Abstract
The β-secretase, BACE1, and the α-secretase, ADAM10, are known to competitively cleave amyloid precursor protein (APP) in the amyloid cascades of Alzheimer’s disease. Cleavage of APP by BACE1 produces a 99-residue C-terminal peptide (APP-C99) that is subsequently cleaved by γ-secretase to form amyloid-β (Aβ) protein, whereas cleavage of APP by ADAM10 is nonamyloidogenic. It has been speculated that ADAM10/APP and BACE1/APP interactions are regulated by colocalization within and outside of liquid-ordered membrane domains; however, the mechanism of this regulation and the character of the proteins’ transmembrane domains are not well understood. In this work, we have developed and characterized minimal congener sequences for the transmembrane domains of ADAM10 and BACE1 using a multiscale modeling approach combining both temperature replica exchange and conventional molecular dynamics simulations based on the coarse-grained Martini2.2 and all-atom CHARMM36 force fields. Our results show that membrane composition impacts the character of the transmembrane domains of BACE1 and ADAM10, adding credence to the speculation that membrane domains are involved in the etiology of Alzheimer’s disease.
Significance
Despite extensive research, the amyloid hypothesis, which posits that Alzheimer’s disease begins with the production and aggregation of β-amyloid peptides, remains controversial. To settle this controversy, we need to better understand the molecular mechanisms governing β-amyloid production. It has been speculated that the localization of amyloid precursor protein, β-secretase, α-secretase, and γ-secretase into different membrane domains controls this process. Using molecular dynamics, we develop and characterize transmembrane domain models of the β-secretase, BACE1, and the α-secretase, ADAM10, in different membrane environments. Future studies can use these models to further develop our understanding of the molecular origins of Alzheimer’s disease.
Introduction
The interplay between the β-secretase (β-site amyloid precursor protein cleaving enzyme 1; BACE1), the α-secretase (a disintegrin and metalloproteinase domain-containing protein 10; ADAM10), and amyloid precursor protein (APP) is a central focus in Alzheimer’s disease (AD) research. According to the amyloid hypothesis of AD, misregulation of the competitive amyloidogenic and nonamyloidogenic APP processing pathways, mediated by BACE1, ADAM10, and γ-secretase, leads to an increase in amyloidogenic β-amyloid (Aβ) peptide production (1,2,3). The oligomerization and fibrilization of amyloidogenic Aβ has been identified as a key factor in the onset of AD (1,2,3).
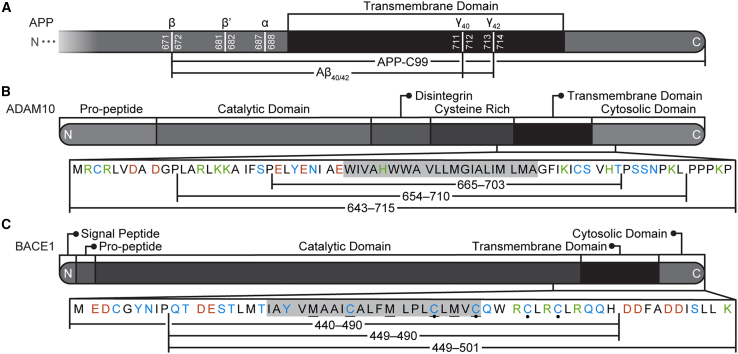
The APP processing pathways begin with cleavage of APP by BACE1 at the β- or β′-sites of APP or by ADAM10 at its α-site (Fig. 1 A) (2). When APP is cleaved at its β-site, a 99-residue C-terminal transmembrane domain (TMD) fragment (APP-C99) is produced (2). Conversely, when cleaved at APP’s β′-site or its α-site, a truncated C-terminal fragment is produced (2). Subsequent cleavage of APP-C99 and the truncated C-terminal APP fragments by γ-secretase produces amyloidogenic Aβ and nonamyloidogenic peptides, respectively (2). Despite extensive research, we do not fully understand what leads to the upregulation of amyloidogenic Aβ.
Figure 1.
Primary sequences of APP, ADAM10, and BACE1. (A) Cleavage sites on APP by β- (β and β′ sites), α- (α site), and γ-secretase ( and sites). (B) ADAM10 and (C) BACE1 domains and the TMD congener sequences tested with REMD. The predicted TMDs are highlighted, acidic residues are orange, basic residues are green, polar residues are blue, and nonpolar residues are black. On BACE1, sulfur belt residues are underlined, and S-palmitoylatable residues are marked with circles.
A possible explanation for the misregulation of these proteins could lie in the observation that high cholesterol is a risk factor for AD (4). It has been shown that membranes phase separate into liquid-ordered phase (Lo) and liquid-disordered phase (Ld) domains, and increases in cholesterol concentration facilitate the formation of the Lo phase (5,6,7). Studies, as described below, exploring the impact of cholesterol concentration on BACE1 activity, ADAM10 activity, and Aβ production have led to the speculation that these proteins could be regulated via partitioning of enzyme and substrate into these domains, with ADAM10 preferring Ld regions and BACE1 and γ-secretase colocalized in Lo domains.
Numerous studies have explored the effect of cholesterol concentration on Aβ production directly. Feeding mice a high cholesterol diet was found to increase Aβ production (8). Small reductions in cholesterol also increased Aβ production, whereas large reductions in cholesterol levels decreased Aβ production (9,10,11,12,13,14). Cholesterol depletion has also been shown to increase ADAM10 processing of APP (15).
Other studies have sought to understand the localization of ADAM10, BACE1, and APP in Lo and Ld membrane domains. Targeting of ADAM10 to lipid rafts with a glycosylphosphatidylinositol-anchor was found to decrease ADAM10 activity (16,17). Additionally, unsaturated fatty acids (typically found in Ld membrane domains) were found to increase membrane fluidity and, in turn, increase ADAM10 activity (18). A consensus exists that BACE1 localization in Lo domains is increased and potentially dependent upon S-palmitoylation of C474, C478, C482, and C485 on the N-terminal side of the TMD and in the N-terminal juxtamembrane domain (JMD). However, the effect of BACE1 localization in Lo domains on amyloidogenic Aβ production is still debated (19,20,21,22,23,24). It was also found that interactions between BACE1 and APP are positively correlated with membrane viscosity (25). Finally, although some studies have pointed to partial localization of APP in Lo and Ld membrane domains, compelling evidence suggests it primarily localizes in the Ld domain near the Ld-Lo boundary (26). Taken together, these results point to the importance of membrane domains in the regulation of APP processing. Nevertheless, further investigation is necessary to develop a detailed mechanism defining the role of membrane domains in APP processing.
Many studies have investigated the localization of BACE1 and ADAM10 in Ld and Lo membrane regions and demonstrated the importance of their respective TMDs for APP processing. Surprisingly, the TMD sequences of BACE1 and ADAM10 have not been clearly defined, and the structural and physicochemical properties of their TMDs remain largely uncharacterized. Experimental and computational work has revealed that BACE1 can form a trimer mediated by a complex between copper and a sulfur-rich motif (M462xxxC466xxxM470xxxC474xM476xC478) within its TMD (27,28,29). However, BACE1’s proposed TMD sequence of 458–478 (Uniprot: P56817) has only been predicted based on sequence and hydrophobicity analysis and not by experimentation or simulation (30). ADAM10’s proposed TMD sequence of 673–693 (Uniprot: O14672) is also only predicted (30). This predictive determination of these TMD sequences does not account for potential changes in membrane composition, and therefore it cannot provide a complete picture of the proteins’ TMDs. Additionally, these predictions do not account for the role of the JMDs in the TMD’s membrane insertion and character, which could be critical when developing a minimal model for TMD characterization.
