Abstract
Much evidence suggests that the major immediate-early (IE) transactivator of human cytomegalovirus (HCMV), IE-2, is likely to be critical for efficient viral replication; however, the lack of an IE-2 mutant HCMV has precluded an experimental test of this hypothesis. As an initial step toward characterizing an IE-2 mutant, we first cloned the HCMV Towne genome as a bacterial artificial chromosome (BAC) and analyzed the ability of transfected Towne-BAC DNA (T-BACwt) to produce plaques following introduction into permissive human fibroblasts. Like Towne viral DNA, transfected T-BACwt DNA was infectious in permissive cells, and the resulting virus stocks were indistinguishable from Towne virus. We then used homologous recombination in Escherichia coli to delete the majority of UL122, the open reading frame encoding the unique portion of IE-2, from T-BACwt. From this deleted BAC, a third BAC clone in which the deletion was repaired with wild-type UL122 was created. In numerous transfections of permissive human foreskin fibroblast cells with these three BAC DNA clones, the rescued BAC and T-BACwt consistently yielded plaques, while the UL122 mutant BAC never generated plaques, even after 4 weeks. Protein and mRNA of other IE genes were readily detected from transfected UL122 mutant BAC DNA; however, reverse transcription-PCR failed to detect mRNA expression from any of five early genes examined. The generalized failure of this mutant to express early genes is consistent with expectations from in vitro assays which have demonstrated that IE-2 transactivates most HCMV promoters. These experiments provide the first direct demonstration that IE-2 is required for successful HCMV infection and indicate that virus lacking IE-2 arrests early in the replication cycle.
Human cytomegalovirus (HCMV; human herpesvirus 5) is a widespread human pathogen that has minor clinical impact on healthy individuals but causes organ disease in immunosuppressed patients and neural damage in fetuses infected in utero (7). Persons infected with the human immunodeficiency virus frequently develop HCMV related pathology as a result of HCMV opportunistic replication, and given the global pandemic nature of human immunodeficiency virus infection (28), HCMV disease can be expected to continue to increase for some time. Transplant recipients undergoing posttransplant immunosuppressive therapy comprise another major class of immunosuppressed patients in which HCMV poses substantial risk. HCMV infection is associated with development of disease and mortality in both solid organ and bone marrow transplant recipients and is a major risk factor for graft-versus-host disease (7). Although recent antiviral agents have reduced the incidence of posttransplant HCMV disease somewhat, transplant frequency is likely to escalate, and thus HCMV infection will continue to pose problems for this group. Of considerable clinical and humanitarian importance is the scope of infant mortality and morbidity produced by congenital HCMV infection. HCMV is transmitted in utero in up to 50% of pregnancies in which the mother has not previously been exposed to the virus (7). In addition to the high rate of transmission, fetal infection is far more likely to produce severe sequelae in HCMV-naive mothers than in those with preexisting HCMV immunity (7). The simple prevalence of HCMV infection in the human population makes fetal HCMV infection the leading viral cause of birth defects (10, 14, 32). In all HCMV disease states, damage is associated with viral replication in the affected tissue that may result from either primary infection or reactivation of latent virus (7).
Primary HCMV infection resolves into a latent, subclinical infection lasting for the life of the host. A thorough understanding of HCMV replication dynamics is therefore central to effective intervention in HCMV disease. HCMV replicative gene expression evolves in the temporal cascade that typifies the herpesvirus family (31). Gene expression initiates from a small number of immediate-early (IE) loci that are expressed without a requirement for prior viral transcription. These IE gene products activate other viral genes and produce changes in the infected cell which aid viral replication. The majority of HCMV IE transcription derives from a single genetic locus, termed the major immediate-early (MIE) region, and produces the IE-1 and IE-2 protein family through complex differential splicing of a primary transcript (24). Other HCMV IE genes include the UL36-38 and US3 families, each of which gives rise to multiple proteins, the partially homologous IRS1 and TRS1 genes encoded within the short repeats, and a 5-kb RNA which appears to be noncoding (24, 30). The greatest amount of experimental data, by far, are available for IE-1 and IE-2; the numerous biochemical activities and interactions with key host cell proteins that have been described for both indicate that the MIE proteins may modulate diverse cellular processes, including transcription, cell cycle control, apoptosis, and subnuclear complex composition (25, 26, 41). Analysis of viral mutants has indicated that UL36, UL37, US3, and IRS1 are all dispensable for replication in cell culture, whereas IE-1 is critical for replication at low multiplicity (4, 18, 25, 27).
Despite the intense research devoted to the HCMV MIE genes and the numerous properties attributed to IE-2 from in vitro studies, there has been no experimental demonstration that IE-2 is critical for HCMV replication. We were unable to purify a mutant of HCMV lacking IE-2 with the classical plaque isolation methods used to recover other HCMV mutants. Our experience suggested that this was primarily due to the difficulty of achieving long-term expression of fully functional IE-2 in cultured cells for the purpose of complementation (A. Marchini and H. Zhu, unpublished observations). A recent report suggesting that IE-2 impedes cell cycle progression may explain why expression is not tolerated in cultured cells (26). Our inability to purify this mutant also suggested, albeit indirectly, that mutants lacking IE-2 are, in fact, impaired for growth. Since this conclusion is only inferred and the experiment neither confirmed the existence of the predicted genotype nor provided any indication of the underlying defect, we chose to create a bacterial artificial chromosome (BAC) of HCMV from which a genome lacking IE-2 could be derived without the need for complementation.
A number of recent reports have established the utility of BAC clones of herpesvirus genomes for the production and purification of genomes containing defined mutations. BAC clones have now been described for herpes simplex virus type 1, pseudorabies virus, varicella-zoster virus, Epstein-Barr virus, and human and murine CMV (6). In addition to obviating any requirement for complementation to create a mutant viral genome, a BAC clone offers several other important advantages over previously used methods for creating recombinant HCMV. First, maintaining the HCMV genome as a BAC clone in Escherichia coli dramatically facilitates mutagenesis by allowing the full power of bacterial genetics to be applied to creation of new mutations. Second, since all mutagenic steps are carried out in E. coli, the time needed to create and isolate new mutants is greatly reduced compared to traditional procedures carried out in eukaryotic cells. Third, in contrast to the strategy of reassembling virus in vivo from overlapping sets of cosmid clones (20), BAC transfection eliminates any requirement for multiple recombination events during reconstitution of virus in eukaryotic cells, which may themselves introduce unwanted mutations, and makes the recovery of recombinant virus much more efficient. Since it remains problematic to complement an IE-2 mutant in cell culture, and considering all of the advantages that a BAC HCMV clone would provide for future experimentation, we decided to generate a BAC clone of HCMV and use it to create an IE-2 deletion mutant. While this approach precluded the isolation of an IE-2 deletion mutant virus stock, transfection experiments using purified BAC DNA allowed preliminary characterization of a HCMV mutant lacking IE-2. The BAC system will also allow rapid and simple rescue analysis to define the functions of IE-2 that are critical for HCMV replication using the extensive collection of in vitro-mutated IE-2 alleles which have been reported.
