Abstract
Endogenous retroviruses are abundant components of mammalian genomes descended from ancient germline infections. In several mammals, the envelope proteins encoded by these elements protect against exogenous viruses, but this activity has not been documented with endogenously expressed envelopes in human. We report that our genome harbors a large pool of envelope-derived sequences with the potential to restrict retroviral infection. To test this, we characterized an envelope-derived protein, Suppressyn. We found that Suppressyn is expressed in preimplantation embryos and developing placenta using its ancestral retroviral promoter. Cell culture assays showed that Suppressyn, and its hominoid orthologs, could restrict infection by extant mammalian type D retroviruses. Our data support a generalizable model of retroviral envelope cooption for host immunity and genome defense.
One-Sentence Summary:
The human genome expresses a vast pool of envelope sequences of retroviral origin one of which can restrict zoonotic viruses.
Viruses pose a persistent threat to human health and may be potent evolutionary drivers of immune adaptation (1–3). It is therefore critical to identify host factors that protect against viral infection. Endogenous retroviruses (ERV) are abundant components of mammalian genomes and are descended from ancient retroviral germline infections (1). ERV sequences are a source of novel genes in evolution (2). ERV-derived envelopes (env) are interesting because they can serve as antiviral factors by binding to and competing for target cell receptors of exogenous viruses (1). Env-derived proteins restricting against contemporary infectious retroviruses have been documented in mouse (Fv4), cat (Refrex-1), and sheep (enJSS6A1) but not in humans (1, 2, 4). The receptor-binding surface domain of an env can be sufficient to restrict infection by this interference mechanism (4). Because the human genome contains thousands of remnant retroviral sequences with coding potential (5), we hypothesized that it includes envelope sequences with the capacity to restrict against modern viruses.
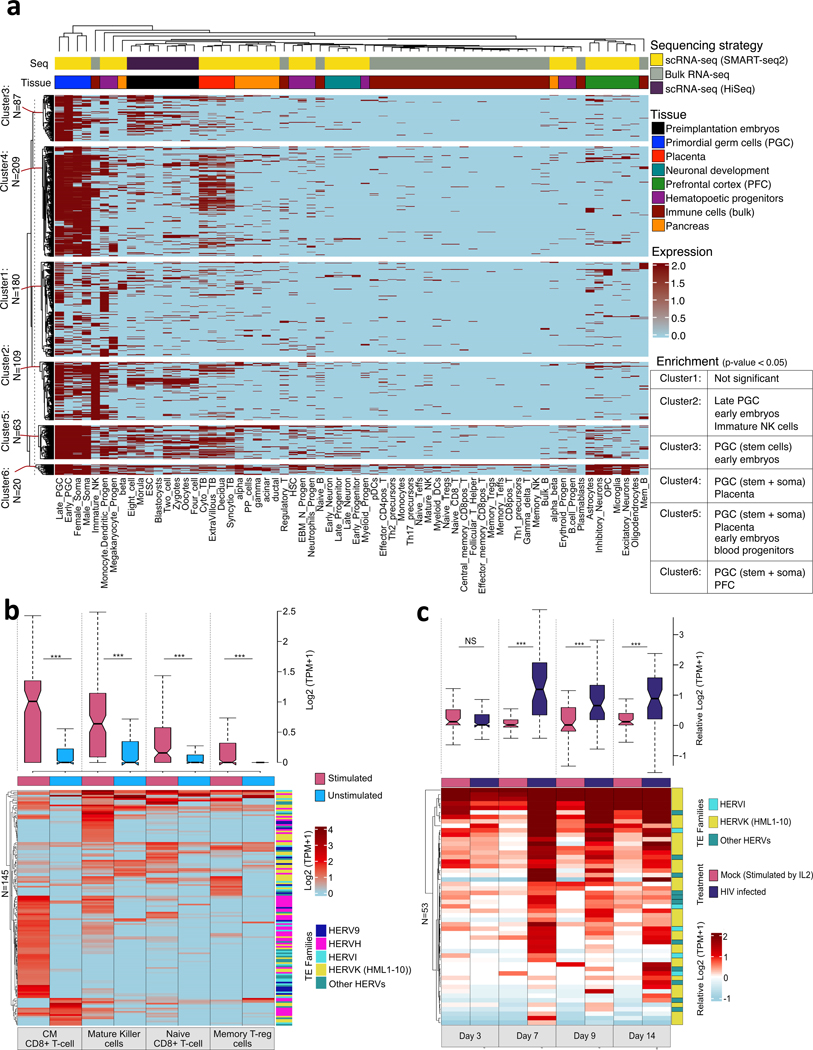
We scanned the human genome for potentially antiviral env-derived open reading frames (envORF) that encode at least 70 aa and are predicted to include the receptor-binding surface domain (see Methods). This search identified 1,507 unique envORFs, including ~20 env-derived sequences currently annotated as human genes such as Supppressyn (SUPYN), Syncytin-1 and Syncytin-2 (6, 7). This number is consistent with a previous genome-wide scan for ERV-derived ORFs predicting 1,731 ORFs with homology to retroviral envelopes (5). Next, we mined transcriptome datasets generated from human embryos and various adult tissues (Table S1) and observed that ~44% of envORFs (668/1507) showed evidence of RNA expression in at least one of the cell types surveyed (Fig. 1A). These analyses revealed three general trends about expressed envORFs: (i) like known env-derived genes, envORFs exhibit tissue-specific expression patterns, but some are widely expressed (HERVK13, ERV3) (Fig. 1A; fig. S1, S2); (ii) the majority of envORFs are expressed during early human development, often in stem and primordial germ cells, and/or in placenta (Fig. 1A; fig. S1); (iii) with the exception of brain, envORFs are rarely expressed in differentiated tissues under normal conditions (Fig. 1A; fig. S1, S2). Since antiviral factors are generally expressed in immune cells and/or induced upon immune stimulation or infection, we also profiled envORF expression using transcriptome datasets generated from various immune cells, including resting, stimulated, and HIV-infected cells (Table S1). These analyses revealed that a substantial fraction of envORFs (145 loci from multiple families) are expressed in immune cells and tend to be induced by immune stimulation (Fig. 1B; fig. S3). Consistent with previous reports (8), we observed that a subset of envORFs are induced upon HIV infection (Fig. 1C; fig. S4). These data suggest that the human genome harbors a vast reservoir of env-derived sequences with coding potential for receptor-binding proteins. Many are expressed in a tissue-specific or infection-inducible fashion, suggesting that they may function as antiviral factors.
Fig. 1. Expression profile of env-derived transcripts over a subset of human cell types.
(A-C) Heatmaps show envORF locus (rows) expression (log2 CPM > 1) in tissues (columns) (A), and stimulated immune (B) or HIV infected CD4+ T-cells (C). Bars above heatmaps denote sequencing strategy (A), tissue (A) or treatment (B, C) with the same color scheme in figures S1, S3, and S4. Rows and columns are ordered by hierarchical clustering of envORF expression. Significant envORF cluster enrichment was calculated by hypergeometric test. (B, C) Boxplots represent distribution of envORF expression levels relative to unstimulated (B) and Mock-infected cells (C). For (C), envORF expression is averaged across donors (n = 3) and normalized to mean expression of unstimulated cells (n = 6) (see fig. S4). (NS: not significant; ***p < 0.01; Wilcoxon rank-sum test)
As a paradigm to test the antiviral activity of an envORF, we focused on SUPYN for two primary reasons. First, SUPYN was reported to be expressed in the developing placenta, as we observe for many envORFs, a tissue that can be vulnerable to viral infection and a barrier against vertical transmission to the fetus (9). Second, SUPYN lacks a transmembrane domain but retains the ability to bind the amino acid transporter ASCT2 (also known as SLC1A5), which is the receptor for a diverse group of retroviruses, including endogenous (e.g. HERV-W) and exogenous viruses such as RD114 in cat and simian retrovirus (SRV) in Old World monkeys (6, 10–12). These viruses are collectively referred to as RD114 and Type D (RDR) retroviruses (12). We hypothesized that SUPYN might restrict RDR retroviruses by virtue of its binding to ASCT2.