In this work, we clearly define the TMD and JMD sequences of ADAM10 and BACE1 and develop congener models for this and future computational and experimental characterization. For each protein, we started with several lengths of peptides including residues in the predicted TMD region and within the flanking JMDs in an all-α-helical structure (Fig. 1 B and C). These peptides were then simulated using temperature replica exchange molecular dynamics (REMD) until a consistent helical region was identified and the conformational ensemble was adequately sampled. Once identified, the optimal length congener models were simulated in various membrane environments using conventional molecular dynamics with all-atom (CHARMM36) and coarse-grained (Martini2.2) force fields. Importantly, these models contain parts of the proteins’ JMDs to preserve any JMD-membrane contacts that may be critical for replicating the properties of the TMDs. Due to the suggested role of membrane domains in mediating the interactions of ADAM10 and BACE1 with APP, we explored the effects of lipid environment on these congener models using simplified membrane models that approximately capture the lipid order of Lo and Ld domains. Dipalmitoylphosphatidylcholine (DPPC):cholesterol (CHOL) mixtures were the first binary mixture observed to produce the Lo lipid phase by Vist and Davis, and they have since been the principal binary lipid mixture used to represent the Lo phase (31). The Lo phase has frequently been used as a reductive representation of the lipid order expected in lipid rafts, which separates from the Ld phase, used to represent the lipid membrane bulk. As such, lipid phase separations that can be observed in models as simple as ternary mixtures of a saturated lipid, unsaturated lipid, and CHOL have frequently been used as simplified models of the formation and separation of lipid rafts from the lipid membrane bulk. In the molecular dynamics simulation literature, DPPC:dilinoleoylphosphatidylcholine (DIPC):CHOL has most-frequently been chosen as a ternary mixture for the study of lipid phase separation (6). As such, in selecting a simplified model of Lo and Ld lipid environments, we have chosen DPPC:CHOL and DIPC:CHOL binary lipid mixtures for characterization of the ADAM10 and BACE1 TMDs and JMDs.
Our results show that the proposed transmembrane congener models for ADAM10 and BACE1 are impacted by their membrane environment. By establishing minimal models for the TMDs of ADAM10 and BACE1, our study sets the stage for future computational and experimental work investigating the impact of membrane composition on ADAM10 and BACE1 structure and partitioning, as well as their interactions with the TMD of the APP substrate.
Materials and methods
Unless otherwise specified, simulation analysis was performed in Python3 with the Numpy, Scipy, Scikit-Learn, and/or MDAnalysis packages (32,33,34,35,36). All molecular visualizations were created using Visual Molecular Dynamics, Stride, and POV-Ray, and figures were created using MatPlotLib (37,38,39,40). Probability density functions were computed using gaussian kernel density estimates with bandwidths optimized by 10-fold K-Fold cross validation (bandwidth values provided in Table S3). Further system and simulation details are provided in Tables S1 and S2.
All-atom proteins in implicit membranes
For each protein, transmembrane congener models with JMDs of varying length were simulated using REMD in implicit membranes with thicknesses of 30, 35, and 40 Å described by the generalized Born with simple switching (GBSW) potential in the CHARMM simulation program (41,42,43). This approach follows that taken in our previous work exploring the structure of APP-C99 (44). The congener models used for BACE1 included residues M440–K501, Q449–K501, and Q449–H490; and the congener models for ADAM10 included residues M643–P715, L654–L710, and E665–T703 (Fig. 1 B–C). Each model was constructed by placing the residues in a completely α-helical structure and centered in the GBSW membrane. The proteins were represented with the CHARMM36m force field (45). The GBSW membranes used a 6-Å smoothing length with 38 angular integration points and a maximum distance for radial integration of 20 Å. The nonpolar surface tension coefficient was set to 0.04 kcal/mol/Å2. The membrane/solvent interface on each side of the membrane was set to 2.5 Å, leaving the hydrophobic cores of the 30-, 35-, and 40-Å membranes as 25, 30, and 35 Å, respectively. Replica exchange was attempted every 50 ps between 16 linearly spaced windows from 310K to 460K. Frames were saved every 10 ps for around 200 ns, and the frames from the last 100 ns from the 310K window were used for analysis.
Coarse-grained simulations
The Martini2.2 coarse-grained protein force field is dependent on a predefined secondary structure (46,47,48). To identify a suitable secondary structure, the final 100 ns of frames from the 310K window of the REMD simulations of the selected congener models were clustered with agglomerative clustering using Ward’s minimum variance method (49). The optimal number of clusters was taken to be that with the highest Silhouette Score (50). This clustering scheme was applied using Julia with the Clustering.jl package and is further described in supporting methods I (51). For each protein and membrane width, the medoid of the largest cluster was mapped to the Martini2.2 model using the martinize.py program with a manually defined secondary structure (46,47,48). The secondary structures of the α-helical residues in the proteins were defined based on the averaged backbone hydrogen bond pattern of the frames in the largest cluster, and for the rest of the residues, they were defined using the DSSP result for the input structure (52).
For each protein, its Martini2.2 congener model was simulated in five single-component lipid membranes: 100 mol% DPPC, 100 mol% DIPC, 100 mol% dilauroylphosphatidylcholine (DLPC), 100 mol% dioleoylphosphatidylcholine (DOPC), and 100 mol% 1-palmitoyl-2-oleoylphosphatidylcholine (POPC). The models were also simulated in DIPC and DPPC membranes with four different concentrations of CHOL: 10 mol% CHOL, 20 mol% CHOL, 30 mol% CHOL, and 40 mol% CHOL. The congener model for each protein and membrane was selected based on the width of a pure lipid membrane composed of the lipid included in the membrane (Table S1). The systems were built using the insane.py program (53).
Five replicates were simulated for each system in GROMACS 2021.5 using the new-rf input parameters recommended by the Martini developers (54,55). Each replicate was first minimized by steepest descent and then equilibrated in the isobaric-isothermal (NPT) ensemble using the velocity rescaling thermostat and a semiisotropic Berendsen barostat with a compressibility of 4.5 10−5 bar−1 and an integration timestep of 20 fs (56). Production simulations were performed for 5 μs in the NPT ensemble using a semiisotropic Parrinello-Rahman barostat with a compressibility of 3.0 10−4 bar−1(57). Coordinates were saved every 1 ns, and the last 4 μs of each simulation was used for analysis. All simulations were performed at 310K with an ion concentration of 0.15 M and with 10% of water represented with Martini’s antifreeze water model.
All-atom proteins in explicit membranes
Final configurations from each 5-μs Martini2.2 replicate simulation of each protein in 7:3 DPPC:CHOL and 9:1 DIPC:CHOL membranes were backmapped to the CHARMM36 force field using the backwards.py program (58,59). Note that due to Martini2.2’s one-to-four bead per atom representation, individual Martini lipids can represent multiple all-atom lipids. Here, the DIPC lipids were backmapped to di-C18:2 PC lipids and the DPPC lipids were backmapped to di-C16:0 PC lipids. Also note that di-C18:2 PC lipids are named DUPC in the CHARMM36 force field, but in this paper, we will continue to refer to them as DIPC for clarity. Backmapping was completed using a revised version of the initram-v5.sh program. The initram-v5.sh program is available with backwards.py on Martini’s web site. The only revisions to initram were to use the Verlet cutoff scheme for neighbor searching and soft-core potentials to make it compatible with our single-precision version of GROMACS. Similar revisions are discussed and utilized in Martini’s backmapping tutorials. An additional 2 nm of water was added along the z axis (perpendicular to the membrane) to the ADAM10 in the 9:1 DIPC:CHOL system using Packmol to prevent interperiodic contact between the JMDs (60). Each system was equilibrated for 5 ns in the NVT ensemble with the velocity rescaling thermostat and a 1-fs timestep and for 10 ns in the NPT ensemble with a 1-fs timestep, the velocity rescaling thermostat, and the semiisotropic Berendsen barostat with a compressibility of 4.5 10−5 bar−1(56,61). Production simulations were performed using the Nose-Hoover thermostat and the semiisotropic Parrinello-Rahmen barostat with a compressibility of 4.5 10−5 bar−1 for 1 μs (57,62,63). Coordinates were saved every 100 ps and the last 900 ns from each simulation was used for analysis. All simulations were performed using GROMACS 2021.2 at 310K with an ion concentration of 0.15 M (54,55).
