Abstract
The nucleocapsid protein (NC) of human immunodeficiency virus type 1 (HIV-1) has two zinc fingers, each containing the invariant metal ion binding residues CCHC. Recent reports indicate that mutations in the CCHC motifs are deleterious for reverse transcription in vivo. To identify reverse transcriptase (RT) reactions affected by such changes, we have probed zinc finger functions in NC-dependent RT-catalyzed HIV-1 minus- and plus-strand transfer model systems. Our approach was to examine the activities of wild-type NC and a mutant in which all six cysteine residues were replaced by serine (SSHS NC); this mutation severely disrupts zinc coordination. We find that the zinc fingers contribute to the role of NC in complete tRNA primer removal from minus-strand DNA during plus-strand transfer. Annealing of the primer binding site sequences in plus-strand strong-stop DNA [(+) SSDNA] to its complement in minus-strand acceptor DNA is not dependent on NC zinc fingers. In contrast, the rate of annealing of the complementary R regions in (−) SSDNA and 3′ viral RNA during minus-strand transfer is approximately eightfold lower when SSHS NC is used in place of wild-type NC. Moreover, unlike wild-type NC, SSHS NC has only a small stimulatory effect on minus-strand transfer and is essentially unable to block TAR-induced self-priming from (−) SSDNA. Our results strongly suggest that NC zinc finger structures are needed to unfold highly structured RNA and DNA strand transfer intermediates. Thus, it appears that in these cases, zinc finger interactions are important components of NC nucleic acid chaperone activity.
Reverse transcription, a critical event in the retrovirus life cycle, consists of a complex series of reactions that culminate in synthesis of a linear, double-stranded DNA copy of the viral RNA genome (27; reviewed in references 4 and 14). This process is catalyzed by the virus-encoded reverse transcriptase (RT) enzyme. However, it is known that in addition to RT, host and other viral factors play important roles in viral DNA synthesis.
One of these accessory factors is the viral nucleocapsid protein (NC), a small basic, single-stranded nucleic acid binding protein, which is tightly associated with genomic RNA in the interior of the mature virus particle (for reviews, see references 14, 16, and 59). Studies on the solution structure of free human immunodeficiency virus type 1 (HIV-1) NC indicated that this protein consists of a flexible polypeptide chain and two rigid zinc-binding domains connected by a short basic peptide linker (55–57, 67, 69, 70). Recently, De Guzman et al. (19) solved the three-dimensional nuclear magnetic resonance structure of NC bound to the SL3 RNA stem-loop in the HIV-1 packaging signal. They showed that the N-terminal basic residues of NC in the complex form a helix that binds to the major groove of the RNA stem largely by nonspecific electrostatic interactions, whereas the zinc fingers are involved in specific interactions with the G residues in the GGAG tetraloop (19).
The zinc fingers are in close spatial proximity in the NC protein (49, 50, 55, 56) and appear to weakly interact with one another (12, 46, 50, 55, 74). Interestingly, the two zinc fingers have similar structures (66). However, they do not have equivalent biological activity: in fact, both zinc fingers are required for HIV-1 replication (22, 30, 32, 34), and the positions of the two fingers cannot be exchanged (22, 30).
NC proteins exhibit an unusual biochemical activity: they can act as nucleic acid chaperones, i.e., they catalyze the folding of nucleic acids into the most thermodynamically stable structures with the maximal number of base pairs (73; for reviews, see references 16, 39, and 59). The nucleic acid chaperone activity of NC is crucial for achieving highly specific and efficient viral DNA synthesis, making it possible for RT to copy structured elements in RNA and DNA templates (24, 36, 40, 42, 44, 48, 78). For example, NC stimulates HIV-1 minus-strand transfer (2, 8, 17, 18, 20, 36, 42, 58, 61, 79). During this step, minus-strand strong-stop DNA [(−) SSDNA] is translocated to the 3′ end of the viral RNA genome, in a reaction facilitated by base-pairing of the repeat (R) sequences at the 3′ ends of the RNA and DNA reactants (reviewed in reference 72). On the basis of studies performed in the absence of RT, You and McHenry (79) proposed that the stimulatory effect of NC on formation of the RNA-DNA hybrid is due to the ability of NC to unfold the TAR stem-loop structures in the R regions; this, in turn, accelerates the rate of annealing. In subsequent work from our group using a reconstituted HIV-1 minus-strand transfer system (36), we reported that NC has another, related function: it blocks RT-catalyzed self-priming reactions induced by the TAR structure. In the absence of NC, synthesis of self-priming products (SP products or SP DNAs) leads to extremely inefficient minus-strand transfer (8, 36) since SP DNAs are dead-end products (36). (Self-priming has also been observed by two other groups [44, 48] in the context of studies on (−) SSDNA synthesis.)
More recently, we developed a reconstituted system that models the reactions associated with HIV-1 plus-strand transfer (77) and have shown that some of these reactions are also stimulated by NC (see below). In this system, RT initially synthesizes plus-strand strong-stop DNA [(+) SSDNA] by copying the minus-strand sequences in the DNA donor template as well as the 18 nucleotides at the 3′ end of the tRNA3Lys primer covalently attached to the donor; this reconstitutes the primer binding site (PBS) in (+) SSDNA. In subsequent steps, the tRNA primer is removed from minus-strand donor DNA so that the 18-nucleotide (nt) complementary PBS sequences at the 3′ ends of (+) SSDNA and the minus-strand acceptor DNA template can anneal. As the plus- and minus-strands are elongated, the donor template is removed by strand displacement. The final product is an 80-bp linear, double-stranded DNA. (A schematic diagram illustrating these reactions is given in Fig. 1 in reference 77). We have demonstrated that the nucleic acid chaperone activity of NC promotes complete tRNA primer removal and annealing of the PBS sequences (77). In addition, it has been shown that DNA strand displacement is enhanced by NC (41a).
A major question that has challenged investigators working on NC concerns the function of the zinc finger structures. There is extensive evidence from mutational studies indicating that the zinc fingers are essential for packaging genomic RNA (reviewed in reference 6). However, there are some mutations in the NC metal ion binding site (CCHC) that alter the zinc coordination center and confer a noninfectious phenotype that cannot be explained by reduced RNA packaging alone (29, 32–34, 51–53, 81). This indicates that the specific native conformation induced by the CCHC zinc coordination center is required for other functions in addition to viral RNA encapsidation. These functions include integration (10), sequence-specific nucleic acid binding (7, 15, 19, 26, 54, 68, 75), and reverse transcription (24, 31, 32, 41a, 60, 62, 71, 78, 80).
In earlier work, we showed that the zinc fingers are important for the ability of NC to reduce RT pausing at a stable stem-loop structure formed by bases in the murine leukemia virus (MuLV) polypurine tract and downstream nucleotides (78). An effect of the zinc fingers on extension of the tRNA3Lys primer (62) and on elongation of HIV-1 minus-strand DNA has also been reported (24). Analysis of viral DNA synthesized by HIV-1 mutant viruses with altered CCHC motifs demonstrated that it was produced at greatly reduced levels (32, 71). In a study of a similar class of MuLV mutants, it was shown that the viral DNA products have severe defects, e.g., degraded viral DNA termini as well as insertions, deletions, and rearrangements in terminal sequences (31). How this phenotype is generated and what steps in reverse transcription are disrupted is not known.
