Abstract
Non-allelic homologous recombination (NAHR), non-homologous end joining (NHEJ), and microhomology-mediated replication-dependent recombination (MMRDR) have all been put forward as mechanisms to explain DNA rearrangements associated with genomic disorders. However, many nonrecurrent rearrangements in humans remain unexplained. To further investigate the mutation mechanisms of these copy number variations (CNVs), we performed breakpoint mapping analysis for 62 clinical cases with intragenic deletions in the human DMD gene (50 cases) and other known disease-causing genes (one PCCB, one IVD, one DBT, three PAH, one STK11, one HEXB, three DBT, one HRPT1, and one EMD cases). While repetitive elements were found in only four individual cases, three involving DMD and one HEXB gene, microhomologies (2–10 bp) were observed at breakpoint junctions in 56% and insertions ranging from 1 to 48 bp were seen in 16 of the total 62 cases. Among these insertions, we observed evidence for tandem repetitions of short segments (5–20 bp) of reference sequence proximal to the breakpoints in six individual DMD cases (six repeats in one, four repeats in three, two repeats in one, and one repeat in one case), strongly indicating attempts by the replication machinery to surpass the stalled replication fork. We provide evidence of a novel template slippage event during replication rescue. With a deeper insight into the complex process of replication and its rescue during origin failure, brought forward by recent studies, we propose a hypothesis based on aberrant firing of replication origins to explain intragenic nonrecurrent rearrangements within genes, including the DMD gene.
Use of array-based comparative genomic hybridization (aCGH) in clinical diagnostic laboratories has accelerated the discovery of several microdeletion and microduplication syndromes (Raedt et al. 2006; Vissers et al. 2009; Mefford et al. 2010; Rosenfeld et al. 2010). These genetic disorders involve a gain, loss, or disruption of a gene(s), referred to as copy number variations (CNVs), resulting from previously undetected submicroscopic genomic rearrangements (Stankiewicz and Lupski 2006). These genomic rearrangements, ranging from simple deletions or duplications to complex rearrangements, are mediated by mutational mechanisms that are not completely understood; however, several mechanisms, including non-allelic homologous recombination (NAHR), non-homologous end joining (NHEJ), fork stalling and template switching (FoSTeS), and microhomology-mediated replication-dependent recombination (MMRDR), have been hypothesized to explain the cause of different genomic rearrangements (Stankiewicz and Lupski 2006; Chen et al. 2010). While NAHR accounts for most recurrent rearrangements, NHEJ, a double-strand break (DSB) repair mechanism, explains the nonrecurrent rearrangements with minimal to no junction homology (Roth et al. 1985; Inoue et al. 2002; Weterings and van Gent 2004).
Recently, breakpoint junction sequencing of several deletion and duplication rearrangements has suggested a role for microhomology in mediating CNVs. Microhomology-mediated mechanisms, collectively referred to as MMRDR, include microhomology-mediated break-induced replication (MMBIR), break-induced serial replication slippage (BISRS), and fork stalling and template switching (FoSTeS) (Lee et al. 2007; Chen et al. 2010). Several studies that provide further evidence to implicate microhomology-mediated replication error mechanisms have been reported (Koszul et al. 2004; Chen et al. 2005a; Hastings et al. 2009a). The above-mentioned microhomology-based mechanisms dovetail with a common hypothesis that upon replication fork stalling, the leading/lagging strand primer/polymerase disengages from its original template, translocates, and then reassociates, probably by using microhomology at the 3′ end, to another available template/replication fork in physical proximity and resumes replication (Chen et al. 2005a, 2010; Lee et al. 2007). These models further postulate that during replication, downstream fork switching (forward invasion) results in a deletion, whereas switching to an upstream fork (backward invasion) results in a duplication, and repeated switches back and forth result in complex rearrangements that can include triplications and inversions (Hastings et al. 2009a). The knowledge of the replication mechanism and its errors that result in CNV formation gained from these hypotheses has added greatly to our understanding of genomic rearrangements. However, a recent large-scale breakpoint junction sequence analysis study demonstrated that microhomology-mediated processes (2–20 bp), including FoSTeS and NHEJ, account for only 28% of observed rearrangements and CNVs (Kidd et al. 2010). Therefore, the mechanisms leading to a majority of genomic rearrangements and associated CNVs remain elusive.
In this study, we performed breakpoint junction sequence analyses of 62 pathogenic nonrecurrent deletion events determined by high-resolution oligonucleotide aCGH in different patients with known genetic disorders. These included 50 Duchenne or Becker muscular dystrophy (DMD/BMD) patients with deletions in the dystrophin (DMD) gene, as well as 12 patients with deletions in different disease-causing genes, including the PCCB, IVD, DBT, PAH, STK11, HEXB, DBT, HRPT1, and EMD genes. We analyzed 124 total breakpoint junction sequences for the presence of microhomology and repetitive elements to understand the putative mechanism involved in deletion causation. Of the 16 cases with insertions, sequence analysis of six particular cases suggested that the template sequences proximal to the mapped deletion breakpoints were repeatedly replicated. This is a novel observation and is not explained by any of the above-mentioned existing mechanisms. We propose a hypothesis based on aberrant firing of replication origins to explain nonrecurrent rearrangements within genes, including the DMD gene, based on the replication rescue model suggested by Doksani et al. (2009), and we suggest that this mechanism may explain other DNA rearrangements in the human genome.