Results and discussion
Development of BACE1 and ADAM10 TMD and JMD models
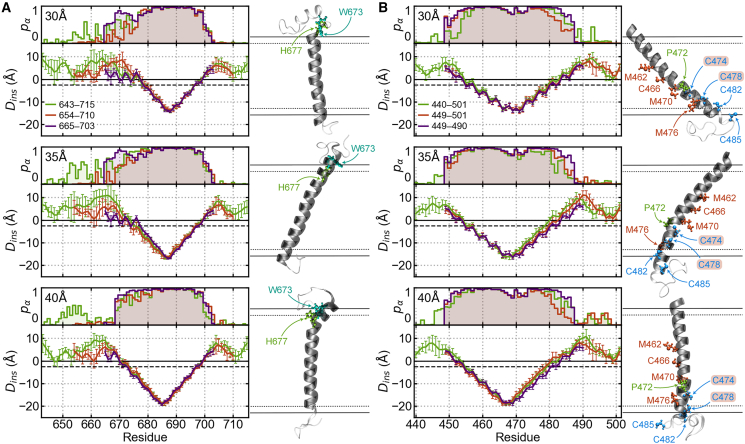
REMD simulations were performed to sample the conformational space of several length congener models of ADAM10 and BACE1 in 30-, 35-, and 40-Å GBSW implicit membranes. These simulations were used to identify minimal length congener sequences having consistent structural features in the three membranes to be used in explicit membrane simulations. Model validation principally focused on two metrics: residue α-helix propensity, , and insertion depth, . Residue α-helix propensity was calculated as the probability that the residue forms an amine-carbonyl backbone hydrogen bond with residue and/or residue . Hydrogen bonds were defined as having a donor-acceptor distance of Å and a donor-hydrogen-acceptor angle . Insertion depths were calculated as the nearest distance from the edge of the GBSW membrane.
For ADAM10, we evaluated model congener sequences including residues M643–P715, L654–L710, and E665–T703. The shortest model (E665–T703) features an extended α-helix and does not include reentrant residues within the N-terminal JMD (Fig. 2 A). We selected the L654–L710 model for further simulation to account for effects of interactions between the JMD and the membrane. In our REMD simulations of the ADAM10 congener models, residues H677 and W673 can form a sidechain-backbone hydrogen bond and induce a hinge in the TMD α-helix. From this point on, the L654–L710 model of ADAM10 will be referred to as ADAM10.
Figure 2.
Structural comparison of congener models in implicit membranes. The residue α-helix propensity, , and insertion depths, , are presented for (A) ADAM10 and (B) BACE1 TMD and JMD congener models in 30-Å (top), 35-Å (middle), and 40-Å (bottom) GBSW membranes. The insertion depth error bars indicate their standard deviations. The medoid structure of the largest cluster from the agglomerative clustering of each simulation is shown to the right of their and results. For both proteins, the residues that induce a hinge/bend in the α-helix are green, and the extracellular JMD is oriented up. For BACE1, S-palmitoylatable residues are blue, and sulfur belt residues are orange.
For BACE1, we evaluated three model congener sequences: M440–K501, Q449–K501, and Q449–H490. Helicity and insertion depths appear consistent across all three models in all membranes (Fig. 2 B). Therefore, the Q449–K501 model is the optimal model sequence, as including the last 11 residues of BACE1’s C-terminal domain provided a more complete model congener sequence with minimal added computational cost. This BACE1 model contains four cysteine residues known to be S-palmitoylated (C474, C478, C482, and C485), five sulfur containing residues believed to contribute to BACE1 trimerization (M462, C466, M470, C474, and C478), and P472, which induces a kink in the α-helix. From this point on, the Q449–K501 model of BACE1 will be referred to as BACE1.
Agglomerative clustering was performed on each REMD simulation, and the largest cluster was used to define the secondary structures of the selected models for Martini2.2 simulations (Figs. 2, 3, and 4). The medoid structures of the largest clusters (Fig. 2) were used as the input structure for mapping from all-atom to Martini representation.
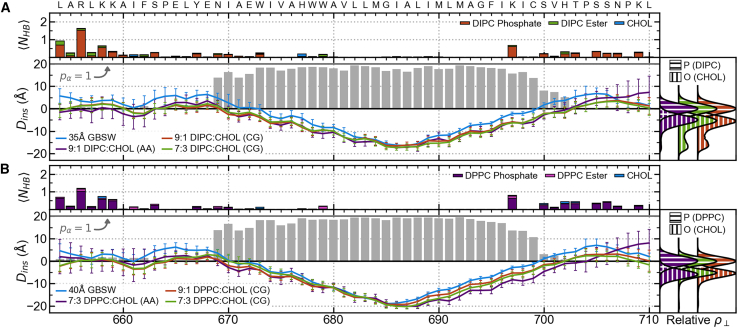
Figure 3.
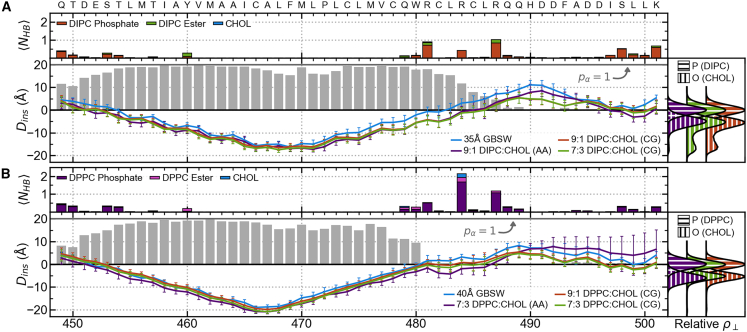
ADAM10 structure and interactions with membrane. Systems with (A) explicit DIPC membranes or a 35-Å implicit membrane and (B) explicit DPPC membranes or a 40-Å implicit membrane. For each set of membranes, residue insertion depths, , are shown with colored lines (error bars show standard deviations), and gray bars indicate the residues’ α-helix propensities, , for the all-atom explicit membrane system (bottom left). Relative lateral atomic densities along the membrane normal, , for lipid phosphate phosphorus atoms and CHOL hydroxyl oxygen atoms (bottom right). Colored bars indicate the average number of hydrogen bonds formed between residues and membrane components () for the all-atom explicit membrane system (top).
Figure 4.
Impact of membrane composition on the orientation of ADAM10. (A) Effect of increase in membrane width on the tilt angle, θ, with confidence ellipses showing the standard deviations. Probability densities of θ and membrane width are shown along their corresponding axes. (B) Tilt angle probability density of all frames from all-atom simulation of ADAM10 in a 7:3 DPPC:CHOL bilayer compared with frames with a hydrogen bond between the sidechain (SC) of H677 and backbone (BB) of W673, and frames with different numbers of hydrogen bonds between the C-terminal JMD and the membrane, NHB(CT-M). (C) Mean orientational angles, , for each residue from all-atom simulations with 9:1 DIPC:CHOL and 7:3 DPPC:CHOL bilayers. (D) Reference graphic depicting θ and angles.
