Abstract
Methylation at the N6-position of either adenosine (m6A) or 2′-O-methyladenosine (m6Am) represents two of the most abundant internal modifications of coding and non-coding RNAs, influencing their maturation, stability and function. Additionally, although less abundant and less well-studied, monomethylation at the N1-position (m1A) can have profound effects on RNA folding. It has been known for several decades that RNAs produced by both DNA and RNA viruses can be m6A/m6Am modified and the list continues to broaden through advances in detection technologies and identification of the relevant methyltransferases. Recent studies have uncovered varied mechanisms used by viruses to manipulate the m6A pathway in particular, either to enhance virus replication or to antagonize host antiviral defenses. As such, RNA modifications represent an important frontier of exploration in the broader realm of virus-host interactions, and this new knowledge already suggests exciting opportunities for therapeutic intervention. In this review we summarize the principal mechanisms by which m6A/m6Am can promote or hinder viral replication, describe how the pathway is actively manipulated by biomedically important viruses, and highlight some remaining gaps in understanding how adenosine methylation of RNA controls viral replication and pathogenesis.
1. Introduction
Methylation at the N6-position of adenosine (m6A) (Fig. 1A) is the most abundant internal modification of eukaryotic polyadenylated RNAs and is installed by an evolutionarily conserved methyltransferase (“writer”) complex consisting of the catalytic enzyme METTL3, an allosteric activator METTL14, and an RNA-binding scaffolding factor WTAP as well as additional RNA-binding regulatory subunits including VIRMA, ZC3H13, RBM15 and RBM15B (Knuckles et al., 2018; Liu et al., 2014; Ping et al., 2014; Wang et al., 2014; Wen et al., 2018). Profiling studies detect m6A in about 25% of mammalian mRNAs (Dominissini et al., 2012; Meyer et al., 2012), long non-coding RNAs (Meyer et al., 2015; Patil et al., 2016) and microRNA precursor RNAs (Alarcón et al., 2015). Predicting m6A sites using computational tools remains challenging. Some 80% of verified methylation events occur at an AC dinucleotide embedded within a degenerate RRACH sequence (where R = A/G, and H = A/C/U), more often referred to as the ‘DRACH motif’ (Harper et al., 1990; Kane and Beemon, 1985). The remaining 20% of sites do not obviously match this sequence (Linder et al., 2015). DRACH-like sequences are abundant in cellular transcriptomes, but only a small fraction are found to be methylated in vivo and these are enriched in the terminal exon especially close to the termination codon or the polyadenylation signal (Ke et al., 2015). Methylation of mRNA and lncRNA precursors is coincident with transcription by RNA polymerase II (RNAPII) and is largely completed before the RNA is released from the chromatin template (Ke et al., 2017). There are two known m6A demethylases (or erasers), the predominantly nuclear ALKBH5 (Zheng et al., 2013), and the more widespread FTO (Jia et al., 2011). Both are α-ketoglutarate-dependent dioxygenases that mediated oxidative demethylation of m6A. Controversy remains as to the in vivo substrate specificity of these demethylases and the extent to which m6A removal occurs through enzymatic demethylation or via RNA turnover (Darnell et al., 2018; Mauer and Jaffrey, 2018; Zhao et al., 2018). As summarized in Fig. 2, the biological functions of m6A are exerted through two general mechanisms: alterations in RNA secondary structure and interactions with RNA binding proteins (or “readers”) that directly or indirectly recognize the methylated adenosine. Readers such as YTHDC1 and YTHDC2, are predominantly nuclear whereas YTHDF1, YTHDF2 and YTHDF3 are predominantly cytoplasmic, although this varies to a large extent by cell type. As evident from their names, many m6A readers include a YT521-B homology (YTH) domain (Liao et al., 2018; Xu et al., 2015), however other unrelated RNA-binding proteins such as IGF2BP1/2/3 (Huang et al., 2018), eIF3 (Meyer et al., 2015), heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2/B1) (Alarcón et al., 2015) and the Tudor domain-containing SND1 (Baquero-Perez et al., 2019) are also capable of m6A recognition. Recent reports demonstrate functional redundancy among readers and suggest that in vivo association with individual methylated RNAs is conferred by differences in relative abundance (Lasman et al., 2020; Zaccara and Jaffrey, 2020). This is an extremely active area of investigation not only because of roles in normal cellular processes but also as the pathogenic basis of human disease (He and He, 2021). For instance, many components of the m6A pathway are dysregulated in cancer (Barbieri and Kouzarides, 2020) or contribute to inflammatory disease (Zong et al., 2021). Intrinsic antiviral defenses are also controlled by RNA m6A modification (Rubio et al., 2018; Winkler et al., 2019), driving some viruses to target m6A modification components or reshape the global cellular m6A modification landscape to benefit virus replication. Lastly, host factors involved in the m6A modification pathway impact virus infection biology through epitranscriptomic changes to both virus and host mRNAs, which upon recognition by m6A reader proteins post-transcriptionally regulate the expression of factors needed for virus replication.
Fig. 1.
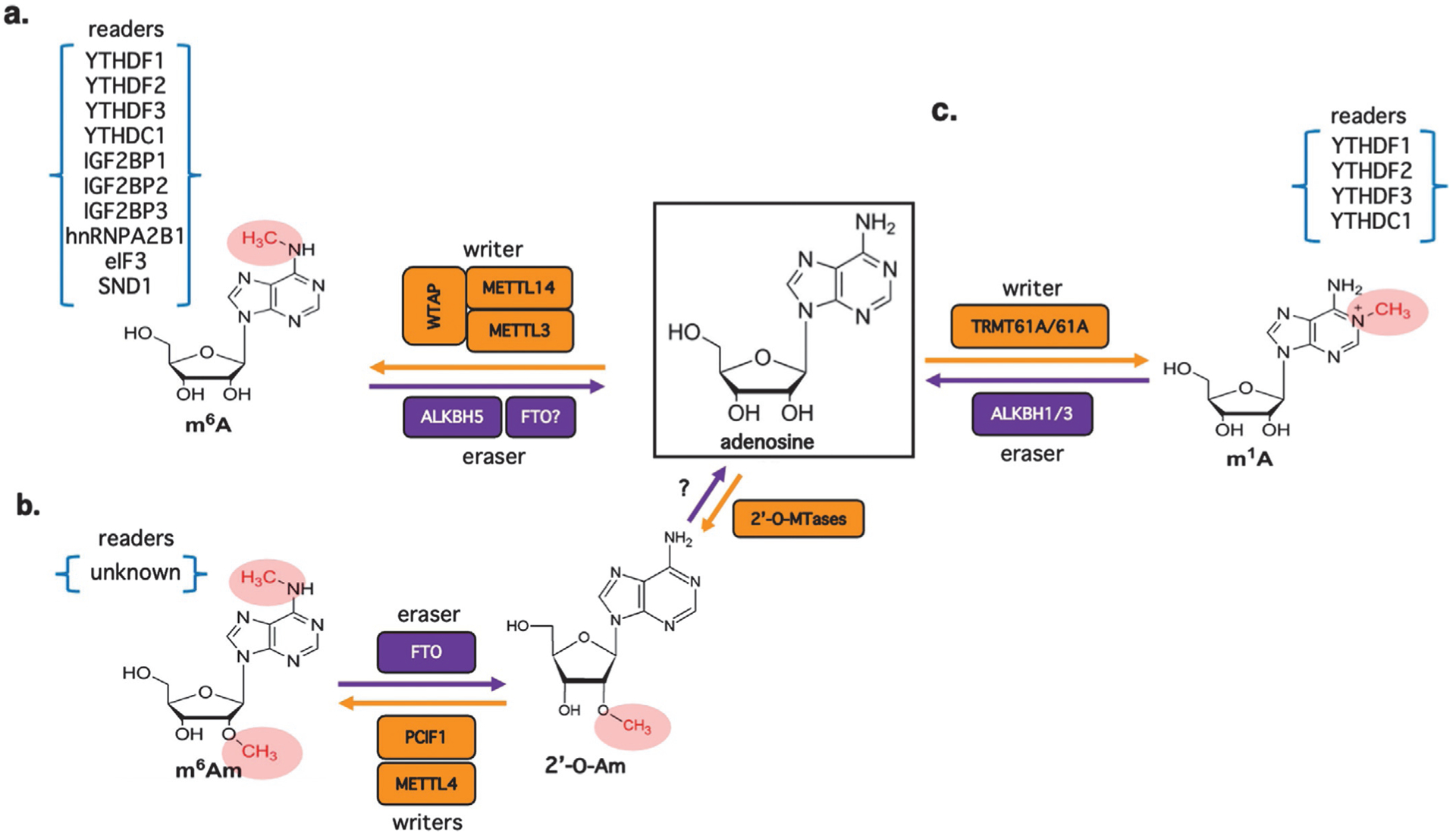
Reversible conversion of adenosine in RNA to m6A, m6Am and m1A. (A) N6-methyladenosine (m6A) is the most abundant internal modification of both mRNA and lncRNA in mammalian cells and is catalyzed by a core writer complex composed of METTL3, METTL14 and WTAP. m6A-modified RNAs can be recognized by a variety of RNA binding proteins (readers), several of which contain a YTH domain. Demethylation is mediated by the ALKBH5 and FTO eraser proteins, although as indicated by the question mark the full extent and relative contribution of FTO in particular, remain uncertain. (B) N6-methylation of cap-adjacent N6,2′-O-dimethyladenosine (m6Am) is catalyzed by PCIF1 (also known as CAPAM) (Akichika et al., 2019). Methylation of the 2′-O-ribose of the first and second transcribed nucleotides is catalyzed by hMTr1 and hMTr2, respectively (Bélanger et al., 2010; Werner et al., 2011). The enzymes responsible for installation of m6A away from the cap structures are less well understood, however internal m6Am methylation of U2 small nuclear RNA (snRNA) is catalyzed by METTL4 at sites of pre-deposited 2’-O-methylation (H. Chen et al., 2020). Note that demethylation of m6Am by FTO generates 2′-O-methyladenosine and it is unclear if or how this can be further converted to the unmethylated ribonucleotide. Specific readers of m6Am have yet to be identified but conceivably they include known m6A readers. (C) Additionally, adenosine can also be monomethylated at the N1 position (m1A) which interferes with the Watson-Crick interface and introduces a positive charge under physiological conditions that together can alter RNA secondary structure. Enzymes including TRMT6/61A, TRMT61B, and TRMT10C have been implicated as m1A writers (Li et al., 2017; Safra et al., 2017), with ALKBH1 and ALKBH3 identified as the erasers (Dominissini et al., 2016; Li et al., 2016). There is evidence that YTHDF1-YTHDF3 and YTHDC1 can act as m1A readers potentially complicating the interpretation of reader depletion studies (Dai et al., 2018; Seo and Kleiner, 2020).
