Abstract
Preaxial polydactyly (PPD) is a common limb malformation in human. A number of polydactylous mouse mutants indicate that misexpression of Shh is a common requirement for generating extra digits. Here we identify a translocation breakpoint in a PPD patient and a transgenic insertion site in the polydactylous mouse mutant sasquatch (Ssq). The genetic lesions in both lie within the same respective intron of the LMBR1/Lmbr1 gene, which resides ≈1 Mb away from Shh. Genetic analysis of Ssq reveals that the Lmbr1 gene is incidental to the phenotype and that the mutation directly interrupts a cis-acting regulator of Shh. This regulator is most likely the target for generating PPD mutations in human.
Keywords: chromosome 7q36‖Ssq‖mouse‖human
Preaxial polydactyly [PPD (MIM190605)] is one of the most frequently observed human congenital limb malformations. Sporadic cases of PPD have been described, but most show an autosomal-dominant mode of inheritance. The limb-specific phenotype varies markedly within families, ranging from a simple addition of a phalanx in triphalangeal thumb to whole digit duplications and tibial aplasia. Using several large families, a PPD locus was mapped to a 450-kb region on chromosome 7q36, and all families described so far are linked to this locus (1–5). Recent reports suggest that PPD constitutes one aspect of a complex disease locus. Acheiropodia (6), complex polysyndactyly (CPS) (7), and acropectoral syndrome (8) are all distinct, limb-specific disorders that map to this region, suggesting that elements essential for limb development are located in this locus.
Sasquatch (Ssq) is a mouse mutation that arose through a transgenic insertion (9). The mutation is semidominant, resulting in supernumerary preaxial (anterior) digits on the hindfeet in the heterozygotes. In homozygotes both fore- and hindlimbs show additional preaxial digits, and in some cases the long bones are shortened such that the limbs appear twisted. The insertion site responsible for the Ssq phenotype is physically linked to within ≈1 Mb of Shh.
Here, we show that Ssq maps to the region on mouse chromosome 5 that corresponds to the human PPD locus. We identify mutations in a PPD patient and in the Ssq mouse. The PPD patient carries a de novo chromosomal translocation. Isolation of the PPD translocation breakpoint and the Ssq transgene insertion site revealed a similar location for these genetic disruptions within the Lmbr1 gene. We provide genetic analysis that shows that the Ssq mutation is not acting locally but in fact interrupts a long-range cis-acting regulator. This regulator operates on Shh residing 1.8 cM away, corresponding to a physical distance of ≈1 Mb. Consequently, disruption of Shh regulation is most likely the basis for PPD in humans.
Materials and Methods
Patient Material.
The translocation patient was clinically examined, and a member of her family was interviewed for family history at the Niigata University Hospital. All studies were approved by the local ethics committee. A member of the family gave written informed consent on behalf of the patient. The PPD families used in this study are unrelated, as described (1, 4).
Fluorescence in Situ Hybridization (FISH) Analysis.
The end sequences of PAC 123K10, BAC 111G19, and cosmid 171G2 were generated to enable alignment with the existing genomic sequence. FISH analysis using YAC, BAC, PAC, and cosmid clones as probes was performed as described (10). Epstein–Barr-virus-mediated lymphoblastoid cell lines established from peripheral blood of the patients were treated with colcemid and harvested according to routine methods. Probes were labeled with digoxigenin-11-dUTP by using a nick translation kit (Roche).
STS Probes and Southern Blot Analysis.
STS probes used for Southern blot analysis were PCR-amplified from human genomic DNA of normal control, using primers specific to the genomic sequence of a BAC clone Rp11-332E22. Probes were labeled with [α-32P]dCTP in a random priming reaction using the RediPrime II DNA labeling system (Amersham Pharmacia Biotech). Southern blot analyses were performed as described (11).
Mutation Analysis and Sequencing.
Primers were designed complementary to intronic DNA around all exons of LMBR1 to cover at least 60 bp on both sides of the exon. PCR products were generated from either genomic DNA cleaned with the Amicon Microcon PCR purification kit (Millipore) or treated with a PCR product presequencing kit (Amersham Pharmacia). Larger fragment sequencing used a transposon-based system (GPS–1 Genome Priming system, New England Biolabs) and plasmid DNA was made using an ABI Prism Miniprep kit (Perkin–Elmer/Applied Biosystems). DNA was sequenced on an ABI-377 automated fluorescence dye sequencer, using Bigdye or dRhodamine chemistry (Perkin–Elmer/Applied Biosystems).
