Abstract
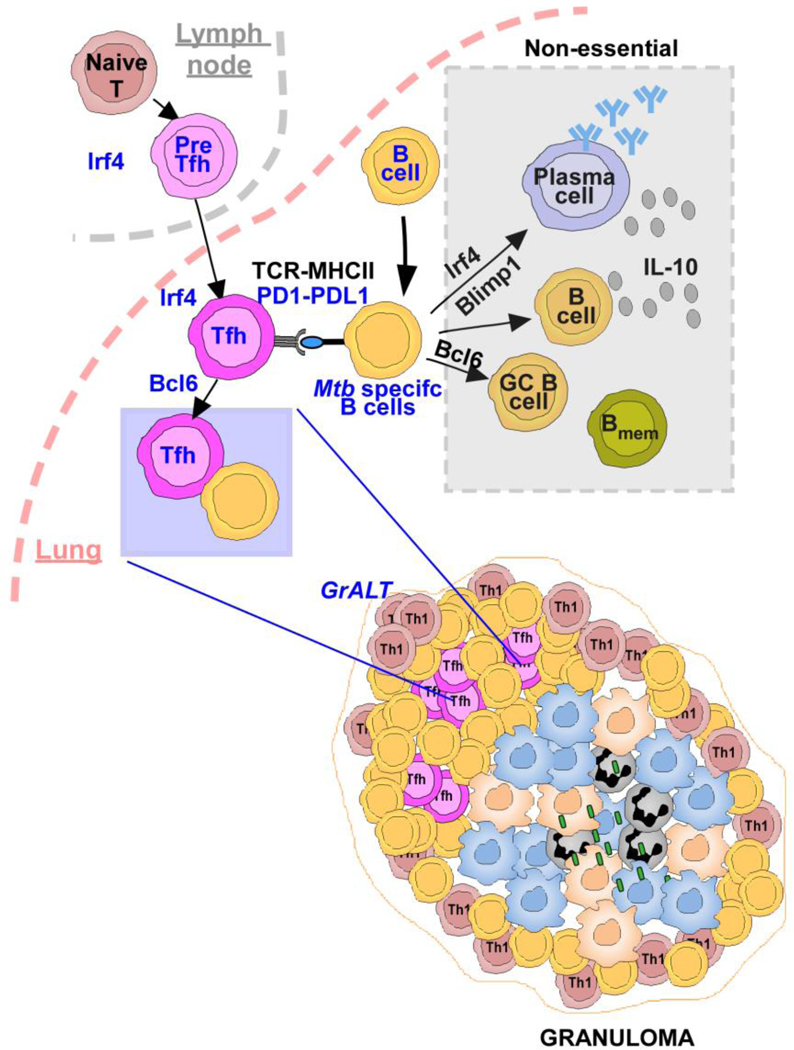
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), is a global cause of death. Granuloma associated lymphoid tissue (GrALT) correlate with protection during TB, but the mechanisms of protection are not understood. During TB, the transcription factor IRF4 in T- but not B cells is required for the generation of Th1, Th17 and Tfh-like cellular responses. A population of IRF4+ T cells co-express the transcription factor BCL6 during Mtb infection, and deletion of Bcl6 (Bcl6fl/fl) in CD4+ T cells (CD4cre) resulted in reduction of Tfh-like cells, impaired localization within GrALT, and increased Mtb burden. In contrast, the absence of germinal center (GC) B cells, MHC Class II expression on B cells, antibody-producing plasma cells, or IL-10-expressing B cells, did not increase Mtb susceptibility. Indeed, antigen-specific B cells enhance cytokine production and strategically localize Tfh-like cells within GrALT via PD-1 and PD-L1 interactions, and mediate Mtb control in both mice and macaques.
Introduction
Tuberculosis (TB) is caused by Mycobacterium tuberculosis (Mtb) results in 1.5 million deaths annually1. TB exists as a spectrum of disease, with variable clinical outcomes potentially linked to the granuloma, a hallmark of TB. Myeloid and neutrophilic granulomas are associated with progression to active pulmonary TB (ATB)2, whilst more organized granulomas associated with lymphoid follicles (GrALT) within tertiary lymphoid organs (TLO) are associated with Mtb control3, 4 and TB latency (LTBI). Here, GrALT is used to describe the lymphoid follicles localized near or within TB granulomas, which are less organized in mouse models3 but highly organized in the diversity outbred mouse model of TB5 and macaques with LTBI4, 5. GrALT facilitates Mtb control by orchestrating optimal interactions between lymphocytes (T- and B cells), and myeloid cells (macrophages and dendritic cells)2, 3, 4, and is poorly organized within human TB granulomas3. Recent studies showed that antibodies from individuals that control Mtb infection display unique profiles, are functionally distinct from those of individuals with ATB6, and can discriminate between individuals with LTBI and ATB7. B cell deficient mice are more susceptible to infection with clinical Mtb strains8, yet the specific functions of B cells involved in Mtb control remains unclear. By contrast, the impact of T cell-mediated immunity, particularly CD4+ T helper cells, on Mtb control is more established9. The co-localization and interaction of antigen-specific B cells and CXCR5-expressing CD4+ T cells within GrALT is critical in mediating protection, but the molecular interactions that generate efficient pulmonary immunity during TB is unknown3, 10. Therefore, the current study was designed to delineate the specific T- and B cell protective functions within the GrALT that mediate Mtb control.
High-throughput transcriptomic analysis across multiple species5, highlighted IRF4 as a potential key regulator of Mtb control. Here we show that IRF4, a critical transcription factor for B- and T cell differentiation, which is also expressed by DCs, is downregulated during progression to ATB in humans and animal models that express a spectrum of TB disease. Irf4 expression in T cells is essential to support lung Th1, Th17 and Tfh-like cell responses, GrALT formation and Mtb control. Importantly, IRF4+ T cells co-express BCL6 during Mtb infection and Bcl6 deficiency in T- and B cells (lacking Tfh-like cells or germinal center [GC] B cells) are required for GrALT formation and Mtb control. In addition, while Mtb-specific B cells are needed for Mtb control and GrALT formation, important B cell effector mechanisms such as MHC-II presentation, antibody production, or IL-10 production by B cells is not required for either control of Mtb or GrALT formation. Upon Mtb infection, B cells upregulated PD-L1 expression and interactions with PD-1-expressing lung Tfh-like cells was required to enhance the differentiation from pre-Tfh to mature Tfh-like cells and localization within GrALT to mediate Mtb control. Thus, our results reveal a critical protective function for Tfh-like cells localized within GrALT, and support the contribution of B cells in the strategic localization of Tfh-like cytokine producing cells within the GrALT for Mtb control.
Results
IRF4 expression is regulated in CD4+ T cells during ATB
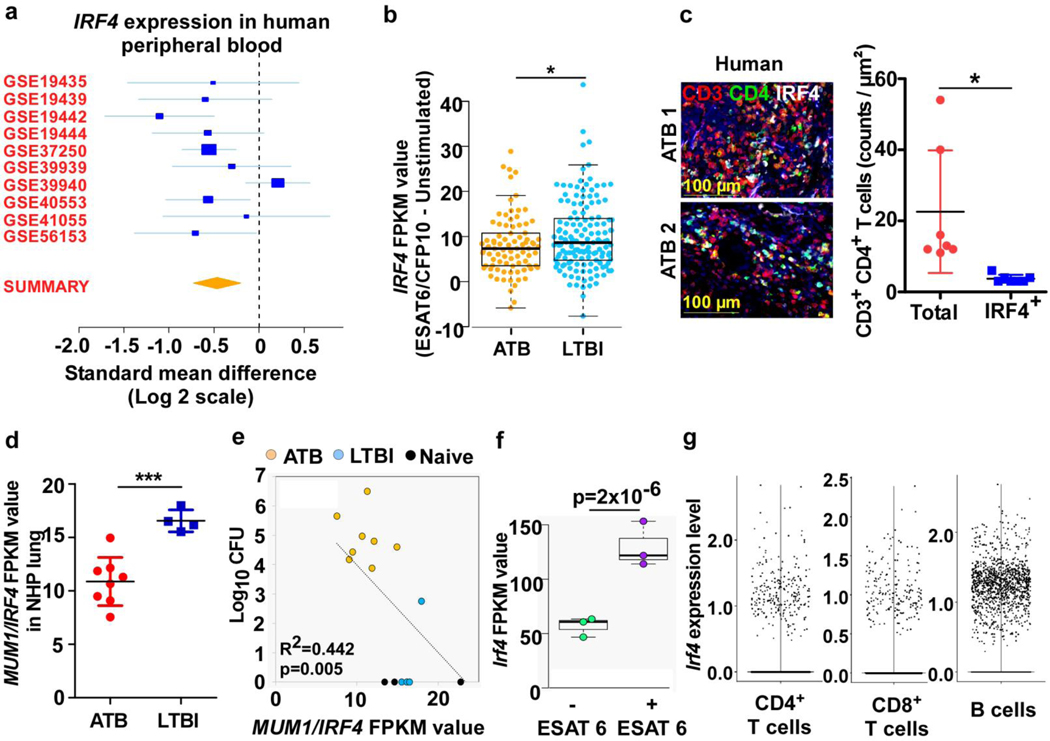
Naive T cells express low levels of the transcription factor IRF4, which is upregulated upon TCR stimulation for differentiation into T helper subsets11. To measure IRF4 mRNA levels during TB disease in humans, 10 human whole blood transcriptome datasets (Supplementary Table 1) from asymptomatic individuals with or without Mtb infection, or with microbiologically-confirmed ATB disease were identified12. Meta-analysis showed down-regulation of IRF4 mRNA expression in ATB, compared to healthy individuals or those who controlled Mtb infection (Fig 1a; Supplementary Table 1). Ex-vivo stimulated PBMCs from Mtb-infected humans, who either remained asymptomatic (LTBI) or progressed to ATB, with Mtb antigen ESAT6/CFP10 peptide pool, showed higher IRF4 levels in individuals with LTBI compared with ATB (Fig 1b). Importantly, only a small proportion of total CD3+CD4+ T cells expressed IRF4 within lung granulomas in human ATB patients (Fig 1c). Also, IRF4 (MUM1) mRNA expression was significantly downregulated in the lungs of Mtb-infected NHP with ATB, when compared with NHP with LTBI (Mtb-infected NHPs with asymptomatic TB) (Fig 1d), and correlated with increased Mtb burden in ATB (Fig 1e). Lung CD4+ T cells isolated from Mtb-infected C57BL/6 mice also showed increased expression of Irf4 after in vitro ESAT-6 stimulation, compared to unstimulated cells (Fig 1f). Single cell RNA sequencing (scRNA-seq) of lung cells from uninfected and Mtb-infected C57BL/6 mice at 50 and 100 days post infection (dpi)13 revealed activated T- and B cell populations13 as cell types that expressed Irf4 (Fig 1g). Thus, IRF4 expression is induced in T- and B cells cells upon antigen/Mtb exposure, but is downregulated in ATB disease in humans and macaques.
Figure 1. Analysis of IRF4 expression in animal models and in human TB.
(a) Forest plot showing IRF4 expression in whole blood transcriptome of human active TB patients (n = 10) in comparison to healthy controls with (n = 8) or without (n = 2) Mtb infection (Table S1). (b) Swarm plot showing relative IRF4 mRNA expression following ESAT6/CFP10-stimulation of purified human T cells from ATB and LTBI donors, data is normalized to unstimulated values (n = 85 ATB and 112 LTBI). Data was analyzed using two-sided t-test, unequal variance. p-values represent significance of differential expression according to DEseq. (c) Representative IF images of lung lesion from patients with ATB (n = 1 male; 72 years of age and 1 female; 51 years of age) show IRF4+CD3+CD4+ T cells; CD3-red, CD4-green, IRF4-white. Graph shows total number of CD3+CD4+ T cells counted and the number of IRF4+ cells among them. Data represents mean ± SD, with two-sided unpaired t-test used to compare groups. (d) MUM1/IRF4 (ID: ENSMUSG00000020156) mRNA levels in NHP lungs (ATB, n=8 and LTBI, n= 4) determined via bulk RNA-seq analysis. (c-d) Data represents mean ± SD, with two-sided unpaired t-test used to compare groups. (e) Correlation between log10 CFU vs MUM1/IRF4 mRNA levels in the lungs of NHP ATB (n = 8), LTBI (n = 4) and naïve (n=4). (f) IRF4 expression by CD4+ T cells sorted from lungs of Mtb-infected C57BL/6 mice, incubated with or without ESAT6/CFP10 peptide (n = 3). FPKM normalization was performed in order to plot the data. DESeq2 was used to calculate significance of differential expression between each of the sample groups. The significance of the Pearson correlation values was tested using a two-tailed t-distribution test with degrees of freedom equal to n – 2. (g) scRNA-seq of cells isolated from the lungs of B6 mice, either uninfected (n = 2), or Mtb-infected at 50 dpi (n = 3) or 100 dpi (n = 3), swarm plot showing expression of Irf4 in CD4+, CD8+ T and B cells. Swarm plot was prepared using the “boxplot” R function and the “beeswarm” R package. The boxed area indicates the interquartile range (IQR) of the data; values below are the lowest 25% of data points, and values above are the highest 25% of data points. The thick black line in the center indicates the median value, and the whiskers extend to the minima and maxima data values, excluding outliers. Outliers are defined as any points 1.5 x IQR above the 3rd quartile or 1.5 x IQR below the 1st quartile (b and f). *, p≤0.05; **,p≤0.005; ***, p≤0.0005.
Irf4 in T cells is indispensable for Mtb control
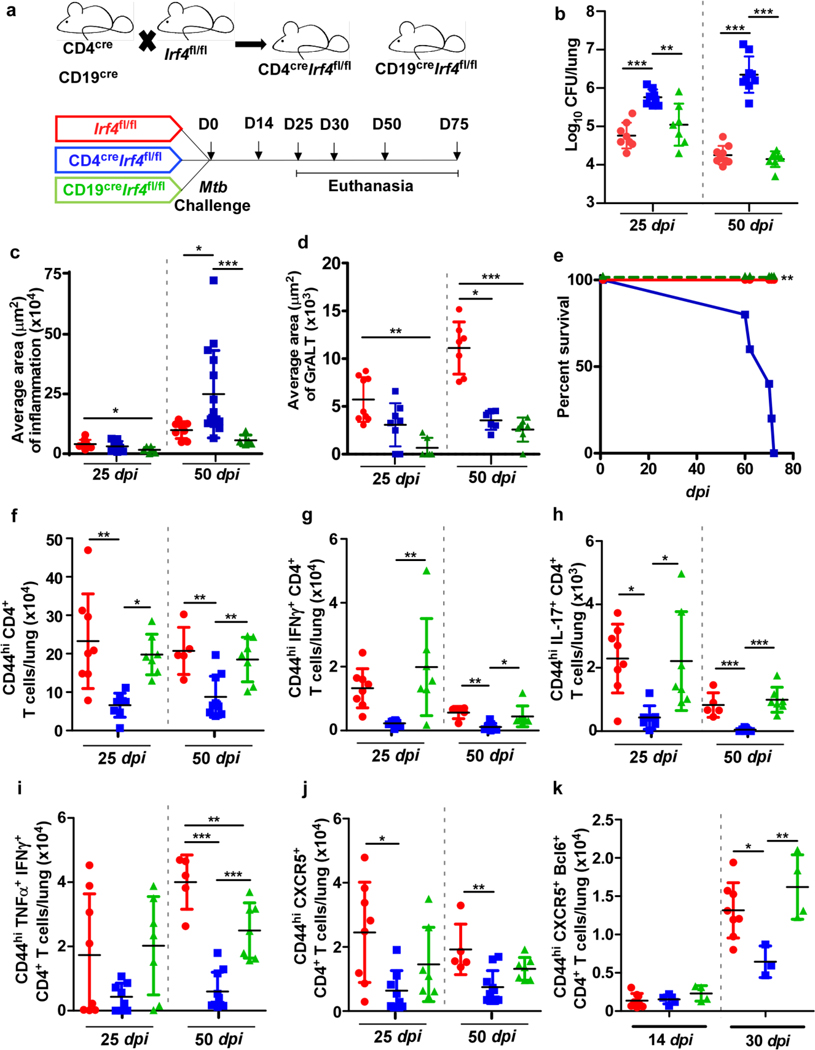
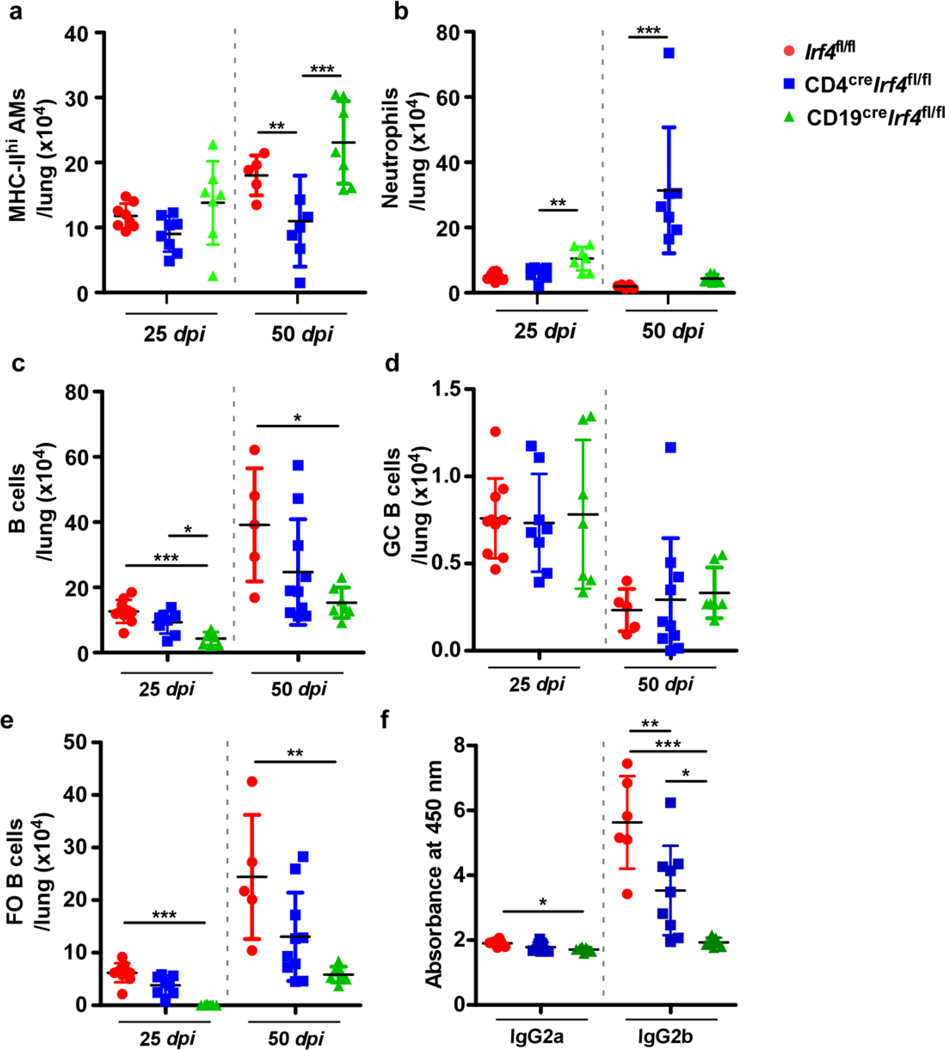
We crossed the Irf4fl/fl mice14 with CD4cre mice or CD19cre mice to generate CD4creIrf4fl/fl (Irf4 deletion in T cells) and CD19creIrf4fl/fl (Irf4 deletion in B cells) mice (Fig 2a) and aerosol infected with Mtb HN878. Mice lacking Irf4 in T cells showed significantly elevated Mtb CFU in the lungs at 25 dpi with further increase at 50 dpi. In contrast, Irf4fl/fl and CD19creIrf4fl/fl mice controlled Mtb burden at both dpi (Fig 2b). The increased lung bacterial burden coincided with exacerbated lung inflammation in CD4creIrf4fl/fl mice, while CD19creIrf4fl/fl mice had significantly lower inflammation than Irf4fl/fl mice (Fig 2c). CD4creIrf4fl/fl also exhibited reduced MHC-II expressing alveolar macrophages (AMs), enhanced neutrophil recruitment to the lungs of CD4creIrf4fl/fl mice, when compared with Irf4fl/fl mice (Extended Data Fig 1a and b). GrALT was significantly smaller in CD4creIrf4fl/fl mice, which were highly susceptible to Mtb infection, but also in CD19creIrf4fl/fl mice, which were not susceptible (Fig 2d). CD4creIrf4fl mice exhibited signs of increased mortality and succumbed to infection by 80 dpi (Fig 2e), compared with Irf4fl/fl and CD19creIrf4fl/fl mice. Thus, while Irf4 expression in both T- and B cells is required for GrALT, our results show that only Irf4 expression in T cells is critical for Mtb control.
