Abstract
The metabolism of glucose and lipids is essential for energy production in the body, and dysregulation of the metabolic pathways of these molecules is implicated in various acute and chronic diseases, such as type 2 diabetes, Alzheimer’s disease, atherosclerosis (AS), obesity, tumor, and sepsis. Post-translational modifications (PTMs) of proteins, which involve the addition or removal of covalent functional groups, play a crucial role in regulating protein structure, localization function, and activity. Common PTMs include phosphorylation, acetylation, ubiquitination, methylation, and glycosylation. Emerging evidence indicates that PTMs are significant in modulating glucose and lipid metabolism by modifying key enzymes or proteins. In this review, we summarize the current understanding of the role and regulatory mechanisms of PTMs in glucose and lipid metabolism, with a focus on their involvement in disease progression associated with aberrant metabolism. Furthermore, we discuss the future prospects of PTMs, highlighting their potential for gaining deeper insights into glucose and lipid metabolism and related diseases.
Keywords: Post-translational modification, Glucose metabolism, Lipid metabolism, Metabolic disease
Introduction
Glucose and lipid metabolism, the main source of energy, is critical for the physiological functions of all tissues and organs (Chen et al. 2019). Dysregulation of glucose and lipid metabolism is a risk factor for many acute and chronic diseases, such as type 2 diabetes, Alzheimer’s disease (AD), atherosclerosis (AS), obesity, tumor, and sepsis (Cheng et al. 2016; Garcia et al. 2023; Gasbarrino et al. 2023; Takeuchi et al. 2023; Yassine et al. 2022). Glucose and lipid metabolism in the body is regulated by various proteins, including key enzymes. Any factor that affects these proteins may influence the metabolic processes. Recently, studies have confirmed that post-translational modifications (PTMs) participate in the metabolic processes of glucose and lipids and have a critical impact on diseases arising from aberrant glucose and lipid metabolism (Sawant et al. 2022; Stocks and Zierath 2022).
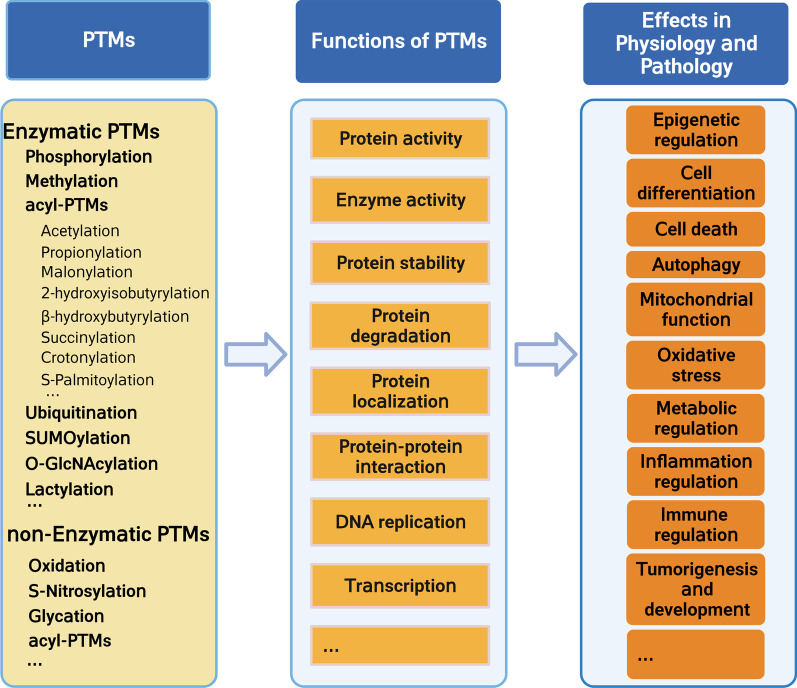
PTM refers to the reversible or irreversible covalent processing of some proteins after translation, which occurs at the amino acid side chains, C-terminus, or N-terminus (Ramazi and Zahiri 2021). Based on their biochemical origin, PTMs are divided into enzymatic (ePTMs) and non-enzymatic (nPTMs) (Jennings et al. 2022; Wold 1981). The effects of ePTMs are precisely controlled by PTM enzyme readers, writers, and erasers, which can add or remove modifications (Jennings et al. 2022). In contrast, nPTMs usually occur spontaneously between nucleophilic or redox-sensitive amino acid side chains and reactive metabolites (Harmel and Fiedler 2018). Approximately two-thirds of proteins in vivo undergo PTMs; these modifications include phosphorylation, acetylation, ubiquitination, methylation, and glycosylation. Emerging evidence reveals that PTMs can expand the diversity of proteins by influencing their functions via altering protein structure, localization, and activity. Ultimately, PTMs play a vital role in various physiological and pathophysiological processes, such as cell replication, cell death, transcription regulation, translation regulation, cellular signal transduction, and immune regulation (Fig. 1) (Meng et al. 2021; Patwardhan et al. 2021; Yu et al. 2022), and are also involved in glucose and lipid metabolism. For example, Lorendeau et al. focused on the metabolic regulation of signaling and transcriptional regulation of mammalian target of rapamycin (mTOR), AMP-activated protein kinase (AMPK), and p53, and discussed functional consequences of PTMs on these enzymes (Lorendeau et al. 2015).
Fig. 1.
Functions and effect of post-translational modifications in physiology and pathology
In this review, we summarize the types and roles of PTMs and illustrate their molecular mechanisms in regulating glucose and lipid metabolism. Additionally, we highlight the roles and mechanisms of PTMs in diseases associated with aberrant glucose and lipid metabolism. Our review aims to provide insights into the treatment of diseases associated with dysregulated glucose and lipid metabolism.
Common types of PTMs
PTMs are complex processes that play extremely important roles in almost all cellular activities. Exploring the regulatory processes of PTMs is of great significance for understanding the molecular mechanisms or finding new biomarkers for various diseases. There are currently more than 400 known PTMs. The most common modifications of proteins associated with glucose and lipid metabolism include phosphorylation, acetylation, ubiquitination, SUMOylation, lactylation, methylation, S-glutathionylation, and glycosylation (Table 1).
Table 1.
Characterizations of several common PTMs
| Type of PTMs | Chemical Structure | Modified Amino Acid Residues | Added | Removed | References |
|---|---|---|---|---|---|
| Phosphorylation |
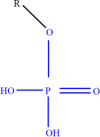
|
Ser, Thr, Tyr, His, Asp, Pro, Glu, Cys, Arg, and Lys (Jennings et al. 2022) | Enzymatic (kinases) | Enzymatic (phosphatases) | Jennings et al. (2022) |
| Ubiquitination |
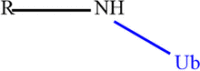
|
Lys, Cys, Ser, and Thr, (Kelsall 2022) | Enzymatic [ubiquitinactivating (E1), -conjugating (E2), and -ligase (E3)] (Reyes-Turcu et al. 2009) | Enzymatic [deubiquitinating enzymes (DUBs)] (Reyes-Turcu et al. 2009) | Kelsall (2022); Reyes-Turcu et al. (2009) |
| Methylation |
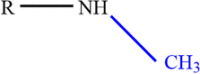
|
Lys (mono-, di-, and tri-methylation) (Bhat et al. 2021), Arg (mono- and di-methylation of Arg) (Xu and Richard 2021) | Enzymatic (Lys methyltransferase and Protein Arg methyltransferases) (Bhat et al. 2021) | Enzymatic (Lys demethylases, Lys-specific histone demethylase 1and JmjC domain-containing 6) (Bhat et al. 2021; Manni et al. 2022) | Bhat et al. (2021; Xu and Richard (2021) |
| O-GlcNAcylation |
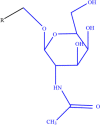
|
Ser and Thr (Yang and Qian 2017) | Enzymatic (O-GlcNAc transferase) (Wang et al. 2023) | Enzymatic (O-GlcNAcase) (Wang et al. 2023) | Wang et al. (2023); Yang and Qian (2017) |
| Acetylation |
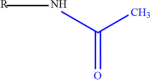
|
Lys, N-terminus of proteins, thiol group of Cys | Enzymatic (acetyltransferases) and non-enzymatic(S-to-N acyl transfer mechanism) | Enzymatic (deacetylases and sirtuins) | James et al. (2018); Jennings et al. (2022) |
| SUMOylation |

|
Lys (Chang and Yeh (2020) | Enzymatic [SUMO activating (E1), -conjugating (E2), and -ligase (E3)] (Yeh 2009) | Enzymatic [deSUMOylating enzyme (SENP)] (Yeh 2009) | Chang et al. (2020); Du et al. (2021); Yeh (2009) |
| Succinylation |
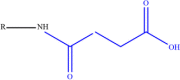
|
Lys | Enzymatic (KAT2A/GCN5) (Wang et al. 2018b) and non-enzymatic | Enzymatic (SIRT5, SIRT7) (Li et al. 2016; Rardin et al. 2013) | Li et al. (2016); Rardin et al. (2013); Wang et al. (2018b) |
| Lactylation |
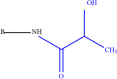
|
Lys (Chen et al. 2021a) | Enzymatic [p300/CBP (Zhang et al. 2019b)] and non-enzymatic(lactyl-glutathione) (Chen et al. 2021a) | Unknown | Chen et al. (2021a) |
| Propionylation |
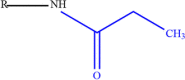
|
Lys (Tang et al. 2022) | Enzymatic (p300/CBP (Chen et al. 2007), GCN5 (Kebede et al. 2017), PCAF (Kebede et al. 2017), MOF (Han et al. 2018), BRPF1-KAT6 (Yan et al. 2020)) and non-enzymatic (high concentrations of propionyl-CoA) (You et al. 2019) | Enzymatic [SIRT1 (Cheng et al. 2009)] | Chen et al. (2007); Cheng et al. (2009); Han et al. (2018); Kebede et al. (2017); Tang et al. (2022); Yan et al. (2020); You et al. (2019) |
| Malonylation |
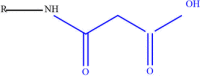
|
Lys (Zou et al. 2023) | Enzymatic (unknown (Zou et al. 2023), donor of malonylation: Malonyl-CoA) | Enzymatic (SIRT5) (Baek et al. 2023; Wang et al. 2022b) | (Baek et al. 2023; Wang et al. (2022b); Zou et al. 2023) |
| S-Palmitoylation |
|
Cys (Liu et al. 2022b) | Enzymatic (zDHHC palmitoylase family) (Zhou et al. 2023) |
Enzymatic (APT1/2, PPT1/ 2, ABHD17A/B/ C) (Zhou et al. 2023) |
Liu et al. (2022b); Zhou et al. (2023) |
| S-Glutathionylation |
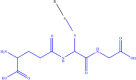
|
Cys (Rashdan et al. 2020) |
Enzymatic (Glutathione S-transferases (GST) ) and non-enzymatic (Martinez-Ruiz and Lamas 2007; Rashdan et al. 2020) |
Enzymatic (Glutaredoxins (Grx), SRX, TRX) (Rashdan et al. 2020) | Martinez-Ruiz and Lamas (2007); Rashdan et al. (2020) |
| Crotonylation |
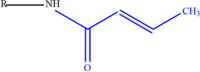
|
Lys (Tan et al. 2011), Ser (Liao et al. 2020) |
Enzymatic (Crotonyltransferases:p300/CBP (Sabari et al. 2015), KAT8 /MOF (Liu et al. 2017), KAT2B /PCAF (Xu et al. 2017), Gcn5 and Esa1 (Kollenstart et al. 2019)) |
Enzymatic [HDAC1–3 (Kelly et al. 2018; Madsen and Olsen 2012) and SIRT1-3 (Bao et al. 2014)] |
Bao et al. (2014); Kelly et al. (2018); Kollenstart et al. (2019); Liao et al. (2020); Liu et al. (2017); Madsen and Olsen (2012); Sabari et al. (2015); Tan et al. (2011); Wang et al. (2021); Xu et al. (2017) |
| 2-hydroxyisobutyrylation |
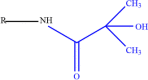
|
Lys (Huang et al. 2018a) |
Enzymatic [p300 (Huang et al. 2018b) and TIP60 (Wang et al. 2022c)] |
Enzymatic [HDAC2 and HDAC3 (Huang et al. 2018a)] | Huang et al. (2018a; b); Wang et al. (2022c) |
| β-hydroxybutyrylation |
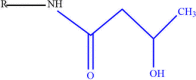
|
Lys (Huang et al. 2021) | Enzymatic [p300/CBP (Huang et al. 2021)] | Enzymatic [HDAC1-3 (Huang et al. 2021), SIRT1-2 (Huang et al. 2021), SIRT3 (Zhang et al. 2019c)] | Huang et al. (2021); Zhang et al. (2019c) |
Phosphorylation
Phosphorylation is the process by which phosphate groups bind to substrates and thus regulate protein activity and interactions under the regulation of protein kinases (Cohen 2002). Phosphorylation affects at least one-third of eukaryotic proteins (Cohen 2000). It is widely involved in regulatory processes, including membrane transport, protein degradation, regulation of enzyme activity (activation or inhibition), and protein interactions. Thus, phosphorylation plays a vital role in regulating cell apoptosis, mitochondrial function, inflammatory response, oxidative stress, cellular signaling, translocation, and autophagy (Carlson et al. 2020; Hepowit et al. 2022; Liu et al. 2021a; Peng et al. 2017; Ross et al. 2023; Zhang et al. 2022). Protein phosphorylation is one of the most common and important PTMs (Sacco et al. 2012). It is a reversible process that is regulated by protein kinases and phosphatases.
The most common phosphorylation sites are in the amino acid side chains of serine (Ser), threonine (Thr), and tyrosine (Tyr) residues (Seok 2021). Overall, phosphorylated Ser is the most abundant (86%), followed by Thr (12%) and Tyr (2%) (Olsen et al. 2006). Phosphorylation plays critical regulatory roles in glycolipid metabolism. For example, phosphorylation of the Ser473 site of protein kinase B (Akt) inhibits the activity of GSK3β, thereby activating glycogen synthase to reduce blood sugar in HepG2 cells (Gao et al. 2018). Additionally, Galectin-3 can mediate cardiac remodeling due to impaired glycolipid metabolism by inhibiting Akt phosphorylation at Thr308/Ser473 (Sun et al. 2021b). AMPK phosphorylation by cellular repressor of E1A stimulated genes 1 (CREG1) can lead to glucose uptake in skeletal muscle cells (Goto et al. 2023). Therefore, phosphorylation sites may have the potential to serve as biomarkers for glucose and lipid metabolism diseases or even as possible therapeutic targets.
Acetylation
Acetylation, a type of PTM that has been extensively explored, refers to the process of acetyl group transfer from acetyl coenzyme A to lysine or other amino acid residues of target proteins. It is catalyzed by acetyltransferases and regulates gene transcription and signal transduction (Drazic et al. 2016). It is a reversible process that is regulated mainly by lysine acetyltransferases (KATs) and lysine deacetylases. Acetylation is classified as Nα-acetylation, Nε-acetylation, and O-acetylation, depending on the addition of acetyl groups to different amino acids and at different sites (Lee et al. 2010). One of the first modifications of histone discovered was acetylation, wherein an acetyl group is added to lysine residues at the N terminus of histone protein, which regulates gene transcription by affecting the binding of DNA to histones (Verdone et al. 2005). Notably, KAT-mediated histone acetylation affects epigenetic processes (He et al. 2018). For example, the activation of Toll-like receptors can promote histone acetylation and thus regulate Myeloid differentiation primary response 88 (MyD88) and Toll/Interleukin-1 receptor-domain-containing adapter-inducing interferon-β (TRIF) signaling, leading to the activation of adenosine triphosphate (ATP)-citrate lyase and thereby promoting energy metabolism (Lauterbach et al. 2019). Recruitment of hypoxia-inducible factor 1 alpha (HIF1α) to hypoxia-responsive elements induces glucose uptake through its interaction with p300-dependent histone acetylation (Anand et al. 2023). Recently, several non-histone acetylations have been identified that mainly affect gene transcription, DNA damage repair, protein folding, cell division, signal transduction, autophagy (Narita et al. 2019). Moreover, acetylation is important for the regulation of metabolism (Zhao et al. 2010). For instance, Zhang et al. discovered that histone deacetylase 8 could alter the glucose metabolism of hepatocellular carcinoma cells by controlling the acetylation of the PKM2 protein at the K62 site, leading to a predominant utilization of glucose through the pentose phosphate pathway (Zhang et al. 2020a). Thus, the balance between acetylation and deacetylation is crucial, and disruption of this balance may lead to disease development.
Ubiquitination and SUMOylation
Ubiquitination, a common PTM, refers to the process in which ubiquitin covalently binding to target proteins and is catalyzed by a three-enzyme cascade, composed of E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases. Ubiquitin (Ub), a highly conserved 76-amino acid protein, contains seven lysine residues (K6, K11, K27, K29, K33, K48, and K63), each of which is ubiquitinated to form distinctive forms of polyubiquitin chains (Swatek and Komender 2016). Various lengths and types of ubiquitinated chains determine the fate of substrate proteins and mediate different signaling pathways. Although ubiquitination mainly regulates the degradation of proteins, studies have found that ubiquitination also plays vital roles in regulating protein activity, protein–protein interactions, subcellular localization, and signal transduction (Komander and Rape 2012; Rajalingam and Dikic 2016). For instance, K48-linked chains are responsible for targeting substrate proteins for proteasomal degradation, while K63-linked chains are involved in several nonproteolytic functions, such as nuclear factor (NF)-κB activation and DNA damage repair (Emmerich et al. 2016; Liu et al. 2018; Yu et al. 2021). Numerous studies have confirmed that ubiquitination is widely involved in various physiological and pathological processes, such as transcriptional regulation, cell proliferation, cell apoptosis, DNA damage repair, and immune regulation (Roberts et al. 2022; Zhong et al. 2022). Notably, ubiquitination is a dynamic and reversible process, and can be counteracted by deubiquitinases (Mevissen and Komander 2017). A dynamic balance between ubiquitination and deubiquitination is necessary to maintain protein homeostasis and function, and abnormalities in the ubiquitin system are associated with the occurrence of many diseases, including neurodegenerative diseases, immune diseases, and cancers (Cockram et al. 2021; Liu et al. 2022a).
