Abstract
The primary sequence of the long unique region L-DNA (L for low GC) of rhesus monkey rhadinovirus (RRV) isolate 26-95 was determined. The L-DNA consists of 130,733 bp that contain 84 open reading frames (ORFs). The overall organization of the RRV26-95 genome was found to be very similar to that of human Kaposi sarcoma-associated herpesvirus (KSHV). BLAST search analysis revealed that in almost all cases RRV26-95 coding sequences have a greater degree of similarity to corresponding KSHV sequences than to other herpesviruses. All of the ORFs present in KSHV have at least one homologue in RRV26-95 except K3 and K5 (bovine herpesvirus-4 immediate-early protein homologues), K7 (nut-1), and K12 (Kaposin). RRV26-95 contains one MIP-1 and eight interferon regulatory factor (vIRF) homologues compared to three MIP-1 and four vIRF homologues in KSHV. All homologues are correspondingly located in KSHV and RRV with the exception of dihydrofolate reductase (DHFR). DHFR is correspondingly located near the left end of the genome in RRV26-95 and herpesvirus saimiri (HVS), but in KSHV the DHFR gene is displaced 16,069 nucleotides in a rightward direction in the genome. DHFR is also unusual in that the RRV26-95 DHFR more closely resembles HVS DHFR (74% similarity) than KSHV DHFR (55% similarity). Of the 84 ORFs in RRV26-95, 83 contain sequences similar to the recently determined sequences of the independent RRV isolate 17577. RRV26-95 and RRV17577 sequences differ in that ORF 67.5 sequences contained in RRV26-95 were not found in RRV17577. In addition, ORF 4 is significantly shorter in RRV26-95 than was reported for RRV17577 (395 versus 645 amino acids). Only four of the corresponding ORFs between RRV26-95 and RRV17577 exhibited less than 95% sequence identity: glycoproteins H and L, uracil DNA glucosidase, and a tegument protein (ORF 67). Both RRV26-95 and RRV17577 have unique ORFs between positions 21444 to 21752 and 110910 to 114899 in a rightward direction and from positions 116524 to 111082 in a leftward direction that are not found in KSHV. Our analysis indicates that RRV26-95 and RRV17577 are clearly independent isolates of the same virus species and that both are closely related in structural organization and overall sequence to KSHV. The availability of detailed sequence information, the ability to grow RRV lytically in cell culture, and the ability to infect monkeys experimentally with RRV will facilitate the construction of mutant strains of virus for evaluating the contribution of individual genes to biological properties.
Kaposi's sarcoma-associated herpesvirus (KSHV) is associated with classical and AIDS-related Kaposi's sarcoma (4, 6–8, 23), as well as with primary effusion lymphoma (25, 36) and multicentric Castleman's disease (14, 31, 38). Sequence analysis of the KSHV genome (34) indicated that it is more closely related to herpesvirus saimiri (HVS) than to other herpesviruses and thus is assigned to the gamma-2 or rhadinovirus subgroup of the herpesvirus family (33). Investigation of the role of individual KSHV genes in replication and disease has been limited by the lack of a permissive cell culture system (13, 19, 32) and of an appropriate animal model.
A herpesvirus that can be grown lytically in cell culture was recently isolated from the peripheral blood of a rhesus monkey (animal 26-95) at the New England Regional Primate Research Center (12). Sequencing and BLAST search analysis of a 10.6-kbp fragment of virion DNA revealed sequences corresponding to KSHV open reading frame (ORF) 7; intact genes for glycoprotein B (ORF 8), DNA polymerase (ORF 9), ORF 10, ORF 11, and viral interleukin-6 (vIL-6; ORF R2); and a partial gene for thymidylate synthetase (TS; ORF 70) (12). Based on these similarities in gene order and sequences this virus was assigned to the rhadinovirus subgroup and named rhesus monkey rhadinovirus (RRV) (12).
Recently, the primary sequence of an independent RRV isolate (isolate 17577) has been determined. RRV17577 sequences over the same 10.6-kbp stretch were found to be colinear and closely related to those from the original RRV26-95 isolate (37). Furthermore, the genome organization of RRV17577 was closely but not entirely colinear with that of KSHV (37).
In this report, we reveal the complete primary sequence of the long unique region (L-DNA) of RRV26-95. The genomic organization of RRV26-95 and sequence of individual ORFs contained within the L-DNA are compared to KSHV and RRV17577 sequences.
MATERIALS AND METHODS
Virus purification.
RRV26-95 was grown in primary rhesus monkey fibroblasts in Dulbecco's modified Eagle's medium (Gibco, Grand Island, N.Y.) supplemented with 20% fetal bovine serum (Sigma, St. Louis, Mo.), 2 mM l-glutamine, and penicillin-streptomycin (50 IU and 50 μg/ml, respectively), as previously described (12). Procedures for the purification of virus have been previously detailed (11). Briefly, cells and debris were removed by low-speed centrifugation following complete cell lysis. The supernatant was then filtered through a 0.45-μm-pore-size filter to remove any residual cells and debris. The filtered supernatant was centrifuged for 3 h at 17,000 rpm in a Sorvall type 19 rotor in order to pellet virus. Resuspended virus was fractionated by Sepharose 4B column chromatography, and virus contained in the void volume was used as a source of virion DNA for cloning.
Cloning of RRV26-95 DNA fragments.
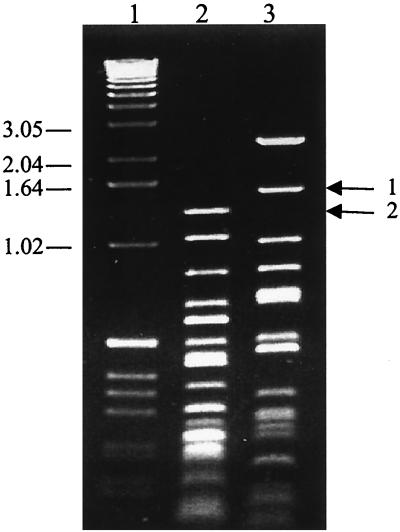
First, 10 μg of purified virion DNA in a 200-μl volume was sonicated for 10 s at 18% capacity in a 550 Sonic Dismembrator (Fischer Scientific, Medford, Mass.). Sonicated DNA was treated with T4 DNA polymerase (New England Biolabs, Beverly, Mass.) to create blunt ends and electrophoresed through 0.8% agarose, and DNA fragments of 0.5 to 1.0 kbp were isolated. Sonicated, purified DNA was inserted into the SmaI site of the pUC18 cloning vector (Pharmacia, Piscataway, N.J.). In addition, unsonicated DNA was digested with KpnI, EcoRI, SmaI, HindIII, and PstI restriction enzymes (New England Biolabs). Then, 5.0- to 10.0-kbp restriction fragments were isolated and cloned into either pUC18 (EcoRI, SmaI, and HindIII) or pSP72 (Promega) (KpnI and PstI) cloning vectors. As a control for the size of a stretch of repetitive DNA between ORF 69 and R13 (Fig. 1), the EcoRI fragment vector (E6) was digested with RsaI and HaeIII enzymes. Restriction fragments were electrophoresed through 0.8% agarose, and 1.5-kbp RsaI and 1.3-kbp HaeIII fragments (Fig. 2) that were predicted to contain the repetitive sequences of interest were isolated and inserted into the SmaI site of the pUC18 cloning vector. The plasmid clones described here were grown in XL-2 Blue MRF′ ultracompetent cells (Stratagene, La Jolla, Calif.).