MATERIALS AND METHODS
Cells, viruses, viral DNA, and cosmid clones.
Primary human foreskin fibroblast (HFF) cells were prepared from tissue samples and grown in Dulbecco's modified Eagle medium plus 10% fetal calf serum. The viral DNA used to create a BAC was purified from total virus particles isolated from HFF cells infected with the Towne strain of HCMV (a gift from T. Shenk) according to established protocols (19). Cosmid subclones comprising the entire AD169 HCMV genome (a gift from B. Fleckenstein and T. Shenk) were used to confirm the structure of the Towne BAC (T-BAC) by Southern blotting.
Production and characterization of a HCMV BAC.
To provide flanking DNA for homologous recombination in eukaryotic cells, two fragments of HCMV DNA were PCR amplified from cosmid clone CM1052, which contains the HindIII K/Q, X, V, and W fragments of AD169 HCMV (12). Amplifications used the following primers (read 5′ to 3′) derived from the published sequence (9) of AD169 HCMV DNA:(i) CCGGATCCCCACCGGGTAGAACC, in which the first cytosine residue following the BamHI site is nucleotide 189941; and (ii) CCAAGCTTGCACAACGGGATGACC, in which the guanosine residue following the HindIII site is the complement of nucleotide 191921. Primers i and ii yielded a 1.98-kb PCR product and introduced restriction sites for BamHI and HindIII at the 5′ and 3′ termini, respectively. A second recombination fragment was amplified similarly, using the following primers: (iii) GAGCCAGAGTATGGG, in which the first residue is nucleotide 200834 and occurs just 5′ to the HindIII site starting at nucleotide 200856; and (iv) CCTATCTACGTGCCC, in which the last cytosine is the complement of nucleotide 204034 and occurs just 3′ to the BamHI site starting at nucleotide 204024. Primers iii and iv yielded a 3.2-kb product including unique BamHI and HindIII sites near the termini.
The F-plasmid vector pMBO1374, a gift from G. Smith (34), was first modified by removal of the unique BamHI site and one of two ClaI sites, giving pMBO1374.1. The two HCMV PCR products (see above) were digested with BamHI and ligated to each other, resulting in a 5.2-kb linear species with the 3′ end of the 3′ homology fragment joined to the 5′ end of the 5′ fragment (Fig. 1A). The 5.2-kb fragment was next digested with HindIII and cloned into the HindIII site of pMBO1374.1, yielding plasmid pUSF-2. A cassette in which the simian virus 40 early promoter and polyadenylation signals control expression of green fluorescent protein (GFP) was PCR amplified from plasmid pGET-07 (a gift from G. Tullis) and cloned into the remaining ClaI site of pUSF-2 via ClaI sites included in the primers. This final construct, pUSF-3, contains the prokaryotic genetic elements necessary to confer maintenance as a BAC in E. coli, HCMV DNA sequences to direct homologous recombination to the unique short (US) region of the viral genome, and the GFP marker to facilitate identification and purification of recombinant HCMV in eukaryotic cells. The flanking DNA deletes 8.9 kb of DNA within the US region of HCMV that has been defined as dispensable for HCMV replication in cell culture (18), truncating IRS1 after amino acid 719 and removing reading frames US1 to US11 plus the carboxy-terminal third of US12.
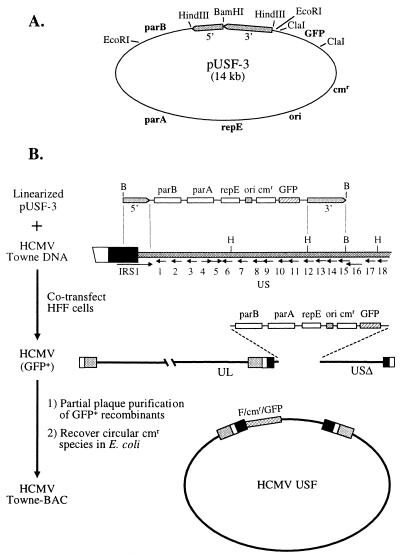
FIG. 1.
Construction of a Towne HCMV BAC. (A) The recombination substrate plasmid for introducing F-plasmid sequences into HCMV, pUSF-3, was derived by modifying pMBO1374 (34). The relative map positions of the replication origin (ori), replication and partition functions (repE, parA, and parB), chloramphenicol resistance (cmr), and the GFP eukaryotic expression cassette (GFP) are indicated in boldface. Two PCR fragments derived from HCMV (gray bars) were inserted into the vector such that BamHI digestion would result in their proper orientation for recombination with the CMV genome (see below). (B) To create a recombinant HCMV with a pUSF-3 insertion, pUSF-3 was digested with BamHI (top) and cotransfected into primary HFF cells with linear, wild-type Towne HCMV DNA. The targeted region of Towne DNA (second line from top) includes a portion of the internal repeat (large bar) and the first 15 open reading frames of the US region of HCMV (numbered arrows). BamHI (B) and HindIII (H) sites present in Towne DNA are indicated above the bar. Recombinant virus having pUSF-3 substituted for US1-12 (third line) was enriched by plaque purification using the GFP marker (flow chart, left). Fresh HFF cells were infected at high MOI with the enriched virus stock for 24 h, after which total genomic DNA was isolated by phenol-chloroform extraction. Aliquots of the total DNA preparation were transformed into Gene Hogs E. coli (Research Genetics), and the transformants were selected for resistance to chloramphenicol. Resistant colonies were expanded, and their circular plasmid DNA was examined by restriction analysis to identify correct recombinants (bottom).