Sequences conserved between mouse and human were also screened for mutations in familial PPD patients. These regions were: 3.7 kb adjacent to exon 5 (downstream); 100 bp at 1 kb upstream of exon 6; 230 bp adjacent to exon 6 (upstream); intron 7; intron 14; 2 kb adjacent to exon 16 (downstream). Database screening and sequence analysis was done using programs from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) and Human Genome Mapping Project (www.hgmp.mrc.ac.uk).
Cloning Ssq Insertion Site.
Ssq/Ssq genomic libraries were made in Lambda FixII and in SuperCos (Stratagene) following the manufacturer's instructions. λ clones (1.6 × 106) were screened using PCR product generated from the transgene (primers GCTTCAGCTCTGTGACATACT and CAGTTTGTCCTTCTTCTGCCC). Subsequent screening identified one λ as containing only one end of the transgene. DNA was made from this (Qiagen Lambda kit) and end sequence obtained. The genomic end fragment was used as a probe on the RPCI21 library (HGMP Resource Centre, Cambridge, U.K.) and PACs identified (RCPI21-542n10 and RCPI21-576b18). FISH and exon trapping (12) were conducted by standard techniques. Complete intron 5 sequence was determine by assembling HTGS draft sequence (GenBank accession no. AC058788) in combination with sequence generated from the integration sites and directed sequence to fill gaps.
Analysis of Lmbr1 Expression by Reverse Transcription (RT)-PCR.
RNA was extracted from E11.5 mouse embryos by using RNAzol (Biogenesis) and cDNA was made using a First-strand cDNA Synthesis Kit (Amersham Pharmacia Biotech). PCR primers used are specific to exons of mouse Lmbr1, except that the exon 6 primer crossreacts with the glycosylphosphatidylinositol-anchored protein homologue and serves as an internal quantification control. PCR was also performed with primers to the mouse HPRT gene as a positive control. Samples were taken after 25, 30, and 35 cycles of PCR, and quantified either by Southern blotting of the gel and probing with an internal oligonucleotide or by addition of [α-32P]dCTP to the reaction and subsequent scanning on a Storm phosphoimager (imagequant package).
Genetic Cis–Trans Test.
A Shh null heterozygous mouse was crossed to a Ssq homozygote mouse to generate Shh null heterozygous mice carrying the Ssq insertion, the F1 generation. The F1s were crossed to CBA wild-type mice to generate the G2 generation. DNA from all mice was analyzed by PCR for the Ssq insertion and for both the Shh wild-type and null alleles. Primers for the Ssq insertion were CTCTGTTTCCTTTTCCTCTATC and GTATGGGATTAATTAAATCTTGTGTC, which generate a 180-bp product. The Shh PCR primers were described by Chiang et al. (13) and generated a 550-bp product for the null allele and 230-bp product for the wild-type allele.
Results
Identification of a PPD Translocation Breakpoint.
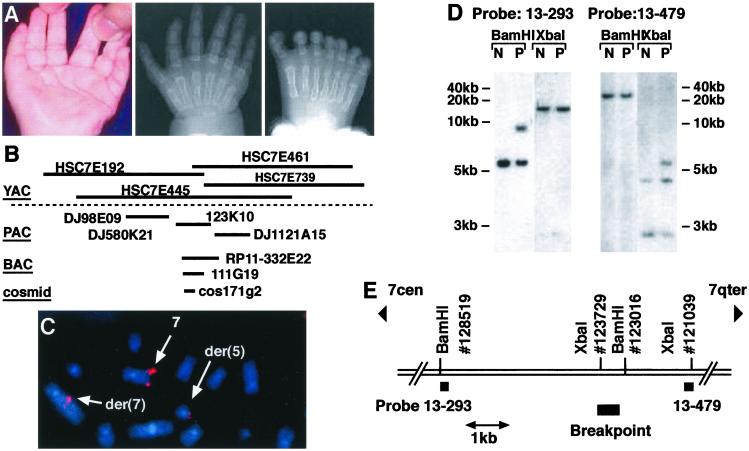
A 3-year-old girl presented with bilateral duplication of the triphalangeal thumb and triplication of the great toe without any associated abnormality (Fig. 1A). She was found to carry a de novo reciprocal translocation t(5,7)(q11,q36). Fine mapping of the translocation breakpoint by use of FISH analysis in the lymphoblastoid cell line derived from the patient (Fig. 1C) identified a BAC clone Rp11-332E22 (GenBank accession no. AC007097) that spanned the breakpoint (Fig. 1B). Using fragments from the BAC, a cosmid clone 171g2 was identified that spanned the breakpoint (Fig. 1B). Sequencing the ends of the cosmid insert identified the genomic fragment as lying inside one of the genes (originally given the designation C7orf2 (5) within the genetically defined critical region for PPD. This gene is the orthologue of the mouse Lmbr1 gene (14). Southern blot analysis of DNA from a lymphoblastoid cell line further refined the site of the breakpoint (Fig. 1D). A number of repeat-free STSs including 13–293 and 13–479 were PCR-amplified and used as probes. This analysis showed that the translocation breakpoint is located in a 714-bp BamHI/XbaI fragment residing between exons 5 and 6 of LMBR1 without an accompanying large deletion (Fig. 1E). This DNA fragment consists of 95% LINES and 5% simple repeated sequence, and seems to neither encode an ORF nor contain known regulatory sequences.