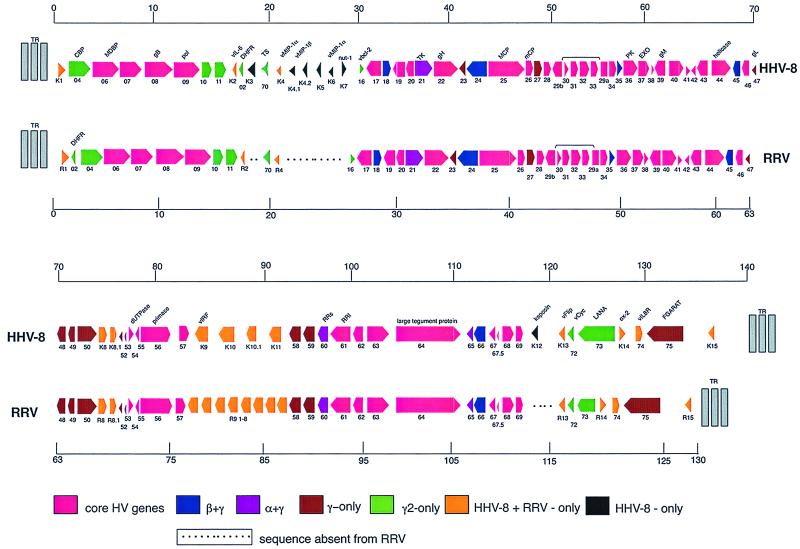
FIG. 1.
Alignment of ORFs of KSHV and RRV26-95. The different colors signify ORFs contained in KSHV and RRV26-95 that are conserved in the indicated herpesvirus subfamilies or subgroups. The square side of the symbol signifies the 5′ end, and the pointed side of the symbol signifies the 3′ end of the depicted ORFs. The ORFs are not drawn to scale.
FIG. 2.
The restriction digestion pattern of the RRV26-95 E6 clone. E6, an 11.0-kbp RRV26-95 restriction fragment clone that contains sequences from ORFs 66 to 71, was independently digested with HaeIII (lane 2) and RsaI (lane 3). Arrow 1 signifies the 1.5-kbp RsaI band (lane 3) and arrow 2 signifies the 1.3-kbp HaeIII band (lane 2) that contains highly repetitive sequences located between ORF 69 and R13 (Fig. 1) of RRV26-95. Lane 1 is a molecular weight marker, and the sizes of the marker bands are indicated at the left in thousands.
Assembly of RRV26-95 sequences.
pUC18 clones were initially screened for insert DNA by HaeII digestion. The sequence of insert DNA from sonication clones or the ends of restriction fragment clones was determined by using M13 forward and reverse primers (Promega, Madison, Wis.) and a Big Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer Cetus, Norwalk, Conn.). The ends of restriction fragment inserts cloned into pSP72 were sequenced by using M13, T7, and SP6 primers (Promega). Sonication clones that contained a high G+C content were sequenced using a dGTP Big Dye Terminator Ready Reaction kit (Perkin-Elmer Cetus). Sequence data were edited using Sequencer 3.0 software (Gene Codes Corporation) and assembled into contiguous overlapping fragments (contigs) by using Sequencer and AssemblyLIGN software (Kodak Lab Research, Rochester, N.Y.). Contig sequences were compared to sequences contained in GenBank by using BLASTX and assigned tentative positions to corresponding KSHV (34) and HVS (1) sequences. Gaps in sequence between contigs were filled by PCR amplification of these sequences by using primers that annealed near the end of contig sequences. The sequence of the majority of these gaps were then determined by using a series of custom primers to walk cloned DNA. For larger gaps, amplified DNA fragments were independently digested with two restriction enzymes with four base recognition sites, AluI and RsaI (New England Biolabs), and the resulting fragments were cloned into the SmaI site of pUC18. The sequence of these fragments was determined by using M13 forward and reverse primers as described above. Using these methods, we acquired the complete double-stranded sequence of the L-DNA of RRV26-95. The sequence presented in this report represents an average of 8.14 readings per base over the entire L-DNA.
Alignment and phylogenetic analysis of RRV26-95, RRV17577, and KSHV sequences.
The positions of ORFs in RRV26-95 sequences were determined by using MacVector 5.0 software (Oxford Molecular Group, Campbell, Calif.). The sequences of these ORFs were aligned to RRV17577 and KSHV ORFs using Gap Analysis software (Wisconsin GCG package, version 9.1; Oxford Molecular Group) and manually adjusted. Phylogenetic trees of RRV26-95, KSHV, and HVS ORF sequences were constructed by using PAUP* 4.0 software. A minimum of 200 bases of open sequence was used to determine putative ORFs in the L-DNA of RRV26-95.
IL-6 rescue assay in B9 cells.
RRV26-95 vIL-6 sequences were PCR amplified and inserted into the multiple cloning site located between the murine leukemia virus (MuLV) long terminal repeat (L) and the simian virus 40 (SV40) early promoter (S) of the vector LXSG (2). Thus, vIL-6 transcription was driven by MuLV sequences, and transcription of the green fluorescent protein (G) was driven by SV40 sequences. The resulting clone and the control LXSG vector were electroporated independently into COS-1 cell cultures. Twenty-four hours later, green fluorescing cells were examined visually and by flow cytometry. Supernatants were collected from vIL-6-expressing and control cultures. These samples were then incubated with the IL-6-dependent B9-cell line (5) for 48 h. At that time the viability of the B9 cells were tested using a cell proliferation kit (Promega).
Nucleotide sequence accession number.
RRV26-95 nucleotide sequence data have been deposited in the GenBank nucleotide sequence database under the accession no. AF210726.
RESULTS
Genomic organization of RRV26-95.
Sequence analysis of RRV26-95 virion DNA revealed 84 ORFs identifiable as herpesvirus genes within the 130,733 kbp that comprise the L-DNA. The L-DNA had a G+C content of 52.1%. We have used a nomenclature for RRV26-95 ORFs that corresponds to that used previously for KSHV ORFs (34). Beginning 513 bp upstream of the start of the R1 ORF, RRV26-95 contains repetitive sequence elements lacking ORFs of significant length (data not shown). These data suggest that the L-DNA of RRV26-95 begins 513 bp upstream of the start of the R1 ORF (10) and suggest that R1 is the leftmost gene in RRV26-95. Sequences 2,863 bp downstream of the RRV26-95 ORF 75 initiate repeating elements absent of any ORFs of significant length (data not shown). These sequences signify the 3′ end of the RRV26-95 L-DNA. R15 is located in this 2,863-bp stretch of sequence and is likely the rightmost gene in RRV26-95. The regions not included in the L-DNA are 65.0% G+C and are likely to represent terminal repeat sequences. Included in the L-DNA sequences of RRV26-95 are 39 genes that are common to all known herpesviruses (Fig. 1). In addition, beta- and gammaherpesvirus-specific ORFs 18, 24, 45, and 66, as well as alpha- and gammaherpesvirus-specific ORFs 21 (thymidine kinase), 60 (ribonucleotide reductase, small), and 65 are contained in RRV26-95 (Fig. 1). Gammaherpesvirus-specific ORFs 27, 48, 49, 50, 52, 58, 59, 67.5, and 75, as well as gamma 2-herpesvirus-specific ORFs 2 (dihydrofolate reductase; DHFR), 4 (complement-binding protein; CBP), 70, 10, 11, and 73, and the TS gene common to gammaherpesviruses and the alphaherpesvirus varicella-zoster virus, are contained in RRV26-95 (Fig. 1).
RRV26-95 contains a highly repetitive region located between R4 and vBcl-2 (ORF 16) (Fig. 1) that is 3,966 bp in length. In this region are two repeating elements: one of 26 bp that is repeated 10 times and is 53.8% G+C and one of 25 bp that is repeated 27 times and is 79.5% G+C (data not shown). The majority of the RRV26-95 sequence in this region was determined from 10.6-kbp PstI (P5) and 10.5-kbp HindIII (H2) restriction fragment clones of virion DNA. The gap in sequence between these two clones was filled using sequences from three clones derived from sonicated virion DNA. These experiments indicated that the size of the gap in sequence between P5 and H2 is 1,274 bp in length. Since we did not have a restriction fragment clone that spanned this region, we amplified these sequences by using PCR primers that annealed to nonrepetitive sequences in P5 and H2 and used the amplified fragment for sizing and for sequencing. The resulting PCR fragment was of a size and sequence (data not shown) that indicated that the sequence represented by continuous restriction and sonication clones accurately represented the RRV26-95 sequence between R4 and ORF 16. We observed one ORF in this region with sequences unique to RRV (see below).