Fig. 2.
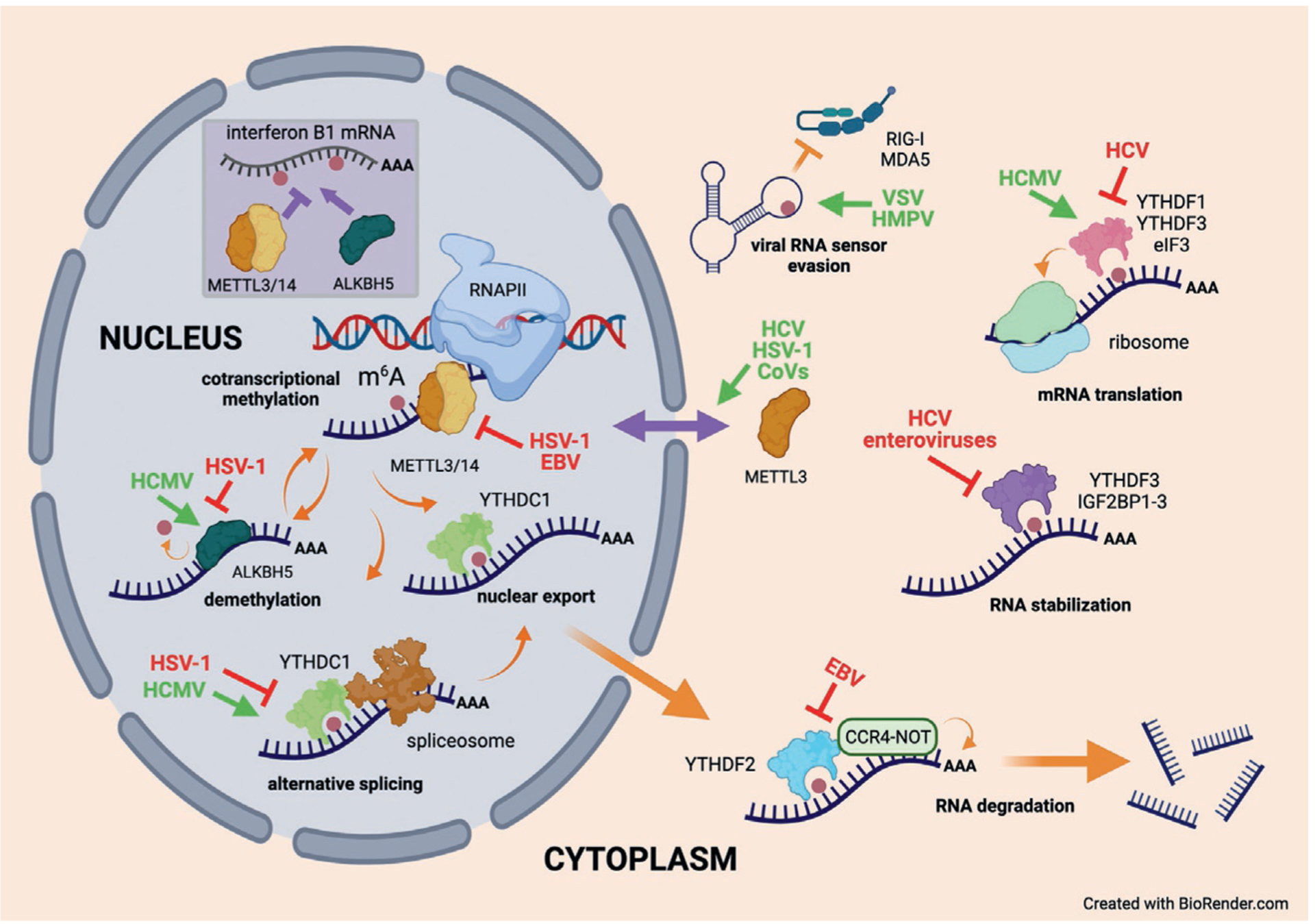
Viruses manipulate the m6A pathway at multiple steps to effect changes in RNA function or fate. During RNA polymerase II (RNAPII) transcription, m6A (red dot) is installed on nascent transcripts by a polymerase-associated complex that includes the essential METTL3 and METTL14 subunits, and is potentially reversed through the actions of the nuclear demethylase ALKBH5. The m6A epitranscriptomic mark is recognized by reader proteins such as YTHDC1 to influence splicing and/or facilitate export into the cytoplasm. Of special relevance to viral infections, the expression of the unspliced IFNβ1 mRNA (inset) is modulated by the competing inhibitory and stimulating activities of METTL3/14 and ALKBH5, respectively. Once in the cytoplasm, m6A-modified polyadenylated RNAs engage with a variety of other reader proteins that recognize m6A directly or indirectly and influence transcript function and fate. Depending on the cell type and possibly the site and density of m6A installation, the binding of YTHDF2 can destabilize mRNAs through polyA tail shortening by the CCR4/NOT complex. Alternatively readers can confer increased stability or can stimulate translation by ribosomes. RNA folding can also be modulated through the presence or absence of m6A and this can limit recognition of virus RNA signatures by innate defense sensors such as RIG-I or MDA5. Many of these steps are manipulated by viruses during infection either to enhance their own gene expression and/or genome replication or to suppress antiviral activities. Here representative viruses that stimulate or upregulate host m6A-associated functions are identified in green, whereas those that repress host functions are shown in red. See text for more discussion.
2. m6A detected in RNAs from a broad spectrum of viruses
Pioneering studies beginning in the mid-1970s established that mRNAs transcribed from DNA viruses including adenovirus type 2, simian virus 40 (SV40), and herpes simplex virus type 1 (HSV-1) are extensively m6A-modified (Canaani et al., 1979; Hashimoto and Green, 1976; Lavi and Shatkin, 1975; Moss et al., 1977; Sommer et al., 1976, 1978). Armed with our current understanding of the pathway and knowing that these viruses use RNAPII to transcribe their genes, and are reliant on many other components of the host RNA processing machinery for transcript maturation, this seem self-evident today but was a remarkable yet under appreciated discovery at the time. Accordingly, coding and non-coding RNAs produced by other herpesviruses such as human cytomegalovirus (HCMV), Epstein–Barr virus (EBV), and Kaposi sarcoma herpesvirus (KSHV) are also extensively m6A-modified (Baquero-Perez et al., 2019; Ye et al., 2017). The genomic RNAs of retroviruses, such as Rous sarcoma virus (Kane and Beemon, 1985), murine leukemia virus (Courtney et al., 2019), and human immunodeficiency virus (HIV) (Kennedy et al., 2017), which are transcribed by RNAPII from chromosomally integrated proviruses exhibit especially high levels of modification. More surprising however is the discovery that many RNA viruses transcribed using their own RNA-dependent RNA polymerase (RdRp) are also m6A-modified. These include negative-sense RNA viruses such as influenza A virus (IAV) which replicate in the nucleus using their own RdRp (Courtney et al., 2017; Narayan et al., 1987), as well as viruses that replicate exclusively in the cytoplasm such as respiratory syncytial virus (RSV) (Xue et al., 2019) and human metapneumovirus (HMPV) (Lu et al., 2020). The growing list of positive-sense RNAs viruses found to be m6A modified include flaviviruses such as Zika virus, dengue virus, yellow fever virus, and West Nile virus (Gokhale et al., 2016; Lichinchi et al., 2016) along with entero-virus 71 (EV71) (Hao et al., 2019), and members of the α- and β-coronavirus subfamilies including the COVID-19 pandemic agent, SARS-CoV-2 (Chen et al., 2020; Burgess et al., 2021; Li et al., 2021; Liu et al., 2021; Zhang et al., 2021). More recently, the double-stranded RNA genome of rotavirus was shown to be m6A-modified (Wang et al., 2022). In many instances, depletion of individual writer, eraser, or reader subunits using RNA interference or gene knockout were found to have both positive and negative impacts on viral replication. The likely mechanisms are discussed below.
