Abstract
GTPases of the Ypt/Rab family play a key role in the regulation of vesicular transport. Their ability to cycle between the GTP- and the GDP-bound forms is thought to be crucial for their function. Conversion from the GTP- to the GDP-bound form is achieved by a weak endogenous GTPase activity, which can be stimulated by a GTPase-activating protein (GAP). Current models suggest that GTP hydrolysis and GAP activity are essential for vesicle fusion with the acceptor compartment or for timing membrane fusion. To test this idea, we inactivated the GTPase activity of Ypt1p by using the Q67L mutation, which targets a conserved residue that helps catalyze GTP hydrolysis in Ras. We demonstrate that the mutant Ypt1-Q67L protein is severely impaired in its ability to hydrolyze GTP both in the absence and in the presence of GAP and consequently is restricted mostly to the GTP-bound form. Surprisingly, a strain with ypt1-Q67L as the only YPT1 gene in the cell has no observable growth phenotypes at temperatures ranging from 14 to 37°C. In addition, these mutant cells exhibit normal rates of secretion and normal membrane morphology as determined by electron microscopy. Furthermore, the ypt1-Q67L allele does not exhibit dominant phenotypes in cell growth and secretion when overexpressed. Together, these results lead us to suggest that, contrary to current models for Ypt/Rab function, GTP hydrolysis is not essential either for Ypt1p-mediated vesicular transport or as a timer to turn off Ypt1p-mediated membrane fusion but only for recycling of Ypt1p between compartments. Finally, the ypt1-Q67L allele, like the wild type, is inhibited by dominant nucleotide-free YPT1 mutations. Such mutations are thought to exert their dominant phenotype by sequestration of the guanine nucleotide exchange factor (GNEF). These results suggest that the function of Ypt1p in vesicular transport requires not only the GTP-bound form of the protein but also the interaction of Ypt1p with its GNEF.
The movement of proteins through the secretory pathway involves their orderly progression through a series of membranous compartments (66). Transport between successive secretory compartments appears to be mediated by vesicles that bud from one compartment and fuse with the next (60, 77). Progress has been made in the past few years in our understanding of the vesicle machinery and the mechanisms regulating the directionality and specificity of vesicle targeting and fusion. Over the last 10 years, the Ypt/Rab family of small GTPases has been shown to play an important role in vesicular trafficking in both yeast and mammalian cells (22, 63, 109). It has been suggested that these proteins act at the different steps of the secretory pathway to ensure the fidelity of vesicular targeting (10, 49, 88, 90). However, the specific mechanism by which Ypt/Rab proteins regulate vesicular trafficking is still unknown.
The ability of Ypt/Rab proteins to cycle between GTP- and GDP-bound forms is thought to be crucial for their function (11, 28, 60, 67). Conversion from the GDP- to the GTP-bound form is achieved by nucleotide exchange, while the shift from the GTP- to the GDP-bound form is accomplished by the endogenous GTPase activity of these proteins. Most GTP-binding proteins have slow intrinsic rates of GTP hydrolysis and nucleotide exchange and thus require accessory factors to stimulate these reactions. Factors that stimulate guanine nucleotide exchange (guanine nucleotide exchange factor [GNEF]) (17, 18, 100, 102) and GTP hydrolysis (GTPase-activating protein [GAP]) (16, 17, 25, 40, 94, 95, 99, 108) have been identified for Rab proteins, but their role in vesicular transport is not clear. In addition, a protein that inhibits GDP dissociation (GDI) has also been identified as a Rab accessory factor. GDI is believed to be involved in recycling of Rab proteins, in their GDP-bound form, between membranes after each round of vesicle fusion (4, 91). Finally, GDI displacement factor (20) has been recently suggested to have a role in Ypt/Rab protein recruitment to the membrane.
The following hypothesis has been advanced to explain how guanine nucleotide exchange and hydrolysis regulate the function of Ypt/Rab proteins: (i) nucleotide exchange stimulated by GNEF is coupled to membrane localization of Rab proteins to the donor (or vesicle) compartment; and (ii) GTP hydrolysis, stimulated by GAP, is important for vesicle fusion with the acceptor compartment (28, 60). At present, there is little evidence for the second part of this hypothesis. A recent alternative suggestion for the role of GTP hydrolysis proposes that the GTPase activity is not required for Ypt/Rab-mediated membrane fusion but rather acts as a timer for this fusion (78). These two alternative models for the role of GTP hydrolysis have arisen from two lines of investigation: the cloning and disruption of GAP genes (see below), and the use of mutations in Ypt/Rab proteins that impair GTP hydrolysis (see Discussion). Our results do not support either of these views but rather suggest a different model in which the GTPase activity of Ypt/Rab proteins is not essential for membrane fusion or its timing but may be required for the recycling of these proteins between compartments.
If the GTPase activity of Ypt/Rab proteins is not crucial for their function, the GAP factors that regulate GTP hydrolysis are also not likely to be essential for Ypt/Rab function. While factors that regulate GTP hydrolysis for Ras and Rho have been identified and characterized (for a review, see reference 54), comparatively little is known about GAPs for Ypt/Rab GTPases. GAP activity was detected in mammalian and yeast cell extracts by using different Ypt/Rab proteins, including Ypt1p and Sec4p (16, 17, 40, 95). In the yeast Saccharomyces cerevisiae, Gyp6p and Gyp7p are GAPs that act on Ypt6p and Ypt7p, respectively (94, 99). Ypt6p is suggested to function in vacuolar protein sorting (94) or transport within the Golgi complex (48), and Ypt7p functions in endocytosis and homotypic vacuole fusion (53, 106). The GYP6 and GYP7 gene products do not have homology with each other or with other GAPs specific for Ras and Rho. Deletion or overexpression of GYP6 has no effect on cell growth. However, since neither YPT6 nor YPT7 is an essential gene, these results leave open the question of whether GAPs are necessary for the function of essential Ypt/Rab proteins.
In this work, we studied the role of GTP hydrolysis in the function of Ypt1p during vesicular transport. Ypt1p, a member of the Ypt/Rab family of GTPases, has an essential function in the regulation of endoplasmic reticulum (ER)-to-Golgi and intra-Golgi transport in the yeast secretory pathway (5, 39, 74, 86, 88). To determine the consequence of preventing GTP hydrolysis by Ypt1p, we created a Q67L mutation; the corresponding residue in Ras is important for GTP hydrolysis (19, 24, 45, 55, 65, 82). To study the effect of this mutation on intrinsic and stimulated GTP hydrolysis, we developed a GTP hydrolysis assay and partially characterized a GAP activity for Ypt1p (75). The ypt1-Q67L mutation causes a severe block in GTP hydrolysis. However, this mutation confers only a minor defect in secretion and cell growth, suggesting that GTP hydrolysis is not essential for Ypt1p function. In addition, using the ypt1-Q67L mutation, we found that being in the GTP-bound form is not sufficient for Ypt1p function in protein transport. We suggest that Ypt1p must interact with its GNEF even if it is loaded with GTP.
MATERIALS AND METHODS
Materials, strains, and plasmids.
All chemical reagents were purchased from Sigma Chemical Co. (St. Louis, Mo.) unless otherwise noted. All DNA restriction endonucleases were from New England BioLabs (Beverly, Mass.) or Boehringer Mannheim (Indianapolis, Ind.). Taq DNA polymerase was from Gibco BRL (Gaithersburg, Md.). The Escherichia coli bacterial strain SURE (Stratagene Inc., La Jolla, Calif.) was used for transformations. Bacteria were transformed by electroporation (8, 21).
Yeast strains used in this study were GPY60 (MATα ura3-52 trp1 leu2 his4 pep4::URA3), NSY125 (MATa, his4-539 lys2-801 ura3-52), NSY126 (MATα his4-539 ade2-ura3-52), NSY406 (see below for construction; MATa his4-539 lys2-801 ura3-52 ypt1-Q67L), DBY1803 (MATa ura3-52 his4-539 lys2-801 ypt1-T40K), NSY161 (MATα his4-539 ura3-52 ypt1-A136D), REE966 (MATa ade2 can1-110r his3-11,15 leu2-3,112 trp1-1 ura3-1). Yeast cells were grown either in YPD (1% yeast extract, 2% Bacto Peptone, 2% dextrose), SD (0.67% nitrogen base with appropriate nutritional supplements, 2% dextrose), or SRaf (0.67% nitrogen base with appropriate nutritional supplements, 2% raffinose) (76). Nutrients omitted for selection purposes are indicated. Yeasts were transformed by the lithium acetate method (27).
Plasmids used in this study were pNS364 (contains GAL1 and GAL10 promoters [41] cloned into pRS316 [89], a CEN URA3-marked vector), pNS326 (42) (contains YPT1 driven by the GAL10 promoter in pNS364), pNS330 (50) (contains ypt1-Q67L in pNS364), pNS327 (42) (YPT1-N121I in pNS364), pNS317 (42) (YPT1-D124N in pNS364), and pNS213 (contains EcoRI-to-BamHI fragment of ypt1-Q67L from pNS330 for integration of ypt1-Q67L allele [see below]).
Mutagenesis, expression, and purification of Ypt proteins.
The Ypt1-Q67L mutant was generated by site-directed mutagenesis as described previously (42). The sequence of the mutagenic oligonucleotide was 5′-TGG GAC ACT GCA GGT CTA GAA CGT TTC CGT ACT-3′. The cloning of wild-type and mutant Ypt1 proteins into pGEX-KT, expression in E. coli, and purification are described elsewhere (42).
GTP hydrolysis assays.
GTP hydrolysis was monitored by the charcoal binding assay (12, 33). Wild-type and Q67L Ypt1 proteins (10 μM) were preloaded with 5 μl of [γ-32P]GTP (2,000 Ci/mmol; Amersham Life Sciences, Arlington Heights, Ill.) in preload buffer (20 mM HEPES [pH 7.2], 20 mM potassium acetate, 5 mM EDTA, 0.5 mg of bovine serum albumin [BSA] per ml, 1 mM dithiothreitol [DTT]) for 15 min at 30°C in a 10-μl volume. Preload reactions were diluted with 50 μl of reaction buffer (20 mM HEPES [pH 7.2], 5 mM magnesium acetate, 300 mM sorbitol, 1 mM DTT) plus 0.5 mg of BSA per ml, and unbound nucleotide was removed at 4°C with two successive acrylamide spin columns (BioSpin6; Bio-Rad Laboratories, Hercules, Calif.) equilibrated with reaction buffer plus BSA. The volume of the flowthrough was adjusted to 250 μl with reaction buffer plus BSA to give a final Ypt1p concentration of 40 nM. GAP-stimulated GTP hydrolysis was measured by incubating 2 nM preloaded Ypt1p with a 5-mg/ml concentration of a P12 (12,000 × g pellet) subcellular fraction (75) in GTP hydrolysis assay buffer (reaction buffer plus 1 mM each GTP, GDP, and ATP) at 30°C. Intrinsic GTP hydrolysis was measured by substituting 5 mg of BSA (a nonspecific protein) per ml for the P12 fraction. Aliquots of 20 μl were removed at the indicated time points and added to 320 μl of ice-cold 5% NoritA activated charcoal in 50 mM NaH2PO4. Samples were vortexed and centrifuged at 1,400 × g for 10 min at 4°C. A 170-μl aliquot of the supernatant was removed and added to 4 ml of scintillation fluid (Ready Protein+; Beckman, Fullerton, Calif.). Samples were quantified in a Beckman scintillation counter.
Construction and analysis of strains.