Results
Deletion confirmation and breakpoint approximation by aCGH
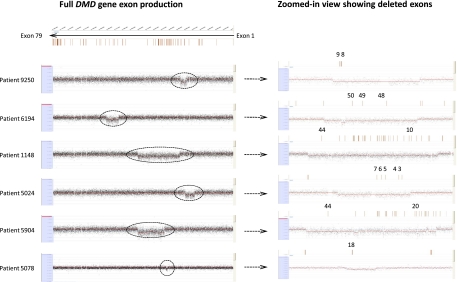
High-resolution oligonucleotide aCGH, custom-designed on a NimbleGen 385K or OGT 44K platform (Oxford Gene Technology) to target 450 genes associated with various genetic disorders, was used to detect deletions and duplications. A log2 ratio of −0.6 was set as the threshold value for deletion calls, and each call was generated from a DLR (deviation log ratio) of segments with at least four probes. DMD is the largest known gene of the human genome (2.4 Mb) and has a high mutation frequency for intragenic deletions (60%–65% of observed mutations) (Den Dunnen et al. 1989; Nobile et al. 1995) and duplications (5%–15% of all mutations) (Oudet et al. 1992; Oshima et al. 2009), making it an excellent model for understanding CNV mechanisms. We found evidence for simple deletions, as well as interrupted deletions in which stretches of deleted DNA were punctuated by DNA with no change in copy number (Figs. 1, 2). A total of 50 DMD cases confirmed to have deletions by aCGH as well as all observed non-DMD deletion cases (12) were processed for further analysis and breakpoint determination (Tables 1, 2). Non-DMD cases included in the study were patients with deletions in the PCCB (one), PAH (three), HRPT1 (one), IVD (one), DBT (three), STK11 (one), HEXB (one), and EMD (one) genes. Distribution of the observed DMD deletions along the gene is shown in Supplemental Data S1. Approximate breakpoint coordinates for the observed deletions were obtained by NimbleScan and CytoSure software and used for subsequent breakpoint sequence analysis.
Figure 1.
(Left) Comparative genomic hybridization data for the DMD gene locus (chrX:31,137,345–33,229,673) for DMD patients using a custom-designed 385K high-density array from NimbleGen. (Right) The zoomed-in view of the corresponding array on the left, with the deleted exons highlighted in red.
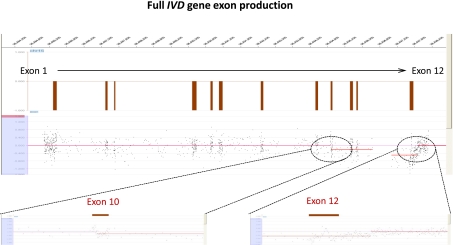
Figure 2.
Comparative genomic hybridization data for the IVD gene locus (chr15:40,697,686–40,713,512) in patient 4551 using a custom-designed 385K high-density array from NimbleGen. The zoomed-in view of the corresponding array highlights the breakpoints for a patient with a deletion mutation encompassing exons 10–12 with breakpoints in intron 9 and the 3′ UTR.
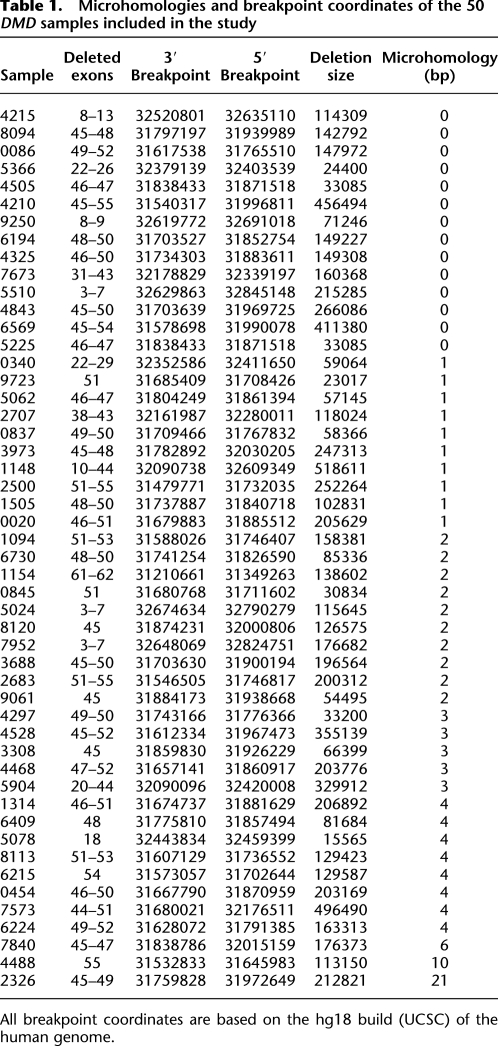
Table 1.
Microhomologies and breakpoint coordinates of the 50 DMD samples included in the study
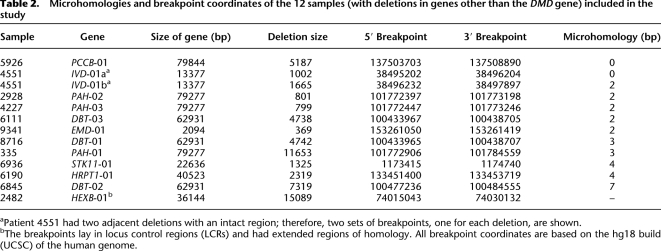
Table 2.