Figure 1.
Phenotype and identification of the breakpoint in the t(5 7)(q11,q36)translocation patient. (A) Left hand (Left, ventral view) of the PPD translocation patient showing duplication of the triphalangeal thumb. Radiogram of the right hand (Center) showing bilateral duplications and of the right foot (Right) showing triplication of the great toe. (B) The YACs, PACs, BACs, and a cosmid used to define the translocation breakpoint. (C) FISH analysis of metaphase chromosomes from a lymphoblastoid cell line from the patient, using cosmid clone171G2 as a probe. Hybridization signals are marked by arrows on the wild-type chromosome 7 and on the products of the translocation, the derived chromosome 5 (der 5) and derived chromosome 7 (der 7). (D) Southern blot analysis with genomic DNA from the lymphoblastoid cell line from the patient (P) and a normal control (N). DNA was digested with restriction endonucleases BamHI and XbaI. STS probe 13–293 (Left) and STS probe 13–479 (Right) were used. The relative positions of the STS probes, restriction sites, and the translocation breakpoint are indicated (E).
Subsequently, we performed mutation analysis on the LMBR1 gene in patients. Analysis of the exons and the intron/exon boundaries of the LMBR1 gene in five unrelated PPD families (1, 4) uncovered no pathogenic mutations (data not shown). Therefore, in the PPD families mutations interrupting either the coding region or the structure of the transcript are not apparent.
Isolation of the Ssq Insertion Site.
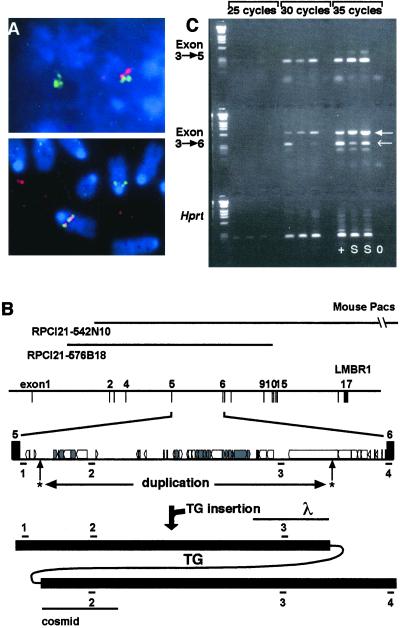
To isolate the transgenic insertion site from the Ssq mutant mouse, a λ genomic library was made from Ssq/Ssq mice. Clones were isolated using the transgene as a probe, one of which contained a junction of the transgene insertion site (Fig. 2B). The genomic end fragment of this λ clone (Fig. 2B) was used to identify mouse PAC clones which, by chromosomal FISH analysis, were shown to co-localize with the transgene (Fig. 2A). Subsequent exon trapping (12) of the PAC RPCI21-542N10 identified a surrounding gene. The complete sequence of the corresponding mouse cDNA was determined and is identical to mouse Lmbr1 (14).
Figure 2.