Characterization of the ADAM10 TMD and JMD
The ADAM10 congener model was simulated in a variety of coarse-grained and all-atom lipid and lipid/CHOL bilayers. Insertion depths and α-helix propensities were calculated to determine the TMD residues. Insertion depths were calculated as the distance along the normal of the nearest leaflet between a residue’s α-carbon (or BB bead for Martini) and the leaflet (supporting methods II). The position and normal of each leaflet were determined using the lipid phosphate phosphorus atoms within 2 nm of the any protein atom. Residue α-helix propensities were calculated in the same manner as with the REMD simulations. Here, we define the TMD as the continuous chain of residues passing through the bilayer with and the TMD α-helix as the continuous chain of residues with .
To understand how the ADAM10 TMD and JMDs interact with and anchor to the membrane, the average number of hydrogen bonds, , formed between each residue and each kind of membrane component were computed for each of the all-atom explicit membrane simulations. Potential hydrogen bond acceptors were identified as atoms whose charge , potential hydrogen atoms involved in hydrogen bonds were identified as those with a charge , and potential hydrogen bond donors were inferred from the potential hydrogens. Like with backbone hydrogen bonds, protein-membrane hydrogen bonds were defined as having a donor-acceptor distance of Å and a donor-hydrogen-acceptor angle .
For ADAM10 in a 9:1 DIPC:CHOL all-atom bilayer, the TMD extends from residues I670 to H702, and the α-helix includes residues N669–C699 (Fig. 3 A). In a 7:3 DPPC:CHOL bilayer, the ADAM10 TMD extends from residues I670 to P704, and residues N669–C699 make up the the α-helix. Similar results were found for the TMD in coarse-grained simulations with DIPC and DPPC membranes with various CHOL concentrations and in pure DOPC, DLPC, and POPC membranes (Fig. S5). These results show that the length of the ADAM10 TMD and TMD α-helix are not substantially impacted by membrane lipid and CHOL concentrations.
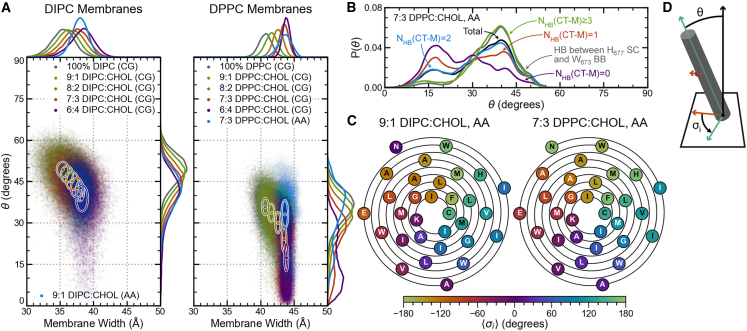
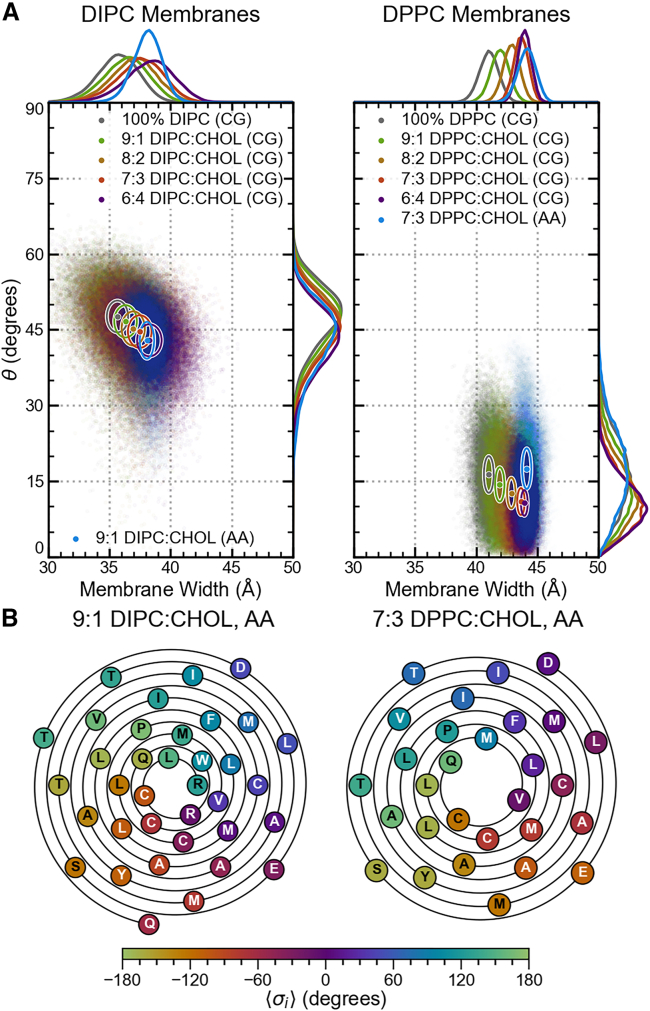
The tilt angle, θ, between ADAM10’s α-helix axes and the membrane normal and the residues’ mean orientational angles, , were calculated for coarse-grained DIPC:CHOL and DPPC:CHOL systems with 0%, 10%, 30%, and 40% CHOL and for the all-atom systems with 9:1 DIPC:CHOL and 7:3 DPPC:CHOL membranes. The angles describe the orientation of a helical residue’s α-carbon in the membrane about the axis of the α-helix, and the θ angle is measured as the angle between the α-helix axis and the membrane normal (Fig. 4 D). Further details about the θ and calculations are provided in supporting methods II.
For ADAM10 in DIPC membranes, the tilt angle decreases linearly with increasing membrane width. In contrast, for DPPC membranes with 30% and 40% CHOL (consistent with concentrations found in Lo membrane domains), ADAM10’s tilt angle distribution becomes bimodal, indicating two distinct configurational states (Fig. 4 A). This bimodality also appears in all-atom 7:3 DPPC:CHOL membrane simulations of ADAM10 and can accounted for by the number of hydrogen bonds between ADAM10’s C-terminal JMD and membrane components and the presence of a hydrogen bond between the sidechain of H677 and the backbone of W673 (Fig. 4 B). Higher tilt angles tend to be associated with configurations with more than two hydrogen bonds between the C-terminal JMD and the membrane () and/or with a sidechain-backbone hydrogen bond between H677 and W673 (). Lower tilt angles are most strongly associated with configurations lacking any hydrogen bonds between the C-terminal JMD and the membrane (). We observe the impact of hydrogen bonds between the protein and the membrane on the tilt angle in the 7:3 DPPC:CHOL membrane but not in the 9:1 DIPC:CHOL membrane. This does not appear to be a product of hydrogen bonding with CHOL as, relative to lipid-protein hydrogen bonds, few of these interactions occur in either membrane (CHOL-protein hydrogen bonds account for only 5.7% and 9.6% of membrane-protein hydrogen bonds in the 9:1 DIPC:CHOL and 7:3 DPPC:CHOL membranes, respectively). Despite the observed change in tilt angle between DIPC and DPPC membranes, the difference in from the 9:1 DIPC:CHOL all-atom to the 7:3 DPPC:CHOL all-atom membrane is unchanged (10 36°; Fig. 4 C). On the N-terminus side, the TMD is anchored to the membrane primarily by residues L654, R656, and K658 (Fig. 3). On the C-terminus side, the TMD is anchored to the membrane by K697 and nonspecifically by various residues in the C-terminal JMD (Fig. 3).