One approach to this problem is to determine whether there is a requirement for the zinc finger structures of NC in well-defined in vitro RT systems. The reconstituted HIV-1 plus- and minus-strand transfer systems that we developed appeared to be especially useful for this type of study. Here, we have begun our investigation of zinc finger function by examining the effects of a mutation that changes the CCHC motifs in both zinc fingers to SSHS. This mutation essentially eliminates the zinc coordination sites necessary for the zinc finger conformation (35) with only a minimal change of six S atoms to six O atoms. The mutant protein is referred to as SSHS NC. Our results indicate that the zinc fingers facilitate maximal removal of the tRNA primer in plus-strand transfer. In addition, the zinc fingers contribute to the dramatic stimulatory effect of NC on minus-strand transfer and are required for efficient annealing of the complementary R regions and for blocking self-priming reactions.
MATERIALS AND METHODS
Materials.
RNA and DNA-RNA oligonucleotides were purchased from Oligos, Etc. (Wilsonville, Oreg.). DNA oligonucleotides for strand transfer assays were obtained from Lofstrand (Gaithersburg, Md.) or from Oligos, Etc. DNA oligonucleotides for site-directed mutagenesis were obtained from either Operon Technologies, Inc. (Alameda, Calif.) or Life Technologies (Rockville, Md.) An RNase H-minus HIV-1 RT having a point mutation changing the active site residue Glu478→Gln (63) was the generous gift of Stuart Le Grice (HIV Drug Resistance Program, NCI-Frederick Cancer Research and Development Center, Frederick, Md.). The MacVector program was purchased from the Oxford Molecular Company (Oxford, England).
Preparation of wild-type and SSHS NC proteins. (i) Wild-Type NC.
Recombinant wild-type NC (55-amino-acid form) was expressed and purified as described previously (78).
(ii) SSHS NC.
The Cys15→Ser Cys18→Ser Cys28→Ser Cys36→Ser Cys39→Ser Cys49→Ser (SSHS/SSHS) mutant, in the context of the pNL4-3 sequence (1; GenBank accession no. M19921) was prepared as follows. A subclone (pRB581, SSHC/SSHC) was prepared by ligating the 503-bp SpeI-ApaI fragment (containing the Cys15→Ser Cys18→Ser mutations) and the 419-bp ApaI-BclI fragment (containing the Cys36→Ser Cys39→Ser mutations) into the SpeI and SalI restriction sites of the pBluescript KS(+) phagemid vector (Stratagene, La Jolla, Calif.). The inserted fragments were derived from the Cys15→Ser Cys18→Ser and Cys36→Ser Cys39→Ser full-length proviral clones that were reported previously (34). pRB581 was mutated in two successive steps using the Quick Change site-directed mutagenesis system (Stratagene) changing Cys28→Ser and then Cys49→Ser. The Cys28→Ser mutation was performed using oligonucleotides 2549 (5′ACA TAG CCA AAA ATT CCA GGG CAC CTA G-3′; the 5′ end corresponds to nt 1988 in the sequence of pNL4-3 [1]) and Z3414D08 (5′-CTA GGT GCC CTG GAA TTT TTG GCT ATG T-3′; the 5′ end corresponds to nt 2015 of the complementary sequence of the pNL4-3 [1]). The Cys49→Ser mutation was introduced using oligonucleotides Z3414D06 (5′-CCA AAT GAA AGA TTC TAC TGA GAG ACA GG-3′; the 5′ end corresponds to nt 2052 in the sequence of the pNL4-3 [1]) and Z3414D07 (5′-CCT GTC TCT CAG TAG AAT CTT TCA TTT GG-3′; the 5′ end corresponds to nt 2080 of the complementary sequence of the pNL4-3 [1]). The resulting plasmid subclone, termed pDB589, containing the SSHS/SSHS mutation was used to generate the recombinant NC expression plasmid as follows. The gene coding for the mutant NC protein (55-amino-acid form) was amplified by PCR from pDB589 using the sense primer 4658-333 and the antisense primer 4658-356; the PCR product was cloned into the pET32a vector (Novagen, Madison, Wis.), and the NC protein was isolated and purified as described previously (10). All mutations were verified using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction kit and an ABI 373 automated sequencer with the upgraded Big Dye filter wheel (PE Applied Biosystems, Foster City, Calif.).
Construction of the full-length HIV-1 clone containing the SSHS/SSHS mutation in NC.
For preparation of the full length HIV-1 proviral clone containing coding regions for the SSHS/SSHS NC zinc fingers, pDB589 (see above) was cut with the restriction endonucleases SpeI and SalI. The 4,278-bp SpeI/SalI fragment containing the sequences coding for the SSHS/SSHS mutation in the zinc fingers was then subcloned into the homologous sites of pNL4-3 (1) to create the plasmid pDB653.
Analysis of HIV-1 virions containing the SSHS/SSHS mutation in the NC domain of Gag.
The pNL4-3 and pDB653 plasmid constructs were transfected into 293T cells as previously described (29, 32). Virus-containing supernatant fluids were clarified by low-speed centrifugation. Samples were taken for RT assays and activities were determined as described earlier (32). The procedures for Western and Northern blot analyses are given in reference 28. Infectivity was determined using either CD4-producing HeLa cells containing a long terminal repeat (LTR)–β-galactosidase construct (HCLZ) (30) or AA2-clone 5 cells (32). Viral DNA was isolated from AA2-clone 5 cells infected with the mutant and wild-type viruses as described earlier (3). PCR analysis of 2-LTR-circularized viral DNA species was performed as described previously (25, 32).
Analysis of complete primer removal.
The assay system models the step in which RNase H cleavage has already removed one base from the 3′ terminus of tRNA3Lys, leaving a 3′ rA attached to the 5′ end of minus-strand DNA (11) and measures formation of the plus-strand 80-nt transfer DNA product (77). Assay conditions, separation of the DNA products by gel electrophoresis, and PhosphorImager analysis of the gel data are described in detail by Wu et al. (77). Note that the chimeric DNA-RNA oligonucleotide and the 17-nt RNA were added in fivefold excess of the amount of (+) SSDNA to ensure complete annealing of (+) SSDNA to these two reactants (77); wild-type or SSHS NC was then added to reaction mixtures prior to addition of wild-type or RNase H-minus (63) RT (77). Reactions were initiated by addition of MgCl2 and deoxyribonucleotide triphosphates (77). Both the plus- and minus-strand 80-bp DNA was synthesized; however, only the plus-strand was detected, since 5′ 32P-labeled (+) SSDNA was used for the reaction.
Annealing reactions.