SUMOylation is an essential PTM similar to ubiquitination. During SUMOylation, a small ubiquitin-like modifier (SUMO) protein covalently binds to target proteins on lysine residues, which is mediated by a specific SUMO E1 activating enzyme, SUMO E2 conjugating enzyme, and SUMO E3 ligase. Unlike ubiquitination, SUMOylation mainly mediates the localization and functional regulation of target proteins instead of promoting degradation (Vertegaal 2022; Zhao 2018). Notably, SUMOylation is reversible, and deSUMOylation is mediated by SUMO-specific proteases, predominantly of the Sentrin/SUMO-specific proteases (SENPs) family. Imbalances in SUMOylation and deSUMOylation have been observed in the progression of various diseases (Mustfa et al. 2017; Zheng et al. 2020) including metabolism-related diseases (Sadeghi et al. 2023; Sapir 2020; Zhu et al. 2022b). Notably, SUMOylation and deSUMOylation have been found to be important in regulating glucose and lipid metabolism. For example, Zheng et al. discovered that adipose lipid storage in mice decreased when SUMO-specific protease 2 (Senp2) was specifically knocked out in adipose tissues (Zheng et al. 2018). Senp2 could regulate adipose lipid storage by suppressing Setdb1 function via the de-SUMOylation of Setdb1, suggesting that Senp2-mediated deSUMOylation regulates lipid metabolism in adipose tissues. In addition, guanosine triphosphate binding protein 4 (GTPBP4) was found to induce dimeric pyruvate kinase M2 SUMOylation and dimer formation through the UBA2-SUMO1 axis, thus promoting aerobic glycolysis in hepatocellular carcinoma (Zhou et al. 2022b). Therefore, further studies on the homeostasis of SUMOylation and deSUMOylation may provide new insights into the diagnosis and treatment of these diseases.
Glycosylation
Protein glycosylation is one of the most abundant and diverse types of PTMs, in which glycan moieties are added to proteins (Reily et al. 2019). By modulating the structure, stability, and function of proteins, glycosylation plays a profound role in various pathological and physiological processes (Bangarh et al. 2023; Pradeep et al. 2023; Reily et al. 2019). There are two major kinds of protein glycosylation in eukaryotes: N-linked (N-glycosylation) and O-linked (O-glycosylation). N-glycosylation involves the attachment of N-glycans (N-acetylglucosamine/GlcNAc) to the amino group of the Asn residue at the sequence Asn–X–Ser/Thr (where X represents any amino acid except for Pro); it initiates in the endoplasmic reticulum (ER) and then further modifications occur in the Golgi apparatus (Pradeep et al. 2023; Schjoldager et al. 2020). O-glycosylation is more complicated, and refers to the covalent addition of diverse glycans (such as N-acetylgalactosamine (GalNAc), fucose, glucose, xylose, and mannose) to the hydroxyl group of Ser/Thr residues, and also on tyrosine, hydroxylysine, and hydroxyproline; it mostly occurs in the Golgi apparatus (Joshi et al. 2018; Li et al. 2022a, b, c; Schjoldager et al. 2020). Notably, O-linked N-acetylglucosamine modification (O-GlcNAcylation), a unique type of O-glycosylation in which O-linked N-acetylglucosamine (O-GlcNAc) is added to Ser and Thr residues of proteins located in the cytoplasm, nucleus, and mitochondria, has received increasing attention in recent years (Gonzalez-Rellan et al. 2022; Yang and Qian 2017). O-GlcNAcylation is mediated by O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA). OGT catalyzes the addition, whereas OGA reversibly removes protein modifications (Gao et al. 2001; Shafi et al. 2000). Research has indicated that O-GlcNAcylation tunes the functions of protein in various ways, including protein cellular localization, protein stability, and protein–protein interaction (Chang et al. 2020). Interestingly, O-GlcNAcylation and phosphorylation have been shown to participate in extensive crosstalk with each other, as they can both occur on Ser and Thr residues of proteins (Hart et al. 2011). O-GlcNAcylation, which is sensitive to cellular metabolic states, has been proposed to function as a “nutrient and stress sensor” in cells (Bond and Hanover 2013; Ruan et al. 2013). Notably, emerging evidence has shown that glycosylation plays a pivotal role in metabolic diseases. For example, Nishimura et al. discovered that suppression of O-GlcNAcylation in the intestine reduced glucose absorption via inhibiting SGLT1 expression, suggesting that regulating O-GlcNAcylation in the intestine may provide a novel strategy for treating absorption disorders, obesity, and diabetes (Nishimura et al. 2022). In addition, Yung et al. revealed that ER stress-mediated perturbation of placental protein glycosylation could lead to the maladaptation of maternal hepatic glucose metabolism, which may be a new mechanism of maternal metabolic disorders (Yung et al. 2023). Thus, considering the intimate relationship between glycosylation and metabolic state, studies targeting the regulatory roles of glycosylation may provide novel insights into the treatment of diseases associated with aberrant metabolism.
Methylation
Methylation, mediated by methyltransferase, is a widespread phenomenon in both eukaryotes and prokaryotes. The substrates of methylation can be DNA, RNA, and proteins. Among these, protein methylation is a common PTM which occurs in both histone and non-histone proteins (Dai et al. 2021). The most common modification sites of methylation are lysine and arginine residues (Jambhekar et al. 2019). Based on the substrates involved, protein methyltransferases can be divided into categories such as protein lysine methyltransferases (PKMTs) and protein arginine methyltransferases (PRMTs) (Bhat et al. 2021; Dai et al. 2021; Xu and Richard 2021). PKMTs can cause monomethylation, bimethylation, or trimethylation of lysines on their substrates (Bhat et al. 2021). Methylation modifications in arginine include monomethylated arginine, asymmetric dimethylarginine, and symmetric dimethylarginine (Blanc and Richard 2017; Dai et al. 2021). Protein methylation plays an essential role in various intracellular processes, including glucose and lipid metabolism, via regulating the function of target proteins (Biggar and Li 2015; Dilworth et al. 2019; Li et al. 2019; Malecki et al. 2022). For example, Han et al. discovered that PRMT6 could mediate asymmetric dimethylation of multiple arginine residues of cAMP-response element binding protein (CREB)-regulated transcriptional coactivator 2 (CRTC2), which enhanced the interaction of CRTC2 with CREB on the promoters of gluconeogenic enzyme-encoding genes and thus played a vital role in hepatic glucose metabolism (Che et al. 2021; Han et al. 2014; Jia et al. 2020). In addition, Jia et al. found that PRMT5 regulates fatty acid metabolism and lipid droplet biogenesis in white adipose tissues, and that Prmt5AKO mice (the Prmt5 gene is specifically present in adipocytes) exhibit sex- and depot-dependent progressive lipodystrophy (Jia et al. 2020). Mechanistically, Prmt5 can not only methylate and release the transcription elongation factor SPT5 from the Berardinelli-Seip congenital lipodystrophy 2 (Bscl2 encodes Seipin, which can mediate lipid droplet biogenesis) promoter but also methylate Sterol Regulatory Element-Binding Transcription Factor 1a (SREBP1a) and promote lipogenic gene expression. Thus, further studies on protein methylation may supply potential therapeutic targets for diseases involving dysregulated glucose and lipid metabolism.
Other PTMs
In addition to the aforementioned PTMs, other PTMs such as lactylation, methylation, S-glutathionylation, N-glycosylation, and palmitoylation have also been observed to participate in glucose and lipid metabolism-related diseases. S-glutathionylation is the formation of mixed disulfides between glutathione and cysteine residues in proteins, which can lead to enhanced or suppressed protein activity (Dalle-Donne et al. 2009). A study performed by Dong et al. found that S-glutathionylation of the AMPK-α catalytic subunit could activate AMPK to improve glucose transportation and degradation while inhibiting glycogen synthesis and maintaining redox balance under a low reactive oxygen species microenvironment, providing new insights into diabetes treatment (Dong et al. 2016). However, many other PTMs have not been studied for their role in glucose and lipid metabolism. Given the extensive regulatory roles of PTMs in protein function, future studies should investigate the regulatory roles and mechanisms of various PTMs in glucose and lipid metabolism to provide potential targets for treatment and diagnosis.
In conclusion, PTMs play important roles in protein functions and participate in various biological processes. Exploring their regulatory roles in glucose and lipid metabolism may provide the basis for clinical diagnosis and therapy.
Roles of PTMs in glucose and lipid metabolism
Roles of PTMs in glucose metabolism
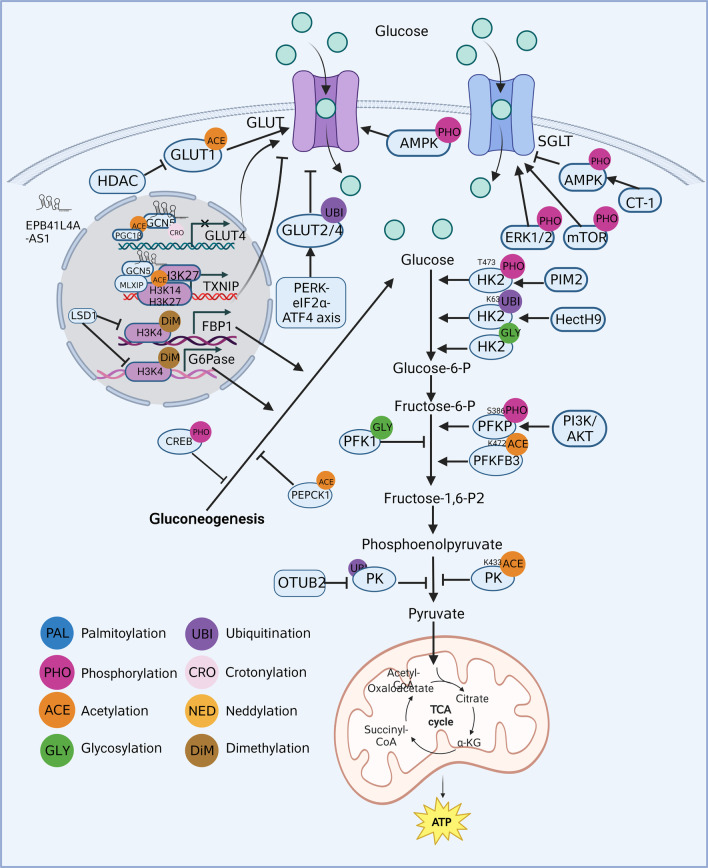
Cell cycle, growth, apoptosis, and energy metabolism are critically affected by glucose metabolism (Mulukutla et al. 2010). Glycolysis (involving three irreversible reactions) and gluconeogenesis (involving four irreversible reactions) are the central processes of glucose metabolism (Chandel 2021). Disorders in glucose metabolism primarily involve disruptions in energy and substance metabolism, and they participate in various pathological processes. For example, dysregulated glucose metabolism is involved in diabetes mellitus and AD (Cao et al. 2022; Huang et al. 2023). The reprogrammed glucose metabolism in the enhanced Warburg effect (or aerobic glycolysis) is considered a hallmark of cancer (Povero 2023). A better understanding of the regulation and molecular mechanisms involved in glucose metabolism can help us to understand the basis of many metabolic disorders. Recent studies have found that rate-limiting enzymes in glucose metabolism, such as facilitated-diffusion glucose transporters (GLUT), phosphofructokinase (PFK), and phosphoenolpyruvate carboxykinase (PEPCK), are tightly regulated by several PTMs, including phosphorylation, acetylation, ubiquitination, glycosylation, crotonylation, and dimethylation (Fig. 2) (Ahmed et al. 2023; He et al. 2022; Yi et al. 2012). For example, glycosylation inhibits PFK1 activity and redirects the flux of glucose from glycolysis through the pentose phosphate pathway (Yi et al. 2012). PEPCK is an important enzyme in the gluconeogenic pathway that catalyzes the conversion of oxaloacetate to phosphoenolpyruvate, thereby participating in glucose synthesis. For example, phosphorylation of both AMPK and forkhead box transcription factor O1 (FoxO1) results in a downregulation of PEPCK and G6Pase, thereby promoting glucose uptake and inhibiting glucose production (Ahmed et al. 2023). PEPCK acetylation, which can occur at various amino acid residues, is emerging as an important regulatory mechanism of its activity and is linked to metabolic diseases (Marin-Hernandez et al. 2022; Xiong et al. 2011; Zhang et al. 2018). In addition to PTMs of PEPCK, other rate-limiting enzymes have also been shown to play important roles. LINC00930 can recruit the retinoblastoma binding protein 5 and general control nonderepressible 5 complex to the promoter of PFKFB3, increasing H3K4 trimethylation and H3K9 acetylation levels and transactivating PFKFB3, thereby promoting glycolytic flux (He et al. 2022). In addition, we found that crotonylation and dimethylation are also involved in glucose metabolism. Crotonylation is a type of acylation that regulates gene expression and metabolic homeostasis, and a recent study has shown its involvement in the regulation of energy metabolism (Gowans et al. 2019). Dimethylation is a process that adds two methyl groups to proteins and is known to regulate gene expression (Jackson et al. 2004); it can also affect glucose metabolism (Pan et al. 2013).
Fig. 2.
Roles of post-translational modifications in glucose metabolism
PTMs in glucose transport
The initial and limiting step in glucose metabolism is glucose transport through the cell membrane via glucose transport proteins. There are two families of cellular glucose transporters: GLUT and sodium-dependent glucose transporters (SGLTs) (Navale and Paranjape 2016). Studies have shown that PTMs regulate GLUT and SGLT in glucose transport. PTMs directly reflect GLUT activity. For example, GLUT 1 and 4 are upregulated following histone deacetylase inhibition, accompanied by an increase in GLUT1 acetylation (Chen et al. 2015a). ER stress-mediated ubiquitination of GLUT-2 and GLUT-4 during hyperglycemia reduces glucose uptake in the liver, exacerbating diabetic pathophysiology (Kumar et al. 2022). Moreover, PTMs indirectly reflect GLUT levels. For instance, phosphorylation of AMPK increases glucose uptake in myocytes for ATP production by mediating the expression and translocation of GLUT-4 protein (Zhang et al. 2019a). Additionally, Liao et al. found that the lncRNA EPB41L4A-AS1 increases histone H3K27 crotonylation in the GLUT-4 promoter region and non-histone PGC1-β acetylation, which inhibits GLUT-4 transcription and suppresses glucose uptake in muscle cells (Liao et al. 2022). Insulin promotes AKT phosphorylation and thus increases GLUT-1 at the plasma membrane in adipocytes to facilitate glucose uptake (Shimamoto et al. 2019). Thioredoxin Interacting Protein (TXNIP) also participates in glucose transport. It is a negative regulator of cellular glucose uptake, reducing glucose influx by promoting GLUT1 endocytosis. It also serves as a direct substrate of AKT, mediating AKT-dependent acute glucose influx, and functions as an adaptor for basal endocytosis of GLUT4 in vivo (Waldhart et al. 2017). SGLTs are also regulated by PTMs. For instance, inhibition of extracellular signal-regulated protein kinase (ERK1/2) and mTOR phosphorylation reduces SGLT-1-mediated glucose uptake (Di Franco et al. 2017). Furthermore, Cardiotrophin-1 inhibits intestinal sugar absorption by reducing SGLT-1 levels through AMPK (Lopez-Yoldi et al. 2016). Overall, PTMs regulate GLUT and SGLT via direct and indirect mechanisms that affect glucose transport. These findings demonstrate that PTMs, such as phosphorylation and acetylation, participate in glucose transport. GLUT and SGLT can serve as interesting therapeutic targets for combating abnormal glucose metabolism-related diseases.
PTMs in glycolysis
Glycolysis is the first step in the breakdown of glucose to produce high-energy molecules ATP and NADH. This process rapidly generates energy by breaking down glucose into pyruvate in the cytosol (Baker and Rutter 2023). Three crucial rate-limiting enzymes, hexokinase (HK), phosphofructokinase (PFK), and pyruvate kinase (PK), control the flux of glycolysis. The activity and protein content of rate-limiting enzymes have essential effects on glucose metabolism. Investigation of the regulatory mechanisms may provide novel insights into therapies for diseases associated with dysregulated glucose metabolism.