Microhomologies and breakpoint coordinates of the 12 samples (with deletions in genes other than the DMD gene) included in the study
Mapping deletion breakpoints reveals the resolution of aCGH
Since the deletion breakpoints determined by aCGH technology are only approximate and are dependent on the spacing of the designed probes, exact breakpoints were further mapped by a PCR approach. Target-specific primer pairs for long-range PCR (LR-PCR) were designed at least 2–3 kb from the approximate breakpoints returned by aCGH (discussed above). Upon electrophoresis and visualization on agarose gels, patient DNAs yielded products of expected size (2–6 kb spanning the breakpoints), while wild-type DNA yielded either no product or a much larger product (based on the size of the deletion). These products were gel-purified and used for DNA walk-in to obtain smaller products amenable to sequencing. Use of gel-purified LR-PCR products compared with genomic DNA for further walk-in by multiplex PCR yielded more specific products and cleaner sequences. Multiplex PCR performed using a mixture of different primer pairs (designed at regular intervals from aCGH-determined breakpoint coordinates) returned several products as visualized on agarose gels. Sequencing the shortest expected band (smallest breakpoint junction product) and aligning the sequence against the human reference human genome by BLAT mapped the exact deletion breakpoints. These breakpoints were on average ±500 bp away from the breakpoints approximated by aCGH analysis (Supplemental Data S2). This distance was consistent across most breakpoints, indicating the high resolution of deletion and duplication detection (as close as <50 bp to exact breakpoints in several cases) achieved by custom-designed oligonucleotide aCGH.
Signatures of known mechanisms at deletion breakpoint junctions
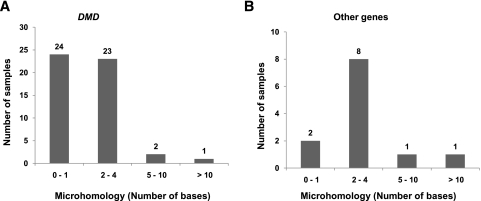
The 2.4-Mb dystrophin gene is composed of 79 exons (of 150–175 bp each), which account for only 0.6% of the gene, interspersed with very large introns ranging from 20 kb to 170 kb (Koenig et al. 1987). Compared with DMD, other genes involved in the study, namely, PCCB , PAH, HRPT1, IVD, DBT, STK11, HEXB, and EMD, are much smaller in size (about 1/30th), ranging from 13 kb to 79 kb (with 10–15 exons), with smaller exons (100–125 bp) and introns (2–17 kb). Sequences across all of the 124 mapped breakpoints, including those of 50 DMD and 12 non-DMD cases, were analyzed for any sequence signatures corresponding to known mutational mechanisms. Repetitive elements were found within 75 bp of mapped breakpoints in only four individual cases, including three DMD and one HEXB. These included a Charlie2 (MER1 family) in case 6224, a LTR16A (LTR) in 9723, a MER5A (MER1) in 4297, and a SINE (Alu) in 2482. Furthermore, microhomology of ≥2 bp was observed in 26 cases (52%), while there was no homology or only 1-bp homology in 24 cases, accounting for 48% of the DMD cases studied (Table 1). Similar observations were made in previous studies involving genomewide human germline CNV breakpoints, where only 30%–40% of cases had any significant microhomology (Conrad et al. 2010; Kidd et al. 2010). Sequence alignment at both distal and proximal breakpoints as returned by BLAT and the presence of microhomology at junctions is represented for each case (see Supplemental Data S3). Microhomologies of 2–4 bp were also observed in a majority of the non-DMD cases (nine out of 12), as shown in Table 2. A graphical representation of the occurrence of microhomologies at breakpoint junctions is shown (Fig. 3). The presence of repetitive elements, microhomologies, and occasional small insertions suggested possible involvement of the known mechanism, namely, NAHR, NHEJ, and MMRDR, in mediating the observed deletions. However, a significant number of cases (33% of total cases) lacked any of these sequence signatures and remained unexplained.
Figure 3.
The presence of microhomologies at breakpoint junctions. The graphs represent the extent of microhomologies at the deletion breakpoint junction sequences in DMD (A) and other genes (B). The x-axis represents the number of bases homologous in both junction sequences of a deletion. The y-axis represents the number of samples that have been observed to have the indicated microhomology.
Junction sequences reveal repeat replications
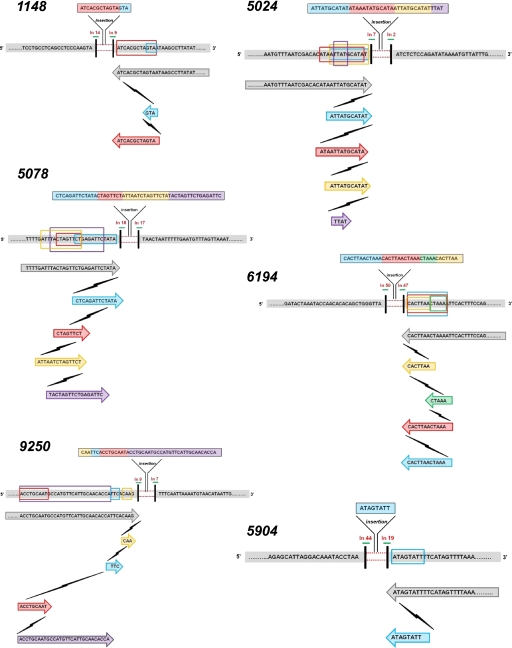
Each of the 62 junction sequences was aligned with the reference human genome sequence deposited in GenBank (Mar. 2006; NCBI36/hg18) using the BLAT tool (http://genome.ucsc.edu/cgi-bin/hgBlat?command=start) to validate their nature and/or origin. Occasionally, part of the sequence did not match with the reference and was inferred to be an insertion. These insertions ranged from 1 to 48 bp and were observed in 16 of the total 62 cases. The majority of these insertions (except for that in IVD) returned no matches to the human reference genome when analyzed individually by the UCSC BLAT tool. Of the 13 DMD cases with insertions, six (5024, 5078, 9250, 5904, 1148, and 6194) were of particular interest. Each of these six sequence insertions was found upon manual alignment to contain short segments (5–20 bp in length), each identical to the reference sequence proximal to the deletion breakpoint (Fig. 4). The number and length of the sequence segments in each of the six cases were:
5024 (…..TAATCGACAC[ATA{/A!TTAT!GCATA]T/}…..),
6194 (…..AAAGTGAAT[/!TTTAG]{TTAAGTG!/}…..),
5078 (…..TTCAAAGGCA!AAT!TTTG/ATT/|TA[\CTAGTT{CT]\GAGATTC|TATA}…..),
1148 (…..[TAGTGCGAT/CAT/]TATTCGGAA.....),
5904 (…..AACTATGAA{AATACTAT}…..), and
9250 (…..C{TTG}T!GAA!/TGGTGTTGCAATGAACATGGC[ATTGCAGGT]/ATCC…..),
varied as shown; each such sequence stretch is shown inclusive of a pair of braces. Note that while the complete sequence shown above in each case (ignoring the braces and special characters) is a reference sequence, the sequences enclosed in a pair of similar braces or special characters is a repetition or re-replication of the respective reference sequence that forms part of the observed insertion. Additionally, a 29-bp insertion observed in the IVD gene (case 4551) upon alignment using the UCSC Genome Browser matched the reference sequence within the deletion breakpoints. This suggests that what appeared as a simple deletion in aCGH data (Fig. 2) was, indeed, two adjacent deletions (or one large deletion) with a 29-bp intact region (in the same orientation as the template).