Identification of the mouse Ssq insertion site. PAC RPCI21-542N10 was isolated as containing the transgene integration site. (A) FISH analysis confirms the colocalization of the PAC with the transgene. PAC RPCI21-542N10 is labeled in green and the human placental alkaline phosphatase reporter gene (HPAP)-containing transgene is labeled in red. (Upper) Colocalization in an interphase nucleus; (Lower) colocalization in a metaphase spread. (B) The regions of the Lmbr1 gene covered by the two PAC clones (RPCI21 542N10 and 576B18) isolated. The position of the intron in the Lmbr1 gene (exon/intron gene structure from human sequence) and the intronic repeat structure (boxes) derived from the complete sequence between exons 5 and 6 is shown. Unique sequence DNA was predicted and four probes were generated (positions shown below intronic structure numbered 1–4). The positions of transgene integration (arrows with asterisks) and the area of intron duplication (below asterisks) are indicated. The resolved structure after integration is shown (bottom line), indicating intronic duplication as defined by probes 1–4. The positions of the λ and cosmid genomic clones (lines underneath) that defined the integration sites are marked. (C) RT-PCR samples were removed after 25, 30, and 35 cycles. RNA is from 11.5-day embryos of wild-type (+) and Ssq/Ssq (S) genotypes; a no RNA control (0) is also included. Top and Middle make use of primer pairs specific to Lmbr1; the first is from exon 3 to exon 5 and examines expression levels 5′ of the transgene integration. Comparable levels are detected in wild-type and Ssq/Ssq embryos. Middle uses primers that cross the integration site spanning exons 3 and 6. With these, at 30 cycles a product is only detected in the wild-type sample. The primers used in this experiment cross-hybridize with the mouse glycosylphosphatidylinositol-anchored protein homologue (closed arrowhead), which serves as positive control for the experiment and to normalize the samples. (Bottom) primers from the HPRT locus were used as positive control for RNA integrity and controls for reverse transcription.
Subsequently, a cosmid library was made from Ssq genomic DNA and additional clones were isolated to more fully elucidate the integration site. The transgene/genomic junction from the other end of transgenic integration was identified (Fig. 3B). Comparison of sequence from this cosmid clone with the complete intron 5 sequence we additionally assembled defined the position of this integration. Multiple copies of the transgene have integrated, lying wholly within intron 5 of the Lmbr1 gene (genomic sites of insertion are marked by asterisks in Fig. 2B). The process of integration has resulted in a duplication of approximately 20 kb of the intron with the transgene residing between the duplication endpoints (lower line in Fig. 2B). The duplication was confirmed by Southern blot analysis using unique sequence probes (probes numbered 1–4 in Fig. 2B). The surrounding DNA including exons 5 and 6 was apparently unaffected by the integration.
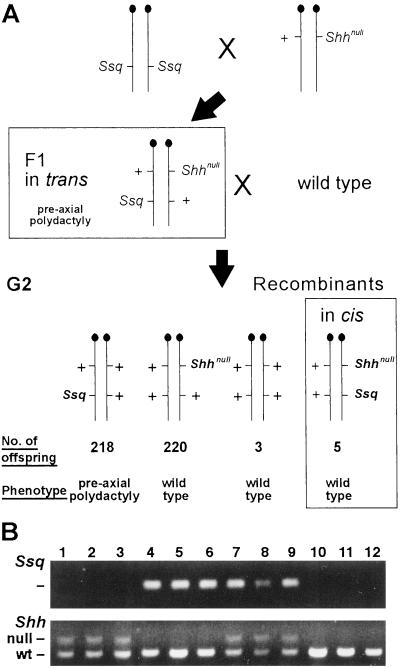
Figure 3.
Cis–trans genetic test. A shows the genetic cross used. A homozygous Ssq mouse was crossed to a Shhnull heterozygote. The F1 mice that carry both mutations on opposite chromosomes, all of which exhibit preaxial polydactyly, were crossed to wild-type mice. The bottom row depicts the outcome of the cross. The majority of G2 progeny were, as expected, found to be heterozygous for either the Ssq allele or the Shhnull (218 and 220 mice, respectively). The mice of interest were those identified as carrying a chromosome 5 that has recombined between the Shh locus and the region of the Ssq insertion. Three had inherited the wild-type alleles at both loci and five carried the Ssq allele and the Shhnull allele in cis. These mice show no additional digits and have wild-type paws. This cross results in a genetic distance of 1.8 cM between Shh and Ssq. B shows examples of the genotyping PCRs used to analyze the G2 progeny. Lanes 1–3 are mice heterozygous for the Shhnull allele. Lanes 4–6 are wild type for Shh but carry the Ssq insertion. Mice in lanes 7–12 carry the recombinant chromosomes; 7–9 show G2s with both Shhnull and Ssq, whereas 10–12 are wild type at both loci.