Numerous studies have shown that PTMs regulate glycolytic processes by regulating the translocation, content, and stability of rate-limiting enzymes. Yang et al. found that phosphorylation of hexokinase 2 (HK2) (T473) increased its activity, ultimately enhancing glucose consumption and lactate production (Yang et al. 2018). HectH9-mediated K63-linked ubiquitination is selective for HK2 regulation, and HectH9 works through HK2 to regulate glycolysis (Lee et al. 2019). Baldini et al. found that O-GlcNAc cycling in HK in hepatocytes is a novel way to regulate HK expression and increase glucose entry into liver cells (Baldini et al. 2016), supporting the crucial roles of PTMs in the glycolytic process. In addition, PTMs play a vital role in glycolysis by regulating PFK. Jeon et al. found that phosphorylation of PFKP (S386) mediated by PI3K/AKT could promote the Warburg effect (Jeon et al. 2021). The Warburg effect is characterized by increased glycolysis and lactate production regardless of oxygen availability (Vander et al. 2009). Furthermore, Li et al. found that acetylation of PFKFB3 (K472) impaired the activity of the nuclear localization signal and resulted in PFKFB3 accumulation in the cytoplasm, leading to PFKFB3 activation and enhanced glycolysis (Li et al. 2018a). O-GlcNAcylation of PFK1 (S529) inhibits its activity and regulates the glycolytic pathway through the pentose phosphate pathway (Yi et al. 2012). Moreover, PTMs regulate the final step of glycolysis by influencing the oligomeric state, subcellular localization, and biological activity of PKs. For instance, phosphorylation of pyruvate kinase muscle isozyme M2 (PKM2) (Y105) has been suggested to facilitate the Warburg effect and tumor cell growth (Kalaiarasan et al. 2014). Furthermore, acetylation of PKM2 (K433) was associated with the degradation of PKM2 and decreased PK activity (Jin et al. 2020). Wang et al. found that ubiquitin aldehyde binding 2 regulates PKM2 stability and nuclear repositioning by inhibiting its ubiquitination and blocking the interaction between PKM2 and its ubiquitin E3 ligase, thereby enhancing PKM2 activity and promoting glycolysis (Wang et al. 2022a). Chaiyawat et al. found that lower O-GlcNAcylation levels led to decreased PKM2 expression but induced higher PKM2-specific activity (Chaiyawat et al. 2015). In glycolysis, carbohydrate response element binding protein (ChREBP) binds to the promoter of liver-type pyruvate kinase and promotes the conversion of phosphoenolpyruvate to PK (Uyeda and Repa 2006). ChREBP is primarily expressed in the liver and adipose tissue and is responsible for the transcriptional control of genes involved in glucose utilization and storage (Ortega-Prieto et al. 2019). The regulation of ChREBP is complex and involves various PTMs, including phosphorylation. Under conditions of high glucose availability, ChREBP is phosphorylated by PKA in the cytoplasm. This phosphorylation event prevents ChREBP from entering the nucleus, and inhibits its transcriptional activity (Davies et al. 2008). However, under conditions of low glucose levels or increased cellular energy demand, ChREBP is dephosphorylated, allowing it to translocate into the nucleus and activate the transcription of target genes involved in glucose metabolism (Nakagawa et al. 2013). The phosphorylation status of ChREBP is tightly controlled by cellular glucose levels and energy status, allowing for fine-tuning of glucose homeostasis in response to changing metabolic demands. Regulation of glycolysis is not only exerted by the expression of glycolytic genes and interactions of glycolytic proteins within their environment but also by PTMs and transcriptional regulation. Various PTMs participate in glycolysis by regulating protein functions and may serve as significant therapeutic targets in diseases involving abnormal glucose metabolism.
PTMs in gluconeogenesis
Gluconeogenesis is a process by which non-carbohydrate precursor molecules are converted to glucose. There are two mechanisms that regulate gluconeogenesis metabolic pathways: direct regulation through rate-limiting enzymes and indirect regulation through non-rate-limiting enzymes. The key enzymes involved in regulating the rate of gluconeogenesis include PEPCK, glucose 6-phosphatase (G6Pase), pyruvate carboxylase (PC), and fructose-1,6-bisphosphatase (FBP-1). PTMs regulate gluconeogenesis by controlling enzyme activity. For example, PEPCK acetylation stimulates its interaction with E3 ubiquitin ligase (Ubiquitin Protein Ligase E3 Component N-Recognin 5) leading to PEPCK degradation in a proteasome-dependent manner (Jiang et al. 2011). Moreover, an increased level of acetylation and a decreased level of ubiquitination in PEPCK protein in mouse hepatocytes blocks PEPCK protein degradation and enhances hepatic glucose production (Wang et al. 2022d). Additionally, increased gluconeogenesis and decreased intracellular glycogen content result from increased H3K4 dimethylation at the G6Pase promoter (Pan et al. 2013). However, in a previous study, it was found that during gluconeogenesis, even though PEPCK expression was reduced by 90% in the liver after the targeted deletion of the PEPCK gene in mice, there was only a 40% reduction in gluconeogenic flux (Johanns et al. 2016). This indicates that the regulation of non-rate-limiting enzymes by PTMs also greatly influences glucose metabolism, as demonstrated in other studies (Gonzalez-Rellan et al. 2022; He et al. 2023; Li et al. 2023; Sun et al. 2020). For example, glucose starvation decreases histone acetylation at multiple sites on H3 (K9, K18, K23, and K27) to activate gluconeogenic and fat metabolism genes (Hsieh et al. 2022). Moreover, phosphorylation of CREB-regulated transcription coactivator 2 and histone deacetylase 5 by AMPK inhibits glucose production (Hunter et al. 2018). Overall, through direct or indirect effects, various PTMs play a vital role in glyconeogenesis, and may be a potential target for its regulation.
In summary, PTMs regulate glucose metabolism, including glucose transport, glycolysis, and gluconeogenesis. They also regulate the activity and protein content of enzymes in these metabolic pathways, which in turn affect glucose metabolism. Therefore, investigating the regulatory mechanisms of PTMs may provide new insights into the development of therapies for diseases associated with dysregulated glucose metabolism.
Roles of PTMs in lipid metabolism
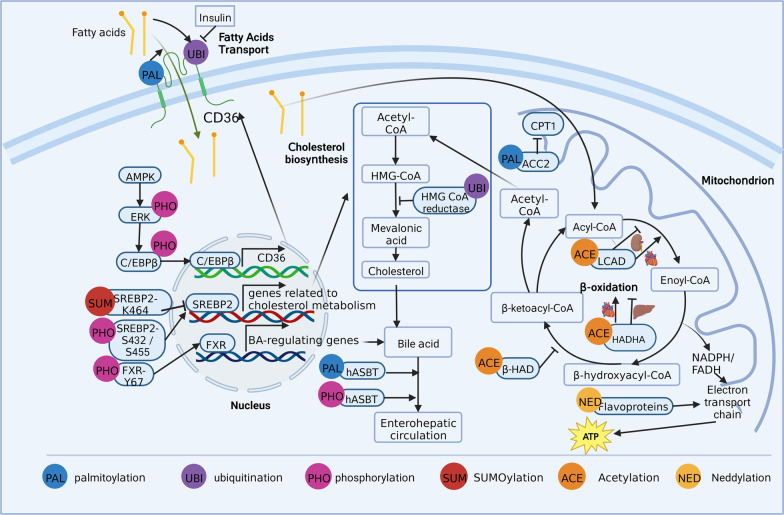
Lipids are hydrophobic molecules that include triacylglycerol, cholesterol, cholesterol esters, phospholipids, glycolipids, and lipoproteins. Lipid metabolism influences various biological processes, including energy metabolism, signal transduction, and the biosynthesis of membrane lipids (Bian et al. 2021; Marx 2022). Aberrant lipid metabolism is closely related to many diseases. For example, insufficient fatty acid uptake and utilization lead to malnutrition, whereas excessive lipid storage and hyperlipidemia are involved in atherosclerosis, obesity, and non-alcoholic fatty liver disease (Liu et al. 2021b; Lu et al. 2021). A better understanding of the factors that regulate lipid metabolism may provide new potential therapeutic strategies. Notably, a growing body of research has found that PTMs play vital roles in lipid metabolism by affecting key proteins at pivotal steps (Fig. 3).
Fig. 3.
Roles of post-translational modifications in lipid metabolism
PTMs in fatty acid transport
When the body requires energy, triglycerides stored in the adipose tissue are mobilized and decomposed into free fatty acids and glycerol, which are released into the blood and transported to tissues requiring energy. This process involves various transporters, including fatty acid translocase (FAT, also named cluster of differentiation 36, CD36), fatty acid transport proteins (FATP), and fatty acid-binding protein (FABP) (Li et al. 2022a). CD36, a key mediator of lipid transport, facilitates the transport and uptake of long-chain fatty acids (Li et al. 2022c). Several PTMs have been shown to affect CD36-mediated lipid transport (Luiken et al. 2016). For instance, Zeng et al. discovered that inhibition of CD36 palmitoylation could increase its localization to mitochondria and enhance its interaction with long-chain acyl-CoA synthetase 1, ultimately enhancing hepatic fatty acid β-oxidation (Zeng et al. 2022). In addition, ubiquitination regulates CD36 levels. However, Smith et al. discovered that fatty acids could strongly enhance the ubiquitination of CD36 and reduce CD36 protein levels, whereas insulin could reduce CD36 ubiquitination and increase CD36 protein levels (Smith et al. 2008). This opposing regulation of CD36 may modulate fatty acid uptake (Smith et al. 2008). Notably, PTMs can also affect CD36 indirectly by modifying upstream kinases and transcription factors. Choi et al. demonstrated that phosphorylation of AMPK by berberine could induce the phosphorylation of ERK1/2 and subsequently cause CCAAT/enhancer-binding protein β (C/EBPβ) binding to the C/EBP-response element in the CD36 promoter, ultimately leading to increased CD36 expression in hepatocytes (Choi et al. 2017).
Studies on PTMs of FATP and FABP are limited. Insulin receptor is a receptor tyrosine kinase. Nielsen et al. discovered that in myocytes and mammary epithelial cells, FABP was phosphorylated in response to insulin stimulation in the presence of tyrosine phosphatase inhibitors, indicating that these phosphorylated FABPs might serve as an intermediary in signal transduction pathways between the insulin receptors and lipid metabolism (Nielsen et al. 1994, Nielsen and Spener 1993). However, no phosphorylation was found in FABP from rat soleus muscle (M-FABP) upon insulin stimulation, suggesting that tyrosine phosphorylation of M-FABP was not an important physiological phenomenon (Prinsen et al. 1994). Thus, the effects of phosphorylation on FABPs from different tissues might be diverse and require further study. Further research is also needed on the role of other PTMs in FABP, and in various PTMs in FATP.
In summary, several PTMs can affect fatty acid transport by modulating the key transporter protein, CD36. Nevertheless, further research is required to explore the effects of other PTMs on CD36 and various PTMs on FATP and FABP to provide novel insights into the regulation of fatty acid transport.
PTMs in fatty acid oxidation (FAO)
Fatty acids, via their oxidation, serve as the primary energy sources in humans and mammals. Normal FAO is essential in maintaining many biological processes, whereas dysregulated FAO is associated with many diseases. FAO is a complex process and can be modulated by many mechanisms. Accumulating evidence has shown that PTMs play vital roles in FAO.
Long-chain acyl-CoA dehydrogenases (LCAD), β-hydroxyacyl-CoA dehydrogenase (β-HAD), hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit α (HADHA), and acetyl-CoA carboxylase 2 (ACC2) are key FAO enzymes, and PTMs have been known to regulate their activity. GCN5L1 is an acetylase that counteracts the deacetylation function of SIRT3 (Lv et al. 2019). Lv et al. reported that LCAD and β-HAD under the control of GCN5 general control of amino acid synthesis 5-like 1 (GCN5L1) led to decreased enzymatic activity and impaired FAO rate in a dyslipidemia-induced kidney injury model. Similarly, Thapa et al. demonstrated that acetylation of HADHA by GCN5L1 decreased its activity in HepG2 cells (Thapa et al. 2018). However, another study (Thapa et al. 2017) reported opposing results in the heart; acetylation of LCAD and HADHA by GCN5L1 enhanced FAO. These discrepancies may be caused by differences in the tissues controlling the acetylation status and FAO or tissue-specific acetylated sites of the enzymes, which should be explored in future studies. Acetyl-CoA carboxylase (ACC) is involved in FAO by inhibiting a key rate-limiting enzyme, carnitine palmitoyl transferase (CPT-I), through malonyl-CoA. A study conducted by O’Neill et al. using ACC2 S212A knock-in mice found that phosphorylation of ACC2 at S221 (S212 in mice) by AMPK regulates skeletal muscle FAO and insulin sensitivity (O’Neill et al. 2014). Neddylation is a ubiquitin-like PTM, in which the ubiquitin-like protein neural precursor cell expressed, developmentally downregulated protein 8 (NEDD8) binds to the target protein by three enzymes: the activating enzyme, conjugating enzyme, and ligase (Zhu et al. 2022a). By affecting the stability, conformation, subcellular localization, and activity of target proteins, neddylation plays critical roles in diverse biological processes including metabolism, immunity, and tumorigenesis (Zou and Zhang 2021). Zhang et al. found that hepatic neddylation could stabilize flavoproteins, thus promoting FAO in neonatal mouse livers and preventing fasting-induced steatosis in adult mice (Zhang et al. 2020b). Flavoproteins are components of the electron transport chain in the mitochondria and are essential for energy metabolism.
In summary, various PTMs regulate FAO, which may provide an extremely promising insight for treating diseases associated with dysregulated FAO. However, it is notable that the function of a type of PTM may vary when it acts on different tissues or modifies different sites of the same protein. Thus, in the future, experimental studies are needed to explore the roles and mechanisms of various PTMs in regulating genes related to FAO in different sites, tissues, or organs, which may provide a basis for developing precise treatments.
PTMs in cholesterol metabolism
Cholesterol and cholesterol esters determine the composition of the plasma membrane, act as precursors of steroid hormones and bile acids, and regulate various cellular functions (Duan et al. 2022). However, excess cholesterol is harmful and can lead to many diseases, such as cardiovascular diseases. For instance, defects in cholesterol biosynthesis cause Smith–Lemli–Opitz syndrome, a neurological and developmental disorder characterized by multiple developmental defects (Tomita et al. 2022). Excessive cholesterol levels are also associated with atherosclerosis (Baumer et al. 2020). Thus, the balance between cholesterol biosynthesis, uptake, transport, and secretion is of great importance for maintaining cholesterol homeostasis. Notably, emerging evidence has indicated that PTMs are vital in regulating cholesterol metabolism.
Several key enzymes and proteins involved in cholesterol metabolism are reportedly regulated by PTMs (Byun et al. 2018; Johnson and DeBose-Boyd 2018; Shimano and Sato 2017). For example, 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMG-CoA reductase) can be ubiquitinated and then degraded through endoplasmic reticulum-associated degradation (Johnson and DeBose-Boyd 2018). HMG-CoA reductase, which catalyzes the synthesis of mevalonic acid, is the rate-limiting enzyme in cholesterol biosynthesis. The regulation of its levels may lead to elevated levels of cholesterol and its precursors, inhibiting cholesterol synthesis and regulating cholesterol homeostasis (Johnson and DeBose-Boyd 2018). PTMs can also participate in cholesterol metabolism by regulating the activity of sterol regulatory element binding protein (SREBP)-2, an isoform of the transcription factor family SREBP, which is involved in regulating the transcription of genes related to cholesterol metabolism (Shimano and Sato 2017). The transactivation capacity of SREBP-2 was shown to decrease when it was modified by SUMO-1 at Lys464 (Hirano et al. 2003). In contrast, phosphorylation of SREBP-2 at Ser-432 and Ser-455 reportedly increased its transactivation capacity (Kotzka et al. 2004). These results indicate that the activity of SREBP-2 is commonly regulated by PTMs, which could provide novel insights into maintaining cholesterol metabolism. Notably, PTMs are also involved in the regulation of bile acid metabolism, which is the main pathway for cholesterol utilization. The Farnesoid X receptor (FXR), which transcriptionally regulates genes involved in bile acid metabolism, is essential in maintaining bile acid homeostasis. Byun et al. discovered that, in response to postprandial FGF19, phosphorylation of FXR by Src was critical for its transcriptional regulation of bile acid levels and may be a potential therapeutic target for treating bile acid-related diseases (Byun et al. 2018). In addition, phosphorylation and palmitoylation of the human apical sodium-dependent bile acid transporter (hASBT), responsible for the reclamation of bile acids from the intestinal lumen, reportedly regulate membrane expression, function, and stability of hASBT, ultimately influencing bile acid enterohepatic circulation and metabolism (Ayewoh et al. 2021; Chothe et al. 2019). Taken together, PTMs, by influencing the expression and activity of key enzymes and transcription factors, may participate in cholesterol metabolism. Considering the importance of cholesterol metabolism balance in maintaining normal physiological processes, these results may provide novel insights for the treatment of diseases caused by dysregulated cholesterol metabolism.
In conclusion, PTMs influence various processes of lipid metabolism via the modification of key proteins. PTMs also influence lipid storage (Qian et al. 2017) and adipogenesis (Su et al. 2022). Considering their importance in regulating target protein expression, activity, and location, exploring the roles of various PTMs in lipid metabolism may lead to the treatment of diseases caused by dysregulated lipid metabolism. However, since the same modification at various sites or various modifications at the same site may lead to different effects on the target protein, the detailed roles of PTMs require further investigation for precise clinical treatments.
Roles of PTMs in diseases associated with dysregulated glucose and lipid metabolism
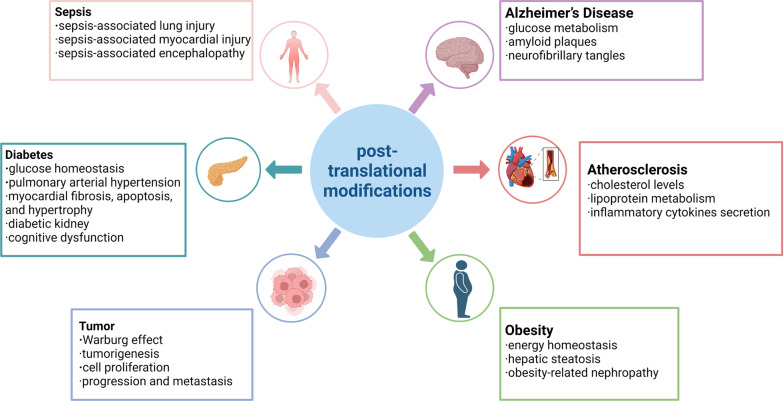
Dysregulated glucose and lipid metabolism are associated with several acute and chronic metabolic diseases, including diabetes mellitus, Alzheimer’s disease, atherosclerosis (AS), obesity, tumor and sepsis. Recently, an increasing number of studies have shown that PTMs play a vital role in these metabolic diseases by regulating glucose and lipid metabolism (Fig. 4).
Fig. 4.