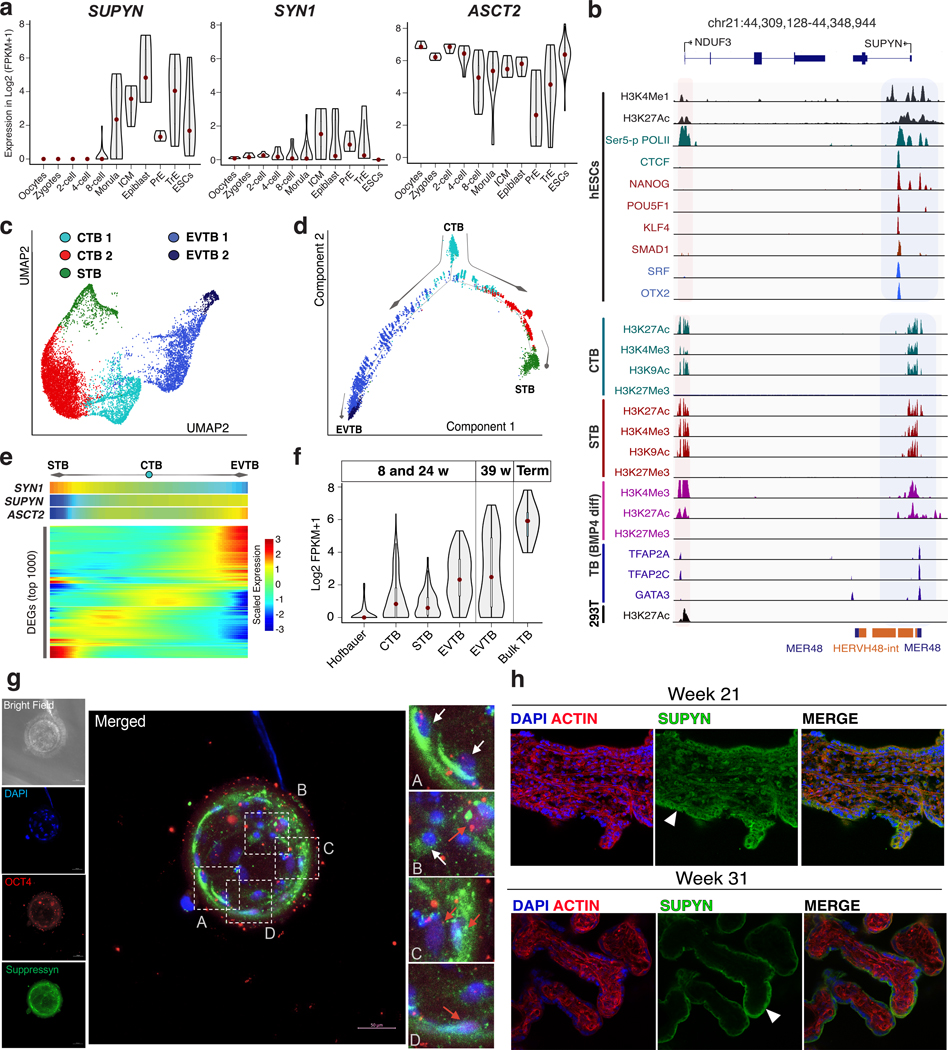
To obtain a more detailed view of SUPYN expression and regulation in human development and tissues, we analyzed publicly available bulk and single-cell RNA-seq, ATAC-seq, DNase-seq, and ChIP-seq datasets generated from adult tissues, preimplantation embryos, human embryonic stem cell lines (hESC), in vitro trophoblast (TB) differentiation models, and placenta explants isolated at multiple stages of pregnancy (Table S1). Apart from testis and cerebellum, SUPYN expression is generally low (<1 log2 (TPM+1)) or absent in adult tissues (fig. S2). SUPYN is expressed at high levels in preimplantation embryos from the 8-cell stage of development (Fig. 2A). Accordingly, the SUPYN promoter region, which corresponds to the long terminal repeat (LTR) of the ancestral HERVH48 provirus, is marked by several peaks of accessible chromatin from 8-cell to blastocyst stages (fig. S5B, C). In hESCs, SUPYN RNA is abundant (Fig. 2A, fig. S5A, C) and its promoter region is marked by histone modifications characteristic of transcriptionally active chromatin (H3K4me1, H3K4Me3, H3K27ac) and bound by core pluripotency (OCT4, NANOG, KLF4, SMAD1) and self-renewal (SRF, OTX2) transcription factors (Fig. 2B, fig. S5C). During hESC to TB differentiation, pluripotency factors (NANOG, OCT4) occupying the SUPYN promoter region are replaced by TB-specific transcription factors (TFAP2A, GATA3); and active chromatin marks (H3K27ac, H3K4me3, H3K9ac) are maintained across all analyzed TB lineages (Fig. 2B). These observations indicate SUPYN expression persists through the TB differentiation process. We next mined single-cell RNA-seq data generated from placenta at multiple developmental stages. After classifying cell clusters based on known markers, we found SUPYN and ASCT2 expression was relatively high in cytotrophoblasts (CTB) and extra-villous trophoblasts (EVTB), but also detectable in syncytiotrophoblasts (STB) (Fig. 2C–F, fig. S6). To confirm these transcriptomic observations, we performed immunostaining of SUPYN in preimplantation embryos as well as second and third trimester placentas. In early embryos, SUPYN is primarily detectable in the trophectoderm, which will give rise to placenta, and in some OCT4-expressing cells, which mark pluripotent stem cells of blastocysts (Fig. 2G, fig. S7, S8). In placenta, SUPYN is widely expressed in STB and potentially CTB of second trimester placental villi (Fig. 2H, fig. S9). The combined transcriptome, genome regulatory data and immunohistochemical staining establish that SUPYN is expressed throughout human fetal development (Fig. 2, fig. S5–9). These analyses also indicate that SUPYN transcription consistently initiates from its ancestral HERVH48 provirus (Fig. 2B, fig. S5B, C, S13).
Fig. 2. Pluripotency and placentation regulatory factors drive SUPYN expression during fetal development.
(A) Violin plots represent single-cell SUPYN, SYN1 and ASCT2 expression in human preimplantation embryo (PrE: primitive endoderm, TrE: trophectoderm) and ESC. (B) Genome browser view of regulatory elements surrounding the SUPYN locus in hESCs, CTB, STB, BMP4-differentiated TB, and 293T cells. ChIP-seq profiles show indicated transcription factors and histone modifications. Shaded area highlights regions of active chromatin. (C) UMAP visualization of TB cell clusters (see fig S6). (D) Monocle2 pseudotime analysis of cell clusters in (C) illustrates developmental trajectory of CTBs to STB and EVTB. (E) Heatmap represents top 1000 differentially expressed genes (row) of single cells (column). Cells are ordered according to pseudotime progression in (D). SYN1, SUPYN, and ASCT2 were fetched from heatmap below. (F) Violin plots denote single-cell SUPYN expression in placental-cell lineages throughout indicated pregnancy stages. (G, H) Confocal microscopy of human preimplantation embryo (G) and placental villus explants (H) stained for SUPYN (green), OCT4 (red), ACTIN (red) and DAPI (blue). Trophectoderm (white arrows) and OCT4-expressing (red arrows) cells are highlighted in (G) subpanels (A-C). STBs are marked by arrowheads (H).
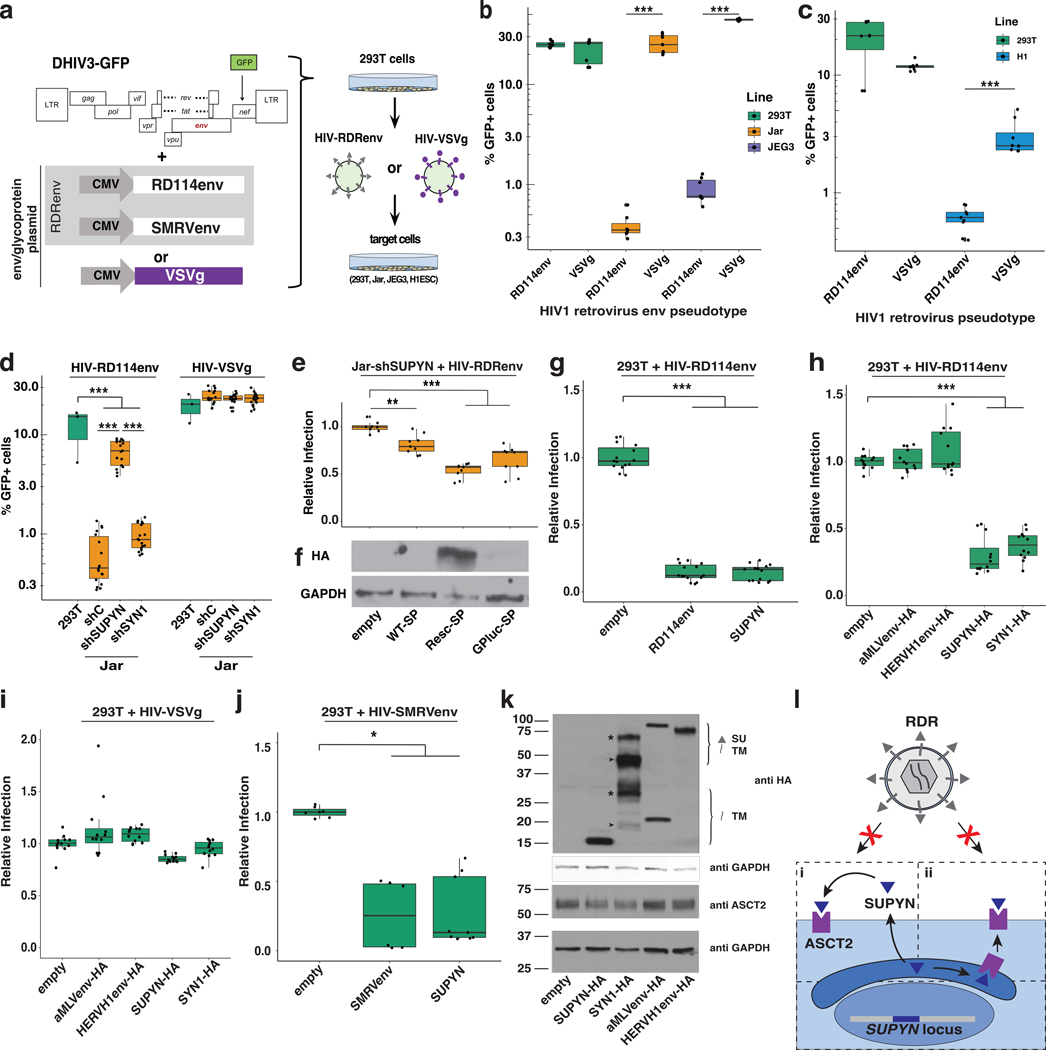
The expression profile of SUPYN is consistent with the hypothesis that it could protect the developing embryo and germline against RDR infection. We first examined whether human placenta-derived Jar and JEG3 cell lines, and H1 hESC, which robustly express SUPYN and ASCT2 (fig. S10, 11A) (10) are resistant to RDR env-mediated infection. We generated HIV-EGFP viral particles pseudotyped with either RD114 env (HIV-RD114) or vesicular stomatitis virus glycoprotein G (HIV-VSVg), which uses the LDL receptor (13), to monitor the level of infection in cell culture based on EGFP expression (Fig. 3A, fig S12). These experiments revealed that Jar, JEG3, and H1 cells were susceptible to HIV-VSVg, as previously reported (14, 15), but resistant to HIV-RD114 infection (Fig. 3B, C). By contrast, 293T cells, which do not express SUPYN (fig. S5A), were similarly susceptible to infection by HIV-RD114 and HIV-VSVg (Fig. 3B, C). To test whether SUPYN contributes to the HIV-RD114 resistance phenotype, we repeated these infection experiments in Jar cells engineered to stably express short hairpin RNAs (shRNA) depleting ~80% of SUPYN mRNA (fig S11A). Depletion of SUPYN in Jar cells resulted in a significant increase in susceptibility to