The ypt1-Q67L mutant allele was integrated into the genomes of yeast strains NSY125 (S288C genetic background) and REE966 (W303 genetic background) at the YPT1 locus by gene replacement as described elsewhere (87). For this purpose, the EcoRI-to-BamHI fragment of ypt1-Q67L was subcloned into the integrating plasmid pRS406 (89) to create pNS213. pNS213 was linearized by partial digestion with NcoI and transformed into yeast. Transformants were selected on media lacking uracil. Recombinants that had lost the plasmid sequence including the URA3 marker were then selected by using 5-fluoroorotic acid (American Bioorganics, Inc., Niagara Falls, N.Y.), which selects against colonies that express the product of the URA3 gene. 5-Fluoroorotic acid-resistant colonies were screened for the presence of the ypt1-Q67L allele by PCR amplification of the YPT1 gene, using yeast genomic DNA as a template (prepared as described in reference 35), and digestion of the PCR product with XbaI, a restriction site created by the ypt1-Q67L mutation. The haploid strain containing the ypt1-Q67L allele was backcrossed twice with NSY126. Diploids were sporulated, and tetrads were dissected. Tetrads were diagnosed for the presence of the ypt1-Q67L allele by PCR amplification of genomic DNA and restriction digestion of the amplified product with XbaI as described above. Tetrads gave rise to four live spores that germinated and grew at similar rates. The chromosomal copy of the ypt1-Q67L allele was sequenced after backcrossing. Genomic DNA was prepared as described above and PCR amplified in two independent reactions with primers flanking the ypt1-Q67L open reading frame (NSOL1 [5′-GGG CCC GCA TGC GCA CCA GTT TTG AGG AGG-3′] and NSOL2 [5′-GGG CCC GGA TCC GAT AAG GAA GAA TG-3′]). The two PCR products were cut with both EcoRV and BamHI and subcloned into the vector pRS306 (89) in independent ligation reactions. DNA was prepared from six independent transformants, equivalent quantities of DNA from each were pooled, and the entire coding region of the ypt1-Q67L gene was sequenced.
The growth phenotype of the ypt1-Q67L strain (NSY406) was tested by spotting 10-fold serial dilutions onto YPD or SD plates. Cells were grown at 10, 14, 26, 30, and 37°C for up to 2 weeks. Plates were photographed when large colonies were visible in the wild-type control strain.
To test for dominance of the ypt1-Q67L allele, yeast strain NSY125 was transformed with pNS330, a CEN plasmid containing the URA3 gene and the ypt1-Q67L coding region under the control of the galactose-inducible GAL10 promoter. For controls, NSY125 was transformed with pNS364 (GAL1/10 vector control), pNS326 (wild-type YPT1), or pNS327 (containing the dominant negative YPT1-N121I allele) (42). Tenfold serial dilutions of cells were inoculated onto SD-Ura (repressing medium), SRaf-Ura (noninducing medium), or SRaf-Ura-plus-2% galactose (inducing medium) plates and grown at 14, 30, or 37°C.
In vivo orthophosphate labeling and isolation of nucleotide associated with Ypt1p.
Cells were grown to an optical density at 600 nm (OD600) of 0.5 to 1. Cells (37.5 OD600 units per immunoprecipitation) were harvested, resuspended to 5 OD600 units/ml in spheroplasting medium (YEP; 0.1% glucose, 1.4 M sorbitol, 50 mM KPi [pH 7.5], 50 mM 2-mercaptoethanol, zymolyase 100T [1 U/OD unit; Seikagaku America, Rockville, Md.]), and incubated at 30°C for 1 h with gentle rotation. Spheroplasts were pelleted, resuspended to 7.5 OD units/ml in low-phosphate medium (low-phosphate YEP [105], 0.1% glucose, 1 M sorbitol), and incubated at 30°C for 30 min. Cells were pelleted and resuspended with 250 μl of low-phosphate medium per 37.5 OD units; 0.5 μCi of [32P]orthophosphate (150 mCi/ml in H2O; Dupont NEN, Boston, Mass.) was added per 37.5 OD600 units of cells in screw-cap 1.5-ml microcentrifuge tubes. Cells were incubated with label for 1 h at 30°C with occasional agitation. Cells were pelleted at 12,000 × g for 1 min in a microcentrifuge and washed twice with 1 ml of low-phosphate medium. Labeled spheroplasts were lysed by the addition of 150 μl of 20 mM HEPES [pH 7.5] and 5 mM MgCl2 and vortexed 20 times with 3-s pulses at a setting of 6 on an S/P vortexer. A 10× stock of buffer 88 (1× buffer 88 is 20 mM HEPES [pH 6.8], 150 mM potassium acetate, 5 mM magnesium acetate, and 250 mM sorbitol [6]) and a 500× stock of protease inhibitor cocktail (1× protease inhibitor cocktail is 1 μg each of leupeptin, chymostatin, pepstatin, antipain, and aprotinin per ml [30]) were added to 1×. When Triton X-114 phase partitioning was performed (see below), an equal volume of 2% Triton X-114 in buffer 88 was added, samples were treated as described below, and aqueous and detergent phases were recovered separately. Ten percent Triton X-100 was added to a final concentration of 1%, and samples were incubated on ice for 15 min. Tubes were centrifuged for 10 min at top speed in a microcentrifuge. Supernatants were collected. A 150-μl mixture of 5 μl of anti-Ypt1p immunoglobulin G (88) or control preimmune serum, 7% protein A-Sepharose (Zymed, San Francisco, Calif.), 1% Triton X-100, and buffer 88 was added to each sample. Immunoprecipitation was carried out for 2 h at 4°C. Samples were pelleted and washed five times with buffer 88 plus 1% Triton X-100 and then once with buffer 88. To the protein A-Sepharose pellet, 20 μl of 50 mM Tris-HCl (pH 7.5)–40 mM EDTA–2% sodium dodecyl sulfate (SDS) was added. Samples were heated at 70°C for 5 min. The Sepharose beads were pelleted in a microcentrifuge, and 5 μl of supernatant was spotted onto polyethyleneimine (PEI)-cellulose thin-layer chromatography (TLC) plates (Aldrich Chemical Co., Milwaukee, Wis.). Plates were developed in water until the front rose 2 cm above the origin, dried, and then developed completely in 1 M LiCl (97). Radioactivity was quantified with a radioanalytic imager (AMBIS Systems QuantProbe 3.0; Ambis Inc., San Diego, Calif.).
Pulse-chase analysis.
For pulse-chase analysis, the secretory marker proteins carboxypeptidase Y (CPY) and invertase were assayed as previously described (39). Cells were grown overnight at 30°C in SD (2% dextrose) minus methionine and cysteine to an OD600 of between 0.5 and 1. For invertase induction, cells were harvested, washed twice, resuspended in SD (0.1% dextrose) minus methionine and cysteine, and grown for 1 h at 30°C. Cells were then incubated for 6 min in medium containing Tran35S-label (50 μCi/OD unit; ICN, Irvine, Calif.). Chase was initiated by the addition of excess unlabeled methionine and cysteine. Aliquots were removed at the indicated time points and added to ice-cold 10 mM sodium azide. Cells were washed and lysed with glass beads. CPY or invertase was immunoprecipitated from cell lysates and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on 8% gels (anti-CPY antibodies were from T. Stevens; anti-invertase antibodies were from C. Kaiser). Gels were soaked with Amplify (Amersham) to enhance the radioactive signal, dried, and exposed to film.
Electron microscopy.
Samples from wild-type (NSY125) and ypt1-Q67L (NSY406) cells were prepared for electron microscopy as described previously (44). In all cases, cells were grown to an OD600 of between 0.5 and 1 in YPD medium at 30°C. Thirty-two cell sections of the ypt1-Q67L strain and 20 sections of the wild-type strain were examined for phenotypes.
Immunofluorescence microscopy.
Immunofluorescence experiments were performed as previously described (72), with some modifications. Cells were grown to an OD600 of between 0.5 and 1, pelleted, resuspended in 0.1 M KPO4 (pH 6.5)–3.7% formaldehyde, and incubated at 26°C for 2 h with rotation for fixation. After being pelleted and washed three times in SP (1.2 M sorbitol, 0.1 M KPO4 [pH 7.5]), cells were resuspended in SP to a final concentration of 5 OD600/200 μl. Recombinant β-glucanase (20,000 U/ml) and 5 mM DTT were added for 1 h at 37°C to spheroplast cells. Spheroplasts were harvested and washed once with SP, and 20 μl was spotted onto coverslips precoated with 0.1% poly-l-lysine. Samples were washed and blocked with Tris-buffered saline (TBS)-BSA (10 mM Tris [pH 7.5], 100 mM NaCl, 2 mg of immunoglobulin G-free BSA per ml). For Ypt1p and Och1p staining, all washes and antibody incubations contained 1% octylglucoside. Primary antibody was added at the appropriate dilution (affinity-purified anti-Ypt1p antibodies, 1:400 [88]; anti-Sec7p serum, preabsorbed to fixed yeast cells, 1:500 [23]; anti-hemagglutinin [HA] antibodies, 1:1,000 [BAbCo, Richmond, Calif.]) and incubated for 1 h. Cells were then washed 10 times with TBS-BSA and incubated with secondary antibody at the appropriate dilution (rhodamine-conjugated anti-rabbit FAb, 1:150; fluorescein isothiocyanate-conjugated anti-mouse antibody, 1:200) for 1 h. After 10 washes with TBS-BSA, coverslips were mounted onto slides by using fluorescein isothiocyanate guard (Vectashield; Vector Laboratories, Burlingame, Calif.). Cells were visualized with a fluorescence microscope (model Axioskop; Carl Zeiss Inc., Thornwood, N.Y.).
Several Golgi markers were examined. Endogenous Sec7p was visualized with anti-Sec7p antibodies (23). Pmr1p, Och1p, and Kex2p were visualized as HA-tagged proteins expressed by plasmids pL161 (3), pOH (32), and pSN218 (59), respectively. The triple-HA-tagged Och1p and single HA-tagged Kex2p were expressed under the control of their own promoters on CEN URA3-marked plasmids. Single-HA-tagged Pmr1p was expressed on a 2μm URA3-marked vector under the control of its own promoter. For protein expression, NSY125 and NSY406 strains were transformed with the appropriate constructs or with the corresponding empty vector as a control.
Cell fractionation and Western blotting.
For detection of Pmr1 protein, NSY125 and NSY406 were transformed with a 2μm plasmid containing HA-tagged PMR1 (pL161 [3]). To determine the distribution of proteins in cell fractions, cells were grown to an OD600 of between 0.5 and 1, pelleted, washed in buffer 88, resuspended in buffer 88 with 1 mM phenylmethylsulfonyl fluoride, and lysed with acid-washed glass beads. Lysates were cleared of unbroken cells by centrifugation at 500 × g. Lysates were then spun at 100,000 × g to generate a supernatant (S100) and pellet (P100). Equivalent quantities of protein were loaded onto 11% gels and separated by SDS-PAGE. Samples were transferred to polyvinylidene difluoride nylon membranes (Immobilon; Millipore, Bedford, Mass.) at 90 V for 1 h at 4°C in 20 mM Tris–150 mM glycine–20% (vol/vol) methanol (15). Membranes were blocked in TBST (20 mM Tris-HCl [pH 7.5], 140 mM NaCl, 1% [vol/vol] Tween 20) plus 5% milk for 1 h at room temperature or overnight at 4°C. Blots were washed once with TBST for 15 min at room temperature and probed with anti-Ypt1p antibody (diluted 1:1,000 in TBST plus 1% milk) or anti-HA monoclonal antibody (diluted 1:1,000; BAbCo) for 1 h at 26°C or with affinity-purified anti-Mnn1p antibody (diluted 1:400; [29]) overnight at 4°C. After four 5-min washes with TBST, membranes were incubated for 1 h at 26°C with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5,000 in TBST plus 1% milk; Amersham) for detection of anti-Ypt1 and anti-Mnn1 antibodies or with horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:15,000; Cappel, Durham, N.C.) for detection of anti-HA antibody. Membranes were washed with TBST four times for 5 min each, developed for chemiluminescence as described by the manufacturer (Amersham), and exposed to film. Bands were quantified by densitometry (Molecular Dynamics apparatus).