To understand whether specific interactions with CHOL, other than just hydrogen bonding, could impact the character of ADAM10 in Lo domains, we analyzed residue-CHOL contacts with a residence time of greater than 25 ns in the all-atom 9:1 DUPC:CHOL and 7:3 DPPC:CHOL systems (Figs. S10A and S11). Within the C-terminal side leaflet, strong interactions occur between CHOL and F695. It is possible that in CHOL-rich Lo domains, these interactions lead to an unfavorable decrease in interactions between the C-terminal JMD and lipids and cause the previously described low-tilt state. Meanwhile in the N-terminal leaflet, interactions between H677, W678, and W679 and CHOL could impact the H677–W673 sidechain-backbone hydrogen bond, and interactions between F662 and CHOL could cause the slight decrease in hydrogen bonding between N-terminal JMD residues L654 and R656 observed in the 7:3 DPPC:CHOL system compared with the 9:1 DUPC:CHOL system.
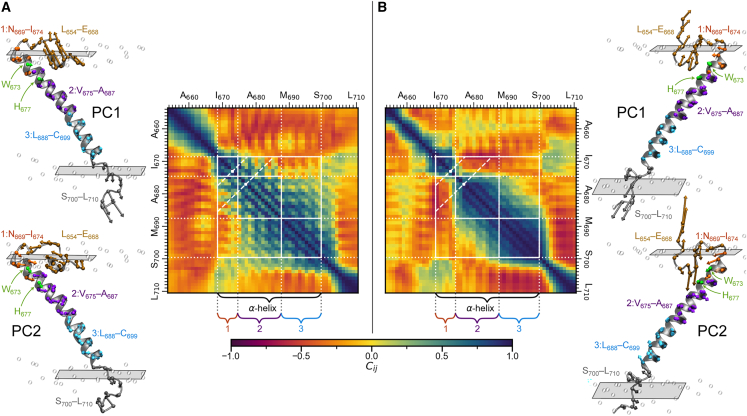
Principal component analyses were performed for the all-atom 9:1 DIPC:CHOL and 7:3 DPPC:CHOL membrane simulations using the RMSD-fitted α-carbon cartesian positions of ADAM10. Principal component analyses have been applied for similar systems to identify critical global protein motions and can be easily applied using the Prody python package and other programs (64). Here, we instead used in-house code described in supporting methods III (65,66,67). Cross correlation matrices were constructed for each membrane using enough principal components to account for 90% of the cumulative variance. For the 9:1 DIPC:CHOL membrane and the 7:3 DPPC:CHOL membrane, the cumulative variances of the first 11 principal components are 90.8% and 91.2%, respectively. The cross correlation matrices of these principal components and the first two principal components for each system are shown in Fig. 5.
Figure 5.
ADAM10 principal components and cross correlation matrices. The first (top) and second (bottom) principal components and principal component cross correlation matrices are shown for (A) 9:1 DIPC:CHOL and (B) 7:3 DPPC:CHOL all-atom membranes. The principal components are shown on the structure closest to the mean structure in PC1 and PC2 space. The lengths of the arrows are proportional to the magnitude of the principal components. N-terminus JMD residues are brown, C-terminus JMD residues are gray, W673 and H677 are green, and residue groups 1, 2, and 3 are orange, purple, and cyan, respectively. The cross correlation matrices were constructed using the first 11 principal components for each system to account for over 90% cumulative variance. Gridlines indicate the α-helix and residue groups 1, 2, and 3. Diagonal dashed lines mark the locations of W673 and H677.
In both 9:1 DIPC:CHOL and 7:3 DPPC:CHOL membranes, the principal component analysis reveals strong correlations between helical residues and residues one helical turn later and a break in the helix between residues W673 and H677. This kink in the helix is more significant in the 7:3 DPPC:CHOL membrane than in the 9:1 DIPC:CHOL membrane. Further, in the 9:1 DIPC:CHOL membrane, residues in helical groups 2 (V675–A687) and 3 (L688–C699) are well correlated, whereas in the 7:3 DPPC:CHOL membrane, no significant correlation is observed. Finally, the principal component analysis further substantiates our previous TMD α-helix definition for ADAM10 of N669–C699 in both membranes.
Characterization of the BACE1 TMD and JMD
Following the protocol applied for ADAM10, the BACE1 congener model was simulated in a variety of coarse-grained and all-atom lipid and lipid/CHOL bilayers, and residue insertion depths and α-helix propensities were calculated. We define the TMD as the continuous chain of residues passing through the bilayer with . The TMD α-helix was defined as the continuous chain of residues with . To understand how BACE1’s TMD and JMDs interact with the membrane, the average number of hydrogen bonds between each residue and each kind of membrane component were computed for each all-atom simulation. Hydrogen bonds were identified in the same manner as with ADAM10.
For BACE1 in a 9:1 DIPC:CHOL all-atom bilayer, the TMD extends from residues E452–C485 and the α-helix is made up of residues Q449–R484 (Fig. 6 A). In a 7:3 DPPC:CHOL all-atom bilayer, residues D451–Q479 make up the α-helix, and the TMD includes residues E452–R481 (Fig. 6 A). In this membrane, residues C482–L486 are also a part of the continuous chain of residues passing through the membrane with . However, as they are not a part of the α-helix and lie flat along the lipid headgroups, they are better classified as part of the C-terminal JMD. These all-atom results are in agreement with corresponding coarse-grained simulations with DIPC and DPPC membranes containing 0%, 10%, 20%, 30%, and 40% CHOL and in pure DOPC, DLPC, and POPC membranes (Fig. S6), suggesting that the length of BACE1’s TMD and α-helix is dependent upon membrane composition due to structural changes in the membrane rather than specific interactions with individual membrane components.
Figure 6.
BACE1 structure and interactions with membrane. Systems with (A) explicit DIPC membranes or a 35-Å implicit membrane and (B) explicit DPPC membranes or a 40-Å implicit membrane. For each set of membranes, residue insertion depths, , are shown with colored lines (error bars show standard deviations), and gray bars indicate the residues’ α-helix propensities, , for the all-atom explicit membrane system (bottom left). Relative lateral atomic densities along the membrane normal, , are shown for lipid phosphate phosphorus atoms and CHOL hydroxyl oxygen atoms (bottom right). Colored bars indicate the average number of hydrogen bonds formed between residues and membrane components () for the all-atom explicit membrane system (top).
The tilt angle, θ, and residue orientational angles, , were calculated for BACE1 in 9:1 DIPC:CHOL and 7:3 DPPC:CHOL all-atom bilayers, and θ was calculated for coarse-grained DIPC and DPPC bilayers with a variety of CHOL concentrations. For BACE1, the tilt angle decreases linearly with increasing membrane width for both DIPC and DPPC membranes (Fig. 7 A). BACE1’s shorter TMD α-helix in DPPC membranes leads to a more pronounced decrease in tilt angle compared with DIPC membranes than what was observed for ADAM10 (Fig. 4 A). In both membranes, BACE1 is anchored to the membrane by hydrogen bonds between the membrane and R481, R484, and R487, belonging to an RCLRCLR motif. However, compared with in the 9:1 DIPC:CHOL membrane, hydrogen binding propensity with the membrane is shifted from R481 to R484 in the 7:3 DPPC:CHOL membrane (Fig. 6). This change in anchoring residue provides a plausible explanation for the shorter TMD α-helix observed in the wider 7:3 DPPC:CHOL membrane and results in a change in the residue orientational angles (Fig. 7 B). The average change in from the 9:1 DIPC:CHOL membrane to the 7:3 DPPC:CHOL membrane is −58 13°, corresponding to the helical twist between the anchoring residues R481 and R484.
Figure 7.