Figure 2. Irf4 expression in T cells is indispensable for Mtb control.
CD4creIrf4fl/fl, CD19creIrf4fl/fl and Irf4fl/fl mice were infected with Mtb HN878 and were euthanized at indicated timepoints. (a) Experimental scheme. (b) Bacterial burden was determined in the lungs of CD4creIrf4fl/fl, CD19creIrf4fl/fl, and Irf4fl/fl mice. Formalin-fixed lung lobes were sectioned and stained with H&E and immunofluorescence to analyze (c) inflammation (average area) and (d) average area of GrALT. (e) CD4creIrf4fl/fl, CD19creIrf4fl/fl and Irf4fl/fl mice were monitored over 75 dpi for disease severity and to enumerate (f) activated CD4+ T cells, (g) IFNγ-producing CD4+ T cells, (h) IL-17-producing CD4+ T cells, (i) CD4+ T cells co-producing IFNγ and TNFα, and (j) CXCR5+ CD4+ T cells by flow cytometry. (k) The mice lungs were assessed for the presence of CXCR5+Bcl6+CD4+ T cells at 14 and 30 dpi. Data represents the mean ± SD, analysis was performed using one-way ANOVA with Tukey’s multiple comparison test (b and f to k), and Kruskal-Wallis ANOVA with Dunn’s multiple comparison test (c and d). Survival between CD4creIrf4fl/fl, CD19creIrf4fl/fl and Irf4fl/fl mice post-infection were compared using the Mantel-Cox test (e). *, p≤0.05; **,p≤0.005; ***, p≤0.0005; n = 4–10 mice/group (b to j) and 3–4 mice /group (k).
Irf4 mediates Tfh-like cell generation during Mtb infection
Accumulation of activated CD4+ T cells at 25 dpi was significantly compromised in CD4creIrf4fl/fl mice, when compared with both Irf4fl/fl and CD19creIrf4fl/fl mice (Fig 2f). Activated CD4+ T cells expressing IFNγ (Fig 2g), IL-17 (Fig 2h), or co-expressing IFNγ and TNFα (Fig 2i) were reduced in CD4creIrf4fl/fl mice compared to CD19creIrf4fl/fl and Irf4fl/fl mice at 25 dpi, and the reduction was maintained until 50 dpi. Additionally, there was a significant reduction in lung CXCR5+CD4+ T cells (i.e., Tfh-like cells) (Fig 2j) and CXCR5+BCL6+ Tfh-like cells in CD4creIrf4fl/fl, compared with Irf4fl/fl mice (Fig 2k). Consistent with the requirement for Irf4 in the development of GC B cells and FO B cells15, we observed a decrease in total B cells, and FO B cells, but not GC B cells in CD19creIrf4fl/fl mice when compared with Irf4fl/fl mice (Extended Data Fig 1c-e). Indeed, Mtb-specific antibody responses remained lower in CD19creIrf4fl/fl mice when compared with Irf4fl/fl mice (Extended Data Fig 1f), suggesting defective production of plasma cells. Mtb-infected CD4creIrf4fl/fl mice also had reduced total numbers of B cells as well FO B cells (Extended Data Fig 1c and e). Irf4 deficiency in B cells impaired GrALT formation, compromised the accumulation of lung Tfh-like cells (albeit not significantly), but did not affect protective immunity to Mtb infection. In contrast, Irf4 deficiency in T cells also reduced GrALT formation, and significantly reduced the accumulation of Th1, Th17 and Tfh-like cells and increased susceptibility in CD4creIrf4fl/fl mice infection.
Tfh-like cells promote protective GrALT formation during TB
We enumerated IRF4+ expressing T cells within the granulomas of macaques with ATB or LTBI. Although total T cell numbers within granulomas were not significantly different between ATB and LTBI, we found increased accumulation of IRF4+ T cells within the protective GrALT of LTBI macaques (Fig 3a-c). Accordingly, more PD1+Tfh-like cells localized and showed a direct and significant correlation with GrALT size in LTBI lungs (Fig 3d). Fewer IRF4+ PD1+ Tfh-like cells were also found within the ATB human granulomas among the total CD3+ T cells enumerated (Fig 3e to f). Thus, these results together suggest that IRF4+ Tfh-like cells (either PD1+, CXCR5+ or Bcl6+) are generated during protection and associated with enhanced GrALT formation in TB.
Figure 3. IRF4+ BCL6+ T cells increase during Mtb infection.
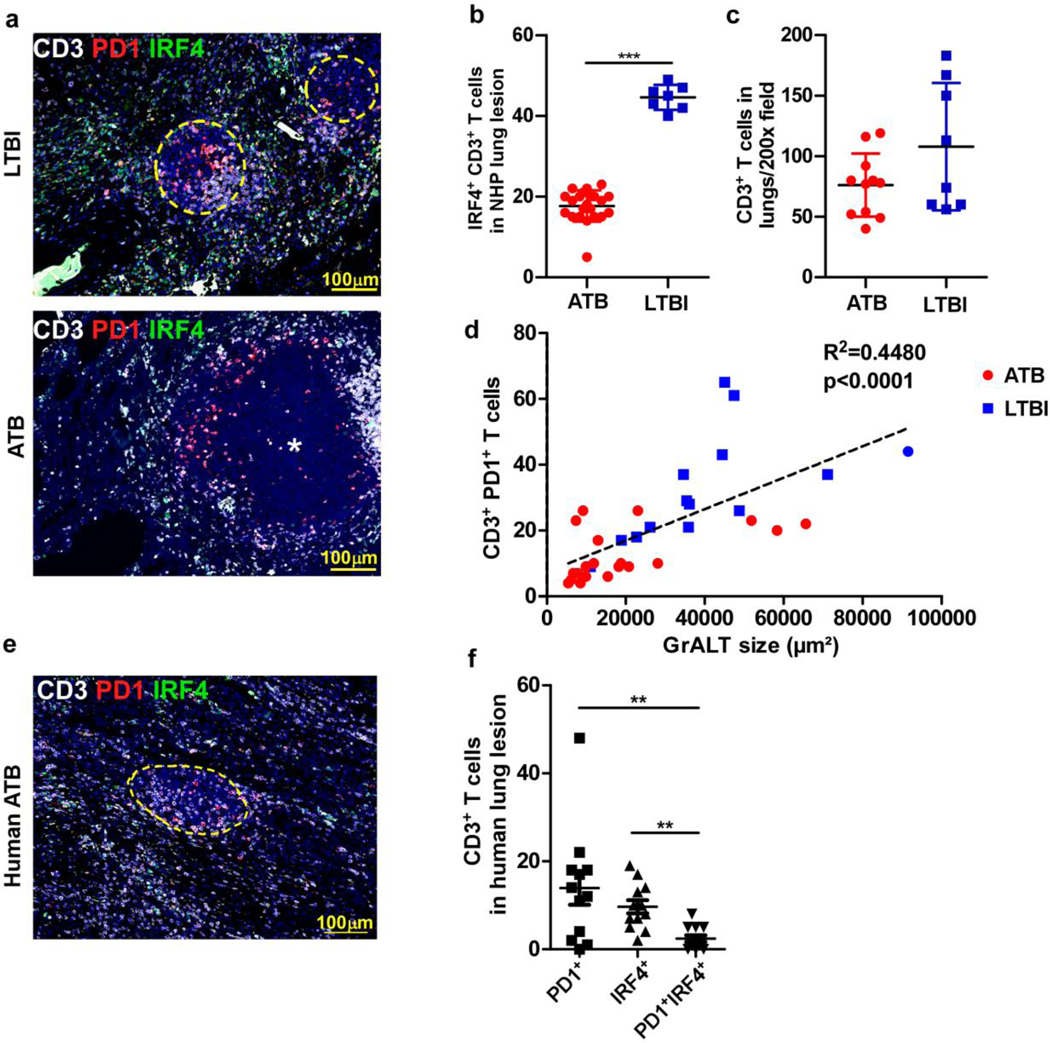
(a) Representative image of inflammatory lesion in lung of NHP LTBI (left) vs ATB (right) stained with antibodies specific for CD3 = white; IRF4 = green; PD1 = red. Circles with yellow dashed lines depict ectopic lymphoid structures and asterix = center of granuloma. The numbers of (b) IRF4+ CD3+ (n = 4 NHPs; 7–21 data points from different fields) and (c) only CD3+ (n=3 NHPs/ group) T cells in NHP ATB vs LTBI lung lesions were estimated by blinded morphometric analysis. (d) Correlation between the number of CD3+CD4+PD1+ Tfh-like cells inside B cell areas with the average area of the GrALT in the lungs of Mtb-infected NHPs (n = 5–6 NHPs/group). (e) Representative human ATB lesion, (f) number of T cells in human lung lesion expressing either PD1, IRF4 or both (n = 3 males and 1 female lung biopsies; age: 29–41 years). Data represents the mean ± SD. Statistical significance was determined with two-sided unpaired t-test (b and c), Pearson’s correlation (d) and Kruskal-Wallis ANOVA with Dunn’s post-hoc test (f). *, p≤0.05; **,p≤0.005; ***, p≤0.0005.
Lung Bcl6+ Tfh-like cells are critical for Mtb control
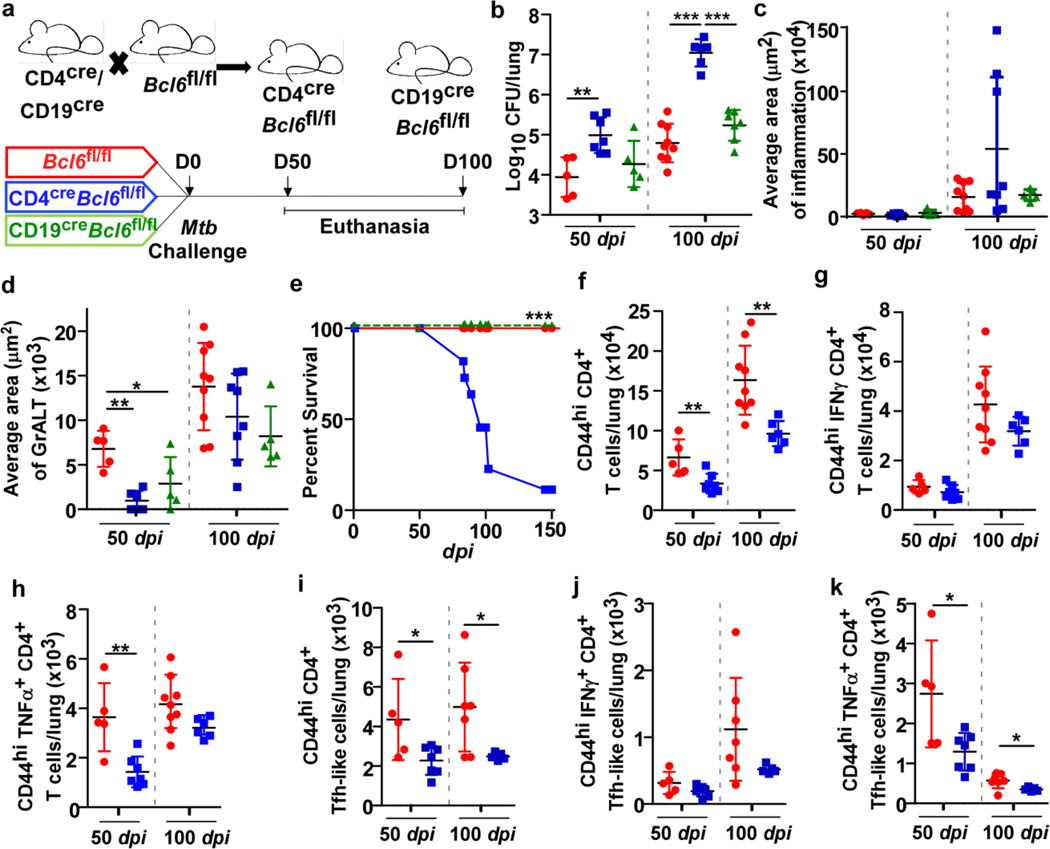
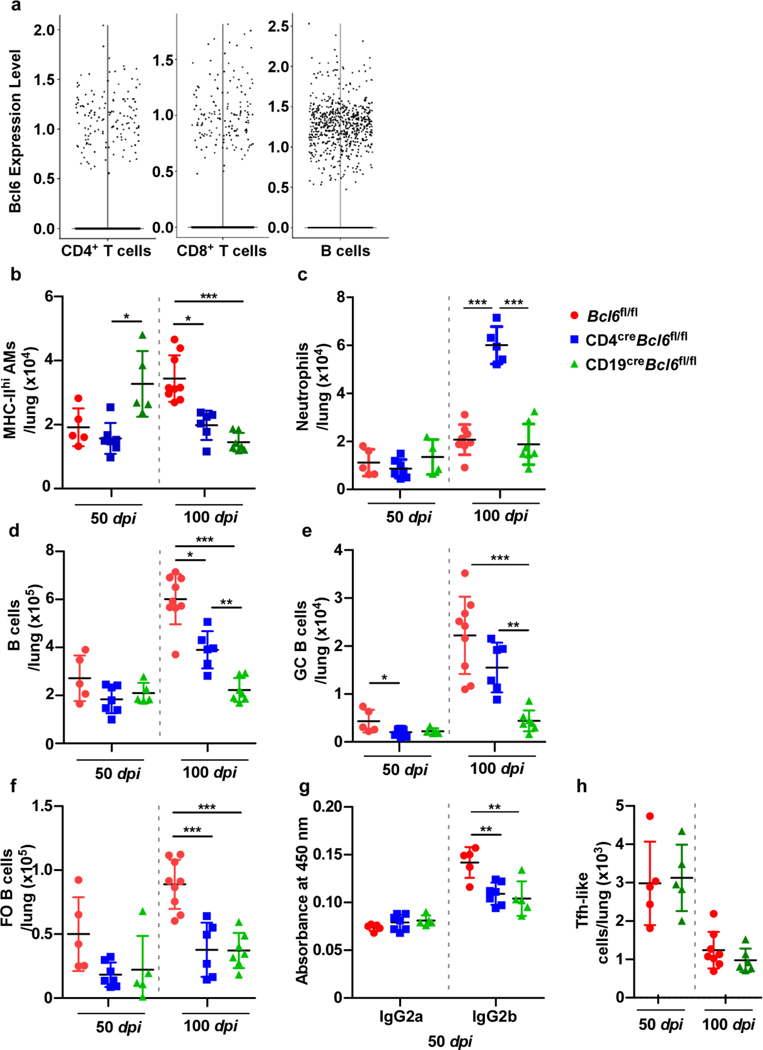
Mouse gene expression dataset of naïve and Mtb-infected DO mice showed that Pou2af1, an upstream regulator of Bcl6 and GC-Tfh development, was the top transcription factor co-expressed with Irf4 (Supplementary Table 2)5. Tfh-like cells were significantly reduced in NHP with ATB and in susceptible CD4creIrf4fl/fl mice following infection. Bcl6 is expressed in both T- and B cells in mouse upon Mtb infection (Extended Data Fig 2a). Therefore, Bcl6 was specifically deleted in T cells, and as Bcl6 is required for the generation of GC B cells, Bcl6 was also eliminated in CD19+ B cells. CD4creBcl6fl/fl and CD19creBcl6fl/fl mice were infected with Mtb (Fig 4a). Notably, lung Mtb CFU (Fig 4b) was significantly higher in CD4creBcl6fl/fl mice compared with Bcl6fl/fl mice at 50 and 100 dpi. Loss of Mtb control was associated with increased lung inflammation (Fig 4c) and smaller GrALT (Fig 4d) at later stages of Mtb infection. CD4creBcl6fl/fl mice, compared with Bcl6fl/fl mice, were highly susceptible to Mtb infection, with a median survival time around 100 dpi (Fig 4e). Similar to CD4creIrf4fl/fl mice (Extended Data Fig 1a), CD4creBcl6fl/fl mice were unable to activate AMs and MHC-II+ expression in lung at 100 dpi, compared with Bcl6fl/fl mice (Extended Data Fig 2b). Additionally, neutrophils were increased within lungs of Mtb-infected CD4creBcl6fl mice (Extended Data Fig 2c).
Figure 4. Tfh-like cells but not GC B cells mediate control of Mtb.