Determination of the prenylation state of Ypt1.
Two methods were used to determine the prenylation state of Ypt1p: Triton X-114 phase partitioning and separation of species by electrophoresis on urea (4 to 8 M)-acrylamide (10 to 15%) gradient gels. Triton X-114 phase partitioning was carried out by the method of Bordier (9), with minor modifications. Briefly, 100 μl of a cell extract (prepared as described above for cell fractionation and Western blotting) was mixed with 100 μl of 2% Triton X-114 in buffer 88 and incubated at 30°C for 5 min. The mixture was layered on top of a cushion of 6% sucrose (wt/vol) and 0.06% Triton X-114 in buffer 88 in a 1.5-ml microcentrifuge tube. Samples were centrifuged at 1,000 × g for 5 min at room temperature. The aqueous phase (remaining above the cushion) was collected. The detergent phase moved through the cushion and was concentrated as a large droplet at the bottom of the tube. The droplet was collected by moving a pipette tip through the cushion and removing the entire droplet as well as some cushion material. The droplet was restored to the original sample volume of 200 μl by dilution with buffer 88. The presence of Ypt1p in total, aqueous, and detergent fractions was assessed by SDS-PAGE and Western blotting as described above.
As an alternative method for detection of prenylated Ypt1p, separation of proteins in cell extracts was performed by SDS-PAGE with gels containing a gradient of 4 to 8 M urea and 10 to 15% acrylamide (80). Proteins were then transferred to polyvinylidene difluoride nylon membranes and processed for Western blotting as described above.
In vitro transport.
In vitro ER-to-Golgi transport assays were carried out as described previously (30). Cell fractions were prepared from strains NSY125 and NSY406 as described previously (30), with minor modifications. An S12 fraction was prepared as described for preparation of an S3 (30) except that lysates were centrifuged at 12,000 × g instead of 3,000 × g. The S12 was substituted for the S3 in transport assays. Buffer 88 was used in place of transport buffer. Ypt1-D124N protein was purified as described previously (42). Briefly, Ypt1p-D124N was expressed in E. coli and purified as a glutathione S-transferase fusion protein on glutathione-agarose beads. The fusion protein on beads was cleaved with thrombin to elute purified Ypt1p-D124N. For inhibition experiments, Ypt1p-D124N was incubated with S12 for 10 min on ice prior to addition of the permeabilized yeast cell fraction.
RESULTS
Ypt1-Q67L mutant protein is restricted to the GTP-bound form in vitro and in vivo.
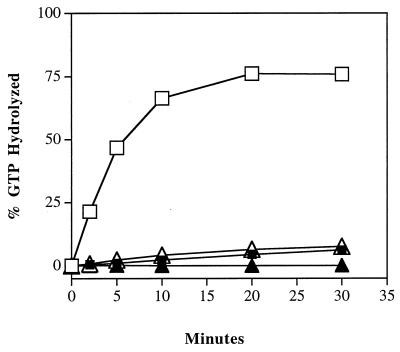
To determine the consequences of blocking hydrolysis of GTP on the ability of Ypt1p to mediate vesicular transport, we created a novel ypt1 mutation by site-directed mutagenesis. This mutation, ypt1-Q67L, is in a residue that is conserved among all Ras-like proteins and has been suggested to coordinate the water molecule that is required for GTP hydrolysis in Ras (19, 24, 45, 55, 65, 82). The analogous mutation was shown to generate proteins defective in the ability to hydrolyze GTP in many members of the Ras superfamily, including a number of Ypt/Rab proteins (14, 34, 93, 103). We tested whether the Ypt1-Q67L protein shows a defect in intrinsic and stimulated GTP hydrolysis. Stimulated GTP hydrolysis was tested in the presence of a yeast cell fraction containing a Ypt1p GAP activity (75). Intrinsic hydrolysis was determined by substituting the yeast cell fraction with an equivalent concentration of BSA, a nonspecific protein. For hydrolysis assays, Ypt1 proteins were expressed and purified from E. coli and preloaded with [γ-32P]GTP. GTP hydrolysis was measured by a charcoal binding assay (12, 33). Wild-type Ypt1p hydrolyzes GTP at a low intrinsic rate of 0.002 mol of GTP per mol of Ypt1p per min at 30°C, similar to the previously published rate of 0.006/mol/mol/min (101). In the presence of saturating amounts of Ypt1-GAP activity, this rate is increased 54-fold to 0.108 mol/mol/min. The Ypt1p-Q67L mutant, however, has an intrinsic rate of hydrolysis that is below the level of detection in this assay. Although this mutant is still responsive to the GAP activity, its stimulated rate of hydrolysis (0.004 mol/mol/min) reaches only a level similar to the intrinsic rate of the wild-type protein (Fig. 1). Therefore, like the analogous Ras mutant, Ypt1p-Q67L, is severely defective in GTP hydrolysis in vitro (about 25-fold less active than the wild type) and is expected to be in the GTP-bound form.
FIG. 1.
Ypt1p-Q67L is defective in intrinsic and GAP-stimulated GTP hydrolysis. GTP hydrolysis was monitored by the charcoal binding assay. Wild-type (squares) and Ypt1p-Q67L (triangles) proteins were preloaded with [γ-32P]GTP for 15 min at 30°C. Unbound nucleotide was removed with two successive acrylamide spin columns. GTP hydrolysis assays were performed by incubating 2 nM preloaded Ypt1p with a P12 subcellular fraction (5 mg/ml) prepared from GPY60 cells (GAP-stimulated hydrolysis; open symbols) or without the P12 fraction (intrinsic hydrolysis; closed symbols) at 30°C. Aliquots were removed at the indicated time points and added to ice-cold activated charcoal to stop the reaction. The charcoal was pelleted, and an aliquot of the supernatant was removed and quantified by scintillation counting. The counts measured at time zero were subtracted as background. GTP binding was slightly less efficient for Ypt1p-Q67L, but hydrolysis rates were normalized for the amount of Ypt1p bound to GTP. Data shown are typical of three independent experiments.
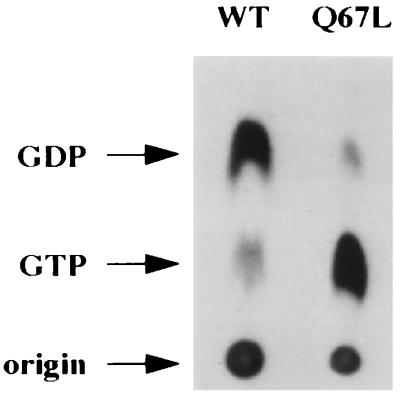
To verify that Ypt1p-Q67L is defective in GTP hydrolysis not only in vitro but also in vivo and is indeed in the GTP-bound form in the cell, we determined the nucleotide-bound state of the wild-type and Q67L Ypt1 proteins in vivo. We constructed a strain containing the mutant ypt1-Q67L allele as the only YPT1 gene (see below), and wild-type and ypt1-Q67L strains were tested by in vivo 32P labeling. Cells were grown to log phase, converted to spheroplasts, and labeled with [32P]orthophosphate. Spheroplasts were then lysed, and Ypt1p was immunoprecipitated. Nucleotides associated with Ypt1p were released by SDS treatment and resolved by PEI-cellulose TLC. Wild-type Ypt1p is primarily associated with GDP, exhibiting a GDP/GTP ratio of 72:28. By contrast, Ypt1p-Q67L is primarily associated with GTP, exhibiting a GDP/GTP ratio of 12:88. While most of the wild-type Ypt1p becomes prenylated in yeast cells, about 30% of the Ypt1p-Q67L was found to be unprenylated (see below). We wanted to verify that the prenylated functional form of Ypt1p-Q67L is predominantly bound to GTP. To examine the nucleotide-bound state of the prenylated pool of Ypt1p-Q67L, a Triton X-114 phase partitioning step was introduced after the in vivo orthophosphate labeling procedure. As seen in Fig. 2, the prenylated pool of Ypt1p-Q67L shows the same GDP/GTP ratio of 12:88 that is observed with the entire cellular pool of Ypt1p-Q67L protein. The GDP/GTP ratio for the prenylated pool of wild-type protein was 81:19, also similar to that in the whole-cell extract. Therefore, even the prenylated pool of Ypt1p-Q67L is predominantly in the GTP-bound state. Thus, the nucleotide state of the mutant protein in vivo is consistent with the observed hydrolysis defect in vitro. The small fraction of Ypt1p-Q67L which is bound to GDP in vivo could arise from residual GTPase activity of the mutant protein. Alternatively, it might represent newly synthesized protein which may bind GDP prior to the first round of nucleotide exchange (2, 85). Together, these results show that the mutant Ypt1p-Q67L is severely impaired in GTP hydrolysis in vitro, and the in vivo GTP/GDP ratio data confirm that hydrolysis by the mutant Ypt1p-Q67L is slower in intact cells.
FIG. 2.
Ypt1p-Q67L is predominantly bound to GTP in vivo. Wild-type (WT; NSY125) and ypt1-Q67L (NSY406) strains were grown to mid-logarithmic phase, spheroplasted, and labeled with [32P]orthophosphate for 1 h at 30°C. Spheroplasts were then lysed osmotically, and lysates were subjected to phase partitioning with 1% Triton X-114. Ypt1p was immunoprecipitated from the detergent phase with anti-Ypt1p antibodies for 2 h at 4°C. Associated nucleotides were released by heating in SDS and resolved by TLC on PEI-cellulose plates. Migration of nucleotides was determined by using unlabeled GDP and GTP standards which were visualized with UV light. Radiolabeled nucleotides were quantified by radioanalytic imaging. The results mentioned in the text are the averages from three independent experiments.
Ypt1-Q67L mutant protein functions efficiently in cell growth and secretion.
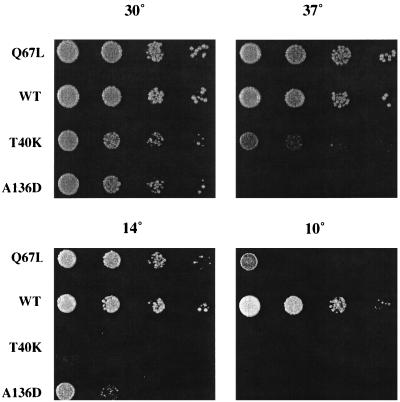
To assess the effects on cell growth and protein transport of substituting wild-type Ypt1p with the hydrolysis-impaired Ypt1p-Q67L mutant, the chromosomal copy of YPT1 was replaced by the ypt1-Q67L mutant allele. Serial dilutions were spotted onto YPD plates and grown at temperatures ranging from 10 to 37°C. Cells carrying the ypt1-Q67L allele as the only copy of the YPT1 gene do not exhibit growth defects at temperatures ranging from 14 to 37°C (Fig. 3). Only when cells are grown at 10°C does a growth defect become apparent. This partial growth defect at 10°C was less pronounced when cell growth was tested on minimal medium (data not shown).
FIG. 3.
ypt1-Q67L mutant cells grow normally at temperatures ranging between 14 and 37°C. Tenfold serial dilutions of the ypt1-Q67L strain (NSY406) were spotted onto YPD plates and grown at the indicated temperatures. Wild type (WT; NSY125), ypt1-T40K (DBY1803), and ypt1-A136D (NSY222) strains were spotted as controls. ypt1-T40K and ypt1-A136D strains are shown to demonstrate that other mutations in YPT1 that deplete Ypt1p function do cause a growth phenotype and that the conditions used in the experiment are effective.
