Abstract
Human monoclonal antibodies are safe, preventive, and therapeutic tools that can be rapidly developed to help restore the massive health and economic disruption caused by the coronavirus disease 2019 (COVID-19) pandemic. By single-cell sorting 4,277 SARS-CoV-2 spike protein-specific memory B cells from 14 COVID-19 survivors, 453 neutralizing antibodies were identified. The most potent neutralizing antibodies recognized the spike protein receptor-binding domain, followed in potency by antibodies that recognize the S1 domain, the spike protein trimer, and the S2 subunit. Only 1.4% of them neutralized the authentic virus with a potency of 1–10 ng/mL. The most potent monoclonal antibody, engineered to reduce the risk of antibody-dependent enhancement and prolong half-life, neutralized the authentic wild-type virus and emerging variants containing D614G, E484K, and N501Y substitutions. Prophylactic and therapeutic efficacy in the hamster model was observed at 0.25 and 4 mg/kg respectively in absence of Fc functions.
Keywords: SARS-CoV-2, COVID-19, monoclonal antibodies, prophylaxis, therapy, emerging variants, Fc functions absence
Graphical abstract
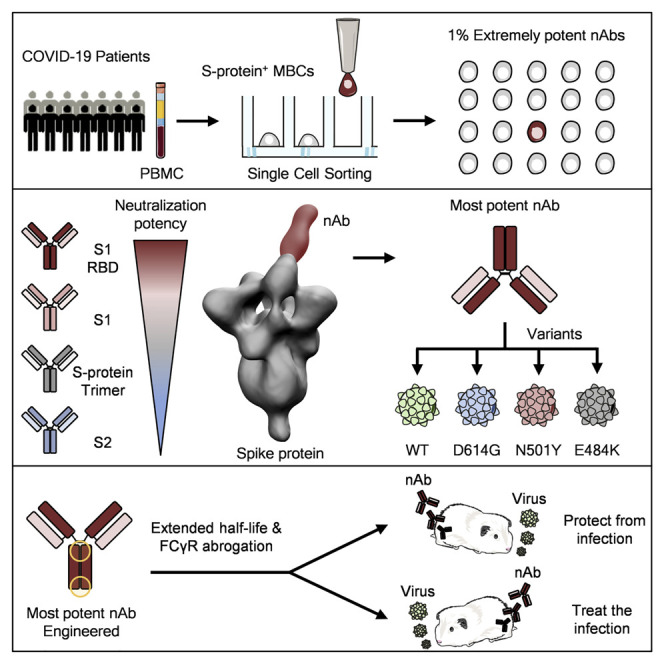
Extremely potent neutralizing human monoclonal antibodies, though rare, are isolated from COVID-19 convalescent patients and suitable for prophylactic and therapeutic interventions of wild-type SARS-CoV-2 as well as emerging variants.
Introduction
The impact of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, with more than 100 million cases, over 2 million deaths, an estimated cost of 16 trillion US dollars to the USA economy (Cutler and Summers, 2020), and 45 million people filing unemployment in the United States alone, is unprecedented (Aratani, 2020).
Vaccines and drugs against SARS-CoV-2 have recently received emergency use authorization (EUA) by the Food and Drug Administration (FDA) for prevention and treatment of coronavirus disease 2019 (COVID-19) (FDA, 2021, 2020).
In spite of this, it is predictable that waves of infection will continue to spread globally, and it is likely to be followed by additional waves over the next few years. This is supported by the emergence of new SARS-CoV-2 variants in the United Kingdom, South Africa, Brazil, and Japan (CDC, 2021).
It is therefore imperative to quickly develop, in parallel to vaccines, therapeutic tools against SARS-CoV-2 and its variants. Among the many therapeutic options available, human monoclonal antibodies (mAbs) can be developed in the shortest time frame. In fact, the extensive clinical experience with the safety of more than 50 commercially available mAbs approved to treat cancer, inflammatory, and autoimmune disorders provides high confidence of their safety (Wellcome and IAVI, 2020). These advantages, combined with the urgency of the SARS-CoV-2 pandemic, support and justify an accelerated regulatory pathway. In addition, the long industrial experience in developing and manufacturing mAbs decreases risks usually associated with technical development of investigational products. Finally, the incredible technical progress in this field allows shortening of conventional timelines and enables a path from discovery to proof-of-concept trials within 5–6 months (Kelley, 2020). A key example is the Ebola case, where mAbs were developed faster than vaccines or other drugs (Kupferschmidt, 2019), becoming the first therapeutic intervention recommended by the World Health Organization (WHO) and approved by the FDA (Mullard, 2020).
During the first months of this pandemic, many groups have been active in isolating and characterizing human monoclonal antibodies from COVID-19 convalescent patients or from humanized mice, and some of them have been progressing quickly to clinical trials for the prevention and cure of SARS-CoV-2 infection (Shi et al., 2020; Hansen et al., 2020; Hsieh et al., 2020; Pinto et al., 2020; Zost et al., 2020a, 2020b; Rogers et al., 2020, Alsoussi et al., 2020). Few of them are already in phase III clinical trials and reported promising preliminary results. Two of them received the EUA from the FDA (Lilly, 2020; Regeneron, 2020).
All these antibodies neutralize SARS-CoV-2 infection by binding to the spike glycoprotein (S protein), a trimeric class I viral fusion protein that mediates virus entry into host cells by engaging with the human angiotensin-converting enzyme 2 (hACE2) and cellular heparan sulfate as receptors (Clausen et al., 2020). The S protein exists in a metastable pre-fusion conformation and in a stable post-fusion form (Wang et al., 2020; Walls et al., 2020; Schäfer et al., 2020). Each S protein monomer is composed of two distinct regions, the S1 and S2 subunits. The S1 subunit contains the receptor-binding domain (RBD), which is responsible for the interaction with hACE2 and heparan sulfate on host cell membranes triggering the destabilization of the prefusion state of the S protein and consequent transition into the post-fusion conformation. This event results in the entry of the virus particle into the host cell and the onset of infection (Wrapp et al., 2020; Walls et al., 2020; Tay et al., 2020; Zou et al., 2020).
As for other mAbs in the field of infectious diseases (Hooft van Huijsduijnen et al., 2020; Sparrow et al., 2017), the dose of mAbs so far used in clinical trials against SARS-CoV-2 is high, ranging from 500 to 8,000 mgs (NCT04411628; NCT04427501; NCT04441918; NCT04425629; NCT04426695; NCT04452318). The high dose poses two important limits to the application of mAbs in the infectious diseases field. First, the high dosage has cost-associated implications, and it only allows for intravenous delivery, making this therapeutic intervention extremely costly and therefore available almost exclusively in high-income countries. Indeed, the high price of this intervention has been a barrier to the global access of mAbs and their use to other fields such as infectious diseases. A solution would be the development of extremely potent mAbs that can be used at lower dosages leading to cost reductions and that can be delivered via intramuscular or subcutaneous injections. A first example is the respiratory syncytial virus (RSV) case, where a potent mAb has recently shown its therapeutic effect in premature infants after only one intramuscular injection of 50 mg (Griffin et al., 2020).
The second limit of mAbs in the field of infectious diseases is the risk of antibody-dependent enhancement (ADE) of disease, which is usually mediated by the binding of the fragment crystallizable (Fc) region portion of the antibody to Fc gamma receptors (FcγRs) expressed by immune cells (Lee et al., 2020). ADE has been clearly demonstrated in the case of SARS-CoV, RSV, and dengue viruses, and the theoretical risk has been raised in the case of SARS-CoV-2 (Lee et al., 2020; Katzelnick et al., 2017; Arvin et al., 2020).
In this work, we pushed the limits of mAb application to fight infectious diseases by selecting extremely potent antibodies with the aim of using them at low dosage to make them affordable and conveniently delivered by intramuscular injection. In addition, we mitigated the risk of ADE by engineering their Fc region. Despite complete lack of Fc-receptor-binding and Fc-mediated cellular activities, engineered mAbs were able to prevent and treat SARS-CoV-2 infection in golden Syrian hamster at a concentration of 0.25 and 4 mg/kg respectively. These antibodies have the potential to globally extend the access and affordability of this important medical tool.
Results
Isolation and characterization of S protein-specific antibodies from SARS-CoV-2 convalescent patients
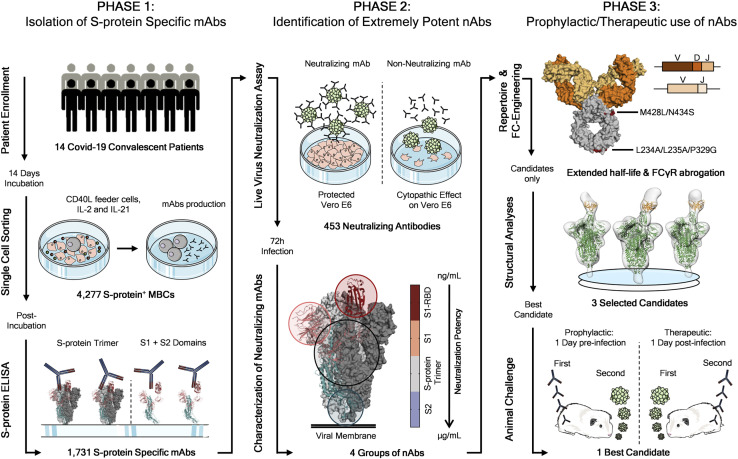
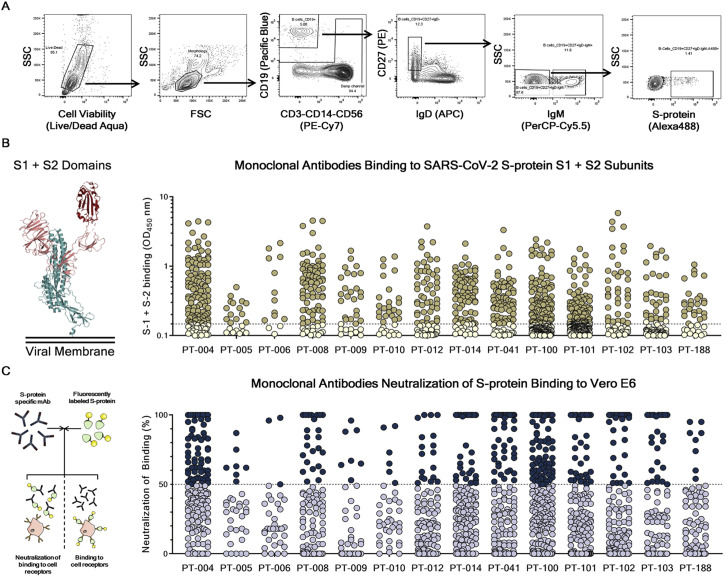
To retrieve mAbs specific for SARS-CoV-2 S protein, peripheral blood mononuclear cells (PBMCs) from fourteen COVID-19 convalescent patients enrolled in this study were collected and stained with fluorescently labeled S protein trimer to identify antigen-specific memory B cells (MBCs). Figure 1 summarizes the overall experimental strategy. The gating strategy described in Figure S1 A was used to single-cell sort, into 384-well plates, IgG+ and IgA+ MBCs binding to the SARS-CoV-2 S protein trimer in its prefusion conformation. The sorting strategy aimed to specifically identify class-switched MBCs (CD19+CD27+IgD−IgM−) to identify only memory B lymphocytes that underwent maturation processes. A total of 4,277 S protein-binding MBCs were successfully retrieved with frequencies ranging from 0.17% to 1.41% (Table S1). Following the sorting procedure, S protein+ MBCs were incubated over a layer of 3T3-CD40L feeder cells in the presence of IL-2 and IL-21 stimuli for 2 weeks to allow natural production of immunoglobulins (Huang et al., 2013). Subsequently, MBC supernatants containing IgG or IgA were tested for their ability to bind either the SARS-CoV-2 S protein trimer in its prefusion conformation or the S protein S1 + S2 subunits (Figure 2 A; Figure S2 B) by enzyme linked immunosorbent assay (ELISA). A panel of 1,731 mAbs specific for the SARS-CoV-2 S protein were identified showing a broad range of signal intensities (Figure 2A; Table S1).
Figure 1.
Workflow and timeline for SARS-CoV-2 neutralizing antibodies identification
The overall scheme shows three different phases for the identification of SARS-CoV-2 neutralizing antibodies (nAbs). Phase 1 consisted in the enrolment of COVID-19 patients (n = 14) from which PBMCs were isolated. Memory B cells were single-cell sorted (n = 4,277), and after 2 weeks of incubation, antibodies were screened for their binding specificity against the S protein trimer and S1/S2 domains. Once S protein-specific monoclonal antibodies (mAbs) were identified (n = 1,731) phase 2 started. All specific mAbs were tested in vitro to evaluate their neutralization activity against the authentic SARS-CoV-2 virus, and 453 nAbs were identified. nAbs showing different binding profiles on the S protein surface were selected for further functional characterization and to identify different neutralizing regions on the antigen. Phase 3 starts with the characterization of the heavy and light chain sequences of selected mAbs (n = 14) and the engineering of the Fc portion of three most promising candidates. The latter were also selected for structural analyses that allowed the identification of the neutralizing epitopes on the S protein. Finally, the most potent antibody was tested for its prophylactic and therapeutic effect in a golden Syrian hamster model of SARS-CoV-2 infection.
Figure S1.
Gating strategy for single-cell sorting and monoclonal antibodies screening for S protein S1 + S2 subunits binding and neutralization of binding (NoB) activity, related to Figure 2
(A) Starting from top left to the right panel, the gating strategy shows: Live/Dead; Morphology; CD19+ B cells; CD19+CD27+IgD-; CD19+CD27+IgD-IgM-; CD19+CD27+IgD-IgM-S-protein+ B cells.
(B) The graph shows supernatants tested for binding to the SARS-CoV-2 S-protein S1 + S2 subunits. Threshold of positivity has been set as two times the value of the blank (dotted line). Darker dots represent mAbs which bind to the S1 + S2 while light yellow dots represent mAbs which do not bind. (B) The graph shows supernatants tested by NoB assay. Threshold of positivity has been set as 50% of binding neutralization (dotted line). Dark blue dots represent mAbs able to neutralize the binding between SARS-CoV-2 and receptors on Vero E6 cells, while light blue dots represent non-neutralizing mAbs.
Figure 2.
Identification of SARS-CoV-2 S protein-specific nAbs
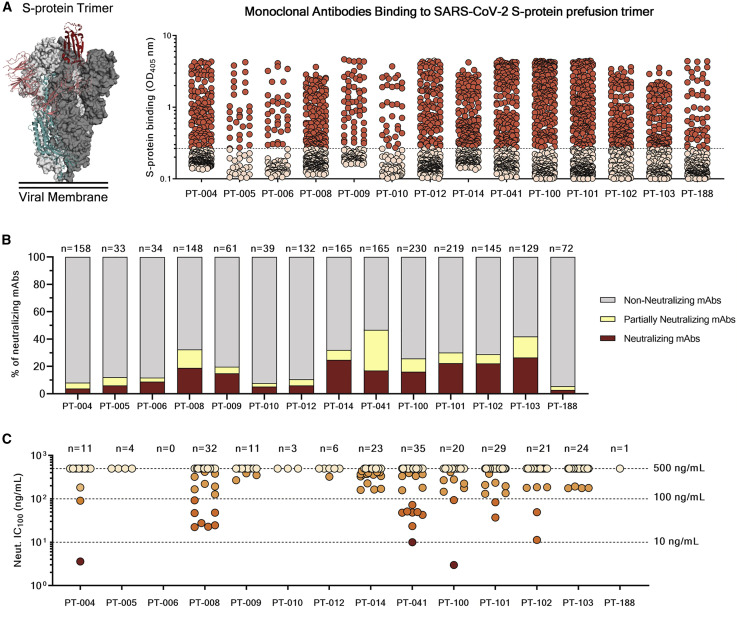
(A) The graph shows supernatants tested for binding to the SARS-CoV-2 S-protein stabilized in its prefusion conformation. Threshold of positivity has been set as two times the value of the blank (dotted line). Red dots represent mAbs that bind to the S protein, while pink dots represent mAbs that do not bind.