Our analysis revealed another highly repetitive region that contained G+C-rich sequences between ORFs 69 and R13 (vFLIP) in RRV26-95 (Fig. 1). We have cloned an EcoRI restriction fragment (E6) of virion DNA that included sequences from ORF 66 to ORF 71, and the size of this fragment (11 kbp) corresponded to the length of the sequence determined for this region from overlapping clones derived from sonicated virion DNA. To further demonstrate that our determined sequence accurately represented RRV26-95 sequences in this region, the E6 clone was digested with HaeIII and RsaI restriction enzymes. Recognition sites for these enzymes flanked the highly repetitive sequence region that is 1,127 bp in length and is 80.3% G+C. These sequences contained two repeating elements: one of 17 to 19 bp that is repeated 44 times and one of 31 bp that is repeated 10 times (data not shown). The HaeIII and RsaI restriction pattern of E6 agreed with the expected pattern based on our determined sequence (Fig. 2). The 1.3-kbp HaeIII and 1.5-kbp RsaI digestion fragments (Fig. 2) were isolated and cloned into the SmaI site of pUC 18. The sequence of these fragments was determined and agreed with our sequence determination from overlapping fragments of sonicated virion DNA (data not shown), indicating that we accurately determined the RRV26-95 sequences in this region. These analyses demonstrate that the sequence between ORF 69 and R13 is 6,106 bp. We observed RRV-unique ORFs in this region in both the rightward and leftward directions without significant similarities to any herpesviral or cellular sequences (see below).
Comparison of RRV26-95 and KSHV genomic organization and sequence.
Our analysis indicates that the genomic organization of RRV26-95 is closely but not entirely colinear with that of KSHV (Fig. 1). In most cases, RRV26-95 genes are in corresponding locations and have the same polarity as corresponding KSHV genes (Fig. 1). However, several ORFs are contained in KSHV sequences that are not contained in RRV26-95 sequences. These are K3 and K5 (bovine herpesvirus-4 immediate-early protein homologues), K7 (nut-1), and K12 (Kaposin) (Fig. 1). RRV26-95 contains only one vMIP-1 homologue compared to three in KSHV (23), and RRV26-95 contains eight homologues of vIRF compared to only four in KSHV (27, 35) (Fig. 1). We have called the RRV26-95 vMIP-1 homologue R4 and the vIRF homologues R9.1-R9.8 (Fig. 1, Table 1). Another difference between RRV26-95 and KSHV is that the DHFR gene is correspondingly located near the left end of the genome in RRV26-95 and the gamma-2 HVS, but in KSHV the DHFR gene is displaced 16,069 nucleotides in a rightward direction in the genome (Fig. 1). In all other cases, RRV26-95 genes are in corresponding locations to and have the same polarity as corresponding KSHV genes (Fig. 1).
TABLE 1.
| ORF | RRV26-95
|
KSHV
|
RRV17577
|
Putative function | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Start position | Stop position | Pol | Size | Size | % Identity | % Similarity | Size | % Identity | % Similarity | ||
| R1 | 513 | 1784 | + | 423 | 289 | 28.0 | 33.0 | 423 | 98.1 | 98.6 | |
| 2 | 2418 | 1852 | − | 188 | 210 | 45.5 | 54.4 | 188 | 99.9 | 99.5 | DHFR |
| 4 | 2595 | 3782 | + | 395 | 550 | 35.7 | 42.3 | 645 | 64.5 | 72.2 | CBP |
| 6 | 4213 | 7611 | + | 1,132 | 1,133 | 63.3 | 71.3 | 1132 | 100.0 | 100.0 | ssDBP |
| 7 | 7636 | 9696 | + | 686 | 695 | 52.2 | 60.8 | 686 | 99.6 | 99.6 | Transport protein |
| 8 | 9683 | 12172 | + | 829 | 845 | 66.4 | 73.9 | 829 | 96.7 | 97.1 | gB |
| 9 | 12290 | 15334 | + | 1,014 | 1,012 | 67.4 | 75.5 | 1,014 | 100.0 | 100.0 | Pol |
| 10 | 15429 | 16679 | + | 416 | 418 | 35.3 | 44.0 | 416 | 100.0 | 100.0 | |
| 11 | 16688 | 17917 | + | 409 | 407 | 33.0 | 42.8 | 409 | 100.0 | 100.0 | |
| R2 | 18712 | 18089 | − | 207 | 204 | 22.6 | 30.6 | 207 | 100.0 | 100.0 | VIL-6 |
| 70 | 19946 | 18945 | − | 333 | 337 | 65.8 | 71.8 | 333 | 98.8 | 99.1 | TS |
| R4 | 20822 | 20466 | − | 118 | 94 | 32.6 | 42.4 | 115 | 94.8 | 95.6 | VMIP |
| 16 | 24788 | 25351 | + | 187 | 175 | 46.0 | 58.0 | 187 | 100.0 | 100.0 | Bcl-2 homolog |
| 17 | 27067 | 25457 | − | 536 | 553 | 45.8 | 52.7 | 536 | 99.4 | 99.6 | Capsid protein |
| 18 | 27066 | 27839 | + | 257 | 257 | 58.0 | 68.1 | 299 | 100.0 | 100.0 | |
| 19 | 29490 | 27847 | − | 547 | 549 | 52.9 | 61.4 | 547 | 99.3 | 99.3 | Tegument protein |
| 20 | 30037 | 28985 | − | 350 | 320 | 46.9 | 53.9 | 350 | 99.4 | 99.4 | |
| 21 | 30036 | 31709 | + | 557 | 580 | 46.4 | 54.9 | 557 | 98.9 | 99.1 | Thymidine kinase |
| 22 | 31696 | 33876 | + | 726 | 730 | 41.2 | 50.8 | 704 | 74.2 | 79.4 | gH |
| 23 | 35081 | 33873 | − | 402 | 404 | 49.0 | 57.5 | 402 | 99.8 | 99.8 | |
| 24 | 37329 | 35131 | − | 732 | 752 | 59.1 | 66.7 | 732 | 99.6 | 99.6 | |
| 25 | 37331 | 41467 | + | 1,378 | 1,376 | 72.3 | 79.8 | 1,378 | 99.6 | 99.6 | MCP |
| 26 | 41499 | 42416 | + | 305 | 305 | 64.3 | 71.8 | 307 | 98.4 | 98.4 | Capsid protein |
| 27 | 42441 | 43250 | + | 269 | 290 | 27.8 | 37.2 | 269 | 100.0 | 100.0 | |
| 28 | 43416 | 43691 | + | 91 | 102 | 26.5 | 30.1 | 91 | 100.0 | 100.0 | |
| 29b | 44787 | 43741 | − | 348 | 351 | 66.4 | 77.8 | 348 | 99.4 | 99.4 | Packaging protein |
| 30 | 44913 | 45143 | + | 76 | 77 | 38.2 | 51.3 | 76 | 98.7 | 98.7 | |
| 31 | 45101 | 45754 | + | 217 | 224 | 46.1 | 57.3 | 217 | 100.0 | 100.0 | |
