferases—that generate the methylation patterns.
Dnmt1 was the first DNA methyltransferase to be identified (Bestor et al., 1988). This enzyme is responsible for maintaining pre-existing methylation patterns during DNA replication (Leonhardt et al., 1992; Liu et al., 1998) and is, therefore, often referred to as the ‘maintenance’ methyltransferase. However, the possibility that Dnmt1 also functions as a de novo methyltransferase cannot be excluded (Vertino et al., 1996; Yoder et al., 1997). Dnmt1 is required for normal development as mice deficient for the Dnmt1 gene are embryonic lethal (Li et al., 1992). Because de novo methylation persists in embryonic stem (ES) cells derived from these mice, the existence of independent de novo methyltransferases was suggested (Lei et al., 1996). Accordingly, two de novo methyltransferases have been discovered recently, namely Dnmt3a and Dnmt3b (Okano et al., 1998). They contain in their C-terminus the highly conserved catalytic domain that is characteristic of DNA methyltransferases (Lauster et al., 1989). Dnmt3a and Dnmt3b also contain an N-terminal non-catalytic domain that shows little sequence similarity between the two proteins except for a cysteine-rich region (or ATRX-homology domain; see below). This region is distinct from the cysteine-rich domain—CXXC motif—of Dnmt1 (Ma et al., 1993; Cross et al., 1997) but shares homology with the conserved plant homeodomain (PHD) fingers, present in many chromatin-associated proteins and thought to mediate protein–protein interactions (Aasland et al., 1995). In particular, the cysteine-rich region of Dnmt3a and Dnmt3b is most closely related to the imperfect PHD (or PHD-like) motif found in ATRX, a putative helicase/ATPase of the SNF2 family (Gibbons et al., 1997). Dnmt3a and Dnmt3b are functional de novo methyltransferases, as shown by various assays (Okano et al., 1998; Hsieh, 1999; Lyko et al., 1999). Their inactivation in mice reveals that they are necessary for genome-wide de novo methylation and are essential for proper development (Okano et al., 1999). Interestingly, this study and experiments using an episomal system in cell lines (Hsieh, 1999) demonstrate that Dnmt3a and Dnmt3b generate a methylation pattern that is non-random and that rather, they have specific and preferred DNA targets. Since DNA methyltransferases show very limited DNA sequence selectivity in vitro (Yoder et al., 1997), other factors are likely to be required to mediate the regional specificity they exhibit.
The establishment of methylation patterns leads to gene silencing. Recent studies indicate that DNA methylation may operate along a common mechanistic pathway, namely the use of histone deacetylase (HDAC) activity to repress transcription. Methyl-CpG binding domain (MBD) proteins, which selectively recognize methylated CpG dinucleotides, have been shown to be components of, or to contact, HDAC complexes. These MBDs can thereby recruit HDACs to methylated DNA, resulting in chromatin remodelling and transcriptional silencing (Jones et al., 1998; Nan et al., 1998; Ng et al., 1999, 2000; Wade et al., 1999; Zhang et al., 1999). Recently, it has been shown that Dnmt1 itself can repress transcription through HDAC activity (Fuks et al., 2000; Robertson et al., 2000; Rountree et al., 2000), suggesting that methylation and deacetylation could act together to potentiate the repressed state. Although these studies have begun to shed light on how methylation silences gene expression, much remains to be discovered to fully understand the details underlying this process.
In the present study, we have addressed the mechanisms by which Dnmt3a functions. We show that Dnmt3a can be targeted to promoters through its association with a sequence-specific transcription factor, RP58. We find that this targeted co-repressor function of Dnmt3a is independent of its de novo methyltransferase activity. In addition, we show that Dnmt3a can repress transcription, using its ATRX-homology domain, by recruiting HDAC activity. Together, these data indicate that Dnmt3a plays a role in transcriptional regulation and also suggest that Dnmt3a may have important functions in processes other than de novo methylation.
Results
Dnmt3a binds the RP58 transcriptional repressor in the yeast two-hybrid system
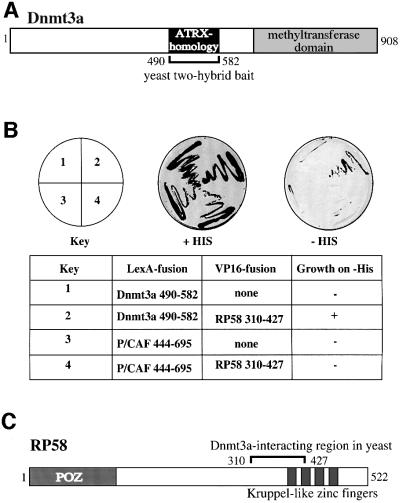
To understand better how patterns of DNA methylation are regulated, we sought to identify proteins that associate with Dnmt3a and might thus target de novo methylation to specific genomic regions. To this end, we employed a yeast two-hybrid screen using as bait the LexA DNA-binding domain fused to Dnmt3a sequences (residues 490–582). We chose this region of Dnmt3a for the screen because it contains the conserved N-terminal cysteine-rich region that shows strong sequence homology with the PHD-like domain of ATRX (Figure 1A; Gibbons et al., 1997). The screen led to the identification of a sequence derived from a RP58 cDNA (amino acids 310–427) as a possible Dnmt3a interaction partner. This interaction is specific since the LexA DNA-binding domain fused to the unrelated P/CAF protein (residues 444–695) failed to bind RP58 310–427–VP16 (Figure 1B). RP58 is a 522-amino-acid protein that contains an N-terminal POZ domain (residues 1–115), belonging to the POZ/zinc finger family (Bardwell and Treisman, 1994), and four sets of Kruppel-type C2H2 zinc finger motifs (residues 372–488) in its C-terminus (Figure 1C). RP58 has been shown to associate with condensed chromatin and to be a sequence-specific DNA-binding protein with transcriptional repression activity (Aoki et al., 1998). Hence, RP58 has features expected of a protein that may recruit Dnmt3a to specific genomic sequences.