(B) The bar graph shows the percentage of non-neutralizing (gray), partially neutralizing (pale yellow), and neutralizing antibodies (dark red) identified per each donor. The total number (n) of antibodies tested per individual is shown on top of each bar.
(C) The graph shows the neutralization potency of each nAb tested once expressed as recombinant full-length IgG1. Dashed lines show different ranges of neutralization potency (500, 100, and 10 ng/mL). Dots were colored based on their neutralization potency and were classified as weakly neutralizing (>500 ng/mL; pale orange), medium neutralizing (100–500 ng/mL; orange), highly neutralizing (10–100 ng/mL; dark orange), and extremely neutralizing (1–10 ng/mL; dark red). The total number (n) of antibodies tested per individual is shown on top of each graph. A COVID-19 convalescent plasma and an unrelated plasma were used as positive and negative control, respectively, in all the assays.
Figure S2.
Characterization and distribution of SARS-CoV-2 S protein-specific nAbs, related to Figure 2
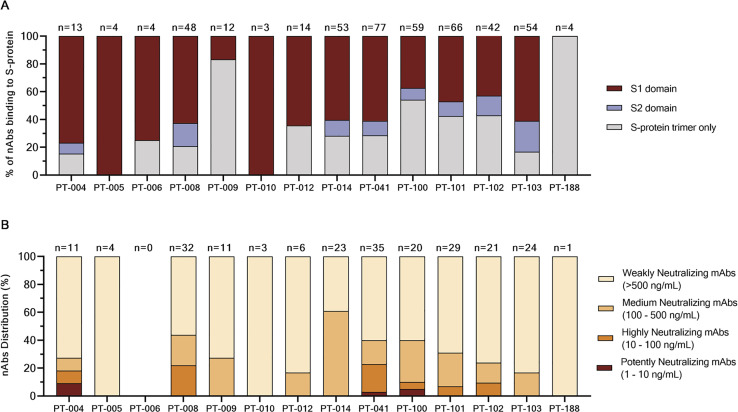
(A) The bar graph shows the distribution of nAbs binding to different S-protein domains. In dark red, light blue and gray are shown antibodies binding to the S1-domain, S2-domain and S-protein trimer respectively. The total number (n) of antibodies tested per individual is shown on top of each bar.
(B) The bar graph shows the distribution of nAbs with different neutralization potencies. nAbs were classified as weakly neutralizing (> 500 ng/mL; pale orange), medium neutralizing (100 – 500 ng/mL; orange), highly neutralizing (10 – 100 ng/mL; dark orange) and extremely neutralizing (1 – 10 ng/mL; dark red). The total number (n) of antibodies tested per individual is shown on top of each bar.
Identification of S protein-specific mAbs able to neutralize SARS-CoV-2
The 1,731 supernatants containing S protein-specific mAbs, were screened in vitro for their ability to block the binding of the streptavidin-labeled S protein to Vero E6 cell receptors and for their ability to neutralize authentic SARS-CoV-2 virus by in vitro microneutralization assay. In the neutralization of binding (NoB) assay, 339 of the 1,731 tested (19.6%) S protein-specific mAbs were able to neutralize the antigen/receptor binding, showing a broad array of neutralization potency ranging from 50% to 100% (Figure S2C; Table S1).
As for the authentic virus neutralization assay, supernatants containing naturally produced IgG or IgA were tested for their ability to protect the layer of Vero E6 cells from the cytopathic effect triggered by SARS-CoV-2 infection. To increase the throughput of our approach, supernatants were tested at a single-point dilution, and to increase the sensitivity of our first screening, a viral titer of 25 50% tissue culture infectious dose (TCID50) was used. For this screening, mAbs were classified as neutralizing, partially neutralizing, and non-neutralizing based on their complete, partial, or absent ability to prevent the infection of Vero E6 cells, respectively. Out of 1,731 mAbs tested in this study, a panel of 453 (26.2%) mAbs neutralized the authentic virus and prevented infection of Vero E6 cells (Table S1). The percentage of partially neutralizing antibodies and neutralizing antibodies (nAbs) identified in each donor was extremely variable ranging from 2.6%–29.7% and 2.8%–26.4% respectively (Figure 2B; Table S2). The majority of nAbs were able to specifically recognize the S protein S1 domain (57.5%; n = 244), while 7.3% (n = 53) of nAbs were specific for the S2 domain, and 35.2% (n = 156) did not recognize single domains but only the S protein in its trimeric conformation (Figure S2A; Table S3). From the panel of 453 nAbs, we recovered the heavy chain (HC) and light chain (LC) variable regions of 220 nAbs, which were expressed as full-length immunoglobulin G1 (IgG1) using the transcriptionally active PCR (TAP) approach to characterize their neutralization potency against the live virus at 100 TCID50. The vast majority of nAbs identified (65.9%; n = 145) had a low neutralizing potency and required more than 500 ng/mL to achieve 100% inhibitory concentration (IC100). A smaller fraction of the antibodies had an intermediate neutralizing potency (23.6%; n = 52) requiring between 100 and 500 ng/mL to achieve the IC100, while 9.1% (n = 20) required between 10 and 100 ng/mL. Finally, only 1.4% (n = 3) of the expressed nAbs were classified as extremely potent nAbs, showing an IC100 lower than 10 ng/mL (Figure 2C; Figure S2B; Table S4).
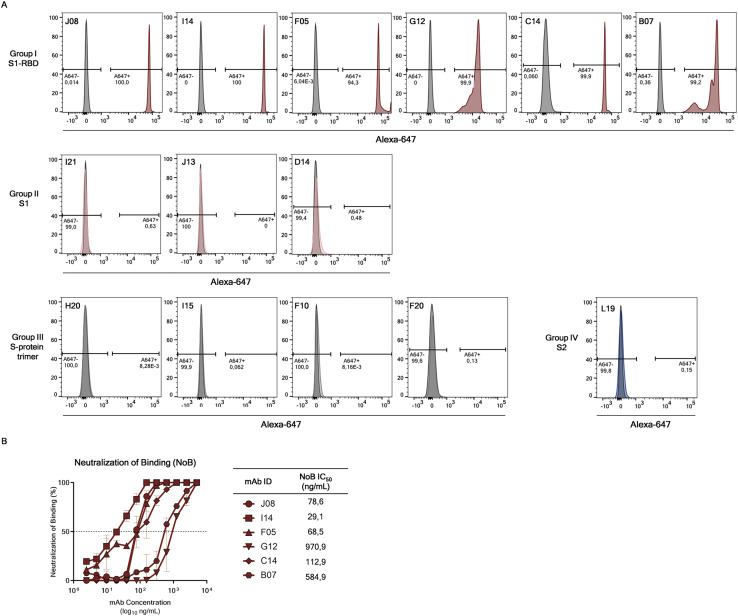
SARS-CoV-2 neutralizing antibodies can be classified into four groups
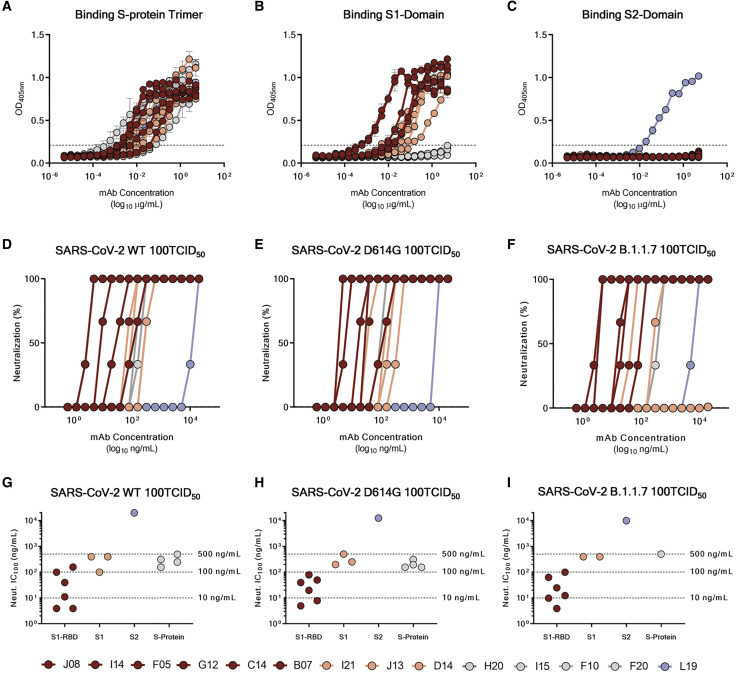
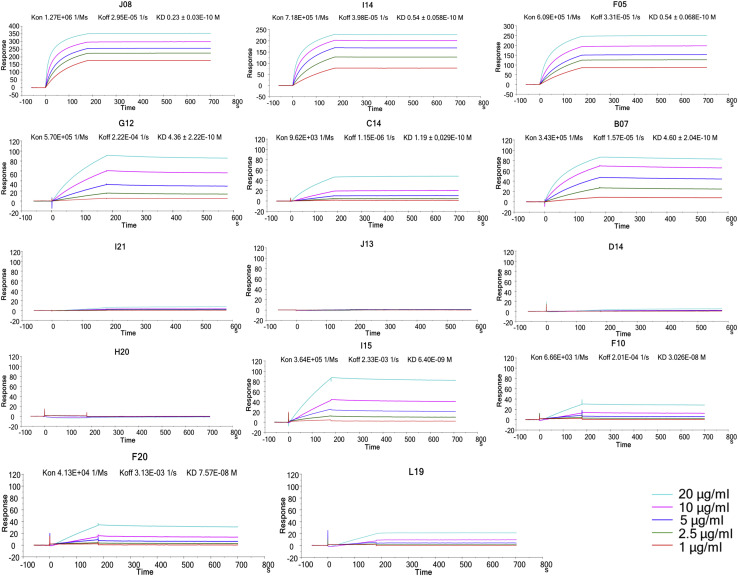
Based on the first round of screening, 14 nAbs were selected for further characterization. All nAbs were able to bind the SARS-CoV-2 S protein in its trimeric conformation (Figure 3 A). The mAbs named J08, I14, F05, G12, C14, B07, I21, J13, and D14 were also able to specifically bind the S1 domain (Figure 3B). The nAbs named H20, I15, F10, and F20 were not able to bind single S1 or S2 domains but only the Sprotein in its trimeric state, while the nAb L19 bound only the S2 subunit (Figures 3B and 3C). Among the group of S1-specific nAbs, only J08, I14, F05, G12, C14, and B07 were able to bind the S1 RBD and to strongly inhibit the interaction between the S protein and Vero E6 receptors, showing a half maximal effective concentration (EC50) at the NoB assay of 78.6, 15.6, and 68.5 ng/mL for J08-MUT, I14-MUT, and F05-MUT, respectively (Figures S3 A and S3B). On the other hand, I21, J13, and D14, despite showing S1 binding specificity, did not show any binding to the RBD and NoB activity (Figure S3A). Based on this description, four different groups of nAbs against SARS-CoV-2 were identified. The first group (Group I) is composed of S1 RBD-specific nAbs (J08, I14, F05, G12, C14, and B07), which showed neutralization potency against the authentic wild type (WT), the D614G variant, and the emerging variant recently isolated in the UK B.1.1.7. S1 RBD-specific nAbs showing a neutralizing potency ranging from 3.9 to 157.5 ng/mL (Figures 3D–3I; Table S5) and picomolar affinity to the S protein with an equilibrium dissociation constant (KD) ranging from 0.2 to 4.6 E−10M (Figure S4 ). In addition to the D614G and the B.1.1.7 variants, the S1 RBD-specific nAb J08 showed also to neutralize SARS-CoV-2 variants containing the E484K mutation (Andreano et al., 2020). The second group (Group II) included S1-specific nAbs that did not bind the RBD (I21, J13, and D14). These antibodies also showed good neutralization potency ranging from 99.2 to 500.0 ng/mL (Figures 3D–3I; Table S5) but inferior to that of S1 RBD-directed nAbs. One antibody from this group was not able to neutralize the B.1.1.7 variant (I21). The third group (Group III) is composed of antibodies able to bind the S-protein only in its whole trimeric conformation (H20, I15, F10, and F20). Antibodies belonging to this group showed lower affinity to the S protein trimer (KD 64.0 E−10M–757.0 E−10M) compared to Group I nAbs and medium neutralization potencies ranging from 155.0 to 492.2 ng/mL against the authentic WT and D614G (Figures 3D–3I; Figure S4; Table S5). On the other hand, only one S protein-specific nAb (D21) showed moderate neutralization activity against the B.1.1.7 with an IC100 of 500.0 ng/mL. Three S protein-specific nAbs (I15, F10, and F20) did not show any functional activity against this latter variant (Figures 3D–3I; Table S5). The fourth and final group (Group IV) is composed of antibodies that exclusively recognized the S2 domain. Different antibodies with similar properties were identified for Group IV, but only the nAb L19 is shown. The Group IV nAb L19 shows the lowest neutralization potency with 19.8 μg/mL for the authentic WT, 12.5 μg/mL against the D614G, and 9.9 μg/mL against the B.1.1.7 variant (Figures 3D–3I; Table S5).
Figure 3.
Functional characterization of potent SARS-CoV-2 S protein-specific nAbs
(A–C) Graphs show binding curves to the S protein in its trimeric conformation, S1 domain, and S2 domain. Mean ± SD of technical triplicates are shown. Dashed lines represent the threshold of positivity.
(D–F) Neutralization curves for selected antibodies were shown as percentage of viral neutralization against the authentic SARS-CoV-2 wild type (D), D614G variant (E), and the emerging variant B.1.1.7 (F). Data are representative of technical triplicates. A neutralizing COVID-19 convalescent plasma and an unrelated plasma were used as positive and negative control, respectively.
(G–I) Neutralization potency of 14 selected antibodies against the authentic SARS-CoV-2 wild type (G), D614G variant (H), and the emerging variant B.1.1.7 (I). Dashed lines show different ranges of neutralization potency (500, 100, and 10 ng/mL). In all graphs, selected antibodies are shown in dark red, pink, gray, and light blue based on their ability to recognize the SARS-CoV-2 S1 RBD, S1 domain, S protein trimer only, and S2 domain, respectively.
Figure S3.
Binding to S protein receptor binding domain (RBD) and NoB activity of S1-RBD antibodies, related to Figure 3
(A) Histograms show the ability of selected antibodies to bind the S-protein RBD. Gray histograms represent the negative control while colored histograms show tested antibodies. Percentage of positive and negative populations are denoted on each graph.
(B) Neutralization of binding (NoB) curves for S1-RBD specific antibodies are shown as percentage of reduction of signal emitted by a fluorescently labled S-protein incubated with Vero E6 cells. Mean ± SD of technical duplicates are shown. Dashed lines represent the threshold of positivity; A neutralizing COVID-19 convalescent plasma and an unrelated plasma were used as positive and negative control, respectively.
Figure S4.
Binding kinetics of SARS-CoV-2 nAbs to the S protein antigen, related to Figure 3
Representative binding curves of selected antibodies to SARS-CoV-2 S-protein trimer. Different curve colors define the spike concentration used in the experiment. Kon, Koff and KD are denoted on each graph.
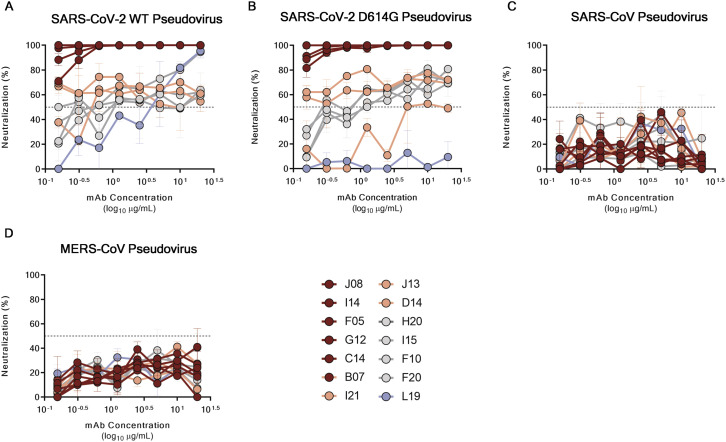
All the antibodies described above were also tested for their ability to cross-neutralize other human coronavirus strains. nAbs were tested against lentiviral pseudotypes expressing the SARS-CoV-2, SARS-CoV-2 D614G, SARS-CoV, and Middle East respiratory syndrome (MERS)-CoV S protein on their viral membrane surface. Neutralization activity was shown against SARS-CoV-2 and D614G pseudotypes, therefore confirming previous data. None of the antibodies reported here were able to cross-neutralize other coronavirus species (Figure S5 ).
