Abstract
Polycystic ovary syndrome (PCOS) is one of the most complicated chronic inflammatory diseases in women of reproductive age and is one of the primary factors responsible for infertility. There is substantial dispute relating to the pathophysiology of PCOS. Consequently, there is a critical need for further research to identify the factors underlying the pathophysiology of PCOS. Three transcriptome profiles of granulosa cells from patients with PCOS and normal controls were obtained from the gene expression integration database. We also obtained relevant microarrays of granulocytes prepared from PCOS patients and normal controls from the gene expression integration database. Then, we used the R package to perform correlations and identify differences between PCOS and normal controls with regard to immune infiltrating cells and functionality. Subsequently, intersecting genes were identified and risk models were constructed. Finally, the results were validated by enzyme linked immunosorbent assay and real-time PCR. We identified 8 genes related to cuproptosis (SLC31A1, PDHB, PDHA1, DLST, DLD, DLAT, DBT, and ATP7A) and 5 genes related to m7G (SNUPN, NUDT16, GEMIN5, DCPS, and EIF4E3) that were associated with immune infiltration. Furthermore, the expression levels of DLAT (P = .049) and NUDT16 (P = .024) differed significantly between the PCOS patients and normal controls, as revealed by multifactorial analysis. Both DLAT and NUDT16 were negatively correlated with immune cell expression and function and expression levels were significantly lower in the PCOS group. Finally, real-time PCR and enzyme linked immunosorbent assay demonstrated that the expression levels of DLAT and NUDT16 were significantly reduced in the granulosa cells of PCOS patients. In conclusion, our findings shed fresh light on the roles of immune infiltration, cuproptosis, and m7G alternations in PCOS. We also provide a reliable biomarker for the pathological classification of PCOS patients.
Keywords: bioinformatics, biomarkers, cuproptosis, immune infiltration, m7G methylation, polycystic ovary syndrome (PCOS)
1. Introduction
Polycystic ovary syndrome (PCOS) affects 5% to 10% of women of reproductive age worldwide and is one of the most common endocrine disorders with clinical manifestations of oligo-ovulation, anovulation, and polycystic ovarian changes. According to some studies, PCOS is an inflammatory disease that exerts effects on ovulation and the ovarian/follicular microenvironment.[1–4] Patients with PCOS have considerably higher serum levels of IL-6, natural killer cells, leukocytes, and the levels of IL-1β and IL-18 in follicular fluid than healthy individuals.[4–8] The inflammatory response of PCOS also exerts significant impact on the proliferation and apoptosis of granulosa cells (GCs).[9] However, few studies have investigated the mechanisms of inflammation in PCOS and there is a clear need to further investigate the specific characteristics of inflammation in PCOS. Recent research discovered that excessive amounts of copper can stimulate protein acylation in the tricarboxylic acid (TCA) cycle, and that this represents a novel trigger of cell death.[10] The TCA cycle is a biochemical pathway that is commonly found in aerobic organisms which is the center of lipid, amino acid, and carbohydrate metabolism and plays an important role in regulating metabolism, cell function, and cell fate.[11] Patients with PCOS exhibit problems with the TCA cycle, glucose, and lipid metabolism.[12] Furthermore, the TCA cycle plays an important role in the proliferation and death of GCs and immune cells, as well as immunological function. Disruption of the TCA cycle results in abnormal amino acid and lipid metabolism, which can lead to accumulation of citric and succinic acids in macrophages, which can promote immune function.[13] Furthermore, lipid oxidation within the TCA cycle is critical for the activity and lifespan of CD8 + T and Treg cells.[14] Previous research found that PCOS patients with a high incidence of cyst formation possessed GCs containing fewer TCA metabolites in their mitochondria, thus resulting in reduced levels of inflammation, thereby increasing cyst rates.[15] Copper ions are intimately associated with the development of PCOS patients who exhibit increased levels of copper in the serum and follicular fluid.[16–18] Collectively, these studies reported evidence that copper ions may have an effect on the development of PCOS and immune infiltration.
In patients with PCOS, low-grade chronic inflammation may also be closely associated with m7G. Studies have increasingly reported that RNA methylation plays a significant role in the functionality of immune cells.[19,20] N7 methyladenosine, also known as m7G RNA methylation, occurs at specific internal sites in tRNA and rRNA molecules in all areas of life. In addition to preventing RNA degradation, m7G RNA methylation also induces RNA processing, modification, and translation factors.[21–23] The activation of T cells can also be triggered by m7G. RNA polymerase (RNMT), also known as m7G-cap methyltransferase, is known to play an important role in mediating T-cell activation and specifically controls the synthesis of ribosomes.[24] The catalytic subunit of m7G methyltransferase, wh, is required for the production of m7G; this is a homolog of Trm82 and wh variants are known to result in the abnormal growth of oocytes.[25,26] Trm82 encodes an essential subunit of the tRNA-encoding gene methyltransferase.[27] Abnormal oocyte quality and a chronic low inflammatory reaction are typical symptoms of PCOS patients. GCs reflect the characteristics of oocytes and are a noninvasive means of evaluating oocyte quality. However, there is a lack of research on the effect of m7G on GCs. Therefore, it is necessary to further explore the effect of m7G on GCs in patients with PCOS.
In summary, immunological response, cuproptosis, and m7G modification, are all known to play important roles in the development of PCOS. Moreover, the etiologies that lead to PCOS are complex and diverse; furthermore, immune factors, cuproptosis, and m7G are all known to exert important effects on the development of PCOS. Therefore, in this study, we investigated key biomarkers that can influence immune factors, cuproptosis, and m7G from multiple perspectives through bioinformatic analysis. Our findings may provide new strategies to study the mechanisms of PCOS and develop new strategies for PCOS.
2. Methods
2.1. Data extraction
Original datasets that compared the gene expression profiles of GCs between PCOS patients and normal controls were downloaded from the gene expression omnibus (GEO) database (Table 1).
Table 1.
The basic backgrounds of the datasets included.
| Platforms | Country | Year | NP | NC | |
|---|---|---|---|---|---|
| GSE34526 | [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | China | 2011 | 7 | 3 |
| GSE80432 | [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array | China | 2021 | 8 | 8 |
| GSE137684 | Agilent-039494 SurePrint G3 Human GE v2 8 × 60K Microarray 039381 | China | 2019 | 8 | 4 |
Note: 1. Number of PCOS sample (NP); 2. number of control sample (NC).
The accession numbers of the 3 datasets were as follows: GSE34526 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE34526); GSE80432 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80432), and GSE137684 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137684). Microarray data from the GSE34526 dataset was based on the GPL570 ([HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array) while the GSE80432 dataset was based on GPL6244. The GSE137684 ([HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version]) was based on GPL17077 (Agilent-039494 SurePrint G3 Human GE v2 8 × 60K Microarray 039381); the chip contained 23 PCOS patients and 15 healthy controls. In the study, we investigated mRNA profiles in ovarian GCs from PCOS patients and healthy controls. The analytic workflow is shown in Figure 1.