Fig. 1. Dnmt3a binds the sequence-specific transcriptional repressor RP58 in a yeast two-hybrid screen. (A) Schematic representation of Dnmt3a. The conserved cysteine-rich region that is closely related to the PHD-like motif of ATRX (residues 490–582), used as bait in the yeast two-hybrid screen, is indicated. The C-terminal methyltransferase domain is shown (numbers indicate amino acid residues). (B) RP58 is a specific Dnmt3a-binding protein in yeast. L40 yeast cells were co-transformed with the plasmids indicated and plated on to –THULL plates containing 35 mM 3-AT in order to examine the specificity of the interaction between bait and prey proteins. (C) Schematic representation of RP58. The N-terminal POZ domain and C-terminal Kruppel-type zinc finger motifs are shown. The region found to interact with Dnmt3a in the yeast two-hybrid screen (residues 310–427) is indicated.
Dnmt3a contacts RP58 directly
To support the yeast two-hybrid data, we first examined the ability of Dnmt3a to bind RP58 in an in vitro glutathione S-transferase (GST) pull-down assay. As shown in Figure 2A (lane 3), full-length RP58 (amino acids 1–522) linked to GST binds efficiently to in vitro translated (IVT) and radiolabelled full-length Dnmt3a. This binding was specific since Dnmt3a failed to bind to the control GST alone (lane 2). When the original fragment of RP58 isolated in the yeast two-hybrid screen (residues 310–427) was fused to GST, it also bound to 35S-labelled Dnmt3a (Figure 2A, lane 4), albeit less strongly than GST–RP58 full length. To provide additional evidence for the association between Dnmt3a and RP58, we asked whether RP58 could purify methyltransferase activity from nuclear extracts. As shown in Figure 2B, RP58 full length (1–522) and 310–427 purified significant methyltransferase activity from nuclear extracts.
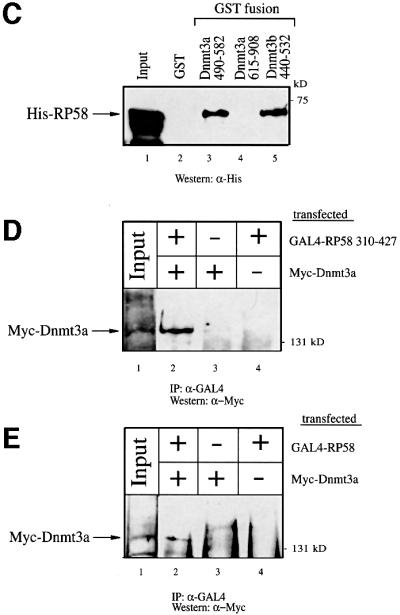
Fig. 2. Dnmt3a interacts with RP58 in vitro and in vivo. (A) Dnmt3a binds RP58 in vitro. The upper panel is a schematic representation of the GST–RP58 fusion proteins, RP58 full length (residues 1–522) and the region of RP58 isolated from the yeast two-hybrid screen (residues 310–427). The indicated GST–RP58 fusion proteins or GST alone were tested in GST pull-down experiments for binding to in vitro translated and 35S-radio labelled full-length Dnmt3a (IVT Dnmt3a). The bound IVT Dnmt3a is indicated by an arrow on the left. Lane 1, IVT [35S]Dnmt3a input (20%). Molecular weight in kDa is indicated on the right. (B) RP58 associates with DNA methyltransferase activity. Equivalent amounts of GST or GST–RP58 fusion proteins bound to Sepharose beads were incubated with HeLa nuclear extract, washed and assayed for methyltransferase activity. Activity is read as c.p.m. of S-adenosyl-l-[methyl-3H]methionine incorporated into an oligonucleotide substrate. Values are normalized to background controls lacking substrate. (C) Dnmt3a binds RP58 directly. GST (lane 2) or the indicated GST fusion proteins of Dnmt3a (residues 490–582, the ATRX-homology region; residues 615–908, encompassing the methyltransferase domain) or GST–Dnmt3b (residues 440–532, the conserved ATRX-homology region) (lanes 3–5) were incubated with histidine-tagged RP58 (His–RP58). Direct binding of RP58 was visualized by western blot analysis using anti-His antibody. Lane 1, His–RP58 input (50%). (D) Dnmt3a co-immunoprecipitates with RP58 310–427 from transfected cell extracts. 293T cells were transiently transfected with 25 µg of pMT3aMyc (expressing Myc-tagged full-length mouse Dnmt3a, Myc–Dnmt3a) and/or 25 µg of pcDNA3GAL4-RP58 310–427 (expressing GAL4-tagged RP58 310–427) as indicated. Whole-cell extracts were then precipitated with anti-GAL4 antibody (5C1) and the presence of Dnmt3a in the immunoprecipitates was visualized by western blot analysis using anti-Myc antibody (9E10). (E) Dnmt3a co-immunoprecipitates with full-length RP58 from transfected cell extracts. 293T cells were transiently transfected as in (D) with Myc–Dnmt3a, and/or pcDNA3GAL4-RP58 (expressing GAL4-tagged RP58 full length) as indicated. Precipitation with the anti-GAL4 antibody was followed by western blot analysis using anti-Myc antibody to detect Dnmt3a.
We next wanted to know whether Dnmt3a contacts RP58 directly. Although the yeast two-hybrid data suggest that the association between LexA–Dnmt3a 490–582 and RP58 310–427–VP16 is direct, we cannot exclude the possibility that binding occurred via an intermediary protein present in yeast. We therefore performed a direct interaction assay by producing both Dnmt3a and RP58 as recombinant proteins in Escherichia coli. As shown in Figure 2C, GST–Dnmt3a 490–582 (containing the ATRX-homology domain) bound histidine-tagged RP58 (lane 3), whereas GST alone (lane 2) or GST–Dnmt3a 615–908 (containing the C-terminal methyltransferase domain; lane 4) did not. Thus, Dnmt3a contacts RP58 in the absence of other proteins and uses the ATRX-homology domain for this interaction. Since this domain of Dnmt3a shows strong sequence conservation with Dnmt3b (Okano et al., 1998), we asked whether the homologous region of Dnmt3b could also bind RP58. Figure 2C (lane 5) indicates that, similarly to Dnmt3a, the ATRX-homology region of Dnmt3b contacts RP58 directly.