Figure S5.
Neutralization activity of selected nAbs against SARS-CoV-2, SARS-CoV, and MERS-CoV pseudotypes, related to Figure 3
(A–D) Graphs show the neutralizing activities of 14 selected nAbs with different SARS-CoV-2 S-protein binding profiles against SARS-CoV-2, SARS-CoV-2 D614G, SARS-CoV and MERS-CoV pseudotypes respectively. Dashed lines represent the threshold of positivity. Mean ± SD of technical duplicates are shown. In all graphs selected antibodies are shown in dark red, pink, gray and light blue based on their ability to recognize the SARS-CoV-2 S1-RBD, S1-domain, S-protein trimer only and S2-domain respectively.
Different pathogen vulnerability regions identified on the S protein
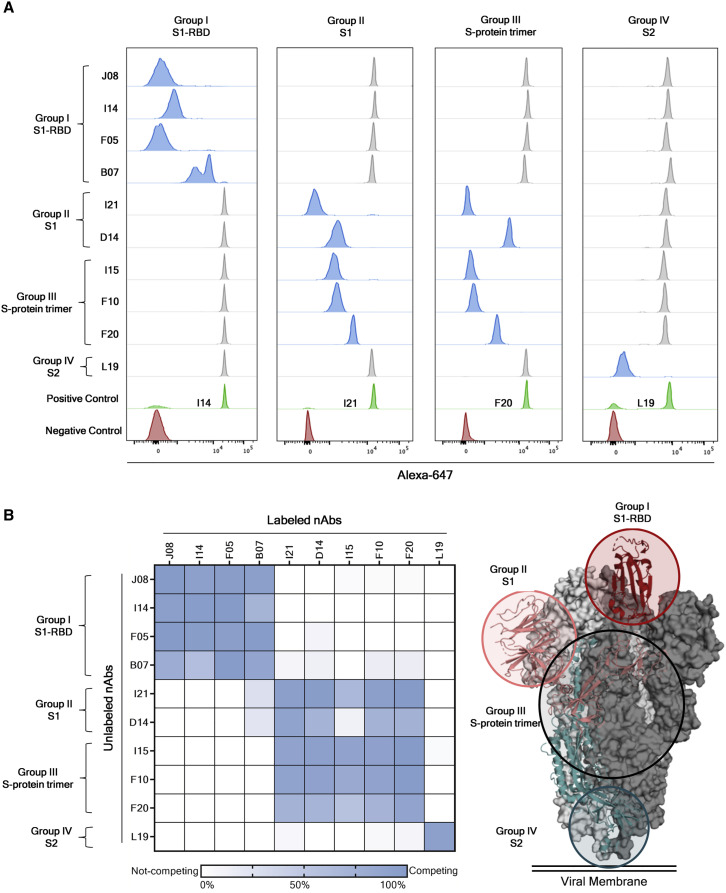
The fourteen selected nAbs were further characterized by a competition assay that allowed speculation on the S protein regions recognized by these antibodies. Briefly, beads were coated with SARS-CoV-2 trimeric S protein and incubated with a primary unlabeled antibody in order to saturate the binding site on the antigen surface. Following the first incubation step, a secondary Alexa-647-labeled antibody was incubated with the antigen/unlabeled-mAb complex. If the secondary labeled antibody did not recognize the same epitope as the primary unlabeled mAb, a fluorescent signal would be detected when tested by flow cytometry. Through this assay, we observed that all Group I nAbs competed among themselves for binding to the S protein RBD, indicating that these antibodies possibly clash against each other and recognize a similar epitope region. All Group II nAbs showed different competition profiles and competed with Group II and Group III nAbs. These results confirmed that Group III antibodies can recognize various regions on the S protein surface as they compete with themselves as well as with antibodies belonging to Group II. Interestingly, nAbs belonging to Group II also competed with the B07 RBD-directed antibody, thereby suggesting that this latter nAb may have a different binding orientation compared to other nAbs included in the Group I. Finally, the Group IV nAb L19 did not compete with any of the other groups identified in this study, suggesting that this class of nAbs recognizes a distant epitope region as compared to Group I, II, and III nAbs (Figures 4A and 4B)
Figure 4.
Identification of four different sites of pathogen vulnerability on the S protein surface
(A) Representative cytometer peaks per each of the four antibody groups are shown. Positive (beads conjugated with only primary labeled antibody) and negative (un-conjugated beads) controls are shown as green and red peaks, respectively. Competing and not-competing nAbs are shown in blue and gray peaks, respectively.
(B) The heatmap shows the competition matrix observed among the 14 nAbs tested. Threshold of competition was set at 50% of fluorescent signal reduction. A speculative representation of the vulnerability sites is shown on the S protein surface.
Genetic characterization of SARS-CoV-2 nAbs
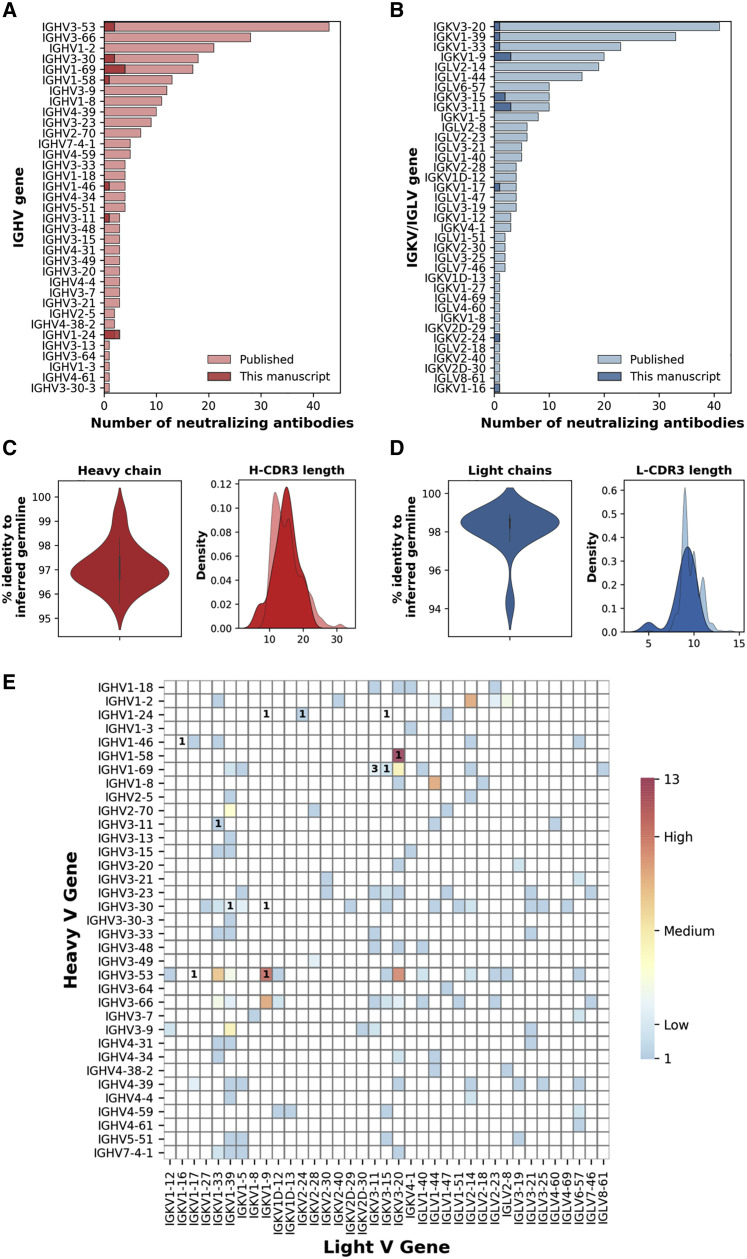
The genes encoding the HCs and LCs of the 14 selected nAbs were sequenced, and their IGHV and IGKV genes were compared with publicly available SARS-CoV-2 neutralizing antibody sequences (Figures 5A and 5B). Four nAbs used one of the most predominant HC V genes for SARS-CoV-2 nAbs (IGHV1-69), while three nAbs used one of the least representative HCV genes (IGHV1-24). Two other nAbs employed the most common germline observed for SARS-CoV-2 nAbs, which is IGHV3-53 (Figure 5A) (Yuan et al., 2020). Interestingly, while IGHV1-69 and IGHV1-24 accommodate IGHJ diversity, nAbs belonging to the IGHV3-53 gene family only showed recombination with the IGHJ6 gene (Table S6). The HC V genes somatic hypermutation level and complementary determining region 3 (H-CDR3) length were also evaluated. Our selected nAbs displayed a low level of somatic mutations when compared to the inferred germlines with sequence identities ranging from 95.6% to 99.3% (Figure 5C left panel; Table S6), confirming what was observed in previous publications (Pinto et al., 2020; Zost et al., 2020b; Rogers et al., 2020; Griffin et al., 2020). The H-CDR3 length spanned from 7 to 21 amino acids (aa) with the majority of the antibodies (n = 6; 42.0%) having a length of 14 to 16 aa that is slightly bigger than previously observed (Figure 5C right panel; Table S6). All of our nAbs used the κ chain, and the majority of them used the common genes IGKV1-9 and IGKV3-11 (n = 6; 42.0%) (Figure 5B; Table S6). The level of IGKV somatic hypermutation was extremely low for LCs showing a percentage of sequence identities ranging from 94.3% to 98.9% (Figure 5D left panel; Table S6). The LC CDR3 (L-CDR3) lengths were ranging from 5 to 10 aa, which is in line with what was previously observed for SARS-CoV-2 nAbs (Figure 5D right panel; Table S6). When paired HC and LC gene analysis was performed, IGHV1-69-derived nAbs were found to rearrange exclusively with IGKV3 gene family, whereas IGHV1-24-derived nAbs accommodate LC diversity (Table S6). Of note, some of our candidates showed unique HC and LC pairing when compared to the public SARS-CoV-2 nAb repertoire. Particularly, five different HC and LC rearrangements not previously described for nAbs against SARS-CoV-2 were identified. These included the IGHV1-24;IGKV1-9, IGHV1-24;IGKV3-15, IGHV1-46;IGKV1-16, IGHV3-30;IGKV1-9, and IGHV3-53;IGKV1-17 (Figure 5E).
Figure 5.
Heavy and light chain analyses of selected nAbs
(A and B) Bar graphs show the heavy and light chains usage for neutralizing antibodies against SARS-CoV-2 in the public repertoire compared to the antibodies identified in this study. Our and public antibodies are shown in dark and light colors, respectively.
(C and D) The heavy and light chain percentage of identity to the inferred germline and amino acidic CDR3 length are shown as violin and distribution plot, respectively.
(E) The heatmap shows the frequency of heavy and light chain pairing for SARS-CoV-2 neutralizing human mAbs already published. The number within the heatmap cells represent the amount of nAbs described in this manuscript showing already published (colored cells) or novel heavy and light chain rearrangements (blank cells).
Fc engineering of candidate nAbs to abrogate Fc receptor binding and extend half-life
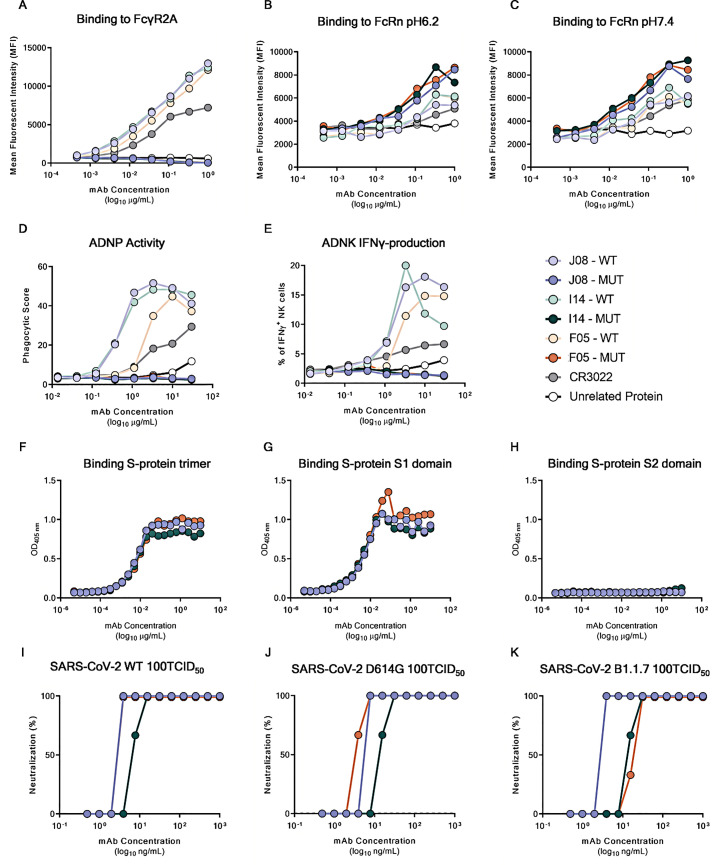
ADE of disease is a potential clinical risk following coronavirus infection (Lee et al., 2020). Therefore, to optimize the suitability for clinical development and reduce the risk of ADE, five different point mutations were introduced in the constant region (Fc) of the three most potent nAbs (J08, I14, and F05), which were renamed J08-MUT, I14-MUT, and F05-MUT. The first two point mutations (M428L and N434S) were introduced to enhance antibody half-life and to increase tissue distribution and persistence (Zalevsky et al., 2010; Gaudinski et al., 2018; Pegu et al., 2017). The remaining three point mutations (L234A, L235A, and P329G) were introduced to reduce antibody dependent functions such as binding to FcγRs and cell-based activities (Schlothauer et al., 2016).
To confirm the lack of FcγR binding as well as the extended half-life, a beads-based Luminex assay was performed. Briefly the beads were coated with SARS-CoV-2 S protein RBD. Antibodies were tested at eight-point dilutions, and the binding was detected with FcγR2A and neonatal Fc receptor (FcRn) at pH6.2 and 7.4. The FcγR2A was selected as it is predominantly expressed on the surface of phagocytic cells (such as monocytes, macrophages, and neutrophils) and is associated with phagocytosis of immune complexes and antibody-opsonized targets (Ackerman et al., 2013). On the other hand, FcRn, which is highly expressed on endothelial cells and circulating monocytes, was selected as it is responsible for the recycling and serum half-life of IgG in the circulation (Mackness et al., 2019). This latter receptor was shown to possess a tighter binding at lower pH (e.g., pH 6.2) compared to a physiological pH (e.g., pH 7.4) (Booth et al., 2018). Results shown in Figure S6 demonstrate that binding to the FcγR2A was completely abrogated for the mutated version of candidate nAbs (J08-MUT, I14-MUT, and F05-MUT) compared to their respective WT versions (J08, I14, and F05) and controls (CR3022 and unrelated protein) (Figure S6A). Furthermore, Fc-engineered antibodies showed increased binding activity to the FcRn at both pH 6.2 and 7.4 compared to their WT counterpart (Figures S6B and S6C). Finally, to evaluate the lack of Fc-mediated cellular activities by our three candidate nAbs, the antibody-dependent neutrophil phagocytosis (ADNP) and antibody-dependent natural killer (ADNK) cell activations were evaluated (Butler et al., 2019; Ackerman et al., 2016; Karsten et al., 2019; Boudreau et al., 2020). For the ADNP assay, primary human neutrophils were used to detect antibody binding to SARS-CoV-2 S protein RBD-coated beads, while ADNK activity was evaluated by using primary human NK cells and detecting the release of the proinflammatory cytokine interferon gamma (IFN-γ). Complete abrogation of both ADNP and ADNK was observed for all three Fc-engineered candidate nAbs compared to their WT versions and control antibody (CR3022), thus confirming the lack of Fc-mediated cellular activities (Figures S6D and S6E).