CD4creBcl6fl/fl and CD19creBcl6fl/fl mice were infected with Mtb HN878 alongside Bcl6fl/fl littermate controls (n = 3–9 mice/group), and lungs were collected and processed at indicated timepoints. (a) Experimental scheme. (b) Bacterial burden was determined in the lungs of CD4creBcl6fl/fl, CD19creBcl6fl/fl, and Bcl6fl/fl mice. Formalin-fixed lung lobes were cut and stained to analyze (c) inflammation (average area) and (d) average area of GrALT. (e) Survival of CD4creBcl6fl/fl, CD19creBcl6fl/fl and Bcl6fl/fl mice were monitored over time. CD4creBcl6fl/fl mice were infected with Mtb HN878 alongside Bcl6fl/fl littermate controls, and flow cytometry was used to enumerate (f) activated CD4+ T cells, (g) IFNγ+ CD4+ T cells, (h) TNFα+ CD4+ cells (i) activated CD4+ Tfh-like cells, (j) IFNγ+ CD4+ Tfh like cells and (k) TNFα+ CD4+ Tfh-like cells in the lungs at the indicated time points. Data represents mean ± SD, and statistical analyses performed using one-way ANOVA with Tukey’s multiple comparison test (b and d), Kruskal-Wallis ANOVA with Dunn’s post-hoc test (c), Mantel-Cox analysis assessing survival distributions between CD4creBcl6fl/fl and Bcl6fl/fl mice post-infection (e), two-sided unpaired t-test (f to k). *, p≤0.05; **,p≤0.005; ***, p≤0.0005.
While activated CD4+ T cells were reduced in Mtb-infected CD4creBcl6fl/fl mice (Fig 4f), accumulation of CD4+ T cells expressing IFNγ was not changed (Fig 4g). There were also early reductions in accumulation of TNFα producing CD4+ T cells (Fig 4h). We previously showed that CXCR5+ Tfh-like cells express cytokines such as IFNγ, TNFα and IL-23. Similar to total activated CD4+ T cells, numbers of IFNγ producing Tfh-like and TNFα-producing Tfh-like cells were reduced in the Mtb-infected lungs of CD4creBcl6fl/fl mice (Fig 4i-k). B cell populations (FO B cells, GC B cells) as well as Mtb-specific antibody response, were also significantly reduced in CD4creBcl6fl/fl and CD19creBcl6fl/fl as compared with Bcl6fl/fl mice (Extended Data Fig 2d-g). Both CD19creBcl6fl/fl and Bcl6fl/fl mice were able to control Mtb (Fig 4b) and displayed similar levels of lung inflammation at 50 and 100 dpi (Fig 4c). Incidentally, CD19creBcl6fl mice, despite maintaining similar inflammation, had smaller GrALT, when compared with Mtb-infected Bcl6fl/fl mice (Fig 4d). Significantly fewer B cells, including GC B cells, FO B cells, as well as Mtb-specific antibody responses, accumulated in the lungs of CD19creBcl6fl/fl mice, compared with Mtb-infected Bcl6fl/fl mice (Extended Data Fig 2d-g). While fewer activated CD4+ T cells and CD4+ T cells expressing IFNγ accumulated during chronic disease in Mtb-infected CD19creBcl6fl/fl mice, there were no differences in Tfh-like cell accumulation (Extended Data Fig 2h). Thus, although both Tfh-like cells and GC B cells participate in GrALT formation, Tfh-like cells are the key component required for pulmonary protective Mtb immunity in mice.
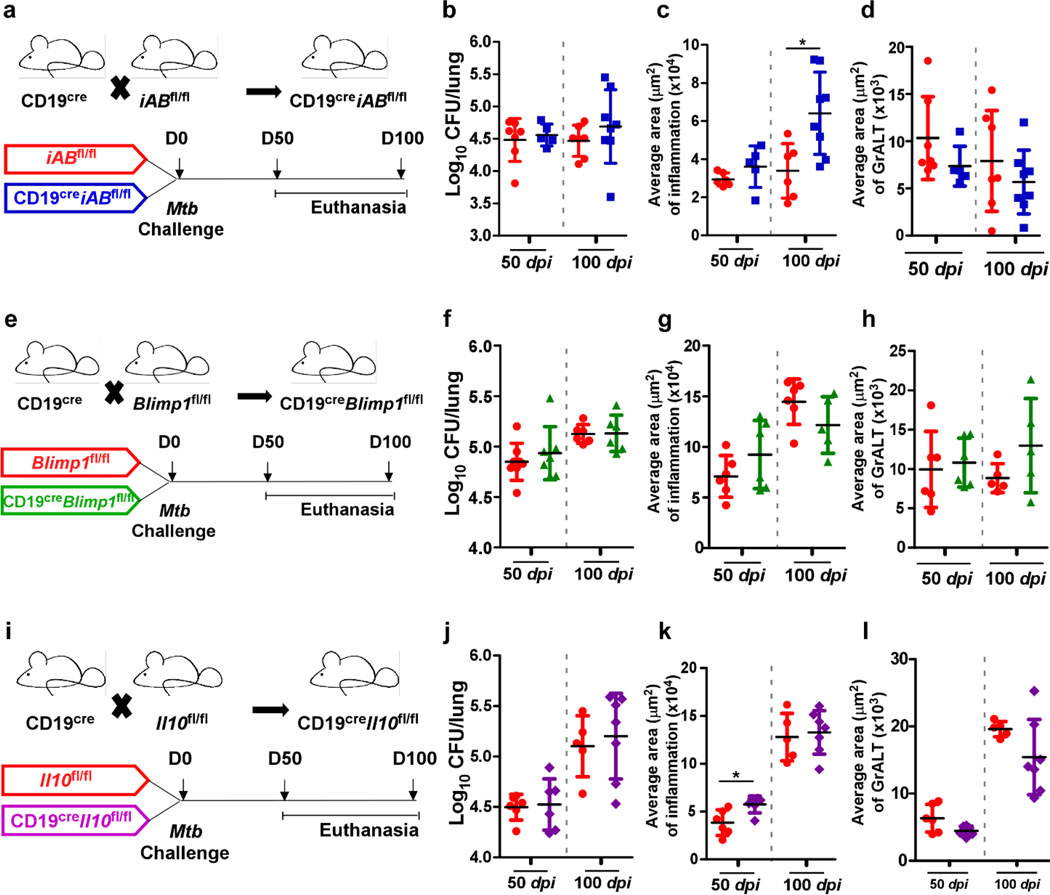
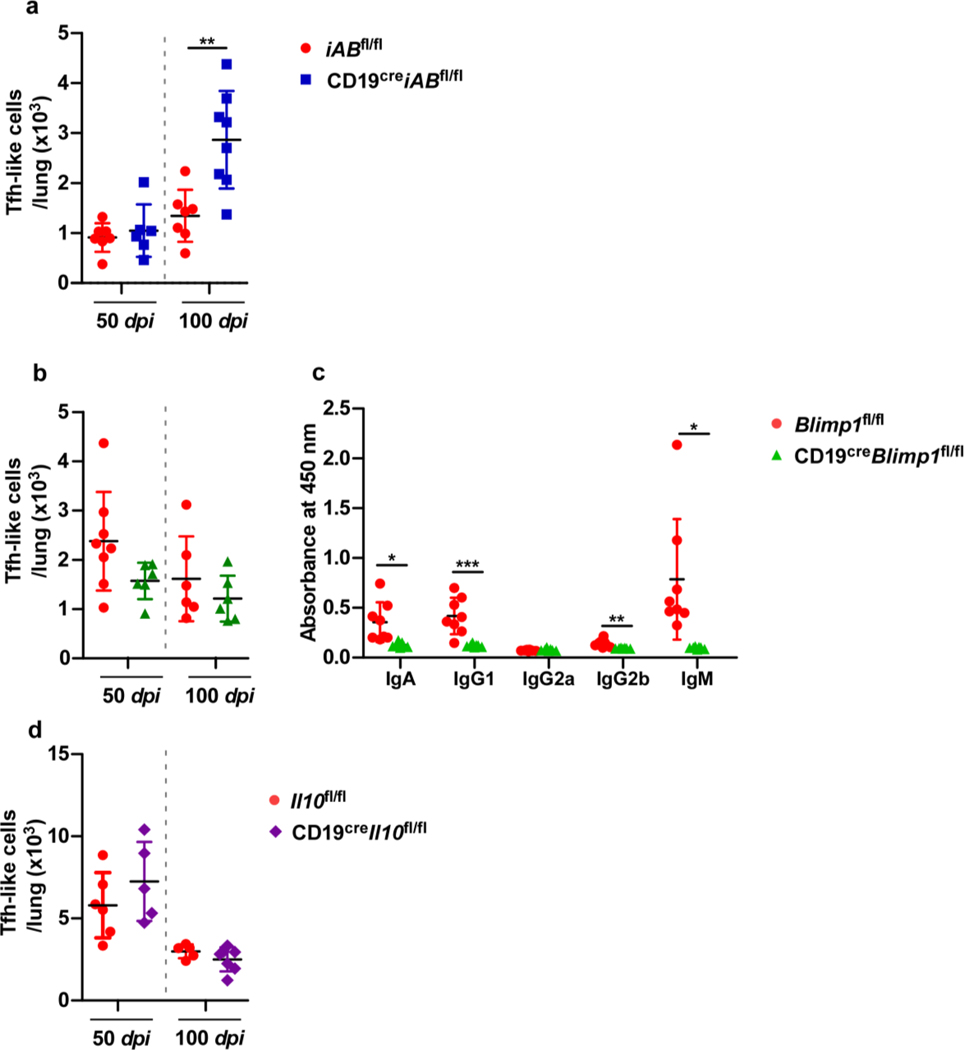
B cells are required for long-term control of clinical strains of Mtb8. Nevertheless, CD19creBcl6fl/fl mice, which lack GC B cells, were still able to control Mtb. Thus, we generated conditional mice deficient for B cell functions including antigen presentation through MHC-II (CD19creiABfl/fl mice) (Fig 5a), plasma cell differentiation and subsequent antibody production (CD19creBlimp1fl/fl mice) (Fig. 5e), and regulatory cytokine IL-10 production (CD19creIl-10fl/fl mice) (Fig 5i). CD19creiABfl/fl mice (Fig 5b), CD19creBlimp1fl mice (Fig 5f), and CD19creIl-10fl/fl mice (Fig 5j) were all able to control Mtb. Early inflammation (Fig 5c, g and k) and accumulation of Tfh-like cells were not impacted in any of the CD19cre mice with compromised B cell effector functions (Extended Data Fig 3a, b and d). CD19creBlimp1fl/fl Mtb-infected mice were severely defective in induction of Mtb-specific antibodies when compared with Blimp1fl/fl Mtb-infected mice (Extended Data Fig 3c). There were no differences observed in GrALT in any of the CD19cre mice during Mtb infection (Fig 5d, h and l). Thus, our results show that B cells participate in Mtb control8, but independent of the generation of GC B cells, MHC-II antigen presentation, antibody production by plasma cells and the modulatory effects of IL-10. Of note, no significant decreases in the Tfh-like cells were observed in any of these B cell deficient mouse models.
Figure 5. Dispensable B cell effector functions in Mtb control.
Conditional B cell knock-out mice and littermate controls were infected with Mtb HN878 (n = 4–10 mice/group). Lungs were collected and processed at the indicated time points. CD19cre mice were crossed with iABfl/fl mice to generate CD19creiABfl/fl mice (MHC-II deficient B cells). (a) Experimental scheme, (b) bacterial burden, (c) average area of inflammation and (d) average area of GrALT are shown. CD19cre mice were crossed with Blimp1fl/fl mice to generate CD19creBlimp1fl/fl mice, and the (e) experimental scheme, (f) bacterial burden, (g) average area of inflammation and (h) average area of GrALT are shown. CD19cre mice were crossed with Il-10fl/fl mice to generate CD19creIl-10fl/fl mice, and the (i) experimental scheme, (j) bacterial burden, (k) average area of inflammation and (l) average area of GrALT are shown. Data represents mean ± SD, and statistical analyses were performed using two-sided unpaired t-test. *, p≤0.05.
Mtb-specific B cells localize Tfh-like cells within GrALT
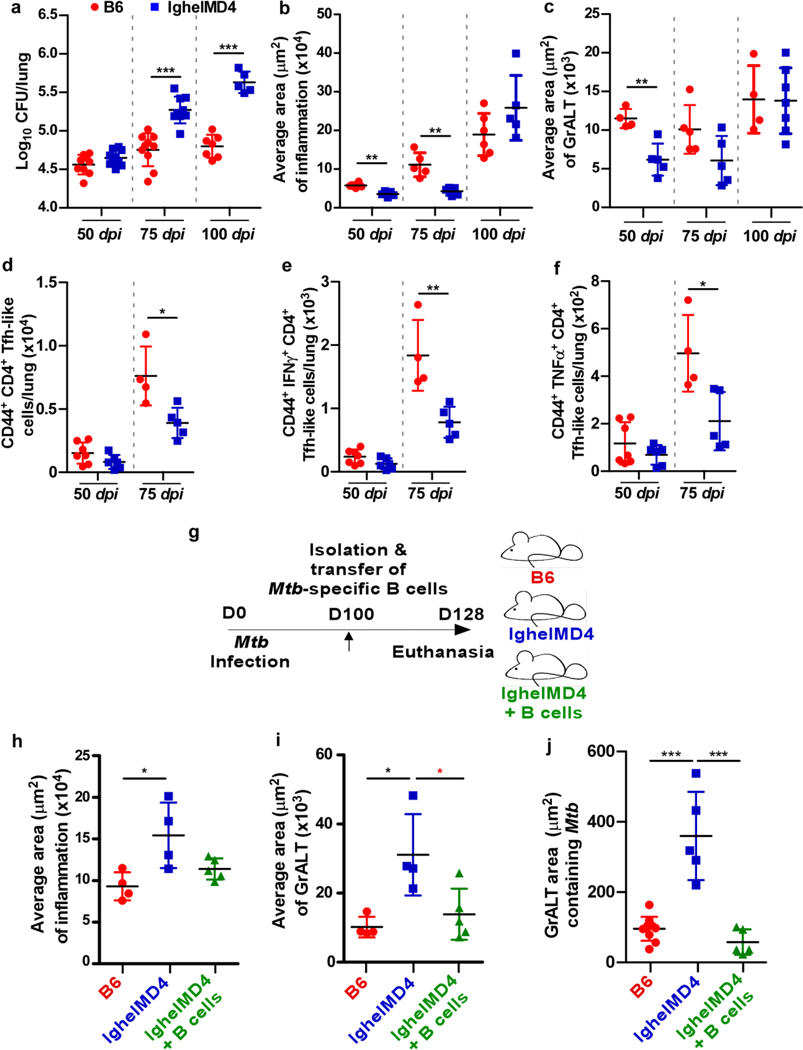
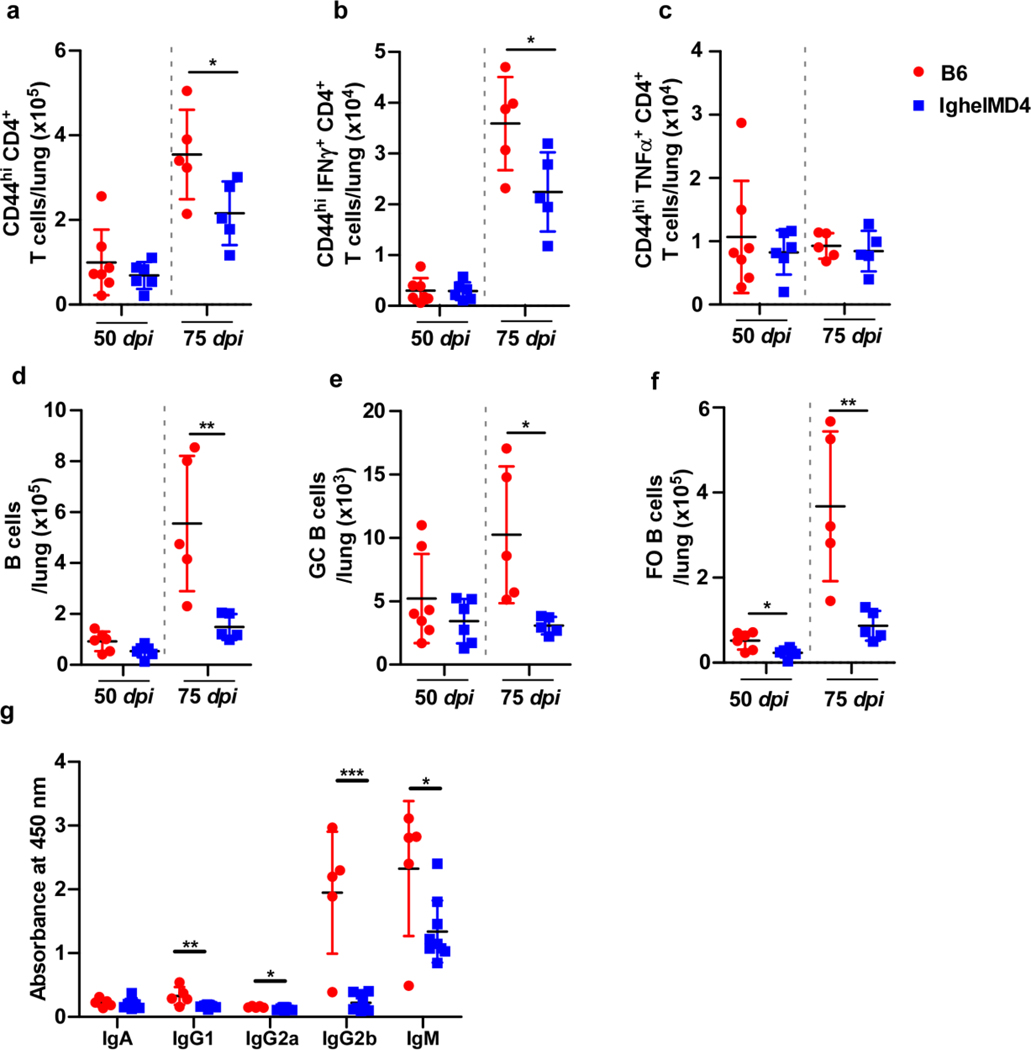
Transgenic IghelMD416 mice, in which the majority (>90%) of B cells recognize hen egg lysozyme and do not recognize Mtb antigens exhibited increased lung Mtb CFU when compared with C57BL/6 Mtb-infected mice during chronic infection (at 75 and 100 dpi), but not at early time points (50 dpi) (Fig 6a). Inflammation did not coincide with the increase in bacterial burden (Fig 6b). While GrALT formation was defective at early time points, non-specific B cells accumulated over time to form large GrALT structures in IghelMD4 Mtb-infected mice (Fig 6c). Fewer total activated CD4+ T cells accumulated in lungs of IghelMD4 mice compared with C57BL/6 mice, but no significant differences were observed in the Th1 cells (i.e., producing cytokines IFNγ or TNFα) (Extended Data Fig 4a-c). Similarly, Tfh-like cells were reduced, and of note, those producing cytokines including IFNγ or TNFα (Fig 6d-f). Additionally, B cell accumulation, including GC B cells and FO B cells, as well as antibody production was reduced in chronically Mtb-infected IghelMD4 mice (Extended Data Fig 4d-g). To address the impact of Mtb-specific B cells on immunopathology in the lungs of Mtb-infected IghelMD4 mice, we adoptively transferred B cells isolated from Mtb-infected C57BL/6 mice into IghelMD4 mice during chronic TB (Fig 6g). Twenty-eight days after transfer (128 dpi), IghelMD4 mice showed higher inflammation and GrALT, whereas inflammation and GrALT were lower in the IghelMD4 mice which received antigen-specific B cells (Fig 6h-i). Notably, significantly lower Mtb were harbored within the GrALT in the IghelMD4 mice that received the B cells (Fig 6j).