The ability of cells to grow well with Ypt1p-Q67L was unexpected, especially since current models suggested a major role for GTP hydrolysis in Ypt/Rab GTPase function, either for membrane fusion or as a timer to turn it off. To rule out the possibility that a compensatory mutation arose in the ypt1-Q67L allele as an intragenic suppressor, the genomic copy of ypt1-Q67L was amplified by PCR in two independent reactions and sequenced. No intragenic reversion was detected. To rule out intergenic suppression, ypt1-Q67L was backcrossed twice and tetrads were analyzed. All four spores of tetrads were viable, with no differences in growth rates, at 30°C. To ensure that the lack of phenotype was not a strain-specific phenomenon, the ypt1-Q67L allele was integrated by gene replacement into two independent genetic backgrounds, S288C and W303, and identical growth phenotypes were observed (data not shown). Most importantly, as shown by the in vivo labeling (see above), close to 90% of the Ypt1p-Q67L mutant is bound to GTP. Together, these results demonstrate that although the Ypt1p-Q67L mutant is severely defective in GTP hydrolysis, it is capable of functioning in the cell to support growth.
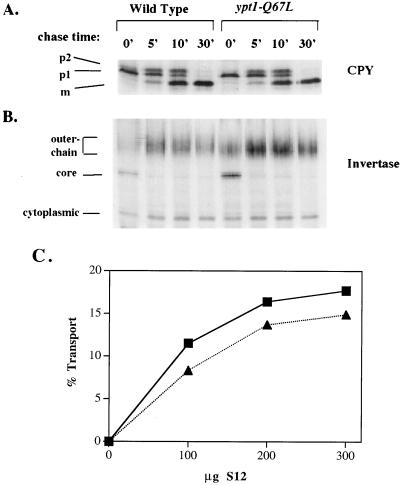
Ypt1p has been shown to mediate ER-to-Golgi transport (5, 74, 86, 88) as well as intra-Golgi transport (39). To determine if GTP hydrolysis by Ypt1p is important for protein transport, the secretory phenotype of the ypt1-Q67L mutant strain was examined. We used two markers: the slowly transported protein CPY, a vacuolar enzyme that traverses the secretory pathway in about 15 to 30 min, and the rapidly transported protein invertase, a secreted enzyme that reaches the periplasmic space in less than 5 min. The kinetics of CPY transport from the ER to the vacuole were determined by pulse-chase analysis (Fig. 4A). Cells were metabolically labeled at 30°C in mid-logarithmic phase of growth with Tran35S-label for 6 min. Chase was initiated by the addition of excess unlabeled methionine and cysteine. At the indicated time points, aliquots were removed, cells were lysed, CPY was immunoprecipitated, and the different modified forms of CPY were separated by SDS-PAGE. During the pulse, there is an accumulation of labeled CPY in its ER form (Fig. 4A, p1). During the chase this form is converted first to the Golgi form (Fig. 4A, p2) by glycosylation and then to the vacuolar form (Fig. 4A, m) by proteolysis, and by 30 min all of the labeled CPY has matured. The kinetics of CPY transport were very similar between wild-type and ypt1-Q67L strains. To detect possible subtle delays in transport in the mutant strain, we also examined invertase secretion. Wild-type and ypt1-Q67L strains were shifted to low-glucose medium at 30°C for 1 h at mid-logarithmic phase to induce the expression and secretion of invertase. Cells were then metabolically labeled, and the label was chased as described above. Aliquots were removed at the indicated time points, and invertase was immunoprecipitated. During the pulse, there is an accumulation of labeled invertase in both the ER from (core glycosylated) and the higher-molecular-weight Golgi form (outer chain mannosylated); within less than 5 min of chase, all of the invertase is converted to the Golgi form. As with CPY, the kinetics of invertase transport were very similar between wild-type and mutant cells (Fig. 4B).
FIG. 4.
Secretory kinetics are normal in ypt1-Q67L mutant cells. (A) In vivo transport of CPY. Wild-type (NSY125) and ypt1-Q67L mutant (NSY406) cells were grown at 30°C in minimal medium without methionine to mid-logarithmic phase. Cells were then pulse-labeled with Tran35S-label for 6 min at 30°C. Samples were chased with excess unlabeled methionine and cysteine for the indicated times (minutes). CPY was immunoprecipitated with anti-CPY antibodies, and modified forms were separated on SDS–8% polyacrylamide gels. p1, ER form; p2, Golgi form; m, mature vacuolar form. (B) In vivo transport of invertase. A pulse-chase experiment was performed as described above for CPY except that invertase was derepressed by switching to low-glucose (0.1%) medium for 1 h at 30°C, and immunoprecipitation was done with anti-invertase antibodies. The constitutive cytoplasmic form, the core ER form, and Golgi outer chain forms are indicated. Rates of transport were determined by quantification of the different forms of CPY and invertase and found to be the same in wild-type and mutant cells. (C) In vitro ER-to-Golgi transport. Transport of 35S-labeled pro-α-factor was assayed in a cell-free reaction (30). Cell fractions were prepared from wild-type (NSY125; squares) or ypt1-Q67L (NSY406; triangles) cells. Permeabilized yeast cells serve as the donor compartment and were incubated with the indicated quantities of S12, the supernatant of a 12,000 × g spin that contributes the acceptor compartment as well as necessary soluble components. Percent transport was calculated as the percentage of ER-modified (core-glycosylated, concanavalin A-precipitable) α-factor that acquired Golgi-specific modifications (anti-α-1,6-mannose-precipitable counts per minute) in 90 min at 20°C. Data shown are typical of four independent experiments.
An alternative assay for the secretory ability of cells is to use them as a source of material for a cell-free protein transport reaction. Some mutants exhibit more severe secretory defects in vitro than in vivo (5, 74). ypt1-Q67L mutant cells were used for the preparation of fractions for the ER-to-Golgi in vitro transport reaction, for which Ypt1p is essential (5, 6, 74, 86). Figure 4C shows that the Ypt1p-Q67L mutant drives this transport reaction almost as efficiently as wild-type Ypt1p. In the case of Rab5, the Q79L mutant protein (analogous to Ypt1-Q67L) stimulated endosome fusion in vitro (7, 34), but no stimulation of ER-to-Golgi transport was seen with Ypt1-Q67L. Together, these data demonstrate that the hydrolysis-impaired Ypt1p-Q67L is able to function efficiently in protein transport both in vivo and in vitro. The slightly lower rate of transport in vitro in the Ypt1p-Q67L-driven reaction (about 20%) is consistent with either a minor role of GTP hydrolysis in transport or a role in recycling of Ypt1p between membranes (see below).
A characteristic phenotype of sec mutants that disrupt protein trafficking is the accumulation of aberrant membranes of secretory compartments that precede the step in which the mutant proteins function (44, 61, 62). Alternatively, abnormally large organelles can arise from increased activity of a secretory regulator, as is the case with the Rab5-Q67L mutant protein (93). It was therefore of interest to examine whether ypt1-Q67L mutant cells accumulate any abnormal membrane structures. Cells from wild-type and ypt1-Q67L strains were fixed, stained, and sectioned for transmission electron microscopy. Micrographs revealed no obvious ultrastructural differences between wild-type and mutant cells (data not shown). Notably, there was no accumulation of small ER-to-Golgi vesicles and large Golgi-to-plasma membrane vesicles, as seen in secretion-defective ypt1 and sec4 mutants, respectively (38, 62, 79), nor was there an exaggeration of ER-like membranes as occurs in mutants blocked in exit from the ER (44, 62). Additionally, there was no occurrence of the Berkeley body structures that form in mutants blocked in exit from the Golgi (38, 61, 62). Thus, by the criterion of maintaining normal organelle structure, Ypt1p-Q67L appears to behave like the wild-type protein.
The Ypt1-Q67L mutant protein exhibits a partial defect in prenylation and localization.
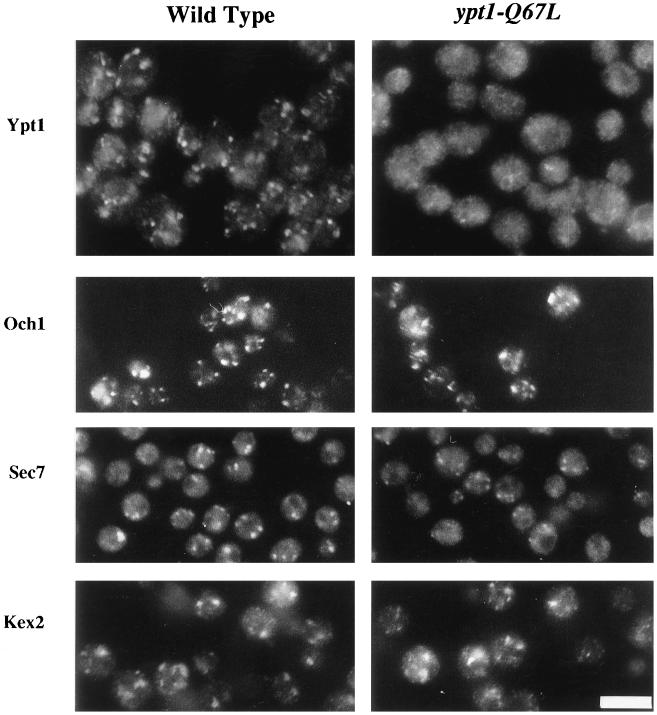
Ypt1p resides on intracellular membranes of the ER, Golgi, and small vesicles, as shown by immunofluorescence, electron microscopy, and cell fractionation studies (6, 56, 71, 88). To assess whether the bound nucleotide influences Ypt1p distribution in cells, we compared the localization of Ypt1p in wild-type and ypt1-Q67L strains. When viewed by indirect immunofluorescence, wild-type Ypt1p localizes to punctate structures throughout the cytoplasm, characteristic of Golgi staining (reference 88 and Fig. 5). The Ypt1p-Q67L mutant staining is much more diffuse, although occasional punctate staining is discernible (Fig. 5). To rule out the possibility that the diffuse staining pattern observed in the mutant strain is due to a disruption of Golgi structure, the localization of resident Golgi proteins was determined. Three markers were examined: HA-tagged Och1p (a cis-Golgi marker [32, 58]), Sec7p (a general Golgi marker [23]), and HA-tagged Kex2p (a trans-Golgi marker [73]). Each of these markers shows a punctate staining pattern that is similar in wild-type and mutant strains, indicating that Golgi structure is not grossly perturbed in the ypt1-Q67L strain. The difference in Ypt1p-Q67L localization is, therefore, not likely to be due to Golgi fragmentation but, as shown below, probably reflects a larger cytosolic pool of mutant protein.
FIG. 5.
In ypt1-Q67L mutant cells, the immunofluorescence staining pattern of Ypt1p-Q67L is abnormal but other Golgi markers are normal. Yeast cells from wild-type (NSY125) or ypt1-Q67L (NSY406) strains were fixed and stained for fluorescence microscopy with affinity-purified anti-Ypt1p antibodies (1:500) or anti-Sec7p antibodies (1:500) as indicated. For visualization of Och1p and Kex2p, the same strains were transformed with plasmids expressing HA-tagged Och1p or Kex2p as indicated, fixed, and stained with anti-HA antibodies (1:1,000 dilution). Bar, 10 μm.