Figure S6.
Characterization of Fc-engineered candidate nAbs, related to Figure 7
(A) the graph shows binding curves of J08, I14 and F05 MUT and WT to the FcγR2A.
(B and C) graphs show binding curves of J08, I14 and F05 MUT and WT to the FcRn at pH 6.2 (B) and 7.4 (C).
(D and E) Graphs show the ADNP and ADNK induced by J08, I14 and F05 MUT and WT versions; all the experiments were run as technical duplicates. In every experiment a control antibody (CR3022) and an unrelated protein were used as positive and negative control respectively.
(F–H) Graphs show binding curves to the S-protein in its trimeric conformation, S1-domain and S2-domain. Mean of technical triplicates are shown.
(I–K) Neutralization curves against the authentic SARS-CoV-2 wild type, the D614G variant and the B.1.1.7 emerging variant for J08-MUT, I14-MUT and F05-MUT shown in blue, green and red respectively. Data are representative of technical triplicates.
Potency and autoreactivity evaluation of Fc-engineered candidates
The three engineered antibodies were tested to confirm their binding specificity and neutralization potency against both the WT, the widespread SARS-CoV-2 D614G mutant and the emerging variant B.1.1.7 (Korber et al., 2020, CDC, 2021) to evaluate their cross-neutralization ability. The three engineered nAbs maintained their S1 domain binding specificity and extremely high neutralization potency with J08-MUT and F05-MUT being able to neutralize both the WT and the D614G variant with an IC100 lower than 10 ng/mL (both at 3.9 ng/mL for the WT and the D614G strains) (Figure S6F – K; Table S5). The antibody J08-MUT also showed extreme neutralization potency against emerging variants as it was able to neutralize the B.1.1.7 with an identical IC100 compared to the WT virus (Figure S6K; Table S5) and has also showed to neutralize variants that include the E484K mutation (Andreano et al., 2020).
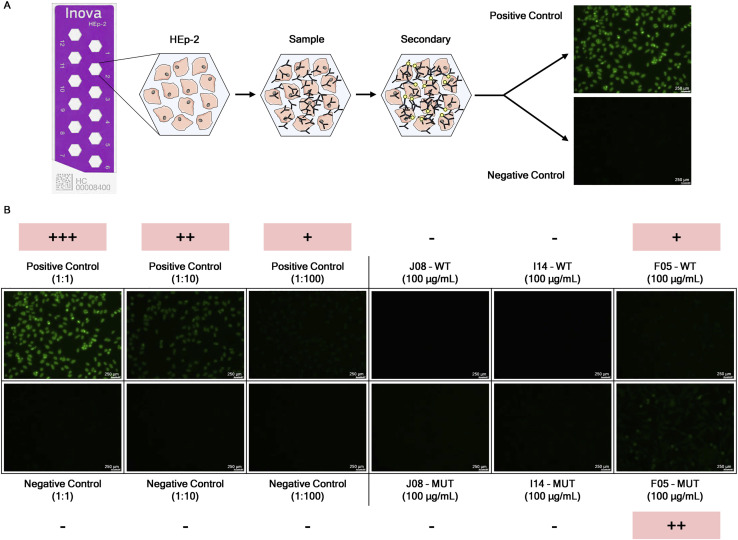
Since it has been reported that SARS-CoV-2 elicited antibodies that can cross-react with human tissues, cytokines, phospholipids, and phospholipid-binding proteins (Zuo et al., 2020; Bastard et al., 2020; Kreer et al., 2020), the three candidate mAbs in both their WT and MUT versions were tested through an indirect immunofluorescent assay against human epithelial type 2 (HEp-2) cells, which expose clinically relevant proteins to detect autoantibody activities (Figure S7 A). As reported in Figure S7B, the positive control presents a different range of detectable signals based on the initial dilution steps (from bright green at 1:1 to very dim green at 1:100). Among all samples tested, only F05 showed moderate level of autoreactivity to human cells, while no signal could be measured for the other antibodies (Figure S7B).
Figure S7.
Autoreactivity assessment of selected SARS-CoV-2 candidate nAbs, related to Figure 7
(A) Schematic representation of the indirect immunofluorescent assay for the screening of autoreactive nAb.
(B) Single figures show the fluorescent signal detected per each sample tested in this assay. Positive and negative controls were used at three different dilutions (1:1, 1:10 and 1:100). Three candidate nAbs were incubated on HEp-2 cells at a concentration of 100 μg/mL. Representative pictures of the scoring system are shown. Autoreactive samples are highlighted in pink. 250 nm scale bar is shown.
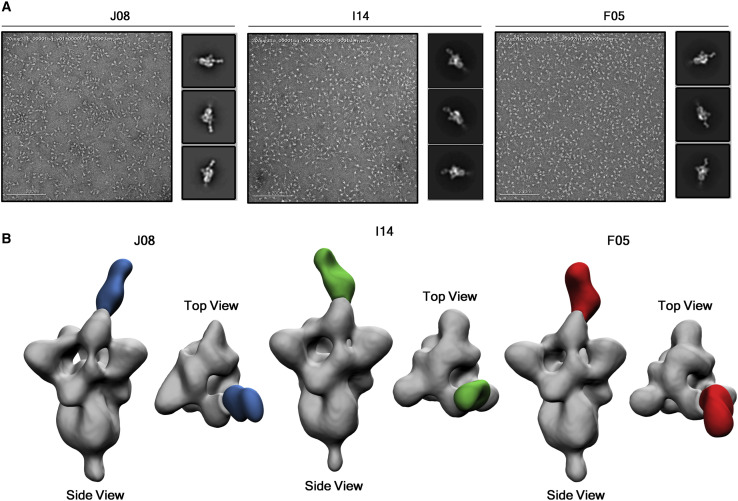
Structural analyses of candidate nAbs
Single-particle negative-stain electron microscopy (nsEM) was used to visualize a stabilized SARS-2-CoV-6P-Mut7 S protein in complex with three separate Fabs: J08, I14, and F05. This recombinant, soluble S protein primarily exhibits 3 RBD’s “down” but can switch to RBD “up” conformation with antibody bound. Inspection of the 2D class averages revealed a mixed stoichiometry of unbound S protein, one Fab bound, and two Fab bound classes, which allowed for 3D refinements of each (Figure 6 A). The three different Fabs bind to the RBD in the “up” conformation, although at different angles and rotations, likely due to the flexibility of the RBD. Model docking of PDB 7BYR (one RBD “up” bound to antibody) shows that the fabs overlap with the receptor-binding motif (RBM) and therefore are positioned to sterically block receptor hACE2 engagement (Figure 6B). To determine the epitope, HC and LC sequences of Fabs J08, I14, and F05 were used to create synthetic models for docking into the nsEM maps. Based on the docking, we predicted that a loop containing residues 477 to 489 (STPCNGVEGFNCY) appeared to be involved in the binding specifically with residue F486 extending into a cavity that is in the middle of the HC and LC of each antibody.
Figure 6.
EM epitope mapping of RBD mAbs
(A) Negative stain for J08, I14, and F05 in complex with the S protein. 200 nm scale bar is shown.
(B) Figures show the binding of J08 (blue), I14 (green), and F05 (red) to the SARS-CoV-2 S protein RBD.
J08-MUT prevents SARS-CoV-2 infection in the golden Syrian hamster
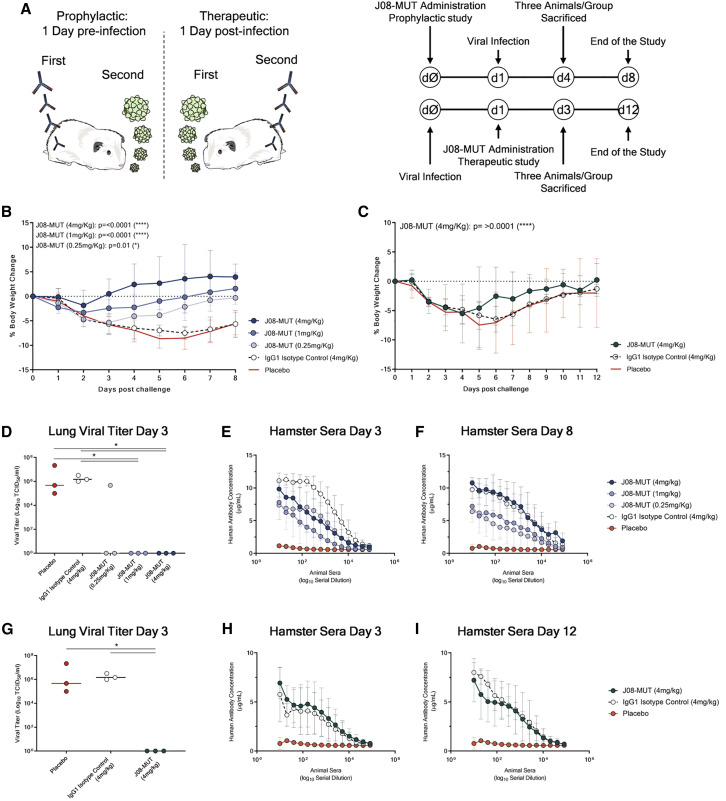
The golden Syrian hamster model has been widely used to assess monoclonal antibody prophylactic and therapeutic activities against SARS-CoV-2 infection. This model has shown to manifest severe forms of SARS-CoV-2 infection mimicking more closely the clinical disease observed in humans (Baum et al., 2020; Imai et al., 2020; Rogers et al., 2020; Sia et al., 2020). We designed a prophylactic study in golden Syrian hamster to evaluate the efficacy of J08-MUT in preventing SARS-CoV-2 infection. For this study, 30 hamsters were divided into five arms (six animals each), which received, J08-MUT at 4, 1, and 0.25 mg/kg via intraperitoneal injection. Placebo and IgG1 isotype control groups were included in the study, which received a saline solution and an anti-influenza antibody at the concentration of 4 mg/kg, respectively. The J08-MUT at 4 mg/kg group and the 1 and 0.25 mg/kg groups were tested in two independent experiments. The IgG1 isotype control group was tested in parallel with the J08-MUT 4 mg/kg group, whereas the placebo is an average of the two experiments. Animals were challenged with 100 μL of SARS-CoV-2 solution (5 × 105 plaque-forming units [PFU]) via intranasal distillation 24 h post-administration of the antibody. Three hamsters per group were sacrificed at 3 days post infection, while the remaining animals were culled at day 8 (Figure 7 A). Body weight change was evaluated daily and considered as a proxy for disease severity. Animals in the control group and those that received the IgG1 isotype antibody lost more than 5% of their original body weight from day 1 to day 6 and then stabilized. These data are in line with previously published data of SARS-CoV-2 infection in a golden Syrian hamster model (Kreye et al., 2020; Liu et al., 2020). In marked contrast, in the prophylactic study, all animals that received J08-MUT were significantly protected from weight loss. Protection was present at all J08-MUT concentrations and was dose dependent (Figure 7B). When J08-MUT was administered at 4 mg/kg, we observed protection from SARS-CoV-2 infection and only a minimal weight loss (average −1.8% of body weight) was noticed 1 day post viral challenge. A higher body weight loss was observed 1 day post infection in hamsters that received J08-MUT at 1 mg/kg (from −1.8% to −3.3%) and 0.25 mg/kg (from −1.8% to −4.7%). In the J08-MUT 4 mg/kg group, all animals quickly recovered and reached their initial weight by day 3. From day 4 on all hamsters gained weight increasing up to 5% from their initial body weight. Hamsters that received the 1 and 0.25 mg/kg dosages completely recovered their initial body weight at day 6 and 8, respectively. Hamsters in the control groups did not recover their initial body weight and at day 8, still showed around 5% of weight loss (Figure 7B). The prophylactic activity of J08-MUT was also reflected in the complete absence of viral titer in the lung tissue at 3 days post infection in all hamsters that received J08-MUT at 4 and 1 mg/kg and also in two out of three hamsters that received J08-MUT at 0.25 mg/kg. On the other hand, hamsters that received the IgG1 isotype control or in the placebo group showed a significantly higher viral titer (Figure 7D).
Figure 7.
Prophylactic and therapeutic efficacy of J08-MUT in the golden Syrian hamster model of SARS-CoV-2 infection
(A) Schematic representation and timelines of prophylactic and therapeutic studies performed in golden Syrian hamster.
(B and C) The figure shows the prophylactic impact of J08-MUT at three different concentrations (4, 1, and 0.25 mg/kg) (B) on body weight loss change (C). The figure shows the therapeutic impact of J08-MUT at 4 mg/kg on body weight loss change. Mean ± SD are denoted in the graphs.
(D–F) The figures show the lung viral titer at day 3 (D) and the detection of human antibodies in hamster sera at day 3 (E) and day 8 (F) in the prophylactic study. Mean ± SD of technical triplicates are shown.
(G–I) The figures show the lung viral titer at day 3 (G) and the detection of human antibodies in hamster sera at day 3 (H) and day 12 (I) in the therapeutic study. Mean ± SD of technical triplicates are shown. Statistical differences were calculated with two-way analysis of variance (ANOVA) for body weight change and with a nonparametric Mann–Whitney t test for the lung viral titer. Significances are shown as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Finally, we performed an ELISA assay to detect the presence of human IgG in hamster sera. All samples that received J08-MUT or the IgG1 isotype control showed detectable human IgGs in the sera in a dose-dependent fashion, while no human IgGs were detected in the placebo group (Figures 7E and 7F). Human IgGs were detected at 3 and up to 7 days post infection (Figures 7E and 7F).
J08-MUT therapy of SARS-CoV-2 infection in the golden Syrian hamster
For the therapeutic study, three groups of six animals each were used to evaluate the ability of J08-MUT to treat SARS-CoV-2 infection in the golden Syrian hamster model. One group received J08-MUT via intraperitoneal injection at 4 mg/kg, and the other two groups received placebo and 4mg/kg IgG1 isotype control, respectively. The experiment was performed in parallel with the initial prophylactic study where J08-MUT was administered at 4 mg/kg and the two control groups. Animals were challenged with 100 μL of SARS-CoV-2 solution (5 × 105 PFU) via intranasal distillation 24 h prior to the administration of the antibody. Three hamsters per group were sacrificed at 3 days post infection while the remaining animals were culled at day 12 (Figure 7A). Despite J08-MUT and control groups showed a similar trend in weight loss in the first 4 days post infection, the treatment group showed a significantly quicker weight recovery (Figure 7C). At day 12, only hamsters that received J08-MUT recovered the initial body weight (Figure 7C). When we analyzed the viral titer in lung tissues, we observed complete absence of the virus at day 3 in all the hamsters treated with J08-MUT at 4 mg/kg, while animals that received the IgG1 isotype control or in the placebo group showed a significantly higher viral titer (Figure 7G). To evaluate the presence of human antibodies in hamster sera, we performed an ELISA assay. All samples that received J08-MUT or the IgG1 isotype control showed detectable human IgGs in the sera in a dose-dependent fashion, while no human IgGs were detected in the placebo group (Figures 7H and 7I). Human IgGs were detected at 3 and up to 11 days post infection (Figures 7H and 7I).
Discussion
This work describes a systematic screening of memory B cells from SARS-CoV-2 convalescent patients to identify extremely potent mAbs against SARS-CoV-2 and their engineering to extend half-life and eliminate the potential risk of ADE. The best antibody neutralized the authentic WT virus and emerging variants at pico molar concentration in vitro and showed prophylactic and therapeutic efficacy in a SARS-CoV-2 hamsters model of infection when used at 0.25 and 4 mg/kg, respectively. The antibody described is a promising candidate for the development of a broadly affordable tool for prevention and therapy of COVID-19.