Having provided evidence for an interaction in vitro between Dnmt3a and RP58, we next wanted to confirm this interaction in vivo. To this end, we used a co-immunoprecipitation approach in which 293T cells were cotransfected with a Myc-tagged full-length Dnmt3a (Myc–Dnmt3a) and a GAL4-tagged RP58 310–427 (GAL4–RP58 310–427). We then lysed the cells and carried out precipitation with anti-Myc antibody followed by western blot analysis using anti-GAL4 antibody. As shown in Figure 2D, Dnmt3a interacted specifically with RP58 310–427 (lane 2) since no precipitate was detected when either Myc–Dnmt3a or GAL4–RP58 310–427 was transfected alone (lanes 3 and 4). As shown in Figure 2E, using this approach we also detect an interaction between Dnmt3a and full-length RP58. A specific interaction is seen between Myc–Dnmt3a and GAL4–RP58 full length (lane 2) and not when either construct was transfected alone (lanes 3 and 4). Taken together, our data indicate that Dnmt3a interacts both in vitro and in vivo with RP58 and that this interaction is direct.
Dnmt3a acts as a transcriptional co-repressor for RP58 in a methylation-independent manner
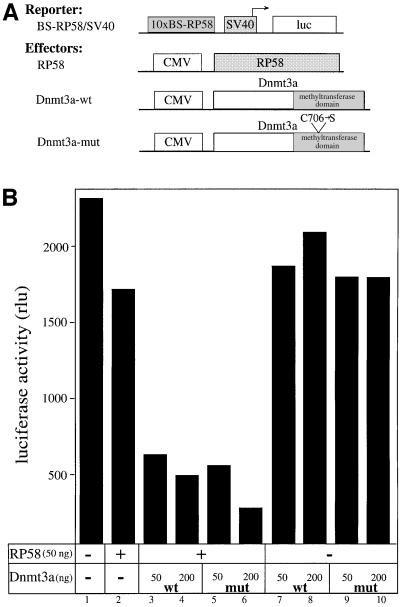
Given that RP58 has been described as a sequence-specific transcriptional repressor (Aoki et al., 1998), our finding that Dnmt3a binds RP58 raises the possibility that Dnmt3a might function as a co-repressor for RP58. To test this idea, we performed transient transfections using a reporter plasmid containing 10 copies of the RP58 DNA-binding site upstream of the SV40 early promoter driving expression of the luciferase gene (BS–RP58/SV40; Figure 3A). This synthetic promoter was chosen, as endogenous RP58-regulated promoters are unknown. As depicted in Figure 3B, cotransfection into 293T cells of the BS–RP58/SV40 reporter, together with a limiting amount of an RP58-expressing vector, repressed transcription only slightly (lane 2). Co-expression of Dnmt3a-wt strongly enhanced RP58 repression (Figure 3B, lanes 3 and 4). This repressive effect required the presence of RP58, since in the absence of the DNA-binding protein no Dnmt3a effect was seen (Figure 3B, lanes 7 and 8). The co-repression function of Dnmt3a is not mediated via its ability to methylate DNA since a point mutant that abolishes its enzymatic activity still represses transcription (lanes 5 and 6) in an RP58-dependent manner (lanes 9 and 10). Bisulphite sequencing of the BS–RP58/SV40 construct after transfection with RP58 and Dnmt3a-wt did not detect any de novo methylation (data not shown), supporting the dispensability of DNA methylase activity in repression by Dnmt3a-wt.
Fig. 3. Dnmt3a cooperates with RP58 to repress transcription in a methylation-independent manner. (A) A schematic representation of the reporter and effector constructs used. The reporter construct BS–RP58/SV40 contains 10 copies of the RP58 DNA-binding site upstream of the SV40 promoter driving expression of the luciferase gene. Constructs expressing full-length RP58, full-length wild-type Dnmt3a (Dnmt3a-wt) and catalytically inactive Dnmt3a (Dnmt3a-mut) are indicated. Dnmt3a-mut contains a single mutation in its methyltransferase catalytic domain that has been shown in vivo to abolish its enzymatic activity (Hsieh, 1999). (B) Dnmt3a is recruited to a promoter via RP58 to repress transcription independently of its methyl transferase activity. 293T cells were transiently transfected with 1 µg of the BS–RP58/SV40 reporter and 50 ng of RP58 in the absence (–) or presence (+) of increasing amounts of Dnmt3a-wt (wt) or Dnmt3a-mut (mut) (50 ng, lanes 3 and 5, and 200 ng, lanes 4 and 6). Absolute luciferase activity is given in rlu. (C and D) RP58 represses transcription independently of methylation. Increasing amounts of RP58 (100, 500 and 1000 ng) were transfected into 293T cells together with 1 µg of unmethylated (C) or SssI-in vitro methylated (D) BS–RP58/SV40 reporter.
As an alternative approach to test whether de novo methylation is required for RP58 to repress transcription, we sought to compare the ability of RP58 to inhibit the activity of an in vitro methylated BS–RP58/SV40 reporter. We reasoned that if methylation is part of the mechanism by which RP58 functions as a repressor, in vitro methylation of the reporter prior to transfection would reduce RP58-mediated repression. Cotransfection of unmethylated (Figure 3C) or in vitro methylated BS–RP58/SV40 reporter (Figure 3D) along with RP58 led to similar dose-dependent transcriptional repression. This suggests that RP58 does not require de novo methylation to repress transcription. Taken together, the data above indicate that Dnmt3a acts as a transcriptional co-repressor when bound to RP58 and that its de novo methyltransferase activity is dispensable for this function.