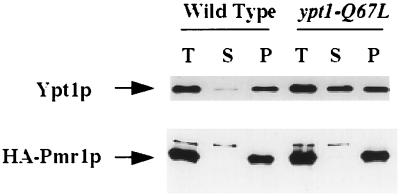
The localization of Ypt1p was verified by cell fractionation. Wild-type and ypt1-Q67L mutant cells were lysed, and cell lysates were centrifuged at 100,000 × g to generate supernatant (S100) and pellet (P100) fractions. Proteins in the cell fractions were resolved by SDS-PAGE, and Ypt1p was visualized by Western blot analysis with anti-Ypt1p antibodies. Ypt1p was found to be more abundant in the S100 fraction in ypt1-Q67L cells than in the wild-type cells (Fig. 6). Specifically, 50% of the total protein is found in the S100 fraction of ypt1-Q67L cells versus 5% in wild-type cells. The fractionation of Pmr1p and Mnn1p was examined to determine how resident Golgi proteins fractionate under these conditions. Pmr1p (Fig. 6 [3, 84]) and Mnn1p (data not shown [29]) were found exclusively in the P100 fraction. Therefore, the Ypt1p-Q67L found in the S100 fraction is likely to be there due to its residence in the cytosol. Together, the cell fractionation and the immunofluorescence results suggest that while most of the wild-type Ypt1p is bound to membranes, about half of the mutant Ypt1p-Q67L mislocalizes to the cytoplasm.
FIG. 6.
Mislocalization of mutant Ypt1p-Q67L to the cytosol as assessed by cell fractionation. Wild-type and ypt1-Q67L mutant cells were lysed with glass beads and centrifuged at 100,000 × g to generate supernatant (S) and pellet (P) fractions (T, total cell lysate). Proteins were resolved by SDS-PAGE, transferred to nylon membranes, and processed for Western blot analysis with affinity purified anti-Ypt1p antibodies (upper panel). For visualization of the Golgi marker Pmr1p, cells were transformed with a plasmid expressing HA-tagged Pmr1p. Cells were fractionated as above, and proteins were resolved by SDS-PAGE on 8% gels and processed for Western blot analysis with anti-HA antibodies (lower panel).
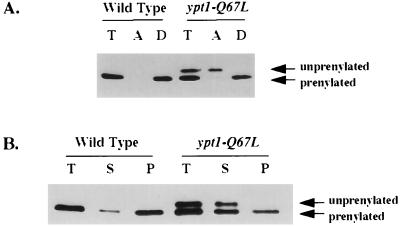
Ypt/Rab proteins require geranylgeranylation for function and for membrane association (56, 104). The machinery that transfers the geranylgeranyl group onto Ypt/Rab proteins has been well characterized. The GDP-bound form of Rab proteins is preferred over the GTP-bound form as a substrate for modification by a factor of 10- to 50-fold (81, 85). Because Ypt1p-Q67L is primarily in the GTP-bound form in vivo (see above), it was possible that ypt1-Q67L cells contain a higher proportion of unprenylated Ypt1p, thereby explaining the observed accumulation in the S100 fraction. Two methods were used to determine the prenylation state of Ypt1p: Triton X-114 phase partitioning (9) and electrophoretic separation on urea-acrylamide gradient gels (80). When cell fractions from the wild-type and ypt1-Q67L mutant cells were subjected to phase partitioning, more Ypt1p was found in the aqueous phase in the mutant strain (20 to 30%, versus 1 to 5% for the wild type) (Fig. 7A; compare lanes 2 and 5). Partitioning into the aqueous phase could occur either because of a prenylation defect or because the Ypt1p-Q67L is complexed with another protein that masks the prenyl group. Therefore, the prenylation state was verified by electrophoretic separation. On urea-acrylamide gradient gels, the prenylated form of Ypt1p migrates faster than the unprenylated form. The aqueous phase of the Triton X-114 extraction was examined with this gel system, and it was found to contain exclusively the lower-mobility form, while the detergent phase contained only the higher-mobility form (Fig. 7A; compare lanes 5 and 6). The unprenylated protein was found exclusively in the S100 fraction, not in the P100 membrane fraction, supporting the observation that prenylation is required for membrane association (Fig. 7B). A striking result evident in Fig. 7B is that in addition to the accumulation of unprenylated protein, there is an accumulation of prenylated Ypt1p-Q67L in the S100 fraction (the total amounts of prenylated Ypt1p in mutant and wild-type cells are similar). In contrast, in wild-type cells, when 81% of the Ypt1p is bound to GDP (see above), most of the prenylated Ypt1p is attached to the membrane. Therefore, the presence of some prenylated Ypt1p-Q67L in the cytosol suggests that the prenylated GTP-bound form is not the preferred form for association with the membrane. This result agrees with the estimate that about 50% of the mutant Ypt1p-Q67L mislocalizes to the cytoplasm, even though only about 25% of it is unprenylated. Together, these data show that Ypt1p-Q67L mutant has partial defects in both prenylation and localization to the membrane and that these two phenotypes are probably at least partially linked. However, it seems that these partial defects do not affect the function of the mutant Ypt1p in protein transport, perhaps because the concentration of Ypt1p in the cell is not limiting.
FIG. 7.
Mutant Ypt1p-Q67L is partially defective in prenylation. (A) A smaller fraction of the mutant Ypt1p-Q67L than of wild-type Ypt1p is prenylated, as determined by Triton X-114 phase partitioning and urea-acrylamide gradient gel electrophoresis. Wild-type (NSY125) and ypt1-Q67L mutant (NSY406) total cell lysates were subjected to phase partitioning with 1% Triton X-114. Total (T), aqueous (A), and detergent (D) phases were electrophoresed on 4 to 8 M urea–10 to 15% acrylamide gels and processed for Western blot analysis with anti-Ypt1p antibodies. Note that the aqueous phase contains all of the unprenylated form and the detergent phase contains all of the prenylated Ypt1p-Q67L. (B) Unprenylated and some prenylated mutant Ypt1p-Q67L is mislocalized to the cytoplasm (S100 fraction). Wild-type and ypt1-Q67L mutant cells were lysed with glass beads and centrifuged at 100,000 × g to generate supernatant (S) and pellet (P) fractions (T, total cell lysate). Proteins were resolved by SDS-PAGE on 4 to 8 M urea–10 to 15% acrylamide gradient gels, transferred to nylon membranes, and processed for Western blot analysis with affinity-purified anti-Ypt1p antibodies. The upper form of Ypt1p is unprenylated; the lower form is prenylated. Quantification indicates that there is the same amount of prenylated Ypt1p in wild-type and mutant strains. Data are typical of three independent experiments.
The ypt1-Q67L mutation does not exert dominant effects on cell growth and secretion.
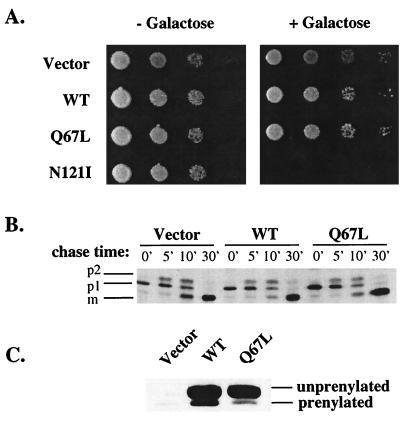
Overexpression of the GTP-bound, activated form of many Ras-like GTPases results in dominant phenotypes ranging from stimulated endocytic rates for Rab5 expression, to oncogenic transformation of cells by Ras, to rearrangements of the actin cytoskeleton by Rho-type GTPases (19, 47, 93). It was therefore of interest to determine the consequences of overexpression of Ypt1p-Q67L. For this purpose, a wild-type strain was transformed with the ypt1-Q67L mutation under the control of the inducible GAL10 promoter on a CEN plasmid. Empty vector, wild-type YPT1, and the dominant mutant YPT1-N121I (42, 83) were transformed as controls. Tenfold serial dilutions were spotted on plates with or without 2% galactose and grown at 30°C (Fig. 8A). Colony size and abundance were identical between cells expressing YPT1 or ypt1-Q67L and those transformed with the vector control, while expression of YPT1-N121I was lethal as reported previously (42, 83). Similar results were observed at both 37 and 14°C (data not shown). Therefore, overexpression of Ypt1p-Q67L does not seem to affect cell growth.
FIG. 8.
ypt1-Q67L is not dominant for growth or secretion when overexpressed. (A) Growth phenotypes. Wild-type YPT1 (WT; pNS326), ypt1-Q67L (pNS330), and YPT1-N121I (pNS327) were expressed from the galactose-inducible GAL10 promoter on a CEN URA-marked plasmid in the strain NSY125. Tenfold serial dilutions of cells were spotted onto SRaf-Ura or SRaf-Ura-plus-2% galactose plates and grown at 30°C. (B) CPY transport. The strains were grown overnight in SRaf-Ura minus methionine at 30°C to mid-logarithmic phase and then switched to inducing media (SRaf-Ura minus methionine plus 2% galactose) for 3 h at 30°C. Cells were harvested and pulse-labeled with Tran35S-label for 6 min at 30°C and chased for the indicated times (minutes). Cells were then lysed, and CPY was immunoprecipitated with anti-CPY antibodies and separated on SDS–8% polyacrylamide gels. p1, ER form; p2, Golgi form; m, mature vacuolar form. (C) Western blot analysis of Ypt1p expression. Cells used for the CPY assay were tested for Ypt1p expression. Ypt1p expression was induced with 2% galactose for 3 h at 30°C. Cells were lysed, and equivalent quantities of extract were run on 4 to 8 M urea–10 to 15% acrylamide SDS-containing gels. Proteins were transferred to nylon membranes and processed for Western blotting with anti-Ypt1p antibodies. The unprenylated and prenylated forms of Ypt1p are indicated.
As a more sensitive measure of Ypt1p function, the rate of secretion in cells overexpressing Ypt1p-Q67L or wild-type Ypt1p was examined. CPY was used as a marker, and its transport in cells containing the empty vector was examined as a control. Using the relatively slowly transported CPY marker should allow us to detect acceleration, as well as delay, in protein transport. Cells were grown to mid-logarithmic phase in medium without galactose. They were then shifted to medium with 2% galactose for 3 h to induce the expression of Ypt1p. Pulse-chase analysis was performed as described above but in medium containing 2% galactose. No difference in the rate of CPY transport was observed between control cells and those overexpressing wild-type Ypt1p or Ypt1p-Q67L (Fig. 8B).
To confirm that Ypt1p is indeed overexpressed in these cells after 3 h of induction, Western blot analysis was performed. Cells were lysed with glass beads, and extracts normalized for cell breakage were loaded on a urea-acrylamide gradient gel. This gel system enables the separation of prenylated and unprenylated forms of Ypt1p (see above). In control cells, just the prenylated form of Ypt1p is visible. In cells overexpressing wild-type Ypt1p or Ypt1p-Q67L, two bands, a highly abundant form that migrates at the same size as the bacterially expressed unprenylated protein and a less abundant prenylated form, are evident. The overexpressed wild-type prenylated protein is 40-fold more abundant than the endogenous Ypt1p. The prenylated Ypt1p-Q67L is 10-fold more abundant than the endogenous protein (Fig. 8C). Therefore, a 10-fold overexpression of prenylated Ypt1p-Q67L does not have a dominant effect on cell growth or the rate of protein transport.
Interaction with GNEF is important even for the Ypt1p-Q67L GTP-bound form.
As shown above, Ypt1-Q67L is primarily in the GTP-bound form. One question that we asked is whether Ypt1p-Q67L requires the activity of the GNEF for its function. In the case of Ras, a hydrolysis-defective mutant, Ras2-G19V, bypasses the requirement for the GNEF (encoded by the CDC25 gene) (13). In addition, mutants of H-Ras that have lost the ability to bind GNEF can be rendered functional with a second mutation, G12V, that reduces GTP hydrolysis (57). It has been hypothesized from this result that this G12V Ras mutant is able to bind GTP at some low rate without the exchange factor and that with impaired GTP hydrolysis activity, there is a sufficient quantity of GTP-bound Ras to sustain function.