In the search for potent antibodies, we found that approximately 10% of the total B cells against the S protein isolated produce neutralizing antibodies, and these can be divided into four different groups recognizing the S1 RBD, S1 domain, S2 domain, and the S protein trimer. Most potently neutralizing antibodies are extremely rare and recognize the RBD, followed in potency by antibodies recognizing the S1 domain, the trimeric structure and the S2 subunit. From these data we can conclude that in COVID-19 convalescent patients, most of the observed neutralization titers are likely mediated by antibodies with medium-high neutralizing potency. Indeed, the extremely potent antibodies and the antibodies against the S2 subunit are unlikely to contribute to the overall neutralizing titers because they are respectively too rare and too poor neutralizers to be able to make a difference. We and others found that the antibody repertoire of convalescent patients is mostly germline-like. This may be a consequence of the loss of Bcl-6-expressing follicular helper T cells and the loss of germinal centers in COVID-19 patients, which may limit and constrain the B cell affinity maturation (Kaneko et al., 2020). It will be therefore important to perform similar studies following vaccination as it is likely that the repertoire of neutralizing antibodies induced by vaccination may be different from the one described here.
Out of the 453 neutralizing antibodies that were tested and characterized, one antibody (J08) showed extremely high neutralization potency against both the WT SARS-CoV-2 virus isolated in Wuhan and emerging variants containing the D614G, E484K, and N501Y variants. During the last few months, several groups reported the identification, 3D structure and passive protection in animal models of neutralizing antibodies against SARS-CoV-2. Most of these studies, with few exceptions, reported antibodies that require from 20 to several hundred ng/mL to neutralize 50% of the virus in vitro. While these antibodies are potentially good for therapy, they will require a high dosage, which is associated with elevated cost of goods, low production capacity, and delivery by intravenous infusion.
The extremely potent mAb described in our study is likely to allow the use of lower quantities of antibodies to reach prophylactic and therapeutic efficacy and as a consequence, decrease the cost of goods and enable sustainable development and manufacturability. This solution may increase the number of doses produced annually and therefore increase antibodies availability in high-income countries as well as low-and middle-income countries. Therefore, our antibodies have the potential to meet the expectations of the call to action to expand access to mAb-based products, recently published by the Wellcome Trust and supported by the WHO and the Coalition for Epidemic Preparedness Innovations (Wellcome and IAVI, 2020).
A potential issue associated with the use of human mAbs against viral pathogens is the potential selection of escape mutants. This is usually addressed by using a combination of antibodies directed against non-overlapping epitopes. While this is an ultimate clear solution, it increases the complexity of development, costs of production, drug availability, and affordability. In our case, we believe that selection of escape mutants upon treatment with a single mAb may be quite difficult as the SARS-CoV-2 RNA-dependent polymerase possesses a proofreading machinery (Romano et al., 2020), and the epitope recognized by the antibodies herein described overlaps with the region necessary to bind the hACE2 receptor. In this regard, it took more than 70 days of continuous co-culture of the virus in presence of the antibodies before we were able to detect the first emergence of escape mutants of the WT SARS-CoV-2 (data not shown).
Finally, a peculiar part of our approach consisted in depleting possible antibody Fc-mediated functions of the antibodies to avoid the risk of ADE. While there is no evidence of ADE in SARS-CoV-2, and most vaccines and mAbs tested so far seem to be safe, it is too early to make definitive conclusions. In addition, two recently published reports suggested that we need to continue to monitor the potential risk of ADE. The first report showed that severe SARS-CoV-2 patients are characterized by an increased proinflammatory signature mediated by the Fcγ receptors triggered by afucosylated IgG1 antibodies (Chakraborty et al., 2020). The second report described that one antibody was associated with worse clinical outcomes when administered to hospitalized patients requiring high-flow oxygen or mechanical ventilation (Lilly, 2020). Therefore, we believe it is important to develop and test antibodies where Fc-mediated functions have been eliminated in the clinical practice. Since the Fc portion contributes significantly to the in vivo potency of the antibodies (Schäfer et al., 2020), removing Fc functions may be a problem for mAbs with low neutralization potency because they may no longer be effective when tested in clinical settings, as already described in other contexts (DiLillo et al., 2014). The extremely high potency shown by our antibodies allowed us to remove Fc functions while maintaining in vivo potency at minimal dosage.
Limitations of the study
While we believe that our antibodies are extremely potent when compared to most of those described in literature, we acknowledge that in most cases, direct comparison was not performed, and we rely on published data.
The second limitation of the study is that in vitro neutralization and in vivo protection in the SARS-CoV-2 hamster model of infection cannot be fully predictive of the behavior of the same antibody in humans, and therefore the real benefit of described antibodies can only be assessed in clinical studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies and fluorophores | ||
| CD19 V421 | BD Biosciences | Cat# 562440; RRID:AB_11153299 |
| IgM PerCP-Cy5.5 | BD Biosciences | Cat# 561285; RRID:AB_10611998 |
| CD27 PE | BD Biosciences | Cat# 340425; RRID:AB_400032 |
| IgD-A700 | BD Biosciences | Cat# 561302; RRID:AB_10646035 |
| CD3 PE-Cy7 | BioLegend | Cat# 300420; RRID:AB_439781 |
| CD14 PE-Cy7 | BioLegend | Cat# 301814; RRID:AB_389353 |
| Streptavidin-PE | Thermo Fisher | Cat#12-4317-87 |
| Goat Anti-Human IgA-UNLB | Southern Biotech | Cat# 2050-01; RRID:AB_2795701 |
| Goat Anti-Human IgA-Alkaline Phosphatase | Southern Biotech | Cat# 2050-04; RRID:AB_2795704 |
| Goat Anti-Human IgG-UNLB | Southern Biotech | Cat# 2040-01; RRID:AB_2795640 |
| Bacterial and virus strains | ||
| SARS-CoV-2 wild type | EVAg | GenBank: MT066156.1 |
| SARS-CoV-2 D614G | EVAg | GenBank: MT527178.1 |
| SARS-CoV-2 B.1.1.7 | INMI | GISAID accession number: EPI_ISL_736997 |
| Biological samples | ||
| PBMCs and IgGs of donor PT-004 | This paper | N/A |
| PBMCs and IgGs of donor PT-005 | This paper | N/A |
| PBMCs and IgGs of donor PT-006 | This paper | N/A |
| PBMCs and IgGs of donor PT-008 | This paper | N/A |
| PBMCs and IgGs of donor PT-009 | This paper | N/A |
| PBMCs and IgGs of donor PT-010 | This paper | N/A |
| PBMCs and IgGs of donor PT-012 | This paper | N/A |
| PBMCs and IgGs of donor PT-014 | This paper | N/A |
| PBMCs and IgGs of donor PT-041 | This paper | N/A |
| PBMCs and IgGs of donor PT-100 | This paper | N/A |
| PBMCs and IgGs of donor PT-101 | This paper | N/A |
| PBMCs and IgGs of donor PT-102 | This paper | N/A |
| PBMCs and IgGs of donor PT-103 | This paper | N/A |
| PBMCs and IgGs of donor PT-188 | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Fetal Bovine Serum (FBS) Hyclone | Sigma-Aldrich | Cat#D2650 |
| DMSO | Sigma-Aldrich | Cat#D2650 |
| RNaseOUT Recombinant Ribonuclease Inhibitor | Thermo Fisher | Cat#10777-019 |
| SuperScript IV Reverse Transcriptase | Thermo Fisher | Cat#18091200 |
| DEPC-Treated water | Thermo Fisher | Cat#AM9916 |
| dNTP Set (100 mM) | Thermo Fisher | Cat#10297018 |
| MgCl2 Magnesium Chloride 25mM | Thermo Fisher | Cat#AB0359 |
| Kapa Long Range Polymerase | Sigma-Aldrich | Cat#KK3005 |
| NEBuilder® HiFi DNA Assembly Master Mix | New England BioLabs | Cat#E2621X |
| Q5® High-Fidelity DNA Polymerases | New England BioLabs | Cat#M0491L |
| Expi293™ Expression Medium | Thermo Fisher | Cat#A1435101 |
| ExpiFectamine™ 293 Transfection Kit | Thermo Fisher | Cat#A14524 |
| Ultra Pure Bovine serum albumin (BSA) | Thermo Fisher | Cat#AM2618 |
| DMEM high Glucose | Thermo Fisher | Cat#11965092 |
| Ficoll-Paque™ PREMIUM | Sigma-Aldrich | Cat#GE17-5442-03 |
| MycoZap Plus-PR | Lonza | Cat#VZA2022 |
| IMDM with GlutaMAX | Thermo Fisher | Cat# 31980048 |
| Benzonase Nuclease | Sigma-Aldrich | Cat#70664-3 |
| IL-2 Recombinant Human Protein | Thermo Fisher | Cat#PHC0023 |
| IL-21 Recombinant Human Protein | Thermo Fisher | Cat#PHC0211 |
| Strep-Tactin DY488 | IBA lifesciences | Cat#2-1562-050 |
| Slide-A-Lyzer™ Dialysis Cassettes | Thermo Fisher | Cat#66003 |
| HiTrap Protein G HP column | Cytiva | Cat#17040503 |
| HisTrap FF Crude column | Cytiva | Cat#17528601 |
| SARS Coronavirus Spike Glycoprotein (S1) | The Native Antigen Company | Cat#REC31809 |
| SARS Coronavirus Spike Glycoprotein (S2) | The Native Antigen Company | Cat#REC31807 |
| Tween-20 | VWR | Cat#A4974.0250 |
| SARS Coronavirus Spike Glycoprotein (S1) | The Native Antigen Company | Cat#REC31806-500 |
| SARS Coronavirus Spike Glycoprotein (S2) | The Native Antigen Company | Cat#REC31807-500 |
| Alkaline Phosphatase Yellow (pNPP) Liquid Substrate System | Sigma-Aldrich | Cat#P7998 |
| Goat Anti-Human IgG-UNLB | SouthernBiotech | Cat#2040-01 |
| Critical commercial assays | ||
| NOVA Lite Hep-2 ANA Kit | Inova Diagnostics / Werfen | Cat#066708100 |
| ELISA Starter Accessory Kit | Bethyl Laboratories | Cat#E101 |
| APEX Alexa Fluor 647 Antibody Labeling Kit | Thermo Fisher | Cat#A10475 |
| Pierce BCA Protein Assay Kit | Thermo Fisher | Cat#23227 |
| Deposited data | ||
| Cloned and tested SARS-CoV-2-neutralizing antibodies | This paper | Patent Application |
| Experimental models: cell lines | ||
| VERO E6 cell line | ATCC | Cat#CRL-1586 |
| Expi293F™ cells | Thermo Fisher | Cat#A14527 |
| 3T3-msCD40L Cells | NIH AIDS Reagent Program | Cat#12535 |
| Oligonucleotides | ||
| Single cell PCR Primer | This paper | N/A |
| Random Hexamer Primer | Thermo Fisher | Cat#SO142 |
| TAP forward primer (TTAGGCACCCCAGGCTTTAC) | This paper | N/A |
| TAP forward primer (AGATGGTTCTTTCCGCCTCA) | This paper | N/A |
| Recombinant DNA | ||
| Human antibody expression vectors (IgG1, Igl, Igk) | (Tiller et al., 2008) | N/A |
| Plasmid encoding SARS-CoV-2 S ectodomain (amino acids 1-1208 of SARS-CoV-2 S; GenBank: MN908947) | (Wrapp et al., 2020) | N/A |
| Plasmid encoding SARS-CoV-2 RBD (amino acids 319 - 591 of SARS-CoV-2 S; GenBank: MN908947) | Jason McLellan Lab | N/A |
| pCDNA3.1+-SARS-CoV-2 Spike from Wuhan-Hu-1 isolate (GenBank MN908947.3) codon optimized | This paper | pCDNA-S2 |
| pCAGGS-SARS-CoV-2 Spike from Wuhan-Hu-1 isolate (GenBank MN908947.3) encoding D614G mutation and codon optimized | This paper | pCAGGS-S2 D614G |
| pCAGGS-SARS1 Spike protein codon optimized | (Carnell et al., 2017) | pCAGGS-S1 |
| pCAGGS-MERS Spike protein codon optimized | (Grehan et al., 2015) | pCAGGS-MERS |
| pCSFLW Firefly luciferase encoding plasmid | (Carnell et al., 2015) | pCSFLW |
| p8.91 HIV Gag/Pol-encoding plasmid | (Carnell et al., 2015) | p8.91 |
| Software and algorithms | ||
| Prism 8 | GraphPad | https://www.graphpad.com/ |
| FlowJo 10.5.3 | FlowJo, LLC | https://www.flowjo.com |
| FastQC | Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| MultiQC 1.9 | MultiQC | https://multiqc.info/ |
| Trimmomatic 0.39 | USADELLAB | http://www.usadellab.org/cms/?page=trimmomatic |
| MiXCR | MI Lanoratory | https://mixcr.readthedocs.io/en/master/index.html |
| NumPy | NumPy | https://numpy.org/ |
| Python 3.7.4 | Python Software Foundation | https://www.python.org/ |
| Other | ||
| BD FACS Aria III Cell Sorter | BD Biosciences | https://www.bdbiosciences.com |
| BD FACS Canto II | BD Biosciences | https://www.bdbiosciences.com |
| Leica DMI-microscope | Leica Biosystem | https://www.leica-microsystems.com |
| LUNA-II Automated Cell Counter | Logo Biosystems | https://logosbio.com |
| Qubit Fluorometric Quantification | Thermo Fisher | https://www.thermofisher.com |
| ÄKTA go | Cytiva Lifesciences | https://www.cytivalifesciences.com |
| GloMax Luminometer | Promega | https://ita.promega.com |
| Varioskan LUX multimode microplate reader | Thermo Fisher | https://www.thermofisher.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Rino Rappuoli (rino.r.rappuoli@gsk.com).
Materials availability
Reasonable amounts of antibodies will be made available by the Lead Contact upon request under a Material Transfer Agreement (MTA) for non-commercial usage.
Data and code availability
Nucleotide and amino acidic sequences of all SARS-CoV-2-neutralizing antibodies were deposited in the Italian patent applications n. 102020000015754 filed on June 30th 2020 and 102020000018955 filed on August 3rd 2020. The accession number for the nucleotide sequences of all SARS-CoV-2-neutralizing antibodies reported in this paper is GenBank: MW_598287 - MW_598314.
Experimental model and subject details
Enrollment of SARS-COV-2 convalescent donors and human sample collection
This work results from a collaboration with the National Institute for Infectious Diseases, IRCCS – Lazzaro Spallanzani Rome (IT) and Azienda Ospedaliera Universitaria Senese, Siena (IT) that provided samples from SARS-CoV-2 convalescent donors, of both sexes, who gave their written consent. The study was approved by local ethics committees (Parere 18_2020 in Rome and Parere 17065 in Siena) and conducted according to good clinical practice in accordance with the declaration of Helsinki (European Council 2001, US Code of Federal Regulations, ICH 1997). This study was unblinded and not randomized.
Method details
Single cell sorting of SARS-CoV-2 S-protein+ memory B cells from COVID-19 convalescent donors
Blood samples were screened for SARS-CoV-2 RNA and for antibodies against HIV, HBV and HCV. Peripheral blood mononuclear cells (PBMCs) were isolated from heparin-treated whole blood by density gradient centrifugation (Ficoll-Paque PREMIUM, Sigma-Aldrich). After separation, PBMC were stained with Live/Dead Fixable Aqua (Invitrogen; Thermo Scientific) in 100 μL final volume diluted 1:500 at room temperature RT. After 20 min incubation cells were washed with PBS and unspecific bindings were saturated with 50 μL of 20% normal rabbit serum (Life technologies) in PBS . Following 20 min incubation at 4°C cells were washed with PBS and stained with SARS-CoV-2 S-protein labeled with Strep-Tactin®XT DY-488 (iba-lifesciences cat# 2-1562-050) for 30 min at 4°C. After incubation the following staining mix was used CD19 V421 (BD cat# 562440), IgM PerCP-Cy5.5 (BD cat# 561285), CD27 PE (BD cat# 340425), IgD-A700 (BD cat# 561302), CD3 PE-Cy7 (BioLegend cat# 300420), CD14 PE-Cy7 (BioLegend cat# 301814), CD56 PE-Cy7 (BioLegend cat# 318318) and cells were incubated at 4°C for additional 30 min. Stained MBCs were single cell-sorted with a BD FACS Aria III (BD Biosciences) into 384-well plates containing 3T3-CD40L feeder cells and were incubated with IL-2 and IL-21 for 14 days as previously described (Huang et al., 2013).