The consequence of the Ssq transgenic insertion on Lmbr1 transcription was analyzed. The transgene containing Lmbr1 gene encodes full-length transcripts, further supporting the proposal that arrangement of exons is unaffected by the insertional mutation. However, Lmbr1 transcription in Ssq embryos does show a high level of premature termination. The 5′ end of the mutant Lmbr1 transcript (Fig. 2C Top) is produced at wild-type levels, but transcription of exons 3′ of the insertion site is detectably lower, representing about 10–30% of wild-type levels (Fig. 2C Middle). The transgene insertion apparently leads to termination of transcription between exons 5 and 6.
Furthermore, we investigated for possible premature transcriptional termination in the familial PPD patients. We amplified LMBR1 transcripts by reverse transcription (RT)-PCR from RNA from lymphoblastoid cell lines of Dutch and Cuban patients, and sequenced the products. Polymorphisms in a Cuban (in exons 13 and 17) and two related Dutch (one in exon 5 and the other in exon 14) patients indicated that both LMBR1 alleles, at the 5′ end and the 3′ end, were capable of being expressed. Furthermore, an overlapping series of three PCR products covering the whole LMBR1 transcript showed no detectable difference in band size between patients and controls (data not shown), indicating that no exons are skipped in patients. These data argue that mutations disrupting LMBR1 transcriptional capacity are not commonly associated with the PPD phenotype.
Cis–Trans Genetic Analysis.
We previously showed (9) an intriguing relationship between the Ssq mutation and Shh expression. The reporter gene contained within the Ssq transgene insertion has acquired a limb-specific, Shh-like expression pattern; not only in the appropriate posterior ZPA (15), but also at an ectopic site at the anterior margin of the limb bud. It is unclear from the present data whether the mechanism for Shh misexpression involves disruption of the Lmbr1 gene and is therefore indirect or, as suggested (9), direct because of disruption of a long range Shh regulatory element. Long range regulation of Shh has been previously postulated for a translocation identified in a holoprosencephaly patient in which the breakpoint resides 250 kb upstream (16).
To examine the basis for preaxial polydactyly we devised a cis–trans genetic test. We predicted that an Shh regulator would function in a cis-acting manner—i.e., that the Ssq mutation would affect only chromosomally linked Shh. In contrast, disruption of a gene that secondarily affected Shh expression would operate on both Shh alleles and, therefore, in trans. A mouse cross was devised to derive a recombinant chromosome 5 in which the Ssq mutation was located in cis to an easily distinguishable Shh allele (Fig. 3A). Toward this end, we crossed Ssq/Ssq male mice with females carrying the mutant Shh null allele (Shhnull) (13) to produce the F1 generation. The Shhnull allele provided a straightforward assay for both the genotype (Fig. 3B) and the phenotype of each offspring. In the analysis of the phenotype, mice carrying the recombinant chromosome would exhibit extra preaxial toes if Ssq is acting on Shh in trans and wild-type feet—i.e., suppression of the Ssq phenotype—if acting in cis.
The F1 generation (n = 14) carrying Ssq and Shhnull on opposing chromosomes showed complete penetrance of the Ssq mutant phenotype. (Shhnull shows no phenotype in the heterozygous state.) The second generation (the G2) was produced by mating the F1 mice to wild type with the aim of generating the recombinant chromosome 5. Each G2 mouse was assessed by PCR for the presence of the Ssq transgene insertion (Fig. 3B Upper) and the neor gene associated with Shhnull (Fig. 3B Lower). In this cross we produced 446 G2 offspring of which eight were recombinants, representing a recombination frequency of 1.8 ± 0.6%. Three of the recombinants were uninformative because they contained the wild-type alleles (Fig. 3A). The other five recombinants carried both the Ssq insertion and Shhnull allele situated in cis. Analysis of the limb phenotype in these five recombinants showed no preaxial polydactyly or other detectable limb phenotypes. In addition, we bred two males carrying the recombinant chromosome 5 (Ssq and Shhnull) with wild-type females and showed that the next generation had no limb abnormalities (21 mice carrying the recombinant chromosome of 37 offspring). On some genetic backgrounds the Ssq heterozygous mutation is not fully penetrant. In this cross 7% of the Ssq/+ mice generated showed no detectable limb phenotype. Two of these Ssq/+ males, which displayed no limb phenotype, were bred further and were shown to transmit the phenotype (7 with additional preaxial digits of 19 offspring). Thus the data demonstrate that the Shh null allele inactivates the effects of the Ssq mutation when located in cis (but not in trans) on chromosome 5. It follows that the Ssq insertion is a dominant acting mutation that interferes with the limb-specific expression of Shh. The data are consistent with a long range limb-specific regulator of Shh residing within or near the Lmbr1 gene.