3. Impact of m6A on the maturation, function and fate of viral RNAs
Studies in mammals, insects and plants have clearly demonstrated that presence or absence of m6A can significantly influence the fate and function of cellular RNAs and it is now becoming clear that many of these processes also apply to animal virus RNAs or are directly manipulated by viruses as part of their infection strategies. Consequently, there is a growing interest in using viruses as tractable systems to explore these processes in more detail. Below we will discuss the major processes from a viral perspective.
3.1. Control of viral transcript diversity
To maximize the coding potential of their relatively small genomes, many viruses have evolved complicated transcriptomes with extensive overlap between transcription units (Depledge et al., 2019). For instance, the adenoviral late mRNAs are produced through an elaborate pattern of alternative splicing combined with cleavage and polyadenylation site (CPAS) readthrough. Using a multifaceted approach that brought together meRIP-seq, direct RNA sequencing, and METTL3 disruption or depletion, it has been shown that m6A is installed at many sites within adenoviral precursor RNAs and that the nuclear m6A reader YTHDC1 regulates alternative splice site usage to enhance the production of the critical late mRNAs (Price et al., 2020). Along similar lines, overexpression or depletion of YTHDC1 was found to have little impact on HIV-1 mRNA nuclear export or stability but did alter the complex pattern of alternative splicing characteristic of HIV-1 mRNAs (Tsai et al., 2021). Interestingly, the sites of YTHDC1 binding show limited overlap with the known splicing signals and as such the underlying mechanism is still unclear. Because these findings mirror the influence of YTHDC1 of splice site usage in cellular transcript, continued studies of the impact of m6A on HIV alternative splicing will be instructive.
3.2. Post-transcriptional control of gene expression
Arguably the most broadly applicable function of m6A is in the control of RNA turnover and there are now a number of examples involving both the stabilization and the destabilization of viral RNAs. Infection of susceptible cells by EBV or KSHV, members of the γ-herpesvirus subfamily, can result in specific cancers. METTL14 was found to be an important factor in EBV-induced oncogenesis and steady-state levels are elevated in EBV-positive cancers (Lang et al., 2019). Modification of the EBV transcriptome by m6A stimulates the expression of viral latent genes through mRNA stabilization but also repressed productive cycle (lytic) gene expression via YTHDF1-mediated RNA decay (Lang et al., 2019). Likewise KSHV productive cycle transcripts are detectably m6A-modified (Baquero-Perez et al., 2019; Hesser et al., 2018; Tan et al., 2018; Ye et al., 2017). Interestingly, depletion of the m6A machinery had differential pro- and anti-viral impacts depending on the infection model being tested (Hesser et al., 2018). In a KSHV-infected renal carcinoma line (iSLK.219), depletion of either METTL3 or YTHDF2 impaired virus reproduction due to reduced accumulation of the ORF50 mRNA encoding the essential viral trans-activator RTA. Surprisingly, YTHDF2 depletion in a latently-infected B-cell line (TREX-BCBL-1) had the opposite outcome.
A more recent study found that YTHDF2 binds to multiple, overlapping sites on the HIV genome to increase viral RNA stability (Tsai et al., 2021). These examples are surprising because YTHDF2 binding to cellular mRNAs, including mRNAs produced by endogenous retroviruses (Chelmicki et al., 2021), results in their destabilization (Wang et al., 2014; Park et al., 2019; Zaccara and Jaffrey, 2020) and implies that the context of YTHDF2 m6A sequence recognition is an RNA stability determinant (Tsai et al., 2021). In line with the KSHV studies discussed above, it is also possible that cell type-dependent differences in the availability of different m6A readers may govern the functional outcomes of transcript methylation. Using a different approach, the cellular Tudor SND1 protein was found to act as an m6A reader that recognizes an m6A-modified hairpin within the KSHV RTA/ORF50 mRNA (Baquero-Perez et al., 2019). The association of SND1 with ORF50 mRNA was dependent on the presence of m6A at that site and resulted in the stabilization of the ORF50 mRNA. Consequently, SND1 was found to be essential for KSHV early gene expression and productive replication.
YTHDF2 can act as a proviral factor in other contexts. In permissive monkey kidney established cell lines, overexpression of YTHDF2 resulted in a fivefold increase in SV40 replication and two-fold increase in plaque size (Tsai et al., 2018). Similarly, CRISPR-Cas9 deletion of YTHDF2 decreased plaque size by approximately 40% and both YTHDF2 or METTL3-deletion reduced viral late protein accumulation. Ablation of m6A acceptor sites resulted in slower replication and reduced plaque size, consistent with m6A installation in cis regulating SV40 late gene expression (Tsai et al., 2018).
Although the above examples implicate m6A in the stabilization of viral RNAs, this is not always the case. As part of the antiviral-response the product of interferon-stimulated gene 20 (ISG20) promotes degradation of viral RNAs though its 3′−5′ exonuclease activity (Espert et al., 2003; Nguyen et al., 2001; Zhou et al., 2011). During infection by hepatitis B Virus (HBV) this destabilizing activity is enhanced by m6A methylation at position 1907 in the epsilon stem loop of the pregenomic RNA (Imam et al., 2018). This site is present in two copies and is recognized by YTHDF2. HBV RNA levels were increased in cells depleted for YTHDF2 or the METTL3/14 subunits of the methyltransferase or by mutagenesis of the methylation sites within the epsilon loops (Imam et al., 2020).
Host m6A-modification functions can stimulate reproduction of at least two RNA viruses that replicate in the nucleus. While HIV replication is stimulated by the METTL3/14 m6A writer complex, m6A erasers ALKBH5 and FTO suppress it (Kennedy et al., 2017; Lichinchi et al., 2016; Tirumuru et al., 2016). Various mechanisms including differential nuclear export, protein accumulation, and virus genome replication have been suggested to account for this. The METTL3 methyltransferase likewise promotes IAV replication and m6A sites in the viral HA segment confer proper protein expression and are needed for pathogenicity in mice (Courtney et al., 2017).
Overexpression of m6A readers or writers and knockdown of demethylases likely resulting from changes in host and virus gene expression stimulate HMPV protein expression and replication (Lu et al., 2020). Recombinant HMPV mutants with altered m6A acceptor sites in G-protein coding regions that did not change the amino acid coding displayed reduced replication kinetics and G protein expression (Lu et al., 2020). Similarly, m6A methyltransferase-depletion decreased RSV replication and gene expression whereas m6A demethylase-depletion had the opposite effect (Xue et al., 2019). In addition, RSV recombinants expressing G transcripts that lack particular m6A clusters displayed reduced replication in established cell lines and were attenuated in rodent models.
4. Impact of m6A on host responses to infection
4.1. Shielding viral RNAs from host pathogen sensors
RNA modifications can also impact whether viral nucleic acids are recognized as a pathogen-associated molecular pattern (PAMP) by host sensors such as RIG-I and MDA5, and thereby help virus to avoid cell intrinsic surveillance mechanisms important for immunity and host defense (Durbin et al., 2016). Along these lines HMPV virion RNA deficient in m6A was recognized better by RIG-I resulting in greater interferon (IFN) expression, suggesting that removing m6A from viral RNA genomes may be a strategy for attenuation (Lu et al., 2020). This may help explain why translocation of METTL3 from the nucleus to the cytoplasm in vesicular stomatitis virus-infected cells served to increase viral RNA m6A modification and decrease RIG-I mediated sensing (Qiu et al., 2021).
Adenosine to inosine (A to I) RNA editing catalyzed by the ISG-encoded adenosine deaminase acting on RNA (ADAR) is also regulated by m6A modification (Eisenberg and Levanon, 2018). ADAR1–2 specifically bind to the PAMP dsRNA where the resulting A to I editing can alter mRNA splicing, primary protein sequence, and RNA secondary structure, including dsRNA PAMP accumulation (Xiang et al., 2018). ADAR1 accumulation in response to IFN treatment is stimulated by m6A modification of its cognate mRNA, which is subsequently recognized by YTHDF1 (Terajima et al., 2021). YTHDF1-depletion reduced levels of IFN-induced A-to-I RNA editing, which in turn activated dsRNA-sensing and enhanced expression of ISGs in several, but not all established cell lines examined. The suppression of VSV replication by YTHDF1-depletion was partially relieved by overexpression of wild-type, but not catalytically inactive ADAR1, indicating that ADAR1 A to I RNA editing activity contributes to YTHDF1-dependent IFN responses. Since ADAR1 editing is known to preclude self activation of the cytosolic PAMP sensor MDA5 by endogenous dsRNA (Liddicoat et al., 2015; Pestal et al., 2015), which induces type I IFN antiviral immunity, m6A modification of ADAR1 mRNA maintains immune homeostasis by restricting self-RNA sensing.