Expression and purification of SARS-CoV-2 S-protein prefusion trimer and receptor binding domain
Plasmid encoding SARS-CoV-2 S-2P construct (Wrapp et al., 2020) and S-protein RBD (generously provided by Jason S. McLellan) were transiently transfected at 1 μg/mL culture into Expi293F cells (Thermo Fisher) using ExpiFectamine 293 Reagent. Cells were grown for six days at 37°C with 8% CO2 shaking 125 rpm according to the manufacturer’s protocol (Thermo Fisher); ExpiFectamine 293 Transfection Enhancers 1 and 2 were added 16 to 18 h post-transfection to boost transfection, cell viability, and protein expression. Cell cultures were harvested three and six days after transfection. Cells were separated from the medium by centrifugation (1,100 g for 10 min at 24°C). Collected supernatants were then pooled and clarified by centrifugation (3,000 g for 15 min at 4°C) followed by filtration through a 0.45 μm filter. Chromatography was conducted at room temperature using the ÄKTA go purification system from GE Healthcare Life Sciences. Expressed proteins were purified by using an immobilized metal affinity chromatography (FF Crude) followed by dialysis into final buffer. Specifically, the filtrated culture supernatant was purified with a 5 mL HisTrap FF Crude column (GE Healthcare Life Sciences) previously equilibrated in Buffer A (20 mM NaH2PO4, 500 mM NaCl + 30 mM imidazol pH 7.4).
The flow rate for all steps of the HisTrap FF Crude column was 5 mL/min. The culture supernatant of spike and RBD cell culture was applied to a single 5 mL HisTrap FF Crude column. The column was washed in Buffer A for 4 column volumes (CV) with the all 4 CV collected as the column wash. Recombinant proteins were eluted from the column applying a first step elution of 4 CV of 50% Buffer B (20 mM NaH2PO4, 500 mM NaCl + 500 mM imidazol pH 7.4) and a second step elution of 2 CV of 100% Buffer B. Elution steps were collected in 1 fractions of 1 mL each. Eluted fractions were analyzed by SDS-PAGE and appropriate fractions containing recombinant proteins were pooled. Final pools were dialyzed against phosphate buffered saline (PBS) pH 7.4 using Slide-A-Lyzer G2 Dialysis Cassette 3.5K (Thermo Scientific) overnight at 4°C. The dialysis buffer used was at least 200 times the volume of the sample.
The final protein concentration was determined by measuring the A520 using the Pierce BCA protein assay kit (Thermo Scientific). Final protein was dispensed in aliquots of 0.5 mL each and stored at −80°C.
ELISA assay with S1 and S2 subunits of SARS-CoV-2 S-protein
The presence of S1- and S2-binding antibodies in culture supernatants of monoclonal S-protein-specific memory B cells was assessed by means of an ELISA implemented with the use of a commercial kit (ELISA Starter Accessory Kit, Catalogue No. E101; Bethyl Laboratories). Briefly, 384-well flat-bottom microtiter plates (384 well plates, Microplate Clear, Greiner Bio-one) were coated with 25 μL/well of antigen (1:1 mix of S1 and S2 subunits, 1 μg/mL each; The Native Antigen Company, Oxford, United Kingdom) diluted in coating buffer (0.05 M carbonate-bicarbonate solution, pH 9.6), and incubated overnight at 4°C. The plates were then washed three times with 100 μL/well washing buffer (50 mM Tris Buffered Saline (TBS) pH 8.0, 0.05% Tween-20) and saturated with 50 μL/well blocking buffer containing Bovine Serum Albumin (BSA) (50 mM TBS pH 8.0, 1% BSA, 0.05% Tween-20) for 1 h (h) at 37°C. After further washing, samples diluted 1:5 in blocking buffer were added to the plate. Blocking buffer was used as a blank. After an incubation of 1 h at 37°C, plates were washed and incubated with 25 μL/well secondary antibody (horseradish peroxidase (HRP)-conjugated goat anti-human IgG-Fc Fragment polyclonal antibody, diluted 1:10,000 in blocking buffer, Catalogue No. A80-104P; (Bethyl Laboratories) for 1 h at 37°C. After three washes, 25 μL/well TMB One Component HRP Microwell Substrate (Bethyl Laboratories) was added and incubated 10–15 min at RT in the dark. Color development was terminated by addition of 25 μL/well 0.2 M H2SO4. Absorbance was measured at 450 nm in a Varioskan Lux microplate reader (Thermo Fisher Scientific). Plasma from COVID-19 convalescent donors (Andreano et al., 2020) and unrelated plasma were used as positive and negative control respectively. The threshold for sample positivity was set at twice the optical density (OD) of the blank.
ELISA assay with SARS-CoV-2 S-protein prefusion trimer and S1 – S2 subunits
ELISA assay was used to detect SARS-CoV-2 S-protein specific mAbs and to screen plasma from SARS-CoV-2 convalescent donors. 384-well plates (384 well plates, microplate clear; Greiner Bio-one) were coated with 3 μg/mL of streptavidin (Thermo Fisher) diluted in coating buffer (0.05 M carbonate-bicarbonate solution, pH 9.6) and incubated at RT overnight. Plates were then coated with SARS-CoV-2 S-protein, S1 or S2 domains at 3 μg/mL and incubated for 1 h at RT. 50 μL/well of saturation buffer (PBS/BSA 1%) was used to saturate unspecific binding and plates were incubated at 37°C for 1 h without CO2. For the first round of screening, supernatants were diluted 1:5 in PBS/BSA 1%/Tween20 0.05% in 25 μL/well final volume and incubated for 1 h at 37°C without CO2. For purified antibodies, and to assess EC50, mAbs were tested at a starting concentration of 5 μg/mL and diluted step 1:2 in PBS/BSA 1%/Tween20 0.05% in 25 μL/well final volume for 1 h at 37°C without CO2. 25 μL/well of alkaline phosphatase-conjugated goat anti-human IgG (Sigma-Aldrich) and IgA (Southern Biotech) were used as secondary antibodies. Wells were washed three times between each step with PBS/BSA 1%/Tween20 0.05%. pNPP (p-nitrophenyl phosphate) (Sigma-Aldrich) was used as soluble substrate to detect SARS-CoV-2 S-protein, S1 or S2 specific mAbs and the final reaction was measured by using the Varioskan Lux Reader (Thermo Fisher Scientific) at a wavelength of 405 nm. Plasma from COVID-19 convalescent donors (Andreano et al., 2020) and unrelated plasma were used as positive and negative control respectively. Samples were considered as positive if OD at 405 nm (OD405) was twice the blank.
SARS-CoV-2 virus and cell infection
African green monkey kidney cell line Vero E6 cells (American Type Culture Collection [ATCC] #CRL-1586) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) - high glucose (Euroclone, Pero, Italy) supplemented with 2 mM L- Glutamine (Lonza, Milano, Italy), penicillin (100 U/mL) - streptomycin (100 μg/mL) mixture (Lonza, Milano, Italy) and 10% Foetal Bovine Serum (FBS) (Euroclone, Pero, Italy). Cells were maintained at 37°C, in a 5% CO2 humidified environment and passaged every 3-4 days.
Wild type SARS-CoV-2 (SARS-CoV-2/INMI1-Isolate/2020/Italy: MT066156), D614G (SARS-CoV-2/human/ITA/INMI4/2020, clade GR, D614G (S): MT527178) and B.1.1.7 (INMI-118 GISAID accession number EPI_ISL_736997) viruses were purchased from the European Virus Archive goes Global (EVAg, Spallanzani Institute, Rome)or received from the Spallanzani Institute, Rome. For virus propagation, sub-confluent Vero E6 cell monolayers were prepared in T175 flasks (Sarstedt) containing supplemented D-MEM high glucose medium. For titration and neutralization tests of SARS-CoV-2, Vero E6 were seeded in 96-well plates (Sarstedt) at a density of 1.5 × 104 cells/well the day before the assay.
Neutralization of Binding (NoB) Assay
To study the binding of the SARS-CoV-2 S-protein to cell-surface receptor(s) we developed an assay to assess recombinant S-protein specific binding to target cells and neutralization thereof. To this aim the stabilized S-protein was coupled to Streptavidin-PE (eBioscience # 12-4317-87, Thermo Fisher) for 1 h at RT and then incubated with Vero E6 cells. Binding was assessed by flow cytometry. The stabilized S-protein bound Vero E6 cells with high affinity (data not shown). To assess the content of neutralizing antibodies in sera or in B cell culture supernatants, two microliters of SARS-CoV-2 Spike-Streptavidin-PE at 5 - 10 μg/mL in PBS-5%FCS were mixed with two microliters of various dilutions of sera or B cell culture supernatants in U bottom 96-well plates. After incubation at 37°C for 1 h, 30 × 103 Vero E6 cells suspended in two microliters of PBS 5% FCS were added and incubated for additional 45 min at 4°C. Non-bound protein and antibodies were removed and cell-bound PE-fluorescence was analyzed with a FACS Canto II flow cytometer (Becton Dickinson). Data were analyzed using the FlowJo data analysis software package (TreeStar, USA). The specific neutralization was calculated as follows: NoB (%) = 1 – (Sample MFI value – background MFI value) / (Negative Control MFI value – background MFI value). Plasma from COVID-19 convalescent donors (Andreano et al., 2020) and unrelated plasma were used as positive and negative control respectively.
Single cell RT-PCR and Ig gene amplification
From the original 384-well sorting plate, 5 μL of cell lysate was used to perform RT-PCR. Total RNA from single cells was reverse transcribed in 25 μL of reaction volume composed by 1 μL of random hexamer primers (50 ng/μL), 1 μL of dNTP-Mix (10 mM), 2 μL 0.1 M DTT, 40 U/μL RNase OUT, MgCl2 (25 mM), 5x FS buffer and Superscript IV reverse transcriptase (Invitrogen). Final volume was reached by adding nuclease-free water (DEPC). Reverse transcription (RT) reaction was performed at 42°C/10’, 25°C/10’, 50°C/60’ and 94°/5¢. Heavy (VH) and light (VL) chain amplicons were obtained via two rounds of PCR. All PCR reactions were performed in a nuclease-free water (DEPC) in a total volume of 25 μL/well. Briefly, 4 μL of cDNA were used for the first round of PCR (PCRI). PCRI-master mix contained 10 μM of VH and 10 μM VL primer-mix, 10mM dNTP mix, 0.125 μL of Kapa Long Range Polymerase (Sigma), 1.5 μL MgCl2 and 5 μL of 5x Kapa Long Range Buffer. PCRI reaction was performed at 95°/3¢, 5 cycles at 95°C/30’’, 57°C/30’’, 72°C/30’’ and 30 cycles at 95°C/30’’, 60°C/30’’, 72°C/30’’ and a final extension of 72°/2’. All nested PCR reactions (PCRII) were performed using 3.5 μL of unpurified PCRI product using the same cycle conditions. PCRII products were then purified by Millipore MultiScreen® PCRμ96 plate according to manufacture instructions. Samples were eluted with 30 μL nuclease-free water (DEPC) into 96-well plates and quantify by.
Cloning of variable region genes and recombinant antibody expression in transcriptionally active PCR
Vector digestions were carried out with the respective restriction enzymes AgeI, SalI and Xho as previously described (Tiller et al., 2008, Wardemann and Busse, 2019). Briefly, 75 ng of IgH, Igλ and Igκ purified PCRII products were ligated by using the Gibson Assembly NEB into 25 ng of respective human Igγ1, Igκ and Igλ expression vectors. The reaction was performed into 5 μL of total volume. Ligation product was 10-fold diluted in nuclease-free water (DEPC) and used as template for transcriptionally active PCR (TAP) reaction which allowed the direct use of linear DNA fragments for in vitro expression. The entire process consists of one PCR amplification step, using primers to attach functional promoter (human CMV) and terminator sequences (SV40) onto the fragment PCRII products. TAP reaction was performed in a total volume of 25 μL using 5 μL of Q5 polymerase (NEB), 5 μL of GC Enhancer (NEB), 5 μL of 5X buffer,10 mM dNTPs, 0.125 μL of forward/reverse primers and 3 μL of ligation product. TAP reaction was performed by using the following cycles: 98°/2’, 35 cycles 98°/10’’, 61°/20’’, 72°/1’ and 72°/5¢ as final extention step. TAP products were purified under the same PCRII conditions, quantified by Qubit Fluorometric Quantitation assay (Invitrogen) and used for transient transfection in Expi293F cell line using manufacturing instructions.
Flask expression and purification of human monoclonal antibodies
Expi293F cells (Thermo Fisher) were transiently transfected with plasmids carrying the antibody heavy chain and the light chains with a 1:2 ratio. Cells were grown for six days at 37°C with 8% CO2 shaking at 125 rpm according to the manufacturer’s protocol (Thermo Fisher); ExpiFectamine 293 transfection enhancers 1 and 2 were added 16 to 18 h post-transfection to boost cell viability and protein expression. Cell cultures were harvested three and six days after transfection. Cells were separated from the medium by centrifugation (1,100 g for 10 min at 24°C). Supernatants collected were then pooled and clarified by centrifugation (3000 g for 15 min, 4°C) followed by filtration through a 0.45 μm filter. Chromatography was conducted at room temperature using the ÄKTA go purification system from GE Healthcare Life Sciences. Affinity chromatography was used to purify expressed monoclonal antibodies using an immobilized protein G column able to bind to Fc region. Specifically, filtrated culture supernatants were purified with a 1 mL HiTrap Protein G HP column (GE Healthcare Life Sciences) previously equilibrated in Buffer A (0.02 M NaH2PO4 pH 7). The flow rate for all steps of the HiTrap Protein G HP column was 1 mL/min. The culture supernatant for every monoclonal antibody cell culture was applied to a single 1 mL HiTrap Protein G HP column. The column was equilibrated in Buffer A for at least 6 column volumes (CV) which was collected as column wash. Each monoclonal antibody was eluted from the column applying a step elution of 6 CV of Buffer B (0.1 M glycine-HCl, pH 2.7). Elution steps were collected in 1 fractions of 1 mL each. Eluted fractions were analyzed by non-reducing SDS-PAGE and fractions showing the presence of IgG were pooled together. Final pools was dialyzed in PBS buffer pH 7.4 using Slide-A-Lyzer G2 Dialysis Cassette 3.5K (Thermo Scientific) overnight at 4°C. The dialysis buffer used was at least 200 times the volume of the sample. For each antibody purified the concentration was determined by measuring the A520 using Pierce BCA Protein Assay Kit (Thermo Scientific). All the purified antibodies were aliquoted and stored at −80°C.
Viral propagation and titration
The SARS-CoV-2 virus was propagated in Vero E6 cells cultured in DMEM high Glucose supplemented with 2% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin. Cells were seeded at a density of 1x106 cells/mL in T175 flasks and incubated at 37°C, 5% CO2 for 18 - 20 h. The sub-confluent cell monolayer was then washed twice with sterile Dulbecco’s PBS (DPBS). Cells were inoculated with 3,5 mL of the virus properly diluted in DMEM 2% FBS at a multiplicity of infection (MOI) of 0.001, and incubated for 1 h at 37°C in a humidified environment with 5% CO2. At the end of the incubation, 50 mL of DMEM 2% FBS were added to the flasks. The infected cultures were incubated at 37°C, 5% CO2 and monitored daily until approximately 80%–90% of the cells exhibited cytopathic effect (CPE). Culture supernatants were then collected, centrifuged at 4°C at 1,600 rpm for 8 min to allow removal of cell debris, aliquoted and stored at −80°C as the harvested viral stock. Viral titers were determined in confluent monolayers of Vero E6 cells seeded in 96-well plates using a 50% tissue culture infectious dose assay (TCID50). Cells were infected with serial 1:10 dilutions (from 10−1 to 10−11) of the virus and incubated at 37°C, in a humidified atmosphere with 5% CO2. Plates were monitored daily for the presence of SARS-CoV-2 induced CPE for 4 days using an inverted optical microscope. The virus titer was estimated according to Spearman-Karber formula (Kundi, 1999) and defined as the reciprocal of the highest viral dilution leading to at least 50% CPE in inoculated wells.