For plus-strand annealing, the reactants were 5′ 32P-labeled (+) SSDNA (50 nt) and minus-strand acceptor DNA (48 nt). Annealing of the complementary 18-nt PBS sequences in each of the reactants was performed in the presence of NC, as described in reference 77 for annealing alone. Unannealed labeled (+) SSDNA and the labeled DNA duplex were separated by electrophoresis in 6% polyacrylamide gels, and the amounts of the unannealed and annealed DNAs were quantified by using a PhosphorImager (Molecular Dynamics) and ImageQuant software. Minus-strand annealing reaction mixtures containing 5′ 32P-labeled (−) SSDNA (131 nt) and acceptor RNA (148 nt) were incubated with NC and analyzed according to procedures given in Guo et al. (37), except that 7.5% polyacrylamide gels rather than agarose gels were used to separate unannealed labeled (−) SSDNA and the labeled RNA-DNA hybrid.
Complete minus-strand transfer system.
The components present in reaction mixtures, incubation conditions, and analysis of DNA products are given in Guo et al. (36). Note that the short DNA oligonucleotide, which primes synthesis of (−) SSDNA in this system, was labeled with 32P at its 5′ end, as described in reference 38.
RESULTS
SSHS mutant NC protein.
NC plays an essential role in HIV-1 minus-strand (2, 8, 17, 18, 20, 36, 42, 58, 61, 79) and plus-strand (5, 77) DNA transfer. To investigate the question of NC zinc finger function in these reactions, we constructed a clone expressing an NC protein in which the three cysteines in each zinc finger were changed to serine, i.e., the six S atoms in the zinc coordination sites in NC were replaced by six O atoms (see Materials and Methods). This is the smallest change that can be made in NC that will essentially destroy the metal ion binding sites, while still allowing retention of neighboring residues such as certain basic and aromatic amino acids, which are also important for HIV-1 NC nucleic acid binding (13, 15, 64, 74, 76). This mutation also prevents disulfide bond formation, which can occur when less than six cysteine residues are modified (12).
Effect of wild-type and SSHS NC proteins on complete removal of the tRNA3Lys primer from minus-strand donor DNA.
In previous work with a reconstituted HIV-1 plus-strand transfer system, we demonstrated that synthesis of the 80-bp strand transfer product is dependent on the removal of 9 nt from the 3′ end of tRNA3Lys: (i) initial RNase H cleavage of the 3′ rA and (ii) the removal of an additional 8 nt by secondary RNase H cleavage and/or the nucleic acid chaperone activity of NC (77). To investigate the effect of replacing the cysteine residues in the two zinc fingers of NC on complete primer removal, we used a model system that mimics the step in which RNase H has already cleaved the 3′ rA (references 11 and 77 and references therein) from the 18 nt sequence at the 3′ end of tRNA3Lys (Fig. 1A) (77). In this system, the readout is synthesis of the 80-bp transfer product: primer removal is followed by annealing of (+) SSDNA to acceptor DNA (Fig. 1A) and subsequent elongation of plus- and minus-strand DNA. As described in Materials and Methods, 5′ 32P-labeled (+) SSDNA was annealed to the chimeric DNA-RNA oligonucleotide and the 17-nt RNA; wild-type or mutant NC was then added, followed by the addition of either wild-type or RNase H-minus (63) RT. The amount of labeled plus-strand 80-nt transfer product was quantified by PhosphorImager analysis of gel data. [Since only (+) SSDNA is labeled in this experiment, the minus-strand 80-nt transfer product is not detected.]
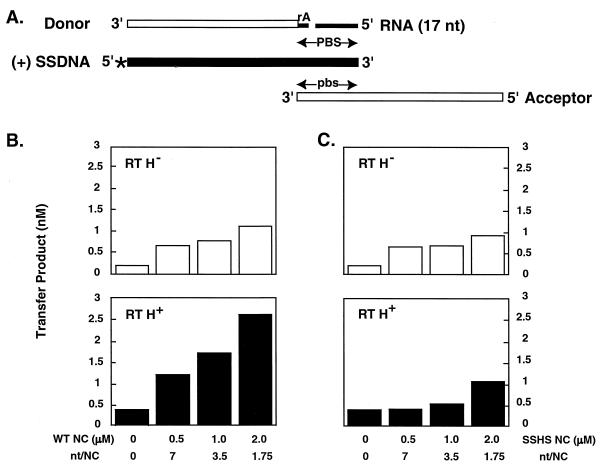
FIG. 1.
Effect of HIV-1 wild-type and SSHS NC proteins on complete removal of the tRNA primer from (−) strand DNA in reaction mixtures containing wild-type or RNase H-minus (63) RTs. (A) Nucleic acid strand transfer intermediates present in the reaction mixtures. The minus-strand donor DNA template (32 nt) is shown with a single rA attached at its 5′ end; a 17-nt RNA representing the 17 bases remaining at the 3′ end of the tRNA3Lys primer after the initial RNase H cleavage, 5′ 32P-labeled (+) SSDNA (50 nt), with the radiolabel indicated by an asterisk, and the minus-strand acceptor DNA template (48 nt) are also shown. (+) SSDNA and the minus-strand donor and acceptor DNAs are represented by filled and open rectangles, respectively; the RNA segments (rA and the 17-nt oligonucleotide) are indicated by narrow filled rectangles. This portion of the figure is taken from Fig. 7A in reference 77. (B and C) Primer removal in reaction mixtures containing increasing concentrations of wild-type (B) or SSHS (C) NC proteins. The amount of the plus-strand 80-nt transfer product (as determined by PhosphorImager analysis of gel data) is plotted against NC concentration. Reactions with RNase H-minus RT (RT H−) are shown in the upper panels, while those containing wild-type RT (RT H+) are shown in the lower panels. The open and solid bars represent results with RNase H-minus and wild-type RTs, respectively.
Figure 1B shows the results obtained from reactions with wild-type NC and mutant or wild-type RT (upper and lower panels, respectively). In accord with our earlier findings (77), the presence of wild-type NC made it possible to synthesize the strand transfer product in the absence of RNase H activity (upper panel). (In the absence of NC and RNase H activity, strand transfer did not occur [Fig. 1B, upper panel; references 5, 65, and 77]). However, if both wild-type NC and wild-type RT were added to the reaction, in effect mimicking in vivo conditions, the amount of strand transfer was stimulated by severalfold (lower panel).
A rather different picture emerged from analysis of reactions containing SSHS NC and mutant or wild-type RT (Fig. 1C, upper and lower panels, respectively). In the presence of mutant NC, the amount of transfer product made was independent of the ability of RT to catalyze RNase H cleavage. Moreover, the data in Fig. 1C were virtually identical to the results obtained under conditions where wild-type NC was present together with RNase H-minus RT (Fig. 1B, upper panel).
Taken together, the data of Fig. 1 suggest that to maximize complete removal of the tRNA primer, both the presence of the zinc finger structures of NC and the RNase H activity of RT are required.
Effect of wild-type and SSHS NC proteins on the annealing reactions in plus- and minus-strand transfer.