Roles of PTMs in metabolic diseases
Role of PTMs in diabetes mellitus (DM)
Diabetes mellitus (DM) is a chronic metabolic disorder characterized by elevated blood glucose levels resulting from inadequate insulin production. Diabetes can be differentiated into type 1 diabetes mellitus (T1DM) and type 2 diabetes mellitus (T2DM). T1DM is an autoimmune disease resulting from the loss of immune tolerance to beta cell autoantigens. PTMs have been shown to regulate glucose and lipid metabolism and are associated with the pathology of DM; for instance, higher doses of AM-879 inhibit Ser273 phosphorylation to improve insulin sensitivity and glucose disappearance rates (Terra et al. 2023). Moreover, glucose and lipid metabolism disturbances promote myocardial fibrosis, apoptosis, and hypertrophy by inhibiting phosphorylation of Akt (T308 or S473) mediated by galectin-3 (Sun et al. 2021b). Although the expression level of enoyl-CoA-hydratase/3-hydroxyacyl-CoA dehydrogenase decreases, acetylation enhances its activity, thereby overall increasing β-oxidation processes in the kidneys of diabetic individuals (Sas et al. 2016). Artemisia dracunculus L. extract increased the phosphorylation of AMPK (T172), affecting downstream signaling of AMPK, inhibiting ACC and increasing SIRT1 protein levels to improve glucose homeostasis by enhancing insulin action and reducing ectopic lipid accumulation (Vandanmagsar et al. 2021). Recent studies have shown that PTMs are also involved in glucose transport, affecting the development of diabetes. For instance, the inhibition of SGLT1 expression and lack of O-GlcNAcylation in the gut decreased glucose absorption (Nishimura et al. 2022). Yang et al. found that Epigynum auritum increased phosphorylation levels of Akt, AMPK, and GSK-3β, which in turn upregulated the expression of GLUT-2 and GLUT-4, thus exerting a hypoglycemic effect (Yang et al. 2022a). Increased O-GlcNAc levels in diabetes decrease ERα activity, which reduces the brain's ability to utilize glucose, reduces the release of neurotrophic factors, and increases the risk of neuronal oxidative stress (Shi et al. 2021). Furthermore, the phosphorylation of ribosomal protein S6 kinase and the SREBP1 pathway in nearby hepatocytes is influenced by calpain proteolysis in cultured ECs, leading to the induction of de novo lipogenesis (Akasu et al. 2022). The identification and characterization of these PTMs present significant challenges and research on PTMs can provide substantial insights into the biological functions of these proteins.
Role of PTMs in Alzheimer’s disease (AD)
AD, a neurodegenerative disease associated with decreased cognitive abilities, is characterized by dysregulated brain glucose metabolism and the accumulation of abnormal protein deposits called myloid plaques and neurofibrillary tangles (NFTs) (Guillozet et al. 2003). PTMs such as phosphorylation, O-GlcNAcylation, and succinylation play a vital role in AD pathogenesis. Tau, a cytosolic phosphoprotein associated with microtubule assembly, is modified by various PTMs. Hyperphosphorylated tau is a major component of NFTs in AD; this indicates that inhibiting the hyperphosphorylation of tau may be a novel therapeutic target for AD. For instance, Zhou et al. discovered that Sirt2 was involved in tau phosphorylation through ERK activation in vivo and in vitro, providing novel insights for the treatment of AD (Zhou et al. 2022a). Xu et al. demonstrated that electroacupuncture preserves cognition in an AD mouse model. At the molecular level, electroacupuncture enhances glucose metabolism and inhibits abnormal phosphorylation of tau protein via the AKT/GSK3β signaling pathway (Xu et al. 2020). Regarding O-GlcNAcylation, Liu et al. revealed that downregulation of tau O-GlcNAcylation leads to abnormal hyperphosphorylation of tau and neurofibrillary degeneration in AD (Liu et al. 2004, 2009). Using proteomic analysis, Tramutola et al. discovered that proteins with reduced O-GlcNAcylation levels are involved in key pathways in the progression of AD, such as neuronal structure, protein degradation, and glucose metabolism (Tramutola et al. 2018). Pinho et al. found that globally reduced O-GlcNAcylation levels were associated with impaired mitochondrial bioenergetic function, disruption of the mitochondrial network, and loss of cell viability in in vitro models of AD (Pinho et al. 2019). These results provide a better understanding of the role of O-GlcNAcylation in AD. In recent years, other PTMs, such as succinylation, lactylation, glycosylation, palmitoylation, and nitrosylation have also been found to participate in AD (Abrams et al. 2011; Andrew et al. 2017; Bukke et al. 2020; Pan et al. 2022; Yang et al. 2022b). Yang et al. discovered that succinylation of the amyloid precursor protein promoted amyloid plaque formation, and succinylation of tau promoted its aggregation to NFTs, indicating that succinylation may be associated with AD (Yang et al. 2022b). Moreover, dysregulated O-GlcNAcylation and succinylation in AD may be caused by abnormalities in brain glucose metabolism (Liu et al. 2009; Yang et al. 2022b), suggesting that these PTMs may link dysregulated brain glucose metabolism to pathological alterations in AD. In conclusion, various PTMs regulate the pathogenesis of AD and provide insights into potential therapeutic targets for AD.
Role of PTMs in atherosclerosis (AS)
AS is characterized by large and medium arteries, which are caused by metabolic disorders of the arterial vessel wall, and is commonly considered a major contributor to cardiovascular diseases (CVDs), including stroke and myocardial infarction (Meng et al. 2022). PTMs have been associated with the pathology of AS by regulating glucose and lipid metabolism. First, abnormal levels of protein phosphorylation have been found to be closely related to the occurrence and development of AS. FGF19-induced phosphorylation of hepatic FXR is a nuclear receptor that plays an important role in maintaining metabolic homeostasis via the transcriptional control of many genes. Byun et al. reported that FXR could maintain cholesterol levels and thus protect against AS (Byun et al. 2019).
Glycosylation has also been shown to participate in regulating AS. Altered glycosylation of various proteins involved in lipoprotein metabolism, such as apolipoproteins and lipoprotein receptors, can change their expression and/or function, thus affecting AS development (Pirillo et al. 2021). For example, low density lipoprotein receptor (LDLR), a glycoprotein, regulates circulating LDL-C levels by binding to LDLs. Glycosylation of LDLR is essential for its function by maintaining its expression and binding affinity with LDLs and very-low-density lipoproteins (Filipovic 1989; van den Boogert et al. 2019; Wang et al. 2018a). Ye et al. showed that the expression of GalNAc-T4 (GALNT4) and protein O-glycosylation were both increased in plaques in ApoE−/−mice, and GALNT4 could increase O-glycosylation of PSGL-1 via the Akt/mTOR and NF-κB pathways, thus priming adhesion and transmigration of monocytes in AS. These results provide novel insights into the role of O-glycosylation in the pathogenesis of AS, suggesting that GALNT4 may be a potential target for AS treatment (Ye et al. 2022).
Moreover, S-nitrosylation and SUMOylation were found to play important roles in AS (Chen et al. 2015b; Li et al. 2018b; Liu et al. 2020). Hyperhomocysteinemia (HHcy) is an independent risk factor for CVDs, including AS. HHcy may participate in AS by regulating S-nitrosylation. Chen et al. reported that HHcy can promote AS by reducing endothelial or aortic protein S-nitrosylation levels (Chen et al. 2015b). In addition, Li et al. reported that HHcy could also reduce the level of protein S-nitrosylation in T cells, ultimately promoting the secretion of inflammatory cytokines and the proliferation of T cells and AS. Mechanistically, HHcy increased the expression of S-nitrosoglutathione reductase (GSNOR), a key enzyme controlling denitrosylation. These results provide new insights into HHcy-induced AS (Li et al. 2018b). Taken together, PTMs, including phosphorylation, glycosylation, S-nitrosylation, and SUMOylation, are instrumental in regulating AS development; however, detailed functions and mechanisms require further investigation to provide a basis for developing precise treatments.
Role of PTMs in obesity
Obesity, a serious public health problem worldwide, is a significant risk factor for many diseases, including CVDs, T2DM, and non-alcoholic fatty liver disease (Wensveen et al. 2015). The pathogenesis of obesity is complicated and includes genetic factors, environmental factors, and metabolic dysregulation (Cruciani et al. 2023). Recent studies have explored the roles and molecular mechanisms of PTMs in obesity. For example, N-myristoylation is a ubiquitous, generally co-translational modification of newly synthesized proteins that involves attachment of the C14 fatty acid (myristic acid) to N-terminal glycine (Rampoldi et al. 2012). Neopane et al. reported that blocking AMPK β1 myristoylation enhanced AMPK activity and protected mice from high-fat diet-induced obesity and hepatic steatosis (Neopane et al. 2022). AMPK is a cellular energy sensor that can phosphorylate a variety of substrates, including key metabolic proteins and transcription factors, to restore energy homeostasis. Therefore, these results may provide a novel strategy for treating metabolic diseases. As mentioned above, FXR controls the expression of many genes involved in bile acid, lipid, glucose, and amino acid metabolism, and maybe a potential target for diseases associated with metabolic disorders. Numerous studies have shown that FXR can be modified by several PTMs and can affect obesity-related disorders. For example, Kim et al. revealed that a dysregulated acetyl/SUMO switch in FXR could promote obesity. Mechanistically, acetylation of FXR blocks its interaction with the SUMO ligase PIASy and inhibits SUMO2 modification at K277, leading to obesity (Kim et al. 2015). These results provide potential therapeutic and diagnostic targets for obesity-related metabolic disorders. Obesity is also a significant risk factor for kidney damage, namely obesity-related nephropathy (Arabi et al. 2022). Chen et al. showed that IκB kinase could inactivate the deubiquitination activity of cylindromatosis protein by activating its phosphorylation, thus promoting the ubiquitination of Nrf2 and aggravating oxidative stress injury in the kidney in obesity-related nephropathy (Chen et al. 2021b). In summary, PTMs play an important role in obesity and obesity-related diseases and represent a large number of potential therapeutic targets. However, in the future, more experimental studies are needed to explore the roles and mechanisms of various PTMs in regulating the expression of genes related to metabolism and the resulting impact on obesity.
Role of PTMs in tumor
PTMs are crucial for controlling tumor immunity and immunotherapy and offering a potential target for enhancing the effectiveness of immunotherapy. Tumor immune microenvironments and the impact of the immune system, in addition to changes in cancer cells, are the primary factors in tumor initiation and development. PTMs such as phosphorylation, ubiquitination, acetylation, and glycosylation, are thought to be associated with tumorigenesis. For instance, epidermal growth factor receptor phosphorylation by PFKP (Y64) has been known to be involved in AKT activation and AKT-mediated phosphorylation of β-catenin (S552), promoting the glycolytic process in brain tumor growth (Lee et al. 2020). Glycyrrhizin inhibits HK2 by decreasing the phosphorylation level of AKT, suppressing the Warburg effect and cell proliferation in peripheral nerve injury (Sun et al. 2021a). PKC increases the phosphorylation and nuclear translocation of PKM2 to enhance lipogenesis and tumor development in prostate cancer cells (Lai et al. 2023). Moreover, tripartite motif-containing 35 (Trim35) regulates the tetrameric and dimeric leaps of PKM2 through ubiquitin action and affects the malignant biological behavior of breast cancer by regulating the Warburg effect (Wu et al. 2022). Glycosylation of PFK1 at (S529) reduces cancer cell proliferation in vitro and slows tumor development (Yi et al. 2012). PKM2 glycosylation may be a novel target for controlling cancer metabolism and tumorigenesis in colorectal cancer (Chaiyawat et al. 2015). Membrane-associated RING-CH 8 promotes ubiquitination-mediated proteasomal degradation to reduce HK2 protein levels, thereby regulating and repressing glycolysis to promote tumor suppressors in colorectal cancer (Wang et al. 2022e). Cisplatin-acetylated PFKFB3 (K472) causes accumulation of PFKFB3 in the cytoplasm, which facilitates its phosphorylation by AMPK, leading to PFKFB3 activation and enhanced glycolysis (Li et al. 2018a). Epidermal growth factor receptor activation rapidly increases PFKFB3 phosphorylation and expression and increases glycolysis in non-small cell lung cancer cells (Lypova et al. 2019). These studies reveal that cancer development, progression, and metastasis are intimately correlated with PTMs, although the underlying molecular pathways remain poorly understood. Further, PTM-mediated dysfunction of glucose and lipid metabolism, especially its effects on various organs, is closely related to tumorigenesis. Hence, PTMs can be highly relevant in the search for drug targets and diagnostic biomarkers in tumorigenesis.
Role of PTMs in sepsis
Sepsis is defined as the presence of systemic signs of infection, while severe sepsis is defined as sepsis plus sepsis-induced organ dysfunction or tissue hypoperfusion (Singer et al. 2016). PTMs are significantly associated with sepsis-associated lung injury, myocardial injury, and encephalopathy. For instance, modulation of sepsis-enhanced glycolysis with 2-deoxy-d-glucose significantly attenuates sepsis-induced cardiac dysfunction. These mechanisms involve attenuating sepsis-induced pro-inflammatory responses and myocardial apoptosis by decreasing mitogen activated protein kinase 3 phosphorylation (Zheng et al. 2017). In addition, in the LPS-treated human umbilical vein endothelial cell (HUVEC) model, dichloroacetate restored pyruvate dehydrogenase complex function by reversing LPS-induced phosphorylation of pyruvate dehydrogenase E1 (S293 and S300), preventing lactic acid production and HUVEC monolayer barrier dysfunction (Mao et al. 2022). Moreover, inhibition of glycolysis or the prevention of PKM2 nuclear aggregation significantly reduces the phosphorylation and activation of transcription factor 2 (ATF2), thus reducing LPS-induced pyroptosis of microglia (Li et al. 2021). ER stress can increase the phosphorylation of signal transducer and activator of transcription 3 (STAT3) and monoclonal antibody to Suppressor of Mothers against Decapentaplegic (SMAD) family member 3 (Smad3), and also activate UPS‐mediated proteolysis to promote sepsis‐induced muscle atrophy (Zheng et al. 2023). Finally, p53 deacetylation by the deacetylase Sirtuin 1 (Sirt1) through resveratrol/quercetin administration or mutation of the acetylated lysine site in p53 promotes renal tubular epithelial cell autophagy, alleviating sepsis-induced acute kidney injury. Other PTM-mediated dysfunctions of glucose metabolism are closely related to sepsis. For instance, cynaroside inhibited glycolysis-related proteins, including PFKFB3, HK2, and HIF-1α, and glycolysis-related hyperacetylation of high mobility group box 1 (HMGB1) to restore PK activity in the septic liver (Pei et al. 2021). Furthermore, Hwang et al. reported a protective effect of glucosamine on sepsis, potentially through the O-GlcNAcylation of nucleocytoplasmic proteins in sepsis-induced lung injury and inflammation (Hwang et al. 2019). Therefore, PTMs exert various physiological effects in sepsis models by affecting the lung, skeleton, brain, and cardiac muscles. Overall, the most common PTMs involved in glucose metabolism include phosphorylation, acetylation, and ubiquitination.
Further prospects of PTMs in glucose and lipid metabolism
PTMs play essential roles in cellular physiology and pathology, regulate glucose and lipid metabolism, and influence almost all aspects of cell biology and pathogenesis. However, many issues remain to be resolved before PTM sites can be used as promising targets for treating glucose and lipid metabolism disorders. Understanding the molecular mechanisms underlying PTMs could shed light on new therapeutic interventions. Although excellent work on PTMs has been carried out in past decades, PTMs of rate-limiting enzymes in glycosphingolipid biosynthesis need to be considered for future development. Moreover, PTMs have diverse functions and can regulate other PTMs, leading to complex regulatory crosstalk. Interprotein crosstalk between phosphorylation and SUMOylation has been widely reported. For example, S-phase kinase-associated protein 2, an E3 ubiquitin ligase, mediates FBP1 protein ubiquitination and degradation induced by phosphatase and tensin homolog loss and promotes the Warburg effect in prostate cancer cell growth (Song et al. 2022). Enhancing the connection between Akt and HK2 through K63-linked ubiquitination eventually leads to an increase in the phosphorylation of HK2 on Thr473 and mitochondrial localization, which is involved in glycolysis and tumor development (Yu et al. 2019). Thus, these PTMs greatly complicate mechanisms that modulate proteasome activity.
In addition, various regulations of rate-limiting enzymes in glucose and lipid metabolism could improve our understanding of the biological roles of these PTMs and provide a foundation for the research of regulatory mechanisms for these types of PTMs. Different diseases affect the corresponding processes of glycolipid metabolism, thus exerting specific regulatory effects. In diabetes, glucose transport disorders and gluconeogenesis have become the focus of disease intervention, and various glucose-lowering drug treatments have been developed based on the PTMs of SGLT and GLUT. However, in sepsis, PTMs specifically regulate metabolic changes in the septic state by modulating the activity and localization of enzymes, such as glycolytic processes, mainly affecting the Warburg effect, which has become an essential target for sepsis intervention. Angiogenesis and immune escape are important intervention targets for PTMs in tumor development. With the development of genomic, transcriptomic, proteomic, and epigenetic technologies, the prospects of novel drugs targeting PTM sites are promising. PTM sites have been proven to be promising therapeutic sites for treating glucose and lipid metabolism disorders, although further studies are needed to elucidate the mechanisms involved.
Advances in next-generation sequencing and mass spectrometry proteomics technologies have led to an explosion of data on PTM sites and disease-associated glucose and lipid metabolism. To predict succinylation sites, machine-learning-based prediction of protein modification sites, such as DeepSuccinylSite, has become popular (Thapa et al. 2020). In the future, PTM sites may become novel biomarkers and therapeutically-related targets for glucose and lipid metabolism diseases. For example, PTMs of blood-derived alpha-synuclein can act as biochemical markers for Parkinson’s disease (Vicente et al. 2017). Although research on PTMs has increased in recent years, their role in glucose and lipid metabolism disorders requires further investigation.