Figure 6. Mtb-specific B cells control chronic Mtb infection.
IghelMD4 mice were infected with Mtb HN878 alongside controls (n = 3–10 mice/group). Lungs were collected and processed at indicated timepoints. (a) Bacterial burden was determined in the lungs. Formalin-fixed lung lobes were sectioned and stained to analyze (b) average area of inflammation and (c) average area of GrALT. (d) Activated CD4+ Tfh-like cells, (e) IFNγ+CD4+Tfh-like cells, and (f) TNFα+CD4+ Tfh-like cells were enumerated by flow cytometry. IghelMD4 mice were infected with Mtb HN878 alongside controls (n = 3–10 mice/group). B cells were isolated from the lungs of Mtb-infected C57BL/6 mice and transferred to IghelMD4 mice (100 dpi). C57BL/6, IghelMD4 and IghelMD4 mice receiving B cells were euthanized at 128 dpi (28 days after transfer) and the lungs were collected and processed. (g) Experimental scheme. Formalin-fixed lung lobes were sectioned and stained with H&E and immunofluorescence to analyze the (h) average area of inflammation, (i) average area of GrALT, and (j) GrALT area containing Mtb. Data represents the mean ± SD. Statistical significance was calculated with unpaired t-test (a to e), one-way ANOVA with Tukey’s multiple comparison test (j and h), Kruskal-Wallis ANOVA with Dunn’s multiple comparison test (i) and Mann-Whitney U test (f and i (red star)). *, p≤0.05; **,p≤0.005; ***, p≤0.0005.
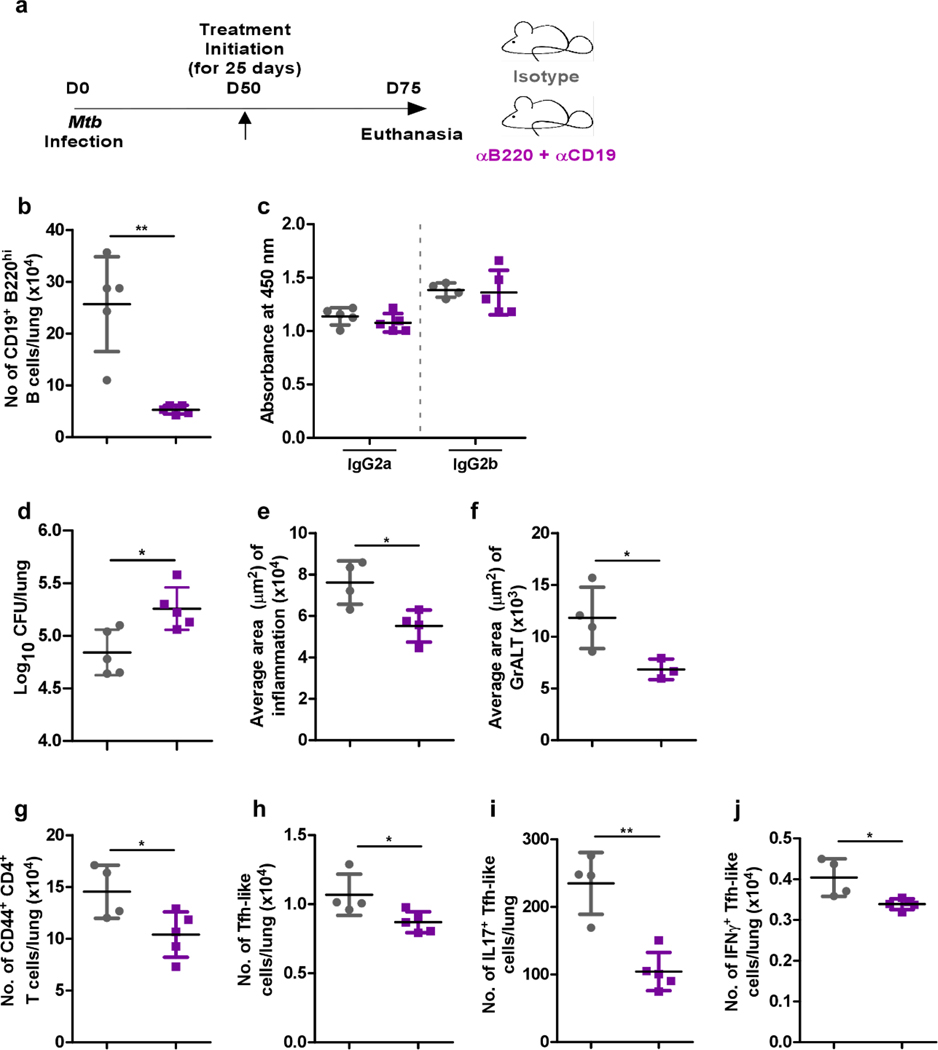
Furthermore, Mtb infection was established in C57BL/6 mice over 50 days, following which α-B220 and α-CD19, or isotype controls were delivered for 25 days (Extended Data Fig 5a). Mtb-infected mice were successfully depleted of B cells in the lung without affecting antibody production (Extended Data Fig 5b and c). Similar to IghelMD4 mice, B cell depleted mice exhibited increased lung Mtb CFU, accumulated fewer activated CD4+ T cells, Tfh-like cells and cytokine-producing Tfh-like cells (Extended Data Fig 5d to j). Therefore, Mtb-specific B cells are required for accumulation of cytokine-producing Tfh-like cells, GrALT formation and local Mtb control.
B cells via PD1-PDL1 axis drive Tfh-like cell localization
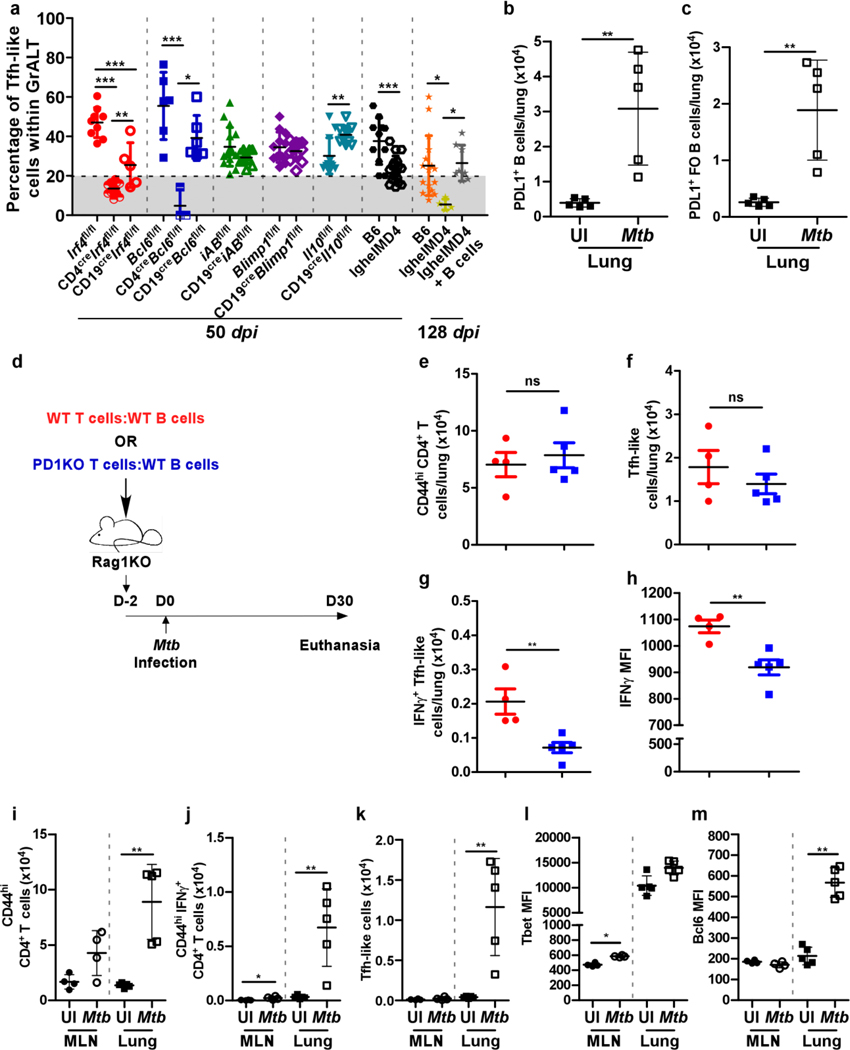
Our previous studies proposed that GrALT is associated with Mtb control and protection3. Yet, in the current study, we found in our conditional deletion models that GrALT formation/organization is not sufficient to explain protection because GrALT was reduced in both CD19creIrf4fl/fl and CD19creBcl6fl/fl mice (Extended Data Fig 6a-b), which were still able to control Mtb. In contrast, CD4creIrf4fl/fl and CD4creBcl6fl/fl mice (Extended Data Fig 6a-b), which were more susceptible to infection, also lack GrALT formation/organization. We therefore evaluated the localization of Tfh-like cells within GrALT in the different CD4 and CD19 Cre mouse models and found that despite its reduced size, the GrALT of Mtb-infected CD19creIrf4fl/fl and CD19creBcl6fl/fl mice still contained a significant localization of Tfh-like cells (Fig 7a, Extended Data Fig 6b). Neither GrALT size nor Tfh-like cell localization within GrALT were impacted in CD19creiABfl/fl, CD19creBlimp1fl/fl, CD19creIl10fl/fl mice (Fig 7a, Extended Data Fig 6a-b). In sharp contrast, in all the susceptible murine models (i.e. CD4creIrf4fl/fl, CD4creBcl6fl/fl and IghelMD4 mice) Tfh-like cells did not localize within GrALT. Thus, there is a threshold of Tfh-like cells within GrALT (Fig 7a), which is needed to mediate Mtb control. We also addressed the localization of Mtb relative to GrALT. Mtb 16sRNA generally localized outside of the GrALT areas during Mtb control (Extended Data Fig 7a). In sharp contrast, in highly susceptible models, such as CD4creIrf4fl/fl and CD4creBcl6fl/fl mice (which lack Tfh cell differentiation and GrALT formation), Mtb localized even within poorly formed GrALT (Extended Data Fig 7a). These results provide novel insights into the dynamic and complex interplay between Tfh-like cells and B cells limiting Mtb within GrALT.
Figure 7. PD1+ Tfh-like cells localize within GrALT for Mtb control.
Irf4fl/fl, CD4creIrf4fl/fl, CD19creIrf4fl/fl, Bcl6fl/fl, CD4creBcl6fl/fl, CD19creBcl6fl/fl, iABfl/fl, CD19creiABfl/fl, Blimp1fl/fl, CD19creBlimp1fl/fl, Il10fl/fl, CD19creIl10fl/fl, C57BL/6 and IghelMD4 mice were infected with Mtb HN878 (n = 3–10 mice/group). Lungs were collected and processed at 50 dpi. Mtb-infected B6, IghelMD4 and IghelMD4 (recipient) mice were euthanized, and lungs were collected and processed at 128 dpi. Formalin-fixed lung lobes were sectioned and stained with immunofluorescent antibodies to determine the (a) percentage of PD1+CD3+ Tfh-like cells within B cell areas. Uninfected or Mtb-infected (30 dpi) C57BL/6 mice (n = 5 mice/group) were euthanized, and lung cells were subjected to flow cytometric analysis for PD-L1 (CD247) expression on (b) total B cells and (c) FO B cells. B cells isolated from the spleens and lymph nodes of C57BL/6 mice and T cells isolated from the spleens and lymph nodes of either C57BL/6 or Pd1KO mice were co-transferred to Rag1KO mice, 1–2 days prior to infection with Mtb HN878 (n = 4–5 mice/group). Mice were euthanized 30 dpi. (d) Experimental scheme. (e) activated CD4+ T cells, (f) CD4+ Tfh-like cells (g) IFNγ+CD4+ Tfh-like cells and (h) IFNγ mean fluorescent intensity in activated CD4+ T cells in the lungs were determined by flow cytometry. Uninfected or Mtb-infected C57Bl/6 mice were euthanized (30 dpi) to collect lung (n = 5 mice/group) and mediastinal lymph node (MLN) (n = 4 mice/group). (i) Activated CD4+ T cells, (j) CD4+IFNγ+ T cells in both MLN and lungs; (k) Tfh like cells; MFIs for transcription factors (l) Tbet and (m) Bcl6 in both MLN and lungs activated CD4+ T cells were determined by flow cytometry. Data represents mean ± SD. Statistical significance was calculated with one-way ANOVA with Tukey’s multiple comparison test for group of three and two-sided unpaired t-test for group of two (a-c and e-h), and two-sided Mann-Whitney U-test (i to m). *, p≤0.05; **,p≤0.005; ***, p≤0.0005.
B cells potentiate IFNγ in protective Tfh-like cells
To delineate the mechanisms by which lung B cells mediate Tfh-like cell localization, we assessed the top genes induced in lung B cells compared with splenic B cells from Mtb-infected mice8. Atf3 (~ 5 fold higher)17, which is rapidly induced upon B cell activation17, Nr4a1(~4.5 fold), Nr4a2 (~3.5 fold), genes induced upon BCR stimulation18, as well as CCR7 (~2 fold higher), which allows B cells to respond and traffic towards T cells19 was induced in lung B cells. These results suggest that BCR stimulation by Mtb antigens is important for the stable downstream interactions with Tfh-like cells. Indeed, Atf3 is a known transcription factor that induces PD-L1 expression17. As such, PD-L1 expression on lungs B cells (Fig 7b) during Mtb infection was increased, particularly in FO B cells (Fig 7c) and not in other subsets (data not shown).
To address if PD-1 and PD-L1 interactions mediate the localization of Tfh-like cells, Rag1KO mice, which lack mature B cells and T cells, received either B cells and T cells from B6 WT mice, or B cells from B6 mice and T cells from Pd1KO mice (1:1 ratio). The Rag1KO mice were infected with Mtb and lung T cell responses were analyzed (Fig 7d). Numbers of total activated lung T cells (Fig 7e) were similar between the two groups, but lung Tfh-like cells were decreased in Rag1KO mice, which received B6 B cells and Pd1KO T cells (Fig 7f). However, IFNγ+ Tfh-like cells (Fig 7g) and IFNγ production on a per cell basis (Fig 7h) was significantly reduced in Mtb-infected Rag1KO, which received B6 B cells and Pd1KO T cells. Finally, in the Rag1KO mice that received B6 B cells and B6 T cells, we observed a reduced Mtb16S rRNA signal and organized GrALT formation. In contrast, an increased Mtb16S rRNA signal, as well as poorly formed GrALT, was detected in Rag1KO mice that received B6 B cells and Pd1KO T cells (Extended Data Fig 7b and c). Thus, the disruption of effective PD1-PDL1 interactions between B- and T cells abrogated the protective effector functions of Tfh-like cells in Mtb-infected mice.
When we compared the T cell responses in the lymph node (LN) vs the lungs from uninfected and Mtb-infected B6 mice, we found that activated T cells (Fig 7i) as well IFNγ-producing T cells (Fig 7j) were significantly increased upon infection in the lungs but not the LNs. Also, accumulation of Tfh-like cells were only found in the lungs but not LNs of Mtb-infected mice (Fig 7k). This was associated with increased T-bet expression (Fig 7l) within Tfh-like cells in LNs, but increased Bcl6 expression (Fig 7m) was observed only in Mtb-infected lungs coinciding with Tfh-like cell accumulation. Following Mtb infection, T cells are primed in the LNs, but reach their full potential, including effector functions and Tfh-like cell phenotype, in the lungs, likely through the PD-1 and PD-L1 interactions with Mtb-specific B cells inside the GrALT.