To generate the recombinant HCMV incorporating the BAC and GFP sequences, pUSF-3 was digested with BamHI and electroporated into HFF cells along with wild-type Towne viral DNA and an expression plasmid for the HCMV tegument protein pp71 (2). One day following transfection, the surviving cells were distributed into 96-well culture dishes, and these were monitored for development of cytopathic effect. When cytopathic effect was uniform in the cultures (10 to 14 days), the supernatants were collected from 12 independent wells in which the majority of cells expressed GFP, and virus carrying the GFP marker was enriched by plaque purification. To recover the recombinant HCMV as covalently closed circular BAC DNA, HFF cells were infected at high multiplicity of infection (MOI) with the enriched stocks for 24 h, after which a crude lysate was prepared from the infected monolayer by lysis at 55°C in Tris-HC1 (50 mM, pH 8.0), EDTA (50 mM), sodium dodecyl sulfate (1%), and proteinase K (100 μg/ml). Aliquots of this crude lysate were transformed, without further purification, into RecA− E. coli DH10B (Research Genetics, Inc.), and transformants resistant to chloramphenicol were selected. As an initial screen for a CMV BAC having the expected structure, Towne viral DNA and BAC DNA from resistant colonies were digested with EcoRI and electrophoresed in agarose, and the fragments were visualized with ethidium bromide. Out of approximately 100 transformants examined, one displayed a restriction pattern nearly identical to that of Towne viral DNA. This BAC clone, designated T-BACwt, was further analyzed by Southern blotting with cosmid probes that spanned the entire AD169 genome.
To characterize the virus which was reconstituted following T-BACwt transfection, cells and supernatant were collected from HFF cultures transfected with either T-BACwt DNA or Towne DNA purified from virions, subjected to two cycles of freeze-thawing, cleared, and titered on fresh HFF cells. To compare the growth kinetics of Towne and T-BACwt viruses, HFF cells were infected with 0.01 PFU/cell at 37°C for 1 h, residual virus was washed out, and the cells were trypsinized and divided among six dishes. Dishes were harvested at various times postinfection, and the progeny virus was titered on fresh HFF cells as described above.
Transfer constructs and conjugative transfer.
The procedure and reagents for conjugative transfer of sequences to BAC DNA in E. coli were generously supplied by G. Smith and L. Enquist and have been described elsewhere (11, 34). To create a transfer vector to mutate UL122 from HCMV, a deletion of approximately 1,230 bp was introduced into an EcoRI-SalI restriction fragment subclone of the Towne MIE region, from the SmaI site at the 5′ end of the UL122 reading frame to the StuI site at the 3′ end (relative to transcription of the gene). This allele was cloned into pGS284, a derivative of the suicide selection vector pCVD442 (34). This allele produces an IE-2 polypeptide with a frameshift after amino acid 135 followed by truncation after an additional 20 unrelated amino acids; it thus contains the 85 residues common to IE-1 and IE-2 plus 50 amino-terminal residues of exon 5 unique to IE-2. A wild-type allele of the IE-2 region was cloned independently into pGS284 and used to rescue the UL122 deletion BAC, also by conjugative transfer. Briefly, to transfer DNA sequences in pGS284 to the HCMV BAC, E. coli S17-λpir containing the GS284 donor plasmid was conjugated with a RecA+ derivative of E. coli DH10B (34) harboring the HCMV BAC DNA. Exconjugates were selected sequentially with antibiotics and sucrose, and the progeny molecules were examined by restriction digestion to identify BAC clones with the intended alteration.
Transfection by electroporation.
Actively dividing HFF cells were trypsinized and suspended in Dulbecco's modified Eagle medium with 10% fetal calf serum. For each transfection, 4 × 106 HFF cells were suspended in 260 μl of medium plus 10% serum and mixed with 2 μg of viral or BAC DNA, 1 μg of plasmid pCMV71 (23) (a gift from J. Baldick), and 1 μg of plasmid pEGFP-N1 (Clontech) in a 0.4-cm cuvette. Following electroporation at 250 V and 960 μF, the cells were plated in a 10-cm-diameter tissue culture plate. Typically, 1 to 2 days after transfection the surviving cells were approaching confluence and were split between 1:2 and 1:4 into new dishes. The passaged cells were cultured at 37°C for plaque outgrowth or harvested for analysis. Plaques were scored on the criteria of visual morphology or GFP expression within multiple adjacent cells in the monolayer. The pattern and spread of GFP fluorescence to neighboring cells in this assay are similar to those observed by IE-1 or IE-2 staining of cultures infected with wild-type Towne virus (2), although GFP expression is delayed by several days relative to IE gene expression.
Molecular analyses, RT-PCR, and immunofluorescence.
Plasmid cloning, restriction enzyme digestion, gel electrophoresis, PCR, and Southern blotting were performed according to established protocols (1). For reverse transcription-PCR (RT-PCR) analysis, cells were transfected with BAC DNA and plasmids as described above. To maximize our ability to detect HCMV gene expression from transfected BAC DNA, cultures in which at least 10 to 20% of the cells expressed GFP 4 to 6 days after transfection were used for RT-PCR analysis. Total RNA was isolated using TRIZOL reagent (Gibco BRL) and digested with RNase-free DNase (Promega). First-strand cDNA was synthesized from an oligo(dT) primer using PowerScript reverse transcriptase (Clontech), according to the manufacturer's protocol. This cDNA served as the template for PCR (35 cycles) using Advantage cDNA polymerase (Clontech) and primer sets for various HCMV IE and early genes as indicated in text and figure legends. PCR products were analyzed by agarose gel electrophoresis.
Expression of HCMV IE proteins in transfected cultures was analyzed by immunofluorescence using monoclonal antibodies 1B12 and 3H9, specific for the unique domains of IE-1 and IE-2, respectively (Marchini and Zhu, unpublished). Briefly, cultures were fixed 3 days after transfection in 4% paraformaldehyde, permeabilized, and blocked with bovine serum albumin. The fixed cells were reacted with primary monoclonal hybridoma culture supernatants followed by secondary antibody conjugated to Alexa-568 (Molecular Probes). Stained cultures were examined by inverted epifluorescence microscopy.
RESULTS
Production of a Towne HCMV BAC.