Figure 4.
Template slippages at deletion breakpoints in DMD cases 5024, 9250, 1148, 5904, 5078, and 6194. In all the cases shown in the figure, short segments of the inserted sequence align with the template (reference genome) sequence proximal to the breakpoint as shown (each such segment of the inserted sequence is highlighted in a different color for each case). These indicate repeated slippage of the replication machinery along the template and re-replication. Each lightning symbol shown in the figure represents a slippage event of the replication machinery. Template sequence proximal to the breakpoints on both 5′ and 3′ ends of the deletion (gray bars); deletion (red dotted lines); breakpoints (vertical black bars). Below this, for each case, the gray arrow bars (with sequence enclosed) refer to the normal replication, while template slippages and re-replications leading to insertion are shown by colored arrows. Each cycle of re-replication is represented in a different color. The introns in which the deletion breakpoints lie are shown above each breakpoint. Each re-replicated sequence is also shown enclosed in corresponding colored boxes on the template DNA.
Discussion
Repeat replication at breakpoint junction suggests template slippage and replication fork direction
The short tandem repeats of sequence segments inserted at deletion breakpoints in six DMD cases (5024, 5078, 9250, 5904, 1148, and 6194) suggest serial slippage of the replisome followed by re-replication of these short segments of template, but subsequent failure to continue replication past the observed breakpoint region (Fig. 4). Although similar observations were made in earlier studies involving gross insertions, evidence of such slippage events adjacent to a gross deletion has not been reported (Chen et al. 2005a,b). Microhomology-mediated mechanisms, which include BISRS (Chen et al. 2005a,c), MMBIR (Hastings et al. 2009a; Zhang et al. 2009), and FoSTeS (Lee et al. 2007; Hastings et al. 2009b; Zhang et al. 2009) explain genomic deletions and duplications, as well as complex rearrangements as happening by several template slippage or switch events due to microhomology (of as little as 1 bp). According to these mechanisms, deletions and duplications are a result of occasional dislodging of the replicating polymerase or primer and reengagement at a different template position based on microhomology, to continue replication (Zhang et al. 2009; Chen et al. 2010). Although microhomology seems plausible for short rearrangements involving a few hundred to a thousand bases, extrapolation of the mechanism to large genomic rearrangements is called into question by the recent advances in our understanding of eukaryotic replication (Debatisse et al. 2004; Blow and Dutta 2005; Doksani et al. 2009; Natsume and Tanaka 2010).
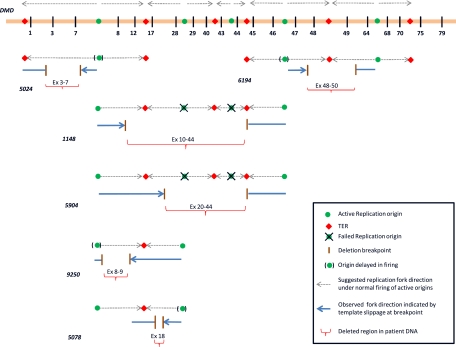
Studies in Escherichia coli (Heller and Marians 2006) and Saccharomyces cerevisiae (Lopes et al. 2006) show that, upon blockage of leading-strand progression by single-strand lesions, replication reinitiates downstream from an unrepaired block, but leaves a lesion or gap of up to 1000 bp. However, when progressing replication forks or active replisomes encounter DSBs, these replisomes do not resume replication, and the downstream template may be replicated (rescue of replication) by the replisomes approaching from adjacent replication forks or origins (Doksani et al. 2009). Invasion of templates at distances several kilobases away from encountered DSBs (as is the case in our current deletions) by stalled polymerases or primers may be less likely in these current DMD cases. Deletions observed in the present study are many kilobases long (70–500 kb) and are more or less the size of a complete replicon or two (Berezney et al. 2000). The large sizes of observed deletions, re-replication events adjacent to these deletions, and lack of microhomology are not explained by a single existing mechanism. One previous study involving replication direction analysis has shown that the entire 2.4-Mb DMD gene in human masculine erythroleukemia cells is replicated by seven active replication origins located upstream of exon 1, between exons 7 and 8, exons 28 and 29, exons 43 and 44, exons 46 and 48, exons 64 and 68, and finally downstream from exon 79 of the DMD gene (Verbovaia and Razin 1997). Replication fork directions under normal firing of all identified origins located within the DMD gene are shown in Figure 5 (indicated by gray arrows along the DMD gene). We believe that the re-replication events observed at the deletion breakpoints in each of the six above-mentioned DMD cases occurred at stalled replication forks during attempts of the replisomes originating from each of the involved origins. Therefore, the end (5′ or 3′) of the deletion at which these slippage and re-replication events were observed indicates the position of the progressing replisome and hence the direction of progression of the replication fork (from the associated origin) prior to the deletion.