SARS-CoV-2 authentic virus neutralization assay
All SARS-CoV-2 authentic virus neutralization assays were performed in the biosafety level 3 (BSL3) laboratories at Toscana Life Sciences in Siena (Italy) and Vismederi Srl, Siena (Italy). BSL3 laboratories are approved by a Certified Biosafety Professional and are inspected every year by local authorities. The neutralization activity of culture supernatants from monoclonal was evaluated using a CPE-based assay as previously described (Manenti et al., 2020). S-protein-specific memory B cells produced antibodies were initially evaluated by means of a qualitative live-virus based neutralization assay against a one-point dilution of the samples. Supernatants were mixed in a 1:3 ratio with a SARS-CoV-2 viral solution containing 25 TCID50 of virus (final volume: 30 μL). After 1 h incubation at 37°C, 5% CO2, 25 μL of each virus-supernatant mixture was added to the wells of a 96-well plate containing a sub-confluent Vero E6 cell monolayer. Following a 2 h incubation at 37°C, the virus-serum mixture was removed and 100 μl of DMEM 2% FBS were added to each well. Plates were incubated for 3 days at 37°C in a humidified environment with 5% CO2, then examined for CPE by means of an inverted optical microscope. Absence or presence of CPE was defined by comparison of each well with the positive control (plasma sample showing high neutralizing activity of SARS-CoV-2 in infected Vero E6 cells (Andreano et al., 2020) and negative control (human serum sample negative for SARS-CoV-2 in ELISA and neutralization assays). Following expression as full-length IgG1 recombinant antibodies were quantitatively tested for their neutralization potency against both the wild type, D614G variant and the B.1.1.7 emerging variants. The assay was performed as previously described but using a viral titer of 100 TCID50. Antibodies were prepared at a starting concentration of 20 μg/mL and diluted step 1:2. Technical triplicates were performed for each experiment.
Production and titration of SARS-CoV-2 pseudotyped lentiviral reporter particles
Pseudotype stocks were prepared by FuGENE-HD (Promega) co-transfection of HEK293T/17 with SARS-CoV-2 spike pcDNA3.1 + expression plasmid, HIV gag-pol p8.91 plasmid and firefly luciferase expressing plasmid pCSFLW in a 1:0.8:1.2 ratio. 2 × 106 cells/cm2 were plated 24 h prior to transfection in 10cm cell culture dishes. 48 and 72 h post transfection, pseudotype-containing culture medium was harvested and filtered thought a 0.45um syringe filter to clear cell debris. Aliquots were stored at −80°C. Titration assays were performed by transduction of HEK293T/17 cells pre-transfected with ACE2 and TMPRRS2 plasmids to calculate the viral titer and infectious dose (PV input) for neutralization assays. SARS-CoV-2 D614G pseudotype was produced using the same procedure as described above. SARS-1 pseudotype was produced in a 1:0.5:0.8 ratio. MERS-pseudotype was produced as previously described (Grehan et al., 2015).
SARS-CoV-2 pseudotyped lentivirus neutralization assay
The potency of the neutralizing mAbs was assessed using lentiviral particles expressing SARS-CoV-2 spike protein on their surface and containing firefly luciferase as marker gene for detection of infection. Briefly, 2 × 106 HEK293T cells were pre-transfected in a 10 cm dish the day before the neutralization assay with ACE2 and TMPRSS2 plasmids in order to be used as optimal target cells for SARS-CoV-2 PV entry. In a 96-well plate mAbs were 2-fold serially diluted in duplicate in culture medium (DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin) starting at 20 μg/mL in a total volume of 100 μL. 1x106 RLU of SARS-CoV-2 pseudotyped lentiviral particles were added to each well and incubated at 37°C for 1 h. Each plate included PV plus cell only (virus control) and cells only (background control). 1 × 104 pre-transfected HEK293T cells suspended in 50 μL complete media were added per well and incubated for 48 h at 37°C and 5% CO2. Firefly luciferase activity (luminescence) was measured using the Bright-Glo assay system with a GloMax luminometer (Promega, UK). The raw Relative Luminescence Unit (RLU) data points were converted to a percentage neutralization value, whereby 100% neutralization equals the mean cell only RLU value control and 0% neutralization equals the mean PV only RLU value control. The normalized data was then plotted using Prism 8 (GraphPad) on a neutralization percentage scale and a NT50 value calculated, using the non-linear regression analysis. Plasma from COVID-19 convalescent donors showing neutralization activity against SARS-CoV-2 (Andreano et al., 2020) were also assessed in this assay.
Characterization of SARS-CoV-2 RBD-Antibodies binding by Flow cytometry
Flow cytometry analysis was performed to define antibodies interaction with S-protein-receptor-binding domain (RBD). Briefly, APEX Antibody Labeling Kits (Invitrogen) was used to conjugate 20 μg of selected antibodies to Alexa fluor 647, according to the manufacturer instructions. To assess the ability of each antibody to bind the RBD domain, 1 mg of magnetic beads (Dynabeads His-Tag, Invitrogen) were coated with 70 μg of histidine tagged RBD, and then 20 μg/mL of each labeled antibody were incubated with 40 μg/mL of beads-bound RBD for 1 h on ice. Then, samples were washed with 200 μL of Phosphate-buffered saline (PBS), resuspended in 150 μL of PBS and assessed with a FACS Canto II flow cytometer (Becton Dickinson). Results were analyzed by FlowJo (version 10).
Flow Cytometry-Based S-protein Competition assay
Antibodies specificity to bind SARS-CoV-2 S-protein and their possible competition was analyzed performing a Flow cytometer-based assay. To this aim, 200 μg of stabilized histidine tagged S-protein were coated with 1 mg of magnetic beads (Dynabeads His-Tag, Invitrogen). 20 μg of each antibody were labeled with Alexa fluor 647 working with the APEX Antibody Labeling Kits (Invitrogen). To test competitive binding profiles of the antibody panel selected, beads-bound S-protein (40 μg/mL) were pre-incubated with unlabeled antibodies (40 μg/mL) for 1 h on ice. Then, each set of the beads-antibody complexes were washed with PBS and separately incubated with each labeled antibody (20 μg/mL) for 1 h on ice. After incubation, the mix Beads-antibodies was washed, resuspended in 150 μL of PBS and analyzed using FACS Canto II flow cytometer (Becton Dickinson). Beads-bound and non-bound S-protein incubated with labeled antibodies were used as positive and negative control, respectively. Population gating and analysis was carried out using FlowJo (version 10).
Antigen-specific FcγR binding
Fluorescently coded microspheres were used to profile the ability of selected antibodies to interact with Fc receptors (Boudreau et al., 2020). The antigen of interest (SARS –CoV-2 S-protein RBD) was covalently coupled to different bead sets via primary amine conjugation. The beads were incubated with diluted antibody (diluted in PBS), allowing “on bead” affinity purification of antigen-specific antibodies. The bound antibodies were subsequently probed with tetramerized recombinant human FcγR2A and FcRN and analyzed using Luminex. The data is reported as the median fluorescence intensity of PE for a specific bead channel.
Antibody-dependent neutrophil phagocytosis
Antibody-dependent neutrophil phagocytosis (ADNP) assesses the ability of antibodies to induce the phagocytosis of antigen-coated targets by primary neutrophils. The assay was performed as previously described (Karsten et al., 2019, Boudreau et al., 2020). Briefly, fluorescent streptavidin-conjugated polystyrene beads were coupled to biotinylated SARS-CoV-2 Spike trimer. Diluted antibody (diluted in PBS) was added, and unbound antibodies were washed away. The antibody:bead complexes are added to primary neutrophils isolated from healthy blood donors using negative selection (StemCell EasySep Direct Human Neutrophil Isolation Kit), and phagocytosis was allowed to proceed for 1 h. The cells were then washed and fixed, and the extent of phagocytosis was measured by flow cytometry. The data is reported as a phagocytic score, which considers the proportion of effector cells that phagocytosed and the degree of phagocytosis. Each sample is run in biological duplicate using neutrophils isolated from two distinct donors. The mAb were tested for ADNP activity at a range of 30 μg/mL to 137.17 ng/mL.
Antibody-dependent NK cell activation
Antibody-dependent NK cell activation (ADNKA) assesses antigen-specific antibody-mediated NK cell activation against protein-coated plates. This assay was performed as previously described (Boudreau et al., 2020). Stabilized SARS-CoV-2 Spike trimer was used to coat ELISA plates, which were then washed and blocked. Diluted antibody (diluted in PBS) was added to the antigen coated plates, and unbound antibodies were washed away. NK cells, purified from healthy blood donor leukopaks using commercially available negative selection kits (StemCell EasySep Human NK Cell Isolation Kit) were added, and the levels of IFN-γ was measured after 5 h using flow cytometry. The data is reported as the percent of cells positive for IFN-γ. Each sample is tested with at least two different NK cell donors, with all samples tested with each donor. The monoclonal antibodies were tested for ADNKA activity at a range of 20 μg/mL to 9.1449 ng/mL.
Affinity evaluation of SARS-CoV-2 neutralizing antibodies
Anti-Human IgG Polyclonal Antibody (Southern Biotech 2040-01) was immobilized via amine group on two flow cells of a CM5 sensor chip. For the immobilization, anti-human IgG Ab diluted in 10mM Na acetate pH 5.0 at the concentration of 25 μg/mL was injected for 360 s over the dextran matrix, which had been previously activated with a mixture of 0.1M 1-ethyl-3(3-dimethylaminopropyl)-carbodiimide (EDC) and 0.4 M N-hydroxyl succinimide (NHS) for 420 s. After injection of the antibody, Ethanolamine 1M was injected to neutralize activated group. 10 μL/min flow rate was used during the whole procedure. Anti-SPIKE protein human mAbs were diluted in HBS-EP+ (HEPES 10 mM, NaCl 150 mM, EDTA 3.4 mM, 0.05% p20, pH 7.4) and injected for 120 s at 10 μL/min flow rate over one of the two flow cells containing the immobilized Anti-Human IgG Antibody, while running buffer (HBS-EP+) was injected over the other flow cell to be taken as blank. Dilution of each mAb was adjusted in order to have comparable levels of RU for each capture mAb. Following the capture of each mAb by the immobilized anti-human IgG antibody, different concentrations of SPIKE protein (20 μg/mL, 10 μg/mL, 5 μg/mL, 2.5 μg/mL and 1 μg/mL in HBS-EP+) were injected over both the blank flow cell and the flow cell containing the captured mAb for 180 s at a flow rate of 80 μL/min. Dissociation was followed for 800 s, regeneration was achieved with a pulse (60 s) of Glycine pH 1.5. Kinetic rates and affinity constant of SPIKE protein binding to each mAb were calculated applying a 1:1 binding as fitting model using the Bia T200 evaluation software 3.1.
Autoreactivity screening test on HEp-2 Cells
The NOVA Lite HEp-2 ANA Kit (Inova Diagnostics) was used in accordance to the manufacturer’s instructions to test antibodies the autoreactivity of selected antibodies which were tested at a concentration of 100 μg/mL. Kit positive and negative controls were used at three different dilutions (1:1, 1:10 and 1:100). Images were acquired using a DMI3000 B microscope (Leica) and an exposure time of 300 ms, channel intensity of 2000 and a gamma of 2.
Genetic Analyses of SARS-CoV-2 S-protein specific nAbs
A custom pipeline was developed for the analyses of antibody sequences and the characterization of the immunoglobulin genes. Raw sequences were stored as ab1 file and transformed into fastaq using Biopython. The reads were then quality checked using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and a report was generated using MultiQC (https://multiqc.info/). The antibody leader sequence and the terminal part of the constant region were removed by trimming using Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic). This latter program was also used to scan and remove low-quality reads using a sliding-window parameter. Once sequences were recovered, germline gene assignment and annotation were perfromed with MiXCR (https://mixcr.readthedocs.io/en/master/index.html), using the single-read alignment parameters, and a CSV-formatted output was generated. Finally, the sequences retrieved from the antibodies described in this manuscript were compared to published neutralizing antibodies against SARS-CoV-2. For this purpose, the Coronavirus Antibody Database, CoV-AbDab (http://opig.stats.ox.ac.uk/webapps/covabdab/) was downloaded and the antibodies with reported neutralization activity against SARS-CoV-2 were extracted. Comparison analysis were performed in Python using NumPy (https://numpy.org/) and, Pandas (https://pandas.pydata.org/) while figures were produced using the Matplotlib tool (https://matplotlib.org/) and Seaborn (https://seaborn.pydata.org/).
Negative-stain electron microscopy
Complexes were formed by incubating SARS-2 CoV-GSAS-6P-Mut7 and respective fabs at a 1:3 (trimer to fab) molar ratio for 30 min at room temperature. After diluting to 0.03 mg/mL in 1X TBS pH 7.4, the samples were deposited on plasma-cleaned copper mesh grids and stained with 2% uranyl formate for 55 s. Automated data collection was made possible through the Leginon software (Suloway et al., 2005) and a FEI Tecnai Spirit (120keV, 56,000x mag) paired with a FEI Eagle (4k by 4k) CCD camera. Other details include a defocus value of −1.5 μm, a pixel size of 2.06 Å per pixel, and a dose of 25 e−/Å2. Raw micrographs were stored in the Appion database (Lander et al., 2009), particles were picked with DoGPicker (Voss et al., 2009), and 2D and 3D classification and refinements were performed in RELION 3.0 (Scheres, 2012). Map segmentation and model docking was done in UCSF Chimera (Pettersen et al., 2004).
Prophylactic and therapeutic passive transfer studies in golden Syrian hamsters
Six- to eight-month-old female Syrian hamsters were purchased from Charles River Laboratories and housed in microisolator units, allowed free access to food and water and cared for under U.S. Department of Agriculture (USDA) guidelines for laboratory animals. For the passive transfer prophylactic experiments, the day prior to SARS-CoV-2 infection six hamsters per group were intraperitoneally administered with 500 μL of a 4, 1 or 0.25 mg/kg dose of J08-MUT mAb. For the passive transfer therapeutic experiments, the day after SARS-CoV-2 infection six hamsters per group were intraperitoneally administered with 500 μL of a 4 mg/kg dose of J08-MUT mAb. Another two groups (n = 6/each) were administered with 500 μL of 4 mg/kg of the anti-influenza virus #1664 human mAb (manuscript in preparation) or PBS only to serve as human IgG1 isotype and mock control groups, respectively. The day after, hamsters were anesthetized using 5% isoflurane, and inoculated with 5 × 105 PFU of SARS-CoV-2 (2019-nCoV/USA-WA1/2020) via the intranasal route, in a final volume of 100 μL. Baseline body weights were measured before infection as well as monitored daily for 7 and 11 days post infection in the prophylactic and therapeutic studies respectively. All experiments with the hamsters were performed in accordance with the NRC Guide for Care and Use of Laboratory Animals, the Animal Welfare act, and the CDC/NIH Biosafety and Microbiological and Biomedical Laboratories as well as the guidelines set by the Institutional Animal Care and Use Committee (IACUC) of the University of Georgia who also approved the animal experimental protocol. All animal studies infection with SARS-CoV-2 were conducted in the Animal Health Research Center (AHRC) Biosafety Level 3 (BSL-3) laboratories of the University of Georgia.