A Towne HCMV BAC was produced and isolated essentially as described by Borst et al. (4), beginning with introduction of BAC sequences into the viral genome via homologous recombination in cultured mammalian cells and then recovery and maintenance of circular genomes in E. coli. To avoid creating a recombinant genome too large to package, we designed a plasmid vector, pUSF-3 (see Fig. 1 and Materials and Methods), that would delete about 9 kb of HCMV DNA from the dispensable portion of the US region (18). Transfer of pUSF-3 to the Towne US target yields a recombinant HCMV containing F-plasmid sequences necessary for episomal maintenance in E. coli, a selectable marker conferring prokaryotic resistance to chloramphenicol, and a cassette for eukaryotic expression of GFP (Fig. 1A).
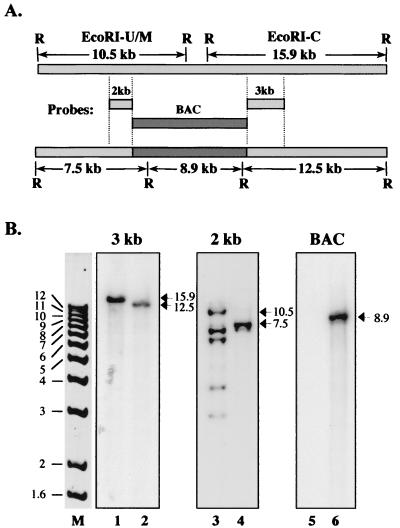
Following electroporation of RecA− E. coli with closed circular DNA from infected HFF cells, BAC DNA was prepared from resistant colonies and examined by restriction digestion. A clone with a restriction pattern comparable to that of Towne viral DNA was examined extensively by Southern blotting (Fig. 2). Initially, blots were probed with each of the HCMV DNA fragments in pUSF-3 to examine the structure of the US region to which the recombination had been targeted (Fig. 2A). A Southern blot probed with the 3-kb pUSF-3 fragment showed the 15.9-kb EcoRI C fragment of Towne replaced by the predicted 12.5-kb fragment in the recombinant (Fig. 2B, lanes 1 and 2). Probing with the 2-kb pUSF-3 fragment showed the predicted reduction of the 10.5-kb EcoRI U/M junction fragment to 7.5 kb (Fig. 2B, lanes 3 and 4). The 2-kb probe contains DNA from the terminal repeat of the US region and thus detected all fragments containing either end of that region. One of these, the 7-kb EcoRI U/Z junction fragment, occurs in both the BAC and viral DNAs (lanes 3 and 4). Consistent with the anticipated lack of linear viral DNA in the BAC, the 2-kb probe detects the 6.5-kb EcoRI M and 3-kb EcoRI Z free-end fragments only in viral DNA (lane 3). The origin of the additional bands of 3.7 and 4.4 kb visible in our Towne viral DNA preparation (lane 3) is unclear; the sizes are consistent with EcoRI-Z fragments that have one and two additional copies, respectively, of the 700-bp terminal a sequence and may arise from differential cleavage between tandem copies of the terminal sequence during cleavage of concatemeric DNA (24). The pMBO1374 BAC vector was also used as a probe and detected an internal 8.9-kb EcoRI fragment from the integrated plasmid in BAC DNA but failed to hybridize to viral DNA (compare lanes 5 and 6). Additional Southern blot analyses with cosmid probes that span the entire HCMV genome further confirmed that this BAC clone contained, without rearrangement, all of the EcoRI fragments present in the parental Towne viral DNA (data not shown). The restriction mapping analysis also indicated that the genome in the BAC clone was in the prototypical isomeric arrangement, consistent with the structure of pUSF-3. As observed with the AD169 HCMV BAC clone, there was no evidence of genome isomerization within E. coli (4). These analyses confirmed all predictions for the structure of a recombinant Towne HCMV-BAC resulting from homologous recombination with pUSF-3; therefore, we examined this clone, designated T-BACwt, further to characterize its biological properties.
FIG. 2.
Genome structure of the Towne HCMV BAC. To ascertain that the T-BAC genome had the correct structure, Towne viral DNA isolated from virions and T-BAC DNA from E. coli were digested with EcoRI and Southern blotted with probes derived from pUSF-3. (A) EcoRI maps of the relevant regions of wild-type Towne (upper bar) and the predicted T-BAC recombinant (lower bar) are shown along with the three pUSF-3 probes (center). The 10.5- and 15.9-kb fragments of wild-type Towne correspond to the EcoRI M/U junction and EcoRI C fragments, respectively, in the prototypic isomer of HCMV DNA (37). (B) Restriction fragment patterns obtained following EcoRI digestion of Towne viral DNA (lanes 1, 3, and 5) and T-BAC DNA (lanes 2, 4, and 6). The probe used (A) is indicated above each set of lanes. The sizes (in kilobases) of 1-kb ladder (Gibco-BRL) standards are shown in lane M.
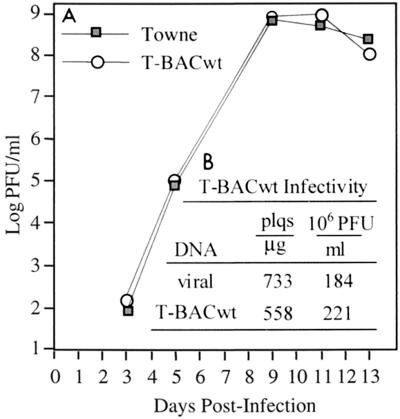
To establish that our T-BAC clone retained the fundamental biological characteristics of Towne viral DNA, T-BACwt DNA purified from E. coli was transfected into HFF cells with an expression plasmid for the HCMV tegument protein pp71 (23). The transfected cells were plated, and the cultures were monitored for development of cytopathic effect and peak virus titer (Fig. 3). Wild-type Towne DNA, purified from virions, and T-BACwt DNA from E. coli both produced plaques following transfection into HFF cells (Fig. 3B). Comparing these two molecules in numerous transfections, we found viral DNA to yield more plaques than the BAC, and vice versa; thus, we feel that the observed difference between viral DNA and T-BACwt is due more to variation inherent to these experiments than to a genuine difference in biological features. The peak titers, of infectious virus particles recovered from transfection supernatants were also similar (Fig. 3B). In addition to characterizing production of virus from transfected BAC DNA, we examined the infectious properties of the resulting HCMV BAC virus stocks following low-MOI infection of HFF cells. Towne and T-BACwt viruses generated very similar yields of infectious progeny (Fig. 3A), a minor difference being that T-BACwt, either as transfected DNA or as infectious T-BAC virus, developed visible cytopathic effect more slowly than its wild-type Towne counterpart (data not shown). We do not know whether this results from the deletion of US genes or from a secondary mutation elsewhere in T-BACwt DNA. Aside from this minor phenotypic difference in cytopathic effect, the additional tests established that the T-BACwt bacterial clone of Towne HCMV retains the essential properties of wild-type Towne virus; therefore, T-BACwt was taken as the starting reagent for subsequent experiments.