HIV-RD114 infection but not HIV-VSVg (Fig. 3D). To confirm that SUPYN expression confers RD114 resistance and control for potential off-target effects of SUPYN-targeting shRNAs, we transfected Jar-shSUPYN cells with two shRNA-resistant, HA-tagged SUPYN rescue constructs and examined their susceptibility to HIV-RD114 infection. The shRNA-targeted SUPYN signal peptide sequence was replaced with a luciferase (SUPYN-lucSP) or modified signal peptide sequence (SUPYN-rescSP) that disrupts shRNA-binding but retains codon identity (see Methods). Transfection with either SUPYN-rescSP or SUPYN-lucSP restored a significant level of resistance to HIV-RD114 infection (Fig. 3E). Western blot analysis of transfected cell lysates showed SUPYN-rescSP was more abundantly expressed than SUPYN-lucSP (Fig. 3F), which may account for the stronger HIV-RD114 resistance conferred by the former (Fig. 3E). These results suggest that SUPYN expression is at least partially responsible for preventing Jar cells from RD114env-mediated infection.
Fig. 3. SUPYN confers resistance to RD114 env-mediated infection.
(A) HIV-glycoprotein reporter virus production and infection assay approach (see Methods). (B-D) Proportion of GFP+ 293T, JEG3, Jar (B), 293T, H1-ESCs (C), and 293T, shSUPYN-Jar (D) cells infected with indicated reporter virus pseudotypes. (E) Relative infection rates of shSUPYN-Jar cells transfected with HA-tagged wild-type (WT-SP), rescue signal peptide (Resc-SP), or luciferase signal peptide (GPluc-SP) encoding SUPYN overexpression constructs. (F) Western Blot analysis (αHA, αGAPDH) of Jar-shSUPYN cell lysates transfected with indicated SUPYN overexpression constructs. (G-J) Relative infection rates of 293T cells transfected with unmodified RD114env (G), SUPYN (G, J), SMRVenv (J) or indicated HA-tagged protein overexpression plasmids and infected with indicated reporter viruses (top). (K) Representative Western blot analysis (αHA, αGAPDH, αASCT2) of 293T cell lysates following transfection with indicated constructs. Arrowheads and asterisks denote unglycosylated and glycosylated protein fragments respectively. (I) Model of SUPYN-dependent RDR infection restriction. SUPYN likely binds ASCT2 following secretion (I) or within the secretory compartment (ii). (n ≥ 3 with ≥ 2 technical replicates; ***adj. p < 0.001; **adj. p < 0.01; *adj. p < 0.05; ANOVA with Tukey HSD)
To test if SUPYN expression alone is sufficient to confer protection against RD114env-mediated infection, we transfected 293T cells with unmodified or HA-tagged SUPYN overexpression constructs and subsequently infected with HIV-RD114 and HIV-VSVg respectively. As a positive control, we also transfected 293T cells with an unmodified RD114env overexpression construct, which is predicted to confer resistance to HIV-RD114. Expression of either SUPYN or RD114env resulted in ~80% reduction in the level of HIV-RD114 infection (Fig. 3G, H) but had no significant effect on HIV-VSVg infectivity (Fig. 3I). Taken together, our knockdown and overexpression experiments indicate SUPYN expression is sufficient to confer resistance to RD114env-mediated infection.
Our RD114env-specific resistance phenotype (Fig. 3G–I) suggests SUPYN restricts viral entry via receptor interference. If so, this protective effect should extend to infection mediated by other RDRenv (11, 16, 17). To test this, we generated HIV-GFP reporter virions pseudotyped with squirrel monkey retrovirus env (HIV-SMRVenv) (11) (Fig. 3J) and infected 293T cells previously transfected with SUPYN, SMRVenv (as a positive control) or an empty vector. Cells expressing SUPYN or SMRVenv showed an ~80% reduction of HIV-SMRVenv infected cells (Fig. 3J), indicating SUPYN can restrict infection mediated by multiple RDRenv.
Another prediction of RDR restriction via receptor interference is that resistance should be conferred by env that bind ASCT2, such as SUPYN, but not those binding other cellular receptors. Consistent with this prediction and previous work with overexpressed SYN1 (18), expressing HA-tagged SUPYN and SYN1 strongly restricted HIV-RD114, while HA-tagged env from amphotrophic murine leukemia virus (aMLV) or human endogenous retrovirus H (HERVH), neither of which are expected to interact with ASCT2 (19–21), had no effect on HIV-RD114 nor HIV-VSVg infection in 293T cells (Fig. 3H, I). All tested env proteins were expressed at comparable levels (Fig. 3K). Furthermore, we observed that SUPYN overexpression led to a decrease in ASCT2 protein level in 293T cells (Fig. 3K, fig. S11E, F). This result suggests that if SUPYN acts by receptor interference, its interaction with ASCT2 leads to partial receptor degradation, which is consistent with some instances of receptor interference (22, 23). In agreement with previous observations (6, 24), we noted that SUPYN knockdown in Jar cells resulted in selective loss of a putative un-glycosylated isoform of ASCT2 (fig. S11B–D). We speculate that the presence of SUPYN-dependent un-glycosylated ASCT2 may result from SUPYN sterically interfering with glycosylation machinery present within the secretory pathway. Collectively, these observations converge on the model that SUPYN restricts against RDR infection through receptor interference by two possible mechanisms that are not mutually exclusive: SUPYN may bind ASCT2 within the secretory pathway or extracellularly following secretion (Fig. 3L).