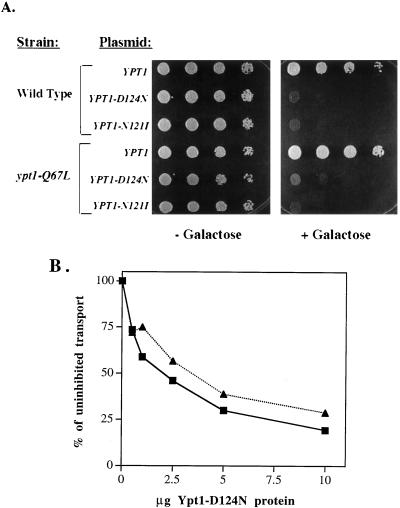
We were interested in determining whether Ypt1p-Q67L requires GNEF function. For the Ras experiment, GNEF activity was blocked by deletion of the CDC25 gene (13). Because the GNEF for Ypt1 has not been identified, we used an alternative strategy. We have previously demonstrated that the D124N and N121I Ypt1p mutant proteins block nucleotide exchange on wild-type Ypt1p (42). In vivo, these mutants exhibit a dominant lethal phenotype. Addition of these mutant proteins to an otherwise wild-type in vitro ER-to-Golgi transport assay blocked vesicle fusion. We tested whether these inhibitors of the Ypt1p GNEF would block Ypt1p-Q67L function both in vivo and in vitro. For in vivo analysis, YPT1-D124N and YPT1-N121I were expressed under the control of the inducible GAL10 promoter in wild-type and ypt1-Q67L strains. When cells were grown in the absence of galactose (noninducing conditions), no growth defect was observed. However, when cells were grown on galactose-containing medium (inducing conditions), lethality correlated with expression of the dominant mutants in both the wild-type and ypt1-Q67L strains (Fig. 9A).
FIG. 9.
Mutant Ypt1p-Q67L requires GNEF for function both in vivo and in vitro. (A) ypt1-Q67L cell growth is inhibited by expression of the dominant nucleotide-free alleles of YPT1. Wild-type (NSY125) and ypt1-Q67L (NSY406) cells were transformed with plasmids carrying the wild-type YPT1 gene (pNS326), YPT1-D124N (pNS317), or YPT1-N121I (pNS327) under the inducible GAL10 promoter. Yeast strains carrying these plasmids express Ypt1p when grown on medium containing galactose. Tenfold serial dilutions were spotted on SRaf-Ura medium without galactose (− Galactose) or with 2% galactose (+ Galactose). (B) In vitro transport in a ypt1-Q67L mutant cell reaction is inhibited by the nucleotide-free Ypt1p-D124N. In vitro transport cellular fractions from wild-type (squares) and ypt1-Q67L mutant (triangles) cells were prepared, and the reactions were performed as for Fig. 4C. Reactions were supplemented with the indicated quantities of purified mutant Ypt1p-D124N. Transport is expressed as percentage of uninhibited transport (reactions with no Ypt1p-D124N). Data shown are typical of four independent experiments.
It was difficult to confirm that the mutant Ypt1p-Q67L has bound GTP when GNEF is inhibited in vivo, since overexpression of the dominant negative mutant proteins interferes with the loading of Ypt1p with labeled GTP (data not shown). However, we could determine the effect of these dominant proteins in vitro. For these experiments, we used fractions prepared from wild-type and ypt1-Q67L mutant cells that do not express the dominant negative proteins and thus contain approximately 19 and 88% prenylated GTP-bound Ypt1p, respectively (see above). Purified Ypt1p-D124N, a dominant negative inhibitor of Ypt1p GNEF, was added to the ER-to-Golgi transport reactions. Ypt1p-D124N blocked transport in the ypt1-Q67L reaction with the same dose dependence and degree of inhibition as in the wild-type reaction (Fig. 9B). As with Ras and elongation factor Tu (EF-Tu) (31, 36, 37, 46, 69, 70), the nucleotide-free mutant Ypt1 proteins do not block the other known regulators of Ypt1p, GAP and GDI (43). However, it is a formal possibility that the dominant mutant proteins affect other, yet unknown aspects of Ypt/Rab function, although there is no precedent for this from analogous mutations in other GTPases as unrelated as Ras and EF-Tu. These results suggest that the presence of GTP-bound wild-type or Q67L mutant Ypt1p is necessary, but that Ypt1p function also requires interaction with Ypt1-GNEF. The proof awaits the identification of the gene encoding Ypt1-GNEF to test that the expected lethality of its deletion cannot be suppressed by the ypt1-Q67L mutation.
DISCUSSION
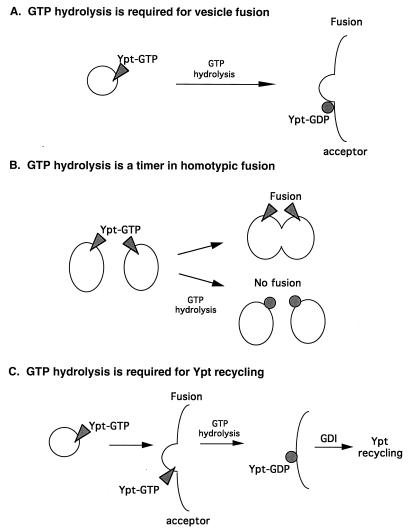
One current view in the field assigns a crucial role to GTP hydrolysis in Ypt/Rab-mediated vesicular transport (1, 28, 60, 63) (Fig. 10A). This view, which stems from a comparison with the role that EF-Tu plays in protein translation, suggests that both the GDP- and the GTP-bound forms of Ypt/Rab proteins are important for their function (10). Support for this idea comes from the study of GTPase-defective mutants of Ypt/Rab proteins. First, the sec4-Q79L allele, which encodes a protein partially defective in GTP hydrolysis, confers partial cold sensitivity for growth and a minor secretory defect at the nonpermissive temperature (103). However, since both the biochemical and the physiological effects of the Q79L mutation on Sec4p are mild, a mutation with a more severe biochemical defect is needed to test this hypothesis. Second, overexpression of two Rab proteins result in inhibition of protein transport: Rab2-Q65L potently inhibits ER-to-Golgi transport (although Rab1-Q67L does not) (96); Rab6, wild type or Q72L, reduces the rate of transport between the cis/medial Golgi and the late Golgi (52) and causes redistribution of Golgi resident proteins into the ER (51). This overexpression approach leaves unresolved the question of whether the endogenous Ypt/Rab protein could function when mutated. A second model suggests that GTP hydrolysis has a role as a timer that turns off Ypt/Rab-mediated membrane fusion (78) (Fig. 10B). This model suggests that GTP hydrolysis is required not for membrane fusion but rather for its prevention (see below).
FIG. 10.
Three models for the role of GTP hydrolysis in Ypt/Rab-mediated vesicular transport. (A) GTP hydrolysis is required for vesicle-membrane fusion (28, 60). (B) GTP hydrolysis is required to turn off Ypt/Rab-mediated homotypic membrane fusion (78). (C) GTP hydrolysis is required for Yptp recycling between membranes (this report). In this model, GTP hydrolysis is not required either for Yptp-mediated membrane fusion or for turning off Yptp function in heterotypic membrane fusion but rather is required for Yptp recycling between membranes.
In this report, we suggest that GTP hydrolysis does not play an important role in Ypt1p-mediated vesicular transport in vivo or in vitro. While Ypt1p function is essential for ER-to-Golgi transport (5, 6, 39, 74, 86, 88), cells expressing only the mutant Ypt1p-Q67L, which is severely impaired in its intrinsic and stimulated GTP hydrolysis, exhibit only a minor defect in cell growth. In addition, Ypt1p-Q67L is almost as efficient as the wild-type protein at supporting vesicular transport in vitro. We calculated that while it takes a wild-type Ypt1p molecule about 9 min in vitro to hydrolyze one molecule of GTP in the presence of saturating amounts of GAP, Ypt1p-Q67L needs about 4.2 h for this reaction. Therefore, because protein transport from the ER to the plasma membrane takes only 5 to 15 min in yeast cells, Ypt1p-Q67L probably exerts its function without the need for GTP hydrolysis. The possibility remains that, since Ypt1p levels are probably not limiting, the residual GTP hydrolysis of Ypt1p-Q67L may support efficient vesicular transport. However, support for the argument that GTP hydrolysis is not required comes from a recent report in which it was shown that Rab5 protein can support endosome fusion in vitro in the presence of a slowly hydrolyzable nucleotide analog, XTPγS (78). In that report, it was suggested that the GTPase activity of Rab5 serves as a timer, regulating the kinetics of membrane docking and fusion (78). Consistent with this hypothesis are the observations that overexpression of the wild-type or Q79L allele of rab5 causes the accumulation of large early endosomes and increased rates of transferrin internalization in vivo (93) and that addition of purified wild-type Rab5 or Rab5-Q79L protein stimulates endosome fusion in vitro (7, 34). Furthermore, depletion of the TSC2 gene product, which encodes a Rab5 GAP, results in increased rates of fluid phase endocytosis (108). In contrast to these results, our in vivo study shows that GTP-bound Ypt1p-Q67L has no effect on secretion even when overexpressed; and results with the Q67L analogous mutations of Sec4p, Rab2, and Rab6 show inhibition of protein transport (see above). The different effects of the GTP hydrolysis defective mutants in Rab5 versus Ypt1p, Sec4p, Rab2, and Rab6 may arise from a difference between the regulation of homotypic and heterotypic membrane fusion. While homotypic membrane fusion could continue if the Ypt/Rab protein stays in its GTP-bound form, in heterotypic membrane fusion, the Ypt/Rab protein, even if it is in its activated GTP-bound form, might not be able to promote membrane fusion since the other components of the targeting/fusion machinery are not present. We suggest that the only role of GTP hydrolysis by Ypt/Rab proteins that function in the exocytic pathway is to generate the GDP-bound form for efficient GDI-mediated recycling between membranes (Fig. 10C). The diverse phenotypes observed after mutating the various exocytic Ypt/Rab proteins may reflect differences in their requirements for efficient recycling.
GDI-mediated recycling of Ypt/Rab proteins involves their reversible association with membranes (68, 107), and GDI function is essential for yeast cell viability and protein transport (26). However, if the role of GTP hydrolysis by Ypt/Rab proteins is to allow efficient GDI-mediated recycling between membranes, it seems that the recycling of Ypt1p is not essential for either protein transport or cell growth. This suggestion is consistent with the observation that Ypt1p is still functional when permanently fixed to membranes via a membrane anchor (64). Thus, recycling of Ypt/Rab proteins can probably also occur via a GDI-independent mechanism that does not involve their detachment from membranes and therefore may not require GTP hydrolysis. Alternatively, it is possible that recycling of Ypt1p is not important for its function, since there is excess Ypt1p in the cell. This conclusion is based on the observation that mutant cells with a 90% reduction in the level of Ypt1p show no growth or secretion defects at the permissive temperature (39, 75a).
The roles of cycling between the GTP- and GDP-bound forms of Ypt/Rab proteins and of the factors that regulate this cycling are still unresolved. Current models suggest that GAPs regulate Ypt/Rab proteins at the acceptor compartment and that GTP hydrolysis and GAP activity are essential for vesicle fusion with this compartment. In these models, GNEF is predicted to function at the donor compartment (or on the vesicles) in the recruitment of Ypt/Rab GTPases to the membrane (60, 92, 98). Our data suggest an alternative model for Ypt/Rab mode of action. Our combined information from studies of a GTPase-defective mutant Ypt1p and of GAP localization suggest that GTP hydrolysis is not essential for vesicle fusion (this study) and that GAP for Ypt1p does not function at the acceptor compartment (75). We have previously shown that nucleotide exchange is essential for Ypt1p-mediated vesicular transport (42). Therefore, in our model the shift from the GDP- to the GTP-bound form is the event that is crucial for vesicle targeting and/or fusion, while GTP hydrolysis and the GDP-bound form are needed for the recycling of Ypt/Rab proteins. We propose the following hypothesis for the roles of nucleotide cycling of Ypt/Rab proteins and for the localization of their accessory proteins: (i) nucleotide exchange, GNEF, and the GTP-bound form are essential for vesicle targeting or fusion, and GNEF functions at the acceptor compartment; (ii) GTP hydrolysis, GAP, and the GDP-bound form do not have a direct role in vesicle fusion but rather function in GDI-mediated recycling of Ypt/Rab proteins between membranes, a process that is not essential for Ypt1p function.
