Summary
Limited information exists about the types of immune cells present in Kaposi sarcoma (KS) lesions, especially in KS in the gastrointestinal tract. Using previously reported RNA-sequencing results from Kaposi sarcoma lesions in skin and gastrointestinal tract with normal matched tissues from the same patients at the same time, we investigated changes in lymphocytes in these tissues. We employed a computational method that determines changes in cell type distributions using KS lesion transcriptome data compared to a reference set of RNA expression patterns of purified immune cells. Since secreted cytokines and chemokines from KSHV-infected cells may influence the microenvironment of Kaposi sarcoma lesions, we performed cytokine profiling of conditioned media from KSHV-infected primary human dermal lymphatic endothelial cells. We also measured how this conditioned media altered the differentiation of macrophages in cell culture assays. These results suggested that factors in conditioned media from KSHV-infected endothelial cells promoted differentiation of a promonocytic cell line to proinflammatory macrophages.
Keywords: KSHV, cibersort, RNA-sequencing
Introduction
Kaposi sarcoma (KS) is caused by the Kaposi sarcoma herpesvirus (KSHV or HHV8). Besides KS, KSHV also causes primary effusion lymphoma (PEL), multicentric Castleman disease (MCD), and KSHV inflammatory cytokine syndrome (KICS). These conditions can occur alone or together and vary in prognosis (1). In the US, KS mainly affects people living with HIV (PWH) and is known as epidemic KS. Despite advances in HIV treatment reducing its incidence, KS remains a common HIV-associated tumor in the US and Southern Africa, where it is a significant health issue. KS commonly occurs in PWH with low CD4+ T cell counts but can also arise from other immune dysregulation. Standard KS treatment for PWH includes antiretroviral therapy (ART). For extensive or ART-resistant disease, systemic therapies like liposomal doxorubicin, paclitaxel, or pomalidomide are used (1). KS may recur despite remission and well-controlled HIV, requiring long-term treatment. The reasons for recurrence are unclear.
Little is known about cellular and viral RNA transcripts in KS lesions and their relation to the microenvironment. We previously aimed to understand the changes in human gene expression and viral transcripts in both skin and GI KS. KS tissue was compared to normal tissue using matched patient samples (2). KS typically manifests as skin lesions but may also occur in visceral organs including the respiratory and gastrointestinal (GI) tracts in severe cases. Information on their relationships between clinical characteristics, inflammation, immunity and KSHV expression is scarce. We also explored variations in the immune microenvironment based on the lesion location and clinical characteristics. Macrophages play a crucial role in the human immune response to viral infections. As key components of the innate immune system, these versatile cells act as first responders, identifying and engulfing pathogens through phagocytosis. Macrophages then present viral antigens on their surface to prime the adaptive immune response, facilitating the production of specific antibodies and viral-specific T cells. They also release a variety of cytokines and chemokines that recruit and activate other immune cells, such as T cells and natural killer cells, to the site of infection. In solid tumors, tumor associated macrophages (TAMs) form, with aberrant expression of cytokines and chemokines, immunosuppressive tumor microenvironment. KSHV was reported to modulate macrophages directly (3) or indirectly (4).
To investigate specific cellular genes linked to KS pathogenesis or KSHV infection, we conducted in vitro studies using KSHV-infected lymphatic endothelial cells (LECs) to study cytokine production and their effects on macrophage gene expression.
Materials and Methods
Cell culture, reagents, nucleofection, and KSHV infection
Human dermal lymphatic endothelial cells (HDLECs) were obtained from Promocell and passaged in EGM2 medium (Lonza) for up to 5 passages, with passages 3 to 5 used for experiments. THP-1 cells (American Type Culture Collection TIB-202) were cultured in RPM1640 media (Gibco) supplemented with 10% Fetal Bovine Serum, Penicillin, and Streptomycin (Gibco). HDLEC de novo infections were carried out using KSHV-BAC16 (concentrated by ultracentrifugation), diluted in EGM2 medium, at multiplicity of infection of 0.25, 0.5, and 1 infectious units (I.U.). 1.0 I.U. is the amount of virus that is sufficient to infect all cells if virus will be distributed equally, as determined by GFP ectopic marker. Polybrene was added at 8 μg/mL. Uninfected samples were included as negative controls. After 16 hours of incubation, cells were washed and overlaid with fresh media.