Impact of membrane composition on the orientation of BACE1. (A) Effect of increase in membrane width on the tilt angle, θ, with confidence ellipses showing the standard deviations. Probability densities of θ and membrane width are shown along their corresponding axes. (B) Mean orientational angles, , for each residue from all-atom simulations with 9:1 DIPC:CHOL and 7:3 DPPC:CHOL bilayers.
To further understand how CHOL impacts the character of BACE1, we examined specific residue-CHOL contacts lasting longer than 25 ns in the all-atom 9:1 DUPC:CHOL and 7:3 DPPC:CHOL systems (Figs. S10B and S12). Within the N-terminal leaflet, these contacts appear to be facilitated by Y460, whose α-carbon is positioned on the side of the helix pointing slightly into the membrane in the 9:1 DUPC:CHOL system ( −109°) and more significantly into the membrane in the 7:3 DPPC:CHOL system ( −161°). On the C-terminal side of the helix, protein-CHOL contacts appear to be facilitated by F469 and W480. In the 9:1 DUPC:CHOL membrane, F469 ( 100°) and W480 ( 107°) are both on the side of the helix pointing slightly toward the outside of the membrane. In the 7:3 DPPC:CHOL system, F469 points in toward the center of the membrane ( 35°), and W480 becomes a part of the C-terminal JMD. It is possible that the interactions between these residues and CHOL drive the reorientation of BACE1 in Lo domains.
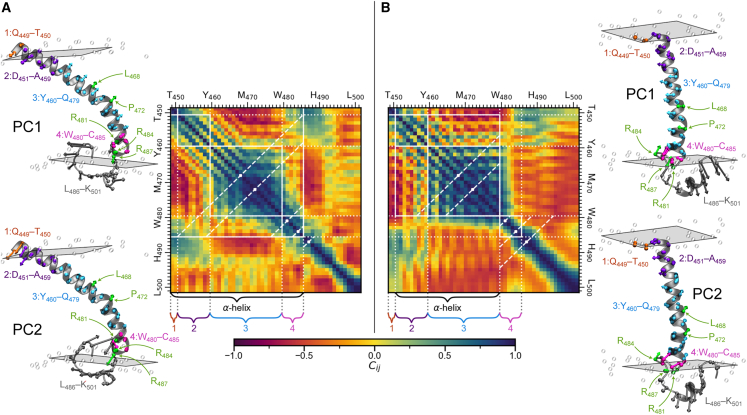
Principal component analyses were performed for both all-atom systems using the RMSD-fitted α-carbon cartesian positions of BACE1. Cross correlation matrices were constructed using the first 11 principal components from the 9:1 DIPC:CHOL system (90.5% cumulative variance) and the first 10 principal components from the 7:3 DPPC:CHOL system (90.5% cumulative variance). The cross correlation matrices and the first two principal components for each system are shown in Fig. 8. The cross correlation matrices reaffirm our previous observation of a shortened helical structure in the 7:3 DPPC:CHOL bilayer compared with the 9:1 DIPC:CHOL bilayer. In the 7:3 DPPC:CHOL bilayer, group 1 (Q449 and T450) and group 4 (W480–C485) lack the helical character seen in the 9:1 DIPC:CHOL bilayer. In the 7:3 DPPC:CHOL bilayer, the essential motions of the residues from L483 to Q488 are better correlated with one another than they are in the 9:1 DIPC:CHOL membrane. This is a consequence of having a membrane anchor within the C-terminal side of the TMD α-helix and another within the C-terminal JMD.
Figure 8.
BACE1 principal components and cross correlation matrices. The first (top) and second (bottom) principal components and principal component cross correlation matrices are shown for (A) 9:1 DIPC:CHOL and (B) 7:3 DPPC:CHOL all-atom membranes. The principal components are shown on the structures closest to the mean structure in PC1 and PC2 space. The lengths of the arrows are proportional to the magnitude of the principal components. C-terminus JMD residues are gray, L468, P472, R481, R484, and R487 are green, and residue groups 1, 2, 3, and 4 are orange, purple, cyan, and pink, respectively. To account for over 90% cumulative variance, the cross correlation matrices for 9:1 DIPC:CHOL and 7:3 DPPC:CHOL were constructed using the first 11 and 10 principal components, respectively. Gridlines indicate the α-helix, and residue groups 1, 2, 3, and 4. Diagonal dashed lines mark the locations of L468, P472, R481, R484, and R487.
Conclusions
Our findings show that ideal transmembrane congener models of ADAM10 and BACE1 include TMD and JMD residues L654–L710 and Q449–K501, respectively. These models were used in our characterization of each protein’s TMD and JMDs and are aptly suited for future computational and experimental characterization.
In both a 9:1 DIPC:CHOL bilayer and a 7:3 DPPC:CHOL bilayer, ADAM10’s TMD α-helix includes residues N669–C699. ADAM10’s TMD is found to be consistently bound to the membrane by hydrogen bonds between lipids and residue K697 on the C-terminal side of its TMD and residues L654, R656, and K658 on the N-terminal side of its TMD. The ADAM10 congener model also interacts with the membrane through protein-CHOL contacts, involving aromatic residues H677, W678, W679, F662, and F695, which are observed to increase in the CHOL-rich 7:3 DPPC:CHOL membrane. With a consistent α-helix definition and anchoring residues across different membranes, the TMD’s orientation in the membrane about its α-helix axis did not differ between the 9:1 DIPC:CHOL membrane and the 7:3 DPPC:CHOL membrane. In membranes with compositions consistent to those found in Ld membrane domains, the tilt of the α-helix decreases linearly with increasing membrane width. However, a second configurational state with a lower tilt angle arises in Lo membranes. This configurational state tends to lack a backbone-sidechain hydrogen bond between W673 and H677 and typically has few hydrogen bonds binding the C-terminal JMD to the membrane. This behavior in Lo membranes could explain the speculated localization and enzymatic activity of ADAM10 in Ld domains, but this requires further investigation.
In both 9:1 DIPC:CHOL and 7:3 DPPC:CHOL bilayers, BACE1 is primarily bound to the membrane by an RCLRCLR motif on the C-terminal side of the TMD and in the C-terminal JMD. However, in the 9:1 DIPC:CHOL membrane, R481 and R487 are the primary anchors, whereas in the 7:3 DPPC:CHOL bilayer, R484 is the primary anchor. Despite having a wider membrane width, this results in a shorter TMD α-helix in the 7:3 DPPC:CHOL bilayer (D451–Q479) than in the 9:1 DIPC:CHOL bilayer (E449–C484). This shorter α-helix results in a more substantial decrease in tilt angle in DPPC membranes than in DIPC membranes compared with what was seen with ADAM10. The BACE1 congener model also interacts with the membrane through protein-CHOL contacts, involving aromatic residues Y460, W480, and F469, which are observed to increase in the CHOL-rich 7:3 DPPC:CHOL membrane. The change in anchoring residues causes a rotation of the TMD α-helix about its axis in the membrane. Together, these factors could contribute to BACE1’s partitioning, homodimerization, and enzymatic activity. It is important to note that our simulations were not of palmitoylated BACE1, which may be necessary for proper partitioning and activity.