Figure 1.
The analytic workflow of the present study.
2.2. Data preprocessing
The 3 raw datasets were preprocessed by the “limma” and “sva” packages in R (version 4.1.3). Preprocessing included background correction, normalization, and “log2” transformation. The receiver operating characteristic (ROC) curves are plotted using the R package “survival ROC” to verify the accuracy of the prediction model.
2.3. Immune infiltration analysis
First, we searched for correlations between the expression levels of hub genes and immune infiltration in PCOS. By applying ssGSEA in the R environment, we were able to analyze the infiltration of 29 immune cells and immune functions that were identified by GSEA (https://www.gsea-msigdb.org/gsea/index.jsp). The ssGSEA score was set to standard and visualized by heat maps generated by the “pheatmap” package in R. The “corrplot” package was then used to investigate the correlation between immune cells and regulators of immune cell functions. Differences between immune cells and immune cell functions between patients with PCOS and normal controls were visualized by the “ggpubr” package in R. Then, we analyzed the original data and applied a cutoff threshold of P < .05 and |logFC|>|0.5 for screening.
2.4. Bioinformatics analysis of curoptosis and m7G
We identified 29 m7G-related genes and 13 curoptosis-related genes from previous systematic reviews.[10,25,28] The correlations between the immune-related gene matrix, the curoptosis-repeated gene matrix, and the m7G-related gene matrix were analyzed by the “ggcorrplot” and “ROCR” packages in R. Then, we constructed a clinical prediction model and created a correlation heat map.
2.5. Functional enrichment analyses
Following data downloads, we applied the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://bioinfo.org/kobas/) and Gene Ontology Resource (GO) (https://david.ncifcrf.gov/tools.jsp). Differences were considered statistically significant at P < .05. Subsequently, the “ggplot2” tool in R was used to generate enrichment maps.
2.6. Experimental validation
The Sichuan Jinxin Xi’nan Women’s and Children’s Hospital Chengdu provided follicular fluid for further validation; in total, 3 patients with PCOS and 3 healthy volunteers provided signed and informed consent. Genes showing the largest changes in expression between the 2 groups were identified by real-time PCR (RT-PCR) and following centrifugation of the GCs in follicular fluid. This study was approved by the Ethics Committee of Chengdu Xi’nan Women’s Hospital.
2.6.1. Inclusion criteria
Patients were included if they were aged 20 to 35 years, needed artificial assisted reproductive technology and underwent an GnRH antagonist protocol. The PCOS group needed to meet the Rotterdam diagnostic criteria of PCOS,[29] including clinical and/or biochemical hyperandrogenism, polycystic ovaries, and/or oligo-/anovulation. For the normal controls, we selected patients with blocked fallopian tubes and normal ovarian function.
2.6.2. Exclusion criteria
Patients were excluded if they refused to provide informed consent or if they had diseases related to the cardiovascular, immune, or nervous system, or had diseases that affected the ovaries, such as cancer or cysts.
2.6.3. Granular cells extraction
Staff at the Sichuan Jinxin Xi’nan Women’s and Children’s Hospital Chengdu collected follicular fluid from eligible patients undergoing controlled ovulation induction. Samples were first centrifuged at 2000 rpm for 10 minutes to separate the supernatant. The remaining cell precipitate was prepared as a suspension with phosphate buffer solution, and the cell suspension was slowly added to the lymphocyte separation solution at a ratio of 1:1; then, the granular cell layer, containing a small amount of erythrocytes, was aspirated by centrifugation at 2000 r/min for 30 minutes, and the erythrocytes were washed and lysed with erythrocyte lysate; finally, the GCs were obtained by washing with phosphate buffer solution to remove the erythrocyte lysate.
2.6.4. Real-time PCR
Subsequently, total RNA was extracted from the GCs by the TRIzol method using reagents from Beijing Baori Medical Biotechnology Co., Ltd (Beijing, China). Total RNA was then reverse transcribed into cDNA using a template of 1 μg of RNA; the reaction conditions were 40 °C for 60 minutes, 25 °C for 5 minutes, and 75 °C for 5 minutes. GAPDH was utilized as an internal control. For PCR 1 μL of cDNA used as a template and the amplification settings were 95 °C for 3 minutes, 40 cycles at 95 °C for 5s, and 60 °C for 32s. The primers were as follows: NUDT16 (forward 5′- TCCCAATTTCCTCTTCTCCCAAAGC -3′, reverse 5′- GATCACGATCCCAACTCCACCAAG -3′), and DLAT (forward 5′- GGGTTATTGCACAGCGATTAAT -3′, reverse 5′- GAAGAATTTGCTTCGGGAACTT -3′). After amplification, the relative expression levels were determined by the 2‐ΔΔCt method.
2.6.5. Enzyme Linked Immunosorbent Assay
Before preparing for enzyme linked immunosorbent assay (ELISA), the supernatants from the follicular fluid of PCOS and normal groups were centrifuged and frozen at ‐80 °C. Subsequently, the concentrations of DLAT and NUDT16 in follicular fluid were measured by reference to the ELISA kit for human DLAT and NUDT16 (Hefei Bomei Biotechnology Co., Ltd., Hefei, China) method at a measurement wavelength of 450 nm, and a reference wavelength of 550 nm.
2.7. Statistical analysis
All statistical analyses were performed in the R Programming language. All statistical tests were two-sided and P < .05 was considered statistically significant.