B-cell depletion reduces control in Mtb-infected macaques
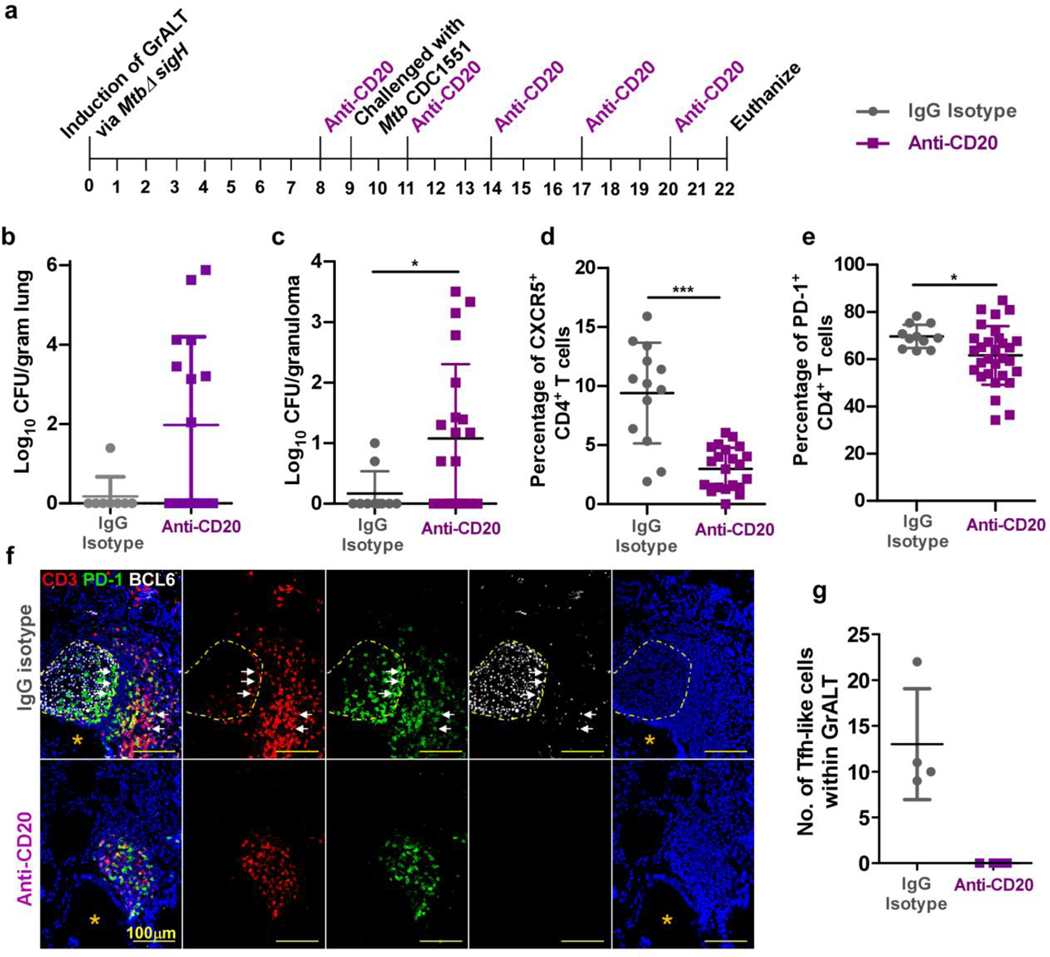
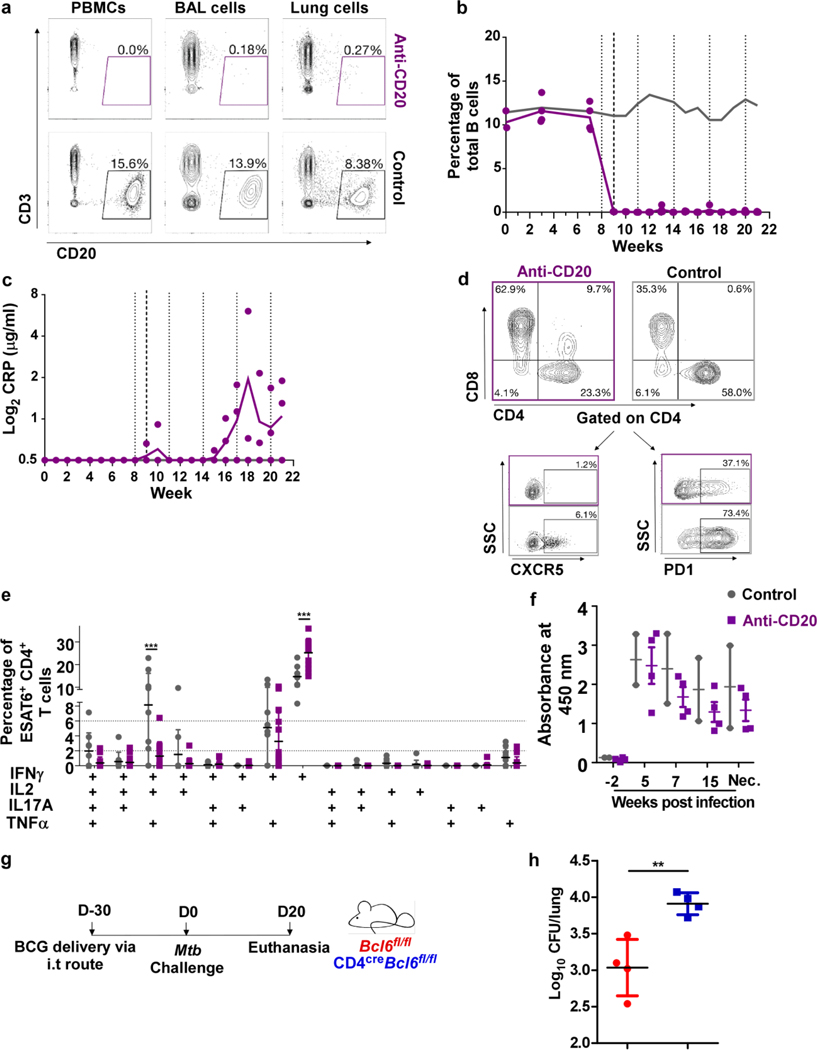
Vaccination of macaques with MtbΔsigH increased GrALT and controlled infection upon challenge with lethal dose of Mtb10. To address the role of B cells in Mtb control, macaques were aerosol vaccinated with 1000 CFU of MtbΔsigH and divided into two groups. The treatment group received intravenous administration of 2B81R (anti-CD20) eight weeks post-vaccination and every three weeks thereafter. The control group received control IgG antibody at the same time points (Fig 8a). B cells, at every time point after the initial treatment, were significantly reduced in the lung when compared to control animals (p<0.05) (Extended Data Fig 8a-b). One week after the first administration of CD20-depleting or control antibodies, animals were challenged with a high-dose of Mtb CDC1551 (Fig 8a), which leads to 100% lethality in susceptible rhesus macaques10. After the B cell depletion and high-dose challenge, 50% of treated animals displayed detectable levels of C-reactive protein (CRP) (Extended Data Fig 8c), a marker of systemic inflammation that is highly indicative of active TB in macaques10. To determine if B cells aid in controlling bacterial growth, either directly or indirectly, we analyzed Mtb burdens at necropsy. B cell depleted animals had significantly higher bacterial burden in randomly collected lung sections (Fig 8b), as well as in individual granulomas (Figure 8c). B cell depleted macaques also showed reduced accumulation of Tfh-like cells expressing CXCR5 (Fig 8d; Extended Data Fig 8d) or PD-1 (Fig 8e; Extended Data Fig 8d). These results were further confirmed by co-staining for BCL6 and PD-1. In contrast to isotype control treated vaccinated macaques, B cell-depleted animals exhibited little to no well-organized GrALT, and lacked localization of CD3+PD-1+BCL6+ Tfh-like cells (Fig 8f, g). Granulomas from control animals retained more polyfunctional cytokine producing CD4+ T cells (IFNγ+TNFα+IL-2+), while B cell depleted granulomas contained more monofunctional, IFNγ-producing CD4+ T cells (Extended Data Fig 8e). As expected, B cell depleted macaques had similar Mtb-specific IgG response to control macaques (Extended Data Fig 8f). Furthermore, B cells may play a direct role in recruiting or retaining CD4+ Tfh-like cells, which may reside in the GrALT.
Figure 8. B cells mediate control in MtbΔsigH-vaccinated macaques.
Macaques were aerosol exposed to MtbΔsigH prior to challenge with virulent Mtb CDC1551 and received either CD20 depleting (n = 4 NHP) or IgG isotype control antibodies (n = 2 NHP). Clinical samples were collected, and macaques were monitored for clinical signs of disease throughout the study. (a) Experimental scheme. Bacterial burden was determined in (b) bulk lung tissue and (c) individual granulomas at necropsy. (d) CXCR5+CD4+ T cells, and (e) PD1+CD4+ T cells were detected by flow cytometry at the time of necropsy. (f) Representative images of a B cell follicle in lungs of a macaque which received IgG control isotype antibodies (top panels) and images of lung tissue from a macaque which received CD20 depleting antibodies (bottom panels); CD3-red, PD1-green, BCL6-white; dotted yellow line indicate germinal center, white arrows indicate CD3+BCL6+PD1+ Tfh-like cells, orange asterix indicates the bronchus. (g) The number of BCL6+ Tfh-like cells with the GrALT. Data represents mean ± SD. Statistical significance was calculated with tow-sided Mann-Whitney U-test (b to e); no statistics calculated as anti-CD20 treated NHP had all ‘zero’ values (g). *, p≤0.05, ***, p≤0.0005.
To confirm the contribution of Tfh-like cells in vaccine-induced immunity to Mtb infection, we vaccinated Bcl6fl/fl and CD4creBcl6fl/fl with BCG by the intratracheal route, followed by rest and subsequent challenge with Mtb HN878 (Extended Data Fig 8g). We found that BCG-vaccinated CD4creBcl6fl/fl mice showed significantly increased Mtb CFU when compared with Bcl6fl/fl mice (Extended Data Fig 8h). These results further confirm a functional role for Tfh-like cells in protective immunity to Mtb challenge after vaccination.
Discussion
TB continues to be a global health threat and the immune parameters that mediate protection or pathology are not fully delineated. Our results here answer long-standing questions regarding the role of B cells in impacting Mtb control through the differentiation and localization of Tfh-like cells within GrALT.
IRF4 is expressed in innate and adaptive immune cells, including macrophages, and dendritic cells, as well as T- and B cells20. Absence of IRF4 in CD4+ T cells affects the differentiation of Th2, Th9, Th17 and Tfh cells. However, the role for IRF4 in generation of Th1 cells is less clear. IRF4 also controls peripheral CD8+ T cells differentiation following viral infection, and the expression of effector molecules such as IFNγ and granzyme B is reduced in CD8+ T cells lacking IRF411. Consistent with these studies, both T cells (including CD4+, CD8+ T cells) and B cells predominantly expressed Irf4 mRNA in the Mtb-infected mouse lung. IRF4 promotes CD8+ T cell exhaustion during chronic viral infection21 or represses CD4+ T cell exhaustion during allograft rejection22. Thus, the decreased levels of IRF4 mRNA in the lungs of Mtb-infected macaques and mice, and in the blood of individuals with ATB, may be a reflection of diminished T cell activation23 and/or depletion of IRF4-expressing T cells. In T cells, IRF4 is induced within hours following TCR stimulation, and its expression depends on TCR affinity24. These results are confirmed by the increased expression of IRF4 mRNA in antigen-specific CD4+ T cells (ESAT6/CFP10 stimulated) from Mtb-infected individuals and mice. At the early time points post Mtb infection, IRF4 expression is up regulated in antigen-specific T- and B cells, and is required for differentiation of T cells into effector subsets and B cells, respectively.
Using Mtb-infected CD4creIrf4fl/fl, CD19creIrf4fl/fl and Irf4fl/fl mice, we show that Irf4 expression in T cells is necessary for effective control of Mtb, results which are similar to those seen in CD4−/− mice25. Consistent with the known role for IRF4 in B cell differentiation11, a reduction in lung FO B cells in Mtb-infected CD19creIrf4fl/fl mice, which coincided with reduced lung inflammation and impaired GrALT formation in the lungs. We did not find any differences in GC cells in the lung, but did observe defective Mtb-specific antibody responses in the CD19creIrf4fl/fl mice. Thus, GC and antibody responses are not directly involved in protection, but Tfh-like localization within GrALT (independent of GrALT size) can still lead to efficient control of Mtb infection.
The formation of well-organized GrALT in the lung is associated with protection and Mtb control in mouse and NHP models3, 4, 10. Our initial results show that IRF4+ T cells localize within GrALT and strategic positioning of Tfh-like cells within GrALT correlates with Mtb control, suggesting a feedback loop where Tfh-like cells provide positive signals to B cells to enhance GrALT formation. These findings led us to experimentally address the role of Bcl6 in T cells and the specific role of lung Tfh-like cells in the mouse model of Mtb infection. Tfh differentiation occurs following dendritic cell (DC) priming, CD4+ T cell activation and engagement of peptide:MHC, co-stimulatory molecules and cytokines. Activated CD4+ T cells differentiate into Bcl6+ pre-Tfh cells or Blimp1+ CD4+ T cells (non-Tfh cells). There is a well-known reciprocal antagonistic relationship between Bcl6 and Blimp1 (encoded by Prdm1), where Bcl6 represses Blimp1, and Blimp1 represses Bcl626. Consistent with the participation of Bcl6 in the generation of Tfh-like cells, we found that Bcl6 deletion in T cells caused a significant reduction in cytokine-producing lung Tfh-like cells, impaired GrALT formation, thereby leading to increased susceptibility to Mtb infection. Deficiencies of Irf4 or Bcl6 in CD4+ T cells resulted in enhanced Mtb susceptibility, albeit CD4creBcl6fl/fl mice succumbed to TB disease at a later timepoint, likely because Bcl6 functions downstream of Irf4. While Bcl6 deficiency in B cells did not affect susceptibility to Mtb infection, GrALT was significantly smaller, likely because the recruitment and transition to GC B cells was affected thus impacting the formation of lymphoid follicles. Despite reduced GrALT, Mtb control was still maintained because Tfh-like cells still efficiently localized (albeit at lower levels than controls but still above a threshold mediating protection) within GrALT. These results unequivocally show a role for Tfh-like cells and Bcl6 in T cells in mediating protective immunity against TB, and a dispensable role for GC B cells in Mtb infection.
While the participation of CD4+ T cell in Mtb control is well-recognized, the contribution of B cells is much less understood. Recent studies showed that there were distinctive B cell subpopulations in the periphery of ATB patients27. Mtb-specific B cells proliferate and are present in human granulomas3 and animal models3, 4, 10 and are located in close proximity to CXCR5-expressing CD4+ T cells and Mtb-infected macrophages3, 10. A novel TB vaccine, MtbΔsigH, which provides protection against Mtb challenge in NHPs, was associated with enhanced GrALT formation, consisting of CD20+ B cells in granulomas10. Also, B cell deficient mice are susceptible to infection with clinical Mtb strains8. Thus, while the protective functions of B cells in Mtb control are suggestive, the exact functional mechanism of action thus far has been elusive. Our data show that Mtb-specific B cells are important for controlling the infection, by mediating effective generation and localization of lung Tfh-like cells, independent of widely recognized B cell functions, including GC B cell differentiation, MHC-II-dependent antigen presentation, plasma cell differentiation/antibody production, or IL-10 synthesis. Recent literature surrounding the role of antibodies during Mtb infection has found distinct differences antibodies isolated from individuals with asymptomatic vs symptomatic TB disease6, 7, differences based on isotype and production site28, and antibodies against Mtb lipids29, although there is no clear consensus on what role antibodies are actually playing during infection. We show that antibodies themselves do not have a direct protective function, as CD19creBlimp1fl/fl mice did not exhibit increased Mtb susceptibility. However, antibodies may have a potential role in preventing Mtb dissemination, which occurs because of impaired cellular immunity30.
Our results validate a role for Mtb-specific B cells in protective immunity, as IghelMD4 mice are more susceptible to Mtb infection, showed impaired accumulation of cytokine-producing Tfh cell and poorly formed GrALT. Lung resident memory B cells, which are distinct from peripheral B cells, acquire effector functions early after challenge, as their strategic placement allows for local antigen encounter at the site of infection31. Absence of MHC-II on CD19 cells or lack of GC B cells is not required for expansion of lung Tfh-like cells and Mtb control. There is no consensus in the literature with several papers highlighting a non-redundant role for MHC-II driven antigen presentation32 or independent of CD40:CD154 interaction. However Rodriguez-Pinto et al33 instead highlight that the CD40:CD154 interaction is required for optimal CD4+ T cell priming by B cells. IL-10 is an anti-inflammatory and inhibitory cytokine that limits protective responses to Mtb infection34. Mtb has previously been shown to utilize IL-10 to evade the immune system. Previous studies indicated that activated effector T and B cells are a major source for IL-10. Consistent with previous data, we did not find a protective or pathological role for IL-10 expression in B cells up to the time points tested.
Lung B cells isolated from Mtb-infected mice expressed heightened markers of BCR activation, such as Atf3 and Nr4a1/Nr4a2. ATF3 is a highly induced gene in lung B cells, which can transcriptionally regulate PD-L1 by directly binding to the PD-L1 promoter region17 or can upregulate PD-L1 expression on marginal zone B cells in an Atf3-dependent manner35. Furthermore, PD-L1 on B cells can interact with PD-1 on T cells, inhibit T cell recruitment into the follicle and concentrate T cells toward the GC. PD-1 and PD-L1 interactions between individual Tfh-like cells and B cells optimized B cell competition and affinity maturation, thus controlling tissue positioning and function of Tfh cells in the spleen36. Our results show that activation of B cells, and upregulation of PD-L1 on B cells may allow interaction with PD-1 on T cells to drive the differentiation of cytokine-producing Tfh-like cells within the GrALT. PD-1 expressing CD4+ T cells accumulate in the lung following Mtb infection, proliferate and are not exhausted, but can differentiate into cytokine-producing cells and may play a central role in maintenance of antigen-specific effector T cells during chronic Mtb infection37. Upon aerosol infection with Mtb, PD-1 deficient mice (and to lesser extent PD-L1 deficient mice) and macaques treated with anti-PD1 antibody are highly susceptible to disease due to exacerbation of T cell responses and lung pathology5, 38, 39. Adoptive transfer studies demonstrate that PD-1-expressing CD4+ T cells limit TB lung pathology by regulating CD4+ T cell responses38. Additionally, PD-1 deficient mice have CD4+ T cells that express lower Tbet levels, despite higher IFNγ expression, while IFNγ and TNFα producing cells were reduced38. Our results show that PD-1 deficient cells when co-transferred with B6 B cells into Rag1KO mice mediated reduced accumulation of Tfh-like cells and impaired IFNγ production. Recent studies have shown that in the absence of PD-1 dependent inhibitory signals, migratory CD4+ T cells are less differentiated and result in increases in the lung parenchymal effector CD4+ T cell population with propensity to over-produce IFNγ and mediate pathology40. These results suggest a multifaceted regulatory role for PD-1 in Mtb infection. On one hand, PD-1 signals could be required to restrain overall CD4+ T cell activation and limit terminal differentiation, while on the other hand, these signals specifically in Tfh-like cells may be driving cytokine production and along with interactions with PD-L1 on B cells may facilitate the localization of Tfh-like cells within the GrALT to mediate Mtb control. Generating mice with Tfh-like cells deficient in PD-1 should allow us dissect out this dichotomy in future studies. PD-L1KO mice are not as susceptible to Mtb infection as PD-1 deficient mice38; thus, it is also possible that other PD-1 ligands are involved in the localization of Tfh-like cells and should be explored in future studies.
The formation and organization of GrALT is a correlate of protection in mouse models3, 41 and in macaques2, 3, 4, 42. However, absence of Irf4 and Bcl6 in T- and B cells both result in similarly defective GrALT organization with differing outcomes in Mtb control. By using a combination of conditionally deleted mice, transgenic mouse strains and adoptive transfer experiments, we show that the protective features of GrALT are derived from the strategic localization of cytokine-producing Tfh-like cells at the site of infection. Clearly, Tfh-like cells and B cells function together to mediate the protective GrALT formation with Tfh-like cell playing a fundamental primary protective role and B cells playing an indirect role. In Mtb-infected CD19creIrf4fl/fl and CD19creBcl6fl/fl mice, reduced size of GrALT is associated with lower accumulation of Tfh-like cells within GrALT. This is also clear in Mtb-infected IghelMD4 mice where absence of Mtb-specific B cells during acute infection is not required for GrALT formation or Mtb control, but during the chronic stages of infection, the poor accumulation of Tfh-like cells within GrALT resulted in increased susceptibility. The feedback interactions between Mtb-specific B- and T cells are present during chronic infection (100 dpi), where Mtb-infected IghelMD4 mice show a larger GrALT composed of non-specific B cells and increased inflammation, which was reversed by adoptive transfer of Mtb-specific B cells. Phuah et al43 depleted B cells in NHPs, and describe increased bacterial burden, dysregulated cytokine producing T cells, and a lower antibody response. Using a model where GrALT is critical in protection i.e., mucosal vaccine strain, MtbΔsigH, and Mtb challenge, we show that despite presence of antibodies, B cell depletion results in increased Mtb CFU, a decrease in T cells that produce multiple cytokines, a reduction in the accumulation of lung Tfh-like cells and GrALT size. We were able to replicate these findings in a mouse model of B cell depletion during Mtb infection, and mice deficient in Tfh-like cells also showed increased susceptibly in vaccine Mtb challenge models, highlighting the importance of targeting the B cell-Tfh-like cell axis in vaccine strategies against Mtb.