Dnmt3a associates with HDAC1 through its ATRX-homology domain
We and others have recently shown that Dnmt1 interacts with the HDACs and represses transcription, at least partly, through HDAC activity (Fuks et al., 2000; Robertson et al., 2000; Rountree et al., 2000). The finding that Dnmt3a has repressive capacity prompted us to investigate whether Dnmt3a may, as is the case for Dnmt1, repress transcription through recruitment of HDAC activity. To test this idea, we first conducted in vitro GST pull-down assays and tested the ability of Dnmt3a to bind HDAC1. Figure 4A shows that GST–Dnmt3a 490–582 (encompassing the ATRX-homology region) bound specifically to full-length HDAC1 translated in vitro (lane 3), whereas GST alone (lane 2) or GST–Dnmt3a 615–908 (lane 4) did not. GST–Rb, a known HDAC1-interacting partner (Brehm et al., 1999), was used as a positive control (Figure 4A, lane 5). Using various HDAC1 fragments fused to GST and in vitro translated full-length Dnmt3a, we found that the N-terminal portion including the catalytic domain contains several binding sites for HDAC1 (Figure 4B).
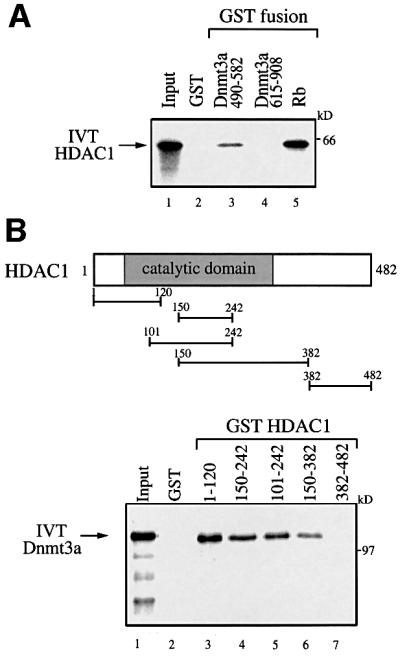

Fig. 4. Dnmt3a interacts with the histone deacetylase HDAC1 in vitro and in vivo. (A) Dnmt3a binds HDAC1 through its ATRX-homology domain in vitro. GST fusion proteins of Dnmt3a (residues 490–582 and residues 615–908) were tested for interaction with HDAC1 in a GST pull-down assay. In vitro translated and 35S-labelled full-length HDAC1 (IVT HDAC1) was incubated with equivalent amounts of bacterially expressed GST (lane 2) or GST fusions of Dnmt3a (lanes 3 and 4) or, as a positive control, GST–Rb (lane 5), and analysed by SDS–PAGE. Lane 1, IVT [35S]HDAC1 input (25%). (B) Dnmt3a binds specific regions of HDAC1 in vitro. The upper panel is a schematic representation of the HDAC1 deacetylase with its catalytic domain depicted by a grey box. The GST–HDAC1 fusions indicated were tested in GST pull-down experiments (lower panel, lanes 3–7), using in vitro translated full-length Dnmt3a (IVT Dnmt3a). Lane 1, IVT [35S]Dnmt3a input (20%). (C) Dnmt3a co-immunoprecipitates with HDAC1 from transfected cell extracts. 293T cells were transiently transfected with 30 µg of pMT3aMyc (expressing Myc-tagged full-length mouse Dnmt3a, Myc–Dnmt3a) and/or 10 µg of pcDNA3-HDAC1-F (expressing Flag-tagged full-length HDAC1, HDAC1-F) as indicated. Whole-cell extracts were then precipitated with anti-Myc antibody (9E10) and the presence of HDAC1-F in the immuno precipitates was visualized by western blot analysis using anti-Flag antibody (M2). (D) Dnmt3a 490–582 co-immunoprecipitates with HDAC1 from transfected cell extracts. 293T cells were transiently transfected with 30 µg of pcDNA3GAL4–Dnmt3a 490–582 (expressing GAL4-tagged Dnmt3a residues 490–582) and/or 10 µg of HDAC1-F as indicated. Whole-cell extracts were then precipitated with anti-GAL4 antibody (5C1) and the presence of HDAC1-F in the immuno precipitates was visualized by western blot analysis using anti-Flag antibody (M2).
The association between Dnmt3a and HDAC1 was also observed using a co-immunoprecipitation approach. 293T cells expressing Myc-tagged full-length Dnmt3a (Myc–Dnmt3a) along with Flag-tagged full-length HDAC1 (HDAC1-F) were lysed, immunoprecipitated with an anti-Myc antibody and western blotted with a Flag antibody. Figure 4C indicates that Dnmt3a interacts specifically with HDAC1 in vivo. Consistent with the in vitro binding data presented in Figure 4A showing that Dnmt3a binds HDAC1 via its ATRX-homology region, immunoprecipitation of GAL4–Dnmt3a 490–582 using a GAL4-specific antibody immunopurifies Flag–HDAC1 (Figure 4D).
Dnmt3a associates with HDAC activity
To determine whether Dnmt3a associates with deacetylase activity, we tested whether antibodies against Dnmt3a could precipitate deacetylase activity from untransfected cells. As shown in Figure 5A (lane 2), immunoprecipitation of endogenous Dnmt3a from HeLa nuclear extracts with anti-Dnmt3a antibodies purified significant amounts of HDAC activity. Control immunoprecipitations with preimmune serum [P.I.(3a), lane 1] or irrelevant antibodies [estrogen receptor (ER; lane 5) and GAL4 (lane 6)] gave background activity (Figure 5A). Using the same assay, we also found that anti-Dnmt3b antibodies precipitated HDAC activity efficiently and specifically (Figure 5A, lane 4). These data demonstrate that native Dnmt3a is associated with HDAC activity, a property that is shared by Dnmt3b.
Fig. 5. Dnmt3 proteins associate with HDAC activity. (A) Endogenous Dnmt3a and Dnmt3b associate with HDAC activity. HeLa nuclear extracts were immunoprecipitated with either preimmune serum to Dnmt3a or Dnmt3b [lanes 1, P.I. (3a), and 3, P.I. (3b)], anti-Dnmt3a and anti-Dnmt3b sera (lanes 2 and 4) or irrelevant antibodies for ER and GAL4 (lanes 5 and 6, respectively). After washing, the immune complexes were tested for HDAC activity. HDAC activity is given as radioactivity (c.p.m.) of [3H]acetate released from an acetylated histone H4 peptide. IP ab, immunoprecipitation antibody. (B) Dnmt3a 490–582 associates with HDAC activity. Equivalent amounts of GST (lane 1) or GST–Dnmt3a 490–582 (lane 2) bound to Sepharose beads were incubated with HeLa nuclear. The beads were then washed and assayed for HDAC activity as described above.