3. Results
3.1. Integration of corrected genes and immune infiltration analysis
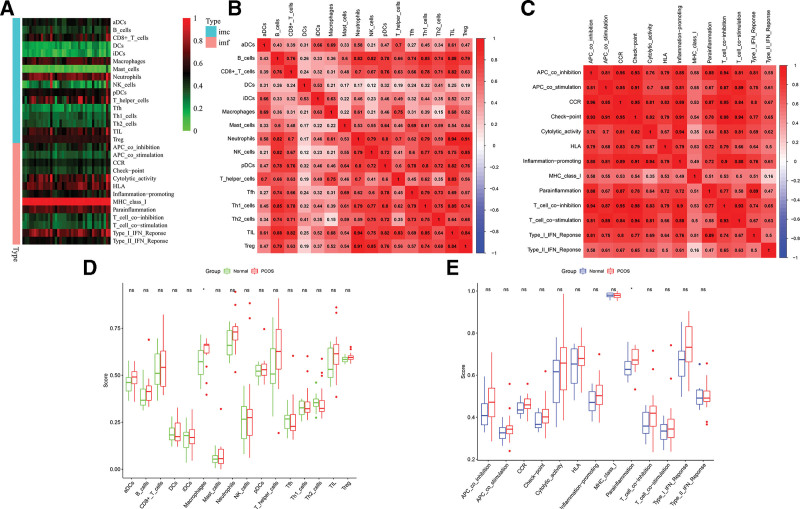
The “limma” and “sva” packages in R were employed to consolidate relevant data from GSE34526, GSE80432, and GSE137684 datasets into a new dataset. Subsequently, immune cells and immune functions were correlated with corrected genes using the “ssGSEA” package, as depicted in the heatmap (Fig. 2A). The analysis revealed heightened expression of CD8 + T cells, macrophages, neutrophils, T helper cells, tumor-infiltrating lymphocytes, and Treg among immune cells, indicating a pronounced immune response in PCOS patients. Additionally, immune functions such as cytolytic activity, human leukocyte antigen, major histocompatibility complex type I, parainflammation, and type I interferon response exhibited increased prominence. Correlation analysis using the “corrplot” package in R demonstrated positive associations among all immune cells, indicating synergistic effects with the activation of immune cells in PCOS patients. Notably, neutrophils exhibited the most significant correlation with Treg cells and tumor-infiltrating lymphocytes (correlation coefficient > 0.9), highlighting their potential role in modulating immune responses in PCOS. There were evident correlations in various aspects, such as APC co-inhibition and CCR, checkpoint, T cell co-dependence of aggression; APC co-stimulation and checkpoint relevance; checkpoint and CCR, inflammation-promoting, T cell co-inhibition, T cell co-directional correlation of simulation; inflammation-promoting and cytologic correlation of activities; CCR and T cell co-stimulation, T cell co-stimulation (correlation coefficients > 0.9) (Fig. 2B and C).
Figure 2.
Immunoinfiltration analysis. (A) Classification heat map of immune cells and immune function. (B) Immunocyte correlation analysis. (C) Correlation analysis of immune function. (D) Difference of immune cell expression between PCOS group and normal group. (E) Difference of immune function between PCOS group and normal group. (1 Immune cells (imc); 2 immune function (imf); 3 ns. P ≥ .05, *P < .05; **P < .01; ***P < .001.)
Upon comparing immune cell expression and immune function between the PCOS and normal groups, we observed a significantly higher expression of macrophages in the PCOS group (P < .05). Additionally, parainflammation in the PCOS group was significantly elevated, indicating the prevalence of long-term low-grade inflammation in this group. This may represent a crucial immunological mechanism contributing to PCOS (Fig. 2D and E).
3.2. Correlation analysis between differential genes and immune infiltration
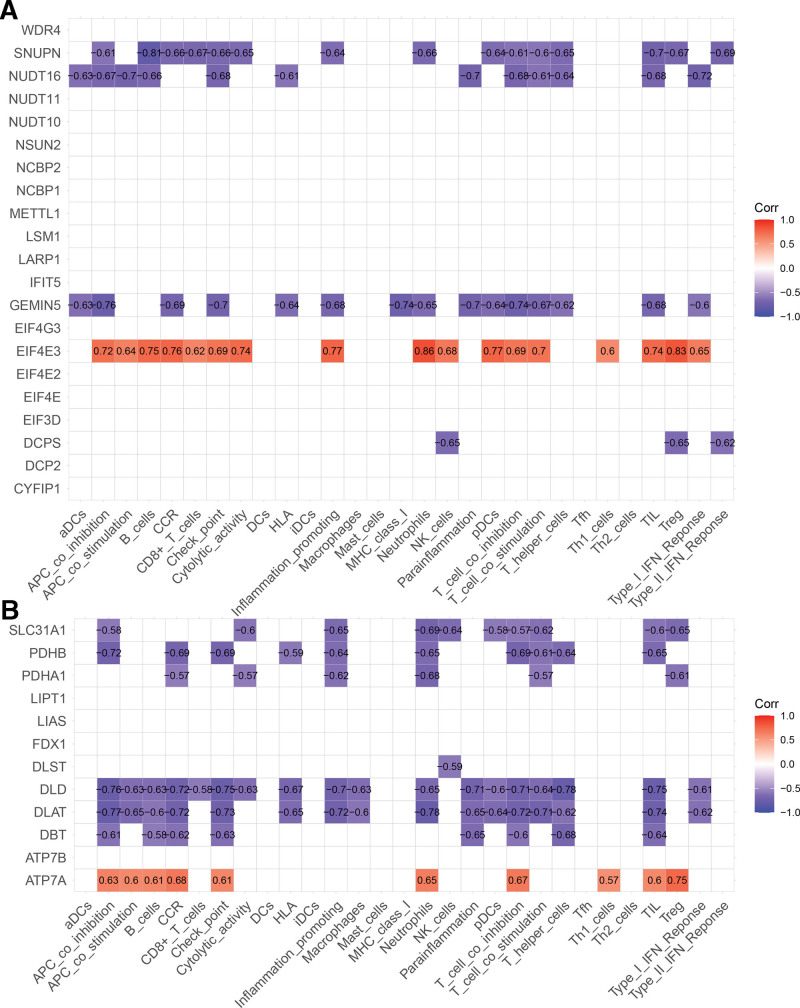
Using the “ggcorrplot” package in R, we analyzed correlations between cuproptosis-related genes, m7G-related genes, and immune-related genes. Ultimately, 8 cuproptosis-related genes correlated with immune-related genes (CIGs), and 5 m7G-related genes correlated with immune-related genes (MIGs) were identified (P < .05). SNUPN, NUDT16, GEMIN5, and DCPS exhibited negative correlations with immune cells and immune function within the MIGs, while EIF4E3 displayed a positive correlation (Fig. 3A). In the CIGs, SLC31A1, PDHB, PDHA1, DLST, DLD, DLAT, DBT, and ATP7A showed negative correlations, while ATP7A exhibited a positive correlation (Fig. 3B).
Figure 3.
Correlation analysis with immune cells and immune function. (A) Correlation analysis of m7G-related genes with immune cells and immune function. (B) Correlation analysis of cuproptosis-related genes with immune cells and immune function.