Based on the multiple lines of evidence in mouse and macaque models, we propose that Mtb-specific B cells likely provide help to lung Tfh-like cells (independent of MHC-II-TCR interactions) and downstream generation and strategic localization of cytokine producing Tfh-like cells within the GrALT for efficient Mtb control. The relationship between Tfh-like cells and B cells is thought to be important because of a reciprocal dependency played out during the generation of affinity-matured antibody. This cognate interaction occurs in germinal centers that form within B cell follicles of secondary lymphoid organs after antigen exposure. However, there is evidence that B cell dependence is not completely required for Tfh-like cells that can develop without B cell help, if T cells get adequate stimulation from peptide antigen-MHC-II complexes displayed on other antigen-presenting cells44. Additionally, B cells are dispensable for the priming of CD4+ T cells and the initial acquisition of a Tfh-like cell phenotype, but B cells might play a more important role in the maintenance, amplification of cytokine responses and function of lung Tfh-like cells. Recent evidence demonstrated that TLOs with a high presence of B cells led to protective CD4+ T cell responses in the lungs. While our results suggest that Mtb-specific B cells are needed for the generation of lung Tfh-like cells, it occurs independently of MHC-II expression on B cells, presence of GC B cells or antibody producing B cells at the site of the infection. Whether this reflects a difference between the Tfh-like cells in the SLOs and the pulmonary TLOs is an area that needs to be investigated in the future.
In conclusion, we show a critical contribution of the transcription factors IRF4 and BCL6 in CD4+ T cells in the generation of cytokine-producing Tfh-like cells, which localize within protective GrALTs to mediate Mtb control. Our results strongly suggest that Mtb-specific B cells are needed for orchestrating Tfh cell differentiation and accumulation in the GrALT, inducing cytokine production via PD1, and influencing GrALT organization. These results together answer long-standing questions regarding the contribution of Tfh-like cells and B cells in the generation of a protective GrALT involved in the generation of local immunity to control Mtb infection and provide novel targets and immune pathways for improving the design of TB vaccines.
Methods
Reanalysis of clinical meta-data
Ten human whole blood transcriptome datasets from healthy and Mtb-infected individuals were identified from the NCBI database Gene Expression Omnibus12. All datasets were downloaded as pre-processed by the original authors and multi-cohort analysis was performed on the summarized log2-transformed gene-level intensities using the MetaIntegrator R Package45. HIV-infected and post treatment samples were excluded from the studies, which profiled the transcriptional response during Mtb infection and treatment response, respectively.
Animals and infection
Mouse experiments.
C57BL/6, CD4cre, CD19cre, Irf4fl/fl, Bcl6fl/fl, iABfl/fl, IghelMD4, Rag1KO and Pd1KO mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Blimp1fl/fl mice were obtained from J. Gommerman (University of Toronto, Canada) and Il10fl/fl mice were kindly provided by A. O’Garra (Imperial College London, UK). All mice were maintained and bred at Washington University in St. Louis. Mice of both sexes between the ages of 6–8 weeks were used in accordance with the approved Institutional Animal Care and Use Committee (IACUC) guidelines at Washington University in St. Louis. CD4creIrf4fl/fl and CD19creIrf4fl/fl mice were obtained by crossing Irf4fl/fl strain with CD4cre and CD19cre, respectively. Similarly, CD4creBcl6fl/fl and CD19creBcl6fl/fl mice, as well as CD19creiABfl/fl mice, CD19creBlimp1fl/fl mice, and CD19creIl10fl/fl mice were generated.
BCG vaccination in mice:
BCG Pasteur (~5×105 CFU) was delivered intratracheally 30 days prior to infection with Mtb. Mice were euthanized at 20 dpi and lungs were harvested, homogenized, and serial dilutions of tissue homogenates plated on 7H11 agar plates supplemented with 2 μg/mL 2-thiophenecarboxylic acid hydrazide to determine Mtb CFU, and to selectively differentiate Mtb CFU from BCG CFU.
B cell depletion in mice.
C57BL/6 mice were infected with Mtb, and infection at 50 dpi, mice received B cell depleting antibodies (150 μg anti-mouse B220 (clone: RA3.3A1/6.1 (TIB-146), BioXCell) and 150 μg anti-mouse CD19 (clone: 1D3, BioXCell) or isotype control antibodies (150 μg IgG, BioXCell) via the intratracheal route every 3 – 4 days, over a 25 day period. At 75 dpi, mice were euthanized, and lungs harvested, homogenized and processed.
Aerosol Infection in mice.
Mtb strain HN878 was cultured and stocked in Proskauer-Beck with 0.05% tween 80 and stored at −80°C. Mice were aerosol infected with ~100 colony forming units (CFU), as described previously46. At specific time points post infection, lungs were harvested, and serial dilutions plated on 7H11 agar plates.
NHP infection.
All animal procedures were approved by the Institutional Animal Care and Use Committee of Tulane National Primate Research Center, Covington, Louisiana, USA, and were performed in accordance with NIH guidelines. Indian rhesus macaques of both sexes verified to be free of Mtb infection by tuberculin skin test were obtained from the Tulane National Primate Research Center. The animals were housed in an ABSL3 facility and were exposed to ~1000 CFU of Mtb CDC1551 via the aerosol route. The animals were periodically monitored for their physiological parameters and disease symptoms.
B cell depletion study in NHPs.
All macaques were similarly aerosol-exposed to ~1000 CFU of MtbΔsigH. This strain harbors a knockout in the transcription factor SigH, and aerosol vaccination with it protects against lethal Mtb challenge. Macaques were then challenged, nine-weeks post-vaccination, with ~1000 CFU of parental, wild-type Mtb CDC1551 via the aerosol route. Tissue samples were collected throughout the study and macaques were monitored for clinical signs of disease. To deplete B cells from macaques, we used 2B81R, a simianized CD20-depleting antibody, which was developed by the NIH Non-human Primate Reagents Resource (https://www.nhpreagents.org/) by fusing the FAB chain of clinically approved Rituximab to a rhesus FC chain. 2B81R was given intravenously to the treated animals at 45–50 mg/kg over the course of 45 minutes to monitor animals for possible development of anaphylactic shock. The antibody administration was first piloted in naïve animals and shown to deplete B cells in the periphery and tissue without induction of clinical signs of TB infection (e.g. C-reactive protein). The antibody was administered to the vaccinated animals one week prior to Mtb-infection (week eight), and then every three weeks subsequently. Control animals were given rhesus control IgG at the same dose at each time point treated animals were given 2B81R.
Generation of single-cell suspensions from lung tissues.
Lung single-cell suspensions from uninfected or Mtb-infected mice/macaque were isolated. Lungs were minced and incubated in Collagenase/DNAse for 30 minutes at 37°C. Lung tissue was pushed through a 70 μm nylon screen to obtain a single-cell suspension. Red blood cells were lysed and the cells were resuspended in suitable media or buffer for further use.
Adoptive transfer in mice.
All mice adoptive transfer studies were performed through the intravenous route. Naïve or Mtb-specific CD4+ T- and/or B cells were isolated from lymph nodes and spleen or lungs of mice via magnetic sorting. Single cell suspensions were prepared from lymph nodes and spleens by passing through 70 μm cell strainer (BD Biosciences) and for lungs as detailed above. The cells were subjected to MACS for isolating CD4+ T cells (Cat # 130–049-201; Miltenyi) and pan-B cells (Cat # 130–104-443; Miltenyi), as per manufacturer protocol. Sorting efficiency was analyzed to about 85–90% and ~1×107/cells in PBS was injected into recipient mice.
In vitro stimulation and transcriptomic analyses of sorted CD4+ T cells.
We reanalyzed RNA-seq datasets from a study that aimed to identify a signature for risk of TB from Mtb-infected participants of the South African Adolescent Cohort Study (ACS)47, 48. 1×106 live PBMC were stimulated with Early Secreted Antigenic Target 6kDa (ESAT-6) and Culture Filtrate Protein 10 (CFP-10). PBMCs, stimulated in media with 0.27 % DMSO, were used as negative controls. After 12 hours of incubation, T cells were purified from PBMC and were lysed in RNeasy RLT buffer (Qiagen). RNA was extracted from sorted cell subsets using RNeasy Plus Micro kit (Qiagen) and subjected to bulk RNA sequencing. For murine studies, data was obtained from a previously published study. Briefly, CD4+ T cells from Mtb-H37Rv infected lungs were isolated using magnetic CD4+ beads (clone GK1.5, Miltenyi Biotec), according to the manufacturer’s instructions. The cells were incubated in the presence or absence of ESAT-6 peptide (5 μg/ml).
Bulk RNA-sequencing
In vitro stimulations.
Both in vitro stimulated human and murine cells were processed for total RNA isolation using RNeasy kits (QIAGEN), RNA-sequenced and analyzed as described previously47, 48.
Mouse or macaque lung tissues.
As described by Ahmed et al5, RNA was extracted and sequencing libraries were generated. RNA-seq reads were aligned to the respective species genome assemblies49 using HISAT2 v2.1.050. FPKM (fragments per kilobase of gene length per million reads mapped) normalization was performed.
Single cell RNA library generation, sequencing, data processing and analyses
As described by Akter et al13 lung single cell suspensions from uninfected and Mtb-infected mice, at 50 and 100 dpi, were enriched for live cells using dead cell depletion kit (Miltenyi Biotec) and were subjected to droplet-based massive parallel single-cell RNA sequencing using Chromium Single Cell 5’ (v1) Reagent Kit in the BSL-3 laboratory as per manufacturer’s instructions (10x Genomics). Data processing was performed as described in Akter et al13.
Flow cytometry staining
Murine studies.
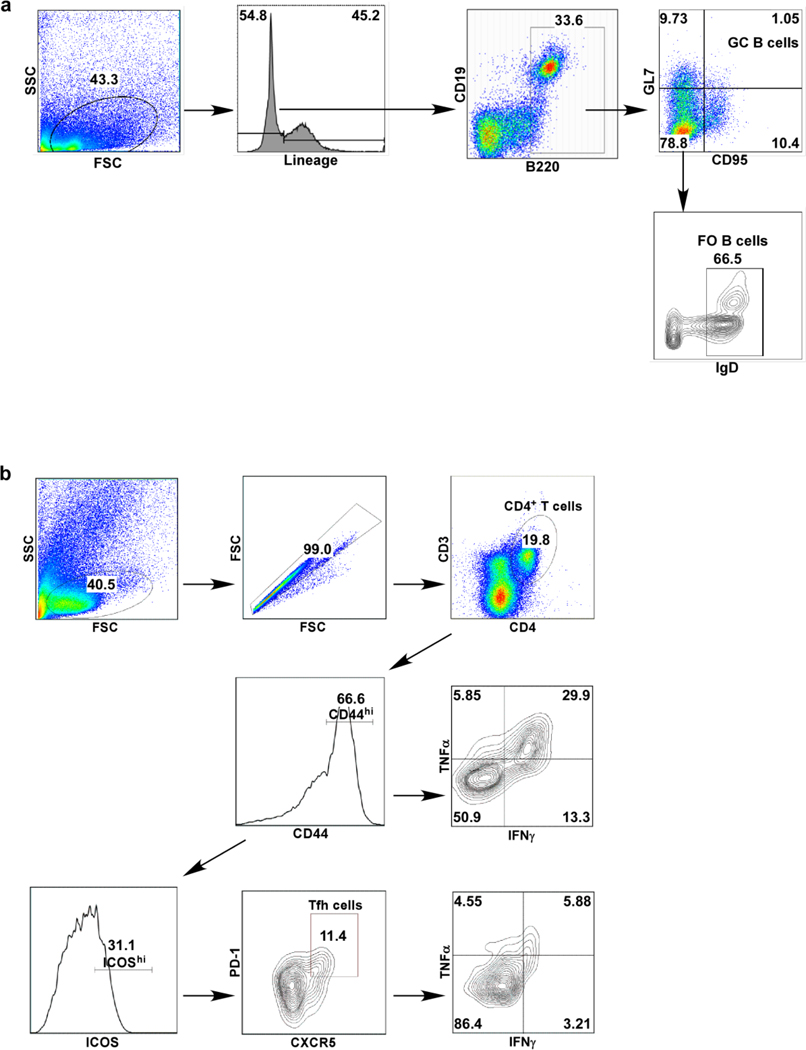
Lung single-cell suspensions were prepared from Mtb-infected mice, as described previously. For flow cytometric analysis, cells were either stained immediately, or stimulated with Phorbol myristate acetate (PMA-50 ng/ml) and ionomycin (750 ng/ml; Sigma Aldrich) in the presence of Golgistop (BD Pharmingen). The following fluorochrome conjugated antibodies were used for myeloid cell surface staining, CD11b APC (clone: M1/70, Tonbo Biosciences), CD11c PE-Cy7 (clone: HL3, BD Pharmingen), GR-1 PerCP-Cy5.5 (Clone: RB6–8C5, BD Pharmingen) and MHC class II FITC (Clone: M5/114.15.2, Tonbo Biosciences). CD19 PE-Cy5 (clone: eBio1D3, eBiosciences), B220 PE-Cy7 (clone: RA3–6B2, BD Pharmingen), GL7 FITC (clone: GL7, BD Pharmingen), CD95 PE (clone: 15A7, eBiosciences), CD138 BV605 (clone: 281–2, BioLegend), and IgD BV421 (clone: 11–26c.2a, BioLegend) were used for B-cell surface staining. CD3 AF700 (clone: 500A2, Biolegend), CD4 Pacific blue (clone: RM4.5, BD Pharmingen), CD44 PE-Cy7 (clone: IM7, Invitrogen), CD8 APC-Cy7 (clone: 53–6.7, BD Pharmingen), CXCR5 PerCP Cy5.5 (clone: 2G8, BD Pharmingen), PD1 PE (clone: J43, BD Pharmingen), and ICOS PE-Cy5 (clone: 7E.17G9, eBiosciences), were used for T cell surface staining. For intracellular staining, fixation/permeabilization concentrate and diluent (BD biosciences) were used to fix and permeabilize lung cells following manufacturer’s instructions. Intracellular staining with IFNγ APC (clone: XMG1.2, Tonbo Biosciences), TNFα FITC (Clone: MP6-XT22, BD Biosciences), IL-17A PE (clone: TC11–18H10, BD Pharmingen), Irf4 FITC (Clone: 3E4, eBiosciences) and Bcl6 PE (Clone: IG191E/A8, Biolegend), or the respective isotype control antibodies (APC rat IgG1κ, FITC rat IgG1κ and PE rat IgG1κ isotype, BD Pharmingen) was performed for 30 min at 4ºC. Samples were acquired on a BD LSRFortessa X-20 Cell Analyzer and the analysis was performed using FlowJo versions 7 and 10 (Treestar). Alveolar macrophages were gated on CD11c+CD11b−, neutrophils were defined as CD11b+CD11c−Gr-1hi cells, monocytes were defined as CD11b+CD11c−Gr-1med cells and recruited macrophages were defined as CD11b+CD11c−Gr-1low cells. GC B-cells as B220hiCD19+GL7+CD95+, and follicular (FO) B cells as B220hiCD19+GL7−CD95−IgD+, and total B cells: combination of all subsets (Extended Data Fig 9a). Activated T-cells were identified as CD3+CD4+CD44hi T-cells and showed expression and/or co-expression of IFNγ, TNFα, and IL-17 cytokines (Extended Data Fig 9b). Tfh-like cells were gated as CD3+CD4+CD44hi ICOShiCXCR5+PD1+ cells (Extended Data Fig 9b). Total numbers of cells within each gate were back calculated based on cell counts / individual lung samples.
NHP studies.
Flow cytometry was performed on tissue samples, including blood, lung, and individual granulomas, from animals throughout the study and at the time of euthanasia10. Where appropriate, single cells from tissue or granulomas were stimulated for 12 hours in the presence of brefeldin A with and without ESAT6/CFP10 peptide stimulation. All samples were stained with fixable live/dead stain (Fixable Viability Stain 520, BD Biosciences), incubated with surface antibodies for 20 minutes at room temperature, and subsequently fixed. For identification of Th1 and Tfh like cells, samples were stained with surface antibodies CD3 Alexa 700 (clone SP34–2, BD Biosciences), CD4 Brilliant violet 605 (clone L200, BD Biosciences), CD8 APC (clone RPA-T8, BD Biosciences), PD1 Brilliant violet 421 (clone EH12.2H7, Biolegend), and CXCR5 PE (clone MU5UBEE, eBioscience). Cells were permeabilized to detect intracellular cytokines with antibodies specific for IFNγ Brilliant violet 510 (clone B27, BD Biosciences), TNFα PE (cloneMAb11, BD Biosciences), IL-2 Brilliant violet 650 (clone MQ1–17H12, BD Biosciences), IL-17A Brilliant violet 711 (clone BL168, Biolegend). Samples were incubated for 2–3 hours at 4ºC. Samples were acquired using a BD LSR Fortessa and data was analyzed using FlowJo v. 7 and 10 software (Treestar).
Histological analyses
Visualization of IRF4 protein expression by CD4+ T cells in human TB granulomas.
Formalin fixed paraffin embedded (FFPE) lung lesions from six ATB patients, both male (4) and female (2), between 29–72 years of age, from the Tuberculosis Outpatient Clinic at the National Institute of Respiratory Diseases (INER) in Mexico City, before anti-Mtb treatment were obtained through informed consent that was approved by the institutional review board of INER. No compensation was provided to patients. Slides were stained with antibodies specific for anti-human CD3ε (Clone: CD3–12, BioRad), PD1 (Cat. No. 10377-MM23, Sino Biological) and IRF4 (Cat. No. LS-C344072, LifeSpan Biosciences). Total CD3+ T cells as well as CD3+PD1+IRF4+ T cells were counted in a blinded manner in the lung lesions.
NHP studies.