It is notable that both ADAM10 and BACE1 have arginine residues within their TMD and JMD sequences, especially BACE1 with its RCLRCLR anchoring motif. Arginine-rich sequences are also observed in helical cell-penetrating peptides that can readily insert into membranes and be internalized by the cell (68). It is possible that the prevalence of arginine in and near the TMDs of ADAM10 and BACE1 facilitates their membrane insertion and trafficking. Despite a large body of research focusing on ADAM10 and BACE1, we still do not know whether the proteins cleave APP in Lo domains or in the Ld domain. The principal impact of lipid composition on the structure and orientation of the BACE1 and ADAM10 TMDs results from variation in bilayer thickness, membrane ordering, and specific interactions with CHOL. In the case of BACE1, this impacts the length of the TMD α-helix and the tilt and orientation of the helix in the membrane. Thanks to its RCLRCLR motif, BACE1 is able to more readily adapt to the wider Lo membranes than ADAM10. For ADAM10, the wider membranes only cause a change in its tilt angle, and in Lo membranes, only part of its ensemble adopts a substantially lower tilt conformation. This suggests ADAM10 may be less suited for localization in Lo domains than BACE1. These effects of membrane composition could impact dimerization of the proteins with APP, preventing or augmenting their APP-cleaving activity. Although this work does not address the question of where ADAM10 and BACE1 cleave APP directly, it provides insight into the influence of membrane composition on the structures and fluctuations of the TMDs of ADAM10 and BACE1. With this foundation of knowledge, the TMD models developed here can be used in future studies exploring their partitioning between Lo and Ld membrane domains and detailed interactions with APP believed to be essential elements of the biogenesis of Aβ and the amyloid cascades of AD. Frames from the all-atom explicit membrane systems for each protein congener model are provided in the Data S1. A coordinate file containing the ADAM10 model in a 7:3 DPPC:CHOL bilayer, Data S2. A coordinate file containing the ADAM10 model in a 9:1 DIPC:CHOL bilayer, Data S3. A coordinate file containing the BACE1 model in a 7:3 DPPC:CHOL bilayer, Data S4. A coordinate file containing the BACE1 model in a 9:1 DIPC:CHOL bilayer for the reader’s convenience.
Author contributions
This research was designed by C.B.A., L.X., G.A.P., and J.E.S. Implicit membrane with all-atom protein REMD simulations were performed by L.X. and G.A.P. and analyzed by L.X., G.A.P., and C.B.A. Coarse-grained and all-atom conventional MD simulations were performed and analyzed by C.B.A. The article was written by C.B.A., G.A.P., and J.E.S.
Acknowledgments
This research was generously supported by the National Institutes of Health (grant R01 GM107703). All simulations were completed on Boston University’s Shared Computing Cluster (SCC) and on the Pittsburg Supercomputing Center’s Bridges-2 computer. The use of Bridges-2 was made possible through a generous allocation grant from the National Science Foundation’s ACCESS (previously XSEDE) program. We thank Ayan Majumder, Seulki Kwon, Jonathan Harris, and Harrison Reiter for our many helpful scientific discussions and Yeol Kyo Choi for guidance on backmapping the systems from Martini2.2 to CHARMM36 representation. We are grateful to our reviewers for their thoughtful consideration of our manuscript and their valuable suggestions that helped us strengthen this article.
Declaration of interests
The authors declare no competing interests.
Editor: Chris Neale.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.08.025.
Supporting material
References
- 1.Chen G.-F., Xu T.-H., et al. Xu H.E. Amyloid beta: structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017;38:1205–1235. doi: 10.1038/aps.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X., Zhou X., et al. Song W. Modifications and trafficking of APP in the pathogenesis of Alzheimer’s disease. Front. Mol. Neurosci. 2017;10:294. doi: 10.3389/fnmol.2017.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tolar M., Abushakra S., Sabbagh M. The path forward in Alzheimer’s disease therapeutics: Reevaluating the amyloid cascade hypothesis. Alzheimers Dement. 2020;16:1553–1560. doi: 10.1016/j.jalz.2019.09.075. [DOI] [PubMed] [Google Scholar]
- 4.Feringa F.M., Van der Kant R. Cholesterol and Alzheimer’s disease; from risk genes to pathological effects. Front. Aging Neurosci. 2021;13:690372. doi: 10.3389/fnagi.2021.690372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baoukina S., Mendez-Villuendas E., et al. Tieleman D.P. Computer simulations of the phase separation in model membranes. Faraday Discuss. 2013;161:63–75. doi: 10.1039/c2fd20117h. [DOI] [PubMed] [Google Scholar]
- 6.Pantelopulos G.A., Straub J.E. Regimes of complex lipid bilayer phases induced by cholesterol concentration in MD simulation. Biophys. J. 2018;115:2167–2178. doi: 10.1016/j.bpj.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veatch S.L., Keller S.L. Seeing spots: complex phase behavior in simple membranes. BBA-Mol. Cell. Res. 2005;1746:172–185. doi: 10.1016/j.bbamcr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Refolo L.M., Malester B., et al. Pappolla M.A. Hypercholesterolemia accelerates the Alzheimer’s amyloid pathology in a transgenic mouse model. Neurobiol. Dis. 2000;7:321–331. doi: 10.1006/nbdi.2000.0304. [DOI] [PubMed] [Google Scholar]
- 9.Abad-Rodriguez J., Ledesma M.D., et al. Dotti C.G. Neuronal membrane cholesterol loss enhances amyloid peptide generation. J. Cell Biol. 2004;167:953–960. doi: 10.1083/jcb.200404149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Refolo L.M., Pappolla M.A., et al. Duff K.E. A cholesterol-lowering drug reduces β-amyloid pathology in a transgenic mouse model of Alzheimer’s disease. Neurobiol. Dis. 2001;8:890–899. doi: 10.1006/nbdi.2001.0422. [DOI] [PubMed] [Google Scholar]
- 11.Fassbender K., Simons M., et al. Hartmann T. Simvastatin strongly reduces levels of Alzheimer’s disease β-amyloid peptides Aβ42 and Aβ40 in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2001;98:5856–5861. doi: 10.1073/pnas.081620098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehehalt R., Keller P., et al. Simons K. Amyloidogenic processing of the Alzheimer β-amyloid precursor protein depends on lipid rafts. J. Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simons M., Keller P., et al. Simons K. Cholesterol depletion inhibits the generation of β-amyloid in hippocampal neurons. Proc. Natl. Acad. Sci. USA. 1998;95:6460–6464. doi: 10.1073/pnas.95.11.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abramov A.Y., Ionov M., et al. Duchen M.R. Membrane cholesterol content plays a key role in the neurotoxicity of β-amyloid: implications for Alzheimer’s disease. Aging Cell. 2011;10:595–603. doi: 10.1111/j.1474-9726.2011.00685.x. [DOI] [PubMed] [Google Scholar]
- 15.Kojro E., Gimpl G., et al. Fahrenholz F. Low cholesterol stimulates the nonamyloidogenic pathway by its effect on the α-secretase ADAM 10. Proc. Natl. Acad. Sci. USA. 2001;98:5815–5820. doi: 10.1073/pnas.081612998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kojro E., Füger P., et al. Postina R. Statins and the squalene synthase inhibitor zaragozic acid stimulate the non-amyloidogenic pathway of amyloid-β protein precursor processing by suppression of cholesterol synthesis. J. Alzheimers Dis. 2010;20:1215–1231. doi: 10.3233/JAD-2010-091621. [DOI] [PubMed] [Google Scholar]
- 17.Harris B., Pereira I., Parkin E. Targeting ADAM10 to lipid rafts in neuroblastoma SH-SY5Y cells impairs amyloidogenic processing of the amyloid precursor protein. Brain Res. 2009;1296:203–215. doi: 10.1016/j.brainres.2009.07.105. [DOI] [PubMed] [Google Scholar]
- 18.Reiss K., Cornelsen I., et al. Bhakdi S. Unsaturated fatty acids drive disintegrin and metalloproteinase (ADAM)-dependent cell adhesion, proliferation, and migration by modulating membrane fluidity. J. Biol. Chem. 2011;286:26931–26942. doi: 10.1074/jbc.M111.243485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjannet S., Elagoz A., et al. Seidah N.G. Post-translational processing of β-secretase (β-amyloid-converting enzyme) and its ectodomain shedding: the pro-and transmembrane/cytosolic domains affect its cellular activity and amyloid-β production. J. Biol. Chem. 2001;276:10879–10887. doi: 10.1074/jbc.M009899200. [DOI] [PubMed] [Google Scholar]