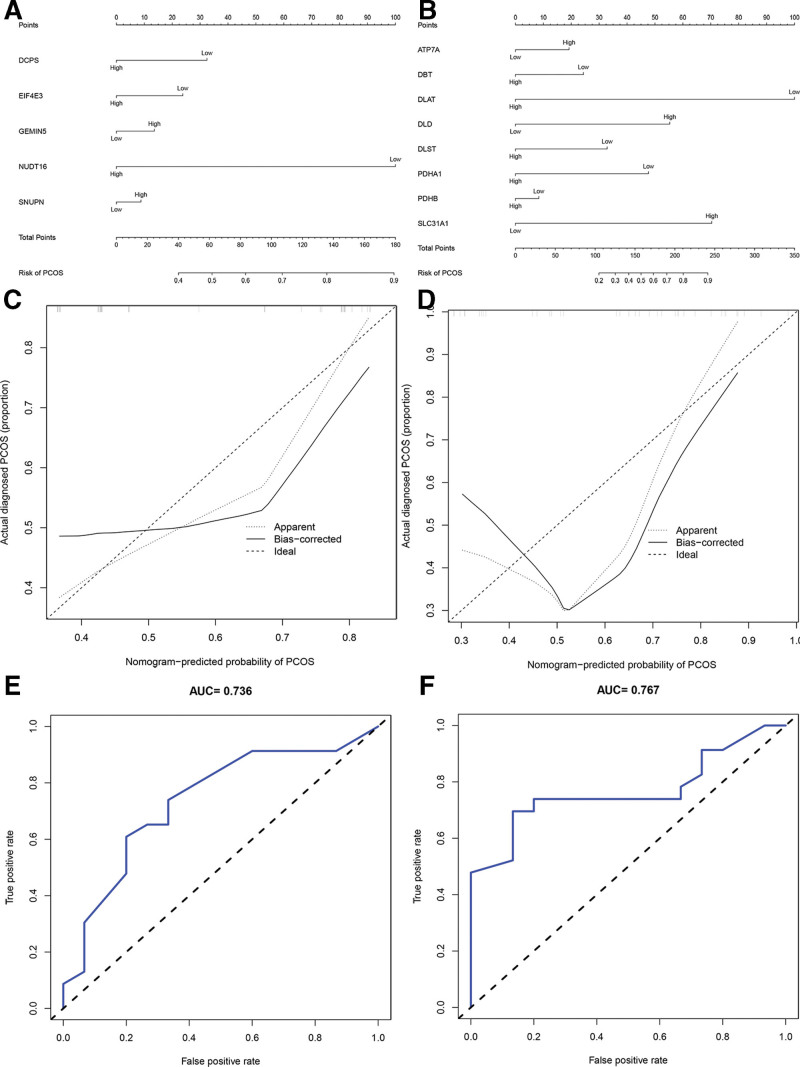
3.3. Development and validation of m7G-related genes and cuproptosis-related genes prognostic signature
Predictive models for PCOS were generated using the most significantly differentially expressed genes with correlation coefficients > 0.5 with immune cells and immune function. This included 5 MIGs (SNUPN, NUDT16, GEMIN5, DCPS, and EIF4E3) and 8 CIGs (SLC31A1, PDHB, PDHA1, DLST, DLD, DLAT, DBT, and ATP7A). ROC analysis demonstrated the predictive ability of MIGs and CIGs. The nomogram, incorporating various prognostic genes, assessed the probability of pathological factors acting individually. To predict the pathogenic factors and therapeutic targets of PCOS patients, a nomogram was constructed. Notably, DLAT and NUDT16 had the most notable impact on the probability of developing PCOS in a multifactorial correlation study. The median risk value of all genes was 1.5. Comparing the 2 groups, genes above the median expression value were considered highly expressed, while genes below the median expression value were considered expressed at low levels. The PCOS group was considered high risk, while the normal group was considered low risk. In the prognostic signature constructed by MIGs, the expression levels of NUDT16 were significantly different between the PCOS group and the normal group (P = .024). The odds ratio (OR) of NUDT16 was 0.15, indicating low expression in PCOS and high levels in normal controls. In the prognostic signature constructed by CIGs, DLAT showed a significant difference between the PCOS group and the normal group (P = .049). The OR of DLAT was 0.07, indicating low expression in PCOS but high levels in normal controls (Table 2).
Table 2.
The prognostic signature of coefficients and OR.
| Gene ID | Coefficients (P) | OR |
|---|---|---|
| DCPS | 0.533 | 0.55 |
| EIF4E3 | 0.693 | 0.64 |
| GEMIN5 | 0.769 | 1.29 |
| NUDT16 | 0.024* | 0.15 |
| SNUPN | 0.881 | 1.18 |
| ATP7A | 0.559 | 1.66 |
| DBT | 0.491 | 0.53 |
| DLAT | 0.049* | 0.07 |
| DLD | 0.299 | 4.27 |
| DLST | 0.311 | 0.42 |
| PDHA1 | 0.2011 | 0.29 |
| PDHB | 0.817 | 0.80 |
| SLC31A1 | 0.083 | 6.33 |
P < .05.
The nomogram effectively predicted the pathological factors in patients with PCOS. And we discovered that DLAT and NUDT16 had the most notable impact on the probability of developing PCOS in a multifactorial correlation study. Specifically, our analysis revealed a noteworthy association between the suppression of DLAT and NUDT16 expression and an increased likelihood of PCOS occurrence. These findings suggest that lower expression levels of DLAT and NUDT16 may correlate with a higher incidence of PCOS (Fig. 4A and B). The C index of the nomograms was 0.736 for the prognostic signature of MIGs and 0.767 for the prognostic signature of CIGs, thereby affirming the favorable predictive capability of the nomograms. Calibration curves demonstrate a close alignment between the observed pathologic factors in PCOS and the anticipated values (Fig. 4C and D). The area under the curve (AUC) was 0.767 for the prognostic signature featuring CIGs and 0.736 for the prognostic signature featuring MIGs, suggesting good prediction performances for each nomogram (Fig. 4E and F).
Figure 4.
Establishment of nomogram and ROC and their forecast performance. (A) Nomogram for predicting pathogenic factors of MIGs. (B) Nomogram for predicting pathogenic factors of CIGs. (C) Calibration curves of nomogram to ascertain the prediction of MIGs. (D) Calibration curves of nomogram to ascertain the prediction of CIGs. (E) ROC curve shows the risk of disease forecast probability through MIGs in detail. (F) ROC curve shows the risk of disease forecast probability through CIGs in detail.
4. Functional enrichment of analysis
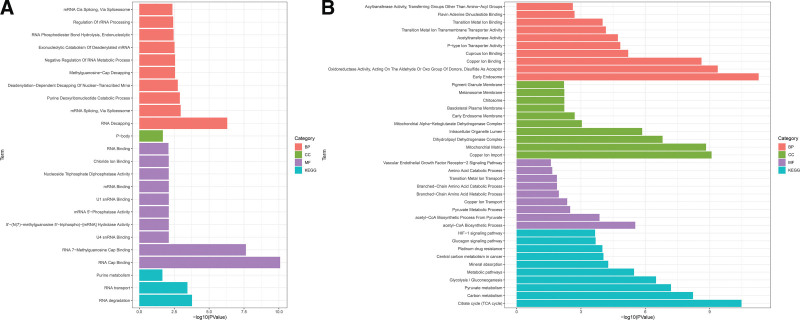
Next, we obtained functional enrichment information for the MIGs and CIGs by GO and KEGG enrichment analysis. The KEGG enrichment analysis highlighted significant enrichment of MIGs in key pathways, including RNA degradation, RNA transport, and purine metabolism. These pathways play essential roles in intracellular nucleic acid metabolism and RNA biology, crucial for maintaining normal cellular function and the physiological state of the organism. In terms of biological processes, a significant enrichment was observed in differential genes related to the regulation of translation. Additionally, the cellular components terms showed significant enrichment in P-bodies. Biological processes (BP) included RNA decapping, mRNA splicing via spliceosome, purine deoxyribonucleotide catabolic process, deadenylation-dependent decapping of nuclear-transcribed mRNA, methylguanosine-cap decapping, negative regulation of RNA metabolic process, exonucleolytic catabolism of deadenylated mRNA, RNA phosphodiester bond hydrolysis, endonucleolytic, regulation of rRNA processing, and mRNA cis splicing via spliceosome. Regarding molecular functions, significant enrichment was detected in RNA cap binding, RNA 7-methylguanosine cap binding, U4 snRNA binding, 5’-(N(7)-methylguanosine 5’-triphospho)-[mRNA] hydrolase activity, mRNA 5’-phosphatase activity, U1 snRNA binding, mRNA binding, nucleoside triphosphate diphosphatase activity, chloride ion binding, and RNA binding. The selected 5 genes were found to be associated with the regulation of translation and RNA biology, as indicated by the enrichment analysis of the MIGs (Fig.5A).