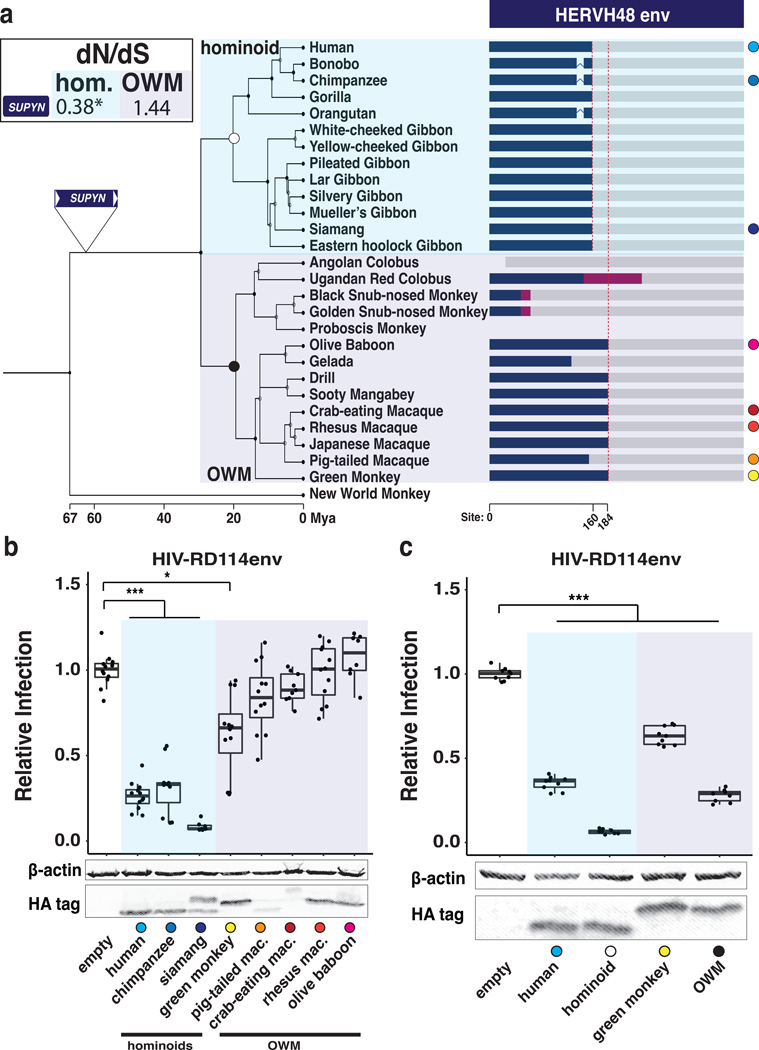
To gain insights into the evolutionary emergence of SUPYN antiviral activity, we used comparative genomics to investigate the origin and functional constraint acting on SUPYN. We found that the HERVH48 provirus from which SUPYN is derived is present at orthologous position across the genomes of all available hominoids and most Old World monkeys (OWM), but is absent in other primate lineages (Fig. 4A, fig. S13). Thus, the provirus that gave rise to SUPYN inserted in the genome of a catarrhine primate ancestor ~27–38 million years ago (25) (Fig. 4A). We next examined whether SUPYN ORFs have evolved under functional constraint during primate evolution. The SUPYN ORF is almost perfectly conserved in length across hominoids, but not in OWM where some species display frameshifting and truncating mutations (Fig. 4A, fig. S14, S15), suggesting SUPYN evolved under different evolutionary regimes in hominoids and OWMs. To test this idea, we analyzed the ratio (𝜔) of nonsynonymous (dN) to synonymous (dS) substitution rates using codeml (26), which provides a measure of selective constraint acting on codons. Log-likelihood ratio tests comparing models of neutral evolution with selection indicate SUPYN evolved under purifying selection in hominoids (ω = 0.38; p = 1.47E-02) but did not depart from neutral evolution in OWMs (ω = 1.44; p = 0.29) (Fig. 4A). These results indicate that SUPYN antiviral activity may be conserved across hominoids but not in OWM. To assess this, we transfected 293T cells with HA-tagged overexpression constructs for the orthologous SUPYN sequences of two hominoid species (chimpanzee, siamang), and five OWM species (African green monkey, pigtailed macaque, crab-eating macaque, rhesus macaque, olive baboon) and challenged these cells with HIV-RD114. Both chimpanzee and siamang SUPYN proteins displayed antiviral activity with potency comparable to or greater than human SUPYN, respectively (Fig. 4B). By contrast, of the five OWM, only African green monkey SUPYN exhibited a modest but significant level of antiviral activity (Fig. 4B, C). The lack of restriction activity for some OWM proteins may be attributed to their relatively low expression level in these human cells (Fig. 4B) and/or their inability to bind the human ASCT2 receptor due to SUPYN sequence divergence (fig. S14, S15). To further trace the evolutionary origins of SUPYN antiviral activity, we expressed ancestral SUPYN sequences phylogenetically reconstructed for the hominoid and OWM ancestors (Fig. S15) and assayed their antiviral activity in 293T cells. Both ancestral proteins were expressed at levels comparable to human and African green monkey SUPYN and exhibited strong restriction activity (Fig. 4C). These data indicate that SUPYN antiviral activity against RDRenv-mediated infection is an ancestral trait preserved over ~20 million years of hominoid evolution, but apparently lost in several OWM lineages.
Fig. 4. SUPYN emerged in a Catarrhine ancestor and has conserved antiviral activity in Hominoids.
(A) Consensus primate phylogeny with cartoon representation of SUPYN ORFs (blue box). Magenta boxes represent SUPYN ORF frameshifts. Red dashed lines denote conserved premature stop codon positions. Grey bars represent degraded HERVH48env sequence. Labeled triangle denotes ancestral lineage where HERVH48env was acquired. Colored circles indicate species used in (B) and (C). SUPYN dN/dS values are shown in box (*p < 0.05; LRT). (B, C) Relative infection rates and Western Blot of 293T cells transfected with primate (B) or ancestral (C) SUPYN-HA constructs and infected with HIV-RD114env. Relative infection rates were determined by normalizing GFP+ counts to empty vector. (n ≥ 3 with ≥ 2 technical replicates ***adj. p < 0.001; *adj. p < 0.05; ANOVA with Tukey HSD)
Because envORFs are predicted to encode proteins with the ability to bind receptors used by their ancestral retroviruses and possibly many contemporary exogenous viruses, we propose that they constitute a reservoir of potential antiviral factors. The prominent expression of envORF, including SUPYN, in the developing germ line suggests that they may have repeatedly acted as a barrier against the vertical propagation of cognate endogenous retroviruses by reinfection (27). This may explain why most HERV families appear to have propagated in the genome primarily via retrotransposition rather than reinfection (28). Under this model, we predict the adaptive benefits of envORFs to be generally transient, unless they have broader antiviral activity or gain additional cellular function. This conjecture may explain why SUPYN has been preserved by natural selection throughout hominoid evolution; not only may SUPYN have shielded the early embryo and nascent germline from the persistent threat of RDRs but also against the adverse effects of SYN1-mediated infections and syncytalization of the developing placenta (6, 10, 29). Our genome may hold many other retrovirus-derived proteins with protective effects against viral infection.
Materials and Methods
Genome-wide search for ERV-derived envelope open reading frames
Candidate envelope open reading frames (envORF) were identified by performing tBLASTn (31) searches of the hg19 human genome assembly using envelope amino acid sequences, obtained from Repbase (32) and published retroviral envelope sequences, as a queries. Each hit was used in reiterative tBLASTn searches, yielding an initial hit list of 82715 candidate envORF sequences. This list of candidates was filtered using the following criteria: (1) EnvORFs must have a length of ≥70 aa. (2) Hits starting at position ≥300 aa were removed because such open reading frames are predicted to encode a portion of the envelope transmembrane domain, which is not expected to play a role in receptor binding. (3) After these processing steps, our list was further condensed to only include unique (non-redundant) open reading frame sequences (n=1507) in the hg19 genome assembly.
RNA-seq analyses
We mined published single cell transcriptome datasets (scRNA-seq) of human pre-implantation embryos isolated at developmental stages ranging from oocyte to blastocyst (33), primordial germ cells (34), human placenta (35–37), neuronal differentiation (38), hematopoietic stem cells (39), pancreas (40), prefrontal cortex (41). Prior to analyzing the transcriptomic datasets, we first recompiled the reference genome annotations using known gene (gencode V19) (42), and envORF sequences to generate a reference transcriptome in fasta format. To guide the transcriptome assembly, we converted this updated genome annotation to gtf format, which was subsequently utilized for our expression quantifications. Reads were mapped to the curated human genome (hg19) with STAR (43) using the following settings --alignIntronMin 20 --alignIntronMax 1000000 --chimSegmentMin 15 --chimJunctionOverhangMin 15 --outFilterMultimapNmax 20. Only uniquely mapped reads were considered for expression calculations. Gene level counts were obtained using featureCounts (44) run with RefSeq annotations. Gene expression levels were calculated at Transcript Per Million (TPM) from counts mapped over the entire gene (defined as any transcript located between Transcription Start Site (TSS) and Transcription End Site (TES)). Only genes and cells that met the following criteria were included in this analysis: (1) each cell must express at least 5,000 genes; (2) each gene must be expressed in at least 1% of cells; (3) each gene must be expressed with log2 TPM >1. We clustered cells meeting these criteria using the default parameters of the Seurat (45) package (v3.1.1) implemented in R (v3.6.0). Seurat applies the most variable genes to get top principal components that are used to discriminate cell clusters in tSNE or UMAP plots. We set Seurat to use 10 principal components in this cluster analysis. For the placental scRNAseq data (Fig. 2, fig. S5), the 2000 most differentially expressed genes were used to define cell clusters. Major clusters corresponding to CTB, STB, EVTB, macrophages, and stromal cells were identified based on the expression of known marker genes. Monocle2 (46) was used to perform single-cell trajectory analysis and cell ordering along an artificial temporal continuum. The top 500 differentially expressed genes were used to distinguish between CTB, STB and EVTB cell populations. The transcriptome from each single cell represents a pseudo-time point along an artificial time vector that denotes the progression of CTB to STB or EVTB respectively.
Data generated on the 10X Genomics scRNA-seq platforms were processed in the following way. Normalized counts and cell-type annotations were used as provided by the original publications. Seurat was used for filtering, normalization, and cell-type identification. The following data processing steps were performed: (1) Cells were filtered based on the criteria that individual cells must have between 1,000 and 5,000 expressed genes with a count ≥1; (2) cells with more than 5% of counts mapping to mitochondrial genes were filtered out; (3) data was normalized by dividing uniquely mapping read counts (defined by Seurat as unique molecular identified (UMI)) for each gene by the total number of counts in each cell and multiplying by 10,000. These normalized values were then natural-log transformed. Cell types were defined by using the top 2000 variable features expressed across all samples. Clustering was performed using the “FindClusters” function with largely default parameters; except resolution was set to 0.1 and the first 20 PCA dimensions were used in the construction of the shared-nearest neighbor (SNN) graph and the generation of 2-dimensional embeddings for data visualization using UMAP. Cell types were assigned based on the annotations provided by the original publication.