If the GTP-bound form of Ypt/Rab proteins is the active form of these proteins and is crucial for executing their functions, while the GDP-bound form is inactive, these proteins are more similar to heterotrimeric G proteins and Ras than to EF-Tu. One question that arises is, why does the Q67L mutation not confer a dominant phenotype with Ypt1p as it does with Ras (19)? In other words, if the Ypt1-Q67L mutant protein is constitutively active, why does it not confer either accelerated secretion and/or uncontrolled membrane fusion that might lead to growth defects? Our explanation is that the nature of the process regulated by the active form of Ypt/Rab proteins is different from that regulated by G proteins and Ras. Thus, active Ypt/Rab proteins promote vesicle targeting and fusion when present on the vesicle. In heterotypic membrane fusion, after the vesicle fuses they are not in the right place or in the right context to stimulate such an event. It is reasonable to suppose that the regulators responsible for stimulating Ypt/Rab proteins to promote membrane targeting and fusion are their GNEFs. This suggestion is supported by our finding that nucleotide-free mutant Ypt1 proteins, which inhibit the Ypt1-GNEF, also block the action of GTP-bound Ypt1p-Q67L. Thus, Ypt/Rab proteins have a unique mode of regulation: like Ras, they are active in the GTP-bound form, but unlike Ras, they seem to require interaction with the GNEF to be biologically active.
ACKNOWLEDGMENTS
We are grateful to B. Glick and A. Turkewitz for helpful discussions and critical reading of the manuscript. We thank T. Stevens, T. Graham, A. Franzusoff, and C. Kaiser for generous gifts of antibodies, and we thank the Electron Microscopy Lab at the University of Chicago and Yi-mei Chen for excellent help with electron microscopy.
Support was provided by training grants 5T32 GM 07151-20 (C.J.R.) and 5T32 HD 07009 (R.J.L.). This research was supported by grant GM45444 from NIH to N.S.
REFERENCES
- 1.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J, editors. Molecular biology of the cell. New York, N.Y: Garland Publishing, Inc.; 1994. pp. 643–644. [Google Scholar]
- 2.Alexandrov K, Horiuchi H, Steele-Mortimer O, Seabra M, Zerial M. Rab escort protein-1 is a multifunctional protein that accompanies newly prenylated rab proteins to their target membranes. EMBO J. 1994;13:5262–5273. doi: 10.1002/j.1460-2075.1994.tb06860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antebi A, Fink G. The yeast Ca2+-ATPase homologue, PMRI, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol Biol Cell. 1992;3:633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araki S, Kikuchi A, Hata Y, Isomura M, Takai Y. Regulation of reversible binding of smg p25A, a ras p21-like GTP-binding protein, to synaptic plasma membranes and vesicles by its specific regulatory protein, GDP dissociation inhibitor. J Biol Chem. 1990;256:13007–13015. [PubMed] [Google Scholar]
- 5.Bacon R A, Salminen A, Ruohola H, Novick P, Ferro-Novick S. The GTP-binding protein Ypt1 is required for transport in vitro: the Golgi apparatus is defective in ypt1 mutants. J Cell Biol. 1989;109:1015–1022. doi: 10.1083/jcb.109.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker D, Wuestehube L, Schekman R, Botstein D, Segev N. GTP-binding Ypt1 protein and Ca2+ function independently in a cell-free transport reaction. Proc Natl Acad Sci USA. 1990;87:355–359. doi: 10.1073/pnas.87.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbieri M A, Guangpu L, Mayorga L, Stahl P. Characterization of Rab5:Q79L-stimulated endosome fusion. Arch Biochem Biophys. 1996;326:64–72. doi: 10.1006/abbi.1996.0047. [DOI] [PubMed] [Google Scholar]
- 8.Becker D M, Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- 9.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 10.Bourne H R. Do GTPases direct membrane traffic in secretion? Cell. 1988;53:669–671. doi: 10.1016/0092-8674(88)90081-5. [DOI] [PubMed] [Google Scholar]
- 11.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 12.Brandt D, Asano T, Pedersen S, Ross E. Reconstitution of catecholamine-stimulated guanosinetriphosphatase activity. Biochemistry. 1983;22:4357–4362. doi: 10.1021/bi00288a002. [DOI] [PubMed] [Google Scholar]
- 13.Broek D, Toda T, Michaeli T, Levin L, Birchmeier C, Zoller M, Powers S, Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987;48:789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- 14.Brondyk W H, McKiernan C J, Burstein E S, Macara I G. Mutants of Rab3A analogous to oncogenic Ras mutants. J Biol Chem. 1993;268:9410–9415. [PubMed] [Google Scholar]
- 15.Burnette W N. “Western Blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- 16.Burstein E, Linko-Stentz K, Lu Z, Macara I. Regulation of the GTPase activity of the ras-like protein p25rab3A. J Biol Chem. 1991;266:2689–2692. [PubMed] [Google Scholar]
- 17.Burstein E S, Macara I G. Characterization of a guanine nucleotide-releasing factor and a GTPase-activating protein that are specific for the ras-related protein p25rab3A. Proc Natl Acad Sci USA. 1992;89:1154–1158. doi: 10.1073/pnas.89.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burton J, Roberts D, Montaldi M, Novick P, De Camilli P. A mammalian guanine-nucleotide-releasing protein enhances function of yeast secretory protein Sec4. Nature. 1993;361:464–467. doi: 10.1038/361464a0. [DOI] [PubMed] [Google Scholar]
- 19.Der C, Finkel T, Cooper G. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell. 1986;44:167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- 20.Dirac-Svejstrup A, Sumizawa T, Pfeffer S. Identification of a GDI displacement factor that releases endosome Rab GTPases from Rab-GDI. EMBO J. 1997;16:465–472. doi: 10.1093/emboj/16.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- 23.Franzusoff A, Redding K, Crosby J, Fuller S R, Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J Cell Biol. 1989;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frech M, Darden T, Pedersen L, Foley C, Charifson P, Anderson M, Wittinghofer A. Role of glutamine-61 in the hydrolysis of GTP by p21 H-ras: an experimental and theoretical study. Biochemistry. 1994;33:3237–3244. doi: 10.1021/bi00177a014. [DOI] [PubMed] [Google Scholar]
- 25.Fukui K, Sasaki T, Imazumi K, Matsuura Y, Nakanishi H, Takai Y. Isolation and characterization of a GTPase activating protein specific for the Rab3 subfamily of small G proteins. J Biol Chem. 1997;272:4655–4658. doi: 10.1074/jbc.272.8.4655. [DOI] [PubMed] [Google Scholar]
- 26.Garrett M D, Zahner J E, Cheney C M, Novick P J. GDI1 encodes a GDP dissociation inhibitor that plays an essential role in the yeast secretory pathway. EMBO J. 1994;13:1718–1728. doi: 10.1002/j.1460-2075.1994.tb06436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gietz D, St. Jean A, Woods R, Schiestl R. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1625. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goud B, McCaffrey M. Small GTP-binding proteins and their role in transport. Curr Opin Cell Biol. 1991;3:626–633. doi: 10.1016/0955-0674(91)90033-u. [DOI] [PubMed] [Google Scholar]
- 29.Graham T, Seeger M, Payne G, MacKay V, Emr S. Clathrin-dependent localization of alpha1,3 mannosyltransferase to the Golgi complex in Saccharomyces cerevisiae. J Cell Biol. 1994;127:667–678. doi: 10.1083/jcb.127.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groesch M, Rossi G, Ferro-Novick S. Reconstitution of endoplasmic reticulum to Golgi transport in yeast: in vitro assay to characterize secretory mutants and functional transport vesicles. Methods Enzymol. 1992;219:137–153. doi: 10.1016/0076-6879(92)19016-y. [DOI] [PubMed] [Google Scholar]
- 31.Haney S A, Broach J R. Cdc25p, the guanine nucleotide exchange factor for the Ras proteins of Saccharomyces cerevisiae, promotes exchange by stabilizing ras in a nucleotide-free state. J Biol Chem. 1994;269:16541–16548. [PubMed] [Google Scholar]
- 32.Harris S, Waters G. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J Cell Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higashijima T, Ferguson K, Smigel M, Gilman A. The effect of GTP and Mg++ on the GTPase activity and the fluorescent properties of Go. J Biol Chem. 1987;262:757–761. [PubMed] [Google Scholar]
- 34.Hoffenberg S, Sanford J, Liu S, Daniel D S, Tuvin M, Knoll B, Wessling-Resnick M, Dickey B. Biochemical and functional characterization of a recombinant GTPase, Rab5, and two of its mutants. J Biol Chem. 1995;270:5048–5056. doi: 10.1074/jbc.270.10.5048. [DOI] [PubMed] [Google Scholar]
- 35.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 36.Hwang Y-W, Sanchez A, Miller D L. Mutagenesis of bacterial elongation factor Tu at lysine 136. J Biol Chem. 1989;264:8304–8309. [PubMed] [Google Scholar]
- 37.Hwang Y W, Zhong J M, Poullet P, Parmeggiani A. Inhibition of SDC25C-domain-induced guanine-nucleotide exchange by guanine ring binding domain mutants of v-H-ras. J Biol Chem. 1993;268:24692–24698. [PubMed] [Google Scholar]
- 38.Jedd G, Mulholland J, Segev N. Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment. J Cell Biol. 1997;137:563–580. doi: 10.1083/jcb.137.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jedd G, Richardson C J, Litt R J, Segev N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J Cell Biol. 1995;131:583–590. doi: 10.1083/jcb.131.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jena B P, Brennwald P, Garrett M D, Novick P, Jamieson J D. Distinct and specific GAP activities in rat pancreas act on the yeast GTP-binding proteins Ypt1 and Sec4. FEBS Lett. 1992;309:5–9. doi: 10.1016/0014-5793(92)80727-x. [DOI] [PubMed] [Google Scholar]
- 41.Johnston M, Davis R W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones S, Litt R J, Richardson C J, Segev N. Requirement of nucleotide exchange for Ypt1 GTPase mediated protein transport. J Cell Biol. 1995;130:1051–1061. doi: 10.1083/jcb.130.5.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones, S., and N. Segev. Unpublished observations.