Figure 5.
GO analysis and KEGG analysis. (A) GO and KEGG analysis of MIGs. (B) GO and KEGG analysis of CIGs.
Regarding BP of CIGs, there was significant enrichment of differential genes in acetyl-CoA biosynthetic process, acetyl-CoA biosynthetic process from pyruvate, pyruvate metabolic process, copper ion transport, branched-chain amino acid metabolic process, branched-chain amino acid catabolic process, transition metal ion transport, amino acid catabolic process, vascular endothelial growth factor receptor-2 signaling pathway, and copper ion import. The significantly enriched molecular function included oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor, copper ion binding, cuprous ion binding, P-type ion transporter activity, acetyltransferase activity, transition metal ion transmembrane transporter activity, transition metal ion binding, flavin adenine dinucleotide binding, and acyltransferase activity, transferring groups other than amino-acyl groups. Enriched cellular components included the mitochondrial matrix, dihydrolipoyl dehydrogenase complex, intracellular organelle lumen, mitochondrial alpha-ketoglutarate dehydrogenase complex, early endosome membrane, basolateral plasma membrane, chitosome, melanosome membrane, pigment granule membrane, and early endosome. The CIGs were mainly enriched in the TCA cycle, carbon metabolism, pyruvate metabolism, glycolysis/gluconeogenesis, metabolic pathways, mineral absorption, central carbon metabolism in cancer, platinum drug resistance, glucagon signaling pathway, and HIF-1 signaling pathway (Fig. 5B). The KEGG pathways associated with the CIGs were predominantly related to the metabolism of sugars, amino acids, fats, and other nutrients, along with the TCA cycle pathway (Fig.5B).
5. The expression of potential biomarkers, as determined by RT-PCR and ELISA
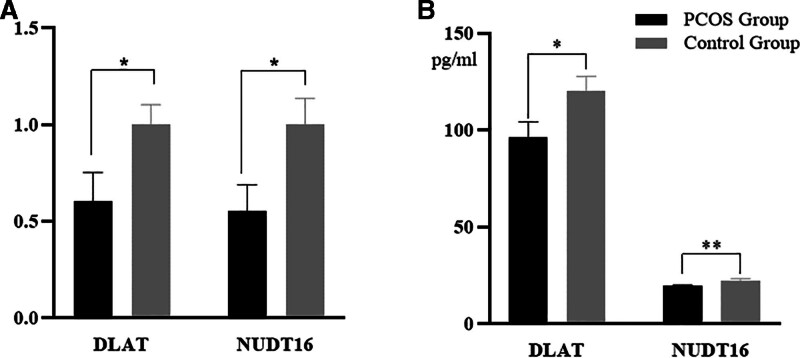
We discovered 5 genes that were strongly associated with m7G, immune cells, and immunological function using a clinical prediction model and identified 8 genes that were strongly connected with cuproptosis, immune cells, and immune function. Therefore, the development of PCOS disease is closely related to the metabolism of various substances, including sugars, amino acids, ribonucleic acids, and oxygen metabolism. DLAT and NUDT16 were identified by “ROCR” package as the genes showing the most significant differences of the 13 genes identified. DLAT (dihydrolipoamide acetyltransferase) is one of the lipid acylation products in cuproptosis that binds directly to copper.[10] Nudix hydrolase 16 (NUDT16) is a member of the NUDIX hydrolase family and is involved in cellular metabolism, homeostasis and mRNA processing; NUDT16 also hydrolyzes m7GppG.[30] Next, the filtered biomarkers, including DLAT and NUDT16, were verified in PCOS samples by RT-PCR and ELISA. The RT-PCR and ELISA results further showed that the expression levels of DLAT (P < .05) and NUDT16 (P < .05) in the PCOS group were significantly lower than those in the control group (Fig. 6).
Figure 6.
Experimental verification of NUDT16 and DLAT. (A) RT-PCR of NUDT16 and DLAT. (B) ELISA of NUDT16 and DLAT.
6. Discussion
To the best of our knowledge, this is the first research investigation of the immunological correlation between cuprotosis, m7G, and PCOS. Fifteen normal samples and 23 PCOS samples from GEO datasets were included in the immune infiltration analysis, and correlations between the cuproptosis-related genes, m7G-related genes, and immune-related genes were then performed. Next, we established 2 different clinical PCOS prediction models using gene clusters composed of MIGs and CIGs (AUC = 0.736 for MIGs and AUC = 0.767 for CIGs). By merging immune-related genes with m7G-related genes and cuproptosis-related genes, we demonstrated the reliability of our bioinformatics findings. We identified 8 immune invasion genes related to cuproptosis and 5 m7G-related immune invasion genes. DLAT and NUDT16 showed the most significant differences in terms of immune infiltration and m7G and cuproptosis, respectively, when compared between the PCOS and normal groups. RT-PCR further confirmed our analysis and showed that the expression levels of DLAT and NUDT16 were significantly lower in the PCOS group.
Immune infiltration analysis identified significant differences in macrophage expression and paraneoplastic response between the PCOS and normal groups. Macrophages play an important role in tissue development (by shaping tissue architecture), the immune response to pathogens (by generating and resolving inflammatory responses), the monitoring and surveillance of tissue changes (by acting as sentinels and effector cells), and especially in maintaining tissue homeostasis (by removing apoptotic or senescent cells, and remodeling and repairing tissue).[31] In addition, hyperandrogenemia in PCOS has been shown to induce chemin-mediated follicular growth arrest by modulating monocyte-macrophage migration and function.[32] Furthermore, macrophage recruitment is a key factor in maintaining paraneoplastic inflammation.[33] When a parainflammatory state or chronic low-grade inflammatory state predominates and this state is not properly controlled or resolved, the immune system’s anti-inflammatory response is similarly dysregulated and unable to suppress inflammatory episodes in a timely and effective manner.[34] In contrast, insulin resistance (IR) and obesity in PCOS can lead to the persistence of a chronic low-grade inflammatory state; the chronic inflammatory process is the most important evidence for the pathogenesis of PCOS.[35] This study further validated the effect of macrophages and parainflammation in PCOS by perfoming bioinformatic analysis of 3 different data sets.