Bulk RNAseq datasets generated from the GTEx consortium (47), placenta (48), 293T (49, 50), immune (51, 52) cells, and hESCs (53, 54) were processed as described above. Briefly, reads were mapped with STAR and uniquely mapped reads were counted with featureCounts. We restricted the visualization to only envORFs that were expressed (Log2 TPM > 1) in at least one of the analyzed or compared samples (see the codes submitted on GitHub).
ChIP-seq, DNAse-seq and ATAC-seq data analysis
Various ChIP-seq datasets representing histone modifications and transcription factors in human embryonic stem cells and their differentiation were retrieved from (55, 56). We obtained the H3K4Me3 for hESC (57), H3K27Ac for CTB to STB primary culture (58), H3K4Me1 for trophoblasts (59), H3K4Me3 and H3K27Me3 for differentiated trophoblasts (60), and GATA2/3, TFAP2A/C (60) ChIP-seq datasets in raw fastq format. DNAse-seq and ATAC-seq datasets were retrieved from references (61) and (54) respectively.
Reads from the above-described datasets were aligned to the hg19 human reference genome using Bowtie2 (62) run in --very-sensitive-local mode. All reads with MAPQ < 10 and PCR duplicates were removed using Picard and samtools (63). Peaks were called by MACS2 (64) (https://github.com/macs3-project/MACS) with the parameters in narrow mode for TFs and broad mode for histone modifications keeping FDR < 1%. ENCODE-defined blacklisted regions (65) were excluded from called peaks. We then intersected these peak sets with repeat elements from hg19 repeat-masked coordinates using bedtools intersectBed (66) with a 50% overlap. To visualize over Refseq genes (hg19) using IGV (67), the raw signals of ChIP-seq were obtained from MACS2, using the parameters: -g hs -q 0.01 -B. The conservation track was visualized through UCSC genome browser (68) under net/chain alignment of given non-human primates (NHPs) and merged beneath the IGV tracks.
Cell culture
293T (provided by Dr. Nels Elde; available from ATCC: CRL-3216) cells were cultured in DMEM (GIBCO, 11995065) containing 10% Fetal Bovine Serum (FBS) (GIBCO, 10438026). Jar cells (provided by Dr. Carolyn Coyne; available from ATCC: HTB-144) were cultured in RPMI (GIBCO, 11875093) containing 10% FBS. JEG3 cells (provided by Dr. Carolyn Coyne; available from ATCC: HTB-36) were cultured in MEM (GIBCO, 11095080) containing 10% FBS. Culture medium for these cell lines was supplemented with sodium pyruvate (GIBCO, 11360070), glutamax (GIBCO, 35050061), and Penicillin Streptomycin (GIBCO, 15140122) according to manufacturer specifications. H1-ESCs (69) (obtained from WiCell: WA01) were grown on Matrigel (Corning, 356277) coated plates in MTESR+ (Stemcell, 05825) growth-media and sub-cultured using Accutase (Innovative Cell Technologies, AT-104) and MTESR+ supplemented with CloneR (Stemcell, 05888). All cell lines were cultured at 37°C and 5% CO2.
Human embryos
Prior to starting the project (documentation available upon request), all the procedures to obtain human pre-implantation embryos were approved by local regulatory authorities and the Spanish National Embryo steering committee. After the signing of informed consent documents by parents, all human pre-implantation embryos were donated and fully anonymized. The embryos used in this study were cryopreserved sibling embryos after successful In Vitro Fertilization pregnancies.
Culturing of human pre-implantation embryos
Human pre-implantation embryos at several stages of development (from +2 to +6 days post-fertilization) were thawed and allowed to grow. Only embryos reaching the blastocyst stage and showing viability in most individual cells were selected for further analyses. Cryopreserved human embryos were thawed using thawing media from Vitrolife (G-1 v.5 and G-2 v.5 media), and were classified based on morphological criteria using inverted Hoffman optical microscopy. More than 50% of thawed embryos showed signs of cell division.
Vector cloning
DHIV3-GFP, phCMV-RD114env, psi(−)-amphoMLV plasmids were provided by Vicente Planelles (University of Utah). pCGCG-SMRVenv plasmid was provided by Welkin Johnson (Boston University). psPAX2 and pVSVg plasmids were provided by John Lis (Cornell University). The following cloning approaches were performed using primers and constructs described in Supplementary Table 3. HERVH1env ORF was PCR-amplified using Q5 polymerase (NEB, M0491L) from HeLa and 293T genomic DNA respectively and cloned into a TOPO vector (ThermoFisher, 450245).
To generate stable SYN1 and SUPYN knock-down cell lines, pHIV lentiviral constructs containing shRNAs targeting SYN1 and SUPYN respectively were cloned using the following strategy. The shRNA encoded in pHIV7-U6-shW3, generously provided by Lars Aagaard (Aarhus University), targets SYN1 (70). SUPYN-targeting shRNAs were designed using siRNA sequences employed by Jun Sugimoto4 as a template. pHIV7 lentiviral constructs were cloned using the pHIV7-U6-shW3 plasmid (70) as a template. pHIV7-U6-shSup-cer, pHIV7-U6-shSup-puro, pHIV7-U6-shC-cer, pHIV7-U6-shC-puro, pHIV7-U6-shSyn1-cer, pHIV7-U6-shSyn1-puro were generated using a Gibson assembly approach. To replace the native GFP marker of pHIV7-U6-shW3 with a Cerulean reporter or puromycin resistance marker, we digested pHIV7-U6-shW3 with NheI (NEB, R3131S) and KpnI (NEB, R3142S). This digest resulted in the production of three DNA fragments: pHIV7 backbone, GFP-, and WPRE-containing fragments. We separately PCR amplified each selection marker and WPRE containing pHIV7 fragment.
InFusion cloning was then used to ligate the digested pHIV7 backbone to the Cerulean or puromycin cassette and WPRE containing PCR product. shRNAs were cloned into the pHIV7-Cerulean/puromycin transfer construct previously digested with NotI (NEB, R0189S) and NheI. U6-promoter containing shRNA cassettes and the CMV promoter driving marker cassette expression were PCR amplified and subsequently InFusion cloned into the NotI/NheI digested pHIV7-cerulean/puromycin backbone.
All pHCMVenv and SUPYN expression constructs, described in this study, were generated as follows: HA-tagged and untagged ORFs with pHCMV homologous overhanging sequence were either PCR amplified using Q5 polymerase (NEB, M0491S) or synthesized (IDT) (see Table S2), and cloned into EcoRI (NEB, R3101T) digested pHCMV backbones using the InFusion cloning kit (Takara Bio, 638920). To generate siRNA-resistant SUPYN rescue constructs, we replaced the native signal peptide sequence4 (which is targeted by siRNAs used in this study) with (1) a Gaussia princeps luciferase SP (SUPYN-lucSP) (71, 72) and (2) a shSUPYN resistant SUPYN rescue construct (SUPYN-rescSP) in which the codons were modified to retain the codon identity but disrupt siRNA binding.
Antibodies
All antibodies used in this study are commercially available. α-GAPDH (D4C6R, D16H11), α-βactin (D6A8), α-HA (C29F4), α-ASCT2 (V501) primary antibodies were purchased from Cell Signaling Technology. α-SUPYN and αOCT4 primary antibodies were purchased from Phoenix Pharma (H-059–052) and Santa Cruz Biotechnology respectively. α-Mouse (#7076) and α-Rabbit (#7074) HRP conjugated secondary antibodies were purchased from Cell Signaling Technology. IRDye secondary antibodies were purchased from Licor (925–32211, 925–68072, 925–32210, 925–68073). Alexa-fluor conjugated secondary antibody was purchased from Invitrogen.
Western Blot
Whole cell extracts from cultured cell lines were prepared using 1x GLO lysis buffer (Promega, E266A). One third volume of 4x Laemli buffer was added to one volume whole cell extract samples, then incubated at 95°C for 5 minutes, and sonicated for 15 minutes at 4°C (amplitude 100; pulse interval 15 seconds on, 15 seconds off). Approximately 30 ug of protein were separated by SDS-PAGE (BioRad, 1610175), transferred to PVDF membrane (BioRad, 1620177), blocked according to antibody manufacturers specification, and incubated overnight in appropriate primary antibody then incubated in IRDye or peroxidase conjugated goat anti-mouse or anti-rabbit secondary antibodies for 1 hour at room temperature. Protein was then detected using ECL reagent (BioRad, 1705061) or the Licor Odyssey imaging system.