FIG. 3.
Biological characterization of T-BACwt. (A) HFF cells were infected with 0.01 PFU/cell of the indicated virus for 1 h at 37°C and nascent virus was titered on fresh HFF cells on the indicated days following infection. Virus yield is averaged from three independent infections with wild-type Towne or T-BACwt virus. (B) HFF cells were transfected with wild-type Towne DNA isolated from virions or T-BACwt DNA purified from E. coli, plus expression vectors for GFP and the HCMV pp71 tegument protein (2). Plaques were counted 7 to 10 days after transfection by visible or fluorescence microscopy, and the yield was expressed as plaques (plqs) per microgram of transfected DNA. When cytopathic effect reached 100% in transfected cultures, nascent virus was titered on fresh HFF cells (indicated as millions of PFU per milliliter of culture supernatant).
Derivation of a deletion mutant of IE-2.
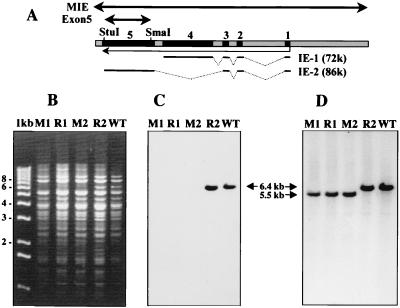
Our primary motivation for creating an HCMV BAC was a long-standing interest in genetic analysis of the MIE gene IE-2 (pp86). Our inability to isolate an IE-2 deletion mutant free of contaminating wild-type virus by plaque purification on normal HFF cells suggested first that this mutant was impaired for growth (Marchini and Zhu unpublished) and additionally that available means for providing complementing IE-2 were inadequate. The IE-1 and IE-2 transcripts share three small 5′ exons and then splice differentially to either of two unrelated exons that encode the majority of each protein (Fig. 4A). To create a mutation which would have maximal effect on IE-2 without directly affecting IE-1, the majority of the main exon unique to IE-2, encoded by the UL122 reading frame, was deleted from a 6.7-kb EcoRI-SalI subclone of the Towne MIE region. The deleted allele with flanking sequences for recombination was cloned into pGS284, a derivative of the positive suicide selection vector pCVD442 (see Materials and Methods), and conjugated into RecA+ E. coli harboring T-BACwt. Exconjugates were selected, and the T-BAC DNA was examined by Southern blotting. Approximately half of the exconjugate T-BAC clones now had a 5.5-kb EcoRI-SalI MIE fragment that was detected by a wild-type MIE probe but failed to hybridize to the deleted SmaI-StuI exon 5 fragment, as would be expected from a T-BAC clone containing the UL122 deletion (clones M1 and M2 in Fig. 4B to D). Two of these were selected for further use and were designated T-BACΔ122. To control for mutations at unrelated loci that might have been introduced during production of the T-BACΔ122 genome, the two T-BACΔ122 clones were each rescued to the wild-type state by repeating the conjugative mating protocol with a wild-type UL122 donor allele. Again, about half of the clones examined following selection had the UL122 locus repaired, as evidenced by the return of the EcoRI-SalI MIE fragment to the 6.7-kb wild-type size, and restored hybridization to the SmaI-StuI exon 5 probe (representative clone R2 in Fig. 4B to D). As is typical with conintegrate resolution, those genomes that did not incorporate the mutation retained the structure of the target BAC. An example of this is the rescue clone labeled R1 in Fig. 4. The two original T-BACΔ122 clones and one repaired derivative of each (designated T-BACΔ122R) were used for the experiments described below.
FIG. 4.
Derivation of UL122 deletion and rescued BAC genomes. (A) Schematic representation of the MIE region of HCMV showing the positions of the five productive cycle exons (numbered black bars) and the splicing patterns (broken lines) of the two most abundant IE proteins, IE-1 and IE-2. Transcription is indicated by the light arrow below the bar. The SmaI and StuI restriction sites at the exon 5 deletion endpoints and the probes used for Southern blotting (heavy arrows) are indicated above the bar. (B to D) Wild-type Towne HCMV DNA (WT) or representative T-BACΔ122 (M1 and M2) and T-BACΔ122R (R1 and R2) DNAs were digested with EcoRI plus SalI and electrophoresed in agarose. The gel was stained with ethidium bromide to record the position of the marker fragments (Gibco 1-kb ladder) (B) and then blotted to nitrocellulose and probed sequentially with probes for UL122 exon 5 (C) and a 6.7-kb EcoRI-SalI fragment containing the entire MIE region of Towne HCMV (D). Positions of the wild-type 6.7-kb fragment and the deleted 5.5-kb derivative are noted.
A UL122 deletion BAC is not infectious.
To begin assessing the biological properties of the UL122 mutant, we electroporated purified T-BACwt, T-BACΔ122, and T-BACΔ122R DNAs into permissive HFF cells and monitored the transfected cultures for plaque development. The transfection experiments were repeated many times with multiple viral DNA and BAC DNA preparations to minimize the chance that negative results were due to defects in the BAC DNA introduced during passage in E. coli or DNA preparation. After 7 to 10 days, nascent plaques were identified by GFP expression, and the total plaque yield from each transfected DNA was scored. Selected representative results are presented in Table 1. Experimental variation, due primarily to differences in transfection efficiency and the quality of individual BAC DNA preparations, produced a wide range of plaque yields for T-BACwt and T-BACΔ122R, from a few to several hundred per transfection; thus, a critical quantitative interpretation of these data is of limited value. Nonetheless, qualitative examination shows the clear difference between these substrates and T-BACΔ122 DNA, which never yielded a recognizable plaque either in the six independent transfections reported in Table 1 or in 15 additional experiments (not shown).
TABLE 1.