- 20.Vetrivel K.S., Meckler X., et al. Thinakaran G. Alzheimer Disease Aβ production in the absence of S-palmitoylated-dependent targeting of BACE1 to lipid rafts. J. Biol. Chem. 2009;284:3793–3803. doi: 10.1074/jbc.M808920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsons R.B., Austen B.M. Protein lipidation of BACE. Biochem. Soc. Trans. 2005;33:1091–1093. doi: 10.1042/BST20051091. [DOI] [PubMed] [Google Scholar]
- 22.Parsons R.B., Austen B.M. Protein–protein interactions in the assembly and subcellular trafficking of the BACE (β-site amyloid precursor protein-cleaving enzyme) complex of Alzheimer’s disease. Biochem. Soc. Trans. 2007;35:974–979. doi: 10.1042/BST0350974. [DOI] [PubMed] [Google Scholar]
- 23.Motoki K., Kume H., et al. Araki W. Neuronal β-amyloid generation is independent of lipid raft association of β-secretase BACE1: analysis with a palmitoylation-deficient mutant. Brain Behav. 2012;2:270–282. doi: 10.1002/brb3.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrew R.J., Fernandez C.G., et al. Thinakaran G. Lack of BACE1 S-palmitoylation reduces amyloid burden and mitigates memory deficits in transgenic mouse models of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2017;114:E9665–E9674. doi: 10.1073/pnas.1708568114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Díaz M., Fabelo N., et al. Marín R. Biophysical alterations in lipid rafts from human cerebral cortex associate with increased BACE1/AβPP interaction in early stages of Alzheimer’s disease. J. Alzheimers Dis. 2015;43:1185–1198. doi: 10.3233/JAD-141146. [DOI] [PubMed] [Google Scholar]
- 26.Capone R., Tiwari A., et al. Kenworthy A.K. The C99 domain of the amyloid precursor protein resides in the disordered membrane phase. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munter L.M., Sieg H., et al. Multhaup G. Model peptides uncover the role of the β-secretase transmembrane sequence in metal ion mediated oligomerization. J. Am. Chem. Soc. 2013;135:19354–19361. doi: 10.1021/ja410812r. [DOI] [PubMed] [Google Scholar]
- 28.Liebsch F., Aurousseau M.R.P., et al. Multhaup G. Full-length cellular β-secretase has a trimeric subunit stoichiometry, and its sulfur-rich transmembrane interaction site modulates cytosolic copper compartmentalization. J. Biol. Chem. 2017;292:13258–13270. doi: 10.1074/jbc.M117.779165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bittner H.J., Guixà-González R., Hildebrand P.W. Structural basis for the interaction of the beta-secretase with copper. BBA-Biomembranes. 2018;1860:1105–1113. doi: 10.1016/j.bbamem.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 30.UniProt Consortium UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023;51:D523–D531. doi: 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vist M.R., Davis J.H. Phase Equilibria of Cholesterol/Dipalmitoylphosphatidylcholine Mixtures: 2H Nuclear Magnetic Resonance and Differential Scanning Calorimetry. Biochemistry. 1990;29:451–464. doi: 10.1021/bi00454a021. [DOI] [PubMed] [Google Scholar]
- 32.Van Rossum G., Drake F.L. CreateSpace; 2009. Python 3 Reference Manual. [Google Scholar]
- 33.Harris C.R., Millman K.J., et al. Oliphant T.E. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Virtanen P., Gommers R., et al. van Mulbregt P., SciPy 10 Contributors SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedregosa F., Varoquaux G., et al. Duchesnay E. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 36.Michaud-Agrawal N., Denning E.J., et al. Beckstein O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 38.Frishman D., Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 39.Persistence of Vision Pty. Ltd . 2004. Persistence of Vision Raytracer.http://www.povray.org/ [Google Scholar]
- 40.Hunter J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- 41.Im W., Lee M.S., Brooks C.L., III Generalized born model with a simple smoothing function. J. Comput. Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 42.Im W., Feig M., Brooks C.L., III An implicit membrane generalized born theory for the study of structure, stability, and interactions of membrane proteins. Biophys. J. 2003;85:2900–2918. doi: 10.1016/S0006-3495(03)74712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brooks B.R., Brooks C.L., III, et al. Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pantelopulos G.A., Straub J.E., et al. Sugita Y. Structure of APP-C991–99 and implications for role of extra-membrane domains in function and oligomerization. BBA-Biomembranes. 2018;1860:1698–1708. doi: 10.1016/j.bbamem.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang J., Rauscher S., et al. MacKerell A.D. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monticelli L., Kandasamy S.K., et al. Marrink S.-J. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theor. Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 47.de Jong D.H., Singh G., et al. Marrink S.J. Improved parameters for the martini coarse-grained protein force field. J. Chem. Theor. Comput. 2013;9:687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 48.Marrink S.J., Risselada H.J., et al. De Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 49.Ward J.H., Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963;58:236–244. [Google Scholar]
- 50.Rousseeuw P.J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. [Google Scholar]
- 51.Bezanson J., Edelman A., et al. Shah V.B. Julia: A fresh approach to numerical computing. SIAM Rev. 2017;59:65–98. [Google Scholar]
- 52.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 53.Wassenaar T.A., Ingólfsson H.I., et al. Marrink S.J. Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J. Chem. Theor. Comput. 2015;11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- 54.Lindahl E., Abraham M.J., et al. van der Spoel D. 2021. GROMACS Source Code [Manual] [DOI] [Google Scholar]
- 55.Abraham M.J., Murtola T., et al. Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 56.Berendsen H.J.C., Postma J.P.M., et al. Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 57.Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 58.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wassenaar T.A., Pluhackova K., et al. Tieleman D.P. Going backward: a flexible geometric approach to reverse transformation from coarse grained to atomistic models. J. Chem. Theor. Comput. 2014;10:676–690. doi: 10.1021/ct400617g. [DOI] [PubMed] [Google Scholar]
- 60.Martínez L., Andrade R., et al. Martínez J.M. PACKMOL: A package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 2009;30:2157–2164. doi: 10.1002/jcc.21224. [DOI] [PubMed] [Google Scholar]
- 61.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126 doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 62.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984;52:255–268. [Google Scholar]
- 63.Hoover W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 64.Zhang S., Krieger J.M., et al. Bahar I. ProDy 2.0: increased scale and scope after 10 years of protein dynamics modelling with Python. Bioinformatics. 2021;37:3657–3659. doi: 10.1093/bioinformatics/btab187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ichiye T., Karplus M. Collective motions in proteins: a covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins. 1991;11:205–217. doi: 10.1002/prot.340110305. [DOI] [PubMed] [Google Scholar]
- 66.Kitao A., Hirata F., Gō N. The effects of solvent on the conformation and the collective motions of protein: normal mode analysis and molecular dynamics simulations of melittin in water and in vacuum. Chem. Phys. 1991;158:447–472. [Google Scholar]
- 67.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr., Sect. A. 1976;32:922–923. [Google Scholar]
- 68.Hao M., Zhang L., Chen P. Membrane Internalization Mechanisms and Design Strategies of Arginine-Rich Cell-Penetrating Peptides. Int. J. Mol. Sci. 2022;23:9038. doi: 10.3390/ijms23169038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.