IF microscopy
For placenta:
Human second trimester placental tissue that resulted from elective terminations was obtained from the University of Pittsburgh Health Sciences Tissue Bank through an honest broker system after approval from the University of Pittsburgh Institutional Review Board (IRB) and in accordance with the University of Pittsburgh’s tissue procurement guidelines. Tissue was excluded in cases of fetal anomalies or aneuploidy. Third trimester placental tissue was obtained through the Magee Obstetric Maternal & Infant (MOMI) Database and Biobank after approval from the University of Pittsburgh IRB. Women who had previously consented for tissue donation and underwent cesarean delivery were included. Placental tissues used in this study were obtained at 18, 23, and unknown weeks gestation with a sex of male, female, and female respectively. Placental tissues were fixed in 4% PFA (in 1x PBS) for 30 minutes, permeabilized with 0.25% Triton X-100 for 30 minutes (on a rocker), washed with 1x PBS and then incubated with primary anti-Suppressyn antibody at 1:200 in 1x PBS for 2–4 hours at room temperature. These samples were incubated with Alexa-fluor conjugated secondary antibody (Invitrogen) diluted 1:1000 and counterstained with actin. DAPI was included in our PBS and then mounted in Vectashield mounting medium with DAPI (Vector Laboratories, H-1200).
For embryo:
Pre-implantation embryos at the blastocyst stage were washed with 1x PBS (Invitrogen) and fixed for 15 minutes using fresh 4% (w/v) paraformaldehyde (Sigma) at room temperature. After fixation, embryos were washed twice on a petri dish using 1x PBS containing 0.1% (v/v) Triton X-100 (Sigma). Next, embryos were permeabilized on a lo-bind 2ml Eppendorf tube using 1x PBS containing 0.5% (v/v) Triton X-100 for 24 hours at 4°C. After permeabilization, embryos were incubated for 24 hours at 4°C in blocking solution [1x PBS containing 0.1% (v/v) Triton X-100, 1% (v/v) Tissue Culture Grade DMSO (Sigma), and 5% (v/v) Normal Goat Serum (Abcam)]. Next, embryos were incubated with the primary antibodies [1:200 anti-Rabbit Suppressyn (Phoenix Pharmaceuticals, Inc), and 1:200 anti-Goat OCT4 (Santa Cruz Biotechnology, Inc)] in fresh blocking solution for 24 hours at 4°C. After 30 minutes at room temperature, embryos were washed twice for 5 minutes using 1x PBS containing 0.1% (v/v) Triton X-100, and subsequently incubated with secondary antibodies in blocking solution containing DAPI (Thermo Scientific) [1:1000 anti-Rabbit Alexa 488 (Life Technologies), and 1:1000 anti-Goat Alexa 555 (Life Technologies)] for 24 hours at 4°C in the dark. Next, embryos were washed twice for 5 minutes using 1x PBS containing 0.1% (v/v) Triton X-100 and mounted in a μ-Slide 8 well (Ibidi®) using 1xPBS and avoiding the generation of air bubbles.
A confocal Zeiss LSM 710 device (Genyo, Spain) was used to process the slides, using a 63X objective, zoom 0.6x, with a 1024×1024 pixels resolution (corresponding to 224.70 μm x 224.70 μm). Fluorescence channels were recorded independently to avoid artifacts, and microscope images shown are maximum projections of several images (2–3 or more). As negative controls, embryos were also incubated with only the secondary antibody.
Virus production
Low passage 293T cells were used to produce lentiviral particles. DHIV3-GFP and env-expression plasmids were co-transfected at a mass ratio of 2:1 using lipofectamine 2000 (ThermoFisher, 11668030). shRNA encoding lentiviral particles were produced by co-transfecting pHIV7, psPAX2, pVSVg according to BROAD institute lentiviral production protocol (https://portals.broadinstitute.org/gpp/public/resources/protocols) using Lipofectamine 2000. Growth media was replaced on transfected cells after overnight incubation. At 72 hours post-transfection, virus containing supernatant was harvested, centrifuged to remove cell debris, filtered through a 0.45 um pore filter, and stored at −80°C.
Infection Assays
293T cells were transfected with env-overexpression constructs using Lipofectamine 2000 and incubated 24 hours. Transfected cells were infected with reporter virus by applying virus (HIV-RD114env, HIV-VSVg, HIV-SMRVenv) stocks in the presence of polybrene (Santa Cruz Bio, sc-134220) at a final concentration of 4 ug/mL. After 6–8 hours, virus stock was replaced with fresh growth media. Infected cells were maintained for 72 hours, replacing media when necessary, and harvested with trypsin. Detached cells were suspended in fresh growth media, strained and analyzed by flow cytometry. For the H1-ESC infection experiment, relative infection rates were calculated by normalizing the percent GFP+, HIV-RD114env infected cells to the percent GFP+ HIV-VSVg infected cells. For env/SUPYN overexpression experiments, relative infection rates were calculated by normalizing the percent GFP+ env/SUPYN-transfected cells to the percent GFP+ empty vector transfected cells. ANOVA with Tukey HSD tests were implemented in R (v3.6.3).
Placental cell shRNA transduction
Placenta-derived cell lines were treated with pHIV-shRNA-virus-containing supernatant and incubated for 72 hours as described in Infection Assays. Cerulean positive cells were sorted using the BD FACS Aria cytometer. Cells transduced with puroR cassette were treated with Puromycin (GIBCO, A1113802) at a final concentration of 3.5 ug/mL for 7 days, then cultured in regular growth media.
RT-qPCR
RNA was isolated from cultured cells using the RNeasy Mini Kit (Qiagen, 74104) and an on column dsDNAse digestion (Qiagen, 79254) was performed. 1–3 ug of total RNA were used to generate cDNA with the maxima cDNA synthesis with dsDNAse kit (ThermoFisher, K1681). qPCR reactions were performed using the LC480 Instrument with Sybr Green PCR master mix (Roche, 04707516001) according to manufacturer’s protocol and using primers indicated in Supplementary Table 2. Gene expression was then quantified using the ΔΔCT method (72). 18S expression was used as a reference housekeeping gene. Wilcox rank sum tests were performed using R (v3.6.3).
Envelope evolutionary sequence analyses
Orthologous SUPYN, SYN1, and SYN2 sequences were extracted from the 30-species MULTIZ alignment (68) and formatted for sequence alignment using the phast package (74). These and additional syntenic SUPYN and SYN2 open reading frame sequences were validated/identified by BLASTn (75) search with default settings of publicly available Catarrhine primate genomes (ncbi.nih.gov). Mariam Okhovat of the Carbone Lab (Oregon Health and Science University) generously provided BAM files containing read alignment information for SUPYN, SYN1, and SYN2 generated from whole genome sequencing of Hoolock leuconedys (Hoolock Gibbon), Symphalangus syndactylus (Siamang), Hylobates muelleri (Müller’s Gibbon), Hylobates lar (Lar Gibbon), Hylobates moloch (Silvery Gibbon), Hylobates pileatus (Pileated Gibbon), and Nomascus gabriellae (Yellow-cheeked Gibbon) (76). Where multiple individuals were sequenced, a consensus sequence was generated using samtools (63) and JalView (77).
To perform dN/dS analyses, orthologous env sequences (>90bp length) encoding the mature sequence downstream of the signal peptide cleavage site, were aligned using MEGA7 (78) and manually converted to PHYLIP format. A Newick tree was generated based on this alignment using the maximum likelihood algorithm implemented in MEGA7. Codeml, implemented in the PAML package, was run to calculate dN/dS values and log likelihood (LnL) scores generated under models M0, M1, M2, M7 and M8 (27). Chi-square tests comparing LnL scores generated under models of neutral evolution and selection were performed.
We used two approaches to reconstruct ancestral hominoid and OWM SUPYN sequences. First, we reconstructed ancestral SUPYN sequences using the majority rule consensus sequence (calculated in JalView) of the hominoid and OWM clade respectively. At positions where nucleotide identity was ambiguous, the dominant nucleotide identity in the neighboring clade was used as a tiebreaker. These sequences were used for our infection assays shown in Fig. 4H. We also employed a maximum likelihood approach using the baseml program, implemented in PAML (27). We reconstructed ancestral SUPYN sequences using the hominoid species, shown in Fig. 1, and the 6 OWM monkeys with the most complete SUPYN-coding open reading frame (olive baboon, drill, crab-eating macaque, rhesus macaque, japanese macaque, green monkey) as our input sequences. Because PAML requires a Newick tree as an input, the MEGAX maximum likelihood algorithm was used to generate a Newick tree with the above described SUPYN sequences (79–81). Baseml was run using models 3–7 (F84, HKY85, T92, TN93, REV). As shown in fig. S16, both the consensus- and maximum likelihood reconstructions were identical for the OWM SUPYN sequences. The consensus-based hominoid sequence reconstruction differed from our maximum likelihood-based reconstruction by two amino acids. These two positions are unlikely to affect the function of the resulting protein because these sites are identical to siamang SUPYN, which restricts RD114env-mediated infection (Fig. 4A, B).
Supplementary Material
Acknowledgments:
We thank Dr. Vicente Planelles for sharing reporter virus plasmids and training; Dr. Welkin Johnson for sharing his SMRVenv expression plasmid; Dr. Lars Aagaard for sharing shRNA transduction constructs; Dr. John Lis for providing lentiviral packaging construct; Dr. Amnon Koren and Rita Rebello for providing embryonic stem cell cultures and technical support respectively; Dr Lucia Carbone and Dr. Mariam Okhovat for sharing gibbon genome sequences. We thank Dr. Ray Malfavon-Borja for producing an initial list of envelope open reading frames in the human genome. We thank Maia Clare for her contribution to evolutionary sequence analyses. We thank Dr. Akiko Iwasaki for her critical reading of this manuscript. We thank members of the Feschotte lab for helpful advice and discussion throughout the project.