Human cytokine analysis
At three days post infection, conditioned media was collected, cleared by centrifugations (500g, 10 minutes, then 2,000g, 20 minutes), and 100 μL for each sample was used for measuring specific cytokines with V-PLEX Proinflammatory Panel 1 Human Kit (Meso Scale Diagnostics). Assays were performed by Clinical Support Laboratory (NCI-Frederick).
THP-1 differentiation assay
2 × 105 THP-1 cells were seeded in 6 well plates in 2 mL complete RPMI media. 10 ng/mL of 12-O-tetradecanoylphorbol-l3-acetate (PMA, Sigma) was added, and cells were incubated for 24 h. 1 mL of media was replaced with 1ml of conditioned media from KHSV-infected HDLEC cells and THP-1 cells were incubated for 24 hours. THP-1 cells were washed with Phosphate-buffered saline (Gibco) and total RNAs were extracted with the Direct-zol RNA miniprep kit (Zymo Research). As a read-out, quantitative reverse transcription-PCR (RT-qPCR) was performed using 500 ng RNA and ReverTra Ace reverse transcription kit (Toyobo), Thunderbird Probe qPCR master mix (Toyobo), and TaqMan probe (CXCL10 Assay ID: Hs00171042_m1, Applied Biosystems) using the ABI StepOnePlus real-time PCR system (Applied Biosystems). Relative mRNA levels were computed using the threshold cycle (ΔΔCt) method with RPS13 (Assay ID: Hs01011487_g1, Applied Biosystems) as a reference gene according to manufacturer's protocol.
Bioinformatics analysis
We analyzed our previous RNA-sequencing data (Gene Expression Omnibus accession number GSE241095) of KS lesion specimens (2). CIBERSORTx software predictions were based on TPM values using the leukocyte signature matrix (LM22) (5). CIBERSORTx was executed using the docker container provided by the authors of CIBERSORTx on their website (https://cibersortx.stanford.edu), with default arguments (i.e. --absolute FALSE, --rmbatchBmode FALSE and - rmbatchSmode FALSE) results in cell-type proportions relative to the reference cell types in the signature matrix. Cell types with values from at least five patient samples were included for further analysis.
Results
As our previous bulk transcriptome analysis of 22 KS lesion samples showed (2) heightened expression in markers associated with activated immune cells, we applied the computational tool, CIBERSORTx (5), to interrogate changes in immune cell types in the KS lesions. This method imputes the expression profile of specific marker genes from each cell type and estimates proportions of different cell types in the original cell mixtures. We calculated the relative amounts of cell type specific signatures in KS lesions compared to their matched normal samples (Figure 1, A-B). Heterogeneity in the immune subtype signatures was noted across both skin and GI KS lesions. Follicular helper T cells and regulatory T cells were decreased in many GI samples, but this observation was not noted in the skin KS samples. There were elevated levels of resting memory CD4+ T cells in 5 of 12 GI KS samples as compared to the paired normal tissue and in 1 skin KS sample as compared to the paired normal tissue. Increased signatures of M1 macrophages were observed in the majority (7 of 10 samples, p = 0.056) of skin KS samples.
Figure 1.