Infectivity of transfected T-BAC DNAsa
| Genotype | No. of exptsb | No. of plaques/transfection
|
|
|---|---|---|---|
| Range | Mean | ||
| Wild type | 7 | 4–642 | 326 |
| Δ122 | 6 | NAc | 0 |
| Δ122R | 7 | 10–648 | 223 |
HFF cells were transfected with T-BACwt, T-BACΔ122, or T-BACΔ122R and monitored for plaque development (see Materials and Methods).
Data are from the indicated number of representative experiments out of a larger number of trials using multiple preparations of viral and BAC DNA over a 6 month period.
NA, not applicable (no preparation of T-BACΔ122 DNA yielded plaques which could be recognized either morphologically or by GFP expression, even after 4 weeks of culture).
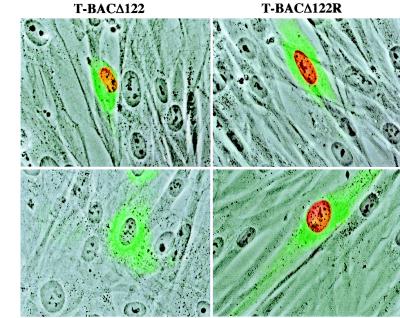
To confirm that the mutation gave the expected phenotype at the protein level, cultures transfected with BAC DNA were fixed and stained using monoclonal antibodies specific for IE-1 or IE-2 expression. With GFP expression from a cotransfected plasmid as a guide to identify transfected cells, expression of IE-1 was readily detected from either T-BACΔ122 or T-BACΔ122R genomes 4 to 5 days after transfection (Fig. 5, top row). In contrast, expression of IE-2 was observed only in cultures transfected with T-BACΔ122R DNA; no IE-2 protein was found in any cells transfected with T-BACΔ122 DNA (Fig. 5, bottom row). The monoclonal antibody used to detect IE-2 was specific to an epitope within the deletion (A. Marchini, unpublished observations); thus, we do not know whether the truncation protein predicted to be synthesized from the deletion allele (see Materials and Methods) was produced in T-BACΔ122-transfected cells.
FIG. 5.
Immunofluorescence of MIE proteins expressed by T-BACΔ122. HFF cells were transfected with T-BACΔ122 or T-BACΔ122R DNA purified from E. coli as described in the text. Four days after transfection, the monolayers were fixed and stained with monoclonal antibodies specific for IE-1 (top row) or IE-2 (bottom row) (see Materials and Methods), followed by a secondary antibody coupled to Alexa-568 (Molecular Probes). Transfected cells were identified by GFP expression from a cotransfected plasmid.
The failure to recover a plaque from 21 independent transfection experiments strongly suggests that the inability of T-BACΔ122 to produce plaques is not a chance occurrence but rather is due to the deficiency of IE-2. Even when T-BACΔ122-transfected HFF cells were cultured for 4 weeks after transfection, we found no evidence of plaque formation as manifested by either visible cytopathic effect or GFP expression. The possibility that the replication defect is due to mutation at a secondary site within the BAC is effectively ruled out by our ability to rescue the phenotype of the mutant with wild-type UL122 sequences. The all-or-nothing nature of this assay, unfortunately, prevents our making any substantial quantitative statement regarding the degree of impairment relative to the control viruses, and since we have been unable to complement the UL122 deletion mutant BAC, we cannot yet generate stocks of mutant virus with which to perform quantitative virological assessments.
The UL122 deletion mutant is defective for early gene expression.
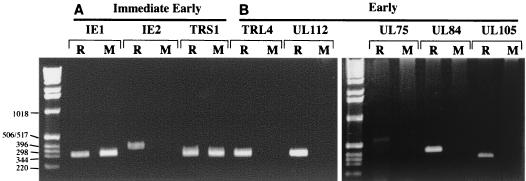
Detection of IE-1 expression from transfected T-BACΔ122 DNA by immunofluorescence indicated that aspects of infection prior to IE gene expression were not markedly impaired in the UL122 mutant. We therefore examined the expression of other IE loci and a panel of early genes using RT-PCR to establish whether these were expressed normally in the mutant. HFF cells were transfected with T-BACΔ122 or T-BACΔ122R DNA and cultured for 5 days, after which total RNA was prepared from the cultures and subjected to RT-PCR analysis using primers to three IE genes and five early genes. RNA expression was detected for IE-1, IE-2, and TRS1 in cells transfected with wild-type Towne (data not shown) and T-BACΔ122R DNA and for IE-1 and TRS1 in the T-BACΔ122-transfected cultures (Fig. 6A). The amplification conditions for this analysis were not quantitative, and therefore the results do not rule out the possibility that IE gene expression is altered in the mutant; however, the results clearly indicate that expression of IE genes, other than IE-2, remained detectable from the T-BACΔ122 genome. In contrast, no expression was detected from the mutant genome for any of the five early genes examined (Fig. 6B). Notably, our assay failed to detect mRNA from TRL4, one of the most abundant early transcripts of HCMV (38). These results, together with the plaque outgrowth experiments in Table 1 demonstrate that HCMV DNA lacking UL122 is growth impaired in HFF cells and that this impediment correlates with a failure to activate expression of early genes.
FIG. 6.
RT-PCR analysis of T-BACΔ122 gene expression. HFF cells were transfected with T-BACΔ122 (lanes M) or T-BACΔ122R (lanes R) DNA purified from E. coli as described in the text. Five days after transfection, RNA was isolated from the cultures and used as template for RT-PCR amplification with primers for three IE genes (IE-1, IE-2, and TRS1) and five early genes (UL75, UL84, UL105, UL122, and TRL4). PCR products were separated by agarose electrophoresis and stained with ethidium bromide. The sizes (in base pairs) of relevant marker bands are noted at the left.