Funding:
National Institutes of Health grant R01 GM112972 (CF)
National Institutes of Health grant R35 GM122550 (CF)
CICE-FEDER-P12-CTS-2256 (JLGP)
The Wellcome Trust-University of Edinburgh Institutional Strategic Support Fund ISFF2 (JLGP)
European Research Council ERC-Consolidator grant ERC-STG-2012-309433 (JLGP)
Howard Hughes Medical Institute International Early Career Scientist grant IECS-55007420 (JLGP)
Private donation from Ms Francisca Serrano - Trading y Bolsa para Torpes (JLGP)
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability:
Materials generated for this study (cell lines, plasmids, primers) are available upon request from Dr. Cedric Feschotte. EnvORF annotations are included among supplementary data files. Accessibility information on publicly available high throughput datasets used in this study are presented in Table S1. Code used to process and plot the high throughput datasets have been deposited on Zenodo and is accessible by the following DOI: 10.5281/zenodo.7038957 (30).
References and Notes
- 1.Johnson WE, Origins and evolutionary consequences of ancient endogenous retroviruses. Nature reviews. Microbiology. 72, 5955 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Frank JA, Feschotte C, Co-option of endogenous viral sequences for host cell function. Current Opinion in Virology. 25, 81–89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Virgin HW, The Virome in Mammalian Physiology and Disease. Cell. 157, 142–150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malfavon-Borja R, Feschotte C, Fighting Fire with Fire: Endogenous Retrovirus Envelopes as Restriction Factors. Journal of Virology. 89, 4047–4050 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakagawa S, Takahashi MU, gEVE: a genome-based endogenous viral element database provides comprehensive viral protein-coding sequences in mammalian genomes. Database J Biological Databases Curation. 2016, baw087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugimoto J, Schust DJ, Kinjo T, Aoki Y, Jinno Y, Kudo Y, Suppressyn localization and dynamic expression patterns in primary human tissues support a physiologic role in human placentation. Scientific Reports. 9, 19502–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T, Paleovirology of “syncytins”, retroviral env genes exapted for a role in placentation. Philosophical Transactions of the Royal Society B: Biological Sciences. 368, 20120507–20120507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Kuyl AC, HIV infection and HERV expression: a review. Retrovirology. 9, 6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arora N, Sadovsky Y, Dermody TS, Coyne CB, Microbial Vertical Transmission during Human Pregnancy. Cell Host and Microbe. 21, 561–567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugimoto J, Sugimoto M, Bernstein H, Jinno Y, Schust D, A novel human endogenous retroviral protein inhibits cell-cell fusion. Scientific Reports. 3, 1462–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinha A, Johnson WE, ScienceDirect Retroviruses of the RDR superinfection interference group: ancient origins and broad host distribution of a promiscuous Env gene. Current Opinion in Virology. 25, 105–112 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Scalise M, Pochini L, Console L, Losso MA, Indiveri C, The Human SLC1A5 (ASCT2) Amino Acid Transporter: From Function to Structure and Role in Cell Biology. Frontiers in cell and developmental biology. 6, 96 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkelshtein D, Werman A, Novick D, Barak S, Rubinstein M, LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proceedings of the National Academy of Sciences of the United States of America. 110, 7306–7311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dottori M, Tay C, Hughes SM, Neural development in human embryonic stem cells-applications of lentiviral vectors. Journal of cellular biochemistry. 112, 1955–1962 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Sakata M, Tani H, Anraku M, Kataoka M, Nagata N, Seki F, Tahara M, Otsuki N, Okamoto K, Takeda M, Mori Y, Analysis of VSV pseudotype virus infection mediated by rubella virus envelope proteins. Scientific Reports. 7, 11607 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson WE, Endogenous Retroviruses in the Genomics Era. Annual review of virology. 2, 135–159 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Sommerfelt MA, Weiss RA, Receptor interference groups of 20 retroviruses plating on human cells. Virology. 176, 58–69 (1990). [DOI] [PubMed] [Google Scholar]
- 18.Ponferrada VG, Mauck BS, Wooley DP, The envelope glycoprotein of human endogenous retrovirus HERV-W induces cellular resistance to spleen necrosis virus. Arch Virol. 148, 659–675 (2003). [DOI] [PubMed] [Google Scholar]
- 19.de Parseval N, Casella J-F, Gressin L, Heidmann T, Characterization of the Three HERVH Proviruses with an Open Envelope Reading Frame Encompassing the Immunosuppressive Domain and Evolutionary History in Primates. Virology. 279, 558–569 (2001). [DOI] [PubMed] [Google Scholar]
- 20.van Zeijl M, Johann SV, Closs E, Cunningham J, Eddy R, Shows TB, O’Hara B, A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proceedings of the National Academy of Sciences. 91, 1168–1172 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller DG, Edwards RH, Miller AD, Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proceedings of the National Academy of Sciences. 91, 78–82 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jobbagy Z, Garfield S, Baptiste L, Eiden MV, Anderson WB, Subcellular redistribution of Pit-2 P(i) transporter/amphotropic leukemia virus (A-MuLV) receptor in A-MuLV-infected NIH 3T3 fibroblasts: involvement in superinfection interference. Journal of Virology. 74, 2847–2854 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JW, Cunningham JM, N-linked glycosylation of the receptor for murine ecotropic retroviruses is altered in virus-infected cells. Journal of Biological Chemistry. 268, 16316–16320 (1993). [PubMed] [Google Scholar]
- 24.Sugimoto J, Schust DJ, Yamazaki T, Kudo Y, Involvement of the HERV-derived cell-fusion inhibitor, suppressyn, in the fusion defects characteristic of the trisomy 21 placenta. Sci Rep-uk. 12, 10552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MAM, Kessing B, Pontius J, Roelke M, Rumpler Y, Schneider MPC, Silva A, O’Brien SJ, Pecon-Slattery J, A molecular phylogeny of living primates. PLoS Genetics. 7, e1001342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Blanco-Melo D, Gifford RJ, Bieniasz PD, Co-option of an endogenous retrovirus envelope for host defense in hominid ancestors. eLife. 6, 11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magiorkinis G, Gifford RJ, Katzourakis A, Ranter JD, Belshaw R, Env-less endogenous retroviruses are genomic superspreaders. Proceedings of the National Academy of Sciences of the United States of America. 109, 7385–7390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Y, Woodward BO, Pastor L, George AM, Petrechko O, Nouvet FJ, Haas DW, Jiang G, Hildreth JEK, Endogenous Retroviral Envelope Syncytin Induces HIV-1 Spreading and Establishes HIV Reservoirs in Placenta. Cell reports. 30, 4528–4539.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh M, Manu-1512/Envelopes_Suppressyn: Evolution and antiviral activity of a human protein of retroviral origin (Zenodo, 2022; https://zenodo.org/record/7038957#.Yw-7KC2B2Lc). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gertz EM, Yu Y-K, Agarwala R, Schäffer AA, Altschul SF, Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC biology. 4, 41 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bao W, Kojima KK, Kohany O, Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA. 6, 11–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, Huang J, Li M, Wu X, Wen L, Lao K, Li R, Qiao J, Tang F, Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nature structural & molecular biology. 20, 1131–1139 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, Yong J, Hu Y, Wang X, Wei Y, Wang W, Li R, Yan J, Zhi X, Zhang Y, Jin H, Zhang W, Hou Y, Zhu P, Li J, Zhang L, Liu S, Ren Y, Zhu X, Wen L, Gao YQ, Tang F, Qiao J, The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell. 161, 1437–1452 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Fan X, Wang R, Lu X, Dang Y-L, Wang H, Lin H-Y, Zhu C, Ge H, Cross JC, Wang H, Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell research. 28, 819–832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pavličev M, Wagner GP, Chavan AR, Owens K, Maziarz J, Dunn-Fletcher C, Kallapur SG, Muglia L, Jones H, Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Research. 27, 349–361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park J-E, Stephenson E, Polański K, Goncalves A, Gardner L, Holmqvist S, Henriksson J, Zou A, Sharkey AM, Millar B, Innes B, Wood L, Wilbrey-Clark A, Payne RP, Ivarsson MA, Lisgo S, Filby A, Rowitch DH, Bulmer JN, Wright GJ, Stubbington MJT, Haniffa M, Moffett A, Teichmann SA, Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 563, 347–353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Close JL, Yao Z, Levi BP, Miller JA, Bakken TE, Menon V, Ting JT, Wall A, Krostag A-R, Thomsen ER, Nelson AM, Mich JK, Hodge RD, Shehata SI, Glass IA, Bort S, Shapovalova NV, Ngo NK, Grimley JS, Phillips JW, Thompson CL, Ramanathan S, Lein E, Single-Cell Profiling of an In Vitro Model of Human Interneuron Development Reveals Temporal Dynamics of Cell Type Production and Maturation. Neuron. 93, 1035–1048.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velten L, Haas SF, Raffel S, Blaszkiewicz S, Islam S, Hennig BP, Hirche C, Lutz C, Buss EC, Nowak D, Boch T, Hofmann W-K, Ho AD, Huber W, Trumpp A, Essers MAG, Steinmetz LM, Human haematopoietic stem cell lineage commitment is a continuous process. Nature cell biology. 