Immunofluorescence was performed on FFPE tissue with multiple slides per animal included for each analysis, as described earlier. B cell depletion was assessed by immune-histochemical staining with anti-CD20 (Clone: L26, Agilent Dako) and CD79a (Clone: HM57, BD Biosciences). Images were taken on a Zeiss FP6 confocal or light microscope where applicable. Data was analyzed using Imaris image software. For determination of IRF4 protein expression by T cells in NHPs, lung sections were stained with antibodies specific for CD20 (Clone: L-26, GeneTex), CD3ε (Clone: M-20, Santa Cruz Biotechnology) and IRF4 (Cat. No. LS-C344072, LifeSpan Biosciences). The total number of CD3+ T cells and CD3+IRF4+ T cells was quantified in the lung lesions of LTBI vs ATB NHPs. To determine IRF4 protein expression by Tfh-like cells in NHPs, lung sections were stained with antibodies specific for CD3ε, PD1, BCL6 (Clone: 1951R; Cat# GTX17716, GeneTex, Irvine, CA) and IRF4. Total CD3+PD1+ T cells and CD3+PD1+IRF4+ T cells were blindly enumerated in the lung lesions. To detect Tfh-like cells in B cell depleted macaques or control macaques, lung sections were stained with antibodies specific for CD3ε, IRF4, PD1 (Cat. No. 10377-MM23, Sino Biological), and BCL6.
Murine studies.
The right upper lobe was collected from mice for histological analysis of inflammation. The lobes were infused with 10% neutral buffered formalin and embedded in paraffin. 5 μm of FFPE lung sections were cut with a microtome, stained with hematoxylin and eosin (H&E) and processed for light microscopy. Images were captured using the automated Nanozoomer digital whole slide imaging system (Hamamatsu Photonics). Regions of inflammatory cell infiltration were delineated utilizing the NDP view2 software (Hamamatsu Photonics), and the percentage of inflammation was calculated by dividing the inflammatory area by the total area of individual lung lobes. All scoring was conducted in a blinded manner utilizing n = 5 to 10 mice per group. Lung sections immune-labelled with CD45R/B220 Biotin (Clone: RA3–6B2, BD Biosciences) and Streptavidin Alexa fluor 555 (Cat. No. S32355), CD3ε (Clone: 145–2C11, Santa Cruz) and goat anti-Armenian hamster IgG Alexa fluor 488 (Cat. No. ab173003, ABcam), and counterstained with DAPI were used to measure the lymphoid follicle area. A second set of lung sections were stained with a combination of antibodies against CD3ε (Clone M-20, Santa Cruz Biotechnology), PD-1 (Clone EPR20665, abcam) and CD45R/B220 (Clone RA3–6B2, BD Biopharmingen) to enumerate CD3+PD1+ Tfh-like cells inside B cells areas within GrALT. Enumeration of CD3+PD1+ Tfh-like cells inside B cells areas of Mtb infected lungs were carried out in mouse infected lungs which were stained with antibodies specific for CD3ε (red), PD1(green) and CD45R/B220 (white). First B cells areas were outlined with an automated tool of the Zeiss Axioplan microscope. Next, CD3+PD1+ Tfh-like cells and CD3+ T cells were enumerated inside discrete B cells areas. Finally, the frequency of CD3+PD1+ Tfh-like cells inside B cell areas was determined.
In situ hybridization.
FFPE lung sections were stained with Mtb 16S rRNA probe (cat # 439161; Advanced Cell Diagnostics, Newark, CA) via in situ hybridization (ISH) using the RNAscope 2.5HD Detection Kit (Brown staining) according to the manufacturer’s recommendations (Advanced Cell Diagnostics, Newark, CA) to provide visual evidence of the spatial location of Mtb. The representative pictures were captured at 40X with the Hamamatsu Nanozoomer 2.0 HT system with NDP scan image acquisition software.
Quantitation of the mycobacterial burden in areas of T and B cell infiltration.
FFPE lung sections of mice infected with Mtb were stained with antibodies specific for CD3ε (red), Mtb (green) and CD45R/B220 (white). 2–3 random 200X pictures were taken from areas with CD3+ T cells and CD45R/B220+ B cells in individual lung lobes. Area covered by Mtb signal was quantitated with NIH Image J software.
Collection of serum and ELISA
murine studies.
Blood was collected via cardiac exsanguination at time of death in BD microtainers (BD). MaxiSorp plates (Nunc) were coated with 100 μg Mtb HN878 CFP-10 (BEI Resources NR-14827) overnight, then blocked with 5 % non-fat dry milk in PBST, and incubated with mouse serum diluted 1:15 – 1:30. Detection antibodies (Goat anti-mouse HRP-conjugated-IgA, -IgG1, -IgG2a, -IgG2b, or -IgM; Southern Biotech), diluted 1:1000, were added to the plates, followed by BD OPTeia TMB substrate (BD). The reaction was stopped with 2N H2SO4, and read at 450 nm on Biotek μcount.
Antibody detection in NHP plasma.
Plasma samples were collected throughout the experiment and at necropsy. Microtitre plates, 96-well, (Corning, 2592) were coated with 100 μg Mtb CDC1551 CFP-10 (BEI Resources NR-14826) overnight, then blocked with 5 % non-fat dry milk in PBST, and incubated with NHP plasma samples diluted 1:15 – 1:240. Detection antibody (Rabbit anti-monkey peroxidase-conjugated IgG, Millipore-Sigma) diluted 1:5000) were added to the plates, followed by SureBlue TMB peroxidase substrate. The reaction was stopped by adding 2N H2SO4, and read at 450 nm.
Statistical analysis and reproducibility
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported before5. Data collection and assignment of animals to the different groups was randomized. For RNA-seq, p-values from Fisher’s exact test were adjusted by Benjamini-Hochberg multiple testing correction. DESeq2 (version 1.4.5)51 with default settings, and a minimum p-value significance threshold of 0.05 (after False Discovery Rate [FDR]52) correction for the number of tests was used for all RNA-Seq comparisons of differential expression. Differential expression for the human dataset was calculated using DESeq2 (version 1.24.0)51 with default settings and DESeq2-normalized read counts were used to calculate relative gene expression values (FPKM). For mouse and NHP studies, all data was analyzed using GraphPad Prism 5, 7 or 9 (La Jolla, CA), after testing for normality and using the indicated methodology in each figure legend, such as two-sided, parametric unpaired t-test (2 groups) or ANOVA with Tukey’s multiple distribution test (3 or more groups); or two-sided, non-parametric Mann-Whitney U-test (2 groups) or Kruskal-Wallis with Dunn’s multiple distribution test (3 or more groups). Outlier analysis was used to exclude any data points; except for Fig 7a where experimental validation was used for the exclusion purpose. All data collection analysis except for histological assessments were not performed blind to the conditions of the experiment. Survival analyses were performed using Mantel Cox test. Pearson correlation was used to analyze linear regressions. Significance is denoted on the figure, and analysis methodology indicated in the figure legend.
Extended Data
Extended Data Fig 1. Irf4 expression by CD19+ B cells is required for accumulation of FO B cells in the lung.
Irf4fl/fl, CD4creIrf4fl/fl, and CD19creIrf4fl/fl mice (n = 5–10 mice/group) were infected with Mtb HN878 and euthanized at 25 and 50 dpi. (a) MHC-IIhi AMs, (b) neutrophils, (c) total B cells, (d) GC B cells, and (e) FO B cells were determined by flow cytometry. Sera collected from mice (50 dpi; dilution shown 1:30) was analyzed via ELISA for levels of (f) IgG2a and IgG2b specific for Mtb antigen CFP-10. Data represent mean ± SD, analysis was performed using one-way ANOVA with Tukey’s multiple comparison test (a, c, d, f), and Kruskal-Wallis ANOVA with Dunn’s multiple comparison test (b, e). *, p≤0.05; **,p≤0.005; ***, p≤0.0005.
Extended Data Fig 2. Bcl6 expression by CD19+ B cells is required for optimal infiltration by B cell subsets in the Mtb-infected lung.
scRNA-seq of cells isolated from the lungs of C57BL/6 mice, either uninfected (n = 2 mice), or Mtb-infected at 50 dpi (n = 3 mice) or 100 dpi (n = 3 mice). (a) Swarm plot showing expression of Bcl6, a downstream target of Irf4, in CD4+, CD8+ T and B cells. Bcl6fl/fl, CD4creBcl6fl/fl, and CD19creBcl6fl/fl mice (n = 5–9 mice/group) were infected with Mtb HN878, euthanized at 50 and 100 dpi, and lungs analyzed by flow cytometry for (b) MHC-IIhi AMs, (c) neutrophils, (d) total B cells, (e) GC B cells, and (f) FO B cells. (g) IgG2a and IgG2b antibody specific against Mtb antigen CFP-10 in peripheral blood serum (diluted 1:30) collected at 50 dpi. (h) Tfh-like cells in the lungs of Bcl6fl/fl vs CD19creBcl6fl/fl mice. Data represent mean ± SD, analysis was performed using Kruskal-Wallis ANOVA with Dunn’s multiple comparison test (b), one-way ANOVA with Tukey’s multiple comparison test (c to g), and two-sided unpaired t-test (h). *, p≤0.05; **,p≤0.005; ***, p≤0.0005.
Extended Data Fig 3. Blimp1 deficiency in B cells impairs antibody production in mice.
iABfl/fl, and CD19creiABfl/fl mice (n = 6–8 mice/group) were infected with Mtb HN878, euthanized at 50 and 100 dpi, and lungs analyzed by flow cytometry for (a) Tfh-like cells. Blimp1fl/fl, and CD19creBlimp1fl/fl mice (n = 6–8 mice/group) were infected with Mtb HN878, euthanized at 50 and 100 dpi. Lungs analyzed by flow cytometry for (b) Tfh-like cells. (c) Immunoglobulin specificity against Mtb antigen CFP-10 in peripheral blood serum (diluted 1:30) collected at 50 dpi were detected by ELISA. Il10fl/fl, and CD19creIl10fl/fl mice (n = 6–8 mice/group) were infected with Mtb HN878 and euthanized at 50 and 100 dpi. (d) Tfh-like cells were enumerated by flow cytometry. Data represent mean ± SD and statistical analysis was performed with two-sided unpaired t-test. *, p≤0.05; **,p≤0.005; ***, p≤0.0005.
Extended Data Fig 4. IghelMD4 mice show reduced Mtb-specific antibody production.
B6 and IghelMD4 mice (n = 5–10 mice/group) were infected with Mtb HN878, euthanized at 50, 75 and 100 dpi and organs were collected and processed. (a) CD44hiCD4+ T cells, (b) IFNγ-producing CD4+ T cells, (c) TNFα-producing CD4+ T cells, (d) total B cells, (e) GC B cells, and (f) FO B cells were determined by flow cytometry. (g) Immunoglobulin specificity against Mtb antigen CFP-10 in peripheral blood serum (diluted 1:15) collected at 100 dpi. Data is mean ± SD, analysis was performed using two-sided unpaired t-test (a to f), and two-sided Mann-Whitney U-test (g). *, p≤0.05; **,p≤0.005.
Extended Data Fig 5. B cell depletion in Mtb-infected mice compromises the accumulation of Tfh-like cells in the lung.
C57BL/6 mice (n = 3–5 mice/group) were infected with Mtb, and infection established over 50 days. At 50 dpi, mice either received B cell depleting antibodies (α-B220 and α-CD19) or isotype via the intratracheal route every 3 – 4 days, over a 25 days period. Mice were euthanized at 75 dpi, and the lungs collected and processed. (a) Experimental scheme. Flow cytometry was used to enumerate (b) CD19+ B220hi B cells, and (c) immunoglobulin specificity against Mtb antigen CFP-10 in peripheral blood serum (diluted 1:30) was determined via ELISA. (d) Bacterial burden was determined in the lungs of Mtb-infected mice receiving either isotype or B cell-depleting antibodies. Formalin-fixed lung lobes were cut and stained to analyze (e) inflammation (average area) and (f) average area of GrALT. Flow cytometry was used to enumerate (g) activated CD4+ T cells, (h) activated CD4+ Tfh-like cells, (i) IL17+ CD4+ Tfh-like cells and (j) IFNγ+ CD4+ Tfh-like cells in the lungs. Data represents mean ± SD, and statistical analyses performed using two-sided unpaired t-test. *, p≤0.05; **,p≤0.005.
Extended Data Fig 6. Strategic positioning of PD1+ Tfh-like cells within GrALT structures mediates Mtb control.
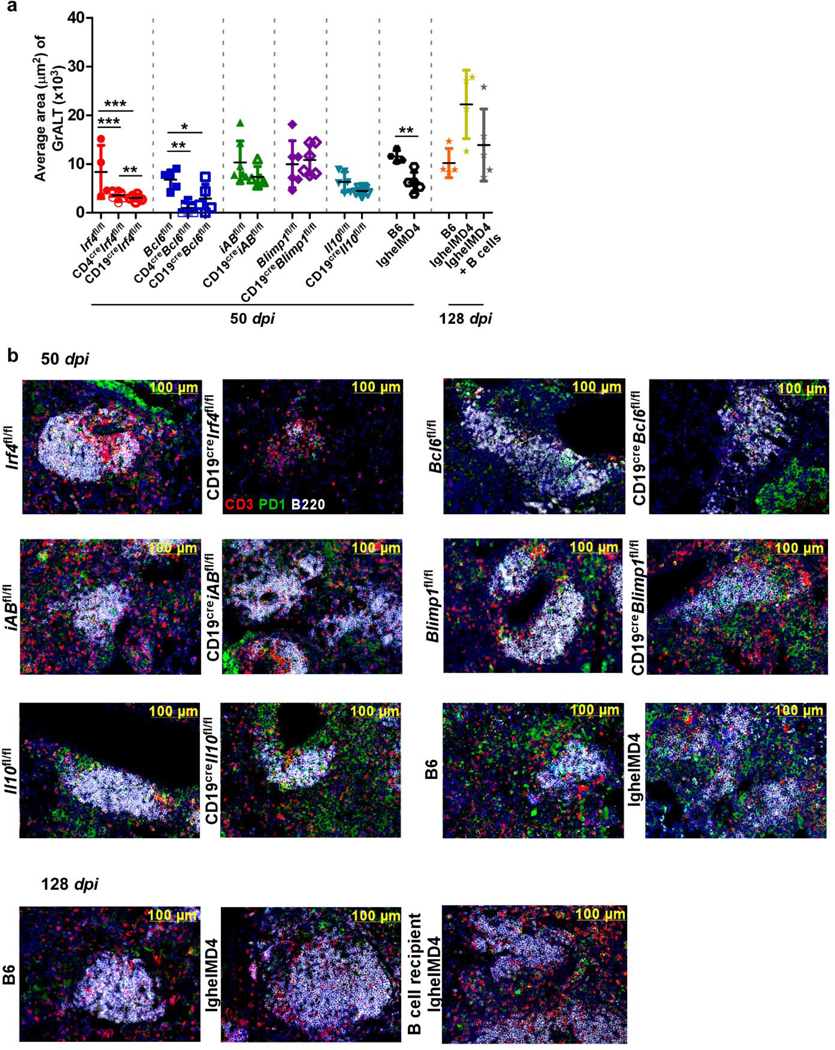
(a) Average area of GrALT collated from all mouse models (n = 3–10 mice/group) used in studies at 50 dpi and 128 dpi. (b) Representative images from FFPE lung sections stained with CD3 (red), PD-1 (green) and CD45R/B220 (white) in indicated models at 50 or 128 dpi. Data represent mean ± SD and statistical analysis was performed using one-way ANOVA with Tukey’s multiple comparison test (3 groups), and two-tailed unpaired t-test (2 groups). *, p≤0.05; **,p≤0.005.
Extended data Fig 7. Mtb 16sRNA is located outside GrALT areas and Pd1+ T cells are critical to control Mtb in mice.
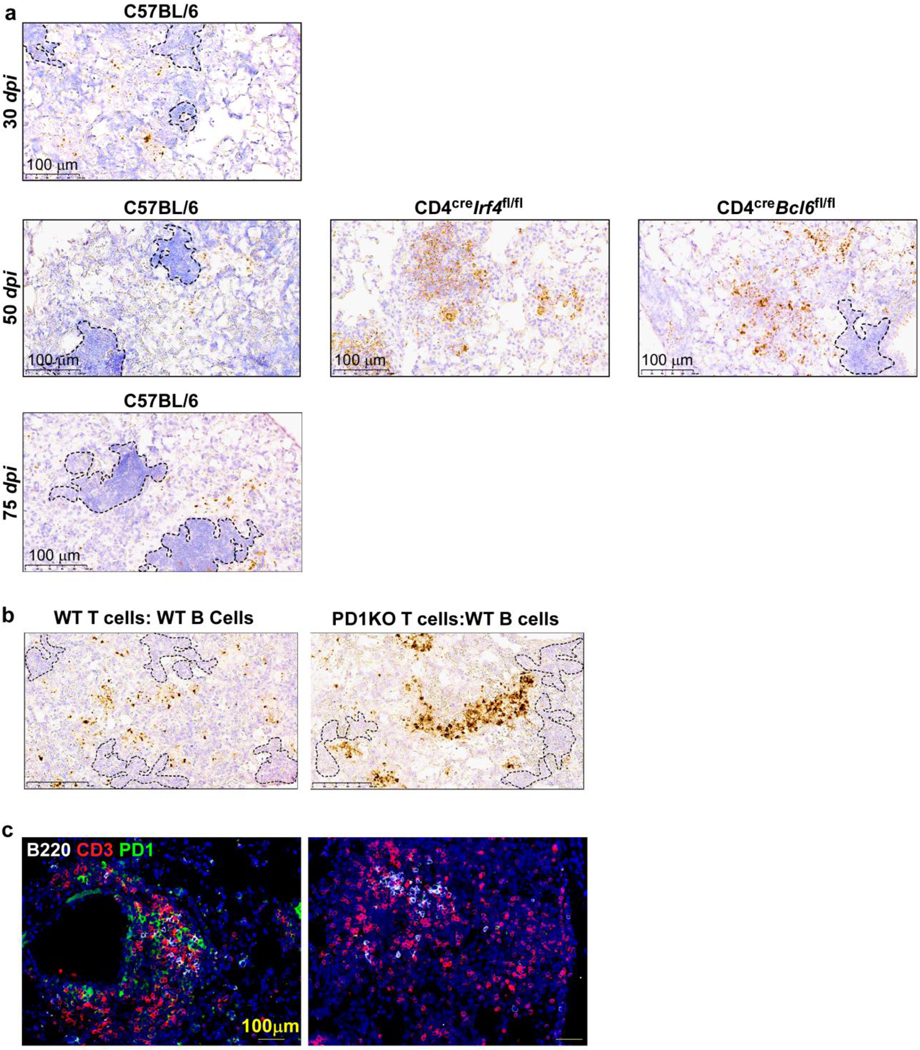
FFPE lungs sections from Mtb-infected B6 and gene deficient mice were stained for the presence of Mtb 16S rRNA via in situ hybridization (ISH) at (a) 30, 50, and 75 dpi. FFPE lungs sections from Rag1KO Mtb-infected mice that either received WT T cells:WT B cells or Pd1KO T cells:WT B cells, were stained for (b) the presence of Mtb 16S rRNA via in situ hybridization (ISH) at 30 dpi, (c) presence of GrALT via immunofluorescence; CD3 (red); B220 (white); PD-1 (green).