Figure 5.
Illustration of the proposed deletion mechanism as explained by failure of replication origins in different patient cases. As shown for each of the six patient cases (5024, 6194, 1148, 5904, 9250, and 5708), failure/delay in origin firing may result in incomplete replication, causing DNA deletion. For example, in patient 5904, failure of origins in introns 28 and 43 may have led to deletions of exons 20–44. The horizontal bar at the top represents the DMD gene with the five mapped replication origins (green circles) and the six termination regions (red rhomboids), along with adjacent numbered exons (vertical black bars). The numbers in bold italics represent the patient sample. For each patient sample illustrated above, we show the template with origins and terminal regions adjacent to the deletion (gray arrowheads showing the direction of replication forks). (Brown horizontal bars) Deletion breakpoints (or replication cease points); (blue bars) patient DNA (or newly replicated DNA); (blue arrowheads) the replication fork direction at breakpoint. The directions of the replication forks are predicted based on the presence of re-replicated sequences at either breakpoint junction. For example, in sample 5024, the template slippage was found at the breakpoint in intron 7, whereas in sample 6194, the template slippage and sequence repetition was found to be at the breakpoint in intron 47. (Red horizontal braces) Corresponding deletion, with the deleted exons mentioned. The crossed-out origins represent replication origins that probably failed to fire during S phase.
To exemplify, in case 5024 (exon 3–7 deletion), template slippage events were observed at the breakpoint in intron 7 as shown in Figure 4, strongly indicating that the replication fork was progressing toward exon 7. Similarly, the replication fork direction in the other five cases was also deduced, represented by blue arrowheads for each deletion case in the figure (Fig. 5). Furthermore, comparison of the assessed replication fork directions in cases 5904 and 9250 suggests that the replisomes have progressed past the termination region (TER) located between exons 12 and 17 and continued replication. The observed progression of replisomes past TER is perhaps an attempt to rescue the template that remained unreplicated due to failure of the origin lying between exons 28 and 29 to fire (Fig. 5). Such rescue of unreplicated template by replisomes approaching from adjacent origins has been reported in yeast (Kitsberg et al. 1993; Doksani et al. 2009). We believe that these deletion events might have occurred during meiotic replication in germ cells, whereby any unreplicated template ends up as a deletion in the haploid gamete cells formed. More convincingly, the paucity of initiation events and the failure of origin firing have been implicated in the fragility of the common FRA3B fragile site in human lymphoblasts (Letessier et al. 2011). The observed fragility of the FRA3B site in lymphoblasts compared with fibroblasts was attributed to cell-type specificity in the number of origins participating in replication. Although our study involves germline deletion events, our observations support the findings of Letessier et al. Similarly, a different study involved the PARK2 and DMD loci, where factors affecting replication timing were reported to contribute to genomic rearrangements equally in both germ cell and somatic cell lines (Mitsui et al. 2010). Furthermore, since DMD is a late replicating gene, it has less time to recover from any stress and complete replication prior to the end of S phase and subsequent chromosome condensation, thus contributing to the high frequency of unrecovered deletion events seen (McAvoy et al. 2007).
Aberrant origin firing and incomplete rescue of replication leads to DNA deletion
Based on our current findings of template slippages and fork reversals, we propose that the observed deletions are a result of aberrant firing of replication origins, coupled with incomplete rescue of replication as found in yeast studies (Branzei and Foiani 2010). Deletions observed in 1148 (exon 10–44 deletion) and 5904 (exon 20–44 deletion) could be due to failure of firing of the two origins between exons 28 and 29 and between exons 43 and 44, as shown in Figure 5 (failed origins are crossed out in the figure). On the other hand, in patients 5024 (exon 3–7 deletion), 5078 (exon 18 deletion), 6194 (exon 48–50 deletion), and 9250 (exon 8–9 deletion), firing of one or more origins appears to have been delayed (origins shown in braces in the figure). The nonrecurrence or the difference in deletion sizes is perhaps explained by the extent of rescue (by adjacent replisomes), which might be dependent on a variety of factors. Hence the observed smaller deletion in 1148 (exon 20–44) compared with that in 5904 (exon 10–44) is perhaps due to a greater rescue of replication. Convincingly enough, TERs have been shown to be polar in yeast, thus stalling the replisome in one direction (nonpermissive end), while allowing it to pass and continue replication in the other direction (permissive end) (Alver and Bielinsky 2010). Therefore, successful rescue of replication is perhaps dependent on such polarity of adjacent TERs, as well. Comparing 1148 and 5904, it appears that the TER between exons 12 and 17 allows the adjacent replisome approaching from the origin in intron 7 to pass and replicate the template of the failed origin between exons 28 and 29 (Fig. 5). However, fork reversal across the TER in 9250 suggests that perhaps this very TER between exon 12 and 17 is not polar at all or is transiently polar, allowing replisomes to pass in either direction. Therefore, at least in humans, both polar and nonpolar TERs may exist. Alternatively, polarity of TERs holding a replication fork at its nonpermissive end could also be transient, persisting only over a period of time during which, if the adjacent fork does not arrive, the polarity may disappear, allowing the held replisome to pass along and replicate the downstream template. Although these alternate explanations need to be explored further, our present study is the first instance in which evidence for such polarity of TERs has been discussed in humans. Nevertheless, additional studies are required to understand the nature of the polarity of TERs.