- 44.Kaiser C A, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- 45.Krengel U, Schlichting I, Scherer A, Schumann R, French M, John J, Kabsch W, Pai E, Wittinghofer A. Three-dimensional structures of the H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 1990;62:539–548. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- 46.Lai C-C, Boguski M, Broek D, Powers S. Influence of guanine nucleotides on complex formation between Ras and CDC25 proteins. Mol Cell Biol. 1993;13:1345–1352. doi: 10.1128/mcb.13.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamarche N, Tapon N, Stowers L, Burbelo P, Aspenström P, Bridges T, Chant J, Hall A. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 48.Li B, Warner J R. Mutation of the Rab6 homologue of Saccharomyces cerevisiae, YPT6, inhibits both early Golgi function and ribosome biosynthesis. J Biol Chem. 1996;271:16813–16819. doi: 10.1074/jbc.271.28.16813. [DOI] [PubMed] [Google Scholar]
- 49.Lian J P, Stone S, Jiang Y, Lyons P, Ferro-Novick S. Ypt1p implicated in v-SNARE activation. Nature. 1994;372:698–701. doi: 10.1038/372698a0. [DOI] [PubMed] [Google Scholar]
- 50.Litt R. The role of the guanine nucleotide exchange factor for Ypt1 GTPase in yeast protein transport. Ph.D. thesis. Chicago, Ill: The University of Chicago; 1997. [Google Scholar]
- 51.Martinez O, Antony C, Pehau-Arnaudet G, Berger E, Salamero J, Goud B. GTP-bound forms of rab6 induce the redistribution of Golgi proteins into the endoplasmic reticulum. Proc Natl Acad Sci USA. 1997;94:1828–1833. doi: 10.1073/pnas.94.5.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez O, Schmidt A, Salamero J, Hoflack B, Roa M, Goud B. The small GTP-binding protein rab6 functions in intra-Golgi transport. J Cell Biol. 1994;127:1575–1588. doi: 10.1083/jcb.127.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayer A, Wickner W. Docking of yeast vacuoles is catalyzed by the Ras-like GTPase Ypt7p after symmetric priming by Sec18p (NSF) J Cell Biol. 1997;136:307–317. doi: 10.1083/jcb.136.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCormick F. The world according to GAP. Oncogene. 1990;5:1281–1283. [PubMed] [Google Scholar]
- 55.Milburn M, Tong L, DeVos A, Brünger A, Yamaizumi A, Nishimura S, Kim S. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 56.Molenaar C M T, Prange R, Gallwitz D. A carboxyl-terminal cysteine residue is required for palmitic acid binding and biological activity of the ras-related yeast YPT1 protein. EMBO J. 1988;7:971–976. doi: 10.1002/j.1460-2075.1988.tb02903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mosteller R, Han J, Broek D. Identification of residues of the H-Ras protein critical for functional interaction with guanine nucleotide exchange factors. Mol Cell Biol. 1994;14:1104–1112. doi: 10.1128/mcb.14.2.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakayama K, Nagasu T, Shimma Y, Kuromitsu J, Jigami Y. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 1992;11:2511–2519. doi: 10.1002/j.1460-2075.1992.tb05316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nothwehr S F, Conibear E, Stevens T H. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J Cell Biol. 1995;132:985–998. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Novick P, Brennwald P. Friends and family: the role of the Rab GTPases in vesicular traffic. Cell. 1993;75:597–601. doi: 10.1016/0092-8674(93)90478-9. [DOI] [PubMed] [Google Scholar]
- 61.Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- 62.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 63.Novick P, Zerial M. The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- 64.Ossig R, Laufer W, Schmitt H D, Gallwitz D. Functionality and specific membrane localization of transport GTPases carrying C-terminal membrane anchors of synaptobrevin-like proteins. EMBO J. 1995;14:3645–3653. doi: 10.1002/j.1460-2075.1995.tb00034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pai E, Krengel U, Petsko G, Goody R, Kabasch W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 67.Pfeffer S. GTP-binding proteins in intracellular transport. Trends Cell Biol. 1992;2:41–46. doi: 10.1016/0962-8924(92)90161-f. [DOI] [PubMed] [Google Scholar]
- 68.Pfeffer S R, Dirac-Svejstrup A B, Soldati T. Rab GDP dissociation inhibitor: putting rab GTPases in the right place. J Biol Chem. 1995;270:17057–17059. doi: 10.1074/jbc.270.29.17057. [DOI] [PubMed] [Google Scholar]
- 69.Powers S, Gonzales E, Christensen T, Cubert J, Broek D. Functional cloning of BUD5, a CDC25-related gene from S. cerevisiae that can suppress a dominant-negative RAS2 mutant. Cell. 1991;65:1225–1231. doi: 10.1016/0092-8674(91)90017-s. [DOI] [PubMed] [Google Scholar]
- 70.Powers S, O’Neill K, Wigler M. Dominant yeast and mammalian RAS mutants that interfere with the CDC25-dependent activation of wild-type RAS in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:390–395. doi: 10.1128/mcb.9.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Preuss D, Mulholland J, Franzusoff A, Segev N, Botstein D. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol Biol Cell. 1992;3:789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pringle J R, Preston R A, Adams A E M, Stearns T, Drubin D, Haarer B K, Jones E. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- 73.Redding K, Holcomb C, Fuller S R. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rexach M F, Schekman R W. Distinct biochemical requirements for the budding, targeting, and fusion of ER-derived transport vesicles. J Cell Biol. 1991;114:219–229. doi: 10.1083/jcb.114.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richardson, C., S. Jones, R. Litt, and N. Segev. Unpublished data.
- 75a.Richardson, C., and N. Segev. Unpublished data.
- 76.Rose M, Winston F, Heiter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 77.Rothman J E. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 78.Rybin V, Ullrich O, Rubino M, Alexandrov K, Simon I, Seabra M, Goody R, Zerial M. GTPase activity of Rab5 acts as a timer for endocytic membrane fusion. Nature. 1996;383:266–269. doi: 10.1038/383266a0. [DOI] [PubMed] [Google Scholar]
- 79.Salminen A, Novick P. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- 80.Sanford J, Foster L, Kapadia Z, Wessling-Resnick M. Analysis of the stoichiometry of Rab protein prenylation. Anal Biochem. 1995;224:547–556. doi: 10.1006/abio.1995.1086. [DOI] [PubMed] [Google Scholar]
- 81.Sanford J, Pan Y, Wessling-Resnick M. Prenylation of Rab5 is dependent on guanine nucleotide binding. J Biol Chem. 1993;268:23773–23776. [PubMed] [Google Scholar]
- 82.Scheffzek K, Ahmadian M R, Kabsch W, Wiesmüller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 83.Schmitt H D, Wagner P, Pfaff E, Gallwitz D. The ras-related YPT1 gene product in yeast: a GTP-binding protein that might be involved in microtubule organization. Cell. 1986;47:401–412. doi: 10.1016/0092-8674(86)90597-0. [DOI] [PubMed] [Google Scholar]
- 84.Schroder S, Schimmoller F, Singer-Kruger B, Riezman H. The Golgi-localization of yeast Emp47p depends on its di-lysine motif but is not affected by the ret1-1 mutation in α-COP. J Cell Biol. 1995;131:895–912. doi: 10.1083/jcb.131.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seabra M. Nucleotide dependence of Rab geranylgeranylation. J Biol Chem. 1996;271:14398–14404. doi: 10.1074/jbc.271.24.14398. [DOI] [PubMed] [Google Scholar]
- 86.Segev N. Mediation of the attachment or fusion step in vesicular transport by the GTP-binding Ypt1 protein. Science. 1991;252:1553–1556. doi: 10.1126/science.1904626. [DOI] [PubMed] [Google Scholar]
- 87.Segev N, Botstein D. The ras-like yeast YPT1 gene is itself essential for growth, sporulation, and starvation response. Mol Cell Biol. 1987;7:2367–2377. doi: 10.1128/mcb.7.7.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Segev N, Mulholland J, Botstein D. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell. 1988;52:915–924. doi: 10.1016/0092-8674(88)90433-3. [DOI] [PubMed] [Google Scholar]
- 89.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;19:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Søgarrd M, Tani K, Ye R R, Geromanos S, Tempst P, Kirchhausen T, Rothman J E, Söllner T. A Rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- 91.Soldati T, Reiderer M A, Pfeffer S R. Rab GDI: a solubilizing and recycling factor for rab9 protein. Mol Biol Cell. 1993;4:425–434. doi: 10.1091/mbc.4.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Soldati T, Shapiro A D, Svejstrup A B D, Pfeffer S R. Membrane targetting of the small GTPase Rab9 is accompanied by nucleotide exchange. Nature. 1994;369:76–78. doi: 10.1038/369076a0. [DOI] [PubMed] [Google Scholar]
- 93.Stenmark H, Parton R G, Steele-Mortimer O, Lütke A, Gruenberg J, Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Strom M, Vollmer P, Tan T J, Gallwitz D. A yeast GTPase-activating protein that interacts specifically with a member of the Ypt/Rab family. Nature. 1993;361:736–739. doi: 10.1038/361736a0. [DOI] [PubMed] [Google Scholar]
- 95.Tan T J, Vollmer P, Gallwitz D. Identification and partial purification of GTPase-activating proteins from yeast and mammalian cells that preferentially act on Ypt1/Rab1 proteins. FEBS Lett. 1991;291:322–326. doi: 10.1016/0014-5793(91)81312-v. [DOI] [PubMed] [Google Scholar]
- 96.Tisdale E J, Bourne J R, Khosravi-Far R, Der C J, Balch W E. GTP-binding mutants of Rab1 and Rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol. 1992;119:749–761. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Trahey M, McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987;238:542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- 98.Ullrich O, Horiuchi H, Bucci C, Zerial M. Membrane association of Rab5 mediated by GDP-dissociation inhibitor and accompanied by GDP/GTP exchange. Nature. 1994;368:157–160. doi: 10.1038/368157a0. [DOI] [PubMed] [Google Scholar]
- 99.Vollmer P, Gallwitz D. High expression cloning, purification, and assay of Ypt-GTPase-activating proteins. Methods Enzymol. 1995;257:118–128. doi: 10.1016/s0076-6879(95)57017-9. [DOI] [PubMed] [Google Scholar]
- 100.Wada M, Nakanishi H, Satoh A, Hirano H, Obaishi H, Matsuura Y, Takai Y. Isolation and characterization of a GDP/GTP exchange protein specific for the Rab3 subfamily small G proteins. J Biol Chem. 1997;272:3875–3878. doi: 10.1074/jbc.272.7.3875. [DOI] [PubMed] [Google Scholar]
- 101.Wagner P, Molenaar C M T, Rauh A J G, Brokel R, Schmitt H D, Gallwitz D. Biochemical properties of the ras-related YPT protein in yeast: a mutational analysis. EMBO J. 1987;6:2373–2379. doi: 10.1002/j.1460-2075.1987.tb02514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Walch-Solimena C, Collins R, Novick P. Sec2 mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Walworth N C, Brennwald P, Kabcenell A K, Garrett M, Novick P J. Hydrolysis of GTP by Sec4 protein plays an important role in vesicular transport and is stimulated by a GTPase-activating protein in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2017–2028. doi: 10.1128/mcb.12.5.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Walworth N C, Goud B, Kabcenell A K, Novick P J. Mutational analysis of SEC4 suggests a cyclical mechanism for the regulation of vesicular traffic. EMBO J. 1989;8:1685–1693. doi: 10.1002/j.1460-2075.1989.tb03560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Warner J. Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. In: Guthrie C, Fink G, editors. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. pp. 423–424. [Google Scholar]
- 106.Wichmann H, Hengst I, Gallwitz D. Endocytosis in yeast: evidence for the involvement of a small GTP binding protein (Ypt7) Cell. 1992;71:1131–1142. doi: 10.1016/s0092-8674(05)80062-5. [DOI] [PubMed] [Google Scholar]
- 107.Wu S, Zeng K, Wilson I, Balch W. Structural insights into the function of the Rab GDI superfamily. Trends Biochem Sci. 1996;21:472–476. doi: 10.1016/s0968-0004(96)10062-1. [DOI] [PubMed] [Google Scholar]
- 108.Xiao G, Shoarinejad F, Jin F, Golemis E, Yeung R. The tuberous sclerosis 2 gene product, tuberin, functions as a Rab5 GTPase activating protein (GAP) in modulating endocytosis. J Biol Chem. 1997;272:6097–6100. doi: 10.1074/jbc.272.10.6097. [DOI] [PubMed] [Google Scholar]
- 109.Zerial M, Stenmark H. Rab GTPases in vesicular transport. Curr Opin Cell Biol. 1993;5:613–620. doi: 10.1016/0955-0674(93)90130-i. [DOI] [PubMed] [Google Scholar]