4.2. Control of IFN mRNA abundance
Depletion of METTL3 or METTL14 in primary human fibroblasts inhibited HCMV reproduction, whereas depletion of ALKBH5 stimulated virus reproduction and spread by nearly 100-fold (Rubio et al., 2018; Winkler et al., 2019). Viral gene expression in adenovirus or RNA virus (VSV, IAV) infected cells were similarly inhibited by METTL3-depletion (Winkler et al., 2019). By contrast, depleting METTL3 or ALKBH5 did not measurably impact replication of other DNA viruses, including HSV-1 or vaccinia (Rubio et al., 2018). The underlying mechanism responsible for regulation of HCMV reproduction by host m6A modification machinery was found to involve differential IFNβ production (Rubio et al., 2018; Winkler et al., 2019). IFNB1 mRNA accumulation was stimulated by METTL14 depletion and restricted by ALKBH5 depletion. Moreover, IFNB1 production in uninfected cells treated with dsDNA was likewise regulated by METTL14 and ALKBH5, and IFNB1 mRNA was enriched in an m6A RNA-containing fraction (Rubio et al., 2018). Finally, genome-wide profiling of cells exposed to dsDNA following ALKBH5 depletion identified differentially expressed genes regulating antiviral immune responses, while METTL14 depletion altered pathways involving metabolic reprogramming, stress responses, and aging (Rubio et al., 2018). In particular IFNB1 mRNA was found to be m6A-modified both within the 3′UTR and coding sequences (Rubio et al., 2018; Winkler et al., 2019). In HCMV-infected cells, IFNB1 mRNA decay was regulated by m6A modification, whereas mRNA decay only modestly contributed to IFNB1 mRNA levels in uninfected cells treated with dsDNA in response to METTL14 or ALKBH5-depletion (Rubio et al., 2018; Winkler et al., 2019). Instead, IFNB1 mRNA levels in uninfected cells exposed to dsDNA were primarily regulated by changes in new RNA synthesis in response to METTL14 or ALKBH5-depletion (Rubio et al., 2018). The effects of interfering with the cellular m6A modification machinery on new RNA synthesis could reflect changes in expression of the host transcription machinery or possibly a more direct effect of m6A modification on chromatin-associated regulatory RNAs (carRNAs), which in turn might impact IFNB1 transcription (Liu et al., 2020). Investigating how the METTL14 m6A methylase subunit and the ALKBH5 demethylase control HCMV replication thus revealed a fundamental mechanism whereby host cell-intrinsic immune responses to dsDNA in uninfected cells are controlled by m6A RNA modification.
In a mechanism reportedly not involving METTL3-dependent m6A installation, the m6A reader YTHDF3 was found to limit ISG expression under homeostatic conditions. VSV reproduction was restricted in YTHDF3-deficient cells and survival of infected mice was enhanced compared to wild-type mice. Together with eIF4G2 and PABPC1, YTHDF3 stimulated translation of FOXO3 mRNA, the protein product of which negatively regulates ISG expression (Zhang et al., 2019). Differential m6A modification of host mRNAs additionally alters cellular metabolism to inhibit VSV replication (Liu et al., 2019). Finally, besides controlling cell intrinsic innate immune responses including type I IFN, specific host transcripts with virus-induced m6A alterations also regulate infection by viruses in the family Flaviviridae including dengue virus, West Nile virus, Zika virus and hepatitis C virus (HCV) (Gokhale et al., 2020).
4.3. Promoting expression of interferon effector proteins
Many ISG encoding mRNAs that are upregulated in response to IFN inducers such as cytoplasmic dsDNA or virus infection (Rubio et al., 2018; Winkler et al., 2019) or presence of type I IFN in the growth medium are also modified by m6A (McFadden et al., 2021). Of these, translation of IFITM1 mRNA was reportedly augmented by METTL3/14-dependent m6A modification within the 3′-UTR (McFadden et al., 2021). Furthermore, overexpression of the m6A reader YTHDF1 was found to increase IFITM1 protein accumulation in response to type I IFN by 25% in an m6A-binding-dependent manner. Accordingly, exposure of METTL3/14-depleted cells to IFNβ increased the percentage of VSV-infected cells by approximately 20%, whereas METTL3/14 overexpression diminished VSV infection to a comparable extent (McFadden et al., 2021). Thus, m6A modification of ISG-encoded mRNAs can intensify the antiviral activity of the type I IFN responsiveness. This report presumed that the reduction in VSV infection does not involve METTL3/14 modification of virus RNAs, since VSV mRNAs were thought to only contain m6Am which is installed by the PCIF1 methyltransferase (discussed later). Others, however, have detected m6A-modified VSV mRNA (Lu et al., 2020; Qiu et al., 2021), potentially complicating this interpretation.
5. Viral manipulation of the m6A pathway
As discussed in the preceding section, the m6A pathway impacts viral replication at multiple levels, often promoting more effective viral replication by enhancing the expression of viral proteins or the stability of viral genomic RNAs. However, there are also situations where the reverse is true and m6A contributes to a more robust antiviral response. As might be anticipated, several viruses have evolved countermeasures to alter the pathway to benefit their replication strategy.
5.1. Altered abundance of m6A factors
In contrast to many viruses, including the evolutionarily related members of the α- and γ-herpesvirus subfamilies, the β-herpesvirus HCMV does not globally impair ongoing host protein synthesis and instead remodels the host translational landscape, thus facilitating productive HCMV replication (McKinney et al., 2014; Tirosh et al., 2015). Unexpectedly, HCMV infection of primary human fibroblasts was found to stimulate the accumulation of m6A methyltransferase subunits METTL3/14, as well as the m6A readers YTHDC1, YTHDF1–3 and the demethylases ALKBH5 and FTO early in the virus lifecycle (Rubio et al., 2018; Winkler et al., 2019). These increases require viral gene expression and are dependent upon mTOR activation in HCMV-infected cells (Rubio et al., 2018).
This contrasts with findings from EBV+ Akata cells undergoing reactivation in which the writer subunits METTL14, WTAP and VIRMA as well as the cytoplasmic YTHDF2 reader are proteolytically cleaved by one or more cellular caspases leading to enhanced EBV replication (Zhang et al., 2021). METTL14 levels are elevated in EBV transformed cells as a response to expression of the latent oncoprotein EBNA3C (Lang et al., 2019). Interestingly, caspase cleavage sites are also present in YTHDF1 and YTHDF3, implying a broad assault on m6A recognition. The caspase 8 mRNA is also m6A modified and bound by YTHDF2, consistent with a feedforward regulatory loop. The likely mechanism for enhanced EBV replication is prevention of YTHDF2-mediated recruitment of the CCR4-NOT deadenylase complex (Du et al., 2016), resulting in the stabilization of EBV productive cycle mRNAs (Xia et al., 2021).
Other viruses similarly target m6A readers for proteolytic degradation. Enteroviruses (EV) encode two cysteine proteases that are used to process the viral polyprotein and also disable host functions through the selective cleavage and degradation of regulatory proteins. An unbiased screen for host targets of the EV 2APRO protease identified the m6A readers YTHDF1–3 as substrates in EV-infected HeLa cells (Kastan et al., 2021). YTHDF3 depletion in EV-infected cells revealed elevated induction of IFN response factor 3 (IRF3) phosphorylation and IFN β/λ 1 mRNAs, while IFN-β/λ 1 release was not significantly altered. However, IFN-stimulated gene (ISG) induction was diminished substantially.
Instead of promoting virus replication, the METTL3/14 m6A methyl-transferase restricted hepatitis C virus (HCV) protein expression and infectious virus production without detectably changing RNA replication, whereas the FTO demethylase stimulated HCV protein expression (Gokhale et al., 2016; Horn and Sarnow, 2017). YTHDF reader proteins are redistributed to virus assembly sites on lipid droplet surfaces and their depletion increased infectious virus production. Furthermore, inactivating m6A acceptor sites within the viral E1 gene similarly increased virus titer without detectably impacting replication (Gokhale et al., 2016). This is consistent with a model whereby m6A regulates infectious virus production and that viral genomes deficient in m6A are preferentially incorporated into virus particles compared with those harboring m6A.
METTL3-depletion also reduced EV71 replication as did silent mutations that destroy m6A acceptor sites in the viral genome without changing coding capacity (Hao et al., 2019; Yao et al., 2020). In contrast, depletion of FTO, YTHDF1–3, or YTHDC1 stimulated EV71 replication. The subcellular distribution of the host m6A modification machinery was also altered in response to EV71 infection, with METTL3 associating with the virus RNA dependent RNA polymerase and colocalizing with viral dsRNA replicative intermediates. While METTL14 was also found to accumulate in the cytoplasm, the cytoplasmic readers YTHDF1 and YTHDF2 were partly redistributed into the nucleus (Hao et al., 2019).
Last but not least, the replication of both seasonal and pandemic β-coronaviruses (CoVs) is suppressed by depletion of METTL3 or the cytoplasmic m6A readers YTHDF1 and YTHDF3 (Burgess et al., 2021; Li et al., 2021; Zhang, Hao, et al., 2021) indicating their positive roles in the viral lifecycle. Moreover, a highly selective small molecule inhibitor of METTL3 catalysis was found to limit CoV RNA synthesis, protein accumulation and replication, providing proof-in-principle that the m6A machinery is a viable anti-viral target (Burgess et al., 2021).