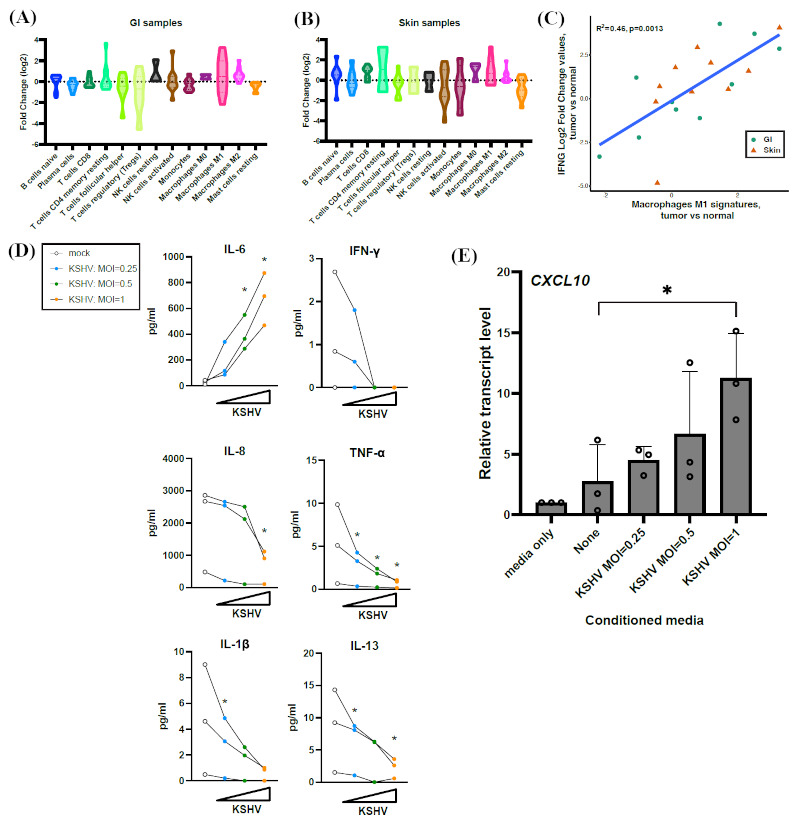
CIBERSORT analysis of KS lesions, cytokine secretion, and macrophage polarization. (A-B) Fold changes of different immune cell types in tumor vs normal samples, were estimated by CIBERSORT. Violin plots show the increased or decreased fold change (log2) of each cell type signature of the KS lesions as compared to normal skin. (C) Correlation analysis of changes in IFNG RNA expression (tumor vs. normal) and changes in M1 macrophage cell signatures (tumor vs. normal). (D) Proinflammatory cytokine levels in conditioned media of KSHV-infected LECs after 3 days were quantitated with an electrochemiluminescence multiplex immunoassay. Each line represents a separate biological replicate. Only cytokines with concentration > 5 pg/mL in any of samples are shown. (E) Macrophage polarization assays with conditioned media and a promonocytic cell line THP-1. THP-1 cells were treated with PMA and then incubated with conditioned media of KSHV-infected LECs (D) for 24 h. Transcript levels of M1 macrophage marker gene CXCL10 was quantitated with RT-qPCR and normalized to RPS13. Multiple paired t-test (two sided) was performed. *p < 0.05.
Macrophages play key roles in antigen presentation, inflammation and phagocytosis. M1-classically activated macrophages are pro-inflammatory and promote cell killing while M2-alternatively activated macrophages are anti-inflammatory and promote tissue healing. IFN-γ signaling promotes M0 macrophage polarization to M1 macrophages (6). Macrophage polarization is widely exploited by viruses through various mechanisms including manipulating cytokines (6). CIBERSORTx analyses predicted differences in M1 and M2 macrophage polarization in the KS tumors compared to normal samples. Within GI KS, RNA expression changes of IFN-γ varied widely as compared to their matched normal samples (2), and M1 macrophage signatures appeared variable in both tissues (Figure 1A). Interestingly, there was a strong positive correlation between IFN-γ expression and M1 macrophage signatures in all tissues (p = 0.0013, Figure 1C).
To further characterize the relationship between cytokines, macrophage polarization and the potential role of directing specific cell types in the KS tissue microenvironment, changes in cytokine levels following KSHV infection of endothelial cells were investigated. An initial question was whether IFN-γ expression levels are changed by KSHV infection of LECs or are the changing levels of IFN-γ expression due to other cell types (e.g. T and NK cells) in KS lesions. LECs were infected with KSHV in cell culture experiments and the conditioned media from these cells was collected. Changes in cytokine secretion from mock-infected and KSHV-infected LECs were measured at 3- and 5-days post-infection. Conditioned media of KSHV-infected LECs contained higher levels of IL-6 in a multiplicity of infection (MOI)-dependent manner (Figure 1D). This increase in IL-6 levels in KSHV-infected endothelial cells was consistent with previous reports (7,8). Other pro-inflammatory cytokines such as IL-8 and IL-1β were reduced in the conditioned media from KSHV-infected cells. Macrophage polarization to the anti-tumor M1 macrophage type is dependent on IFN-γ and TNF-ɑ, and polarization to the pro-tumor M2 macrophage type depends on IL-4 and IL- 13. Secretion of all these cytokines was reduced upon infection. IL-4 concentration was less than 5 pg/mL in all samples and was not shown. To assess the effect of conditioned media from KSHV-infected endothelial cells on macrophage polarization, we differentiated a promonocytic cell line THP-1 with conditioned media of infected LECs. THP-1 cells can be differentiated to M1 or M2 types. Increasing the amount of KSHV in infections polarized THP-1 differentiation more to a M1 profile as measured by an M1 macrophage marker gene, CXCL10 (Figure 1E). KSHV infection in LECs thus regulates secretion of pro-inflammatory cytokines and may promote M1 macrophage polarization. The M1 polarization observed in KS lesions was unlikely due to changes in IFN-γ secretion from KSHV-infected endothelial cells since IFN-γ secretion levels only decreased with KSHV infection in LECs (Figure 1D). These cell culture experiments (Figure 1, D-E) showing an increase in an M1 macrophage marker was consistent with the increased M1 cell signatures observed in skin KS (Figure 1B), compared to the matched normal skin tissue.