Determination of viral load by TCID50 assay
Lung tissues were homogenized in 1 mL of DMEM containing 1% fetal bovine serum (FBS) and 1% penicillin/streptomycin. The lung homogenate supernatant was diluted 10-fold (10° to 106) and used to determine median tissue culture infection dose (TCID50) in Vero E6 cells as previously described (Jang and Ross, 2020).
Human IgG detection in hamster sera
ELISA assay was used to detect the human IgG J08-MUT in hamster sera. 384-well plates (384 well plates, Microplate Clear; Greiner Bio-one) were coated with 2 μg/mL of unlabled goat anti-human IgG (SouthernBiotech) diluted in sterile PBS and incubated at 4°C overnight. 50 μL/well of saturation buffer (PBS/BSA 1%) was used to saturate unspecific binding and plates were incubated at 37°C for 1 h without CO2. Hamster sera were diluted in PBS/BSA 1%/Tween20 0.05% at a starting dilution of 1:10. Fourteen reciprocal dilutions were performed. Alkaline phosphatase-conjugated goat anti-human IgG (Sigma-Aldrich) was used as secondary antibody and pNPP (p-nitrophenyl phosphate) (Sigma-Aldrich) was used as soluble substrate. Wells were washed three times between each step with PBS/BSA 1%/Tween20 0.05%. The final reaction was measured by using the Varioskan Lux Reader (Thermo Fisher Scientific) at a wavelength of 405 nm. Samples were considered as positive if OD at 405 nm (OD405) was twice the blank.
Acknowledgments
We wish to thank Fondazione Toscana Life Sciences in the persons of Dr. Fabrizio Landi and Dr. Andrea Paolini and the whole Administration for their help and support. In particular, we would like to thank Mr. Francesco Senatore, Mrs. Laura Canavacci, and Mrs. Cinzia Giordano for their support in preparing all the documents needed for the ethical approval of the clinical studies carried out within this project. We wish to thank the National Institute for Infectious Diseases, IRCCS, Lazzaro Spallanzani Rome (IT), and the Azienda Ospedaliera Universitaria Senese, Siena (IT) for providing blood samples from COVID-19 convalescent donors under studies approved by local ethic committees. We also wish to thank all the nursing staff who chose to cooperate for blood withdrawal and all the donors who decided to participate in this study. We would like to thank the whole GSK Vaccines Pre-clinical Evidence Generation and Assay – Immunolgy function led by Dr. Oretta Finco for their availability and support as well as Mrs. Simona Tavarini, Mrs. Chiara Sammicheli, Dr. Monia Bardelli, Dr. Michela Brazzoli, Dr. Elisabetta Frigimelica, Dr. Erica Borgogni, and Dr. Elisa Faenzi for sharing their expertise, extreme availability, and technical support. We would also like to thank Dr. Mariagrazia Pizza and Dr. Simone Pecetta for initial insightful advice and discussions on this project. We would like to thank Dr. Jason McLellan and his team for generously providing the SARS-CoV-2 S protein stabilized in its prefusion conformation used in this study. Furthermore, we would like to thank Dr. Daniel Wrapp and Dr. Nianshuang Wang for the precious information and suggestions. The authors wish to thank Seromyx for their support in assessing the functionality of the antibodies Fc portion. The authors would like to thank the University of Georgia Animal Resource staff, technicians, and veterinarians for animal care as well as the staff of the University of Georgia AHRC BSL-3 facility for providing biosafety and animal care. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2 Isolate USA-WA1/2020, NR-52281. This work was funded by the European Research Council (ERC) advanced grant agreement no. 787552 (vAMRes). The Laboratory is also supported by the Wellcome Trust. This publication was supported by funds from the “Centro Regionale Medicina di Precisione” and by all the people who answered the call to fight with us the battle against SARS-CoV-2 with their kind donations on the platform ForFunding (https://www.forfunding.intesasanpaolo.com/DonationPlatform-ISP/nav/progetto/id/3380). This publication was supported by the European Virus Archive goes Global (EVAg) project, which has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 653316. This publication was supported by the COVID-2020-12371817 project, which has received funding from the Italian Ministry of Health. This work was funded, in part, by the University of Georgia (UGA) (UGA-001) and a contract by Fondazione Toscana Life Sciences. In addition, T.M.R. is supported by the Georgia Research Alliance as an Eminent Scholar. We wish to thank AchilleS Vaccines and the EU Malaria Fund for funding and managing the development of the human monoclonal antibody for clinical studies.
Author contributions
E.A., I.P., P.P., N.M., E.P., G.P., A.M., L. Benincasa, M.T., F.V., A.K., J.B., L.D., C.D.G., H.J., G.S., J.L.T., G.O., C.D.S., and D.C. conceived, performed experiments, and analyzed data. E.N., C.A., C.C., F.M., A.E., and M.F. enrolled patients and isolated PBMCs. R.R. and E.A. wrote the manuscript. A.B.W., E.M., N.T., T.M.R., M.R.C., G.I., L. Bracci, C.S., and R.R. coordinated the project.
Declaration of interests
R.R. is an employee of GSK group of companies. E.A., A.K., D.C., C.D.S, I.P., N.M., E.P., P.P., C.S., M.T., F.V., and R.R. are listed as inventors of full-length human monoclonal antibodies described in Italian patent applications no. 102020000015754 filed on June 30, 2020 and no. 102020000018955 filed on August 3, 2020.
Published: February 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.02.035.
Supplemental information
References
- Ackerman M.E., Dugast A.-S., McAndrew E.G., Tsoukas S., Licht A.F., Irvine D.J., Alter G. Enhanced phagocytic activity of HIV-specific antibodies correlates with natural production of immunoglobulins with skewed affinity for FcγR2a and FcγR2b. J. Virol. 2013;87:5468–5476. doi: 10.1128/JVI.03403-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman M.E., Mikhailova A., Brown E.P., Dowell K.G., Walker B.D., Bailey-Kellogg C., Suscovich T.J., Alter G. Polyfunctional HIV-Specific Antibody Responses Are Associated with Spontaneous HIV Control. PLoS Pathog. 2016;12:e1005315. doi: 10.1371/journal.ppat.1005315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsoussi W.B., Turner J.S., Case J.B., Zhao H., Schmitz A.J., Zhou J.Q., Chen R.E., Lei T., Rizk A.A., McIntire K.M., et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. 2020;205:915–922. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Monego S.D., Pantano E., Manganaro N., Manenti A., et al. SARS-CoV-2 escape in vitrofrom a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aratani L. 2020. Jobless America: the coronavirus unemployment crisis in figures. The Guardian, May 28, 2020.https://www.theguardian.com/business/2020/may/28/jobless-america-unemployment-coronavirus-in-figures [Google Scholar]
- Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth B.J., Ramakrishnan B., Narayan K., Wollacott A.M., Babcock G.J., Shriver Z., Viswanathan K. Extending human IgG half-life using structure-guided design. MAbs. 2018;10:1098–1110. doi: 10.1080/19420862.2018.1490119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau C.M., Yu W.-H., Suscovich T.J., Talbot H.K., Edwards K.M., Alter G. Selective induction of antibody effector functional responses using MF59-adjuvanted vaccination. J. Clin. Invest. 2020;130:662–672. doi: 10.1172/JCI129520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A.L., Fallon J.K., Alter G. A Sample-Sparing Multiplexed ADCP Assay. Front. Immunol. 2019;10:1851. doi: 10.3389/fimmu.2019.01851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell G.W., Ferrara F., Grehan K., Thompson C.P., Temperton N.J. Pseudotype-based neutralization assays for influenza: a systematic analysis. Front. Immunol. 2015;6:161. doi: 10.3389/fimmu.2015.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell G.W., Grehan K., Ferrara F., Molesti E., Temperton N. An Optimized Method for the Production Using PEI, Titration and Neutralization of SARS-CoV Spike Luciferase Pseudotypes. Bio Protoc. 2017;7:e2514. doi: 10.21769/BioProtoc.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2021. Emerging SARS-CoV-2 Variants.https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html [Google Scholar]
- Chakraborty S., Gonzalez J., Edwards K., Mallajosyula V., Buzzanco A.S., Sherwood R., Buffone C., Kathale N., Providenza S., Xie M.M., et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 2020;22:67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.A., McVicar R.N., et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020;183:1043–1057.e15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler D.M., Summers L.H. The COVID-19 Pandemic and the $16 Trillion Virus. JAMA. 2020;324:1495–1496. doi: 10.1001/jama.2020.19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D.J., Tan G.S., Palese P., Ravetch J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . 2020. Coronavirus (COVID-19) Update: FDA Authorizes Monoclonal Antibodies for Treatment of COVID-19.https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-monoclonal-antibodies-treatment-covid-19 [Google Scholar]
- FDA . 2021. COVID-19 Vaccines.https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccines [Google Scholar]
- Gaudinski M.R., Coates E.E., Houser K.V., Chen G.L., Yamshchikov G., Saunders J.G., Holman L.A., Gordon I., Plummer S., Hendel C.S., et al. VRC 606 Study Team Safety and pharmacokinetics of the Fc-modified HIV-1 human monoclonal antibody VRC01LS: A Phase 1 open-label clinical trial in healthy adults. PLoS Med. 2018;15:e1002493. doi: 10.1371/journal.pmed.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grehan K., Ferrara F., Temperton N. An optimised method for the production of MERS-CoV spike expressing viral pseudotypes. MethodsX. 2015;2:379–384. doi: 10.1016/j.mex.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin M.P., Yuan Y., Takas T., Domachowske J.B., Madhi S.A., Manzoni P., Simões E.A.F., Esser M.T., Khan A.A., Dubovsky F., et al. Nirsevimab Study Group Single-Dose Nirsevimab for Prevention of RSV in Preterm Infants. N. Engl. J. Med. 2020;383:415–425. doi: 10.1056/NEJMoa1913556. [DOI] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft van Huijsduijnen R., Kojima S., Carter D., Okabe H., Sato A., Akahata W., Wells T.N.C., Katsuno K. Reassessing therapeutic antibodies for neglected and tropical diseases. PLoS Negl. Trop. Dis. 2020;14:e0007860. doi: 10.1371/journal.pntd.0007860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Doria-Rose N.A., Longo N.S., Laub L., Lin C.L., Turk E., Kang B.H., Migueles S.A., Bailer R.T., Mascola J.R., Connors M. Isolation of human monoclonal antibodies from peripheral blood B cells. Nat. Protoc. 2013;8:1907–1915. doi: 10.1038/nprot.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H., Ross T.M. Dried SARS-CoV-2 virus maintains infectivity to Vero E6 cells for up to 48 h. Vet. Microbiol. 2020;251:108907. doi: 10.1016/j.vetmic.2020.108907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N., Kuo H.-H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J., et al. Massachusetts Consortium on Pathogen Readiness Specimen Working Group Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell. 2020;183:143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten C.B., Mehta N., Shin S.A., Diefenbach T.J., Slein M.D., Karpinski W., Irvine E.B., Broge T., Suscovich T.J., Alter G. A versatile high-throughput assay to characterize antibody-mediated neutrophil phagocytosis. J. Immunol. Methods. 2019;471:46–56. doi: 10.1016/j.jim.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzelnick L.C., Gresh L., Halloran M.E., Mercado J.C., Kuan G., Gordon A., Balmaseda A., Harris E. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358:929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley B. Developing therapeutic monoclonal antibodies at pandemic pace. Nat. Biotechnol. 2020;38:540–545. doi: 10.1038/s41587-020-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreer C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K., et al. Longitudinal Isolation of Potent Near-Germline SARS-CoV-2-Neutralizing Antibodies from COVID-19 Patients. Cell. 2020;182:843–854.e12. doi: 10.1016/j.cell.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.-C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.D., et al. A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundi M. One-hit models for virus inactivation studies. Antiviral Res. 1999;41:145–152. doi: 10.1016/s0166-3542(99)00008-x. [DOI] [PubMed] [Google Scholar]
- Kupferschmidt K. Successful Ebola treatments promise to tame outbreak. Science. 2019;365:628–629. doi: 10.1126/science.365.6454.628. [DOI] [PubMed] [Google Scholar]
- Lander G.C., Stagg S.M., Voss N.R., Cheng A., Fellmann D., Pulokas J., Yoshioka C., Irving C., Mulder A., Lau P.-W., et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- Lilly . 2020. Lilly’s neutralizing antibody bamlanivimab (LY-CoV555) receives FDA emergency use authorization for the treatment of recently diagnosed COVID-19.https://investor.lilly.com/news-releases/news-release-details/lillys-neutralizing-antibody-bamlanivimab-ly-cov555-receives-fda [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Mackness B.C., Jaworski J.A., Boudanova E., Park A., Valente D., Mauriac C., Pasquier O., Schmidt T., Kabiri M., Kandira A., et al. Antibody Fc engineering for enhanced neonatal Fc receptor binding and prolonged circulation half-life. MAbs. 2019;11:1276–1288. doi: 10.1080/19420862.2019.1633883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manenti A., Maggetti M., Casa E., Martinuzzi D., Torelli A., Trombetta C.M., Marchi S., Montomoli E. Evaluation of SARS-CoV-2 neutralizing antibodies using a CPE-based colorimetric live virus micro-neutralization assay in human serum samples. J. Med. Virol. 2020;92:2096–2104. doi: 10.1002/jmv.25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. FDA approves antibody cocktail for Ebola virus. Nat. Rev. Drug Discov. 2020;19:827. doi: 10.1038/d41573-020-00197-8. [DOI] [PubMed] [Google Scholar]
- Pegu A., Hessell A.J., Mascola J.R., Haigwood N.L. Use of broadly neutralizing antibodies for HIV-1 prevention. Immunol. Rev. 2017;275:296–312. doi: 10.1111/imr.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Regeneron . 2020. Regeneron’s casirivimab and imdevimab antibody cocktail for COVID-19 is first combination therapy to receive FDA emergency use authorization.https://investor.regeneron.com/news-releases/news-release-details/regenerons-regen-cov2-first-antibody-cocktail-covid-19-receive [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.-T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R., et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2020;218:e20201993. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlothauer T., Herter S., Koller C.F., Grau-Richards S., Steinhart V., Spick C., Kubbies M., Klein C., Umaña P., Mössner E. Novel human IgG1 and IgG4 Fc-engineered antibodies with completely abolished immune effector functions. Protein Eng. Des. Sel. 2016;29:457–466. doi: 10.1093/protein/gzw040. [DOI] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow E., Friede M., Sheikh M., Torvaldsen S. Therapeutic antibodies for infectious diseases. Bull. World Health Organ. 2017;95:235–237. doi: 10.2471/BLT.16.178061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Renia L., Macary P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss N.R., Yoshioka C.K., Radermacher M., Potter C.S., Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H., Busse C.E. Expression Cloning of Antibodies from Single Human B Cells. Methods Mol. Biol. 2019;1956:105–125. doi: 10.1007/978-1-4939-9151-8_5. [DOI] [PubMed] [Google Scholar]
- Wellcome and IAVI (2020). Expanding access to monoclonal antibody-based products: a global call to action. https://www.iavi.org/news-resources/expanding-access-to-monoclonal-antibody-based-products-a-global-call-to-action.
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.-C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J., Chamberlain A.K., Horton H.M., Karki S., Leung I.W.L., Sproule T.J., Lazar G.A., Roopenian D.C., Desjarlais J.R. Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 2010;28:157–159. doi: 10.1038/nbt.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y., Estes S.K., Ali R.A., Gandhi A.A., Yalavarthi S., Shi H., Sule G., Gockman K., Madison J.A., Zuo M., et al. Prothrombotic autoantibodies in serum from patients hospitalized with COVID-19. Sci. Transl. Med. 2020;12:eabd3876. doi: 10.1126/scitranslmed.abd3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nucleotide and amino acidic sequences of all SARS-CoV-2-neutralizing antibodies were deposited in the Italian patent applications n. 102020000015754 filed on June 30th 2020 and 102020000018955 filed on August 3rd 2020. The accession number for the nucleotide sequences of all SARS-CoV-2-neutralizing antibodies reported in this paper is GenBank: MW_598287 - MW_598314.