5.2. Reshaping the host m6A landscape
In contrast to HCMV, herpes simplex virus (HSV) radically alters the transcriptome of the host cell such that host protein synthesis is substantially impaired. This involves several mechanisms that predominate at different times in the infection cycle, including the destabilization of mRNAs through endolytic cleavage by the virus host shutoff (vhs) protein, and disruption of 3′-end formation and transcription termination by the ICP27 protein and an overall decline in transcription of host genes by RNAPII (Friedel et al., 2021; Oroskar and Read, 1989; Wang et al., 2021). Intriguingly, the ICP27 protein also brings about a profound redistribution of nuclear m6A factors comprising multiple subunits of the writer complex (METTL3, METTL14, VIRMA, ZC3H13, RBM15 and RBM15B but interestingly not WTAP, which remains nuclear), and the m6A reader YTHDC1 and eraser ALKBH5, all of which accumulate in the cytoplasm (Srinivas et al., 2021). Redistribution is evident by immunofluorescence microscopy and biochemical fractionation and occurs relatively late in the infection cycle. Importantly, this requires expression of ICP27 either from the virus or a transfected plasmid, implicating the dysregulation of the m6A pathways in some or all aspects of host shutoff. Consistent with the radical disassembly and redistribution of the methyltransferase complex, there is a global loss of m6A on newly synthesized host and viral polyadenylated RNAs. Although the expression of viral proteins of all three kinetic classes is sensitive to the depletion of m6A factors early in the replication cycle this requirement is lost as the infection proceeds paralleling the loss of m6A installation (Srinivas et al., 2021).
6. m6Am and m1A modifications
As with cellular transcripts, viral RNAs transcribed in the nucleus by host RNAPII include an m7G cap at the 5′-end that confers stability and allows for the recruitment of additional RNA processing and translation factors (Furuichi and Shatkin, 2000). Cap-proximal 2′-O methylated adenosines—frequently the first one or two transcribed nucleotides—can also be methylated at the N6 position (Fig. 1B), and this modification (m6Am) is installed by the RNAPII-associated methyltransferase PCIF1/CAPAM (Akichika et al., 2019; Sendinc et al., 2019). While multiple studies concur that m6Am controls cellular gene expression post-transcriptionally, the underlying mechanism(s) remain unclear with one transcriptome-wide analysis reporting it promotes capped mRNA translation (Akichika et al., 2019), whereas another found that it represses cap-dependent translation (Sendinc et al., 2019), while others found that PCIF-depletion reduced the stability of a subset of m6Am-annotated mRNAs without substantially impacting mRNA translation (Boulias et al., 2019). Further investigation is required to understand and possibly resolve these differences and of course discern if multiple mechanism of action exist depending upon biological context. Although m6Am is present in this position in almost a third of cellular mRNAs, its influence on viral biology has not been studied extensively (Wei et al., 1975). It has been shown however, that PCIF1 is responsible for the m6Am modification of RNAPII-transcribed mRNAs encoded by nuclear viruses such adenovirus (Wei et al., 1975), and KSHV (Tan et al., 2018). Interestingly m6Am was not detected on HIV genomic RNA in spite of the presence of other chemical modifications including m6A (Kennedy et al., 2016). In fact HIV infection induces a dramatic decrease in m6Am of cellular mRNAs (Zhang et al., 2021). Using PCIF1 depleted T cells, 854 out of 2237 m6Am-modified transcripts were impacted by HIV infection. This alteration of the host epitranscriptome is mediated by viral protein R (Vpr), which can interact with PCIF1 to induce its ubiquitination and degradation. Among the m6Am-regulated genes identified, PCIF1 reportedly inhibited HIV infection by enhancing transcription factor ETS1 (ETS Proto-Oncogene 1, transcription factor) stability, which binds to the HIV promoter and represses viral transcription (Zhang, Kang, et al., 2021).
Unexpectedly, m6Am has been found in mRNAs produced by negative-strand RNA viruses including VSV, measles, and rabies virus that replicate in the cytoplasm (Tartell et al., 2021). Although interfering with PCIF1-dependent modification of the VSV mRNA cap did not have a detectable impact on translation, decay or infectivity, the antiviral effects of IFNβ treatment were significantly enhanced. This implies that the cap-proximal m6Am modification of VSV mRNA represents a strategy to subvert the host IFN response. Future studies will likely shed light on the mechanisms by which cap structures lacking m6Am are recognized and may identify other viruses that are shielded in this way.
Readers should keep in mind that adenosine can also undergo monomethylation at the N1 position (m1A) (Fig. 1C). This modification is less abundant than m6A or m6Am, comprising only 0.02% of adenosine in RNAs from human cells and is found in mRNAs, lncRNAs and tRNAs (Dominissini et al., 2016; Li et al., 2016; Safra et al., 2017). By introducing a positive charge and by disrupting Watson-Crick paring, m1A can have strong effects on RNA secondary structure and folding equilibria (Zhou et al., 2016). Although installed by different methyltransferases that include but are not limited to TRMT6/TRMT61A, m1A-modified RNAs can be recognized by m6A readers YTHDC1 and YTHDF1–3, potentially complicating the interpretation of depletion studies (Dai et al., 2018; Zheng et al., 2020). Profiling by both meRIP-seq and mass spectrometry has detected m1A in influenza virus RNAs (Furuse, 2021) but not in HIV genomic RNA (Šimonová et al., 2019). The extent to which m1A modification is found in RNAs encoded by other viruses, including DNA viruses and RNA viruses that replicate in the cytoplasm, remains unknown and is ripe for future exploration.
7. Future directions
Viral epitranscriptomics, the study of post-transcriptional modifications of viral RNAs, is a burgeoning new field of research (Dang et al., 2019; Gokhale and Horner, 2017; Horn and Sarnow, 2017; Imam et al., 2020; Kennedy, Courtney, et al., 2017; Williams et al., 2019). As illustrated by the examples discussed above, the m6A pathway makes a substantial contribution to the replication and pathogenesis of a broad spectrum of important and well-studied DNA and RNA viruses including many that cause disease in humans. In the near future, studies will likely focus on key mechanistic details such as how and where m6A is installed and how this impacts RNA secondary structures and/or the RNA-protein interactome. For many viral transcriptomes, the locations of m6A installation are poorly resolved both in terms of pinpointing the exact adenosine residues that are modified and more so, determining the frequency with which they are modified in individual RNA molecules. These shortfalls very much reflect current limitations in mapping technologies. The majority of studies have used antibody-based RNA capture followed by RNA fragmentation and short-read sequencing (meRIP-seq). Aside from concerns about antibody specificity, as m6A antibodies widely used for m6Am/m6A mapping do not discriminate between the two functionally distinct modifications (Dominissini et al., 2012; Meyer et al., 2012), the popular meRIP-seq mapping approach identifies regions in the order of 100–300 nucleotides that contain one or more methylated adenosines and thus cannot distinguish single sites from clusters of sites (Grozhik and Jaffrey, 2018; McIntyre et al., 2020; Zheng et al., 2020). Offering close to nucleotide resolution, miCLIP is more technically involved and also vulnerable to the same antibody selectivity issues. Alternative antibody-independent approaches such as SCARLET (site-specific cleavage and radioactive-labeling followed by ligation-assisted extraction and thin-layer chromatography) (Liu et al., 2013) and MAZTER-seq (Garcia-Campos et al., 2019) offer the potential for nucleotide-resolution mapping but so far have seen little application in viral infection systems. The former is technically demanding and the latter cannot detect all sites. Newer approaches such as introduction of a selective in vitro demethylation step prior to RNA immunoprecipitation (m6Am-seq) could help to address the lack of antibody selectivity and have shown that m6Am is installed in the 5′-UTRs of mRNAs encoding stress-response factors during heat shock and hypoxia (Sun et al., 2021). Likewise efforts to harness the ability of long-read or direct RNA sequencing to detect and map modified ribonucleotides with transcript-level precision are slowly being brought to bare on viral systems (Burgess et al., 2021; Campos et al., 2021; Chang et al., 2021; Price et al., 2020; Srinivas et al., 2021). Due to their small genomes and in many cases ease of manipulation, viruses represent an exciting arena in which to study the biological contributions of individual modification sites and also potentially delineate the criteria for site-specific installation and removal by writers and erasers as well as recognition by readers.
7.1. Installation of m6A on viral RNAs in the cytoplasm
Using MeRIP-seq and other profiling techniques, studies from many different laboratories have detected m6A on the genomic RNAs of viruses that replicate exclusively in the cytoplasm. At first blush this seems entirely paradoxical because these RNAs are not synthesized by host RNAPII, but instead by a virus-encoded RdRp. There is ample evidence that METTL3 is the predominant catalytic activity responsible for methylation of cellular coding and non-coding transcripts generated in the nucleus, including RNAs made by dsDNA viruses and at least two studies have reported the association of METTL3 and METTL14 with RNAPII through coimmunoprecipitation (Huang et al., 2019; Liu et al., 2021). However, some METTL3 has been observed in the cytoplasm of lung cancer cells, murine embryonic stem cells (mESC) and HCV-infected cells suggesting a more dynamic subcellular localization than is assumed (Gokhale et al., 2016; Lin et al., 2016; Wen et al., 2018). The precise nature of the trigger(s) that alter METTL3 subcellular distribution in response to some virus infection remain unknown. Whether redistribution of METTL3 in response to virus infection might have any cell type specific requirements is likewise unknown. Conceivably interactions with viral proteins involved in replication, such as the SARS-CoV-2 RdRp subunit nsp16 (Zhang, Hao, et al., 2021), might be capable of recruiting METTL3 alone or in conjunction with METTL14. This is a little visited area of cellular regulation where studies using viruses as the focus might be especially instructive.