As the ATRX-homology domain of Dnmt3a is required for the interaction with HDAC1 (Figure 4), we next asked whether this domain mediated the association with HDAC activity. Figure 5B shows that GST–Dnmt3a 490–582 (lane 2) purifies deacetylase activity from HeLa nuclear extracts, whereas GST alone (lane 1) failed to bind significant levels of activity.
Dnmt3a actively represses transcription in a TSA-sensitive manner
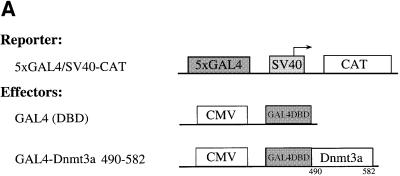
HDAC activity has been shown to be repressive for transcription (Kouzarides, 1999). Having shown that the ATRX-homology region of Dnmt3a binds HDAC1 and associates with deacetylase activity, we set out to establish whether this domain of Dnmt3a has repressive capacity and if so, whether deacetylase activity is required for this repression. To this end, we fused residues 490–582 of Dnmt3a to the GAL4 DNA-binding domain and tested the ability of the fusion protein to repress a reporter bearing five GAL4 binding sites upstream of the SV40 early promoter linked to the CAT gene (5×GAL4/SV40-CAT). Figure 6B shows that transient transfection of 293T cells with increasing amounts of GAL4–Dnmt3a 490–582 efficiently repressed reporter gene expression relative to the GAL4 DNA-binding domain alone. The observed repression was specific as a reporter lacking the GAL4 sites was not affected by GAL4–Dnmt3a 490–582 (data not shown).
Fig. 6. The ATRX-homology region of Dnmt3a actively represses transcription in a TSA-sensitive manner. (A) A schematic representation of the reporter and effector constructs used. The reporter construct 5×GAL4/SV40-CAT contains five binding sites for the yeast transcriptional activator GAL4 upstream of the SV40 early promoter driving the CAT gene. Dnmt3a 490–582, encompassing the ATRX-homology region, is fused to the GAL4 DNA-binding domain. (B) Dnmt3a represses transcription when fused to the GAL4 DNA-binding domain. U2OS cells were transiently transfected with 3 µg of the 5×GAL4/SV40-CAT reporter along with increasing amounts (5 and 10 µg) of GAL4–Dnmt3a 490–582. Whole-cell extracts were used in CAT assays and the results quantified on a phosphoimager. The activity of the reporter in the absence of GAL4 fusion is normalized to a value of 100. (C) Inhibition of HDAC activity relieves repression by Dnmt3a 490–582. U2OS cells were transiently transfected with 3 µg of the 5×GAL4/SV40-CAT reporter along with 5 µg of GAL4–Dnmt3a 490–582. About 10 h after transfection, the cells were treated (+) with the deacetylase inhibitor TSA (100 nM) for 24 h before harvesting, or left untreated (–). CAT assays were performed as above. The activity of the reporter, in the absence of TSA and GAL4 fusion, is normalized to a value of 100.
We next asked whether the repression mediated by GAL4–Dnmt3a (490–582) could be relieved by the addition of the specific HDAC inhibitor trichostatin A (TSA; Yoshida et al., 1995). As shown in Figure 6C, the repressive effect observed with GAL4–Dnmt3a 490–582 on reporter activity was substantially relieved in cells treated with TSA. Collectively, these results indicate that Dnmt3a contains a transcriptional repression domain, encompassing the ATRX-homology region, which recruits deacetylase activity to exert this repressive effect.
Discussion
Here, we show that Dnmt3a is a multi-faceted protein that has the ability to regulate transcription by acting as a co-repressor. Using several protein-based interaction assays, we have identified RP58 as a Dnmt3a-interacting partner. Previous work has shown that RP58 represses transcription through its capacity to function as a sequence-specific DNA-binding factor (Aoki et al., 1998). Significantly, we find that Dnmt3a can repress transcription when recruited to the promoter by RP58. Thus, our findings reveal a role for Dnmt3a as a transcriptional co-repressor for the first time and indicate that Dnmt3a can be targeted to specific regulatory loci through its recruitment by a sequence-specific transcription factor. Since RP58 has been reported to associate predominantly with condensed chromatin (Aoki et al., 1998), the requirement of Dnmt3a for RP58-directed repression may be particularly relevant in controlling heterochromatin-mediated gene inactivation processes. Given that the consensus DNA-binding site for RP58 is found in various promoters of tissue-specific genes (Aoki et al., 1998), an alternative possibility could be that Dnmt3a is targeted by RP58 to these tissue-specific regulated genes and is involved in the control of genes associated with cellular differentiation.
Although we cannot exclude the possibility that the newly identified co-repression function of Dnmt3a may in some instances require its de novo methyltransferase activity, we find that in the context of RP58, this enzymatic activity is dispensable for Dnmt3a to act as a co-repressor. We demonstrate this in three ways. First, a catalytically defective mutant of Dnmt3a still retains the ability to act as a co-repressor for RP58. Secondly, we did not detect de novo methylation within the RP58-responsive promoter using bisulphite sequencing. Thirdly, RP58 could repress a methylated and unmethylated reporter equally well, indicating that RP58 does not require methylation to repress transcription. Although Dnmt3a clearly contributes to the establishment of de novo methylation patterns, as demonstrated by its inactivation in mice (Okano et al., 1999), the above data indicate that de novo methylation is not the sole mechanism by which Dnmt3a functions. It is therefore possible that the developmental defects observed in the Dnmt3a knockout mice are associated not only with its de novo methylation activity (Okano et al., 1998) but also with other functions of Dnmt3a, such as its targeted co-repressor function. Our observation that Dnmt3a can operate to repress transcription independently of its methylation activity is supported by a recent report on Dnmt1, which demonstrates that it can repress transcription from promoters containing E2F-binding sites without the apparent requirement of its methyltransferase activity (Robertson et al., 2000).
