Abstract
In Drosophila, microRNAs (miRNAs) typically guide Argonaute1 to repress mRNA, whereas small interfering RNAs (siRNAs) guide Argonaute2 to destroy viral and transposon RNA. Unlike siRNAs, miRNAs rarely base pair extensively to the mRNAs they regulate. We find that extensive complementarity between a target RNA and an Argonaute1-bound miRNA triggers miRNA tailing and 3′-to-5′ trimming. In flies, Argonaute2-bound small RNAs—but not those bound to Argonaute1—bear a 2′-O-methyl group at their 3′ ends. This modification blocks target-directed small RNA remodeling: in flies lacking Hen1, the enzyme that adds the 2′-O-methyl group, Argonaute2-associated siRNAs are tailed and trimmed. Target-complementarity also affects small RNA stability in human cells. These results provide an explanation for the partial complementarity between animal miRNAs and their targets.
In Drosophila melanogaster, different pathways produce siRNAs and miRNAs (1). Fly siRNAs associate with Argonaute2 (Ago2) to direct cleavage of target RNAs with extensive complementarity to the siRNA guide (RNA interference; RNAi), whereas miRNAs typically act with Argonaute1 (Ago1) to decrease the translation and stability of partially complementary mRNAs (2–5). In plants, both siRNAs and miRNAs bind target mRNAs through extensive base pairing across the entire small RNA guide (6). The reason for the difference between animals and plants in the degree of complementarity between miRNAs and their targets remains enigmatic.
In flies, a key step in the production of a functional siRNA-Ago2 complex, but not a miRNA-Ago1 complex, is the addition of a 2′-O-methyl group to the 3′ end of the small RNA by Hen1 (7–9), an S-adenosylmethionine-dependent methyltransferase (10). Plant Hen1 protects siRNAs and miRNAs alike from 3′-terminal uridylation and degradation (10–12). The function of siRNA methylation in flies is unknown.
miRNA sensors affect steady-state miRNA levels
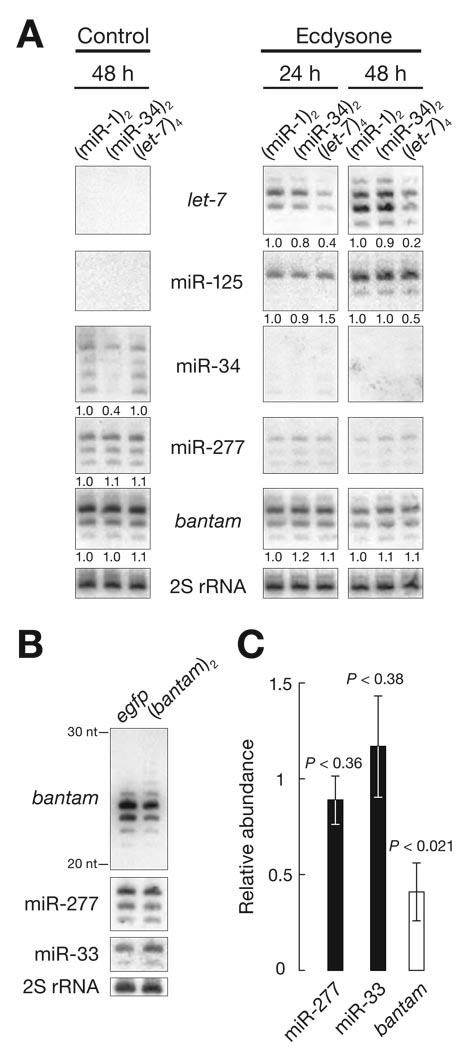
In a set of Drosophila cell lines that expressed reporter transgenes with one or more highly complementary sites for miRNAs (“miRNA sensors”) we noted that the steady-state abundance of the corresponding miRNA was decreased (Fig. 1).
Fig. 1.