We constructed a PCOS risk model using several genes: SLC31A1, PDHB, PDHA1, DLST, DLD, DLAT, DBT, and ATP7A; of these, DLAT may be the m7G gene with the greatest impact on PCOS. SLC31A1, also known as copper transporter protein 1, is a key determinant of intracellular copper distribution.[36] In reaction to bacterial infection, macrophages enhance the expression of SLC31A1, elevate the surface expression of copper transporter protein 1, and accumulate copper within their phagosomes through the involvement of ATP7A. This mechanism plays a pivotal role in the antimicrobial activity of macrophages, contributing to their effective eradication of pathogens.[37] PDHA1 and PDHB are subunits of the pyruvate dehydrogenase complex, both of which catalyze the conversion of pyruvate to acetyl coenzyme A.[38,39] Macrophage SIRT-3 deacetylation at lysine 83 activates PDHA1; this inhibits NLRP3 inflammatory vesicle activation and IL-1β release.[40] Acetyl coenzyme A promotes the expression of inflammatory cytokines by regulating histone acetylation. Deglycosylase DJ-1 connects to PDHB in Tregs and inhibits PDHA phosphorylation to maintain Treg cell differentiation and the functional integrity of T cells.[41] Some studies have confirmed the presence of T-cell and macrophage recruitment in the follicular fluid of PCOS patients and the ovarian tissue of a rat model of PCOS.[32,42] DLD, also known as dihydrolipoamide dehydrogenase, is an important gene that regulates lipoic acid metabolism.[43] Research has confirmed that alpha-lipoic acid reduces inflammatory vesicle activity and the levels of pro-inflammatory cytokines.[44] In addition, alpha-lipoic acid is useful for IR in PCOS.[45] DLST is 1 of the 3 subunits of the α-ketoglutarate dehydrogenase enzyme complex and is essential for maintaining the TCA cycle and cellular energy supply.[46] In CD11bGr1 myeloid cells, DLST exhibits immunosuppressive effects by attenuating T cell activity.[47] Previous research showed that DLAT induces inflammation in the intestine, causing effect on Th17 and also directing bacterial molecular mechanisms that alter the bacterial transcriptome to coordinate metabolomic cycles, further leading to inflammation generation.[48] Dihydrolipoamide branched-chain transacylase is the transacylase subunit of the BCKDH complex and the E2 subunit of the pyruvate dehydrogenase complex which is mainly involved in the TCA cycle and the glycolytic pathway.[49] Previous research showed that macrophages maintain active pyruvate flux via pyruvate dehydrogenase and that when pyruvate dehydrogenase flux is inhibited, macrophage expression is reduced.[50] Other research showed that a rat model of PCOS experienced the inhibition of pyruvate dehydrogenase complex.[51]
We constructed a PCOS risk model using NUDT16, SNUPN, GEMIN5, DCPS, and EIF4E3; of these, the m7G gene NUDT16 may be the gene with the greatest impact on PCOS. Recent research showed that NUDT16 mediates the specific degradation of Rift Valley fever virus mRNA.[52] NUDT16 has a potential role in mRNA degradation and the posttranslational regulation of inflammatory molecules during sepsis.[53] Furthermore, m7G-cap is known to be hypermethylated and imported into the adapter snurportin1 (SNUPN) to recognize m3G-cap; the recognition of snurportin1 by the m3G-cap has important specificity for m7G capped RNA.[54] Abnormal GEMIN5 function will induce extensive defects in mRNA splicing. Cleavage sites in the RNA decapping enzyme are 2 related immunosensors of the retinoic acid-inducible gene I-like receptor family and are also targets of GEMIN5 and play a role in type I interferon response and signaling.[55] Scavenger decapping enzyme (DCPS) is a nucleotide hydrolase, a member of the histidine triad family of pyrophosphatases.[56] Previous research confirmed that dissociation of the histidine triad protein subunit HINT/MITF complex is associated with mast cell activation.[57] EIF4E is a family of cap-binding proteins with the highest affinity for eukaryotic 5′ cap structures. EIF4E1 mediates global translation and its activity is controlled through the PI3K/AKT/mTOR pathway.[58] Meanwhile, the PI3K/AKT/mTOR pathway can be activated by various types of stimuli to play a role the inhibition of apoptosis and autophagy and the promotion of cell growth; furthermore, the autophagic apoptosis of IR and GCs in PCOS is closely related to the PI3K/AKT/mTOR pathway.[59]
Functional enrichment analysis showed that the above genes were significantly enriched in nucleic acid metabolism, copper metabolism, and the HIF signaling pathway. Previous research reported obvious hypoxic reactions in the GCs of PCOS patients, and that the HIF signal pathway was inhibited.[60] The HIF signaling pathway plays a role in hypoxic response and glucose metabolism and also plays a key role in the follicular development and ovulation.[61,62] It is possible that these genes might overregulate cuproptosis and the HIF signaling pathway, thus exerting effect on the inflammatory response, eventually resulting in PCOS.
7. Conclusion and limitations
In this study, we identified 8 genes related to cuproptosis, 5 genes related to m7G and immune infiltration. We generated the first clinical prediction model for PCOS based on cuproptosis, m7G, and immune infiltration, which provides a foundation for subsequent basic research relating to cuproptosis in PCOS immunity.
However, our study has limitations. Firstly, the small sample size we screened by individual GEO datasets may have led to some bias in the results. Second, in the experimental validation stage, we only verified the differences in target gene expression in a small sample size. Furthermore, we did not conduct further mechanistic studies; these issues need to be explored in future research.
Author contributions
Data curation: Yuemeng Zhao, Liying Liu, Laixi Ji, Haijun Wang.
Funding acquisition: Yuxia Cao, Ying Lan, Laixi Ji, Haijun Wang.
Investigation: Yuemeng Zhao.
Methodology: Yuemeng Zhao, Laixi Ji, Haijun Wang.
Project administration: Haijun Wang.
Resources: Haijun Wang.
Validation: Jianheng Hao, Laixi Ji.
Visualization: Haijun Wang.
Writing – original draft: Yuemeng Zhao.
Writing – review & editing: Yuemeng Zhao, Liying Liu, Yuxia Cao, Ying Lan.