19, 271–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enge M, Arda HE, Mignardi M, Beausang J, Bottino R, Kim SK, Quake SR, Single-Cell Analysis of Human Pancreas Reveals Transcriptional Signatures of Aging and Somatic Mutation Patterns. Cell. 171, 321–330.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Gephart MGH, Barres B, Quake SR, A survey of human brain transcriptome diversity at the single cell level. Proceedings of the National Academy of Sciences of the United States of America. 112, 7285–7290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, Barnes I, Berry A, Bignell A, Carbonell Sala S, Chrast J, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, García Girón C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Hunt T, Izuogu OG, Lagarde J, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Ruffier M, Schmitt BM, Stapleton E, Suner M-M, Sycheva I, Uszczynska-Ratajczak B, Xu J, Yates A, Zerbino D, Zhang Y, Aken B, Choudhary JS, Gerstein M, Guigó R, Hubbard TJP, Kellis M, Paten B, Reymond A, Tress ML, Flicek P, GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, gky955- (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England). 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England). 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data. Cell. 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu X, Hill A, Packer J, Lin D, Ma Y-A, Trapnell C, Single-cell mRNA quantification and differential analysis with Census. Nature methods. 14, 309–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gte. Consortium, Genetic effects on gene expression across human tissues. Nature. 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zadora J, Singh M, Herse F, Przybyl L, Haase N, Golic M, Yung HW, Huppertz B, Cartwright JE, Whitley G, Johnsen GM, Levi G, Isbruch A, Schulz H, Luft FC, Müller DN, Staff AC, Hurst LD, Dechend R, Izsvák Z, Disturbed Placental Imprinting in Preeclampsia Leads to Altered Expression of DLX5, a Human-Specific Early Trophoblast Marker. Circulation. 136, 1824–1839 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Antonio MD, Weghorn D, Antonio-Chronowska AD, Coulet F, Olson KM, DeBoever C, Drees F, Arias A, Alakus H, Richardson AL, Schwab RB, Farley EK, Sunyaev SR, Frazer KA, Identifying DNase I hypersensitive sites as driver distal regulatory elements in breast cancer. Nature communications. 8, 436 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, Thi VLD, Shilvock AR, Hoffmann H-H, Rosenberg BR, Rice CM, Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown. Cell. 172, 811–824.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calderon D, Nguyen MLT, Mezger A, Kathiria A, Müller F, Nguyen V, Lescano N, Wu B, Trombetta J, Ribado JV, Knowles DA, Gao Z, Blaeschke F, Parent AV, Burt TD, Anderson MS, Criswell LA, Greenleaf WJ, Marson A, Pritchard JK, Landscape of stimulation-responsive chromatin across diverse human immune cells. Nature Publishing Group. 51, 1494–1505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shytaj IL, Lucic B, Forcato M, Penzo C, Billingsley J, Laketa V, Bosinger S, Stanic M, Gregoretti F, Antonelli L, Oliva G, Frese CK, Trifunovic A, Galy B, Eibl C, Silvestri G, Bicciato S, Savarino A, Lusic M, Alterations of redox and iron metabolism accompany the development of HIV latency. Embo J. 39, e102209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dixon G, Pan H, Yang D, Rosen BP, Jashari T, Verma N, Pulecio J, Caspi I, Lee K, Stransky S, Glezer A, Liu C, Rivas M, Kumar R, Lan Y, Torregroza I, He C, Sidoli S, Evans T, Elemento O, Huangfu D, QSER1 protects DNA methylation valleys from de novo methylation. Science. 372 (2021), doi: 10.1126/science.abd0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J, Xu J, Liu B, Yao G, Wang P, Lin Z, Huang B, Wang X, Li T, Shi S, Zhang N, Duan F, Ming J, Zhang X, Niu W, Song W, Jin H, Guo Y, Dai S, Hu L, Fang L, Wang Q, Li Y, Li W, Na J, Xie W, Sun Y, Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature. 557, 256–260 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Tsankov AM, Gu H, Akopian V, Ziller MJ, Donaghey J, Amit I, Gnirke A, Meissner A, Transcription factor binding dynamics during human ES cell differentiation. Nature. 518, 344–349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barakat TS, Halbritter F, Zhang M, Rendeiro AF, Perenthaler E, Bock C, Chambers I, Functional Dissection of the Enhancer Repertoire in Human Embryonic Stem Cells. Cell stem cell. 23, 276–288.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang WWC, Castillo-Venzor A, Gruhn WH, Kobayashi T, Penfold CA, Morgan MD, Sun D, Irie N, Surani MA, Sequential enhancer state remodelling defines human germline competence and specification. Nat Cell Biol. 24, 448–460 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwak Y-T, Muralimanoharan S, Gogate AA, Mendelson CR, Human Trophoblast Differentiation Is Associated With Profound Gene Regulatory and Epigenetic Changes. Endocrinology. 160, 2189–2203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunn-Fletcher CE, Muglia LM, Pavličev M, Wolf G, Sun M-A, Hu Y-C, Huffman E, Tumukuntala S, Thiele K, Mukherjee A, Zoubovsky S, Zhang X, Swaggart KA, Lamm KYB, Jones H, Macfarlan TS, Muglia LJ, Anthropoid primate-specific retroviral element THE1B controls expression of CRH in placenta and alters gestation length. PLoS biology. 16, e2006337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krendl C, Shaposhnikov D, Rishko V, Ori C, Ziegenhain C, Sass S, Simon L, Müller NS, Straub T, Brooks KE, Chavez SL, Enard W, Theis FJ, Drukker M, GATA2/3-TFAP2A/C transcription factor network couples human pluripotent stem cell differentiation to trophectoderm with repression of pluripotency. Proceedings of the National Academy of Sciences of the United States of America. 114, E9579–E9588 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao L, Wu K, Liu Z, Yao X, Yuan S, Tao W, Yi L, Yu G, Hou Z, Fan D, Tian Y, Liu J, Chen Z-J, Liu J, Chromatin Accessibility Landscape in Human Early Embryos and Its Association with Evolution. Cell. 173, 248–259.e15 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nature methods. 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup, The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England). 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gaspar JM, Improved peak-calling with MACS2. bioRxiv, 496521 (2018). [Google Scholar]
- 65.Amemiya HM, Kundaje A, Boyle AP, The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Scientific Reports. 9, 9354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics (Oxford, England). 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robinson JT, Thorvaldsdóttir H, Winckler W, Integrative genomics viewer. nature.com (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Hinrichs AS, Gonzalez JN, Gibson D, Diekhans M, Clawson H, Casper J, Barber GP, Haussler D, Kuhn RM, Kent WJ, The UCSC Genome Browser database: 2019 update. Nucleic acids research. 47, D853–D858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM, Embryonic Stem Cell Lines Derived from Human Blastocysts. Science. 282, 1145–1147 (1998). [DOI] [PubMed] [Google Scholar]
- 70.Aagaard L, Bjerregaard B, Kjeldbjerg AL, Pedersen FS, Larsson L-I, Rossi JJ, Silencing of endogenous envelope genes in human choriocarcinoma cells shows that envPb1 is involved in heterotypic cell fusions. The Journal of general virology. 93, 1696–1699 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luft C, Freeman J, Elliott D, Al-Tamimi N, Kriston-Vizi J, Heintze J, Lindenschmidt I, Seed B, Ketteler R, Application of Gaussia luciferase in bicistronic and non-conventional secretion reporter constructs. BMC biochemistry. 15, 14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Knappskog S, Ravneberg H, Gjerdrum C, Trösse C, Stern B, Pryme IF, The level of synthesis and secretion of Gaussia princeps luciferase in transfected CHO cells is heavily dependent on the choice of signal peptide. Journal of biotechnology. 128, 705–715 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Schmittgen TD, Livak KJ, Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 74.Hubisz MJ, Pollard KS, Siepel A, PHAST and RPHAST: phylogenetic analysis with space/time models. Briefings in bioinformatics. 12, 41–51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye J, McGinnis S, Madden TL, BLAST: improvements for better sequence analysis. Nucleic acids research. 34, W6–9 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okhovat M, Nevonen KA, Davis BA, Michener P, Ward S, Milhaven M, Harshman L, Sohota, Fernandes JD, Salama SR, O’Neill RJ, Ahituv N, Veeramah KR, Carbone L, Co-option of the lineage-specific LAVA retrotransposon in the gibbon genome. Proceedings of the National Academy of Sciences of the United States of America. 117, 19328–19338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ, Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics (Oxford, England). 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kumar S, Stecher G, Tamura K, MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stecher G, Tamura K, Kumar S, Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol Biol Evol. 37, 1237–1239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar S, Stecher G, Li M, Knyaz C, Tamura K, MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tamura K, Nei M, Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10, 512–526 (1993). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Materials generated for this study (cell lines, plasmids, primers) are available upon request from Dr. Cedric Feschotte. EnvORF annotations are included among supplementary data files. Accessibility information on publicly available high throughput datasets used in this study are presented in Table S1. Code used to process and plot the high throughput datasets have been deposited on Zenodo and is accessible by the following DOI: 10.5281/zenodo.7038957 (30).