Another novel finding of the present study is the close connection we have uncovered between Dnmt3a and histone deacetylation. We show that Dnmt3a binds HDAC1 both in vitro and in vivo, and consistent with this, Dnmt3a associates with HDAC activity in the cell. In addition, we demonstrate that Dnmt3a sequences can actively repress transcription through the recruitment of HDAC activity. We find that the link between Dnmt3a and deacetylation is mediated by the cysteine-rich region of Dnmt3a that is closely related to the PHD–like motif of ATRX. This protein is involved in the ATRX syndrome, which is associated with α-thalassaemia (Gibbons et al., 1997). The finding that two-thirds of the mutations causing the ATRX syndrome lie in its PHD-like motif highlights the biological significance of this motif. The PHD-like domain also shares homology with the PHD motif of Mi2β, a component of the NURD deacetylase complex (Zhang et al., 1998; Xu et al., 1999). In agreement with our observation, Mi2β was found to bind HDAC1 through its PHD zinc fingers (Zhang et al., 1998). Interestingly, the PHD-like motif of Dnmt3a is well conserved with the other Dnmt3 family members, Dnmt3b (Okano et al., 1998) and the recently isolated Dnmt3L (Aapola et al., 2000). This suggests that these proteins are also likely to bind HDAC activity. Indeed, we have demonstrated that endogenous Dnmt3b associates with HDAC activity. It is therefore likely that transcriptional repression through recruitment of deacetylase activity is a conserved and shared feature within the Dnmt3 family.
Collectively, our results provide evidence that Dnmt3a can be targeted to promoters or regions of the genome associated with the RP58 repressor. The targeted co-repressor functions of Dnmt3a are independent of its de novo methylation activity. We find that Dnmt3a has a transcriptional repressor domain that functions by recruiting HDAC activity. This domain may contribute to the ability of Dnmt3a to repress RP58, although this still remains to be established. Thus, our data highlight a novel co-repressor function for Dnmt3a, which is independent of its de novo methylase activity. The identification of Dnmt3L, which lacks a methylase domain but retains the potential to associate with deacetylase activity, highlights the fact that, in some circumstances, the HDAC-associated functions may be working independently of the methylase functions. The challenge in the future is to establish whether there are instances where DNA methylase and associated deacetylase activities have a role to play in the same biological process.
Materials and methods
Constructs
We cloned Dnmt3a 490–582 as a LexA fusion into the yeast expression vector pBTM116. We cloned various domains of HDAC1, RP58, Dnmt3a and Dnmt3b into the pGEX vector (Pharmacia) by PCR using appropriate sets of primers. Full-length wild-type and catalytic-site mutant Dnmt3a (pMT3aMyc and pMT3aMut, respectively; Hsieh, 1999) were a kind gift from C.-L.Hsieh. For the GAL4 fusion experiments, we cloned sequences from Dnmt3a (residues 490–582) or RP58 (residues 310–427 and full length) into the pcDNA3.1-GAL4 vector using PCR. pcDNA3.1-GAL4 has the GAL4 DNA-binding domain (residues 1–147) under the control of a CMV promoter. pcDNA3.1-RP58, RP58-HIS and the reporter construct BS10-pGL2C (BS–RP58/SV40) have been described (Aoki et al., 1998). pcDNA3-HDAC1-F (HDAC1-F), pING14A-HDAC1, pGEX-Rb 379–928 and the reporter construct 5×GAL4/SV40-CAT have been described previously (Morkel et al., 1997; Brehm et al., 1998). We verified all constructs by DNA sequencing.
GST fusion proteins, in vitro translation and pull-down assays
We expressed GST and GST fusion proteins in E.coli XA90 using the pGex (Pharmacia) vector system, and purified protein from crude bacterial lysates according to the manufacturer’s instructions. In vitro transcription–translation was performed using the TNT system (Promega). HDAC1 and Dnmt3a were translated in vitro from pING14A-HDAC1 and pMT3aMyc, respectively. GST pull-downs were performed essentially as described previously (Bannister and Kouzarides, 1995).
Cell culture, transfections, CAT and luciferase assays
Cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum and grown at 37°C, 5% CO2. U2OS cells were transfected using the calcium phosphate coprecipitation method. For TSA experiments, the precipitate was washed off 10 h after transfection and incubated for an additional 24 h, either in the presence or absence of TSA (100 nM; Waco BioProducts) before harvesting. Cells were then washed once in phosphate-buffered saline, resuspended in 100 µl 0.25 M Tris pH 8.0 and lysed by freezing and thawing three times (liquid nitrogen/37°C). Supernatants were clarified by centrifugation (5 min, 12 000 g). We performed CAT assays as described previously (Hagemeier et al., 1993) using 60 µl of the extracts and quantitated the results on a phosphoimager. For luciferase assays, cells were harvested and lysed using the Promega Luciferase Assay System according to the manufacturer’s instructions. Luciferase activity was quantitated on a luminometer (Dynex Technologies) and is expressed as arbitrary light units (rlu). All transfections were done at least in duplicate with a representative experiment shown for each set of transfection.
Immunoprecipitations and western blot analysis
We transiently transfected 293T cells in culture dishes (15 cm diameter) with either 25 µg of pMT3aMyc and/or 25 µg of pcDNA3GAL4-RP58 310–427 (or pcDNA3GAL4-RP58 full length) for Dnmt3a–RP58 co-immunoprecipitations. For Dnmt3a–HDAC1 co-immunoprecipitations, 30 µg of pMT3aMyc or pcDNA3GAL4-Dnmt3a 490–582 and/or 10 µg of pcDNA3-HDAC1-F were transfected. Cells were harvested 36 h post-transfection, lysed in 1 ml of IPH buffer (Bannister and Kouzarides, 1996) at 4°C for 30 min and debris was removed by centrifugation. Immunoprecipitations were then carried out as described previously (Fuks et al., 2000). For Dnmt3a–RP58 co-immunoprecipitations, cells were lysed in 1 ml of modified Z′-buffer (Fuks et al., 2000) containing 300 mM KCl and 0.5% NP-40, which were diluted to 100 mM KCl and 0.1% NP-40 in immunoprecipitations. Antibodies used were anti-Myc (9E10; Roche) or anti-GAL4 (5C1; Santa Cruz). Western blot analysis was carried out according to standard techniques (Martinez-Balbas et al., 2000) using anti-Flag (M2; Kodak) or anti-Myc antibody (9E10; Roche).