The two closely spaced deletions in sample 4551 (exon 10–12 deletion) involving the IVD gene can be explained by delayed firing of the associated origin, resulting in incomplete replication. Interestingly enough, a recent investigation of evolutionary breakpoints in yeast has provided strong evidence for a correlation of replication origins and rearrangement breakpoints irrespective of local genomic architecture (Di Rienzi et al. 2009). In particular, deletion hotspots (between exons 42 and 45) in DMD have been shown to coincide with replication origins (Verbovaia and Razin 1997; Gualandi et al. 2006). Additionally, replication stress hindering origin firing has been shown to induce microdeletions and pathogenic CNVs at the FHIT/FRA3B locus, as well as genomewide (Durkin et al. 2008; Arlt et al. 2009). The template slippages observed at the breakpoints could be an effect of the replication stress undergone by the progressing replisomes while rescuing the otherwise unreplicated template, or they could be a result of the unusual termination of replication occurring at a region other than the TER, because of the final checkpoint activation or cell phase change. Alternatively, these could result from the negative torque accumulated by the replication forks traveling distances longer than usual (past TER), coupled with the long stretches of unwound templates lying ahead due to failure of active origins associated with them.
The dependence of successful replication of DMD (2.4 Mb long) on several active origins and termination junctions, logically because of its size, perhaps explains why there are more intragenic deletions in DMD than any other known disease-causing genes, which are generally much smaller and might have either one or even no intragenic origins. On the contrary, failure of any single origin involving replication of such smaller genes may result in multigenic deletions, rather than intragenic ones.
Aberrant origin firing: A plausible explanation for duplication CNVs
Aberrant replication origin firing may also explain chromosomal duplications as the result of occasional firing of the otherwise dormant or passively replicated origins in response to replication stress or topological changes in the genomic architecture. Firing of dormant origins may cause re-replication of the DNA template, leading to DNA duplications. Plasticity of origin selection during replication stress has been shown to potentiate dormant origin firing and subsequent duplications (Anglana et al. 2003). Convincingly, genotoxic stress conditions in the mouse immunoglobulin heavy chain (IgH) locus or random DSBs in yeast have been shown to induce firing of dormant origins and replication of template, resulting in genomic instability and tumorigenesis (Doksani et al. 2009; Masai et al. 2010; Borowiec and Schildkraut 2011). Studies in fission yeast suggest that such dormant origins are present along the genome at a high frequency of about one every 10 kb, and a proportionately similar frequency could be expected in the human genome, as well (Hayashi et al. 2007). Mammalian replicons are believed to be 30–450 kb in size with the most frequent sizes being 75–150 kb (Berezney et al. 2000). The frequent large sizes of observed duplications (mostly >50 kb as seen in DMD) (A Ankala, unpubl.) that are almost the size of one complete replicon or two (170–500 kb in DMD) further supports this hypothesis.
We believe that, as suggested by Mitsui and others, microhomology at CNV breakpoints may be attributed to a repair mechanism, such as microhomology-mediated end joining (MMEJ) rather than a recombination mechanism (Verkaik et al. 2002; Mitsui et al. 2010). Finally, we provide a new direction for understanding the various genomic rearrangements in the human genome based on aberrant firing of active and dormant origins of replication. We believe our hypothesis can be extended to explain intergenic deletions and duplications as well as complex rearrangements in the human genome that cannot be explained by other established mechanisms, such as NAHR, NHEJ, and MMRDR.
Methods
Samples
From 2007 through 2010, a total of 513 samples for various known and unknown genetic disorders were referred to the Emory Genetics Laboratory for evaluation for the presence of deletions or duplications. The samples were used for research, with informed consent approved by the Institutional Review Board for Human Subject Research at Emory University School of Medicine.
Ultra-high-resolution gene level array comparative genome hybridization (aCGH) design
Gene-targeted high-resolution oligonucleotide CGH array was custom-designed on a NimbleGen 385K platform (till June 2010) or OGT 44K platform (July 2010 onward) to detect deletions and duplications in 450 genes associated with various genetic disorders. The NimbleGen 385K platform used long oligonucleotides (45–60-mer) to achieve isothermal Tm across the array, with repeat sequence masking implemented to ensure greater sensitivity and specificity. The OGT 44K platform has 44,000 unique sequence probes tiled on the array. Both arrays were designed with average spacing of 10 bp within coding regions and 25 bp within promoter, intronic regions, and 3′ UTR, with repeat sequence masking. Use of intronic oligonucleotide probes allows robust detection of dosage changes of the gene within the entire genomic region, as well as determination of approximate breakpoints.
aCGH protocol and analysis
DNA was extracted from patient samples using the Puregene DNA Extraction Kit (Gentra Systems) according to the manufacturer's instructions. Male and female wild-type control DNA was obtained from Promega. Each patient and reference DNA sample was sonicated for 500–2000 base fragments. Patient and reference DNA samples were labeled using Klenow enzyme (NEB) and Cy3 or Cy5 9-mer wobble primers (TriLink BioTechnologies), respectively. After labeling, each sample was purified by isopropanol precipitation and reconstituted in ultra-pure water. To the reference DNA, 13 μg of labeled patient DNA was added, and the products were desiccated in a vacufuge (Savant DNA 120) and then resuspended in appropriate hybridization buffer, along with Cy3 and Cy5 control CPK6 50-mer oligonucleotides. This mixture was hybridized to the array for 16–20 h at 42°C in a MAUI Hybridization System (BioMicro Systems). Arrays were then washed according to the manufacturer's recommendation and immediately scanned on a GenePix 4000 scanner (Molecular Devices). After scanning, we extracted data from images and achieved within-array normalization using the manufacturer-provided software (NimbleScan) and CytoSure. Normalized log2 ratio data were analyzed using two different analysis programs: (SegMNT or DNA copy) NimbleScan (NimbleGen Systems, Inc.) and GLAD (Hupe et al. 2004). Both software programs report breakpoints for predicted deletions or duplications in the patient (or test) sample relative to the reference and also display results graphically in a bar graph, where the y-axis indicates gain or loss of material (1 = gain, 0 = normal, −1 = loss), while the x-axis indicates the position of each feature on the chromosome.