Target-directed destabilization of miRNAs. (A) Northern analysis of total RNA from S2 cells stably expressing egfp mRNA bearing in its 3′ UTR two target sites for miR-1 [(miR-1)2] or miR-34 [(miR-34)2] or four target sites for let-7 [(let-7)4]. Time of ecdysone (right panels) or control treatment (left panel) is indicated. Relative levels of the miRNAs are indicated below the lanes. (B) Representative Northern analysis of total RNA from a clonal S2 cell line expressing egfp mRNA bearing in its 3′ UTR two target sites for bantam [(bantam)2] and a clonal control cell line expressing sole egfp mRNA. (C) Mean ± standard deviation for three biologically independent replicates of the experiment in (B). Levels of each miRNA were first normalized to the 2S rRNA loading control.
For example, a stably transformed S2 cell line constitutively expressing egfp mRNA bearing in its 3′ untranslated region (UTR) two sites fully complementary to miR-34 showed a decrease in miR-34, but not other miRNAs, relative to control egfp cell lines (Fig. 1A and fig. S1). Treatment of S2 cells with ecdysone induces the expression of let-7 and miR-125, among other miRNAs (13). After ecdysone treatment, the steady-state abundance of let-7, but not miR-125, which resides in the same primary transcript (fig. S1), was lower at both 24 and 48 h after induction in a stable S2 cell line expressing egfp bearing in its 3′ UTR four sites fully complementary to let-7, compared to cell lines expressing egfp with two sites for miR-1 or miR-34 (Fig. 1A). Similarly, a clonal cell line expressing an egfp mRNA bearing two sites complementary to the miRNA, bantam, showed reduced levels of bantam (P < 0.001) but not of miR-277 (P < 0.26) or miR-33 (P < 0.38), compared to a clonal cell line expressing egfp without miRNA-complementary sites (Fig. 1B,C, fig. S2 and SOM text). Transient expression of a primary RNA containing both let-7 and miR-1 hairpins in clonal reporter lines producing egfp with sites complementary to miR-1 or let-7 decreased only the miRNA that can bind the complementary target (fig. S3 and SOM text). Finally, miR-34 levels were significantly reduced (P < 0.01) in four independently derived, clonal reporter cell lines expressing egfp bearing two miR-34 complementary sites (fig. S4 and SOM text). Importantly, the expression of mRNAs containing sites perfectly complementary to a miRNA does not influence the detection of those miRNAs by Northern hybridization (fig. S5 and SOM text).