7.2. Virus-directed post-translational modifications
Relatively little is known about the regulation of the protein factors that function within the m6A and m6Am pathways but as has been shown repeatedly for other regulatory processes such as protein phosphorylation, sumoylation and ubiquitylation, viruses can often prove especially useful in uncovering post-translational modifications that control cellular pathways (Song et al., 2021). For instance, METTL3 can be conjugated to SUMO1 at four or more positions, impacting its catalytic activity (Du et al., 2018) and a number of viruses have developed strategies to interfere SUMO-signaling (Zhang et al., 2020). Global m6A levels are reduced during reactive oxygen species (ROS)-induced stress resulting in increased expression of genes involved in of genome integrity and DNA repair (Yu et al., 2021). Mechanistically, ROS induces the phosphorylation of ALKBH5 through activation of ERK/JNK-signaling. This post-translational modification allows the recruitment of the SUMO E3 ligase PIAS4 and addition of SUMO1 which inhibits the m6A demethylase activity.
7.3. Incorporating the methyladenosine pathways into antiviral strategies
Recent development of the highly potent first-in-class small molecule METTL3 inhibitor STM2457 offers the opportunity to selectively extinguish m6A installation dependent upon METTL3 catalytic activity (Yankova et al., 2021). While originally developed for cancer chemotherapy, this bioavailable METTL3 inhibitor demonstrates anti-viral activity against both seasonal and pandemic β-coronaviruses in cultured cells (Burgess et al., 2021). This provides a proof of concept that host RNA modification pathway components can be targeted by small molecule inhibitors and identifies new therapeutic opportunities to control acute viral pathogens. As METTL3 depletion increases IFNB1 production in response to virus infection or dsDNA exposure (Rubio et al., 2018; Winkler et al., 2019), bioavailable METTL3 inhibitors are also likely to augment type I IFN responses and show utility in combination with other anti-virals or perhaps as immune-stimulating adjuvants. Targeting the enzymes responsible for catalytic adenosine methylation may be more effective than targeting readers, which act stoichiometrically, because there is less functional redundancy and more selectivity across the pathways.
Acknowledgments
We apologize to our colleagues whose work was not cited owing to space limitations. Work in the author’s laboratories is supported by grants from the National Institute of Allergy and Infectious Diseases to A.C.W. (AI130618, AI147163) and to I.M. (AI073898, AI152543) and from the National Institute of General Medical Sciences to I.M. (GM056927).
References
- Akichika S, et al. , 2019. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science (New York, NY) 363 (6423), eaav0080. 10.1126/science.aav0080. [DOI] [PubMed] [Google Scholar]
- Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF, 2015. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 162 (6), 1299–1308. 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF, 2015. N6-methyladenosine marks primary microRNAs for processing. Nature 519 (7544), 482–485. 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero-Perez B, Antanaviciute A, Yonchev ID, Carr IM, Wilson SA, Whitehouse A, 2019. The Tudor SND1 protein is an m6A RNA reader essential for replication of Kaposi’s sarcoma-associated herpesvirus. eLife 8, 482. 10.7554/elife.47261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I, Kouzarides T, 2020. Role of RNA modifications in cancer. Nature Reviews Cancer 20 (6), 303–322. 10.1038/s41568-020-0253-2. [DOI] [PubMed] [Google Scholar]
- Bélanger F, Stepinski J, Darzynkiewicz E, Pelletier J, 2010. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. Journal of Biological Chemistry 285 (43), 33037–33044. 10.1074/jbc.m110.155283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulias K, et al. , 2019. Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Molecular cell 75 (3), 631–643.e8. 10.1016/j.molcel.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess HM, et al. , 2021. Targeting the m6A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication. Genes & Development 35 (13–14), 1005–1019. 10.1101/gad.348320.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos JHC, Maricato JT, Braconi CT, Antoneli F, Janini LMR, Briones MRS, 2021. Direct RNA sequencing reveals SARS-CoV-2 m6A sites and possible differential DRACH motif methylation among variants. Viruses 13 (11), 2108. 10.3390/v13112108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaani D, Kahana C, Lavi S, Groner Y, 1979. Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Research 6 (8), 2879–2899. 10.1093/nar/6.8.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JJ-Y, et al. , 2021. Transcriptional and epi-transcriptional dynamics of SARS-CoV-2 during cellular infection. Cell Reports 35 (6), 109108. 10.1016/j.celrep.2021.109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelmicki T, Roger E, Teissandier A, Dura M, Bonneville L, Rucli S, Dossin F, Fouassier C, Lameiras S, Bourc’his D, 2021. m6A RNA methylation regulates the fate of endogenous retroviruses. Nature 17, 1–5. 10.1038/s41586-020-03135-1. [DOI] [PubMed] [Google Scholar]
- Chen J, Jin L, Wang Z, Wang L, Chen Q, Cui Y, Liu G, 2020. N6-methyladenosine regulates PEDV replication and host gene expression. Virology 548, 59–72. 10.1016/j.virol.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, et al. , 2020. METTL4 is an snRNA m6Am methyltransferase that regulates RNA splicing. Cell Research 30 (6), 544–547. 10.1038/s41422-019-0270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG, Chalem A, Bogerd HP, Law BA, Kennedy EM, Holley CL, Cullen BR, 2019. Extensive epitranscriptomic methylation of A and C residues on murine leukemia virus transcripts enhances viral gene expression. mBio 10 (3), 313. 10.1128/mbio.01209-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, Cullen BR, 2017. Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host & Microbe 22 (3), 377–386.e5. 10.1016/j.chom.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Wang T, Gonzalez G, Wang Y, 2018. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Analytical Chemistry 90 (11), 6380–6384. 10.1021/acs.analchem.8b01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Xie Y, Cao P, Xin S, Wang J, Li S, Li Y, Lu J, 2019. N6-methyladenosine and viral infection. Frontiers in Microbiology 10, 417. 10.3389/fmicb.2019.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB, Ke S, Darnell JE, 2018. Pre-mRNA processing includes N6 methylation of adenosine residues that are retained in mRNA exons and the fallacy of “RNA epigenetics”. RNA (New York, NY) 24 (3), 262–267. 10.1261/rna.065219.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depledge DP, Mohr I, Wilson AC, 2019. Going the distance: Optimizing RNA-Seq strategies for transcriptomic analysis of complex viral genomes. Journal of Virology 93 (1), 860. 10.1128/jvi.01342-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, et al. , 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Dominissini D, et al. , 2016. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530 (7591), 441–446. 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L, 2016. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nature Communications 7 (1), 12626. 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, et al. , 2018. SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Research 46 (10), 5195–5208. 10.1093/nar/gky156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin AF, Wang C, Marcotrigiano J, Gehrke L, 2016. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio 7 (5). 10.1128/mbio.00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY, 2018. A-to-I RNA editing—Immune protector and transcriptome diversifier. Nature Reviews Genetics 19 (8), 473–490. 10.1038/s41576-018-0006-1. [DOI] [PubMed] [Google Scholar]
- Espert L, Degols G, Gongora C, Blondel D, Williams BR, Silverman RH, Mechti N, 2003. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. Journal of Biological Chemistry 278 (18), 16151–16158. 10.1074/jbc.m209628200. [DOI] [PubMed] [Google Scholar]
- Friedel CC, et al. , 2021. Dissecting herpes simplex virus 1-induced host shutoff at the RNA level. Journal of Virology 95 (3). 10.1128/jvi.01399-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, Shatkin AJ, 2000. Viral and cellular mRNA capping: Past and prospects. Advances in Virus Research 55, 135–184. 10.1016/s0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse Y, 2021. RNA modifications in genomic RNA of influenza A virus and the relationship between RNA modifications and viral infection. International Journal of Molecular Sciences 22 (17), 9127. 10.3390/ijms22179127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Campos MA, et al. , 2019. Deciphering the “m6A Code” via antibody-independent quantitative profiling. Cell 178 (3), 731–747.e16. 10.1016/j.cell.2019.06.013. [DOI] [PubMed] [Google Scholar]
- Gokhale NS, Horner SM, 2017. RNA modifications go viral. PLoS Pathogens 13 (3), e1006188. 10.1371/journal.ppat.1006188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NS, McIntyre ABR, Mattocks MD, Holley CL, Lazear HM, Mason CE, Horner SM, 2020. Altered m6A modification of specific cellular transcripts affects flaviviridae infection. Molecular Cell 77 (3), 542–555.e8. 10.1016/j.molcel.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NS, et al. , 2016. N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host & Microbe 20 (5), 654–665. 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozhik AV, Jaffrey SR, 2018. Distinguishing RNA modifications from noise in epitranscriptome maps. Nature Chemical Biology 14 (3), 215–225. 10.1038/nchembio.2546. [DOI] [PubMed] [Google Scholar]