We have previously shown that NC stimulates annealing of the complementary PBS sequences in (+) SSDNA and minus-strand acceptor DNA during HIV-1 plus-strand transfer (77). In principle, it is possible that the effect of the zinc fingers observed in the primer removal experiment (Fig. 1) actually reflects an enhancing effect on annealing and not on primer removal per se. (In our system, elongation is not affected by NC [77].) It was therefore of interest to examine the effect of the SSHS mutation on this annealing reaction. The amount of 5′ 32P-labeled (+) SSDNA hybridized to minus-strand acceptor DNA was determined as described in Materials and Methods; the NC concentration was 0.28 μM. Note that the short dashed line at the bottom of Fig. 2A-1 represents annealing in the absence of NC (77).
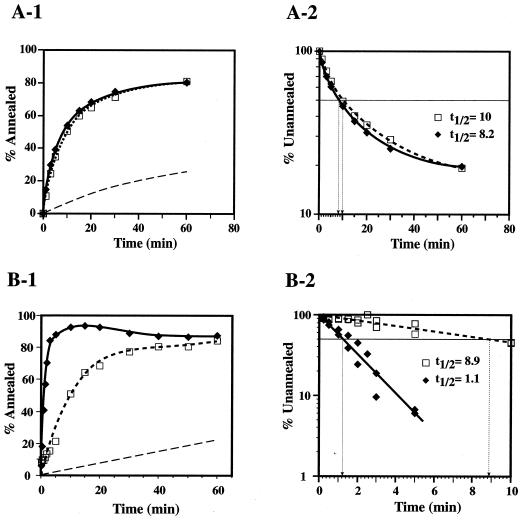
FIG. 2.
Effects of the zinc fingers on the kinetics of NC-facilitated annealing reactions required for HIV-1 plus- and minus-strand transfer. Annealing was performed as described in Materials and Methods. Reactions were incubated for the indicated times with wild-type or SSHS NC proteins. (A) Plus-strand transfer annealing reactions. The concentration of NC in the reaction mixtures was 0.28 μM. (A-1) The percentage of total (+) SSDNA annealed was plotted against the time of incubation. (A-2) The data in panel A-1 were replotted as semilogarithmic plots of the percentage of unannealed (+) SSDNA versus the time of incubation. (B) Minus-strand transfer annealing reactions. (B-1) The NC concentration in the reaction mixtures was 1 μM. The percentage of total (−) SSDNA annealed was plotted against the time of incubation. (B-2) The data in panel B-1 and the data from an independent experiment were replotted as semilogarithmic plots of the percentage of unannealed (−) SSDNA versus the time of incubation. Symbols: solid diamonds, solid lines, wild-type NC; open squares, dashed lines, SSHS NC. The short dashed lines in panels A-1 and B-1 represent annealing in the absence of NC and are based on data in references 77 and 37, respectively.
Both wild-type and mutant NCs had a marked stimulatory effect on the rate and extent of annealing, and the annealing curves were almost superimposable (Fig. 2A-1). Replotting the data in Fig. 2A-1 as semilogarithmic plots (Fig. 2A-2) indicated that the plots show curvature, as previously reported for this system (77) and as is typical for most annealing reactions (reference 21 and references therein). In this experiment, the half-life (t1/2) values for the overall reaction were calculated to be 8.2 and 10 min for the reactions with wild-type and SSHS NC proteins, respectively. Calculation of the initial rates gave even closer t½ values: 6.2 min for wild-type NC and 6.8 min for SSHS NC (data not shown). At a lower NC concentration (0.14 μM), the mutant NC was still as active as the wild-type protein (data not shown).
The data presented in Fig. 2A-1 and A-2 demonstrate that maintaining the CCHC motif in both zinc fingers of HIV-1 NC is not a requirement for promoting the annealing of the complementary PBS sequences in the plus-strand transfer intermediates. Moreover, these findings lead to the conclusion that the results in Fig. 1 reflect a direct effect of the zinc finger structures on the primer removal step.
A similar experiment was performed to determine the effect of the cysteine-to-serine substitution on the annealing reaction that occurs during HIV-1 minus-strand transfer, i.e., annealing of the complementary R sequences in (−) SSDNA and the 3′ end of viral RNA. (Note that in our constructs, R contains the entire TAR sequence [36].) The amount of 5′ 32P-labeled (−) SSDNA hybridized to plus-strand acceptor RNA was determined using the conditions described in Materials and Methods. For comparison, data obtained for annealing in reactions without NC are represented in Fig. 2B-1 by a short dashed line (37).
In the presence of wild-type NC, annealing proceeded much faster than it did with SSHS NC: the wild-type reaction was essentially complete by ∼5 min, whereas, it took ∼40 min for SSHS NC-catalyzed annealing to reach a plateau level. However, the extent of annealing was approximately the same for both proteins (Fig. 2B-1). In this case, the semilogarithmic plots were linear (Fig. 2B-2; references 37 and 79), reflecting pseudo first-order kinetics (79). The t½ values for the wild-type and SSHS reactions, calculated from the plots shown in Fig. 2B-2, were 1.1 and 8.9 min, respectively. This indicates that wild-type NC catalyzed annealing at a rate 8-fold greater than that observed in the reaction with SSHS NC.
Thus, in contrast to the data for plus-strand annealing (Fig. 2A-1 and A-2), the results of Fig. 2B-1 and B-2 demonstrate that the presence of the two CCHC motifs has a major impact on annealing in minus-strand transfer.
Requirement for zinc finger structures to inhibit self-priming in the complete minus-strand transfer system.
The observation that the zinc finger structures facilitate annealing in minus-strand transfer (Fig. 2B) led us to investigate whether zinc coordination plays a role in the ability of NC to reduce self-priming from (−) SSDNA (8, 36, 44). To approach this question, we tested the effects of wild-type and SSHS NC proteins on minus-strand transfer in the complete HIV-1 reconstituted system (36).
Figure 3A illustrates gel analysis of reactions containing increasing concentrations of wild-type and mutant NC proteins. In agreement with previous findings (36), inspection of the gel shows that synthesis of the strand transfer product (T) was greatly stimulated by wild-type NC (lanes 2 to 6) compared to the amount made in the absence of NC (lane 1). In addition, wild-type NC significantly reduced synthesis of SP DNAs. By contrast, SSHS NC promoted low levels of strand transfer and was unable to effectively inhibit self-priming (lanes 7 to 11).
FIG. 3.
Effect of HIV-1 wild-type and SSHS NC proteins on minus-strand transfer and self-priming. (A) Gel analysis. Reaction mixtures were incubated with increasing amounts of wild-type (lanes 2 to 6) or SSHS (lanes 7 to 11) NC proteins for 30 min at 37°C, according to the procedures given in Guo et al. (36) for the complete minus-strand transfer system. The DNA products were separated by gel electrophoresis in a 6% sequencing gel (36). The concentrations of NC were as follows: lane 1, no NC; lanes 2 and 7, 0.4 μM; lanes 3 and 8, 0.8 μM; lanes 4 and 9, 1.6 μM; lanes 5 and 10, 2.4 μM; and lanes 6 and 11, 3.2 μM. The positions of the primer (P), (−) SSDNA, SP products (SP), and the transfer product (T) are indicated. (B and C) PhosphorImager analysis. The gel data shown in panel A were quantified by PhosphorImager analysis, as previously described (36). The concentrations of transfer product (B) and SP products (C) are plotted against NC concentration. Symbols: solid diamonds, solid lines, wild-type NC; open squares, dashed lines, SSHS NC.