Extended data Fig 8. Compromised accumulation of Tfh-like cells in macaques and mice lungs dampens vaccine mediated protection against Mtb.
Macaques were aerosol exposed to MtbΔsigH prior to challenge with virulent Mtb CDC1551 and received either CD20 depleting (n = 4 NHPs) or IgG isotype control antibodies (n = 2 NHPs). Clinical samples were collected throughout the study and at necropsy. (a) FACS profiles for detection of B cells in PBMCs (left panel), BAL (middle panel), and lung (right panels) in CD20-depleted (upper panels) and control macaques (lower panels). (b) The percentage of total peripheral blood B cells (flow cytometry) and (c) Levels of C-reactive protein (CRP) were determined from samples collected throughout the study. (d) FACS profile for detection of T cell subsets in macaque lungs. (e) ESAT6+ CD4+ T cells producing IFNγ, IL2, IL17 and TNFα in lung tissue were enumerated by flow cytometry at the final end-point. (f) CFP-10 specific antibody levels in serum (diluted 1:15) collected at indicated time points. Data represent mean ± SD and statistical analysis was performed with two-sided Mann-Whitney U-test. ***, p≤0.0005. Bcl6fl/fl and CD4creBcl6fl/fl mice (n = 4 to 7 mice/group) were mucosally vaccinated with BCG and rested for thirty days following which mice were infected with Mtb HN878. Mice were euthanized at 20 dpi. (g) Experimental scheme. Lungs were collected and bacterial burden was determined in (h) Bcl6fl/fl and CD4creBcl6fl/fl mice. Data represent mean ± SD and statistical analysis was performed with two-sided unpaired t-test. **, p≤0.005.
Extended data Fig 9. Gating strategy for B cells and T cells flow-cytometric analysis.
Representative FACS plots with gates defining (a) total B cells, GC B cells, and FO B cells, and (b) CD4 T cells, cytokine producing Th1 cells, Tfh-like cells and cytokine producing Tfh-like cells.
Extended data Fig 10. Graphical abstract: Mtb specific B cells and Tfh-like cells interact to mediate protection against Mtb.
B cell effector functions including formation of GC B cells, IL-10 production and antibody/plasma cells have non-essential roles during in vivo Mtb infection. However, Mtb antigen-specific B cells interact with Tfh-like cells likely through PDL1-PD1 to drive differentiation and effective localization of cytokine-producing Tfh-like cells within GrALT to mediate Mtb control (left). Key transcription factors Irf4 and Bcl6 (blue text) are essential for differentiation of Tfh-like cells from naive CD4+ T cells. PD1-PDL1 (blue text) receptor-ligand axis likely drive Tfh-like cell localization and homing into GrALT but this interaction is independent of TCR-MHCII interaction, IL-10 production or antibody response (black text). Thus, Tfh-like cells and B cell interactions play critical roles in mediating Mtb control within GrALT.
Supplementary Material
Acknowledgements
This work was supported by Washington University in St. Louis; NIH grant HL105427, AI111914 to S.A.K.; AI123780 to S.A.K., D.K., T.S., and M.M.; AI134236 to S.A.K and D.K.; R01AI134240, R01AI138587, P51OD011133, P51OD011104, U42OD010442, S10OD028732 and C06OD030079 to D.K.; NIH grant AR069655 (Center for Musculoskeletal Research, University of Rochester); and the Department of Molecular Microbiology, Washington University in St. Louis. We thank Ms. Misty Veschak, Drs. Micah D. Dunlap and Shyamala Thirunavukkarasu of Washington University of St. Louis, Dept. of Molecular Microbiology, and Andrea Seeger and Lynn Reese of University of Rochester, Dept. of Surgical Pathology for technical support and assistance.
Footnotes
Competing interests.
The authors declare no completing interests
Data availability
Bulk RNA-seq data are available in the NCBI Gene Expression Omnibus with accession numbers GSE66578 and SRX960261–267. All macaque bulk RNA-Seq reads are available in NCBI Sequence Read Archive (SRA), Bioproject PRJNA523820. ScRNA-seq data for B6 mice is available GEO with accession number GSE200639.
REFERENCES
- 1.WHO. Global Tuberculosis Report (2020). [Google Scholar]
- 2.Scott NR, Swanson RV, Al-Hammadi N, Domingo-Gonzalez R, Rangel-Moreno J, Kriel BA, Bucsan AN, Das S, Ahmed M, Mehra S, Treerat P, Cruz-Lagunas A, Jimenez-Alvarez L, Muñoz-Torrico M, Bobadilla-Lozoya K, Vogl T, Walzl G, du Plessis N, Kaushal D, Scriba TJ, Zúñiga J, Khader SA S100A8/A9 regulates CD11b expression and neutrophil recruitment during chronic tuberculosis. J Clin Invest 130, 3098–3112 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slight SR, Rangel-Moreno J, Gopal R, Lin Y, Fallert Junecko BA, Mehra S, Selman M, Becerril-Villanueva E, Baquera-Heredia J, Pavon L, Kaushal D, Reinhart TA, Randall TD, Khader SA CXCR5⁺ T helper cells mediate protective immunity against tuberculosis. J. Clin. Invest. 123, 712–726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esaulova E, Das S, Singh DK, Choreño-Parra JA, Swain A, Arthur L, Rangel-Moreno J, Ahmed M, Singh B, Gupta A, Fernández-López LA, de la Luz Garcia-Hernandez M, Bucsan A, Moodley C, Mehra S, García-Latorre E, Zuniga J, Atkinson J, Kaushal D, Artyomov MN, Khader SA The immune landscape in tuberculosis reveals populations linked to disease and latency. Cell Host Microbe 29, 165–178 e168 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed M, Thirunavukkarasu S, Rosa BA, Thomas KA, Das S, Rangel-Moreno J, Lu L, Mehra S, Mbandi SK, Thackray LB, Diamond MS, Murphy KM, Means T, Martin J, Kaushal D, Scriba TJ, Mitreva M, Khader SA Immune correlates of tuberculosis disease and risk translate across species. Sci Transl Med 12, 528 eaay0233 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu LL, Chung AW, Rosebrock TR, Ghebremichael M, Yu WH, Grace PS, Schoen MK, Tafesse F, Martin C, Leung V, Mahan AE, Sips M, Kumar MP, Tedesco J, Robinson H, Tkachenko E, Draghi M, Freedberg KJ, Streeck H, Suscovich TJ, Lauffenburger DA, Restrepo BI, Day C, Fortune SM, Alter G. A. A Functional Role for Antibodies in Tuberculosis. Cell 167, 433–443 e414 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu LL, Das J, Grace PS, Fortune SM, Restrepo BI, Alter G Antibody Fc Glycosylation Discriminates Between Latent and Active Tuberculosis. J Infect Dis 222, 2093–2102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choreño-Parra JA, Bobba S, Rangel-Moreno J, Ahmed M, Mehra S, Rosa B, Martin J, Mitreva M, Kaushal D, Zúñiga J, Khader SA Mycobacterium tuberculosis HN878 Infection Induces Human-Like B-Cell Follicles in Mice. J Infect Dis 221, 1636–1646 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green AM, Difazio R & Flynn JL IFN-gamma from CD4 T cells is essential for host survival and enhances CD8 T cell function during Mycobacterium tuberculosis infection. J Immunol 190, 270–277 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaushal D, Foreman TW, Gautam US, Alvarez X, Adekambi T, Rangel-Moreno J, Golden NA, Johnson AM, Phillips BL, Ahsan MH, Russell-Lodrigue KE, Doyle LA, Roy CJ, Didier PJ, Blanchard JL, Rengarajan J, Lackner AA, Khader SA, Mehra S. Mucosal vaccination with attenuated Mycobacterium tuberculosis induces strong central memory responses and protects against tuberculosis. Nat Commun 6, 8533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mittrücker HW, Matsuyama T, Grossman A, Kündig TM, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi PS, Mak TW Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science 275, 540–543 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Edgar R, Domrachev M & Lash AE Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akter S, Chauhan KS, Dunlap MD, Choreño-Parra JA, Lu L, Esaulova E, Zúñiga J, Artyomov MN, Kaushal D, Khader SA Mycobacterium tuberculosis infection drives a type I IFN signature in lung lymphocytes. Cell Reports 39, 110983 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol 7, 773–782 (2006). [DOI] [PubMed] [Google Scholar]
- 15.De Silva NS, Simonetti G, Heise N & Klein U The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunological Reviews 247, 73–92 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Mason DY, Jones M & Goodnow CC Development and follicular localization of tolerant B lymphocytes in lysozyme/anti-lysozyme IgM/lgD transgenic mice. Internation Immunology 4, 163–175 (1992). [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Kuang X, Zhang Y, Ye Y, Li J, Liang L, Xie Z, Weng L, Guo J, Li H, Ma F, Chen X, Zhao S, Su J, Yang N, Fang F, Xie Y, Tao J, Zhang J, Chen M, Peng C, Sun L, Zhang X, Liu J, Han L, Xu X, Hung MC, Chen X ADORA1 Inhibition Promotes Tumor Immune Evasion by Regulating the ATF3-PD-L1 Axis. Cancer Cell 37, 324–339.e328 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Tan C, Hiwa R, Mueller JL, Vykunta V, Hibiya K, Noviski M, Huizar J, Brooks JF, Garcia J, Heyn C, Li Z, Marson A, Zikherman J NR4A nuclear receptors restrain B cell responses to antigen when second signals are absent or limiting. Nature immunology 21, 1267–1279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reif K, Ekland EH, Ohl L, Nakano H, Lipp M, Förster R, Cyster JG. Balanced responsiveness to chemoattractants from adjacent zones determines B-cell position. Nature. 416, 94–99 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Nam S & Lim JS. Essential role of interferon regulatory factor 4 (IRF4) in immune cell development. Arch Pharm Res 39, 1548–1555 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Man K, Gabriel SS, Liao Y, Gloury R, Preston S, Henstridge DC, Pellegrini M, Zehn D, Berberich-Siebelt F, Febbraio MA, Shi W, Kallies A. Transcription Factor IRF4 Promotes CD8(+) T Cell Exhaustion and Limits the Development of Memory-like T Cells during Chronic Infection. Immunity 47, 1129–1141.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Wu J, Zhang H, Shi X, Xiao X, Fan Y, Minze LJ, Wang J, Ghobrial RM, Xia J, Sciammas R, Li XC, Chen W. Ablation of Transcription Factor IRF4 Promotes Transplant Acceptance by Driving Allogenic CD4(+) T Cell Dysfunction. Immunity 47, 1114–1128.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayaraman P, Jacques MK, Zhu C, Steblenko KM, Stowell BL, Madi A, Anderson AC, Kuchroo VK, Behar SM. TIM3 Mediates T Cell Exhaustion during Mycobacterium tuberculosis Infection. Plos Pathog 12, e1005490 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, Pellegrini M, Belz GT, Smyth GK, Febbraio MA, Nutt SL, Kallies A. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol 14, 1155–1165 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Caruso AM, Serbina N, Klein E, Triebold K, Bloom BR, Flynn JL. Mice deficient in CD4 T cells have only transiently diminished levels of IFN-gamma, yet succumb to tuberculosis. J Immunol 162, 5407–16 (1999). [PubMed] [Google Scholar]
- 26.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 Are Reciprocal and Antagonistic Regulators of T Follicular Helper Cell Differentiation. Science 325, 1006–10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.du Plessis WJ, Keyser A, Walzl G & Loxton AG Phenotypic analysis of peripheral B cell populations during Mycobacterium tuberculosis infection and disease. J Inflamm (Lond) 13, 23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmermann N, Thormann V, Hu B, Köhler AB, Imai-Matsushima A, Locht C, Arnett E, Schlesinger LS, Zoller T, Schürmann M, Kaufmann SH, Wardemann H. Human isotype-dependent inhibitory antibody responses against Mycobacterium tuberculosis. EMBO Mol Med 8, 1325–1339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ordoñez C, Savage HP, Tarajia M, Rivera R, Weeks-Galindo C, Sambrano D, Riley L, Fernandez PL, Baumgarth N, Goodridge A. Both B-1a and B-1b cells exposed to Mycobacterium tuberculosis lipids differentiate into IgM antibody-secreting cells. Immunology 154, 613–623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao M, Valentini D, Poiret T, Dodoo E, Parida S, Zumla A, Brighenti S, Maeurer M. B in TB: B Cells as Mediators of Clinically Relevant Immune Responses in Tuberculosis. Clin Infect Dis 61Suppl 3, S225–234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allie SR, Bradley JE, Mudunuru U, Schultz MD, Graf BA, Lund FE, Randall TD. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat Immunol 20, 97–108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giles JR, Kashgarian M, Koni PA & Shlomchik MJ B Cell-Specific MHC Class II Deletion Reveals Multiple Nonredundant Roles for B Cell Antigen Presentation in Murine Lupus. J Immunol 195, 2571–2579 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez-Pinto D & Moreno J B cells can prime naive CD4+ T cells in vivo in the absence of other professional antigen-presenting cells in a CD154-CD40-dependent manner. Eur J Immunol 35, 1097–1105 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Redford PS, Murray PJ & O’Garra A The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal Immunol 4, 261–270 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Nus M, Sage AP, Lu Y, Masters L, Lam BYH, Newland S, Weller S, Tsiantoulas D, Raffort J, Marcus D, Finigan A, Kitt L, Figg N, Schirmbeck R, Kneilling M, Yeo GSH, Binder CJ, de la Pompa JL, Mallat Z. Marginal zone B cells control the response of follicular helper T cells to a high-cholesterol diet. Nature Medicine 23, 601–610 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Shi J, Hou S, Fang Q, Liu X, Liu X, Qi H. PD-1 Controls Follicular T Helper Cell Positioning and Function. Immunity 49, 264–274.e264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiley WW, Shafiani S, Wittmer ST, Tucker-Heard G, Moon JJ, Jenkins MK, Urdahl KB, Winslow GM, Woodland DL. Distinct functions of antigen-specific CD4 T cells during murine Mycobacterium tuberculosis infection. Proc of the Natl Acad Sci 107, 19408–19413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barber DL, Mayer-Barber KD, Feng CG, Sharpe AH & Sher A CD4 T Cells Promote Rather than Control Tuberculosis in the Absence of PD-1–Mediated Inhibition. J Immunol 186, 1598–607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kauffman KD, Sakai S, Lora NE, Namasivayam S, Baker PJ, Kamenyeva O, Foreman TW, Nelson CE, Oliveira-de-Souza D, Vinhaes CL, Yaniv Z, Lindestam Arleham CS, Sette A, Freeman GJ, Moore R; NIAID/DIR Tuberculosis Imaging Program; Sher A, Mayer-Barber KD, Andrade BB, Kabat J, Via LE, Barber DL. PD-1 blockade exacerbates Mycobacterium tuberculosis infection in rhesus macaques. Sci Immunol 6, eabf3861 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakai S, Kauffman KD, Sallin MA, Sharpe AH, Young HA, Ganusov VV, Barber DL. CD4 T Cell-Derived IFN-γ Plays a Minimal Role in Control of Pulmonary Mycobacterium tuberculosis Infection and Must Be Actively Repressed by PD-1 to Prevent Lethal Disease. PLoS Pathog 12, e1005667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das S, Marin ND, Esaulova E, Ahmed M, Swain A, Rosa BA, Mitreva M, Rangel-Moreno J, Netea MG, Barreiro LB, Divangahi M, Artyomov MN, Kaushal D, Khader SA. Lung Epithelial Signaling Mediates Early Vaccine-Induced CD4+ T Cell Activation and Mycobacterium tuberculosis Control. mBio 12, e01468–01421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed S, Raqib R, Guðmundsson GH, Bergman P, Agerberth B, Rekha RS. Host-Directed Therapy as a Novel Treatment Strategy to Overcome Tuberculosis: Targeting Immune Modulation. Antibiotics (Basel) 9, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phuah J, Wong EA, Gideon HP, Maiello P, Coleman MT, Hendricks MR, Ruden R, Cirrincione LR, Chan J, Lin PL, Flynn JL. Effects of B Cell Depletion on Early Mycobacterium tuberculosis Infection in Cynomolgus Macaques. Infect Immun 84, 1301–1311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deenick EK, Chan A, Ma CS, Gatto D, Schwartzberg PL, Brink R, Tangye SG. Follicular helper T cell differentiation requires continuous antigen presentation that is independent of unique B cell signaling. Immunity 33, 241–253 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References.
- 45.Haynes WA, Vallania F, Liu C, Bongen E, Tomczak A, Andres-Terrè M, Lofgren S, Tam A, Deisseroth CA, Li MD, Sweeney TE, Khatri P. Empowering Multi-Cohort Gene Expression Analysis to Increase Reproducibility. Pac Symp Biocomput. 22, 144–153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, Locksley RM, Haynes L, Randall TD, Cooper AM. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol 8, 369–377 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, Mahomed H, Erasmus M, Whatney W, Hussey GD, Abrahams D, Kafaar F, Hawkridge T, Verver S, Hughes EJ, Ota M, Sutherland J, Howe R, Dockrell HM, Boom WH, Thiel B, Ottenhoff THM, Mayanja-Kizza H, Crampin AC, Downing K, Hatherill M, Valvo J, Shankar S, Parida SK, Kaufmann SHE, Walzl G, Aderem A, Hanekom WA; ACS and GC6–74 cohort study groups. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. The Lancet 387, 2312–2322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahomed H, Hawkridge T, Verver S, Geiter L, Hatherill M, Abrahams DA, Ehrlich R, Hanekom WA, Hussey GD; SATVI Adolescent Study Team. Predictive factors for latent tuberculosis infection among adolescents in a high-burden area in South Africa. Int J Tuberc Lung Dis 15, 331–336 (2011). [PubMed] [Google Scholar]
- 49.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, To JK, Laird MR, Lavidas I, Liu Z, Loveland JE, Maurel T, McLaren W, Moore B, Mudge J, Murphy DN, Newman V, Nuhn M, Ogeh D, Ong CK, Parker A, Patricio M, Riat HS, Schuilenburg H, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Zadissa A, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Cunningham F, Yates A, Flicek P. Ensembl 2018. Nucleic Acids Res 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim D, Langmead B & Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anders S & Huber W Differential expression analysis for sequence count data. Genome Biol 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benjamini Y & Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 57, 289–300 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk RNA-seq data are available in the NCBI Gene Expression Omnibus with accession numbers GSE66578 and SRX960261–267. All macaque bulk RNA-Seq reads are available in NCBI Sequence Read Archive (SRA), Bioproject PRJNA523820. ScRNA-seq data for B6 mice is available GEO with accession number GSE200639.