Discussion
The mapping location of human PPD to a subtelomeric region on 7q36 is well established and the condition has been referred to as both triphalangeal thumb-polysyndactyly (MIM190605) and preaxial polydactyly type II (MIM174500). Heus et al. (5) defined an approximate 400-kb critical region and identified potential genes. One of these genes originally given the genetic designation C7orf2 (5) is the homologue of a gene identified in mouse as Lmbr1 (14).
We show that the breakpoint for a reciprocal t(5,7)(q11,q36) chromosomal translocation in a patient with PPD resides in intron 5 of the LMBR1 gene. In addition we show that the polydactylous Ssq mutation results from a transgene insertion that resides in the corresponding intron of the mouse Lmbr1. These mutational events show that LMBR1/Lmbr1 is central to the generation of extra preaxial digits. Initially, we hypothesized that these mutations would potentially generate truncated LMBR1 protein products with corresponding deleterious effects. Indeed reverse transcription (RT)-PCR analysis of the mouse Lmbr1 transcript showed that the transgene insertion interfered with full-length transcript production. A systematic mutational analysis of PPD families, however, raised doubts that disruption of the LMBR1 protein product was a common cause for the manifestation of PPD. Analysis of lymphoblastoid cell lines from PPD patients revealed no further evidence of truncated LMBR1 transcripts and a comprehensive analysis of the coding region showed no pathogenic mutations.
To determine the mechanism responsible for the production of polydactyly from this locus, we relied on the Ssq mouse for further analysis. Previously we showed that, as occurs in several other preaxial polydactyly mutations in the mouse (17–19), Shh is mis-expressed in an ectopic anterior site in the limb bud directly correlating with extra digits. Interestingly, the expression pattern of the reporter gene contained within the Ssq transgene reflects limb Shh expression in both the normal posterior and the ectopic anterior site (9). These data are consistent with the Ssq locus directly affecting the expression of Shh; the Ssq mutation affecting a gene that resides at least 800 kb (1.6 cM) away. To examine this possibility, we performed a genetic cis–trans test and showed that indeed the Ssq mutation is suppressed in cis by the Shh null mutation. Thus, the data support the hypothesis that long-range disruption of Shh regulation is the mechanism for limb dysgenesis in Ssq and, furthermore, underlies the generation of extra toes in PPD.
In addition to PPD, acheiropodia (20, 21) and complex polysyndactyly (CPS) (7), two other limb-specific defects with distinctly different characteristics, map to the chromosome 7q36 region. Little is known about the molecular etiology of CPS; however, a recent report showed that the autosomal recessive acheiropodia (6) results from a small deletion within the LMBR1 gene. Acheiropodia is a severe limb-specific phenotype and, in contrast to PPD, patients present loss of all bones of the hands and feet, and the tibia is truncated distally and the radius, ulna, and fibula are lost. Affected individuals (who carry the same ancestral haplotype) show deletions in both LMBR1 alleles that remove exon 4 and ≈5–6 kb of surrounding genomic DNA. These data suggest that acheiropodia results from loss of function of the Lmbr1 gene (6). Based on our hypothesis that an Shh regulator resides in the Lmbr1 gene, we propose an alternative hypothesis. The ≈5-kb acheiropodia deletion from within the Lmbr1 gene not only removes exon 4 but may remove surrounding cis-acting regulatory elements essential for limb-specific Shh activity. There are no mouse models for acheiropodia; however, analysis of the limb in the Shh targeted deletion has been reported (22, 23). In accord, although the overall mouse phenotype is complex and severe, in Shh−/− mutant mice the limbs show loss of all bones of the feet and truncations of the long bones. The limb phenotype is similar to that seen in the acheiropodia patients. We suggest that, in contrast to Ssq and PPD, acheiropodia results from a limb-specific loss of Shh expression.
We propose a model (Fig. 4) in which Shh limb-specific regulatory elements arose as a discrete unit and reside at a considerable distance in the Lmbr1 gene. This regulator must detect the target gene at a considerable distance. Depending on the mode of perturbation in this region, the regulatory element is either inactivated as in the recessive acheiropodia condition or modified leading to misexpression as in the Ssq insertion. This model suggests that an element, normally a part of the regulatory domain, drives expression in both the anterior and the posterior limb regions. The anterior expression is actively repressed and it is disruption of this repression that causes PPD. Because PPD is relatively common in the human population (1), it seems likely that a number of independent mutations can interrupt the regulatory element, allowing mis-expression.