| 32 | 45691 | 47085 | + | 464 | 454 | 42.9 | 51.2 | 464 | 100.0 | 100.0 | |
| 33 | 47057 | 48067 | + | 336 | 312 | 42.7 | 52.8 | 336 | 100.0 | 100.0 | |
| 29a | 48968 | 47985 | − | 327 | 312 | 61.2 | 66.7 | 327 | 100.0 | 100.0 | Packaging protein |
| 34 | 48967 | 49950 | + | 327 | 327 | 49.4 | 59.8 | 327 | 99.7 | 99.7 | |
| 35 | 49931 | 50380 | + | 149 | 151 | 37.2 | 49.3 | 149 | 100.0 | 100.0 | |
| 36 | 50286 | 51593 | + | 435 | 444 | 48.7 | 59.1 | 435 | 100.0 | 100.0 | Kinase |
| 37 | 51598 | 53016 | + | 472 | 486 | 63.9 | 72.6 | 480 | 100.0 | 100.0 | Alkaline exonuclease |
| 38 | 52971 | 53180 | + | 69 | 61 | 45.0 | 56.7 | 69 | 100.0 | 100.0 | |
| 39 | 54399 | 53263 | − | 378 | 399 | 59.5 | 73.3 | 378 | 100.0 | 100.0 | gM |
| 40 | 54534 | 55940 | + | 468 | 457 | 33.0 | 42.5 | 468 | 99.8 | 99.8 | Helicase-primase |
| 41 | 55925 | 56536 | + | 203 | 205 | 28.7 | 37.9 | 203 | 99.5 | 99.5 | Helicase-primase |
| 42 | 57351 | 56533 | − | 272 | 278 | 48.0 | 58.7 | 272 | 99.6 | 99.6 | |
| 43 | 59035 | 57305 | − | 576 | 605 | 62.0 | 70.1 | 576 | 100.0 | 100.0 | Capsid protein |
| 44 | 58974 | 61346 | + | 790 | 788 | 66.0 | 74.0 | 790 | 100.0 | 100.0 | Helicase-primase |
| 45 | 62448 | 61387 | − | 353 | 407 | 37.0 | 43.6 | 352 | 99.4 | 99.4 | |
| 46 | 63260 | 62568 | − | 230 | 255 | 56.8 | 68.6 | 255 | 83.5 | 85.2 | UDG |
| 47 | 63727 | 63236 | − | 163 | 167 | 31.2 | 38.8 | 169 | 56.4 | 62.0 | gL |
| 48 | 65154 | 63985 | − | 389 | 402 | 29.6 | 39.1 | 389 | 100.0 | 100.0 | |
| 49 | 66289 | 65384 | − | 301 | 302 | 53.8 | 65.8 | 301 | 99.7 | 99.7 | |
| 50 | 66480 | 68024 | + | 514 | 631 | 44.9 | 54.7 | 514 | 99.7 | 100.0 | Transactivator |
| R8 | |||||||||||
| R8.1 | |||||||||||
| 52 | 70806 | 70387 | − | 139 | 131 | 47.7 | 60.8 | 139 | 100.0 | 100.0 | |
| 53 | 71184 | 70870 | − | 104 | 110 | 49.0 | 53.8 | 104 | 100.0 | 100.0 | |
| 54 | 71260 | 72132 | + | 290 | 318 | 43.1 | 51.0 | 290 | 100.0 | 100.0 | dUTPase |
| 55 | 72825 | 72193 | − | 210 | 227 | 55.2 | 62.8 | 210 | 100.0 | 100.0 | |
| 56 | 72837 | 75323 | + | 828 | 843 | 52.7 | 61.8 | 828 | 100.0 | 100.0 | DNA replication protein |
| 57 | 75564 | 76892 | + | 442 | 275 | 47.1 | 61.4 | 442 | 99.8 | 100.0 | Immediate-early protein |
| R9.1 | 78499 | 77252 | − | 415 | 449 | 28.7 | 60.6 | 415 | 98.3 | 98.8 | vIRF |
| R9.2 | 79919 | 78672 | − | 415 | 449 | 23.2 | 37.2 | 415 | 99.5 | 99.5 | vIRF |
| R9.3 | 81300 | 80245 | − | 351 | 449 | 24.3 | 31.9 | 351 | 100.0 | 100.0 | vIRF |
| R9.4 | 82559 | 81474 | − | 361 | 449 | 25.5 | 34.2 | 253 | 99.6 | 99.6 | vIRF |
| R9.5 | 84193 | 83036 | − | 385 | 449 | 29.5 | 38.4 | 385 | 100.0 | 100.0 | vIRF |
| R9.6 | 85512 | 84340 | − | 390 | 449 | 22.9 | 33.4 | 390 | 100.0 | 100.0 | vIRF |
| R9.7 | 86946 | 85879 | − | 355 | 449 | 25.0 | 34.6 | 355 | 99.4 | 99.4 | vIRF |
| R9.8 | 88201 | 87107 | − | 364 | 449 | 26.8 | 33.6 | 364 | 100.0 | 100.0 | vIRF |
| 58 | 89529 | 88447 | − | 360 | 357 | 38.9 | 46.2 | 360 | 99.7 | 100.0 | |
| 59 | 90724 | 89540 | − | 394 | 396 | 51.8 | 60.2 | 394 | 100.0 | 100.0 | DNA replication protein |
| 60 | 91767 | 90853 | − | 304 | 305 | 70.1 | 78.3 | 314 | 100.0 | 100.0 | Ribonucleotide reductase, small |
| 61 | 94145 | 91779 | − | 788 | 792 | 61.6 | 69.6 | 788 | 99.0 | 99.2 | Ribonucleotide reductase, large |
| 62 | 95143 | 94148 | − | 331 | 331 | 57.6 | 65.4 | 331 | 99.4 | 99.4 | Assembly-DNA maturation |
| 63 | 95142 | 97961 | + | 939 | 927 | 43.5 | 52.9 | 939 | 99.8 | 99.8 | Tegument protein |
| 64 | 97965 | 105611 | + | 2,548 | 2,635 | 41.3 | 51.2 | 2,548 | 99.6 | 99.6 | Tegument protein |
| 65 | 106131 | 105622 | − | 169 | 170 | 45.5 | 55.8 | 169 | 100.0 | 100.0 | Capsid protein |
| 66 | 107483 | 106137 | − | 448 | 429 | 47.3 | 52.7 | 448 | 99.8 | 99.8 | |
| 67 | 108185 | 107571 | − | 222 | 271 | 67.1 | 71.2 | 224 | 87.8 | 87.8 | Tegument protein |
| 67.5 | 108461 | 108201 | − | 86 | 80 | 62.5 | 71.2 | 86 | 98.8 | 98.8 | |
| 68 | 108596 | 109969 | + | 457 | 545 | 49.1 | 58.3 | 457 | 100.0 | 100.0 | Glycoprotein |
| 69 | 109991 | 110884 | + | 297 | 225 | 65.5 | 73.1 | 297 | 100.0 | 100.0 | |
| R13 | 117515 | 116991 | − | 174 | 188 | 33.1 | 40.1 | 174 | 99.4 | 99.4 | FLIP homolog |
| 72 | 118338 | 117574 | − | 254 | 257 | 39.4 | 50.2 | 254 | 99.6 | 99.6 | Cyclin D homolog |
| 73 | 119992 | 118646 | − | 448 | 1,162 | 18.3 | 24.9 | 447 | 99.8 | 97.8 | Latency-associated nuclear antigen |
| R14 | 120646 | 121407 | + | 253 | 348 | 32.8 | 37.6 | 253 | 100.0 | 100.0 | N-CAM Ox-2 homolog |
| 74 | 121704 | 122732 | + | 342 | 342 | 42.0 | 54.7 | 342 | 100.0 | 100.0 | G-protein-coupled receptor |
| 75 | 126733 | 122837 | − | 1,298 | 1,296 | 44.9 | 53.2 | 1,298 | 99.8 | 99.9 | Tegument protein-FGARAT |
| R15 | − | 490 | |||||||||
For RRV ORFs (column 1), we have used a nomenclature that corresponds to that used previously for KSHV ORFs (37). Columns 2 and 3 give the positions of the start and stop codons of the indicated ORFs relative to the 5′ extent of the long unique region. Column 4 indicates the polarity (Pol) of each ORF. Columns 5, 6, and 9 give the sizes (in numbers of amino acids) of each ORF in RRV26-95, KSHV, and RRV17577, respectively. Columns 7 and 8 show the percent identity and similarity, respectively, between RRV26-95 and KSHV ORFs. Columns 10 and 11 are the same as columns 7 and 8 except for RRV26-95 and RRV17577, respectively. Column 12 signifies the putative functions of the indicated ORFs.