Quantification of the gel data by PhosphorImager analysis confirmed these observations. Figure 3B shows that the presence of wild-type NC dramatically increased synthesis of the strand transfer product in a dose-dependent manner (∼32-fold with saturating levels of NC); synthesis reached a plateau at ∼1.6 μM NC (1.75 nt/NC). However, in the presence of SSHS NC, the increase in synthesis of the strand transfer product was ∼6-fold at the highest NC concentration tested (3.2 μM, 0.88 nt/NC). In the linear portion of the curves, e.g., at concentrations of 0.4 and 0.8 μM, stimulation of strand transfer was ∼8-fold greater with wild-type NC. The reactions were also analyzed for the synthesis of SP products (Fig. 3C). As expected (36), increasing concentrations of wild-type NC greatly reduced the amounts of SP DNAs that were made. The SSHS NC protein had almost no effect on self-priming and, in repeated experiments, only a slight reduction in synthesis of SP products was observed.
The results presented above demonstrate that zinc coordination is required for two crucial aspects of NC activity in minus-strand transfer: (i) to promote efficient annealing (Fig. 2B) and (ii) to inhibit self-priming (Fig. 3A and C). Thus, a mutant such as SSHS NC, lacking zinc finger structures, has only a small stimulatory effect on the overall minus-strand transfer reaction (Fig. 3A and B).
Analysis of HIV-1 virions with the SSHS mutation in both zinc fingers.
The results from the in vitro experiments described above suggested that virions with the SSHS/SSHS mutation in the NC domain of Gag would be defective. To investigate this possibility, virus particles were harvested and characterized in a number of different assays. Western blot analysis showed that the protein band patterns of the mutant virus were indistinguishable from those of the wild-type. In both cases, CA protein (p24CA) was clearly visible, indicating that the viral protease was expressed and could catalyze normal cleavage of the Gag precursor (data not shown). Additional results are summarized in Table 1. The ratios of p24CA to RT activity were similar for the mutant and wild-type viruses. The full-length genomic RNA content of the mutant, determined by Northern blot analysis, was <10% of the wild-type virus, a result comparable to findings we reported for similar mutants of HIV-1 (34). In addition, the mutant was replication defective in assays utilizing HCLZ (30) and AA2-clone 5 cells, which measure infectivity in single- and multiple-rounds of replication, respectively. Analysis of 2-LTR-circularized viral DNA species showed that the expected 170-bp PCR product (25, 32) was easily detected in DNA from cells infected with the wild-type virus but was not observed in DNA obtained from cells infected with the mutant.
TABLE 1.
Properties of the SSHS/SSHS mutant and wild-type virusesa
| Sample | RT activity (cpm/ml) | p24CA (ng/ml)b | RNA content (% of WT) | HCLZ titer (foci/ml) | AA2- clone 5 titer (TCID/ml) |
|---|---|---|---|---|---|
| Controlc | 0 | 0 | 0 | <100 | |
| Mutant | 5.2 × 106 | 20 | <10 | 0 | <100 |
| WT | 4.7 × 107 | 276 | 100 | 1.3 × 105 | 106 |
WT, wild type; HCLZ (30); TCID, tissue culture infective dose.
Measured using the p24CA antigen capture kit and procedures obtained from the AIDS Vaccine Program's Biological Products Laboratory, SAIC–Frederick, National Cancer Institute–Frederick Cancer Research and Development Center.
Supernatant from cells transfected with sheared salmon sperm DNA used for mock infection.
These results demonstrate that SSHS/SSHS mutant virions are noninfectious and have defects in synthesis of 2-LTR circular viral DNA as well as in viral RNA packaging.
DISCUSSION
The goal of the present study was to develop an in vitro assay for zinc finger function during HIV-1 DNA synthesis and, in particular, to investigate the question of whether zinc coordination facilitates steps associated with HIV-1 minus- and plus-strand transfer. Our approach was to exploit reconstituted systems that model strand transfer events occurring during virus infection (36, 77) and to compare the activities of wild-type NC and a mutant, termed SSHS NC, having all of the cysteine residues in the zinc fingers changed to serine. This mutation involves a minimal change in NC, since the only sequence change results from replacement of the six S atoms with six O atoms. However, the mutation drastically reduces or eliminates the ability of the protein to bind zinc and effectively destroys the zinc finger structures (35) that are normally part of the wild-type protein. The results demonstrate that catalysis of reactions involving highly structured nucleic acid strand transfer intermediates is dependent on the presence of the zinc coordinating residues in NC.
During HIV-1 plus-strand transfer, initial RNase H cleavage of the rA at the 3′ terminus of the tRNA3Lys primer (11) is not sufficient to achieve complete removal of the tRNA from minus-strand DNA (5, 65, 77). tRNA removal is most efficient when both RNase H and NC activities are present (77). In this report we show that, unlike the situation with wild-type NC, the extent to which SSHS NC promotes primer removal is independent of whether there is concomitant RNase H cleavage and does not reach the level obtained with both RNase H and wild-type NC (Fig. 1). These data lead to the conclusion that the zinc fingers facilitate maximal removal of the tRNA primer.
It has been proposed that RT and NC form a complex that is dependent upon the zinc finger structures in NC (23, 47) and, more specifically, that such a complex may involve zinc-finger dependent (23) NC interactions (9, 58) with the RNase H domain. Under the conditions of our experiments, however, we have not been able to detect changes in RNase H cleavage in either minus-strand (37) or plus-strand (77) transfer in the presence of wild-type NC. In our earlier work, we showed that NC can catalyze destabilization of the 17-nt RNA-DNA hybrid remaining after the initial RNase H cleavage event (77). This hybrid is quite stable: using the MacVector program with specification of our salt conditions and the sequence at the 3′ end of tRNA3Lys to calculate the predicted Tm of the hybrid, we obtained a value of 67.5°C. The present results suggest that to fully unfold the structure formed by the hybrid, NC function must include zinc coordination as well as ionic interactions contributed by the basic residues of the protein.
In addition to facilitating primer removal, NC stimulates annealing of the complementary 18-nt PBS sequences at the 3′ termini of (+) SSDNA and minus-strand DNA during plus-strand transfer (77). Both wild-type and SSHS NC proteins have equivalent activity in the annealing assay and there is no requirement for zinc coordination (Fig. 2A-1 and A-2). In minus-strand transfer, however, the rate of annealing of the complementary R regions in (−) SSDNA and 3′ acceptor RNA is ∼8-fold higher with wild-type NC than it is with SSHS NC (Fig. 2B-1 and B-2). This finding was unexpected. Although it has been reported that the zinc fingers are required for primer tRNA placement in vitro (60), results from other studies (e.g., references 43 and 45) have indicated that the presence or absence of the zinc finger structures or omission of zinc in the reactions (21) has no effect on the ability of NC to stimulate RNA-DNA hybrid or DNA duplex formation in vitro.