Thus far, proteomic studies on PTMs that regulate development have primarily focused on phosphorylation. Frequently, abnormal phosphorylation causes cellular processes to become disorganized, which ultimately results in the onset and progression of illnesses. Consequently, medications often target kinases and phosphatases. Nearly one-third of the pharmaceutical industry's current drug development initiatives focus on PKs, one of the most significant categories of therapeutic targets. For example, the hypoglycemic and anti-obesity characteristics of cardiotrophin-1 (CT-1) may be explained by the fact that CT-1 limits intestinal sugar absorption by lowering SGLT-1 levels through AMPK phosphorylation (Lopez-Yoldi et al. 2016). Additionally, by increasing the activities of HK, glycogen synthase, and the phosphorylation of glycogen synthase kinase 3 (GSK3) protein, whole-grain highland barley enhances glycogen storage in the liver (Deng et al. 2020). Similarly, other PTMs can be used as targets for drug therapy. The PI3-K/Akt-GSK3beta-FBW7 signaling axis was downregulated by xanthohumol, which led to the ubiquitination of c-Myc and inhibition of tumor glycolysis (Yuan et al. 2020). Further multicenter clinical studies are needed to emphasize the role of modifications in clinical applications and confirm their clinical significance.
Conclusion
In this review, we summarized the latest advancements pertaining to PTMs involved in regulating glucose and lipid metabolism. Regulation of rate-limiting metabolic enzymes is essential for controlling cellular metabolic changes. PTMs offer a dynamic way to regulate subcellular localization, stability, and protein interactions and activity. Moreover, PTMs regulate cellular metabolism, especially involving rate-limiting metabolic enzymes. In recent years, PTMs have been shown to participate in nearly all aspects of vital biological processes by regulating protein functions, such as glucose transport, glycolysis, and gluconeogenesis, and aberrant states of PTMs are frequently implicated in diseases involving glucose and lipid metabolism. Hence, PTM sites may become potential therapeutic targets for regulating glucose and lipid metabolism and controlling disease progression. Therefore, in-depth insights into the mechanisms of PTMs in glucose and lipid metabolism may provide a theoretical basis for developing new drugs. Thus, future studies should focus on the following issues:
Molecular mechanisms underlying the role of AKT in PTMs of GLUT and SGLT transporter proteins.
Regulation of PFK by PTMs that affects the Warburg effect and glycolytic pathway in sepsis.
Thorough investigation of the expression of PC and regulation of its activity, as PC, in addition to PEPCK, FBP-1 and G6Pase, is another rate-limiting enzyme in gluconeogenesis.
Clinical research regarding precision medicine and potential therapeutic targets for clinical diagnosis, prognosis, and therapy of PTMs in lipid and glucose metabolism.
Improved understanding of the physiological effects of crosstalk between different PTMs.
Regulation of non-rate-limiting enzymes, in addition to the PTMs of critical enzymes involved in glycolipid metabolism.
Acknowledgements
The authors thank all research team members for their contributions to this work. The figures are created with Biorender.com.
Abbreviations
- ACC
Acetyl-CoA carboxylase
- ATF2
Activation of transcription factor 2
- AD
Alzheimer’s disease
- AMPK
AMP-activated protein kinase
- AS
Atherosclerosis
- ATP
Adenosine triphosphate
- CVDs
Cardiovascular diseases
- CPT-I
Carnitine palmitoyl transferase
- DUBs
Deubiquitinases
- DM
Diabetes mellitus
- ERAD
Endoplasmic reticulum-associated degradation
- FXR
Farnesoid X receptor
- FAO
Fatty acid oxidation
- FAT
Fatty acid translocase
- FATP
Fatty acid transport proteins
- FABP
Fatty acid-binding protein
- GLUT
Glucose transporters
- HIF1α
Hypoxia-inducible factor 1 alpha
- HK
Hexokinase
- HUVEC
Human umbilical vein endothelial cell
- KATs
Lysine acetyltransferases
- KDACs
Lysine deacetylases
- MYD88
Myeloid differentiation primary response 88
- OGT
O-GlcNAc transferase
- OGA
O-GlcNAcase
- PEPCK
Phosphoenolpyruvate carboxykinase
- PFK
Phosphofructokinase
- PFK
Phosphofructokinase
- PTMs
Post-translational modifications
- PC
Pyruvate carboxylase
- PK
Pyruvate kinase
- AKT
Protein kinase B
- SUMO
Small ubiquitin-like modifier
- SGLTs
Sodium-dependent glucose transporters
- TRIF
Toll/Interleukin-1 receptor-domain-containing adapter-inducing interferon-β
Author contributions
Y-HY and RW contributed to the conceptualization, methodology, formal analysis, and original draft preparation. NY contributed to the methodology and visualization. T-NZ and C-FL contributed to the conceptualization, validation, review and editing, supervision, project administration, and funding acquisition. RW and Y-HY contributed equally to this work.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81971810 to Chun-Feng Liu and 82102254 to Tie-Ning Zhang) and the Natural Science Foundation of Liaoning Province (No. 2017225003 and No. 2018108001 to Chun-Feng Liu), the Major Scientific and Technological Special Project of Liaoning Province (No. 2020JH1/10300001), Shenyang’s Science and Technology Program (No. 20-205-4-002), and 345 Talent Project of Shengjing Hospital of China Medical University (M0691).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yu-Hang Yang and Ri Wen contributed equally to this work.
Contributor Information
Tie-Ning Zhang, Email: cmuztn@vip.qq.com.
Chun-Feng Liu, Email: liucf@sj-hospital.org.
References
- Abrams AJ, Farooq A, Wang G. S-nitrosylation of apoe in Alzheimer’s disease. Biochemistry. 2011;50(17):3405–3407. doi: 10.1021/bi200266v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed SA, Sarma P, Barge SR, Swargiary D, Devi GS, Borah JC. Xanthosine, a purine glycoside mediates hepatic glucose homeostasis through inhibition of gluconeogenesis and activation of glycogenesis via regulating the ampk/ foxo1/akt/gsk3beta signaling cascade. Chem Biol Interact. 2023;371:110347. doi: 10.1016/j.cbi.2023.110347. [DOI] [PubMed] [Google Scholar]
- Akasu R, Miyazaki T, Elhussiny MZ, Sugiura Y, Tomitsuka Y, Haraguchi S, et al. Calpain-mediated proteolytic production of free amino acids in vascular endothelial cells augments obesity-induced hepatic steatosis. J Biol Chem. 2022;298(6):101953. doi: 10.1016/j.jbc.2022.101953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S, Khan MA, Zubair H, Sudan SK, Vikramdeo KS, Deshmukh SK, et al. Myb sustains hypoxic survival of pancreatic cancer cells by facilitating metabolic reprogramming. EMBO Rep. 2023;24(3):e55643. doi: 10.15252/embr.202255643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew RJ, Fernandez CG, Stanley M, Jiang H, Nguyen P, Rice RC, et al. Lack of bace1 s-palmitoylation reduces amyloid burden and mitigates memory deficits in transgenic mouse models of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2017;114(45):E9665–E9674. doi: 10.1073/pnas.1708568114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi T, Shafqat A, Sabbah BN, Fawzy NA, Shah H, Abdulkader H, et al. Obesity-related kidney disease: beyond hypertension and insulin-resistance. Front Endocrinol. 2022;13:1095211. doi: 10.3389/fendo.2022.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayewoh EN, Czuba LC, Nguyen TT, Swaan PW. S-acylation status of bile acid transporter hasbt regulates its function, metabolic stability, membrane expression, and phosphorylation state. Biochim Biophys Acta-Biomembr. 2021;1863(2):183510. doi: 10.1016/j.bbamem.2020.183510. [DOI] [PubMed] [Google Scholar]
- Baek J, Sas K, He C, Nair V, Giblin W, Inoki A, et al. The deacylase sirtuin 5 reduces malonylation in non-mitochondrial metabolic pathways in diabetic kidney disease. J Biol Chem. 2023 doi: 10.1016/j.jbc.2023.102960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SA, Rutter J. Metabolites as signalling molecules. Nat Rev Mol Cell Biol. 2023 doi: 10.1038/s41580-022-00572-w. [DOI] [PubMed] [Google Scholar]
- Baldini SF, Steenackers A, Olivier-Van SS, Mir AM, Mortuaire M, Lefebvre T, et al. Glucokinase expression is regulated by glucose through o-glcnac glycosylation. Biochem Biophys Res Commun. 2016;478(2):942–948. doi: 10.1016/j.bbrc.2016.08.056. [DOI] [PubMed] [Google Scholar]
- Bangarh R, Khatana C, Kaur S, Sharma A, Kaushal A, Siwal SS, et al. Aberrant protein glycosylation: implications on diagnosis and immunotherapy. Biotechnol Adv. 2023;66:108149. doi: 10.1016/j.biotechadv.2023.108149. [DOI] [PubMed] [Google Scholar]
- Bao X, Wang Y, Li X, Li XM, Liu Z, Yang T, et al. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. Elife. 2014 doi: 10.7554/eLife.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer Y, Mehta NN, Dey AK, Powell-Wiley TM, Boisvert WA. Cholesterol crystals and atherosclerosis. Eur Heart J. 2020;41(24):2236–2239. doi: 10.1093/eurheartj/ehaa505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Umit KH, Jin J, Gozani O. Epigenetics and beyond: targeting writers of protein lysine methylation to treat disease. Nat Rev Drug Discov. 2021;20(4):265–286. doi: 10.1038/s41573-020-00108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X, Liu R, Meng Y, Xing D, Xu D, Lu Z. Lipid metabolism and cancer. J Exp Med. 2021 doi: 10.1084/jem.20201606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar KK, Li SS. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol. 2015;16(1):5–17. doi: 10.1038/nrm3915. [DOI] [PubMed] [Google Scholar]
- Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65(1):8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Bond MR, Hanover JA. O-glcnac cycling: a link between metabolism and chronic disease. Annu Rev Nutr. 2013;33:205–229. doi: 10.1146/annurev-nutr-071812-161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukke VN, Villani R, Archana M, Wawrzyniak A, Balawender K, Orkisz S, et al. The glucose metabolic pathway as a potential target for therapeutics: crucial role of glycosylation in Alzheimer’s disease. Int J Mol Sci. 2020 doi: 10.3390/ijms21207739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun S, Kim DH, Ryerson D, Kim YC, Sun H, Kong B, et al. Postprandial fgf19-induced phosphorylation by src is critical for fxr function in bile acid homeostasis. Nat Commun. 2018;9(1):2590. doi: 10.1038/s41467-018-04697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun S, Jung H, Chen J, Kim YC, Kim DH, Kong B, et al. Phosphorylation of hepatic farnesoid x receptor by fgf19 signaling-activated src maintains cholesterol levels and protects from atherosclerosis. J Biol Chem. 2019;294(22):8732–8744. doi: 10.1074/jbc.RA119.008360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Bu C, Zhang J, Ren Y, Zhou G, Chen C, et al. Exosomal circular rna hsa_circ_0046060 of umbilical cord mesenchymal stromal cell ameliorates glucose metabolism and insulin resistance in gestational diabetes mellitus via the mir-338-3p/g6pc2 axis. Int J Endocrinol. 2022;2022:9218113. doi: 10.1155/2022/9218113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CR, Asfaha JB, Ghent CM, Howard CJ, Hartooni N, Safari M, et al. Phosphoregulation of phase separation by the SARS-cov-2 n protein suggests a biophysical basis for its dual functions. Mol Cell. 2020;80(6):1092–1103. doi: 10.1016/j.molcel.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiyawat P, Chokchaichamnankit D, Lirdprapamongkol K, Srisomsap C, Svasti J, Champattanachai V. Alteration of o-glcnacylation affects serine phosphorylation and regulates gene expression and activity of pyruvate kinase m2 in colorectal cancer cells. Oncol Rep. 2015;34(4):1933–1942. doi: 10.3892/or.2015.4178. [DOI] [PubMed] [Google Scholar]
- Chandel NS. Carbohydrate metabolism. Cold Spring Harbor Perspect Biol. 2021 doi: 10.1101/cshperspect.a040568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Yeh E. Sumo: from bench to bedside. Physiol Rev. 2020;100(4):1599–1619. doi: 10.1152/physrev.00025.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YH, Weng CL, Lin KI. O-glcnacylation and its role in the immune system. J Biomed Sci. 2020;27(1):57. doi: 10.1186/s12929-020-00648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P, Yu L, Friedman GK, Wang M, Ke X, Wang H, et al. Integrin alphavbeta3 engagement regulates glucose metabolism and migration through focal adhesion kinase (fak) and protein arginine methyltransferase 5 (prmt5) in glioblastoma cells. Cancers. 2021 doi: 10.3390/cancers13051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6(5):812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Du J, Zhao YT, Zhang L, Lv G, Zhuang S, et al. Histone deacetylase (hdac) inhibition improves myocardial function and prevents cardiac remodeling in diabetic mice. Cardiovasc Diabetol. 2015;14:99. doi: 10.1186/s12933-015-0262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu R, Zhang G, Yu Q, Jia M, Zheng C, et al. Hypercysteinemia promotes atherosclerosis by reducing protein s-nitrosylation. Biomed Pharmacother. 2015;70:253–259. doi: 10.1016/j.biopha.2015.01.030. [DOI] [PubMed] [Google Scholar]
- Chen L, Chen XW, Huang X, Song BL, Wang Y, Wang Y. Regulation of glucose and lipid metabolism in health and disease. Sci China Life Sci. 2019;62(11):1420–1458. doi: 10.1007/s11427-019-1563-3. [DOI] [PubMed] [Google Scholar]
- Chen AN, Luo Y, Yang YH, Fu JT, Geng XM, Shi JP, et al. Lactylation, a novel metabolic reprogramming code: current status and prospects. Front Immunol. 2021;12:688910. doi: 10.3389/fimmu.2021.688910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, Hong H, Lei YT, Zou J, Yang YY, He LY. Ikappab kinase promotes nrf2 ubiquitination and degradation by phosphorylating cylindromatosis, aggravating oxidative stress injury in obesity-related nephropathy. Mol Med. 2021;27(1):137. doi: 10.1186/s10020-021-00398-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Tang Y, Chen Y, Kim S, Liu H, Li SS, et al. Molecular characterization of propionyllysines in non-histone proteins. Mol Cell Proteomics. 2009;8(1):45–52. doi: 10.1074/mcp.M800224-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SC, Scicluna BP, Arts RJ, Gresnigt MS, Lachmandas E, Giamarellos-Bourboulis EJ, et al. Broad defects in the energy metabolism of leukocytes underlie immunoparalysis in sepsis. Nat Immunol. 2016;17(4):406–413. doi: 10.1038/ni.3398. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Lee KY, Jung SH, Kim HS, Shim G, Kim MG, et al. Activation of ampk by berberine induces hepatic lipid accumulation by upregulation of fatty acid translocase cd36 in mice. Toxicol Appl Pharmacol. 2017;316:74–82. doi: 10.1016/j.taap.2016.12.019. [DOI] [PubMed] [Google Scholar]
- Chothe PP, Czuba LC, Ayewoh EN, Swaan PW. Tyrosine phosphorylation regulates plasma membrane expression and stability of the human bile acid transporter asbt (slc10a2) Mol Pharm. 2019;16(8):3569–3576. doi: 10.1021/acs.molpharmaceut.9b00426. [DOI] [PubMed] [Google Scholar]
- Cockram PE, Kist M, Prakash S, Chen SH, Wertz IE, Vucic D. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 2021;28(2):591–605. doi: 10.1038/s41418-020-00708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. The regulation of protein function by multisite phosphorylation–a 25 year update. Trends Biochem Sci. 2000;25(12):596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- Cohen P. The origins of protein phosphorylation. Nat Cell Biol. 2002;4(5):E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- Cruciani S, Delitala AP, Cossu ML, Ventura C, Maioli M. Management of obesity and obesity-related disorders: from stem cells and epigenetics to its treatment. Int J Mol Sci. 2023 doi: 10.3390/ijms24032310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Ren T, Zhang Y, Nan N. Methylation multiplicity and its clinical values in cancer. Expert Rev Mol Med. 2021;23:e2. doi: 10.1017/erm.2021.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A. Protein s-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci. 2009;34(2):85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Davies MN, O'Callaghan BL, Towle HC. Glucose activates chrebp by increasing its rate of nuclear entry and relieving repression of its transcriptional activity. J Biol Chem. 2008;283(35):24029–24038. doi: 10.1074/jbc.M801539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng N, Guo R, Zheng B, Li T, Liu RH. Irs-1/pi3k/akt pathway and mirnas are involved in whole grain highland barley (hordeum vulgare l.) Ameliorating hyperglycemia of db/db mice. Food Funct. 2020;11(11):9535–9546. doi: 10.1039/d0fo01990a. [DOI] [PubMed] [Google Scholar]
- Di Franco A, Cantini G, Tani A, Coppini R, Zecchi-Orlandini S, Raimondi L, et al. Sodium-dependent glucose transporters (sglt) in human ischemic heart: a new potential pharmacological target. Int J Cardiol. 2017;243:86–90. doi: 10.1016/j.ijcard.2017.05.032. [DOI] [PubMed] [Google Scholar]
- Dilworth D, Barsyte-Lovejoy D. Targeting protein methylation: from chemical tools to precision medicines. Cell Mol Life Sci. 2019;76(15):2967–2985. doi: 10.1007/s00018-019-03147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong K, Wu M, Liu X, Huang Y, Zhang D, Wang Y, et al. Glutaredoxins concomitant with optimal ros activate ampk through s-glutathionylation to improve glucose metabolism in type 2 diabetes. Free Radic Biol Med. 2016;101:334–347. doi: 10.1016/j.freeradbiomed.2016.10.007. [DOI] [PubMed] [Google Scholar]