Multiplex PCR analysis and breakpoint mapping
Approximate breakpoint coordinates for the various patient samples were acquired from the array results mentioned above. Gene sequences covering 1000 bp upstream of the 5′ breakpoint and 1000 bp downstream from the 3′ breakpoint were downloaded from the UCSC Genome Browser. A combination of three sets of primers spaced 250 bp from each other was used in a multiplex PCR, and the shortest product obtained (absent in control DNA) was sequenced. To achieve combined efficiency and target multiple amplicons in one single reaction, a multiplex PCR was performed in which three sets of forward primers and reverse primers were combined in the same reaction for each single patient sample. Based on the position of the primer, whether it is intact or deleted, single or multiple products were observed on agarose gel electrophoresis. Assessing the size of the amplicon, the appropriate set of primers involved in its amplification was chosen and then sequenced using ABI 3730. These sequences were BLATted in the UCSC Genome Browser, and the breakpoint junction sequences and coordinates were noted. The sequences surrounding the breakpoints were aligned and microhomology sequences noted (Supplemental Data S3). All those samples that failed with initial multiplex PCR were further checked with long-range PCR using a TAKARA kit, and then walked in with a new set of primers. Samples that failed long-range PCR were considered to have complex rearrangements and were not analyzed further. Reference human sequences were obtained from the UCSC Genome Browser (http://genome.ucsc.edu). The ABI2FASTA converter (available online) was used to extract FASTA sequence files from ABI output files (http://www.dnabaser.com).
Custom-developed script to detect repetitive elements
We checked for the presence of repetitive elements and microsatellites using a custom-written Perl script that queries the UCSC database (hg18 build). We queried for nested Repeats, simple Repeats, exapted Repeats, and microsat tables to look for interrupted repeats, simple repeats, exapted repeats, and microsatellites, respectively.
Acknowledgments
This work was supported, in part, by NIH grant 1RC1NS 069541-01 and MDA grant MDA138896 to M.R.H.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.123463.111.
References
- Alver RC, Bielinsky AK 2010. Termination at stop2. Mol Cell 39: 487–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M 2003. Dynamics of DNA replication in mammalian somatic cells: Nucleotide pool modulates origin choice and interorigin spacing. Cell 114: 385–394 [DOI] [PubMed] [Google Scholar]
- Arlt MF, Mulle JG, Schaibley VM, Ragland RL, Durkin SG, Warren ST, Glover TW 2009. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am J Hum Genet 84: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484 [DOI] [PubMed] [Google Scholar]
- Blow JJ, Dutta A 2005. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec JA, Schildkraut CL 2011. Open sesame: Activating dormant replication origins in the mouse immunoglobulin heavy chain (igh) locus. Curr Opin Cell Biol 23: 284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M 2010. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN 2005a. Complex gene rearrangements caused by serial replication slippage. Hum Mutat 26: 125–134 [DOI] [PubMed] [Google Scholar]
- Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN 2005b. Intrachromosomal serial replication slippage in trans gives rise to diverse genomic rearrangements involving inversions. Hum Mutat 26: 362–373 [DOI] [PubMed] [Google Scholar]
- Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN 2005c. Meta-analysis of gross insertions causing human genetic disease: Novel mutational mechanisms and the role of replication slippage. Hum Mutat 25: 207–221 [DOI] [PubMed] [Google Scholar]
- Chen JM, Cooper DN, Ferec C, Kehrer-Sawatzki H, Patrinos GP 2010. Genomic rearrangements in inherited disease and cancer. Semin Cancer Biol 20: 222–233 [DOI] [PubMed] [Google Scholar]
- Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, Hurles ME 2010. Mutation spectrum revealed by breakpoint sequencing of human germline cnvs. Nat Genet 42: 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M, Toledo F, Anglana M 2004. Replication initiation in mammalian cells: Changing preferences. Cell Cycle 3: 19–21 [PubMed] [Google Scholar]
- Den Dunnen JT, Grootscholten PM, Bakker E, Blonden LAJ, Ginjaar HB, Wapenaar MC, Van Paassen HMB, Van Broeckhoven C, Pearson PL, Van Ommen GJB 1989. Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and complementary DNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet 45: 835–847 [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, Collingwood D, Raghuraman MK, Brewer BJ 2009. Fragile genomic sites are associated with origins of replication. Genome Biol Evol 1: 350–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, Bermejo R, Fiorani S, Haber JE, Foiani M 2009. Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell 137: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SG, Ragland RL, Arlt MF, Mulle JG, Warren ST, Glover TW 2008. Replication stress induces tumor-like microdeletions in fhit/fra3b. Proc Natl Acad Sci 105: 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualandi F, Rimessi P, Trabanelli C, Spitali P, Neri M, Patarnello T, Angelini C, Yau SC, Abbs S, Muntoni F, et al. 2006. Intronic breakpoint definition and transcription analysis in DMD/BMD patients with deletion/duplication at the 5′ mutation hot spot of the dystrophin gene. Gene 370: 26–33 [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR 2009a. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327 doi: 10.1371/journal.pgen.1000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G 2009b. Mechanisms of change in gene copy number. Nat Rev Genet 10: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Katou Y, Itoh T, Tazumi A, Yamada Y, Takahashi T, Nakagawa T, Shirahige K, Masukata H 2007. Genome-wide localization of pre-rc sites and identification of replication origins in fission yeast. EMBO J 26: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Marians KJ 2006. Replication fork reactivation downstream of a blocked nascent leading strand. Nature 439: 557–562 [DOI] [PubMed] [Google Scholar]