The amount of viral infection varied among the patient samples, as measured by the KSHV transcript reads in the RNA-sequencing results. The KSHV transcript values were summated together into KSHV transcripts per million (KSHV TPM, Figure 2) as a proxy of virus infection levels. These values reflect the total amount of normalized KSHV viral transcript reads per sample. KSHV TPM and CIBERSORT values for changes between tumor and matched normal samples were analyzed using a non-parametric correlation analysis. This method was used to investigate whether predicted changes in cell types were correlative with each other or with the overall level of KSHV infection (KSHV TPM). All samples were used for the correlative analysis in Figure 2A, which found a strong correlation (R2= 0.85) between the amount of KSHV gene expression (KSHV TPM) and levels of regulatory T cells (Tregs). Since different immune responses may occur between skin and GI locations, additional correlation analyses were conducted separately with only the GI (Figure 2B) and the skin (Figure 2C) samples. In the GI samples, a similar correlation was observed between KSHV infection and levels of Tregs (Figure 2B), which promotes an M2-like TAM formation and immunosuppressive environment. The strongest negative correlation was between Tregs and monocytes in GI samples. In skin samples there was a strong negative correlation between KSHV infection levels and NK resting cells, suggesting that with more KSHV infection, decreased levels of NK resting cells were found in skin KS tumors. A strong positive correlation was also found between changes in M0 macrophages and M2 macrophages, but not with M1, in skin KS. Though CIBERSORTx scores for macrophages are dysregulated both in GI and skin KS compared to normal (Figure 1, A-B), the underlying mechanisms may be distinct.
Figure 2.
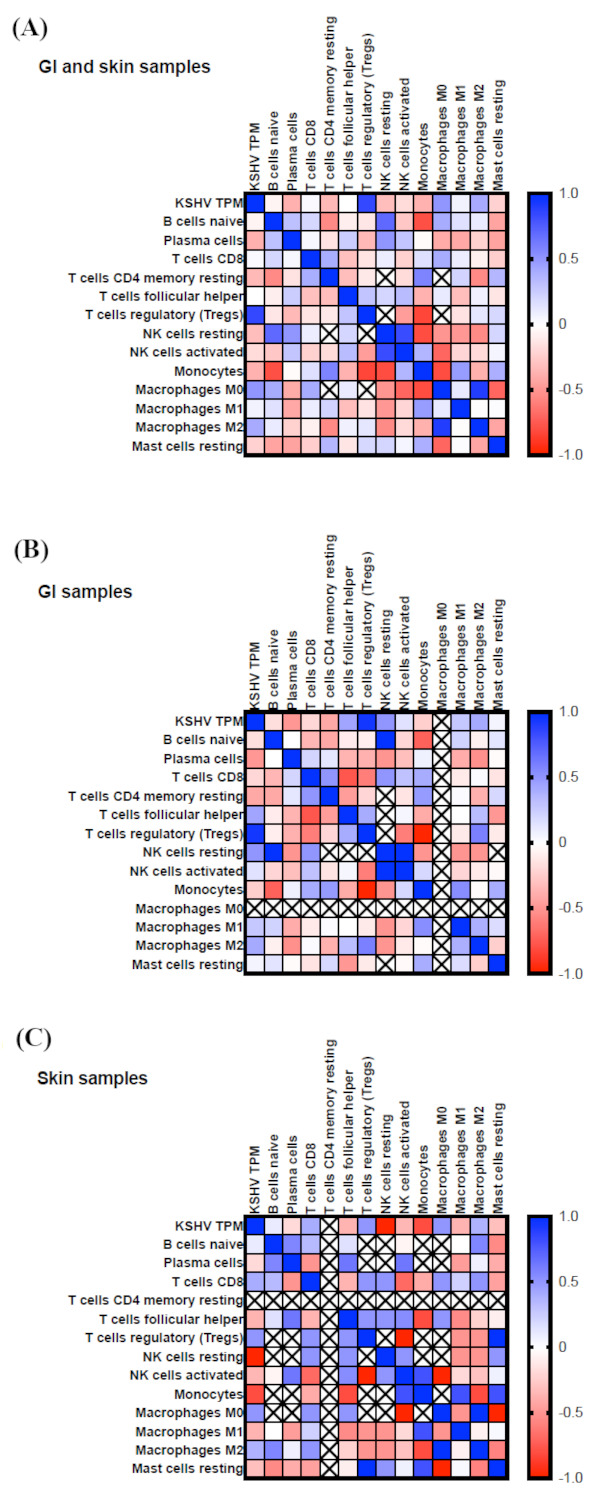
Correlation analysis of CIBERSORT values and KSHV infection. Normalized KSHV transcripts in the RNA-sequencing results were summarized and reported as total KSHV transcripts per million reads (KSHV TPM). Changes in CIBERSORT values were analyzed in combination with KSHV TPM values. Blue colors indicate strong positive correlations and red colors show strong negative correlations in all samples (A), only GI samples (B), and only skin samples (C).
Discussion
Differences in skin and GI lesions gene expression may be associated with immune profiles in respective tumor microenvironments. A recent study using single cell sequencing analysis of KS among participants who were recently diagnosed with HIV and KS who were antiretroviral therapy naïve demonstrated an abundance of T cells and macrophages (9). A previous study of skin KS lesions of participants with endemic and epidemic KS from sub-Saharan Africa demonstrated the distribution of immune cells by infection of KSHV cells. In these analyses, we noted that classically activated M1 macrophages were evenly distributed in KS lesions, whereas alternatively activated macrophages were notable in areas that did not have KSHV infected cells (10). M1 macrophages are often amplified by IFN-γ signaling to promote cell killing (11). This study also highlighted the abundance of CD8+ T cells in KS lesions compared to normal skin but this was not related to KSHV infected cells.