You and McHenry (79) suggested that the unusual pseudo first-order kinetics of the minus-strand annealing reaction (Fig. 2B-2) (37, 79) are due to the fact that NC-catalyzed unfolding of TAR is the rate-limiting step. The current data further suggest that the zinc fingers play a crucial role in kinetic rearrangements of the ternary complex formed by the highly structured RNA and DNA strands with NC; ultimately, this process culminates in formation of a thermodynamically stable RNA-DNA hybrid. Although the 18-nt minus-strand PBS DNA can form a stable hairpin that is destabilized by NC (41), it is not as structured as the TAR stem-loop and has a much lower Tm. This finding may explain why the ability of NC to coordinate zinc does not seem to be necessary for NC-catalyzed annealing of the complementary PBS sequences. Thus, the results of the plus- and minus-strand annealing assays suggest that the contribution of the zinc fingers correlates with the degree of structural complexity of the strand transfer intermediates.
Further evidence for zinc finger function in minus-strand transfer was obtained when we examined the overall reaction in the complete system (36). As expected, we found that wild-type NC dramatically inhibits self-priming from (−) SSDNA (Fig. 3A and C). However, SSHS NC is essentially unable to block these dead-end reactions (Fig. 3A and C). These observations are consistent with the finding that SSHS NC stimulates synthesis of the strand transfer product to a significantly lower extent than wild-type NC (Fig. 3A and B).
Previous reports on zinc finger function in overall HIV-1 minus-strand transfer (17, 39a) and annealing of the complementary R regions (39a, 45) differ from the results presented here (Fig. 3 and Fig. 2B-1 and B-2, respectively). In the earlier work, little (17) or no (39a) reduction in minus-strand transfer could be detected when reactions contained a synthetic 72-amino-acid version of HIV-1 NC that was missing both zinc fingers. Possible differences in the structures of a zinc finger deletion mutant of the 72-amino-acid protein and a 55-amino-acid mutant having only a change in the zinc coordinating residues of NC could contribute to the different findings. It is also possible that differences in the ionic conditions are responsible for the discrepancy in the results. The earlier experiments were conducted in a lower salt environment than the conditions used here. Low-ionic-strength conditions favor protein-nucleic acid interactions dominated by ionic interactions between the basic residues of the protein and the nucleic acid phosphates (74) and would tend to minimize the contributions of the zinc finger structures. At high salt concentrations, the electrostatic nonspecific interactions between NC and a nucleic acid are weakened, while hydrophobic, zinc-finger sequence-dependent interactions become more apparent (26, 74).
In addition to the in vitro studies, we have also examined the phenotype of viral particles carrying the SSHS mutation in both zinc fingers of the NC domain in Gag. As might be anticipated (e.g., see reference 34), the virions were replication defective, packaged low levels of virion RNA, and were unable to synthesize detectable levels of 2-LTR-circularized DNA (Table 1). HIV-1 mutants containing other alterations in the CCHC motifs (changes to CCCC and/or CCHH) that preserve the ability to bind zinc, are (within the limits of detection) noninfectious, despite the fact that some of these mutants package as much as 100% of the wild-type levels of viral RNA (32). Moreover, these mutants have defects in synthesis of full-length viral DNA, but the steps in reverse transcription affected by the mutations are not known. Using our results with the SSHS NC protein as a reference point, it will now be of interest to determine the effect of other recombinant NC mutant proteins with altered CCHC motifs in the in vitro plus- and minus-strand DNA transfer reactions.
In conclusion, we have shown that the ability of NC to coordinate zinc facilitates the tRNA primer removal step in plus-strand transfer as well as annealing, reduction of self-priming, and increased strand transfer efficiency in minus-strand transfer. Our results suggest that the zinc fingers help to unfold highly structured RNA and DNA strand transfer intermediates and imply that in certain cases, zinc finger interactions play a role in the nucleic acid chaperone activity of NC. Finally, by identifying RT reactions dependent on zinc finger function, this work provides an in vitro assay system to study a role for the NC zinc fingers in the infectious process.
ACKNOWLEDGMENTS
Jianhui Guo and Tiyun Wu contributed equally to the work presented in this study.
We thank Stuart Le Grice for his generous gift of an RNase H-minus RT. We are also indebted to Alan Rein for a thoughtful reading of the manuscript.
This work was supported in part by the National Institutes of Health Intramural AIDS Targeted Antiviral Program and in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract NO1-CO-56000.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix J-L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arad U. Modified Hirt procedure for rapid purification of extrachromosomal DNA from mammalian cells. BioTechniques. 1998;24:760–762. doi: 10.2144/98245bm14. [DOI] [PubMed] [Google Scholar]
- 4.Arts E J, Wainberg M A. Human immunodeficiency virus type 1 reverse transcriptase and early events in reverse transcription. Adv Virus Res. 1996;46:97–163. doi: 10.1016/s0065-3527(08)60071-8. [DOI] [PubMed] [Google Scholar]
- 5.Auxilien S, Keith G, Le Grice S F J, Darlix J-L. Role of post-transcriptional modifications of primer tRNALys,3 in the fidelity and efficacy of plus-strand DNA transfer during HIV-1 reverse transcription. J Biol Chem. 1999;274:4412–4420. doi: 10.1074/jbc.274.7.4412. [DOI] [PubMed] [Google Scholar]
- 6.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 7.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 8.Brulé F, Bec G, Keith G, Le Grice S F J, Roques B P, Ehresmann B, Ehresmann C, Marquet R. In vitro evidence for the interaction of tRNA3Lys with U3 during the first strand transfer of HIV-1 reverse transcription. Nucleic Acids Res. 2000;28:634–640. doi: 10.1093/nar/28.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cameron C E, Ghosh M, Le Grice S F J, Benkovic S J. Mutations in HIV reverse transcriptase which alter RNase H activity and decrease strand transfer efficiency are suppressed by HIV nucleocapsid protein. Proc Natl Acad Sci USA. 1997;94:6700–6705. doi: 10.1073/pnas.94.13.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carteau S, Gorelick R J, Bushman F D. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Champoux J J. Roles of ribonuclease H in reverse transcription. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 103–117. [Google Scholar]
- 12.Chertova E N, Kane B P, McGrath C, Johnson D G, Sowder II R C, Arthur L O, Henderson L E. Probing the topography of HIV-1 nucleocapsid protein with the alkylating agent N-ethylmaleimide. Biochemistry. 1998;37:17890–17897. doi: 10.1021/bi980907y. [DOI] [PubMed] [Google Scholar]