DISCUSSION
The findings that IE-2 transactivates gene expression in a promiscuous manner and interacts with a number of important cellular proteins have implied the importance of IE-2 to HCMV growth. Our inability to purify IE-2 mutants in the absence of complementation has further suggested that IE2 is probably critical to HCMV replication, but the consequent inability to produce a pure stock of mutant virus has precluded the formal demonstration of this hypothesis. For this study of UL122, we created a new recombinant Towne HCMV with a substitution of BAC maintenance functions and a GFP marker for a dispensable portion of the US region. This new Towne HCMV-BAC, T-BACwt, had no gross rearrangements to its genome and had a restriction enzyme map identical to the parent Towne genome except for the area targeted for recombination. T-BACwt has retained the originally characterized restriction map through numerous passages in E. coli (H. Zhu, unpublished observations), confirming the previous observations that the HCMV genome appears to be quite stable as a BAC clone and that it also remains in a single isomeric configuration in E. coli (4). Our finding that T-BACwt DNA and Towne viral DNA yield comparable numbers of plaques after transfection into permissive cells provides additional evidence that a single isomer of an HCMV genome is as infectious as a mixture. T-BACwt entirely lacks US1 through US11, from the US region of HCMV, and truncates IRS1 and US12, confirming previous findings that this group of genes is dispensable for replication in cell culture (18, 21). The comparable yield and growth kinetics of virus stocks derived from transfected T-BACwt DNA and Towne virus argue that we may justifiably equate our BAC clone and Towne virus for cell culture experiments. Using T-BACwt to isolate a UL122 mutant genome in E. coli has circumvented the need for plaque purification to generate a genotypically pure stock and thereby provided a critical reagent to begin characterizing an IE-2 mutant. The experiments that we have described using these BAC clones provide the first compelling experimental evidence that HCMV replication is substantially dependent on the function of one or more gene products encoded by sequences in UL122.
Under the conditions that we used, viral DNA lacking UL122 was no longer infectious following transfection into permissive HFF cells. While we have attempted to maximize the specific infectivity of the transfected DNA by expressing the pp71 HCMV tegument protein in the transfected cells (2), this assay ultimately gave an all-or-nothing result. While we cannot state that the UL122 mutant is completely unable to replicate, it is nevertheless clear from our results that T-BACΔ122 is highly defective for growth. Establishing the degree to which replication is impaired in the mutant and whether the phenotype proves to be multiplicity dependent, as observed for an IE-1 mutant (15), must await the development of a complementing system capable of yielding a pure stock of mutant virus. Our results further indicated that the defect in replication of a UL122 mutant involves a failure to activate expression of early HCMV genes. The detection of IE proteins from this mutant indicates that the defect must occur after IE gene expression but before early genes are activated, therefore focusing attention primarily on IE gene functions themselves. Given the large amount of evidence documenting IE-2 transactivation activity, this may not be an unexpected phenotype. It must be noted, however, that a second, 55-kDa IE-2 polypeptide is also substantially encoded by UL122 sequences (37). Very little is known about this minor IE-2 species; however, Baracchini et al. have demonstrated transactivation by this protein (3). Since the T-BACΔ122 deletion is also expected to affect the 55-kDa species, we can not presently ascribe the observed phenotype unambiguously to the 86-kDa IE-2.
Our results also do not indicate the manner in which IE-2 affects early gene expression. Many reports have established that transactivation by IE-2 is not typically dependent on sequence-specific DNA binding but rather may occur more through protein-protein interactions (24). Other work has shown that IE-2 interacts with multiple basal and general transcription factors (8, 17, 22, 33) and known cell cycle regulators (13, 16, 26, 36). An intriguing hypothesis to explain what appears to be a general failure by the UL122 mutant to activate early genes is that interaction of IE-2 with global regulatory factors, such as retinoblastoma protein, CREB, and S1 (22, 39), leads to relief of transcriptional repression that otherwise restricts early gene expression (5, 35, 40). Such a mechanism might account for the observation that IE-2 appears to be able to transactivate most or all HCMV early genes as well as many non-HCMV promoters and also suggests that expression of IE-2 would be a critical element used by the virus to control entry into the lytic cycle from the latent state. It is still unclear which of the various interactions reported for IE-2 are important in transactivation or whether any hierarchy exists; because our mutation is a large deletion, it probably has pleiotropic effects on IE-2 activity. The ability to rapidly recombine new alleles into the deletion mutant genome in the BAC system provides an excellent platform with which to further examine specific aspects of IE-2 transactivation and its relation to HCMV biology.
Because IE-1 and IE-2 share certain amino-terminal sequences, it is not possible to create a simple deletion mutant in which all sequences encoding IE-2 are completely removed without altering the structure of IE-1. Our exon 5 deletion is predicted to express a carboxyl truncation of IE-2 which contains the 85 amino acids shared between IE-1 and IE-2 followed by an additional 55 amino acids from exon 5. Given that transactivator character has been previously ascribed to the shared domain (29), it can be imagined that the truncated IE-2 protein might retain some measure of IE-2 functionality. Formally, therefore, it can be questioned whether the phenotype of T-BACΔ122 DNA that we observed is due not to the loss of IE-2 function but rather to a dominant negative effect on virus replication exerted by the truncated polypeptide. We believe that this is unlikely for the following reason. If HFF cells are cotransfected as described with T-BACwt DNA plus an expression subclone of the MIE locus that should produce a truncation product identical to T-BACΔ122, there is no reduction in plaque yield (Zhu, unpublished). Assuming that the majority of cells transfected with BAC DNA also expressed a truncation protein from the plasmid DNA, this result is inconsistent with dominant negative interference by the residual protein product from the deleted MIE construct. It seems, therefore, simpler to conclude that the deletion of most of UL122 imparts a replication deficiency on HCMV.
The phenotypes associated with deletion of more than half of the HCMV IE genes have now been described for recombinant mutant strains, at least within the context of replication in cultured cells (4, 15, 18, 25, 27). Interestingly, only mutations in the MIE region have so far produced marked effects on virus replication (15). This doubtless reflects the simplicity of cell culture versus the whole organism; however, it also clearly indicates that the MIE locus is a reservoir of functions that are very basic to the mechanism of HCMV replication.
ACKNOWLEDGMENTS
We thank T. Shenk, G. Tullis, G. Smith, L. Enquist, and B. Fleckenstein for advice and reagents and T. Shenk for helpful discussion and critically reading the manuscript.
A.M. received postdoctoral support for this project from the Howard Hughes Medical Institute and fellowship PF-3893 from the American Cancer Society. This work was supported grants from The Foundation of UMDNJ (11-2000) and American Heart Association (9930280T) to H.Z.