By BLAST analysis, the level of similarity between RRV26-95 and KSHV for the 39 herpesvirus core gene sequences ranges from a low of 30.1% for ORF 28 to a high of 75.5% for ORF 9 (DNA polymerase) (Fig. 1, Table 1). The beta-gamma- (ORFs 18, 24, 45, and 66) and alpha-gamma- (ORFs 21, 60, and 65) herpesvirus-specific genes contained in RRV26-95 range in similarity to corresponding KSHV genes from a low of 43.6% (ORF 45) to a high of 78.3% (ORF 60) (Table 1). Furthermore, similarities between RRV26-95 and KSHV gammaherpesvirus-specific genes (ORFs, 27, 48, 49, 50, 52, and 75) range from 37.2% (ORF 27) to 65.8% (ORF 49) (Table 1). In almost all cases, beta-gamma-, alpha-gamma-, and gammaherpesvirus-specific ORF sequences of RRV26-95 are more similar to KSHV than to other herpesviruses (data not shown). DHFR is unusual in that RRV26-95 DHFR more closely resembles HVS DHFR (74% similarity) than KSHV DHFR (55% similarity). With the exception of the DHFR gene, all homologues are correspondingly located in KSHV and RRV26-95.
Our analysis revealed a number of ORFs that are common only to RRV and KSHV. Among these are ORF 1 (termed K1 in KSHV and R1 in RRV), vIL-6, vMIP, a series of interferon regulatory factors (vIRFs), vFlip, vOx-2, and a vIL-8 receptor (Fig. 1). These ORFs range in similarity with corresponding KSHV ORFs from a low of 30.6% (vIL-6) to a high of 42.8% (vMIP) (Table 1). Unlike K1 sequences of some KSHV isolates which can display a high degree of variability (41), the R1 sequences of RRV26-95 and RRV17577 were 98% identical. Short ORFs that correspond in location to the multiply spliced KSHV K8 (18, 40), K8.1 (17, 30) and K15 (9) genes were also observed in RRV26-95 and were called R8, R8.1, and R15. Additional ORFs are located between RRV26-95 ORFs 50 and 52 which are likely to contain additional R8, as well as R8.1, sequences but these gene sequences cannot be determined directly from the primary sequence in the absence of detailed knowledge of splicing patterns. Downstream of ORF 75 sequences, RRV26-95 contains open sequences that correspond to open sequences in RRV17577. These sequences are correspondingly located to the multiply spliced KSHV K15 gene (9, 30) and thus have been termed R15.
Comparison of RRV26-95 and RRV17577 genomic organization and sequence.
Our analysis indicates that RRV26-95 and RRV17577 have colinear genomic organizations (Table 1). Of the 84 ORFs in the RRV26-95 L-DNA, 83 contain corresponding ORFs in RRV17577 and the overall divergence within these sequences is only 2.24%. One exception is that RRV26-95 contains sequences for the herpesvirus core gene ORF 67.5 (Fig. 1) which were not reported in the RRV17577 sequence (37) (Table 1). RRV26-95 ORF 67.5 (Fig. 1) is similar in size (86 amino acids [aa]) (Table 1) to corresponding KSHV sequences (80 aa) and contains several stretches of amino acids that are absolutely conserved between RRV26-95 and KSHV (Fig. 3). Our inspection of the RRV17577 sequence under accession no. AF083501 revealed an ORF 67.5 that was not noted by Searles et al. (37). It exhibited 98.8% identity with ORF 67.5 of RRV26-95 (Table 1). The sequences of RRV26-95 and RRV17577 also differ in a region in the L-DNA containing highly repetitive sequence located between vMIP-1 and vBcl-2 (Fig. 1) which is 288 bases (7%) shorter in RRV26-95 (3,966 bp) than in RRV17577 (4,254 bp). In this region RRV26-95 contains one ORF which is 96% identical to corresponding RRV17577 sequences (see below). The corresponding region in KSHV is 8,313 bp and contains the K4.1, K4.2, K5, K6, and K7 genes (Fig. 1) (26, 34). Thus, both RRV26-95 and RRV17577 (37) do not contain K4.2, K5, K6, and K7 gene sequences. RRV26-95 R4 has limited similarity to KSHV K4.1 but has high similarity to RRV17577 R3, which is the name given to the corresponding gene in this isolate (Table 1) (37). Despite the limited overall similarity, RRV26-95 R4 contains four cysteine residues that are conserved among human MIP homologues, K4, K6 (22, 34), and RRV17577 R3 (37), which are characteristic of β-chemokines (data not shown). Our analyses also demonstrate that the sequences between ORF 69 and R13 are 6,106 bp for RRV26-95 compared to 6,314 bp for RRV17577. This region contains highly repetitive, G+C-rich sequences which are similar to what was observed for RRV17577. The sequences in this region of both isolates do not contain sequences that are detectably similar to K12 (Kaposin) sequences (24) (Fig. 1).
FIG. 3.
Alignment of KSHV and RRV26-95 ORF 67.5. An alignment was constructed of KSHV (accession no. U75698) and RRV26-95 ORF 67.5 amino acid sequences by using CLUSTAL W software. Conserved residues are shaded in black.
Another exception to the overall similarity between RRV26-95 and RRV17577 is that the rhadinovirus-specific CBP sequences are significantly shorter in RRV26-95 than was observed for corresponding sequences from RRV17577 (395 versus 645 aa) (Table 1) (37). RRV26-95 CBP sequences are contained in a 4.2-kbp KpnI restriction fragment clone (K29) of virion DNA that spanned from just upstream of R1 gene sequences to ORF 6 sequences (Fig. 1). This span of virion DNA matched the sequence determined from overlapping clones derived from sonicated virion DNA and thus accurately represents RRV26-95 CBP sequence. We aligned RRV26-95 CBP amino acid sequences with RRV17577, KSHV, and HVS. Our analysis revealed that a region of conservation exists among these sequences that begins at the N′ termini of RRV26-95, KSHV, and HVS sequences which corresponds to amino acid 294 in the reported RRV17577 CBP open sequences (Fig. 4). Included in these sequences are 15 conserved cysteine residues (Fig. 4). The sequences in this region of conservation are most similar between RRV26-95 and RRV17577 sequences (Fig. 4). The N′-terminal 293 aa of the 645-aa RRV17577 CBP do not align with the other rhadinovirus CBP sequences depicted here. KSHV CBP is longer (550 aa) than RRV26-95 (395 aa) and HVS CBP (360 aa) and contains C-terminal sequences downstream of the region of conservation that also do not align with RRV or HVS sequences (Fig. 4).
FIG. 4.
Alignment of rhadinovirus CBP sequences. An alignment was constructed of CBP amino acid sequences from RRV17577 (accession no. AF083501), RRV26-95, KSHV (accession no. U75698), and HVS (accession no. X64346) by using CLUSTAL W software. The first 293 aa of RRV17577 CBP sequences did not align with the other rhadinovirus CBP sequences depicted here. Conserved cysteines are shaded in black. Deletion polymorphisms between sequences starting at amino acid 294 of RRV17577 CBP are indicated with dashes.
Reading frames unique to RRV.