- 13.Cimarelli A, Sandin S, Höglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 15.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darlix J-L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 17.Darlix J-L, Vincent A, Gabus C, De Rocquigny H, Roques B. Trans-activation of the 5′ to 3′ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV 1 RNA. C R Acad Sci Paris Life Sci. 1993;316:763–771. [PubMed] [Google Scholar]
- 18.Davis W R, Gabbara S, Hupe D, Peliska J A. Actinomycin D inhibition of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase and nucleocapsid protein. Biochemistry. 1998;37:14213–14221. doi: 10.1021/bi9814890. [DOI] [PubMed] [Google Scholar]
- 19.De Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 20.DeStefano J J. Human immunodeficiency virus nucleocapsid protein stimulates strand transfer from internal regions of heteropolymeric RNA templates. Arch Virol. 1995;140:1775–1789. doi: 10.1007/BF01384341. [DOI] [PubMed] [Google Scholar]
- 21.Dib-Hajj F, Khan R, Giedroc D P. Retroviral nucleocapsid proteins possess potent nucleic acid strand renaturation activity. Protein Sci. 1993;2:231–243. doi: 10.1002/pro.5560020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorfman T, Luban J, Goff S P, Haseltine W, Göttlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Druillennec S, Caneparo A, de Rocquigny H, Roques B P. Evidence of interactions between the nucleocapsid protein NCp7 and the reverse transcriptase of HIV-1. J Biol Chem. 1999;274:11283–11288. doi: 10.1074/jbc.274.16.11283. [DOI] [PubMed] [Google Scholar]
- 24.Drummond J E, Mounts P, Gorelick R J, Casas-Finet J R, Bosche W J, Henderson L E, Waters D J, Arthur L O. Wild-type and mutant HIV type 1 nucleocapsid proteins increase the proportion of long cDNA transcripts by viral reverse transcriptase. AIDS Res Hum Retrovir. 1997;13:533–543. doi: 10.1089/aid.1997.13.533. [DOI] [PubMed] [Google Scholar]
- 25.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher R J, Rein A, Fivash M, Urbaneja M A, Casas-Finet J R, Medaglia M, Henderson L E. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilboa E, Mitra S W, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 28.Gorelick R J, Benveniste R E, Gagliardi T D, Wiltrout T A, Busch L K, Bosche W J, Coren L V, Lifson J D, Bradley P J, Henderson L E, Arthur L O. Nucleocapsid protein zinc-finger mutants of simian immunodeficiency virus strain Mne produce virions that are replication defective in vitro and in vivo. Virology. 1999;253:259–270. doi: 10.1006/viro.1998.9513. [DOI] [PubMed] [Google Scholar]
- 29.Gorelick R J, Chabot D J, Ott D E, Gagliardi T D, Rein A, Henderson L E, Arthur L O. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J Virol. 1996;70:2593–2597. doi: 10.1128/jvi.70.4.2593-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorelick R J, Fu W, Gagliardi T D, Bosche W J, Rein A, Henderson L E, Arthur L O. Characterization of the block in replication of nucleocapsid protein zinc finger mutants from Moloney murine leukemia virus. J Virol. 1999;73:8185–8195. doi: 10.1128/jvi.73.10.8185-8195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorelick R J, Gagliardi T D, Bosche W J, Wiltrout T A, Coren L V, Chabot D J, Lifson J D, Henderson L E, Arthur L O. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology. 1999;256:92–104. doi: 10.1006/viro.1999.9629. [DOI] [PubMed] [Google Scholar]
- 33.Gorelick R J, Henderson L E, Hanser J P, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc Natl Acad Sci USA. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorelick R J, Nigida S M, Jr, Bess J W, Jr, Arthur L O, Henderson L E, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green L M, Berg J M. Retroviral nucleocapsid protein-metal ion interactions: folding and sequence variants. Proc Natl Acad Sci USA. 1990;87:6403–6407. doi: 10.1073/pnas.87.16.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo J, Wu T, Bess J, Henderson L E, Levin J G. Actinomycin D inhibits human immunodeficiency virus type 1 minus-strand transfer in in vitro and endogenous reverse transcriptase assays. J Virol. 1998;72:6716–6724. doi: 10.1128/jvi.72.8.6716-6724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo J, Wu W, Yuan Z Y, Post K, Crouch R J, Levin J G. Defects in primer-template binding, processive DNA synthesis, and RNase H activity associated with chimeric reverse transcriptases having the murine leukemia virus polymerase domain joined to Escherichia coli RNase H. Biochemistry. 1995;34:5018–5029. doi: 10.1021/bi00015a013. [DOI] [PubMed] [Google Scholar]
- 39.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 39a.Hsu M, Rong L, de Rocquigny H, Roques B P, Wainberg M A. The effect of mutations in the HIV-1 nucleocapsid protein on strand transfer in cell-free reverse transcription reactions. Nucleic Acids Res. 2000;28:1724–1729. doi: 10.1093/nar/28.8.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji X, Klarmann G J, Preston B D. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry. 1996;35:132–143. doi: 10.1021/bi951707e. [DOI] [PubMed] [Google Scholar]
- 41.Johnson P E, Turner R B, Wu Z R, Hairston L, Guo J, Levin J G, Summers M F. A mechanism for plus-strand transfer enhancement by HIV-1 nucleocapsid protein during reverse transcription. Biochemistry. 2000;39:9084–9091. doi: 10.1021/bi000841i. [DOI] [PubMed] [Google Scholar]
- 41a.Kelleher C D, Champoux J J. Characterization of RNA strand displacement synthesis by Moloney murine leukemia virus reverse transcriptase. J Biol Chem. 1998;273:9976–9986. doi: 10.1074/jbc.273.16.9976. [DOI] [PubMed] [Google Scholar]
- 42.Kim J K, Palaniappan C, Wu W, Fay P J, Bambara R A. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J Biol Chem. 1997;272:16769–16777. doi: 10.1074/jbc.272.27.16769. [DOI] [PubMed] [Google Scholar]
- 43.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix J-L. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lapadat-Tapolsky M, Gabus C, Rau M, Darlix J-L. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J Mol Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- 45.Lapadat-Tapolsky M, Pernelle C, Borie C, Darlix J-L. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995;23:2434–2441. doi: 10.1093/nar/23.13.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee B M, De Guzman R N, Turner B G, Tjandra N, Summers M F. Dynamical behavior of the HIV-1 nucleocapsid protein. J Mol Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- 47.Lener D, Tanchou V, Roques B P, Le Grice S F J, Darlix J-L. Involvement of HIV-1 nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J Biol Chem. 1998;273:33781–33786. doi: 10.1074/jbc.273.50.33781. [DOI] [PubMed] [Google Scholar]
- 48.Li X, Quan Y, Arts E J, Li Z, Preston B D, De Rocquigny H, Roques B P, Darlix J-L, Kleiman L, Parniak M A, Wainberg M A. Human immunodeficiency virus type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA3Lys in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. J Virol. 1996;70:4996–5004. doi: 10.1128/jvi.70.8.4996-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mély Y, De Rocquigny H, Morellet N, Roques B P, Gérard D. Zinc binding to the HIV-1 nucleocapsid protein: A thermodynamic investigation by fluorescence spectroscopy. Biochemistry. 1996;35:5175–5182. doi: 10.1021/bi952587d. [DOI] [PubMed] [Google Scholar]