Direct interaction assays using His-tagged proteins
We expressed histidine-tagged RP58 in E.coli XA-90 using the Qiagen vector system. Proteins were purified using nickel chelate chromatogaphy and eluted using 150 mM imidazole. His–RP58 was incubated with GST fusion proteins, washed and resolved as for GST pull-down assays. Gels were blotted on to nitrocellulose membranes and subjected to western blot analysis as described (Martinez-Balbas et al., 2000) using anti-His antibody (H15; Santa Cruz) as primary antibody.
Immunoprecipitation of histone deacetylase activity from cell extracts
We carried out immunoprecipitations as described previously (Martinez-Balbas et al., 2000) with 50 µl of HeLa nuclear extracts (Computer Cell Culture Centre, Belgium) and either Dnmt3a- or Dnmt3b-specific antisera (164A and 157A, respectively, a gift from En Li), preimmune sera (a gift from En Li), anti-ER (HC-20; Santa Cruz) or anti-GAL4 (5C1; Santa Cruz) antibodies. Antibody complexes were collected on protein A/G–Sepharose beads, washed four times in IPH buffer and assayed for HDAC activity. We performed HDAC assays essentially as described previously (Taunton et al., 1996; Brehm et al., 1999) in a volume of 300 µl of IPH buffer containing 250 000 c.p.m. of a tritium-labelled acetylated H4 peptide.
In vitro methylation
The reporter construct BS10–RP58/SV40 was methylated in vitro using SssI methylase (New England Biolabs) for 2 h at 37°C, according to the manufacturer’s instructions. Methylation was verified by protection of the construct from digestion with methylation-sensitive restriction enzymes. Methylated DNA was purified prior to transfection using a High Pure purification kit (Roche).
Methyltransferase assay
Methyltransferase assays were performed essentially as described previously (Fuks et al., 2000). GST fusions were incubated in HeLa nuclear extract, washed and assayed for methyltransferase activity in a 100 µl reaction containing 500 ng of a 33 bp hemimethylated oligonucleotide substrate (Ramchandani et al., 1997), 2 µl S-adenosyl-l-[methyl-3H]methionine (77 Ci/mmol; Amersham), 50 mM Tris–HCl pH 7.5, 5 mM EDTA, 50% glycerol, 5 mM dithiothreitol (DTT) and protease inhibitors. After incubation at 37°C for 2 h, unincorporated nuclides were removed with Biospin chromatography columns (Bio-Rad) and incorporation of radioactivity was determined by liquid scintillation counting. Values were normalized to background controls lacking DNA substrate. As an additional control, proteins were tested for methyltransferase activity without prior incubation in HeLa nuclear extract.
Yeast two-hybrid screen
Dnmt3a 490–582 was cloned into pBTM116 as a LexA fusion. This construct was transformed into L40 yeast together with a mouse 9.5–10.5 d.p.c. cDNA library ligated into pVP16 as described (Lavender et al., 1997). Proteins interacting with Dnmt3a 490–582 were identified by growth on –THULL plates in the presence of 35 mM 3-aminotriazole (3-AT). Plasmids rescued from positive yeast colonies were retransformed with either pBTM116-Dnmt3a 490–582 or pBTM116-P/CAF 444–695 to assess the specificity of the interaction.
Acknowledgments
Acknowledgements
We thank Chih-Lin Hsieh for Dnmt3a wild-type (pMT3aMyc) and mutant (pMT3aMut) constructs, and En Li for Dnmt3a and Dnmt3b immune sera. F.F. was supported by the Wiener-Anspach Foundation and the Fonds National de la Recherche Scientifique (‘Chargé de recherches du FNRS’). W.A.B. was supported by a scholarship from the South African National Research Foundation. The research in the T.K. laboratory is supported by a programme grant from the Cancer Research Campaign.