RRV26-95 was found to contain a number of reading frames not found in KSHV. A 102-aa reading frame is present between positions 21,444 and 21,752. This reading frame which is unique to RRV (RU-1) corresponds in location to K4.1, K4.2, K5, K6, and K7 of KSHV but has no homology to them or to any other KSHV sequences (Fig. 1 and Table 2). A 102-aa reading frame with 96% identity is equivalently located in RRV17577 (Table 2). Similarly, RRV26-95 contains a series of ORFs between positions 110,910 and 114,899 in a rightward direction and between positions 116,625 and 111,082 in a leftward direction that have no similarity with KSHV but are reasonably conserved when compared to RRV17577 (Table 2). These ORFs correspond in location (Fig. 1) but lack detectable similarity to the K12 (Kaposin) gene. As described above, the RRV26-95 sequences in this region contain two G+C-rich repeating elements: one of 17 to 19 bp that is repeated 44 times and one that is 31 bp and is repeated 10 times. The corresponding region of KSHV immediately rightward of the K12 gene contains two distinct 23-bp G+C-rich repeating sequences: DR1, which is 900 bp in length, and DR2, which is 370 bp in length (34, 35). Thus, although RRV lacks a Kaposin gene, it contains sequences in this region that are comparable to KSHV.
TABLE 2.
Identity and similarity of RRV-unique (RU) ORFsa
| ORF | RRV26-95
|
RRV17577
|
|||||
|---|---|---|---|---|---|---|---|
| Start position | Stop position | Pol | Size (aa) | Size (aa) | % Identity | % Similarity | |
| RU1-R | 21752 | 22060 | R | 102 | 102 | 96.0 | 96.0 |
| RU2-R | 111218 | 111451 | R | 77 | 77 | 100.0 | 100.0 |
| RU3-R | 111810 | 112115 | R | 101 | 89 | 100.0 | 100.0 |
| RU4-R | 112944 | 114128 | R | 394 | 92 | 100.0 | 100.0 |
| RU5-R | 114231 | 114494 | R | 87 | 101 | 100.0 | 100.0 |
| RU6-R | 114292 | 114507 | R | 71 | 486 | 78.9 | 78.9 |
| RU7-R | 114782 | 115207 | R | 141 | – | – | – |
| RU8-L | 20251 | 20000 | L | 83 | 70 | 33.3 | 40.4 |
| RU9-L | 111659 | 111390 | L | 89 | 89 | 100.0 | 100.0 |
| RU10-L | 111892 | 111614 | L | 92 | 92 | 100.0 | 100.0 |
| RU11-L | 114492 | 114271 | L | 75 | 75 | 98.6 | 98.6 |
| RU12-L | 115805 | 115584 | L | 73 | 73 | 97.3 | 97.3 |
| RU13-L | 116070 | 115606 | L | 154 | 151 | 87.2 | 87.2 |
| RU14-L | 116832 | 115838 | L | 228 | 228 | 100.0 | 100.0 |
Columns 2 and 3 indicate the position of the start and stop codons of the indicated ORFs relative to the leftmost extent of the long unique region. Column 4 indicates the polarity (Pol) of each ORF (R, rightward; L, leftward). R, positive polarity; L, negative polarity. Columns 5 and 6 indicate the size of each ORF in RRV26-95 and RRV17577, respectively. Columns 7 and 8 show the percent identity and similarity between RRV26-95 and RRV17577 ORFs. Dashes in the RU7 row indicate that RU7 is not observed in RRV17577.
RRV26-95 ORF 73 sequences.
RRV26-95 contains sequences for rhadinovirus-specific ORF 73 sequences (Fig. 1) which are 98% identical to RRV17577 sequences (Table 1). This ORF is significantly shorter (448 aa) than corresponding sequences in KSHV (1,162 aa) (34). The N′ termini of RRV26-95 and KSHV ORF 73 are both proline-rich (Fig. 5). Furthermore, BLAST search analysis revealed detectable similarity in the C′-terminal sequences of these genes (data not shown). Downstream of the proline-rich sequences, KSHV ORF 73 sequences encode a long, highly acidic repetitive domain (Fig. 5B) in contrast to RRV26-95 sequences, which only encode a short stretch of acidic residues in the corresponding region of this gene (Fig. 5A). Thus, the difference in length between ORF 73 of RRV26-95 and KSHV is accounted for by the difference in the length of the acidic sequences.
FIG. 5.
The ORF 73 amino acid sequences of RRV26-95 (A) and KSHV (B) (accession no. U75698). Proline-rich sequences are shaded in black. The repetitive, acidic sequences in KSHV and the short stretch of acidic sequences in RRV26-95 are indicated in boldface.
Phylogenetic analysis of RRV26-95 vIRF sequences.
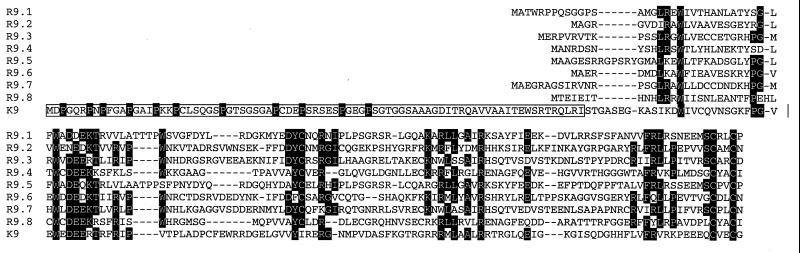
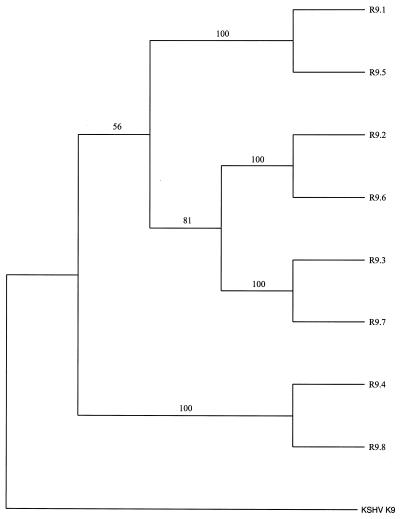
RRV26-95 contains eight copies of vIRF homologues. These sequences have the highest similarity to the KSHV K9 vIRF homologue. To maintain consistency in nomenclature with KSHV, we have called the eight RRV26-95 vIRFs R9.1 to R9.8. K9 contains a proline-rich motif near its N terminus that is necessary for the interaction of K9 with p300 (M. Li, B. Damania, X. Alvarez, V. Ogryzko, K. Ozato, and J. U. Jung, submitted for publication). This interaction leads to the inhibition of the histone acetyltransferase activity of p300. Interestingly, none of the eight vIRF homologues in RRV26-95 contains an N-terminal proline-rich motif (Fig. 6). Our analysis also reveals that four of the R9 ORFs have 100% similarity and four have a minimum similarity of 98.8% with the corresponding vIRF ORFs in RRV17577. A phylogenetic tree was built by using the R9 homologue sequences and was rooted to K9 sequences using PAUP* 4.0 software (Fig. 7). These analyses demonstrate that R9.1 branches with R9.5, R9.2 branches with R9.6, R9.3 branches with R9.7, and R9.4 branches with R9.8. There is very strong support (bootstrap values of 100%) for the branches separating these sequences. These data suggest that R9.5 to R9.8 arose from gene duplication of R9.1 to R9.4 or vice versa. Parsimony analysis produced a tree with an identical topology (data not shown).
FIG. 6.
Alignment of N-terminal RRV26-95 and KSHV vIRF homologue sequences. An alignment was constructed of RRV26-95 R9.1 to R9.8 and KSHV K9 (accession no. U75698) N-terminal amino acid sequences by using CLUSTAL W software. The proline residues within the N-terminal region unique to KSHV K9 are shaded in black. Conserved residues among the K9 and R9 homologues are also shaded in black. Deletion polymorphisms are indicated with dashes.
FIG. 7.
Phylogenetic analysis of vIRF homologue genes of RRV26-95 and KSHV. CLUSTAL W software was used to align full-length R9.1 to R9.8 and K9 (accession no. U75698) amino acid sequences. The neighbor-joining method was used to generate this phylogeny by using PAUP* 4.0 software, with K9 sequences serving as the outgroup. Bootstrap values from 1,000 replications (repeated three times) are shown for each branch point.