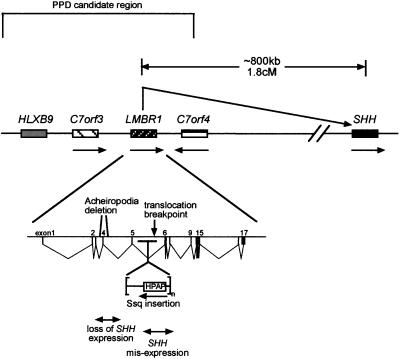
Figure 4.
Diagram of the human PPD critical region on chromosome 7q36 and the relationship with Shh. The corresponding region in mouse contains Lmbr1 surrounded by the orthologues of C7orf3 and C7orf4 and is ≈800 kb from Shh. No other genes are known to lie between Shh and the PPD critical region, suggesting that this is a gene poor region. The arrows indicate the transcriptional orientation. The structure of the 17 exon containing LMBR1 gene is depicted at the bottom and shows the relative position of each mutation. HPAP is the human placental alkaline phosphatase reporter gene contained within the transgene construct that resides at the Ssq insertion site. The double-headed arrows indicate the gene regions responsible for the opposing acheiropodia and PPD (Ssq) phenotypes. It is predicted that mutations between exons 5 and 6 release normal repression of the ectopic anterior expression pattern, whereas deletions around exon 4 inactivate limb-specific expression.
A second polydactylous mouse mutation hemimelic extra toe (Hx) was mapped to the same chromosomal region as the Ssq insertion site, and in accord, Lmbr1 was identified as a candidate gene (14). As in the PPD families, no causal mutation in the coding region was identified for Hx. Recent reports (14, 24) suggest that the Lmbr1 gene itself has a role in Hx. Both Ssq (9) and Hx (17) effect anterior mis-expression of Shh in the early limb bud; thus, the prospect exists that Ssq and Hx are acting by different mechanisms to mis-regulate Shh. If so, these two mutations together may provide insights into a complex locus affecting Shh expression and controlling the pattern of digits in the limb bud. However, a simpler possibility remains that Hx and Ssq are allelic; further genetic evidence is required to determine this.
We have shown that mutational events can have long-range affects. In most cases such a genetic distance would argue against analysis of a gene so far removed from the mutation and would lead to the analysis of the nearest gene carrying the insertion or the translocation breakpoint. In the case of Ssq, however, the reporter gene expression hinted that a Shh regulatory element may be the basis for polydactyly, which we confirm here. This raises the prospect that other situations exist in which genetic analysis has lead to interest in a tightly linked but unaffected gene.
Acknowledgments
We thank Kazutoya Osoegawa and Pieter de Jong and the U.K. Human Genome Mapping Project (HGMP) resource for providing the RPCI21 PAC library and Dr. John West for critically reading the manuscript. Thanks to Muriel Lee for the mouse FISH analysis; V. Ranaldi, L. McKie, and W. Mungel for expert technical assistance; and Sandy Bruce for photographic expertise. We also thank Dr. C. Hayes and P. Glenister (Medical Research Council Mammalian Genetics Unit, Harwell, U.K.) for supplying the Shh mutant mice. Finally, we thank Prof. Dr. Hans Galjaard and the “Stichting Klinische Genetica Rotterdam” for their continuous support. This study was partially supported by grants from the Japan Society for Promotion of Science (Research for the Future, to S.N. and T.H.), the Ministry of Education, Sports, and Culture (to S.N.), and the Netherlands Organization for Scientific Research (NWO) (to P.H.).