- Hao H, et al. , 2019. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Research 47 (1), 362–374. 10.1093/nar/gky1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JE, Miceli SM, Roberts RJ, Manley JL, 1990. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Research 18 (19), 5735–5741. 10.1093/nar/18.19.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto SI, Green M, 1976. Multiple methylated cap sequences in adenovirus type 2 early mRNA. Journal of Virology 20 (2), 425–435. 10.1128/jvi.20.2.425-435.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He PC, He C, 2021. m6 A RNA methylation: From mechanisms to therapeutic potential. The EMBO Journal 139, e105977. 10.15252/embj.2020105977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesser CR, Karijolich J, Dominissini D, He C, Glaunsinger BA, 2018. N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathogens 14 (4), e1006995. 10.1371/journal.ppat.1006995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn SRG, Sarnow P, 2017. Making the mark: The role of adenosine modifications in the life cycle of RNA viruses. Cell Host & Microbe 21 (6), 661–669. 10.1016/j.chom.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, et al. , 2018. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nature Cell Biology 20 (3), 285–295. 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, et al. , 2019. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 567 (7748), 414–419. 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam H, Khan M, Gokhale NS, McIntyre ABR, Kim G-W, Jang JY, Kim S-J, Mason CE, Horner SM, Siddiqui A, 2018. N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proceedings of the National Academy of Sciences of the United States of America 115 (35), 8829–8834. 10.1073/pnas.1808319115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam H, Kim G-W, Mir SA, Khan M, Siddiqui A, 2020. Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified Hepatitis B Virus transcripts. PLoS Pathogens 16 (2), e1008338. 10.1371/journal.ppat.1008338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam H, Kim G-W, Siddiqui A, 2020. Epitranscriptomic(N6-methyladenosine) modification of viral RNA and virus-host interactions. Frontiers in Cellular and Infection Microbiology 10, 584283. 10.3389/fcimb.2020.584283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, et al. , 2011. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature Chemical Biology 7 (12), 885–887. 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane SE, Beemon K, 1985. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: Implications for RNA processing. Molecular and Cellular Biology 5 (9), 2298–2306. 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan JP, Tremblay MW, Brown MC, Trimarco JD, Dobrikova EY, Dobrikov MI, Gromeier M, 2021. Enterovirus 2Apro cleavage of the YTHDF m6A readers implicates YTHDF3 as a mediator of type I interferon-driven JAK/STAT signaling. mBio 12 (2). 10.1128/mbio.00116-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE, Darnell RB, 2017. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes & Development 31 (10), 990–1006. 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, et al. , 2015. A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes & Development 29 (19), 2037–2053. 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Bogerd HP, et al. , 2017. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell host & Microbe 22 (6), 830. 10.1016/j.chom.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Courtney DG, Tsai K, Cullen BR, 2017. Viral epitranscriptomics. Journal of Virology 91 (9). 10.1128/jvi.02263-16. e02263–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, et al. , 2016. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host & Microbe 19 (5), 675–685. 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles P, et al. , 2018. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes & Development 32 (5–6), 415–429. 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang F, Singh RK, Pei Y, Zhang S, Sun K, Robertson ES, 2019. EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PLoS Pathogens 15 (6), e1007796. 10.1371/journal.ppat.1007796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasman L, et al. , 2020. Context-dependent functional compensation between Ythdf m6A reader proteins. Genes & Development 34 (19–20), 1373–1391. 10.1101/gad.340695.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi S, Shatkin AJ, 1975. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proceedings of the National Academy of Sciences of the United States of America 72 (6), 2012–2016. 10.1073/pnas.72.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C, 2016. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nature Chemical Biology 12 (5), 311–316. 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- Li X, et al. , 2017. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Molecular Cell 68 (5), 993–1005.e9. 10.1016/j.molcel.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, et al. , 2021. METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Reports 35 (6), 109091. 10.1016/j.celrep.2021.109091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Sun H, Xu C, 2018. YTH domain: A family of N6-methyladenosine (m6A) readers. Genomics, Proteomics & Bioinformatics 16 (2), 99–107. 10.1016/j.gpb.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, Rana TM, 2016. Dynamics of human and viral RNA methylation during zika virus infection. Cell Host & Microbe 20 (5), 666–673. 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR, 2015. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349 (6252), 1115–1120. 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Choe J, Du P, Triboulet R, Gregory RI, 2016. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Molecular Cell 62 (3), 335–345. 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR, 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nature Methods 12 (8), 767–772. 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Li F, Lin J, Fukumoto T, Nacarelli T, Hao X, Kossenkov AV, Simon MC, Zhang R, 2021. m6A-independent genome-wide METTL3 and METTL14 redistribution drives the senescence-associated secretory phenotype. Nature Cell Biology 23 (4), 355–365. 10.1038/s41556-021-00656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T, 2013. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA (New York, NY) 19 (12), 1848–1856. 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, You Y, Lu Z, Yang J, Li P, Liu L, Xu H, Niu Y, Cao X, 2019. N6-methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science (New York, NY) 365 (6458), 1171–1176. 10.1126/science.aax4468. [DOI] [PubMed] [Google Scholar]
- Liu J, et al. , 2014. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature Chemical Biology 10 (2), 93–95. 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, et al. , 2020. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367 (6477), 580–586. 10.1126/science.aay6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, et al. , 2021. The m6A methylome of SARS-CoV-2 in host cells. Cell Research, 1–11. 10.1038/s41422-020-00465-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, et al. , 2020. N6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nature Microbiology 227, 1–15. 10.1038/s41564-019-0653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Jaffrey SR, 2018. FTO, m6 Am, and the hypothesis of reversible epitranscriptomic mRNA modifications. FEBS Letters 592 (12), 2012–2022. 10.1002/1873-3468.13092. [DOI] [PubMed] [Google Scholar]
- McFadden MJ, McIntyre ABR, Mourelatos H, Abell NS, Gokhale NS, Ipas H, Xhemalçe B, Mason CE, Horner SM, 2021. Post-transcriptional regulation of antiviral gene expression by N6-methyladenosine. Cell Reports 34 (9), 108798. 10.1016/j.celrep.2021.108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre ABR, Gokhale NS, Cerchietti L, Jaffrey SR, Horner SM, Mason CE, 2020. Limits in the detection of m6A changes using MeRIP/m6A-seq. Scientific Reports 10 (1), 6590. 10.1038/s41598-020-63,355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Zavadil J, Bianco C, Shiflett LA, Brown S, Mohr I, 2014. Global reprogramming of the cellular translational landscape facilitates cytomegalovirus replication. Cell Reports 6 (6), 1175. 10.1016/j.celrep.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian S-B, Jaffrey SR, 2015. 5’ UTR m(6)A promotes cap-independent translation. Cell 163 (4), 999–1010. 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR, 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149 (7), 1635–1646. 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B, Gershowitz A, Stringer JR, Holland LE, Wagner EK, 1977. 5’-Terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. Journal of Virology 23 (2), 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan P, Ayers DF, Rottman FM, Maroney PA, Nilsen TW, 1987. Unequal distribution of N6-methyladenosine in influenza virus mRNAs. Molecular and Cellular Biology 7 (4), 1572–1575. 10.1128/mcb.7.4.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LH, Espert L, Mechti N, Wilson DM, 2001. The human interferon- and estrogen-regulated ISG20/HEM45 gene product degrades single-stranded RNA and DNA in vitro. Biochemistry 40 (24), 7174–7179. 10.1021/bi010141t. [DOI] [PubMed] [Google Scholar]
- Oroskar AA, Read GS, 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. Journal of Virology 63 (5), 1897–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, Kim YK, 2019. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Molecular Cell 74 (3), 494–507.e8. 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- Patil DP, Chen C-K, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR, 2016. m(6) A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537 (7620), 369–373. 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB, 2015. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity 43 (5), 933–944. 10.1016/j.immuni.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping X-L, et al. , 2014. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Research 24 (2), 177–189. 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AM, et al. , 2020. Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing. Nature Communications 11 (1), 6016–6017. 10.1038/s41467-020-19,787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, et al. , 2021. N6-methyladenosine RNA modification suppresses antiviral innate sensing pathways via reshaping double-stranded RNA. Nature Communications 12 (1), 1582. 10.1038/s41467-021-21,904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio RM, Depledge DP, Bianco C, Thompson L, Mohr I, 2018. RNA m6 A modification enzymes shape innate responses to DNA by regulating interferon β. Genes & Development 32 (23–24), 1472–1484. 10.1101/gad.319475.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, Erlacher M, Rossmanith W, Stern-Ginossar N, Schwartz S, 2017. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551 (7679), 251–255. 10.1038/nature24456. [DOI] [PubMed] [Google Scholar]