References
- Aapola U. et al. (2000) Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics, 65, 293–298. [DOI] [PubMed] [Google Scholar]
- Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. [DOI] [PubMed] [Google Scholar]
- Aoki K., Meng,G., Suzuki,K., Takashi,T., Kameoka,Y., Nakahara,K., Ishida,R. and Kasai,M. (1998) RP58 associates with condensed chromatin and mediates a sequence- specific transcriptional repression. J. Biol. Chem., 273, 26698–26704. [DOI] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides,T. (1995) CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J., 14, 4758–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Bardwell V.J. and Treisman,R. (1994) The POZ domain: a conserved protein–protein interaction motif. Genes Dev., 8, 1664–1677. [DOI] [PubMed] [Google Scholar]
- Bestor T.H. (2000) The DNA methyltransferases of mammals. Hum. Mol. Genet., 9, 2395–2402. [DOI] [PubMed] [Google Scholar]
- Bestor T., Laudano,A., Mattaliano,R. and Ingram,V. (1988) Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol., 203, 971–983. [DOI] [PubMed] [Google Scholar]
- Brehm A., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- Brehm A., Nielsen,S.J., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1999) The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J., 18, 2449–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross S.H., Meehan,R.R., Nan,X. and Bird,A. (1997) A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nature Genet., 16, 256–259. [DOI] [PubMed] [Google Scholar]
- Fuks F., Burgers,W.A., Brehm,A., Hughes-Davies,L. and Kouzarides,T. (2000) DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nature Genet., 24, 88–91. [DOI] [PubMed] [Google Scholar]
- Gibbons R.J. et al. (1997) Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nature Genet., 17, 146–148. [DOI] [PubMed] [Google Scholar]
- Hagemeier C., Cook,A. and Kouzarides,T. (1993) The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res., 21, 4998–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L. (1999) In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell. Biol., 19, 8211–8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R. (1997) DNA methylation and imprinting: why bother? Trends Genet., 13, 323–329. [DOI] [PubMed] [Google Scholar]
- Jones P.L., Veenstra,G.J., Wade,P.A., Vermaak,D., Kass,S.U., Landsberger,N., Strouboulis,J. and Wolffe,A.P. (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet., 19, 187–191. [DOI] [PubMed] [Google Scholar]
- Kafri T., Ariel,M., Brandeis,M., Shemer,R., Urven,L., McCarrey,J., Cedar,H. and Razin,A. (1992) Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev., 6, 705–714. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (1999) Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev., 9, 40–48. [DOI] [PubMed] [Google Scholar]
- Lauster R., Trautner,T.A. and Noyer-Weidner,M. (1989) Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J. Mol. Biol., 206, 305–312. [DOI] [PubMed] [Google Scholar]
- Lavender P., Vandel,L., Bannister,A.J. and Kouzarides,T. (1997) The HMG-box transcription factor HBP1 is targeted by the pocket proteins and E1A. Oncogene, 14, 2721–2728. [DOI] [PubMed] [Google Scholar]
- Lei H., Oh,S.P., Okano,M., Juttermann,R., Goss,K.A., Jaenisch,R. and Li,E. (1996) De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development, 122, 3195–3205. [DOI] [PubMed] [Google Scholar]
- Leonhardt H., Page,A.W., Weier,H.U. and Bestor,T.H. (1992) A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell, 71, 865–873. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor,T.H. and Jaenisch,R. (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell, 69, 915–926. [DOI] [PubMed] [Google Scholar]
- Liu Y., Oakeley,E.J., Sun,L. and Jost,J.P. (1998) Multiple domains are involved in the targeting of the mouse DNA methyltransferase to the DNA replication foci. Nucleic Acids Res., 26, 1038–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F., Ramsahoye,B.H., Kashevsky,H., Tudor,M., Mastrangelo,M.A., Orr-Weaver,T.L. and Jaenisch,R. (1999) Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nature Genet., 23, 363–366. [DOI] [PubMed] [Google Scholar]
- Ma Q. et al. (1993) Analysis of the murine All-1 gene reveals conserved domains with human ALL-1 and identifies a motif shared with DNA methyltransferases. Proc. Natl Acad. Sci. USA, 90, 6350–6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas M.A., Bauer,U.M., Nielsen,S.J., Brehm,A. and Kouzarides,T. (2000) Regulation of E2F1 activity by acetylation. EMBO J., 19, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M., Boubelik,M. and Lehnert,S. (1987) Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development, 99, 371–382. [DOI] [PubMed] [Google Scholar]
- Morkel M., Wenkel,J., Bannister,A.J., Kouzarides,T. and Hagemeier,C. (1997) An E2F-like repressor of transcription. Nature, 390, 567–568. [DOI] [PubMed] [Google Scholar]
- Nan X., Ng,H.H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenman,R.N. and Bird,A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- Ng H.-H. and Bird,A. (1999) DNA methylation and chromatin modification. Curr. Opin. Genet. Dev., 9, 158–163. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Zhang,Y., Hendrich,B., Johnson,C.A., Turner,B.M., Erdjument-Bromage,H., Tempst,P., Reinberg,D. and Bird,A. (1999) MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nature Genet., 23, 58–61. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Jeppesen,P. and Bird,A. (2000) Active repression of methylated genes by the chromosomal protein MBD1. Mol. Cell. Biol., 20, 1394–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M., Xie,S. and Li,E. (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genet., 19, 219–220. [DOI] [PubMed] [Google Scholar]
- Okano M., Bell,D.W., Haber,D.A. and Li,E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247–257. [DOI] [PubMed] [Google Scholar]
- Ramchandani S., MacLeod,A.R., Pinard,M., von Hofe,E. and Szyf,M. (1997) Inhibition of tumorigenesis by a cytosine-DNA, methyltransferase, antisense oligodeoxynucleotide. Proc. Natl Acad. Sci. USA, 94, 684–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A. (1998) CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBO J., 17, 4905–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K.D., Ait-Si-Ali,S., Yokochi,T., Wade,P.A., Jones,P.L. and Wolffe,A.P. (2000) DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nature Genet., 25, 338–342. [DOI] [PubMed] [Google Scholar]
- Rountree M.R., Bachman,K.E. and Baylin,S.B. (2000) DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nature Genet., 25, 269–277. [DOI] [PubMed] [Google Scholar]
- Sanford J.P., Clark,H.J., Chapman,V.M. and Rossant,J. (1987) Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev., 1, 1039–1046. [DOI] [PubMed] [Google Scholar]
- Surani M.A. (1998) Imprinting and the initiation of gene silencing in the germ line. Cell, 93, 309–312. [DOI] [PubMed] [Google Scholar]
- Taunton J., Hassig,C.A. and Schreiber,S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- Vertino P.M., Yen,R.W., Gao,J. and Baylin,S.B. (1996) De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5)-methyltransferase. Mol. Cell. Biol., 16, 4555–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade P.A., Gegonne,A., Jones,P.L., Ballestar,E., Aubry,F. and Wolffe,A.P. (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nature Genet., 23, 62–66. [DOI] [PubMed] [Google Scholar]
- Xu G.L. et al. (1999) Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature, 402, 187–191. [DOI] [PubMed] [Google Scholar]
- Yoder J.A., Soman,N.S., Verdine,G.L. and Bestor,T.H. (1997) DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol., 270, 385–395. [DOI] [PubMed] [Google Scholar]
- Yoshida M., Horinouchi,S. and Beppu,T. (1995) Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. BioEssays, 17, 423–430. [DOI] [PubMed] [Google Scholar]
- Zhang Y., LeRoy,G., Seelig,H.P., Lane,W.S. and Reinberg,D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell, 95, 279–289. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ng,H.H., Erdjument-Bromage,H., Tempst,P., Bird,A. and Reinberg,D. (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev., 13, 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]