REFERENCES
- 1.Ausubel F M, et al., editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1987. [Google Scholar]
- 2.Baldick C J, Jr, Marchini A, Patterson C E, Shenk T. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol. 1997;71:4400–4408. doi: 10.1128/jvi.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baracchini E, Glezer E, Fish K, Stenberg R M, Nelson J A, Ghazal P. An isoform variant of the cytomegalovirus immediate-early auto repressor functions as a transcriptional activator. Virology. 1992;188:518–529. doi: 10.1016/0042-6822(92)90506-k. [DOI] [PubMed] [Google Scholar]
- 4.Borst E M, Hahn G, Koszinowski U H, Messerle M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J Virol. 1999;73:8320–8329. doi: 10.1128/jvi.73.10.8320-8329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 6.Britt W J. Infectious clones of herpesviruses: a new approach for understanding viral gene function. Trends Microbiol. 2000;8:262–265. doi: 10.1016/s0966-842x(00)01747-9. [DOI] [PubMed] [Google Scholar]
- 7.Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, Knipe D M, Howley P M, editors. Fields Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2523. [Google Scholar]
- 8.Caswell R, Hagemeier C, Chiou C J, Hayward G, Kouzarides T, Sinclair J. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J Gen Virol. 1993;74:2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- 9.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A d, Kouzarides T, Martignetti J A, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 10.Dahle A F, Fowler K B, Wright J D, Boppana S B, Britt W J, Pass R F. Longitudinal investigation of hearing disorders in children with congenital cytomegalovirus. J Am Acad Audiol. 2000;11:283–290. [PubMed] [Google Scholar]
- 11.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 12.Fleckenstein B, Muller I, Collins J. Cloning of the complete human cytomegalovirus genome in cosmids. Gene. 1982;18:39–46. doi: 10.1016/0378-1119(82)90054-3. [DOI] [PubMed] [Google Scholar]
- 13.Fortunato E A, McElroy A K, Sanchez I, Spector D H. Exploitation of cellular signaling and regulatory pathways by human cytomegalovirus. Trends Microbiol. 2000;8:111–119. doi: 10.1016/s0966-842x(00)01699-1. [DOI] [PubMed] [Google Scholar]
- 14.Fowler K B, McCollister F P, Dahle A J, Boppana S, Britt W J, Pass R F. Progressive and fluctuating sensorineural hearing loss in children with asymptomatic congenital cytomegalovirus infection. J Pediatr. 1997;130:624–630. doi: 10.1016/s0022-3476(97)70248-8. [DOI] [PubMed] [Google Scholar]
- 15.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagemeier C, Walker S, Caswell R, Kouzarides T, Sinclair J. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol. 1992;66:4452–4456. doi: 10.1128/jvi.66.7.4452-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones T R, Muzithras V P. A cluster of dispensable genes within the human cytomegalovirus genome short component: IRS1, US1 through US5, and the US6 family. J Virol. 1992;66:2541–2546. doi: 10.1128/jvi.66.4.2541-2546.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones T R, Muzithras V P, Gluzman Y. Replacement mutagenesis of the human cytomegalovirus genome: US10 and US11 gene products are nonessential. J Virol. 1991;65:5860–5872. doi: 10.1128/jvi.65.11.5860-5872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kemble G, Duke G, Winter R, Spaete R. Defined large-scale alterations of the human cytomegalovirus genome constructed by cotransfection of overlapping cosmids. J Virol. 1996;70:2044–2048. doi: 10.1128/jvi.70.3.2044-2048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kollert-Jons A, Bogner E, Radsak K. A 15-kilobase-pair region of the human cytomegalovirus genome which includes US1 through US13 is dispensable for growth in cell culture. J Virol. 1991;65:5184–5189. doi: 10.1128/jvi.65.10.5184-5189.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lang D, Gebert S, Arlt H, Stamminger T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 25.Mocarski E S, Kemble G W, Lyle J M, Greaves R F. A deletion mutant in the human cytomegalovirus gene encoding IE1 (491aa) is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy E A, Streblow D N, Nelson J A, Stinski M F. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J Virol. 2000;74:7108–7118. doi: 10.1128/jvi.74.15.7108-7118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patterson C E, Shenk T. Human cytomegalovirus UL36 protein is dispensable for viral replication in cultured cells. J Virol. 1999;73:7126–7131. doi: 10.1128/jvi.73.9.7126-7131.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piot P. Global AIDS epidemic: time to turn the tide. Science. 2000;288:2176–2178. doi: 10.1126/science.288.5474.2176. [DOI] [PubMed] [Google Scholar]
- 29.Pizzorno M C, Mullen M A, Chang Y N, Hayward G S. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J Virol. 1991;65:3839–3852. doi: 10.1128/jvi.65.7.3839-3852.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plachter B, Traupe B, Albrecht J, Jahn G. Abundant 5 kb RNA of human cytomegalovirus without a major translational reading frame. J Gen Virol. 1988;69:2251–2266. doi: 10.1099/0022-1317-69-9-2251. [DOI] [PubMed] [Google Scholar]
- 31.Roizman B, Sears A E. Herpes simplex viruses and their replication. 3rd Edition ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 32.Scott L L, Hollier L M, Dias K. Perinatal herpesvirus infections. Herpes simplex, varicella, and cytomegalovirus. Infect Dis Clin North Am. 1997;11:27–53. doi: 10.1016/s0891-5520(05)70340-8. [DOI] [PubMed] [Google Scholar]
- 33.Scully A L, Sommer M H, Schwartz R, Spector D H. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J Virol. 1995;69:6533–6540. doi: 10.1128/jvi.69.10.6533-6540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith G A, Enquist L W. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J Virol. 1999;73:6405–6414. doi: 10.1128/jvi.73.8.6405-6414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sommer M H, Scully A L, Spector D H. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J Virol. 1994;68:6223–6231. doi: 10.1128/jvi.68.10.6223-6231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speir E, Modali R, Huang E S, Leon M B, Shawl F, Finkel T, Epstein S E. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 37.Stinski M. Cytomegalovirus and its replication. New York. N.Y: Raven Press; 1991. [Google Scholar]
- 38.Wathen M W, Stinski M F. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J Virol. 1982;41:462–477. doi: 10.1128/jvi.41.2.462-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J, O'Neill J, Barbosa M S. Transcription factor Sp1 mediates cell-specific trans-activation of the human cytomegalovirus DNA polymerase gene promoter by immediate-early protein IE86 in glioblastoma U373MG cells. J Virol. 1998;72:236–244. doi: 10.1128/jvi.72.1.236-244.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H S, Postigo A A, Dean D C. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFβ, and contact inhibition. Cell. 1999;97:53–61. doi: 10.1016/s0092-8674(00)80714-x. [DOI] [PubMed] [Google Scholar]
- 41.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]