- Drazic A, Myklebust LM, Ree R, Arnesen T. The world of protein acetylation. Biochim Biophys Acta. 2016;1864(10):1372–1401. doi: 10.1016/j.bbapap.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Du C, Chen X, Su Q, Lu W, Wang Q, Yuan H, et al. The function of sumoylation and its critical roles in cardiovascular diseases and potential clinical implications. Int J Mol Sci. 2021 doi: 10.3390/ijms221910618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Gong K, Xu S, Zhang F, Meng X, Han J. Regulation of cholesterol homeostasis in health and diseases: from mechanisms to targeted therapeutics. Signal Transduct Target Ther. 2022;7(1):265. doi: 10.1038/s41392-022-01125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich CH, Bakshi S, Kelsall IR, Ortiz-Guerrero J, Shpiro N, Cohen P. Lys63/met1-hybrid ubiquitin chains are commonly formed during the activation of innate immune signalling. Biochem Biophys Res Commun. 2016;474(3):452–461. doi: 10.1016/j.bbrc.2016.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovic I. Effect of inhibiting n-glycosylation on the stability and binding activity of the low density lipoprotein receptor. J Biol Chem. 1989;264(15):8815–8820. doi: 10.1016/S0021-9258(18)81866-X. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic o-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-n-acetylglucosaminidase from human brain. J Biol Chem. 2001;276(13):9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- Gao J, He X, Ma Y, Zhao X, Hou X, Hao E, et al. Chlorogenic acid targeting of the akt ph domain activates akt/gsk3beta/foxo1 signaling and improves glucose metabolism. Nutrients. 2018 doi: 10.3390/nu10101366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MB, Schadler KL, Chandra J, Clinton SK, Courneya KS, Cruz-Monserrate Z, et al. Translating energy balance research from the bench to the clinic to the community: parallel animal-human studies in cancer. CA Cancer J Clin. 2023 doi: 10.3322/caac.21773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasbarrino K, Hafiane A, Gianopoulos I, Zheng H, Mantzoros CS, Daskalopoulou SS. Relationship between circulating adipokines and cholesterol efflux in subjects with severe carotid atherosclerosis. Metabolism. 2023;140:155381. doi: 10.1016/j.metabol.2022.155381. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rellan MJ, Fondevila MF, Dieguez C, Nogueiras R. O-glcnacylation: a sweet hub in the regulation of glucose metabolism in health and disease. Front Endocrinol. 2022;13:873513. doi: 10.3389/fendo.2022.873513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A, Endo Y, Yamashita H. Creg1 stimulates ampk phosphorylation and glucose uptake in skeletal muscle cells. Biochem Biophys Res Commun. 2023;641:162–167. doi: 10.1016/j.bbrc.2022.12.028. [DOI] [PubMed] [Google Scholar]
- Gowans GJ, Bridgers JB, Zhang J, Dronamraju R, Burnetti A, King DA, et al. Recognition of histone crotonylation by taf14 links metabolic state to gene expression. Mol Cell. 2019;76(6):909–921. doi: 10.1016/j.molcel.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillozet AL, Weintraub S, Mash DC, Mesulam MM. Neurofibrillary tangles, amyloid, and memory in aging and mild cognitive impairment. Arch Neurol. 2003;60(5):729–736. doi: 10.1001/archneur.60.5.729. [DOI] [PubMed] [Google Scholar]
- Han HS, Jung CY, Yoon YS, Choi S, Choi D, Kang G, et al. Arginine methylation of crtc2 is critical in the transcriptional control of hepatic glucose metabolism. Sci Signal. 2014;7(314):ra19. doi: 10.1126/scisignal.2004479. [DOI] [PubMed] [Google Scholar]
- Han Z, Wu H, Kim S, Yang X, Li Q, Huang H, et al. Revealing the protein propionylation activity of the histone acetyltransferase mof (males absent on the first) J Biol Chem. 2018;293(9):3410–3420. doi: 10.1074/jbc.RA117.000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmel R, Fiedler D. Features and regulation of non-enzymatic post-translational modifications. Nat Chem Biol. 2018;14(3):244–252. doi: 10.1038/nchembio.2575. [DOI] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between o-glcnacylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Han Z, Liu L, Zheng YG. Chemical biology approaches for investigating the functions of lysine acetyltransferases. Angew Chem Int Ed Engl. 2018;57(5):1162–1184. doi: 10.1002/anie.201704745. [DOI] [PubMed] [Google Scholar]
- He B, Pan H, Zheng F, Chen S, Bie Q, Cao J, et al. Long noncoding rna linc00930 promotes pfkfb3-mediated tumor glycolysis and cell proliferation in nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2022;41(1):77. doi: 10.1186/s13046-022-02282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, B'Nai TA, Yu L, Yao Y, Zhang R, Zahr T, et al. Ppargamma acetylation orchestrates adipose plasticity and metabolic rhythms. Adv Sci. 2023;10(2):e2204190. doi: 10.1002/advs.202204190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepowit NL, Kolbe CC, Zelle SR, Latz E, MacGurn JA. Regulation of ubiquitin and ubiquitin-like modifiers by phosphorylation. FEBS J. 2022;289(16):4797–4810. doi: 10.1111/febs.16101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Murata S, Tanaka K, Shimizu M, Sato R. Sterol regulatory element-binding proteins are negatively regulated through sumo-1 modification independent of the ubiquitin/26 s proteasome pathway. J Biol Chem. 2003;278(19):16809–16819. doi: 10.1074/jbc.M212448200. [DOI] [PubMed] [Google Scholar]
- Hsieh WC, Sutter BM, Ruess H, Barnes SD, Malladi VS, Tu BP. Glucose starvation induces a switch in the histone acetylome for activation of gluconeogenic and fat metabolism genes. Mol Cell. 2022;82(1):60–74. doi: 10.1016/j.molcel.2021.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Luo Z, Qi S, Huang J, Xu P, Wang X, et al. Landscape of the regulatory elements for lysine 2-hydroxyisobutyrylation pathway. Cell Res. 2018;28(1):111–125. doi: 10.1038/cr.2017.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Tang S, Ji M, Tang Z, Shimada M, Liu X, et al. P300-mediated lysine 2-hydroxyisobutyrylation regulates glycolysis. Mol Cell. 2018;70(4):663–678. doi: 10.1016/j.molcel.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhang D, Weng Y, Delaney K, Tang Z, Yan C, et al. The regulatory enzymes and protein substrates for the lysine beta-hydroxybutyrylation pathway. Sci Adv. 2021 doi: 10.1126/sciadv.abe2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CW, Rust NC, Wu HF, Hart GW. Altered o-glcnacylation and mitochondrial dysfunction, a molecular link between brain glucose dysregulation and sporadic Alzheimer’s disease. Neural Regen Res. 2023;18(4):779–783. doi: 10.4103/1673-5374.354515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RW, Hughey CC, Lantier L, Sundelin EI, Peggie M, Zeqiraj E, et al. Metformin reduces liver glucose production by inhibition of fructose-1-6-bisphosphatase. Nat Med. 2018;24(9):1395–1406. doi: 10.1038/s41591-018-0159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JS, Kim KH, Park J, Kim SM, Cho H, Lee Y, et al. Glucosamine improves survival in a mouse model of sepsis and attenuates sepsis-induced lung injury and inflammation. J Biol Chem. 2019;294(2):608–622. doi: 10.1074/jbc.RA118.004638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Johnson L, Jasencakova Z, Zhang X, PerezBurgos L, Singh PB, et al. Dimethylation of histone h3 lysine 9 is a critical mark for dna methylation and gene silencing in arabidopsis thaliana. Chromosoma. 2004;112(6):308–315. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- Jambhekar A, Dhall A, Shi Y. Roles and regulation of histone methylation in animal development. Nat Rev Mol Cell Biol. 2019;20(10):625–641. doi: 10.1038/s41580-019-0151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AM, Smith AC, Smith CL, Robinson AJ, Murphy MP. Proximal cysteines that enhance lysine n-acetylation of cytosolic proteins in mice are less conserved in longer-living species. Cell Rep. 2018;24(6):1445–1455. doi: 10.1016/j.celrep.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings EQ, Fritz KS, Galligan JJ. Biochemical genesis of enzymatic and non-enzymatic post-translational modifications. Mol Aspects Med. 2022;86:101053. doi: 10.1016/j.mam.2021.101053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SM, Lim JS, Park SH, Lee JH. Wnt signaling promotes tumor development in part through phosphofructokinase 1 platelet isoform upregulation. Oncol Rep. 2021 doi: 10.3892/or.2021.8185. [DOI] [PubMed] [Google Scholar]
- Jia Z, Yue F, Chen X, Narayanan N, Qiu J, Syed SA, et al. Protein arginine methyltransferase prmt5 regulates fatty acid metabolism and lipid droplet biogenesis in white adipose tissues. Adv Sci. 2020;7(23):2002602. doi: 10.1002/advs.202002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, et al. Acetylation regulates gluconeogenesis by promoting pepck1 degradation via recruiting the ubr5 ubiquitin ligase. Mol Cell. 2011;43(1):33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Zhang W, Wang Y, Liu J, Hao F, Li Y, et al. Pyruvate kinase m2 promotes the activation of dendritic cells by enhancing il-12p35 expression. Cell Rep. 2020;31(8):107690. doi: 10.1016/j.celrep.2020.107690. [DOI] [PubMed] [Google Scholar]
- Johanns M, Lai YC, Hsu MF, Jacobs R, Vertommen D, Van Sande J, et al. Ampk antagonizes hepatic glucagon-stimulated cyclic amp signalling via phosphorylation-induced activation of cyclic nucleotide phosphodiesterase 4b. Nat Commun. 2016;7:10856. doi: 10.1038/ncomms10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BM, DeBose-Boyd RA. Underlying mechanisms for sterol-induced ubiquitination and er-associated degradation of hmg coa reductase. Semin Cell Dev Biol. 2018;81:121–128. doi: 10.1016/j.semcdb.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi HJ, Narimatsu Y, Schjoldager KT, Tytgat H, Aebi M, Clausen H, et al. Snapshot: o-glycosylation pathways across kingdoms. Cell. 2018;172(3):632. doi: 10.1016/j.cell.2018.01.016. [DOI] [PubMed] [Google Scholar]
- Kalaiarasan P, Subbarao N, Bamezai RN. Molecular simulation of tyr105 phosphorylated pyruvate kinase m2 to understand its structure and dynamics. J Mol Model. 2014;20(9):2447. doi: 10.1007/s00894-014-2447-6. [DOI] [PubMed] [Google Scholar]
- Kebede AF, Nieborak A, Shahidian LZ, Le Gras S, Richter F, Gomez DA, et al. Histone propionylation is a mark of active chromatin. Nat Struct Mol Biol. 2017;24(12):1048–1056. doi: 10.1038/nsmb.3490. [DOI] [PubMed] [Google Scholar]
- Kelly R, Chandru A, Watson PJ, Song Y, Blades M, Robertson NS, et al. Histone deacetylase (hdac) 1 and 2 complexes regulate both histone acetylation and crotonylation in vivo. Sci Rep. 2018;8(1):14690. doi: 10.1038/s41598-018-32927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsall IR. Non-lysine ubiquitylation: doing things differently. Front Mol Biosci. 2022;9:1008175. doi: 10.3389/fmolb.2022.1008175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Xiao Z, Kwon S, Sun X, Ryerson D, Tkac D, et al. A dysregulated acetyl/sumo switch of fxr promotes hepatic inflammation in obesity. EMBO J. 2015;34(2):184–199. doi: 10.15252/embj.201489527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollenstart L, de Groot A, Janssen G, Cheng X, Vreeken K, Martino F, et al. Gcn5 and esa1 function as histone crotonyltransferases to regulate crotonylation-dependent transcription. J Biol Chem. 2019;294(52):20122–20134. doi: 10.1074/jbc.RA119.010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kotzka J, Lehr S, Roth G, Avci H, Knebel B, Muller-Wieland D. Insulin-activated erk-mitogen-activated protein kinases phosphorylate sterol regulatory element-binding protein-2 at serine residues 432 and 455 in vivo. J Biol Chem. 2004;279(21):22404–22411. doi: 10.1074/jbc.M401198200. [DOI] [PubMed] [Google Scholar]
- Kumar PV, Mathur A, Fareed KM, Kakkar P. Endoplasmic reticulum stress induces degradation of glucose transporter proteins during hyperglycemic hepatotoxicity: role of perk-eif2alpha-atf4 axis. Eur J Pharmacol. 2022;926:175012. doi: 10.1016/j.ejphar.2022.175012. [DOI] [PubMed] [Google Scholar]
- Lai X, Liang Y, Jin J, Zhang H, Wu Z, Li G, et al. Protein kinase c epsilon promotes de novo lipogenesis and tumor growth in prostate cancer cells by regulating the phosphorylation and nuclear translocation of pyruvate kinase isoform m2. Exp Cell Res. 2023;422(1):113427. doi: 10.1016/j.yexcr.2022.113427. [DOI] [PubMed] [Google Scholar]
- Lauterbach MA, Hanke JE, Serefidou M, Mangan M, Kolbe CC, Hess T, et al. Toll-like receptor signaling rewires macrophage metabolism and promotes histone acetylation via atp-citrate lyase. Immunity. 2019;51(6):997–1011. doi: 10.1016/j.immuni.2019.11.009. [DOI] [PubMed] [Google Scholar]
- Lee TY, Hsu JB, Lin FM, Chang WC, Hsu PC, Huang HD. N-ace: using solvent accessibility and physicochemical properties to identify protein n-acetylation sites. J Comput Chem. 2010;31(15):2759–2771. doi: 10.1002/jcc.21569. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Li CF, Ruan D, He J, Montal ED, Lorenz S, et al. Non-proteolytic ubiquitination of hexokinase 2 by hecth9 controls tumor metabolism and cancer stem cell expansion. Nat Commun. 2019;10(1):2625. doi: 10.1038/s41467-019-10374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Shao F, Ling J, Lu S, Liu R, Du L, et al. Phosphofructokinase 1 platelet isoform promotes beta-catenin transactivation for tumor development. Front Oncol. 2020;10:211. doi: 10.3389/fonc.2020.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, et al. Sirt7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun. 2016;7:12235. doi: 10.1038/ncomms12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FL, Liu JP, Bao RX, Yan G, Feng X, Xu YP, et al. Acetylation accumulates pfkfb3 in cytoplasm to promote glycolysis and protects cells from cisplatin-induced apoptosis. Nat Commun. 2018;9(1):508. doi: 10.1038/s41467-018-02950-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang Y, Zhang Y, Lu S, Miao Y, Yang J, et al. Gsnor modulates hyperhomocysteinemia-induced t cell activation and atherosclerosis by switching akt s-nitrosylation to phosphorylation. Redox Biol. 2018;17:386–399. doi: 10.1016/j.redox.2018.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Wei X, Jiang DS. Protein methylation functions as the posttranslational modification switch to regulate autophagy. Cell Mol Life Sci. 2019;76(19):3711–3722. doi: 10.1007/s00018-019-03161-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Lu H, Wang X, Duan C, Zhu X, Zhang Y, et al. Pyruvate kinase m2 (pkm2) interacts with activating transcription factor 2 (atf2) to bridge glycolysis and pyroptosis in microglia. Mol Immunol. 2021;140:250–266. doi: 10.1016/j.molimm.2021.10.017. [DOI] [PubMed] [Google Scholar]
- Li H, Herrmann T, Seessle J, Liebisch G, Merle U, Stremmel W, 2022. Role of fatty acid transport protein 4 in metabolic tissues: insights into obesity and fatty liver disease. Biosci Rep. [DOI] [PMC free article] [PubMed]
- Li J, Guo B, Zhang W, Yue S, Huang S, Gao S, et al. Recent advances in demystifying o-glycosylation in health and disease. Proteomics. 2022;22(23–24):e2200156. doi: 10.1002/pmic.202200156. [DOI] [PubMed] [Google Scholar]
- Li Y, Huang X, Yang G, Xu K, Yin Y, Brecchia G, et al. Cd36 favours fat sensing and transport to govern lipid metabolism. Prog Lipid Res. 2022;88:101193. doi: 10.1016/j.plipres.2022.101193. [DOI] [PubMed] [Google Scholar]
- Li S, Feng F, Deng Y. Resveratrol regulates glucose and lipid metabolism in diabetic rats by inhibition of pdk1/akt phosphorylation and hif-1alpha expression. Diabetes Metab Syndr Obes. 2023;16:1063–1074. doi: 10.2147/DMSO.S403893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao P, Bhattarai N, Cao B, Zhou X, Jung JH, Damera K, et al. Crotonylation at serine 46 impairs p53 activity. Biochem Biophys Res Commun. 2020;524(3):730–735. doi: 10.1016/j.bbrc.2020.01.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Xu N, Zhang H, Liao W, Wang Y, Wang S, et al. Persistent high glucose induced epb41l4a-as1 inhibits glucose uptake via gcn5 mediating crotonylation and acetylation of histones and non-histones. Clin Transl Med. 2022;12(2):e699. doi: 10.1002/ctm2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Iqbal K, Grundke-Iqbal I, Hart GW, Gong CX. O-glcnacylation regulates phosphorylation of tau: a mechanism involved in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2004;101(29):10804–10809. doi: 10.1073/pnas.0400348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Shi J, Tanimukai H, Gu J, Gu J, Grundke-Iqbal I, et al. Reduced o-glcnacylation links lower brain glucose metabolism and tau pathology in Alzheimer’s disease. Brain. 2009;132(Pt 7):1820–1832. doi: 10.1093/brain/awp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wei W, Liu Y, Yang X, Wu J, Zhang Y, et al. Mof as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferase-deficient and crotonyltransferase-competent cbp/p300. Cell Discov. 2017;3:17016. doi: 10.1038/celldisc.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Gan W, Su S, Hauenstein AV, Fu TM, Brasher B, et al. K63-linked polyubiquitin chains bind to dna to facilitate dna damage repair. Sci Signal. 2018 doi: 10.1126/scisignal.aar8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YZ, Xiao X, Hu CT, Dai Y, Qu SL, Huang L, et al. Sumoylation in atherosclerosis. Clin Chim Acta. 2020;508:228–233. doi: 10.1016/j.cca.2020.05.033. [DOI] [PubMed] [Google Scholar]