Abbreviations:
- AUC
- area under the curve
- CIGs
- curoptosis-related genes that were correlated with immune-related genes
- ELISA
- enzyme linked immunosorbent assay
- GEO
- gene expression omnibus
- GO
- Gene Ontology Resource
- IR
- insulin resistance
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- MIGs
- m7G-related genes that were correlated with immune-related genes
- PCOS
- polycystic ovary syndrome
- ROC
- receiver operating characteristic
- RT-PCR
- real-time PCR
- TCA
- tricarboxylic acid
This study was supported by grants from the National Natural Science Foundation of China (No. 82074549), the Shanxi Provincial Natural Fund (No. 201901D111399), the Shanxi University of Traditional Chinese Medicine 2020 Cultivation Program for Science and Technology Innovation Capacity (No. 2020PY-LP-03) and the National Natural Science Foundation of China (No. 82105028).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
How to cite this article: Zhao Y, Liu L, Hao J, Wang H, Cao Y, Lan Y, Ji L. Identification and validation of novel genes related to immune microenvironment in polycystic ovary syndrome. Medicine 2024;103:43(e40229).
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Contributor Information
Yuemeng Zhao, Email: zymtcm@163.com.
Liying Liu, Email: 767000032@qq.com.
Jianheng Hao, Email: hjhvvip@163.com.
Haijun Wang, Email: whjdavid@163.com.
Yuxia Cao, Email: cyxzijun@163.com.
Ying Lan, Email: lanyingtcm@163.com.
References
- [1].Li Y, Chen C, Ma Y, et al. Multi-system reproductive metabolic disorder: significance for the pathogenesis and therapy of polycystic ovary syndrome (PCOS). Life Sci. 2019;228:167–75. [DOI] [PubMed] [Google Scholar]
- [2].Patel S. Polycystic ovary syndrome (PCOS), an inflammatory, systemic, lifestyle endocrinopathy. J Steroid Biochem Mol Biol. 2018;182:27–36. [DOI] [PubMed] [Google Scholar]
- [3].Rostamtabar M, Esmaeilzadeh S, Tourani M, et al. Pathophysiological roles of chronic low-grade inflammation mediators in polycystic ovary syndrome. J Cell Physiol. 2021;236:824–38. [DOI] [PubMed] [Google Scholar]
- [4].Liu Y, Liu H, Li Z, et al. The release of peripheral immune inflammatory cytokines promote an inflammatory cascade in PCOS patients via altering the follicular microenvironment. Front Immunol. 2021;12:685724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fulghesu AM, Sanna F, Uda S, Magnini R, Portoghese E, Batetta B. IL-6 serum levels and production is related to an altered immune response in polycystic ovary syndrome girls with insulin resistance. Mediators Inflamm. 2011;2011:389317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].He S, Mao X, Lei H, et al. Peripheral blood inflammatory-immune cells as a predictor of infertility in women with polycystic ovary syndrome. J Inflamm Res. 2020;13:441–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schmidt J, Weijdegard B, Mikkelsen AL, Lindenberg S, Nilsson L, Brännström M. Differential expression of inflammation-related genes in the ovarian stroma and granulosa cells of PCOS women. Mol Hum Reprod. 2014;20:49–58. [DOI] [PubMed] [Google Scholar]
- [8].Benson S, Janssen OE, Hahn S, et al. Obesity, depression, and chronic low-grade inflammation in women with polycystic ovary syndrome. Brain Behav Immun. 2008;22:177–84. [DOI] [PubMed] [Google Scholar]
- [9].Yi W, Li X, Chen K, Li J, Chen K, Pan A. Effect of rNA interference on Oatp3a1 gene expression on biological characteristics and immune factors of ovarian granulosa cells in rats with PCOS. Am J Transl Res. 2020;12:4659–68. [PMC free article] [PubMed] [Google Scholar]
- [10].Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375:1254–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Martinez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun. 2020;11:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sun L, Hu W, Liu Q, et al. Metabonomics reveals plasma metabolic changes and inflammatory marker in polycystic ovary syndrome patients. J Proteome Res. 2012;11:2937–46. [DOI] [PubMed] [Google Scholar]
- [13].O’neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16:553–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Michalek RD, Gerriets VA, Jacobs SR, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kordus RJ, Hossain A, Malter HE, LaVoie HA. Mitochondrial metabolic substrate utilization in granulosa cells reflects body mass index and total follicle stimulating hormone dosage in in vitro fertilization patients. J Assist Reprod Genet. 2020;37:2743–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sun Y, Wang W, Guo Y, et al. High copper levels in follicular fluid affect follicle development in polycystic ovary syndrome patients: Population-based and in vitro studies. Toxicol Appl Pharmacol. 2019;365:101–11. [DOI] [PubMed] [Google Scholar]
- [17].Kanafchian M, Esmaeilzadeh S, Mahjoub S, Rahsepar M, Ghasemi M. Status of serum copper, magnesium, and total antioxidant capacity in patients with polycystic ovary syndrome. Biol Trace Elem Res. 2020;193:111–7. [DOI] [PubMed] [Google Scholar]
- [18].Pokorska-Niewiada K, Brodowska A, Szczuko M. The content of minerals in the PCOS group and the correlation with the parameters of metabolism. Nutrients. 2021;13:2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zha L, Wang J, Cheng X. The effects of RNA methylation on immune cells development and function. FASEB J. 2022;36:e22552. [DOI] [PubMed] [Google Scholar]
- [20].Motorin Y, Helm M. RNA nucleotide methylation. WIREs RNA. 2011;2:611–31. [DOI] [PubMed] [Google Scholar]
- [21].Luo Y, Yao Y, Wu P, Zi X, Sun N, He J. The potential role of N(7)-methylguanosine (m7G) in cancer. J Hematol Oncol. 2022;15:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Galloway A, Cowling VH. mRNA cap regulation in mammalian cell function and fate. Biochim Biophys Acta Gene Regul Mech. 2019;1862:270–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA. 2011;2:277–98. [DOI] [PubMed] [Google Scholar]
- [24].Galloway A, Kaskar A, Ditsova D, et al. Upregulation of RNA cap methyltransferase RNMT drives ribosome biogenesis during T cell activation. Nucleic Acids Res. 2021;49:6722–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tomikawa C. 7-Methylguanosine modifications in transfer RNA (tRNA). Int J Mol Sci . 2018;19:4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu J, Hou JH, Hsieh T-S. A new Drosophila gene wh (wuho) with WD40 repeats is essential for spermatogenesis and has maximal expression in hub cells. Dev Biol. 2006;296:219–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Alexandrov A, Grayhack EJ, Phizicky EM. tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA. 2005;11:821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhang LS, Liu C, Ma H, et al. Transcriptome-wide mapping of internal N(7)-Methylguanosine methylome in mammalian mRNA. Mol Cell. 2019;74:1304–16.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rotterdam ESHRE/ASRM-Sponsored PCOS consensus workshop group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS). Hum Reprod. 2004;19:41–7. [DOI] [PubMed] [Google Scholar]