- 50.Mély Y, Jullian N, Morellet N, De Rocquigny H, Dong C Z, Piémont E, Roques B P, Gérard D. Spatial proximity of the HIV-1 nucleocapsid protein zinc fingers investigated by time-resolved fluorescence and fluorescence resonance energy transfer. Biochemistry. 1994;33:12085–12091. doi: 10.1021/bi00206a011. [DOI] [PubMed] [Google Scholar]
- 51.Méric C, Goff S P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Méric C, Gouilloud E, Spahr P-F. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): deletions of Cys-His boxes. J Virol. 1988;62:3328–3333. doi: 10.1128/jvi.62.9.3328-3333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Méric C, Spahr P F. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J Virol. 1986;60:450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morellet N, Déméné H, Teilleux V, Huynh-Dinh T, de Rocquigny H, Fournié-Zaluski M-C, Roques B P. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC) J Mol Biol. 1998;283:419–434. doi: 10.1006/jmbi.1998.2098. [DOI] [PubMed] [Google Scholar]
- 55.Morellet N, de Rocquigny H, Mély Y, Jullian N, Déméné H, Ottmann M, Gérard D, Darlix J L, Fournie-Zaluski M C, Roques B P. Conformational behaviour of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H NMR. J Mol Biol. 1994;235:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- 56.Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix J-L, Roques B P. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Omichinski J G, Clore G M, Sakaguchi K, Appella E, Gronenborn A M. Structural characterization of a 39-residue synthetic peptide containing the two zinc binding domains from the HIV-1 p7 nucleocapsid protein by CD and NMR spectroscopy. FEBS Lett. 1991;292:25–30. doi: 10.1016/0014-5793(91)80825-n. [DOI] [PubMed] [Google Scholar]
- 58.Peliska J A, Balasubramanian S, Giedroc D P, Benkovic S J. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 59.Rein A, Henderson L E, Levin J G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 60.Remy E, de Rocquigny H, Petitjean P, Muriaux D, Theilleux V, Paoletti J, Roques B P. The annealing of tRNA3Lys to human immunodeficiency virus type 1 primer binding site is critically dependent on the NCp7 zinc fingers structure. J Biol Chem. 1998;273:4819–4822. doi: 10.1074/jbc.273.9.4819. [DOI] [PubMed] [Google Scholar]
- 61.Rodríguez-Rodríguez L, Tsuchihashi Z, Fuentes G M, Bambara R A, Fay P J. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J Biol Chem. 1995;270:15005–15011. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- 62.Rong L, Liang C, Hsu M, Kleiman L, Petitjean P, De Rocquigny H, Roques B P, Wainberg M A. Roles of the human immunodeficiency virus type 1 nucleocapsid protein in annealing and initiation versus elongation in reverse transcription of viral negative-strand strong-stop DNA. J Virol. 1998;72:9353–9358. doi: 10.1128/jvi.72.11.9353-9358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schatz O, Cromme F V, Grüninger-Leitch F, Le Grice S F J. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989;257:311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- 64.Schmalzbauer E, Strack B, Dannull J, Guehmann S, Moelling K. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J Virol. 1996;70:771–777. doi: 10.1128/jvi.70.2.771-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith C M, Smith J S, Roth M J. RNase H requirements for the second strand transfer reaction of human immunodeficiency virus type 1 reverse transcription. J Virol. 1999;73:6573–6581. doi: 10.1128/jvi.73.8.6573-6581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.South T L, Blake P R, Hare D R, Summers M F. C-terminal retroviral-type zinc finger domain from the HIV-1 nucleocapsid protein is structurally similar to the N-terminal zinc finger domain. Biochemistry. 1991;30:6342–6349. doi: 10.1021/bi00239a036. [DOI] [PubMed] [Google Scholar]
- 67.South T L, Blake P R, Sowder III R C, Arthur L O, Henderson L E, Summers M F. The nucleocapsid protein isolated from HIV-1 particles binds zinc and forms retroviral-type zinc fingers. Biochemistry. 1990;29:7786–7789. doi: 10.1021/bi00486a002. [DOI] [PubMed] [Google Scholar]
- 68.South T L, Summers M F. Zinc- and sequence-dependent binding to nucleic acids by the N-terminal zinc finger of the HIV-1 nucleocapsid protein: NMR structure of the complex with the psi-site analog, dACGCC. Protein Sci. 1993;2:3–19. doi: 10.1002/pro.5560020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Summers M F, Henderson L E, Chance M R, Bess J W, Jr, South T L, Blake P R, Sagi I, Perez-Alvarado G, Sowder III R C, Hare D R, Arthur L O. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Surovoy A, Dannull J, Moelling K, Jung G. Conformational and nucleic acid binding studies on the synthetic nucleocapsid protein of HIV-1. J Mol Biol. 1993;229:94–104. doi: 10.1006/jmbi.1993.1011. [DOI] [PubMed] [Google Scholar]
- 71.Tanchou V, Decimo D, Péchoux C, Lener D, Rogemond V, Berthoux L, Ottmann M, Darlix J-L. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J Virol. 1998;72:4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Telesnitsky A, Goff S P. Strong-stop strand transfer during reverse transcription. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 49–83. [Google Scholar]
- 73.Tsuchihashi Z, Brown P O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Urbaneja M A, Kane B P, Johnson D G, Gorelick R J, Henderson L E, Casas-Finet J R. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J Mol Biol. 1999;287:59–75. doi: 10.1006/jmbi.1998.2521. [DOI] [PubMed] [Google Scholar]
- 75.Vuilleumier C, Bombarda E, Morellet N, Gérard D, Roques B P, Mély Y. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry. 1999;38:16816–16825. doi: 10.1021/bi991145p. [DOI] [PubMed] [Google Scholar]
- 76.Wu J Q, Ozarowski A, Maki A H, Urbaneja M A, Henderson L E, Casas-Finet J R. Binding of the nucleocapsid protein of type 1 human immunodeficiency virus to nucleic acids studied using phosphorescence and optically detected magnetic resonance. Biochemistry. 1997;36:12506–12518. doi: 10.1021/bi970676f. [DOI] [PubMed] [Google Scholar]
- 77.Wu T, Guo J, Bess J, Henderson L E, Levin J G. Molecular requirements for human immunodeficiency type 1 plus-strand transfer: analysis in reconstituted and endogenous reverse transcription systems. J Virol. 1999;73:4794–4805. doi: 10.1128/jvi.73.6.4794-4805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu W, Henderson L E, Copeland T D, Gorelick R J, Bosche W J, Rein A, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.You J C, McHenry C S. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J Biol Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]
- 80.Yu Q, Darlix J-L. The zinc finger of nucleocapsid protein of Friend murine leukemia virus is critical for proviral DNA synthesis in vivo. J Virol. 1996;70:5791–5798. doi: 10.1128/jvi.70.9.5791-5798.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]