- Liu T, Lv YF, Zhao JL, You QD, Jiang ZY. Regulation of nrf2 by phosphorylation: consequences for biological function and therapeutic implications. Free Radic Biol Med. 2021;168:129–141. doi: 10.1016/j.freeradbiomed.2021.03.034. [DOI] [PubMed] [Google Scholar]
- Liu YX, Yuan PZ, Wu JH, Hu B. Lipid accumulation and novel insight into vascular smooth muscle cells in atherosclerosis. J Mol Med. 2021;99(11):1511–1526. doi: 10.1007/s00109-021-02109-8. [DOI] [PubMed] [Google Scholar]
- Liu B, Ruan J, Chen M, Li Z, Manjengwa G, Schluter D, et al. Deubiquitinating enzymes (dubs): decipher underlying basis of neurodegenerative diseases. Mol Psychiatry. 2022;27(1):259–268. doi: 10.1038/s41380-021-01233-8. [DOI] [PubMed] [Google Scholar]
- Liu Z, Xiao M, Mo Y, Wang H, Han Y, Zhao X, et al. Emerging roles of protein palmitoylation and its modifying enzymes in cancer cell signal transduction and cancer therapy. Int J Biol Sci. 2022;18(8):3447–3457. doi: 10.7150/ijbs.72244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Yoldi M, Castilla-Madrigal R, Lostao MP, Barber A, Prieto J, Martinez JA, et al. Cardiotrophin-1 decreases intestinal sugar uptake in mice and in caco-2 cells. Acta Physiol. 2016;217(3):217–226. doi: 10.1111/apha.12674. [DOI] [PubMed] [Google Scholar]
- Lorendeau D, Christen S, Rinaldi G, Fendt SM. Metabolic control of signalling pathways and metabolic auto-regulation. Biol Cell. 2015;107(8):251–272. doi: 10.1111/boc.201500015. [DOI] [PubMed] [Google Scholar]
- Lu Q, Guo P, Liu A, Ares I, Martinez-Larranaga MR, Wang X, et al. The role of long noncoding rna in lipid, cholesterol, and glucose metabolism and treatment of obesity syndrome. Med Res Rev. 2021;41(3):1751–1774. doi: 10.1002/med.21775. [DOI] [PubMed] [Google Scholar]
- Luiken JJ, Chanda D, Nabben M, Neumann D, Glatz JF. Post-translational modifications of cd36 (sr-b2): implications for regulation of myocellular fatty acid uptake. Biochim Biophys Acta. 2016;1862(12):2253–2258. doi: 10.1016/j.bbadis.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Lv T, Hu Y, Ma Y, Zhen J, Xin W, Wan Q. Gcn5l1 controls renal lipotoxicity through regulating acetylation of fatty acid oxidation enzymes. J Physiol Biochem. 2019;75(4):597–606. doi: 10.1007/s13105-019-00711-6. [DOI] [PubMed] [Google Scholar]
- Lypova N, Telang S, Chesney J, Imbert-Fernandez Y. Increased 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 activity in response to egfr signaling contributes to non-small cell lung cancer cell survival. J Biol Chem. 2019;294(27):10530–10543. doi: 10.1074/jbc.RA119.007784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen AS, Olsen CA. Profiling of substrates for zinc-dependent lysine deacylase enzymes: hdac3 exhibits decrotonylase activity in vitro. Angew Chem Int Ed Engl. 2012;51(36):9083–9087. doi: 10.1002/anie.201203754. [DOI] [PubMed] [Google Scholar]
- Malecki JM, Davydova E, Falnes PO. Protein methylation in mitochondria. J Biol Chem. 2022;298(4):101791. doi: 10.1016/j.jbc.2022.101791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni W, Jianxin X, Weiqi H, Siyuan C, Huashan S. Jmjd family proteins in cancer and inflammation. Signal Transduct Target Ther. 2022;7(1):304. doi: 10.1038/s41392-022-01145-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Sun M, Chen Z, Zeng Z, Wu J, Chen Z, et al. The pyruvate dehydrogenase complex mitigates lps-induced endothelial barrier dysfunction by metabolic regulation. Shock. 2022;57(6):308–317. doi: 10.1097/SHK.0000000000001931. [DOI] [PubMed] [Google Scholar]
- Marin-Hernandez A, Rodriguez-Zavala JS, Jasso-Chavez R, Saavedra E, Moreno-Sanchez R. Protein acetylation effects on enzyme activity and metabolic pathway fluxes. J Cell Biochem. 2022;123(4):701–718. doi: 10.1002/jcb.30197. [DOI] [PubMed] [Google Scholar]
- Martinez-Ruiz A, Lamas S. Signalling by no-induced protein s-nitrosylation and s-glutathionylation: convergences and divergences. Cardiovasc Res. 2007;75(2):220–228. doi: 10.1016/j.cardiores.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Marx C. Lipid metabolism disorders. Dtsch Arztebl Int. 2022;119(13):229. doi: 10.3238/arztebl.m2022.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Sandow JJ, Czabotar PE, Murphy JM. The regulation of necroptosis by post-translational modifications. Cell Death Differ. 2021;28(3):861–883. doi: 10.1038/s41418-020-00722-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Liu H, Liu J, Pang Y, Liu Q. Advances in immunotherapy modalities for atherosclerosis. Front Pharmacol. 2022;13:1079185. doi: 10.3389/fphar.2022.1079185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen T, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- Mulukutla BC, Khan S, Lange A, Hu WS. Glucose metabolism in mammalian cell culture: new insights for tweaking vintage pathways. Trends Biotechnol. 2010;28(9):476–484. doi: 10.1016/j.tibtech.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Mustfa SA, Singh M, Suhail A, Mohapatra G, Verma S, Chakravorty D, et al. Sumoylation pathway alteration coupled with downregulation of sumo e2 enzyme at mucosal epithelium modulates inflammation in inflammatory bowel disease. Open Biol. 2017 doi: 10.1098/rsob.170024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Ge Q, Pawlosky R, Wynn RM, Veech RL, Uyeda K. Metabolite regulation of nucleo-cytosolic trafficking of carbohydrate response element-binding protein (chrebp): role of ketone bodies. J Biol Chem. 2013;288(39):28358–28367. doi: 10.1074/jbc.M113.498550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Weinert BT, Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol. 2019;20(3):156–174. doi: 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- Navale AM, Paranjape AN. Glucose transporters: physiological and pathological roles. Biophys Rev. 2016;8(1):5–9. doi: 10.1007/s12551-015-0186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neopane K, Kozlov N, Negoita F, Murray-Segal L, Brink R, Hoque A, et al. Blocking ampk beta1 myristoylation enhances ampk activity and protects mice from high-fat diet-induced obesity and hepatic steatosis. Cell Rep. 2022;41(12):111862. doi: 10.1016/j.celrep.2022.111862. [DOI] [PubMed] [Google Scholar]
- Nielsen SU, Spener F. Fatty acid-binding protein from rat heart is phosphorylated on tyr19 in response to insulin stimulation. J Lipid Res. 1993;34(8):1355–1366. doi: 10.1016/S0022-2275(20)36965-0. [DOI] [PubMed] [Google Scholar]
- Nielsen SU, Rump R, Hojrup P, Roepstorff P, Spener F. Differentiational regulation and phosphorylation of the fatty acid-binding protein from rat mammary epithelial cells. Biochim Biophys Acta. 1994;1211(2):189–197. doi: 10.1016/0005-2760(94)90268-2. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fujita Y, Ida S, Yanagimachi T, Ohashi N, Nishi K, et al. Glycaemia and body weight are regulated by sodium-glucose cotransporter 1 (sglt1) expression via o-glcnacylation in the intestine. Mol Metab. 2022;59:101458. doi: 10.1016/j.molmet.2022.101458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- O'Neill HM, Lally JS, Galic S, Thomas M, Azizi PD, Fullerton MD, et al. Ampk phosphorylation of acc2 is required for skeletal muscle fatty acid oxidation and insulin sensitivity in mice. Diabetologia. 2014;57(8):1693–1702. doi: 10.1007/s00125-014-3273-1. [DOI] [PubMed] [Google Scholar]
- Ortega-Prieto P, Postic C. Carbohydrate sensing through the transcription factor chrebp. Front Genet. 2019;10:472. doi: 10.3389/fgene.2019.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Mao C, Wang YX. Suppression of gluconeogenic gene expression by lsd1-mediated histone demethylation. PLoS ONE. 2013;8(6):e66294. doi: 10.1371/journal.pone.0066294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan RY, He L, Zhang J, Liu X, Liao Y, Gao J, et al. Positive feedback regulation of microglial glucose metabolism by histone h4 lysine 12 lactylation in Alzheimer’s disease. Cell Metab. 2022;34(4):634–648. doi: 10.1016/j.cmet.2022.02.013. [DOI] [PubMed] [Google Scholar]
- Patwardhan A, Cheng N, Trejo J. Post-translational modifications of g protein-coupled receptors control cellular signaling dynamics in space and time. Pharmacol Rev. 2021;73(1):120–151. doi: 10.1124/pharmrev.120.000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Le Y, Chen H, Feng J, Liu Z, Zhu J, et al. Cynaroside prevents macrophage polarization into pro-inflammatory phenotype and alleviates cecal ligation and puncture-induced liver injury by targeting pkm2/hif-1alpha axis. Fitoterapia. 2021;152:104922. doi: 10.1016/j.fitote.2021.104922. [DOI] [PubMed] [Google Scholar]
- Peng S, Barba-Bon A, Pan YC, Nau WM, Guo DS, Hennig A. Phosphorylation-responsive membrane transport of peptides. Angew Chem Int Ed Engl. 2017;56(49):15742–15745. doi: 10.1002/anie.201707979. [DOI] [PubMed] [Google Scholar]
- Pinho TS, Correia SC, Perry G, Ambrosio AF, Moreira PI. Diminished o-glcnacylation in Alzheimer’s disease is strongly correlated with mitochondrial anomalies. Biochim Biophys Acta-Mol Basis Dis. 2019;1865(8):2048–2059. doi: 10.1016/j.bbadis.2018.10.037. [DOI] [PubMed] [Google Scholar]
- Pirillo A, Svecla M, Catapano AL, Holleboom AG, Norata GD. Impact of protein glycosylation on lipoprotein metabolism and atherosclerosis. Cardiovasc Res. 2021;117(4):1033–1045. doi: 10.1093/cvr/cvaa252. [DOI] [PubMed] [Google Scholar]
- Povero D. Novel oncometabolites and metabolic checkpoints involved in hepatocellular carcinoma development. J Hepatol. 2023;78(3):463–466. doi: 10.1016/j.jhep.2023.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradeep P, Kang H, Lee B. Glycosylation and behavioral symptoms in neurological disorders. Transl Psychiatry. 2023;13(1):154. doi: 10.1038/s41398-023-02446-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinsen CF, Werten PJ, Maassen JA, Veerkamp JH. No significant tyrosine phosphorylation of muscle fatty acid-binding protein. Biochim Biophys Acta. 1994;1215(1–2):103–108. doi: 10.1016/0005-2760(94)90097-3. [DOI] [PubMed] [Google Scholar]
- Qian H, Chen Y, Nian Z, Su L, Yu H, Chen FJ, et al. Hdac6-mediated acetylation of lipid droplet-binding protein cidec regulates fat-induced lipid storage. J Clin Invest. 2017;127(4):1353–1369. doi: 10.1172/JCI85963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalingam K, Dikic I. Snapshot: expanding the ubiquitin code. Cell. 2016;164(5):1074. doi: 10.1016/j.cell.2016.02.019. [DOI] [PubMed] [Google Scholar]
- Ramazi S, Zahiri J. Posttranslational modifications in proteins: resources, tools and prediction methods. Database. 2021 doi: 10.1093/database/baab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampoldi F, Sandhoff R, Owen RW, Grone HJ, Porubsky S. A new, robust, and nonradioactive approach for exploring n-myristoylation. J Lipid Res. 2012;53(11):2459–2468. doi: 10.1194/jlr.D026997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, et al. Sirt5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18(6):920–933. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashdan NA, Shrestha B, Pattillo CB. S-glutathionylation, friend or foe in cardiovascular health and disease. Redox Biol. 2020;37:101693. doi: 10.1016/j.redox.2020.101693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in health and disease. Nat Rev Nephrol. 2019;15(6):346–366. doi: 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JZ, Crawford N, Longley DB. The role of ubiquitination in apoptosis and necroptosis. Cell Death Differ. 2022;29(2):272–284. doi: 10.1038/s41418-021-00922-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross KE, Zhang G, Akcora C, Lin Y, Fang B, Koomen J, et al. Network models of protein phosphorylation, acetylation, and ubiquitination connect metabolic and cell signaling pathways in lung cancer. PLoS Comput Biol. 2023;19(3):e1010690. doi: 10.1371/journal.pcbi.1010690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan HB, Singh JP, Li MD, Wu J, Yang X. Cracking the o-glcnac code in metabolism. Trends Endocrinol Metab. 2013;24(6):301–309. doi: 10.1016/j.tem.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Tang Z, Huang H, Yong-Gonzalez V, Molina H, Kong HE, et al. Intracellular crotonyl-coa stimulates transcription through p300-catalyzed histone crotonylation. Mol Cell. 2015;58(2):203–215. doi: 10.1016/j.molcel.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco F, Perfetto L, Castagnoli L, Cesareni G. The human phosphatase interactome: an intricate family portrait. FEBS Lett. 2012;586(17):2732–2739. doi: 10.1016/j.febslet.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi M, Dehnavi S, Shohan M, Jamialahmadi T, Sathyapalan T, Sahebkar A. Potential role of sumo and sumoylation in the pathogenesis of diabetes mellitus. Curr Med Chem. 2023;30(14):1623–1637. doi: 10.2174/0929867329666220817142848. [DOI] [PubMed] [Google Scholar]
- Sapir A. Not so slim anymore-evidence for the role of sumo in the regulation of lipid metabolism. Biomolecules. 2020 doi: 10.3390/biom10081154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sas KM, Kayampilly P, Byun J, Nair V, Hinder LM, Hur J, et al. Tissue-specific metabolic reprogramming drives nutrient flux in diabetic complications. JCI Insight. 2016;1(15):e86976. doi: 10.1172/jci.insight.86976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawant DA, Kalhotra P, Novickis AT, Dasgupta S. Regulation of tumor metabolism by post translational modifications on metabolic enzymes. Cancer Gene Ther. 2022 doi: 10.1038/s41417-022-00521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager KT, Narimatsu Y, Joshi HJ, Clausen H. Global view of human protein glycosylation pathways and functions. Nat Rev Mol Cell Biol. 2020;21(12):729–749. doi: 10.1038/s41580-020-00294-x. [DOI] [PubMed] [Google Scholar]
- Seok SH. Structural insights into protein regulation by phosphorylation and substrate recognition of protein kinases/phosphatases. Life-Basel. 2021 doi: 10.3390/life11090957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafi R, Iyer SP, Ellies LG, O'Donnell N, Marek KW, Chui D, et al. The o-glcnac transferase gene resides on the x chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97(11):5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JJ, Liu HF, Hu T, Gao X, Zhang YB, Li WR, et al. Danggui-shaoyao-san improves cognitive impairment through inhibiting o-glcnac-modification of estrogen alpha receptor in female db/db mice. J Ethnopharmacol. 2021;281:114562. doi: 10.1016/j.jep.2021.114562. [DOI] [PubMed] [Google Scholar]
- Shimamoto S, Nakashima K, Kamimura R, Kohrogi R, Inoue H, Nishikoba N, et al. Insulin acutely increases glucose transporter 1 on plasma membranes and glucose uptake in an akt-dependent manner in chicken adipocytes. Gen Comp Endocrinol. 2019;283:113232. doi: 10.1016/j.ygcen.2019.113232. [DOI] [PubMed] [Google Scholar]
- Shimano H, Sato R. Srebp-regulated lipid metabolism: convergent physiology—divergent pathophysiology. Nat Rev Endocrinol. 2017;13(12):710–730. doi: 10.1038/nrendo.2017.91. [DOI] [PubMed] [Google Scholar]
- Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Su X, El-Maghrabi R, Stahl PD, Abumrad NA. Opposite regulation of cd36 ubiquitination by fatty acids and insulin: effects on fatty acid uptake. J Biol Chem. 2008;283(20):13578–13585. doi: 10.1074/jbc.M800008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Zhang J, Liu X, Li M, Wang D, Kang Z, et al. Pten loss promotes warburg effect and prostate cancer cell growth by inducing fbp1 degradation. Front Oncol. 2022;12:911466. doi: 10.3389/fonc.2022.911466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks B, Zierath JR. Post-translational modifications: the signals at the intersection of exercise, glucose uptake, and insulin sensitivity. Endocr Rev. 2022;43(4):654–677. doi: 10.1210/endrev/bnab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Meng C, Xu J, Su Z, Xiao C, Yang D. Histone methyltransferase smyd2 drives adipogenesis via regulating stat3 phosphorylation. Cell Death Dis. 2022;13(10):890. doi: 10.1038/s41419-022-05321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Wei G, Liu H, Xiao D, Huang J, Lu J, et al. Inhibition of xbp1s ubiquitination enhances its protein stability and improves glucose homeostasis. Metabolism. 2020;105:154046. doi: 10.1016/j.metabol.2019.154046. [DOI] [PubMed] [Google Scholar]
- Sun Z, Tan Z, Peng C, Yi W. Hk2 is associated with the warburg effect and proliferation in liver cancer: targets for effective therapy with glycyrrhizin. Mol Med Rep. 2021 doi: 10.3892/mmr.2021.11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Zhang L, Li L, Shao C, Liu J, Zhou M, et al. Galectin-3 mediates cardiac remodeling caused by impaired glucose and lipid metabolism through inhibiting two pathways of activating akt. Am J Physiol-Heart Circul Physiol. 2021;320(1):H364–H380. doi: 10.1152/ajpheart.00523.2020. [DOI] [PubMed] [Google Scholar]
- Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26(4):399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Kameyama K, Miyauchi E, Nakanishi Y, Kanaya T, Fujii T, et al. Fatty acid overproduction by gut commensal microbiota exacerbates obesity. Cell Metab. 2023;35(2):361–375. doi: 10.1016/j.cmet.2022.12.013. [DOI] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Zhan Z, Zhang Y, Huang X. Propionylation of lysine, a new mechanism of short-chain fatty acids affecting bacterial virulence. Am J Transl Res. 2022;14(8):5773–5784. [PMC free article] [PubMed] [Google Scholar]
- Terra MF, Garcia-Arevalo M, Avelino TM, Degaki KY, Malospirito CC, de Carvalho M, et al. Am-879, a ppary non-agonist and ser273 phosphorylation blocker, promotes insulin sensitivity without adverse effects in mice. Metabol Open. 2023;17:100221. doi: 10.1016/j.metop.2022.100221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa D, Zhang M, Manning JR, Guimaraes DA, Stoner MW, O'Doherty RM, et al. Acetylation of mitochondrial proteins by gcn5l1 promotes enhanced fatty acid oxidation in the heart. Am J Physiol-Heart Circul Physiol. 2017;313(2):H265–H274. doi: 10.1152/ajpheart.00752.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa D, Wu K, Stoner MW, Xie B, Zhang M, Manning JR, et al. The protein acetylase gcn5l1 modulates hepatic fatty acid oxidation activity via acetylation of the mitochondrial beta-oxidation enzyme hadha. J Biol Chem. 2018;293(46):17676–17684. doi: 10.1074/jbc.AC118.005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa N, Chaudhari M, McManus S, Roy K, Newman RH, Saigo H, et al. Deepsuccinylsite: a deep learning based approach for protein succinylation site prediction. BMC Bioinformatics. 2020;21(Suppl 3):63. doi: 10.1186/s12859-020-3342-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita H, Hines KM, Herron JM, Li A, Baggett DW, Xu L. 7-dehydrocholesterol-derived oxysterols cause neurogenic defects in smith-lemli-opitz syndrome. Elife. 2022 doi: 10.7554/eLife.67141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramutola A, Sharma N, Barone E, Lanzillotta C, Castellani A, Iavarone F, et al. Proteomic identification of altered protein o-glcnacylation in a triple transgenic mouse model of Alzheimer’s disease. Biochim Biophys Acta-Mol Basis Dis. 2018;1864(10):3309–3321. doi: 10.1016/j.bbadis.2018.07.017. [DOI] [PubMed] [Google Scholar]
- Uyeda K, Repa JJ. Carbohydrate response element binding protein, chrebp, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006;4(2):107–110. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- van den Boogert M, Larsen LE, Ali L, Kuil SD, Chong P, Loregger A, et al. N-glycosylation defects in humans lower low-density lipoprotein cholesterol through increased low-density lipoprotein receptor expression. Circulation. 2019;140(4):280–292. doi: 10.1161/CIRCULATIONAHA.118.036484. [DOI] [PubMed] [Google Scholar]
- Vandanmagsar B, Yu Y, Simmler C, Dang TN, Kuhn P, Poulev A, et al. Bioactive compounds from artemisia dracunculus l. Activate ampk signaling in skeletal muscle. Biomed Pharmacother. 2021;143:112188. doi: 10.1016/j.biopha.2021.112188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander HM, Cantley LC, Thompson CB. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdone L, Caserta M, Di Mauro E. Role of histone acetylation in the control of gene expression. Biochem Cell Biol. 2005;83(3):344–353. doi: 10.1139/o05-041. [DOI] [PubMed] [Google Scholar]
- Vertegaal A. Signalling mechanisms and cellular functions of sumo. Nat Rev Mol Cell Biol. 2022;23(11):715–731. doi: 10.1038/s41580-022-00500-y. [DOI] [PubMed] [Google Scholar]
- Vicente MH, Cassio R, Correia-Guedes L, Gomes MA, Chegao A, Miranda E, et al. Posttranslational modifications of blood-derived alpha-synuclein as biochemical markers for Parkinson’s disease. Sci Rep. 2017;7(1):13713. doi: 10.1038/s41598-017-14175-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldhart AN, Dykstra H, Peck AS, Boguslawski EA, Madaj ZB, Wen J, et al. Phosphorylation of txnip by akt mediates acute influx of glucose in response to insulin. Cell Rep. 2017;19(10):2005–2013. doi: 10.1016/j.celrep.2017.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Mao Y, Narimatsu Y, Ye Z, Tian W, Goth CK, et al. Site-specific o-glycosylation of members of the low-density lipoprotein receptor superfamily enhances ligand interactions. J Biol Chem. 2018;293(19):7408–7422. doi: 10.1074/jbc.M117.817981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Guo YR, Xing D, Tao YJ, Lu Z. Supramolecular assembly of kat2a with succinyl-coa for histone succinylation. Cell Discov. 2018;4:47. doi: 10.1038/s41421-018-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Mu G, Qiu B, Wang M, Yu Z, Wang W, et al. The function and related diseases of protein crotonylation. Int J Biol Sci. 2021;17(13):3441–3455. doi: 10.7150/ijbs.58872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yuan Y, Zou Y, Qi Z, Huang G, Liu Y, et al. Fructose-1,6-bisphosphatase 2 represses cervical cancer progression via inhibiting aerobic glycolysis through promoting pyruvate kinase isozyme type m2 ubiquitination. Anticancer Drugs. 2022;33(1):e198–e206. doi: 10.1097/CAD.0000000000001185. [DOI] [PubMed] [Google Scholar]
- Wang HL, Chen Y, Wang YQ, Tao EW, Tan J, Liu QQ, et al. Sirtuin5 protects colorectal cancer from dna damage by keeping nucleotide availability. Nat Commun. 2022;13(1):6121. doi: 10.1038/s41467-022-33903-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Jiang Y, Peng P, Liu G, Qi S, Liu K, et al. Quantitative proteomics reveals the role of lysine 2-hydroxyisobutyrylation pathway mediated by tip60. Oxidative Med Cell Longev. 2022;2022:4571319. doi: 10.1155/2022/4571319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhou F, Li M, Zhang Y, Li N, Shao L. Mir-34a-5p promotes hepatic gluconeogenesis by suppressing sirt1 expression. Exp Cell Res. 2022;420(1):113336. doi: 10.1016/j.yexcr.2022.113336. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang MM, Geng Y, Ye CY, Zang YS. Membrane-associated ring-ch protein (march8) is a novel glycolysis repressor targeted by mir-32 in colorectal cancer. J Transl Med. 2022;20(1):402. doi: 10.1186/s12967-022-03608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HF, Wang YX, Zhou YP, Wei YP, Yan Y, Zhang ZJ, et al. Protein o-glcnacylation in cardiovascular diseases. Acta Pharmacol Sin. 2023;44(1):8–18. doi: 10.1038/s41401-022-00934-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensveen FM, Valentic S, Sestan M, Turk WT, Polic B. The “big bang” in obese fat: events initiating obesity-induced adipose tissue inflammation. Eur J Immunol. 2015;45(9):2446–2456. doi: 10.1002/eji.201545502. [DOI] [PubMed] [Google Scholar]
- Wold F. In vivo chemical modification of proteins (post-translational modification) Annu Rev Biochem. 1981;50:783–814. doi: 10.1146/annurev.bi.50.070181.004031. [DOI] [PubMed] [Google Scholar]
- Wu H, Guo X, Jiao Y, Wu Z, Lv Q. Trim35 ubiquitination regulates the expression of pkm2 tetramer and dimer and affects the malignant behaviour of breast cancer by regulating the warburg effect. Int J Oncol. 2022 doi: 10.3892/ijo.2022.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Lei QY, Zhao S, Guan KL. Regulation of glycolysis and gluconeogenesis by acetylation of pkm and pepck. Cold Spring Harb Symp Quant Biol. 2011;76:285–289. doi: 10.1101/sqb.2011.76.010942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Richard S. Cellular pathways influenced by protein arginine methylation: implications for cancer. Mol Cell. 2021;81(21):4357–4368. doi: 10.1016/j.molcel.2021.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Wan J, Zhan J, Li X, He H, Shi Z, et al. Global profiling of crotonylation on non-histone proteins. Cell Res. 2017;27(7):946–949. doi: 10.1038/cr.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu A, Zeng Q, Tang Y, Wang X, Yuan X, Zhou Y, et al. Electroacupuncture protects cognition by regulating tau phosphorylation and glucose metabolism via the akt/gsk3beta signaling pathway in Alzheimer’s disease model mice. Front Neurosci. 2020;14:585476. doi: 10.3389/fnins.2020.585476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Rousseau J, Machol K, Cross LA, Agre KE, Gibson CF, et al. Deficient histone h3 propionylation by brpf1-kat6 complexes in neurodevelopmental disorders and cancer. Sci Adv. 2020;6(4):eaax21. doi: 10.1126/sciadv.aax0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Qian K. Protein o-glcnacylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol. 2017;18(7):452–465. doi: 10.1038/nrm.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Ren C, Qiao P, Han X, Wang L, Lv S, et al. Pim2-mediated phosphorylation of hexokinase 2 is critical for tumor growth and paclitaxel resistance in breast cancer. Oncogene. 2018;37(45):5997–6009. doi: 10.1038/s41388-018-0386-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ML, Lu C, Fan ZF, Zhao TR, Cheng GG, Wang YD, et al. Hypoglycemic and hypolipidemic effects of epigynum auritum in high fat diet and streptozotocin-induced diabetic rats. J Ethnopharmacol. 2022;288:114986. doi: 10.1016/j.jep.2022.114986. [DOI] [PubMed] [Google Scholar]
- Yang Y, Tapias V, Acosta D, Xu H, Chen H, Bhawal R, et al. Altered succinylation of mitochondrial proteins, app and tau in Alzheimer’s disease. Nat Commun. 2022;13(1):159. doi: 10.1038/s41467-021-27572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine HN, Self W, Kerman BE, Santoni G, Navalpur SN, Abdullah L, et al. Nutritional metabolism and cerebral bioenergetics in Alzheimer’s disease and related dementias. Alzheimers Dement. 2022 doi: 10.1002/alz.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Guo H, Wang L, Li Y, Xu M, Zhao X, et al. Galnt4 primes monocytes adhesion and transmigration by regulating o-glycosylation of psgl-1 in atherosclerosis. J Mol Cell Cardiol. 2022;165:54–63. doi: 10.1016/j.yjmcc.2021.12.012. [DOI] [PubMed] [Google Scholar]
- Yeh ET. Sumoylation and de-sumoylation: wrestling with life’s processes. J Biol Chem. 2009;284(13):8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WR, et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337(6097):975–980. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You D, Wang MM, Yin BC, Ye BC. Precursor supply for erythromycin biosynthesis: engineering of propionate assimilation pathway based on propionylation modification. ACS Synth Biol. 2019;8(2):371–380. doi: 10.1021/acssynbio.8b00396. [DOI] [PubMed] [Google Scholar]
- Yu X, Wang R, Zhang Y, Zhou L, Wang W, Liu H, et al. Skp2-mediated ubiquitination and mitochondrial localization of akt drive tumor growth and chemoresistance to cisplatin. Oncogene. 2019;38(50):7457–7472. doi: 10.1038/s41388-019-0955-7. [DOI] [PubMed] [Google Scholar]
- Yu HC, Huang KY, Lu MC, Huang TH, Liu SQ, Lai NS, et al. Down-regulation of loc645166 in t cells of ankylosing spondylitis patients promotes the nf-kappab signaling via decreasingly blocking recruitment of the ikk complex to k63-linked polyubiquitin chains. Front Immunol. 2021;12:591706. doi: 10.3389/fimmu.2021.591706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Liu J, Liu C, Liu R, Liu L, Yu Z, et al. Post-translational modifications of cgas-sting: a critical switch for immune regulation. Cells. 2022 doi: 10.3390/cells11193043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Peng G, Xiao G, Yang Z, Huang J, Liu Q, et al. Xanthohumol suppresses glioblastoma via modulation of hexokinase 2 -mediated glycolysis. J Cancer. 2020;11(14):4047–4058. doi: 10.7150/jca.33045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung HW, Zhao X, Glover L, Burrin C, Pang PC, Jones C, et al. Perturbation of placental protein glycosylation by endoplasmic reticulum stress promotes maladaptation of maternal hepatic glucose metabolism. iScience. 2023;26(1):105911. doi: 10.1016/j.isci.2022.105911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S, Wu F, Chen M, Li Y, You M, Zhang Y, et al. Inhibition of fatty acid translocase (fat/cd36) palmitoylation enhances hepatic fatty acid beta-oxidation by increasing its localization to mitochondria and interaction with long-chain acyl-coa synthetase 1. Antioxid Redox Signal. 2022;36(16–18):1081–1100. doi: 10.1089/ars.2021.0157. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yang S, Chen J, Su Z. Unraveling the regulation of hepatic gluconeogenesis. Front Endocrinol. 2018;9:802. doi: 10.3389/fendo.2018.00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DS, Liang GY, Liu DX, Yu J, Wang F. Role of phosphorylated amp-activated protein kinase (ampk) in myocardial insulin resistance after myocardial ischemia-reperfusion during cardiopulmonary bypass surgery in dogs. Med Sci Monitor. 2019;25:4149–4158. doi: 10.12659/MSM.916517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Cao R, Niu J, Yang S, Ma H, Zhao S, et al. Molecular basis for hierarchical histone de-beta-hydroxybutyrylation by sirt3. Cell Discov. 2019;5:35. doi: 10.1038/s41421-019-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Shen M, Wu C, Chen Y, Lu J, Li J, et al. Hdac8-dependent deacetylation of pkm2 directs nuclear localization and glycolysis to promote proliferation in hepatocellular carcinoma. Cell Death Dis. 2020;11(12):1036. doi: 10.1038/s41419-020-03212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang YL, Qiu G, Pian L, Guo L, Cao H, et al. Hepatic neddylation targets and stabilizes electron transfer flavoproteins to facilitate fatty acid beta-oxidation. Proc Natl Acad Sci U S A. 2020;117(5):2473–2483. doi: 10.1073/pnas.1910765117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao M, Gao H, Yu G, Zhao Y, Yao F, et al. Mapk signalling-induced phosphorylation and subcellular translocation of pdhe1alpha promotes tumour immune evasion. Nat Metab. 2022;4(3):374–388. doi: 10.1038/s42255-022-00543-7. [DOI] [PubMed] [Google Scholar]
- Zhao X. Sumo-mediated regulation of nuclear functions and signaling processes. Mol Cell. 2018;71(3):409–418. doi: 10.1016/j.molcel.2018.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Ma H, Zhang X, Tu F, Wang X, Ha T, et al. Enhanced glycolytic metabolism contributes to cardiac dysfunction in polymicrobial sepsis. J Infect Dis. 2017;215(9):1396–1406. doi: 10.1093/infdis/jix138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Cao Y, Chen Y, Wang J, Fan Q, Huang X, et al. Senp2 regulates adipose lipid storage by de-sumoylation of setdb1. J Mol Cell Biol. 2018;10(3):258–266. doi: 10.1093/jmcb/mjx055. [DOI] [PubMed] [Google Scholar]
- Zheng C, Li D, Zhan W, He K, Yang H. Downregulation of senp1 suppresses lps-induced macrophage inflammation by elevating sp3 sumoylation and disturbing sp3-nf-kappab interaction. Am J Transl Res. 2020;12(11):7439–7448. [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Dai H, Chen R, Zhong Y, Zhou C, Wang Y, et al. Endoplasmic reticulum stress promotes sepsis-induced muscle atrophy via activation of stat3 and smad3. J Cell Physiol. 2023 doi: 10.1002/jcp.30950. [DOI] [PubMed] [Google Scholar]
- Zhong T, Lei K, Lin X, Xie Z, Luo S, Zhou Z, et al. Protein ubiquitination in t cell development. Front Immunol. 2022;13:941962. doi: 10.3389/fimmu.2022.941962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Jung CG, Kim MJ, Watanabe A, Abdelhamid M, Taslima F, et al. Insulin deficiency increases sirt2 level in streptozotocin-treated Alzheimer’s disease-like mouse model: increased sirt2 induces tau phosphorylation through erk activation. Mol Neurobiol. 2022;59(9):5408–5425. doi: 10.1007/s12035-022-02918-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Yin Y, Yu M, Gao D, Sun J, Yang Z, et al. Gtpbp4 promotes hepatocellular carcinoma progression and metastasis via the pkm2 dependent glucose metabolism. Redox Biol. 2022;56:102458. doi: 10.1016/j.redox.2022.102458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Hao Q, Liang Y, Kong E. Protein palmitoylation in cancer: molecular functions and therapeutic potential. Mol Oncol. 2023;17(1):3–26. doi: 10.1002/1878-0261.13308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chu F, Zhang M, Sun W, Zhou F. Association between neddylation and immune response. Front Cell Dev Biol. 2022;10:890121. doi: 10.3389/fcell.2022.890121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Gu H, Peng C, Xia F, Cao H, Cui H. Regulation of glucose, fatty acid and amino acid metabolism by ubiquitination and sumoylation for cancer progression. Front Cell Dev Biol. 2022;10:849625. doi: 10.3389/fcell.2022.849625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T, Zhang J. Diverse and pivotal roles of neddylation in metabolism and immunity. FEBS J. 2021;288(13):3884–3912. doi: 10.1111/febs.15584. [DOI] [PubMed] [Google Scholar]
- Zou L, Yang Y, Wang Z, Fu X, He X, Song J, et al. Lysine malonylation and its links to metabolism and diseases. Aging Dis. 2023;14(1):84–98. doi: 10.14336/AD.2022.0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Li H, Herrmann T, Seessle J, Liebisch G, Merle U, Stremmel W, 2022. Role of fatty acid transport protein 4 in metabolic tissues: insights into obesity and fatty liver disease. Biosci Rep. [DOI] [PMC free article] [PubMed]
Data Availability Statement
Not applicable.