- [30].Chrabaszczewska M, Winiewska-Szajewska M, Ostrowska N, et al. Insight into the binding and hydrolytic preferences of hNudt16 based on nucleotide diphosphate substrates. Int J Mol Sci . 2021;22:10929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Italiani P, Boraschi D. From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front Immunol. 2014;5:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lima PDA, Nivet AL, Wang Q, et al. Polycystic ovary syndrome: possible involvement of androgen-induced, chemerin-mediated ovarian recruitment of monocytes/macrophages. Biol Reprod. 2018;99:838–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–35. [DOI] [PubMed] [Google Scholar]
- [34].Rea IM, Gibson DS, Mcgilligan V, McNerlan SE, Alexander HD, Ross OA. Age and age-related diseases: role of inflammation triggers and cytokines. Front Immunol. 2018;9:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rudnicka E, Suchta K, Grymowicz M, et al. Chronic low grade inflammation in pathogenesis of PCOS. Int J Mol Sci . 2021;22:3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lutsenko S. Dynamic and cell-specific transport networks for intracellular copper ions. J Cell Sci. 2021;134:jcs240523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hodgkinson V, Petris MJ. Copper homeostasis at the host-pathogen interface. J Biol Chem. 2012;287:13549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu Z, Yu M, Fei B, Fang X, Ma T, Wang D. miR‑21‑5p targets PDHA1 to regulate glycolysis and cancer progression in gastric cancer. Oncol Rep. 2018;40:2955–63. [DOI] [PubMed] [Google Scholar]
- [39].Li A, Zhang Y, Zhao Z, Wang M, Zan L. Molecular characterization and transcriptional regulation analysis of the bovine PDHB gene. PLoS One. 2016;11:e0157445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wei T, Gao J, Huang C, Song B, Sun M, Shen W. SIRT3 (Sirtuin-3) prevents Ang II (Angiotensin II)-Induced macrophage metabolic switch improving perivascular adipose tissue function. Arterioscler Thromb Vasc Biol. 2021;41:714–30. [DOI] [PubMed] [Google Scholar]
- [41].Danileviciute E, Zeng N, Capelle C, et al. PARK7/DJ-1 promotes pyruvate dehydrogenase activity and maintains Treg homeostasis during ageing. Nat Metab. 2019;4:589–607. [DOI] [PubMed] [Google Scholar]
- [42].Wu R, Fujii S, Ryan NK, et al. Ovarian leukocyte distribution and cytokine/chemokine mRNA expression in follicular fluid cells in women with polycystic ovary syndrome. Hum Reprod. 2007;22:527–35. [DOI] [PubMed] [Google Scholar]
- [43].Solmonson A, Deberardinis RJ. Lipoic acid metabolism and mitochondrial redox regulation. J Biol Chem. 2018;293:7522–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Di Nicuolo F, Castellani R, Ticconi C, et al. α-Lipoic acid and its role on female reproduction. Curr Protein Pept Sci. 2021;22:767–74. [DOI] [PubMed] [Google Scholar]
- [45].Yang Y, Li W, Liu Y, Li Y, Gao L, Zhao J-jun. Alpha-lipoic acid attenuates insulin resistance and improves glucose metabolism in high fat diet-fed mice. Acta Pharmacol Sin. 2014;35:1285–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Diaz-Munoz MD, Bell SE, Fairfax K, et al. The RNA-binding protein HuR is essential for the B cell antibody response. Nat Immunol. 2015;16:415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Udumula MP, Sakr S, Dar S, et al. Ovarian cancer modulates the immunosuppressive function of CD11b(+)Gr1(+) myeloid cells via glutamine metabolism. Mol Metab. 2021;53:101272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Colliou N, Ge Y, Sahay B, et al. Commensal Propionibacterium strain UF1 mitigates intestinal inflammation via Th17 cell regulation. J Clin Invest. 2017;127:3970–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Xie W, Xi P, Liu Y, Zhang Z, Sun T. A comprehensive analysis of the prognostic value and immune infiltration of low expression DBT in clear cell renal cell carcinoma. Front Pharmacol. 2022;13:1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Meiser J, Krämer L, Sapcariu SC, et al. Pro-inflammatory macrophages sustain pyruvate oxidation through pyruvate dehydrogenase for the synthesis of itaconate and to enable cytokine expression. J Biol Chem. 2016;291:3932–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cui P, Hu W, Ma T, et al. Long-term androgen excess induces insulin resistance and non-alcoholic fatty liver disease in PCOS-like rats. J Steroid Biochem Mol Biol. 2021;208:105829. [DOI] [PubMed] [Google Scholar]
- [52].Mcclure C, Brudecki L, Yao ZQ, McCall CE, El Gazzar M. Processing body formation limits proinflammatory cytokine synthesis in endotoxin-tolerant monocytes and murine septic macrophages. J Innate Immun. 2015;7:572–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Huang SSY, Rinchai D, Toufiq M, et al. Transcriptomic profile investigations highlight a putative role for NUDT16 in sepsis. J Cell Mol Med. 2022;26:1714–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Strasser A, Dickmanns A, Luhrmann R, Ficner R. Structural basis for m3G-cap-mediated nuclear import of spliceosomal UsnRNPs by snurportin1. EMBO J. 2005;24:2235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Saiz M, Martinez-Salas E. Uncovering targets of the Leader protease: Linking RNA-mediated pathways and antiviral defense. Wiley Interdiscip Rev RNA. 2021;12:e1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Shao ZQ, Zhang YM, Pan XZ, Wang B, Chen J-Q. Insight into the evolution of the histidine triad protein (HTP) family in Streptococcus. PLoS One. 2013;8:e60116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Weinstein IB, Li H. Mast cells provide a “HINT” to the function of an exotic nucleotide. Immunity. 2004;20:119–20. [DOI] [PubMed] [Google Scholar]
- [58].Weiss B, Allen GE, Kloehn J, Abid K, Jaquier-Gubler P, Curran JA. eIF4E3 forms an active eIF4F complex during stresses (eIF4FS) targeting mTOR and re-programs the translatome. Nucleic Acids Res. 2021;49:5159–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tong C, Wu Y, Zhang L, Yu Y. Insulin resistance, autophagy and apoptosis in patients with polycystic ovary syndrome: Association with PI3K signaling pathway. Front Endocrinol (Lausanne). 2022;13:1091147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Xie Q, Hong W, Li Y, et al. Chitosan oligosaccharide improves ovarian granulosa cells inflammation and oxidative stress in patients with polycystic ovary syndrome. Front Immunol. 2023;14:1086232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Nagao A, Kobayashi M, Koyasu S, Chow CCT, Harada H. HIF-1-Dependent reprogramming of glucose metabolic pathway of cancer cells and its therapeutic significance. Int J Mol Sci . 2019;20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Balamurugan K. HIF-1 at the crossroads of hypoxia, inflammation, and cancer. Int J Cancer. 2016;138:1058–66. [DOI] [PMC free article] [PubMed] [Google Scholar]