Functional analysis of the RRV26-95 vIL-6 homologue.
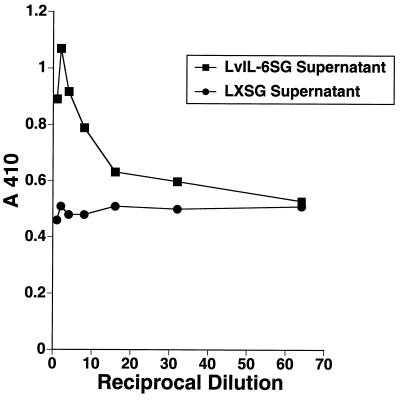
RRV26-95 sequences contain an ORF located immediately downstream of ORF 11 (Fig. 1) that is of similar size (Table 1) and in a corresponding location to the KSHV K2 gene (22, 27). We have termed this corresponding gene in RRV26-95 R2. Despite limited overall similarity to K2 (Table 1), the R2 gene contains four cysteine residues that are conserved in the IL-6 family of cytokines (Fig. 8), and these cysteines have been shown to be important for the proper folding of the protein necessary for interaction with the IL-6 receptor (21). To test functionality, we inserted RRV26-95 vIL-6 sequences into the vector LXSG (2) as described in Materials and Methods. In this construct, vIL-6 transcription was driven by MuLV sequences and transcription of the green fluorescent protein was driven by SV40 sequences. The resulting clone and the control LXSG vector were electroporated independently into COS-1 cell cultures, and green fluorescing cells were subsequently identified by flow cytometry. The parallel cultures were found to have been transfected with comparable efficiency (data not shown). Supernatants from these cultures were collected and incubated with the IL-6-dependent cell line B9 (5). Cell proliferation assays revealed that B9 cells incubated with vIL-6 supernatants were rescued in a dose-dependent manner in comparison to B9 cells incubated with control supernatants (Fig. 9). Similar observations have been made by others for KSHV K2 (22, 27) and RRV17577 R2 sequences (15).
FIG. 8.
Alignment of IL-6 sequences. An alignment was constructed of homologue IL-6 amino acid sequences from macaque (accession no. P51494), human (accession no. P05231), KSHV K2 (accession no. U75698), and RRV26-95 R2 by using CLUSTAL W software. Conserved cysteine residues that have been previously demonstrated to be necessary for IL-6 receptor binding are shaded in black. Deletion polymorphisms are indicated with dashes.
FIG. 9.
B9-cell rescue by RRV26-95 vIL-6. Twofold serial dilutions of supernatants from COS-1 cells transfected with a vector containing RRV26-95 vIL-6 or control vector were incubated with IL-6-dependent B9 cells. The rate of proliferation of B9 cells in the parallel cultures was determined by enzyme-linked immunosorbent assay with a cell proliferation kit.
DISCUSSION
Our analyses of RRV26-95 sequences demonstrate the similarity of this isolate to KSHV in genomic organization (Fig. 1) and in the sequence of individual ORFs (Table 1). BLAST search analysis revealed that in almost all cases RRV26-95 coding sequences have a greater degree of similarity to corresponding KSHV genes than to any other herpesvirus. All of the ORFs present in KSHV have at least one counterpart in RRV26-95 except K3 and K5 (bovine herpesvirus-4 immediate-early protein homologues), K7 (nut-1), and K12 (Kaposin). These data suggest that these KSHV-specific genes were acquired subsequent to the divergence of Asian and African Old World primates. The repetitive, acidic domain in ORF 73 of KSHV (Fig. 5) and the proline-rich domain in K9 (Fig. 6), absent in RRV26-95, may also have been acquired subsequent to this divergence. DHFR is unusual in that it is located near the left end of the genome in RRV26-95 corresponding in location to HVS DHFR and more closely resembles HVS DHFR (74% similarity) than KSHV DHFR (55% similarity). It seems likely that KSHV has rearranged or reacquired the DHFR gene in the course of evolution from Old World primate to human rhadinoviruses.
Overall, RRV26-95 is very similar to RRV17577 both in sequence and in genomic organization. Furthermore, only four of 84 ORFs exhibited less than 95% sequence identity (Table 1). Despite the high overall conservation between the two RRV isolates, some differences were noted between the two. Most notably, the CBP sequences in RRV17577 are significantly longer than in RRV26-95 (Table 1). Alignment of these sequences with those of KSHV and HVS demonstrate that the N′-terminal 293 aa of RRV17577 encoding open CBP sequence are not detectably similar to those of the RRV26-95, KSHV, or HVS CBP coding sequences (Fig. 4), whereas the 3′-most 352 aa are similar to these rhadinovirus CBP coding sequences. Further work will be needed to determine whether the CBP sequences of RRV17577 are unique. Although ORF 67.5 was not noted in the description of Searles et al. (37), it is present in the submitted sequence. These sequences are well conserved among herpesviruses, although their role in viral replication remains undefined. ORF 67.5 sequences are 72% similar between RRV26-95 and KSHV (Table 1).
The EBNA1 gene of Epstein-Barr virus (EBV) contains a long Gly-Ala repeat motif that plays a role in the inhibition of EBNA1-specific major histocompatibility complex class I antigen presentation (16). Recently, it has been shown that the EBNA1 gene of a rhesus lymphocryptovirus (20) contains a Gly-Ala repeat domain that is considerably shorter than corresponding EBV sequence (3). This rhesus homologue does not maintain antigen presentation inhibition activity observed for the human homologue. KSHV ORF 73 sequences contain a large, repetitive acidic motif (Fig. 5B), the function of which remains to be determined. Conversely, RRV26-95 contains only a short stretch of acidic residues in the corresponding region of ORF 73 (Fig. 5A). It will be interesting to determine if the repetitive acidic motif contributes to the function of KSHV ORF 73.
The K12 region is the most abundantly transcribed region in KSHV latent infection (35, 39). Recently, it has been demonstrated that translation of the transcripts in this region is complex (35). The predominant translation product initiates at a CUG codon and does not include K12 sequences but does include the two G+C-rich repeating units (DR1 and DR2) located immediately to the right of the K12 gene (34, 35). In the corresponding region, RRV26-95 also contains two distinct G+C-rich repeating elements that are comparable to the DR1 and DR2 sequences in KSHV but does not contain sequences that are detectably similar to K12 (34, 35). It seems likely that RRV translation will also be complex in this region and may include sequences from the two G+C-rich repeating elements, as well as the 12 ORFs located in this region that are unique to RRV (Table 2).
The association of KSHV with Kaposi's sarcoma and other proliferative abnormalities has led to its intense study by numerous laboratories. To date, these studies have been limited by the lack of a permissive, lytic system for KSHV (13, 19, 32) and by the lack of a direct animal model. This has precluded direct demonstration of the role of individual gene products in KSHV replication, persistence, and disease. We have demonstrated here that RRV26-95 is similar to KSHV both in genomic organization and in the sequence of individual ORFs. Permissive growth of RRV26-95 in rhesus monkey fibroblast cells (12) will facilitate genetic manipulation of RRV sequences, including the engineering of point mutations and deletions, as well as RRV recombinants that contain marker, cellular, or KSHV gene sequences. Study of the biological properties of such mutant and recombinant viruses should provide insight into the relative importance and role of individual genes to biological properties.
ACKNOWLEDGMENTS
We thank Jae Jung for valuable scientific discussions and comments; Yuan Chang for providing the B9 cell line; Kevin Clarke, Michelle Ethier, Susan Czajak, Daniel Silva, and Kim Deary for assistance; and Joanne Newton and Deborah Letourneau for manuscript preparation.
This study was supported by Public Health Service grants AI38131 and RR00168.