Limitations of this study are largely dependent on the clinical samples available and the technical shortcomings. The clinical samples that we analyzed had variations in the concurrent diseases occurring at the time of biopsy and some samples were obtained before or after treatment for KS. Additionally, biopsies were obtained from different areas of the skin and gastrointestinal tract and different tissues had various levels of KSHV gene expression. We used a computational method to query different cell types in these samples, based on RNA expression patterns, but complementary approaches (e.g. flow cytometry, immunohistochemistry) could strengthen the evidence for these cell type differences between patient samples. These complementary approaches were not feasible in our study, due to the limiting size of the biopsy material.
In our analyses, there were differences noted in the skin and GI KS lesions compared to normal counterparts including dysregulated macrophages and higher resting memory CD4+ T cells particularly in GI KS lesions. One hypothesis was that increased IFN-γ expression in KS lesions might be increasing M1 polarization. However, IFN-γ secretion levels dropped with KSHV infection in our experiments using LECs. In our results using conditioned media from KSHV-infected LECs, we did find an increase in an M1 macrophage marker, CXCL10. These results suggest that secreted factors other than IFN-γ in the conditioned media from KSHV-infected cells may be promoting M1 polarization. However, in actual KS lesions, there are infiltrating cells with capability to secrete various cytokines and chemokines beyond KSHV-infected endothelial cells, which may be the overall changes in immune cell population including macrophages. Correlational analysis also demonstrated that such differences may be related to site-specific immune environments between the skin and GI tract rather than broadly associated with KSHV or HIV infection. In GI KS, but not in skin KS, KSHV total TPM positively correlated with regulatory T cells, which promote M2 macrophage differentiation. In contrast, skin KS, only, showed preference to M2 differentiation than M1 in tumor samples compared to matched normal skin. We also note the limitation of this bulk RNA-Seq studies and needs for single cell and spatial analysis to further interrogate the importance of immune cells to regulate tumor microenvironment. Taken together, we showed KSHV's ability to dysregulate immune cells like macrophages in vitro and in clinical specimens.