Abbreviations
- PPD
preaxial polydactyly
- FISH
fluoreqscence in situ hybridization
References
- 1.Heutink P, Zguricas J, van Oosterhout L, Breedveld G J, Testers L, Sandkuijl L A, Snijders P J, Weissenbach J, Lindhout D, Hovius S E, et al. Nat Genet. 1994;6:287–292. doi: 10.1038/ng0394-287. [DOI] [PubMed] [Google Scholar]
- 2.Hing A V, Helms C, Slaugh R, Burgess A, Wang J C, Herman T, Dowton S B, Donis-Keller H. Am J Med Genet. 1995;58:128–135. doi: 10.1002/ajmg.1320580208. [DOI] [PubMed] [Google Scholar]
- 3.Radhakrishna U, Blouin J L, Mehenni H, Patel U C, Patel M N, Solanki J V, Antonarakis S E. Am J Hum Genet. 1997;60:597–604. [PMC free article] [PubMed] [Google Scholar]
- 4.Zguricas J, Heus H, Morales-Peralta E, Breedveld G, Kuyt B, Mumcu E F, Bakker W, Akarsu N, Kay S P, Hovius S E, et al. J Med Genet. 1999;36:32–40. [PMC free article] [PubMed] [Google Scholar]
- 5.Heus H C, Hing A, van Baren M J, Joosse M, Breedveld G J, Wang J C, Burgess A, Donnis-Keller H, Berglund C, Zguricas J, et al. Genomics. 1999;57:342–351. doi: 10.1006/geno.1999.5796. [DOI] [PubMed] [Google Scholar]
- 6.Ianakiev P, van Baren M J, Daly M, Toledo S, Cavalcanti M G, Neto J C, Silveira E L, Freire-Maia A, Heutink P, Kilpatrick M W, Tsipouras P. Am J Hum Genet. 2001;68:38–45. doi: 10.1086/316955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukurov O, Boehmer A, Flynn J, Nicolai J P, Hamel B C, Traill S, Zaleske D, Mankin H J, Yeon H, Ho C, et al. Nat Genet. 1994;6:282–286. doi: 10.1038/ng0394-282. [DOI] [PubMed] [Google Scholar]
- 8.Dundar M, Gordon T M, Ozyazgan I, Oguzkaya F, Ozkul Y, Cooke A, Wilkinson A G, Holloway S, Goodman F R, Tolmie J L. J Med Genet. 2001;38:304–309. doi: 10.1136/jmg.38.5.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharpe J, Lettice L, Hecksher-Sorensen J, Fox M, Hill R, Krumlauf R. Curr Biol. 1999;9:97–100. doi: 10.1016/s0960-9822(99)80022-0. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi E, Hori T, O'Connell P, Leppert M, White R. Hum Genet. 1990;86:14–16. doi: 10.1007/BF00205165. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Nehls M, Pfeifer D, Micklem G, Schmoor C, Boehm T. Curr Biol. 1994;4:983–989. doi: 10.1016/s0960-9822(00)00222-0. [DOI] [PubMed] [Google Scholar]
- 13.Chiang C, Litingtung Y, Lee E, Young K E, Corden J L, Westphal H, Beachy P A. Nature (London) 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 14.Clark R M, Marker P C, Kingsley D M. Genomics. 2000;67:19–27. doi: 10.1006/geno.2000.6225. [DOI] [PubMed] [Google Scholar]
- 15.Pearce R V, Tabin C J. J Exp Zool. 1998;282:677–690. [PubMed] [Google Scholar]
- 16.Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, Mitchell H F, Donis-Keller H, Helms C, Hing AV, et al. Nat Genet. 1996;14:353–356. doi: 10.1038/ng1196-353. [DOI] [PubMed] [Google Scholar]
- 17.Masuya H, Sagai T, Wakana S, Moriwaki K, Shiroishi T. Genes Dev. 1995;9:1845–1653. doi: 10.1101/gad.9.13.1645. [DOI] [PubMed] [Google Scholar]
- 18.Masuya H, Sagai T, Moriwaki K, Shiroishi T. Dev Biol. 1997;182:42–51. doi: 10.1006/dbio.1996.8457. [DOI] [PubMed] [Google Scholar]
- 19.Lettice L, Hecksher-Sørenson J, Hill R E. Development (Cambridge, UK) 1999;126:4729–4736. doi: 10.1242/dev.126.21.4729. [DOI] [PubMed] [Google Scholar]
- 20.Toledo S P A, Saldanha P H. J Genet Hum. 1972;20:253–258. [PubMed] [Google Scholar]
- 21.Fett-Conte A C, Richiere-Costa A. Am J Med Genet. 1990;36:341–344. doi: 10.1002/ajmg.1320360320. [DOI] [PubMed] [Google Scholar]
- 22.Chiang C, Litingtung Y, Harris M P, Simandl B K, Li Y, Beachy P A, Fallon J F. Dev Biol. 2001;236:421–435. doi: 10.1006/dbio.2001.0346. [DOI] [PubMed] [Google Scholar]
- 23.Kraus P, Fraidenraich D, Loomis C A. Mech Dev. 2001;100:45–58. doi: 10.1016/s0925-4773(00)00492-5. [DOI] [PubMed] [Google Scholar]
- 24.Clark R M, Marker P C, Roessler E, Dutra A, Schimenti J, Muenke M, Kingsley D M. Genetics. 2001;159:715–726. doi: 10.1093/genetics/159.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]