REFERENCES
- 1.Albrecht J C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittman S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander L, Lee H, Rosenzweig M, Jung J U, Desrosiers R C. An EGFP-containing vector system that facilitates stable and transient expression assays. BioTechniques. 1997;23:64–66. doi: 10.2144/97231bm12. [DOI] [PubMed] [Google Scholar]
- 3.Blake N W, Moghaddam A, Rao P, Kaur A, Glickman R, Cho Y, Marchini A, Haigh T, Johnson R P, Rickinson A B, Wang F. Inhibition of antigen presentation by the glycine/alanine repeat domain is not conserved in simian homologues of Epstein-Barr virus nuclear antigen 1. J Virol. 1999;73:7381–7389. doi: 10.1128/jvi.73.9.7381-7389.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boshoff C, Whitby D, Hatzioannou T, Fisher C, van der Walt J, Hatzakis A, Weiss R A, Schultz T F. Kaposi's sarcoma associated herpesvirus in HIV-negative Kaposi sarcoma. Lancet. 1995;345:1043–1044. doi: 10.1016/s0140-6736(95)90780-7. [DOI] [PubMed] [Google Scholar]
- 5.Brakenhoff J P, de Hon F D, Fontaine V, ten Boekel E, Schooltink H, Rose-John S, Heinrich P C, Content J, Aarden L A. Development of a human interleukin-6 receptor antagonist. J Biol Chem. 1994;269:86–93. [PubMed] [Google Scholar]
- 6.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 8.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 9.Choi J K, Shim S N, Lee B K, Li M, Jung J U. Identification of the K15 latent gene of Kaposi's sarcoma-associated herpesvirus. J Virol. 2000;74:436–446. [PMC free article] [PubMed] [Google Scholar]
- 10.Damania B, Li M, Choi J-K, Alexander L, Jung J U, Desrosiers R C. Identification of the R1 oncogene and its protein product from the rhadinovirus of rhesus monkeys. J Virol. 1999;73:5123–5131. doi: 10.1128/jvi.73.6.5123-5131.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel M, Letvin N, Sehgal P, Schmidt D, Silva D, Solomon K R, Hodi F S, Jr, Ringler D, Hunt R D, King N W, Desrosiers R C. Prevalence of antibodies to 3 retroviruses in a captive colony of macaque monkeys. Int J Cancer. 1988;41:601–608. doi: 10.1002/ijc.2910410421. [DOI] [PubMed] [Google Scholar]
- 12.Desrosiers R C, Sasseville V G, Czajak S C, Zhang X, Mansfield K G, Kaur A, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foreman K E, Friborg J J, Kong W P, Woffendin C, Polverini P J, Nickoloff B J, Nabel G J. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N Engl J Med. 1997;336:163–171. doi: 10.1056/NEJM199701163360302. [DOI] [PubMed] [Google Scholar]
- 14.Gessain A, Sudaka A, Briere J, Fouchard N, Nicola M A, Rio B, Arborio M, Troussard X, Diebold J, de Thé G. Kaposi sarcoma-associated herpes-like virus (human herpesvirus type 8) DNA sequences in multicentric Castleman's disease: is there any relevant association in non-human immunodeficiency virus-infected patients? Blood. 1996;87:414–416. [PubMed] [Google Scholar]
- 15.Kaleeba J A, Bergquam E P, Wong S W. A rhesus macaque rhadinovirus related to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 encodes a functional homologue of interleukin-6. J Virol. 1999;73:6177–6181. doi: 10.1128/jvi.73.7.6177-6181.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen P M, Klein G, Kurilla M G, Masucci M G. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 17.Li M, MacKey J, Czajak S C, Desrosiers R C, Lackner A A, Jung J U. Identification and characterization of Kaposi's sarcoma-associated herpesvirus K8.1 virion glycoprotein. J Virol. 1999;73:1341–1349. doi: 10.1128/jvi.73.2.1341-1349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin S F, Robinson D R, Miller G, Kung H J. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J Virol. 1999;73:1909–1917. doi: 10.1128/jvi.73.3.1909-1917.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller G, Heston L, Grogan L, Grdoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moghaddam A, Rosenzweig M, Lee-Parritz D, Annis B, Johnson R P, Wang F. An animal model for acute and persistent Epstein-Barr virus infection. Science. 1997;276:2030–2033. doi: 10.1126/science.276.5321.2030. [DOI] [PubMed] [Google Scholar]
- 21.Molden J, Chang Y, You Y, Moore P S, Goldsmith M A. A Kaposi's sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J Biol Chem. 1997;272:19625–19631. doi: 10.1074/jbc.272.31.19625. [DOI] [PubMed] [Google Scholar]
- 22.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 23.Moore P S, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and those without HIV infection. N Engl J Med. 1995;332:1181–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 24.Muralidhar S, Pumfery A M, Hassani M, Sadaie M R, Kishishita M, Brady J N, Doniger J, Medveczky P, Rosenthal L J. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) transforming gene. J Virol. 1998;72:4980–4988. doi: 10.1128/jvi.72.6.4980-4988.1998. . (Erratum, 73:2568, 1999.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nador R G, Cesarman E, Chadburn A, Dawson D B, Ansari M Q, Sald J, Knowles D M. Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi's sarcoma-associated herpes virus. Blood. 1996;88:645–656. [PubMed] [Google Scholar]
- 26.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neipel F, Albrecht J-C, Ensser A, Huang Y-Q, Li J J, Friedman-Kien A E, Fleckenstein B. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. 1997;71:839–842. doi: 10.1128/jvi.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholas J, Ruvolo V R, Burns W H, Sandford G, Wan X, Ciufo D, Hendrickson S B, Guo H G, Haywood G S, Reitz M S. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 29.Poole L J, Zong J C, Ciufo D M, Alcendor D J, Cannon J S, Ambinder R, Orenstein J M, Reitz M S, Hayward G S. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J Virol. 1999;73:6646–6660. doi: 10.1128/jvi.73.8.6646-6660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raab M S, Albrecht J C, Birkmann A, Yaguboglu S, Lang D, Fleckenstein B, Neipel F. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J Virol. 1998;72:6725–6731. doi: 10.1128/jvi.72.8.6725-6731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabkin C S, Janz S, Lash A, Coleman A E, Musaba E, Liotta L, Biggar R J, Zhuang Z. Monoclonal origin of multicentric Kaposi's sarcoma lesions. N Engl J Med. 1997;336:988–993. doi: 10.1056/NEJM199704033361403. [DOI] [PubMed] [Google Scholar]
- 32.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 33.Roizmann B, Desrosiers R C, Fleckenstein B, Lopez C, Minson A C, Studdert M J. The family Herpesviridae: an update. Arch Virol. 1992;123:425–449. doi: 10.1007/BF01317276. [DOI] [PubMed] [Google Scholar]
- 34.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadler R, Wu L, Forghani B, Renne R, Zhong W, Herndier B, Ganem D. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J Virol. 1999;73:5722–5730. doi: 10.1128/jvi.73.7.5722-5730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Said J W, Chien K, Takeuchi S, Tasaka T, Asou H, Cho S K, de Vos S, Cesarman E, Knowles D M, Koeffler H P. Kaposi's sarcoma-associated herpesvirus (KSHV or HHV8) in primary effusion lymphoma: ultrastructural demonstration of herpesvirus in lymphoma cells. Blood. 1996;87:4937–4943. [PubMed] [Google Scholar]
- 37.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M-F, Clauvel J-P, Raphael M, Degos L, Sigaux F. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 39.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu F X, Cusano T, Yuan Y. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J Virol. 1999;73:5556–5567. doi: 10.1128/jvi.73.7.5556-5567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zong J C, Ciufo D M, Alcendor D J, Wan X, Nicholas J, Browning P J, Rady P L, Tyring S K, Orenstein J M, Rabkin C S, Su I J, Powell K F, Croxson M, Foreman K E, Nickoloff B J, Alkan S, Hayward G S. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi's sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol. 1999;73:4156–4170. doi: 10.1128/jvi.73.5.4156-4170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]