- Hupe P, Stransky N, Thiery JP, Radvanyi F, Barillot E 2004. Analysis of array cgh data: From signal ratio to gain and loss of DNA regions. Bioinformatics 20: 3413–3422 [DOI] [PubMed] [Google Scholar]
- Inoue K, Osaka H, Thurston VC, Clarke JTR, Yoneyama A, Rosenbarker L, Bird TD, Hodes ME, Shaffer LG, Lupski JR 2002. Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am J Hum Genet 71: 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, Malig M, Kallicki J, Kaul R, Wilson RK, Eichler EE 2010. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143: 837–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsberg D, Selig S, Keshet I, Cedar H 1993. Replication structure of the human β-globin gene domain. Nature 366: 588–590 [DOI] [PubMed] [Google Scholar]
- Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM 1987. Complete cloning of the Duchenne muscular-dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 50: 509–517 [DOI] [PubMed] [Google Scholar]
- Koszul R, Caburet S, Dujon B, Fischer G 2004. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J 23: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CMB, Lupski JR 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131: 1235–1247 [DOI] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M 2011. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470: 120–123 [DOI] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21: 15–27 [DOI] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You ZY, Yoshizawa-Sugata N, Oda M 2010. Eukaryotic chromosome DNA replication: Where, when and how? Annu Rev Biochem 79: 89–130 [DOI] [PubMed] [Google Scholar]
- McAvoy S, Ganapathiraju S, Perez DS, James CD, Smith DI 2007. DMD and IL1RAPL1: Two large adjacent genes localized within a common fragile site (FRAXC) have reduced expression in cultured brain tumors. Cytogenet Genome Res 119: 196–203 [DOI] [PubMed] [Google Scholar]
- Mefford HC, Shafer N, Antonacci F, Tsai JM, Park SS, Hing AV, Rieder MJ, Smyth MD, Speltz ML, Eichler EE, et al. 2010. Copy number variation analysis in single-suture craniosynostosis: Multiple rare variants including RUNX2 duplication in two cousins with metopic craniosynostosis. Am J Med Genet A 152: 2203–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui J, Takahashi Y, Goto J, Tomiyama H, Ishikawa S, Yoshino H, Minami N, Smith DI, Lesage S, Aburatani H, et al. 2010. Mechanisms of genomic instabilities underlying two common fragile-site-associated loci, PARK2 and DMD, in germ cell and cancer cell lines. Am J Hum Genet 87: 75–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T, Tanaka TU 2010. Spatial regulation and organization of DNA replication within the nucleus. Chromosome Res 18: 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C, Galvagni F, Marchi J, Roberts R, Vitiello L 1995. Genomic organization of the human dystrophin gene across the major deletion hot-spot and the 3′-region. Genomics 28: 97–100 [DOI] [PubMed] [Google Scholar]
- Oshima J, Magner DB, Lee JA, Breman AM, Schmitt ES, White LD, Crowe CA, Merrill M, Jayakar P, Rajadhyaksha A, et al. 2009. Regional genomic instability predisposes to complex dystrophin gene rearrangements. Hum Genet 126: 411–423 [DOI] [PubMed] [Google Scholar]
- Oudet C, Hanauer A, Clemens P, Caskey T, Mandel J-L 1992. Two hot spots of recombination in the DMD gene correlate with the deletion prone regions. Hum Mol Genet 1: 599–603 [DOI] [PubMed] [Google Scholar]
- Raedt TD, Stephens M, Heyns I, Brems H, Thijs D, Messiaen L, Stephens K, Lazaro C, Wimmer K, Kehrer-Sawatzki H, et al. 2006. Conservation of hotspots for recombination in low-copy repeats associated with the NF1 microdeletion. Nat Genet 38: 1419–1423 [DOI] [PubMed] [Google Scholar]
- Rosenfeld JA, Ballif BC, Torchia BS, Sahoo T, Ravnan JB, Schultz R, Lamb A, Bejjani BA, Shaffer LG 2010. Copy number variations associated with autism spectrum disorders contribute to a spectrum of neurodevelopmental disorders. Genet Med 12: 694–702 [DOI] [PubMed] [Google Scholar]
- Roth DB, Porter TN, Wilson JH 1985. Mechanisms of nonhomologous recombination in mammalian-cells. Mol Cell Biol 5: 2599–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR 2006. Well-characterized rearrangement-based diseases and genome structural features at the locus. In Genomic disorders: The genomic basis of disease, pp. 403–406 Humana Press, Totowa, NJ [Google Scholar]
- Verbovaia LV, Razin SV 1997. Mapping of replication origins and termination sites in the Duchenne muscular dystrophy gene. Genomics 45: 24–30 [DOI] [PubMed] [Google Scholar]
- Verkaik NS, Esveldt-van Lange RE, van Heemst D, Bruggenwirth HT, Hoeijmakers JH, Zdzienicka MZ, van Gent DC 2002. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur J Immunol 32: 701–709 [DOI] [PubMed] [Google Scholar]
- Vissers LE, Bhatt SS, Janssen IM, Xia Z, Lalani SR, Pfundt R, Derwinska K, de Vries BB, Gilissen C, Hoischen A, et al. 2009. Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum Mol Genet 18: 3579–3593 [DOI] [PubMed] [Google Scholar]
- Weterings E, van Gent DC 2004. The mechanism of non-homologous end-joining: A synopsis of synapsis. DNA Repair (Amst) 3: 1425–1435 [DOI] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR 2009. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet 41: 849–853 [DOI] [PMC free article] [PubMed] [Google Scholar]