Conclusion
The immune cell composition of KS lesions, particularly in the gastrointestinal tract, remains poorly understood. To address this, we analyzed RNA-sequencing data from KS lesions in both skin and gastrointestinal tissues, using matched normal tissues from the same patients as control samples. We compared the transcriptomes of KS lesions with reference immune cell RNA expression profiles to assess changes in immune cell populations. This analysis found the greatest changes in predicted T cells, monocytes, and macrophages. We conducted cytokine profiling of conditioned media from KSHV-infected human dermal lymphatic endothelial cells, since cytokines and chemokines from KSHV-infected cells may modify the lesion cellular environment. Additionally, we studied how this conditioned media affected the differentiation of macrophages growing in culture. The results showed that factors from the infected cells encouraged the differentiation of promonocytic cells into proinflammatory macrophages (M1). A correlation analysis revealed that as KSHV gene expression increased in tissues, the predicted amount of immune-suppressive T regulatory cells also increased. These studies indicate how KSHV infection and changes in cytokines and chemokines may shape the immune environment of KS lesions.
Funding
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health 1ZIABC011176 (JZ).
Conflict of Interest
The authors have no conflicts of interest to disclose.
References
- 1. Lurain KA, Ramaswami R, Krug LT, Whitby D, Ziegelbauer JM, Wang HW, Yarchoan R. HIV-associated cancers and lymphoproliferative disorders caused by Kaposi sarcoma herpesvirus and Epstein-Barr virus. Clin Microbiol Rev. 2024; e0002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ramaswami R, Tagawa T, Mahesh G, et al. Transcriptional landscape of Kaposi sarcoma tumors identifies unique immunologic signatures and key determinants of angiogenesis. J Transl Med. 2023; 21:653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gilardini Montani MS, Falcinelli L, Santarelli R, Granato M, Romeo MA, Cecere N, Gonnella R, D'Orazi G, Faggioni A, Cirone M. KSHV infection skews macrophage polarisation towards M2-like/TAM and activates Ire1 alpha-XBP1 axis up-regulating pro-tumorigenic cytokine release and PD-L1 expression. Br J Cancer. 2020; 123:298-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhaskaran N, Ghosh SK, Yu X, Qin S, Weinberg A, Pandiyan P, Ye F. Kaposi's sarcoma-associated herpesvirus infection promotes differentiation and polarization of monocytes into tumor-associated macrophages. Cell Cycle. 2017; 16:1611-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, Khodadoust MS, Esfahani MS, Luca BA, Steiner D, Diehn M, Alizadeh AA. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019; 37:773-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sang Y, Miller LC, Blecha F. Macrophage polarization in virus-host interactions. J Clin Cell Immunol. 2015; 6:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Glaunsinger B, Ganem D. Highly selective escape from KSHV-mediated host mRNA shutoff and its implications for viral pathogenesis. J Exp Med. 2004; 200:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Butler LM, Jeffery HC, Wheat RL, Rae PC, Townsend K, Alkharsah KR, Schulz TF, Nash GB, Blackbourn DJ. Kaposi's sarcoma-associated herpesvirus infection of endothelial cells inhibits neutrophil recruitment through an interleukin-6-dependent mechanism: A new paradigm for viral immune evasion. J Virol. 2011; 85:7321-7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Phipps W CD, Xu Y, Kafeero J, Mooka P, Okoche L, Basemera D, Flach B, Towlerton A, Warren EH. Single cell evaluation of Kaposi sarcoma tumors reveals complex immune infiltrate. In: Conference on Retroviruses and Opportunistic Infection (Seattle, Washington, 2019. [Google Scholar]
- 10. Lidenge SJ, Tso FY, Ngalamika O, Kolape J, Ngowi JR, Mwaiselage J, Wood C, West JT. Lack of CD8+ T-cell co-localization with Kaposi's sarcoma-associated herpesvirus infected cells in Kaposi's sarcoma tumors. Oncotarget. 2020; 11:1556-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014; 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]