- Sendinc E, Valle-Garcia D, Dhall A, Chen H, Henriques T, Navarrete-Perea J, Sheng W, Gygi SP, Adelman K, Shi Y, 2019. PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Molecular Cell 75 (3), 620–630.e9. 10.1016/j.molcel.2019.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo KW, Kleiner RE, 2020. YTHDF2 recognition of N1-methyladenosine (m1A)-modified RNA is associated with transcript destabilization. ACS Chemical Biology 15 (1), 132–139. 10.1021/acschembio.9b00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimonová A, et al. , 2019. LC/MS analysis and deep sequencing reveal the accurate RNA composition in the HIV-1 virion. Scientific Reports 9 (1), 8697. 10.1038/s41598-019-45,079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S, Lavi U, Darnell JE, 1978. The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. Journal of Molecular Biology 124 (3), 487–499. 10.1016/0022-2836(78)90183-3. [DOI] [PubMed] [Google Scholar]
- Sommer S, Salditt-Georgieff M, Bachenheimer S, Darnell JE, Furuichi Y, Morgan M, Shatkin AJ, 1976. The methylation of adenovirus-specific nuclear and cytoplasmic RNA. Nucleic Acids Research 3 (3), 749–765. 10.1093/nar/3.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Liu D, Greco TM, Cristea IM, 2021. Post-translational modification control of viral DNA sensors and innate immune signaling. Advances in Virus Research 109, 163–199. 10.1016/bs.aivir.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas KP, Depledge DP, Abebe JS, Rice SA, Mohr I, Wilson AC, 2021. Widespread remodeling of the m6A RNA-modification landscape by a viral regulator of RNA processing and export. Proceedings of the National Academy of Sciences of the United States of America 118 (30). 10.1073/pnas.2104805118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Li K, Zhang X, Liu J, Zhang M, Meng H, Yi C, 2021. m6Am-seq reveals the dynamic m6Am methylation in the human transcriptome. Nature Communications 12 (1), 4778. 10.1038/s41467-021-25,105-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B, et al. , 2018. Viral and cellular N6-methyladenosine and N6,2’-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nature Microbiology 3 (1), 108–120. 10.1038/s41564-017-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartell MA, Boulias K, Hoffmann GB, Bloyet L-M, Greer EL, Whelan SPJ, 2021. Methylation of viral mRNA cap structures by PCIF1 attenuates the antiviral activity of interferon-β. Proceedings of the National Academy of Sciences of the United States of America 118 (29), e2025769118. 10.1073/pnas.2025769118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terajima H, Lu M, Zhang L, Cui Q, Shi Y, Li J, He C, 2021. N6-methyladenosine promotes induction of ADAR1-mediated A-to-I RNA editing to suppress aberrant antiviral innate immune responses. PLoS Biology 19 (7), e3001292. 10.1371/journal.pbio.3001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh O, Cohen Y, Shitrit A, Shani O, Le-Trilling VTK, Trilling M, Friedlander G, Tanenbaum M, Stern-Ginossar N, 2015. The transcription and translation landscapes during human cytomegalovirus infection reveal novel host-pathogen interactions. PLoS Pathogens 11 (11), e1005288. 10.1371/journal.ppat.1005288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L, 2016. N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife 5, e15528. 10.7554/elife.15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Bogerd HP, Kennedy EM, Emery A, Swanstrom R, Cullen BR, 2021. Epitranscriptomic addition of m6A regulates HIV-1 RNA stability and alternative splicing. Genes & Development 35 (13–14), 992–1004. 10.1101/gad.348508.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Courtney DG, Cullen BR, 2018. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathogens 14 (2), e1006919. 10.1371/journal.ppat.1006919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, et al. , 2022. m6A modifications regulate intestinal immunity and rotavirus infection. eLife 11, (e73628). 10.7554/eLife.73628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC, 2014. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nature Cell Biology 16 (2), 191–198. 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. , 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. , 2021. Mechanism and consequences of herpes simplex virus 1-mediated regulation of host mRNA alternative polyadenylation. PLoS Genetics 17 (3), e1009263. 10.1371/journal.pgen.1009263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C-M, Gershowitz A, Moss B, 1975. N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257 (5523), 251–253. 10.1038/257251a0. [DOI] [PubMed] [Google Scholar]
- Wen J, et al. , 2018. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Molecular Cell 69 (6), 1028–1038.e6. 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M, Purta E, Kaminska KH, Cymerman IA, Campbell DA, Mittra B, Zamudio JR, Sturm NR, Jaworski J, Bujnicki JM, 2011. 2′-O-ribose methylation of cap2 in human: Function and evolution in a horizontally mobile family. Nucleic Acids Research 39 (11), 4756–4768. 10.1093/nar/gkr038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GD, Gokhale NS, Horner SM, 2019. Regulation of viral infection by the RNA modification N6-methyladenosine. Annual Review of Virology 6 (1), 235–253. 10.1146/annurev-virology-092818-015559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler R, et al. , 2019. m6A modification controls the innate immune response to infection by targeting type I interferons. Nature Immunology 20 (2), 173–182. 10.1038/s41590-018-0275-z. [DOI] [PubMed] [Google Scholar]
- Xia T-L, et al. , 2021. N(6)-methyladenosine-binding protein YTHDF1 suppresses EBV replication and promotes EBV RNA decay. EMBO Reports 22 (4), e50128. 10.15252/embr.202050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J-F, Yang Q, Liu C-X, Wu M, Chen L-L, Yang L, 2018. N6-methyladenosines modulate A-to-I RNA editing. Molecular Cell 69 (1), 126–135.e6. 10.1016/j.molcel.2017.12.006. [DOI] [PubMed] [Google Scholar]
- Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J, 2015. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. The Journal of Biological Chemistry 290 (41), 24902–24913. 10.1074/jbc.m115.680389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, et al. , 2019. Viral N6-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nature Communications 10 (1), 4595. 10.1038/s41467-019-12,504-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankova E, et al. , 2021. Small molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature, 1–8. 10.1038/s41586-021-03536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, et al. , 2020. N6-methyladenosine modifications enhance enterovirus 71 ORF translation through METTL3 cytoplasmic distribution. Biochemical and Biophysical Research Communications 527 (1), 297–304. 10.1016/j.bbrc.2020.04.088. [DOI] [PubMed] [Google Scholar]
- Ye F, Chen ER, Nilsen TW, 2017. Kaposi’s sarcoma-associated herpesvirus utilizes and manipulates RNA N6-adenosine methylation to promote lytic replication. Journal of Virology 91 (16), 201. 10.1128/jvi.00466-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Wei J, Cui X, Yu C, Ni W, Bungert J, Wu L, He C, Qian Z, 2021. Post-translational modification of RNA m6A demethylase ALKBH5 regulates ROS-induced DNA damage response. Nucleic Acids Research 49 (10), 5779–5797. 10.1093/nar/gkab415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S, Jaffrey SR, 2020. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 181, 1–14. 10.1016/j.cell.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Hao H, Ma L, Zhang Y, Hu X, Chen Z, Liu D, Yuan J, Hu Z, Guan W, 2021. Methyltransferase-like 3 modulates severe acute respiratory syndrome coronavirus-2 RNA N6-methyladenosine modification and replication. mBio, e0106721. 10.1128/mbio.01067-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Kang Y, Wang S, Gonzalez GM, Li W, Hui H, Wang Y, Rana TM, 2021. HIV reprograms host m6Am RNA methylome by viral Vpr protein-mediated degradation of PCIF1. Nature Communications 12 (1), 5543. 10.1038/s41467-021-25,683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Zhang X, Wang J, Ma Y, Zhang L, Cao X, 2019. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proceedings of the National Academy of Sciences of the United States of America 116 (3), 976–981. 10.1073/pnas.1812536116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Zhu C, Robertson ES, Cai Q, 2020. Role of SUMOylation in human oncogenic herpesvirus infection. Virus Research 283, 197962. 10.1016/j.virusres.2020.197962. [DOI] [PubMed] [Google Scholar]
- Zhang K, Zhang Y, Maharjan Y, Sugiokto FG, Wan J, Li R, 2021. Caspases switch off the m6A RNA modification pathway to Foster the replication of a ubiquitous human tumor virus. mBio, e0170621. 10.1128/mbio.01706-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Nachtergaele S, Roundtree IA, He C, 2018. Our views of dynamic N6-methyladenosine RNA methylation. RNA 24 (3), 268–272. 10.1261/rna.064295.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Gan H, Yang F, Yao Y, Hao F, Hong L, Jin L, 2020. Cytoplasmic m1A reader YTHDF3 inhibits trophoblast invasion by downregulation of m1A-methylated IGF1R: Cell. Discovery 6 (1), 12. 10.1038/s41421-020-0144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H-X, Zhang X-S, Sui N, 2020. Advances in the profiling of N6-methyladenosine (m6A) modifications. Biotechnology Advances 45, 107656. 10.1016/j.biotechadv.2020.107656. [DOI] [PubMed] [Google Scholar]
- Zheng G, et al. , 2013. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular Cell 49 (1), 18–29. 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, et al. , 2011. Antiviral activities of ISG20 in positive-strand RNA virus infections. Virology 409 (2), 175–188. 10.1016/j.virol.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, et al. , 2016. m1A and m1G disrupt A-RNA structure through the intrinsic instability of Hoogsteen base pairs. Nature Structural & Molecular Biology 23 (9), 803–810. 10.1038/nsmb.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong X, et al. , 2021. The N6-methyladenosine RNA-binding protein YTHDF1 modulates the translation of TRAF6 to mediate the intestinal immune response. Nucleic Acids Research 49 (10), 5537–5552. 10.1093/nar/gkab343. [DOI] [PMC free article] [PubMed] [Google Scholar]