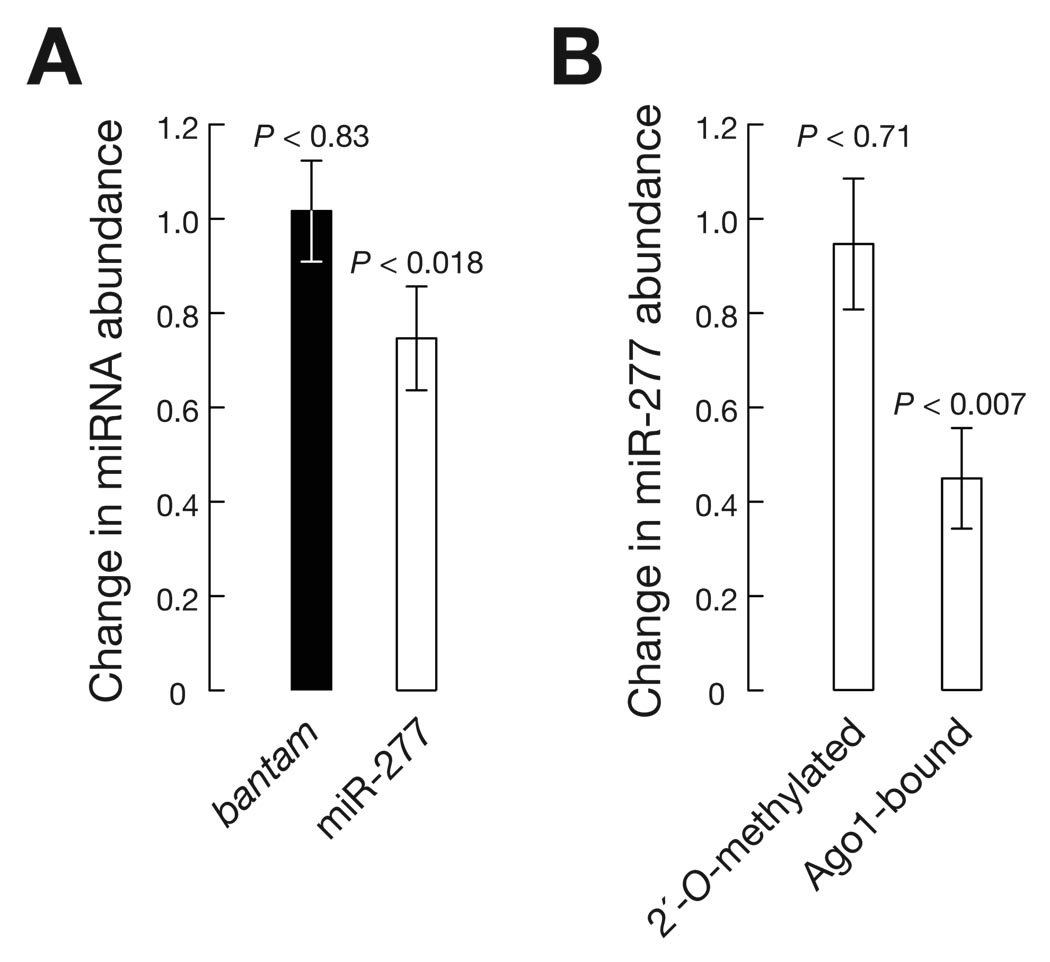
Target-dependent destabilization of miR-277
Drosophila siRNAs and miRNAs partition between Ago1 and Ago2 according to their duplex structure (14–17). Consequently, some miRNAs, including miR-277, partition into both Ago1 and Ago2 (18). Expression of egfp bearing two sites fully complementary to miR-277 caused a small but significant reduction in the abundance of that miRNA (P < 0.018) when compared to an egfp-expressing control cell line (Fig. 2A). We used Ago1-immunoprecipitation as well as oxidation with NaIO4 followed by β-elimination to distinguish between Ago1- and Ago2-loaded miR-277 (Fig. 2B and fig. S6) (8). Ago2-loaded small RNAs bear 2′-O-methyl modified 3′ termini, making them refractory to oxidation; Ago1-loaded miRNAs bear 2′,3′ hydroxy 3′ termini, so that oxidation followed by β-elimination removes their final nucleotide, making them one-nucleotide shorter. Relative to a control reporter, the miR-277–complementary reporter had a significant effect on Ago1-(P < 0.007) but not Ago2-associated miR-277 (P < 0.71; Fig. 2B, fig. S6 and SOM text). This is consistent with earlier observations that Ago2 but not Ago1 silences an egfp reporter with target sites perfectly complementary to miR-277 (18).
Fig. 2.
Target-dependent destabilization of miR-277. (A) Change in miR-277 or bantam abundance in total RNA from a clonal S2 cell line expressing egfp mRNA bearing in its 3´ UTR two target sites for miR-277 [(miR-277)2], relative to a clonal control cell line expressing sole egfp mRNA. Prior to comparison, miRNA levels were normalized to the 2S rRNA loading control. (B) Change in 2′-O-methylated (estimated by resistance to oxidation and β-elimination) and Ago1-associated miR-277 (determined by immunoprecipitation) (see SOM text and fig. S6 for details). In all experiments at least two technical replicates of three biologically independent measurements were used to determine mean ± standard deviation.
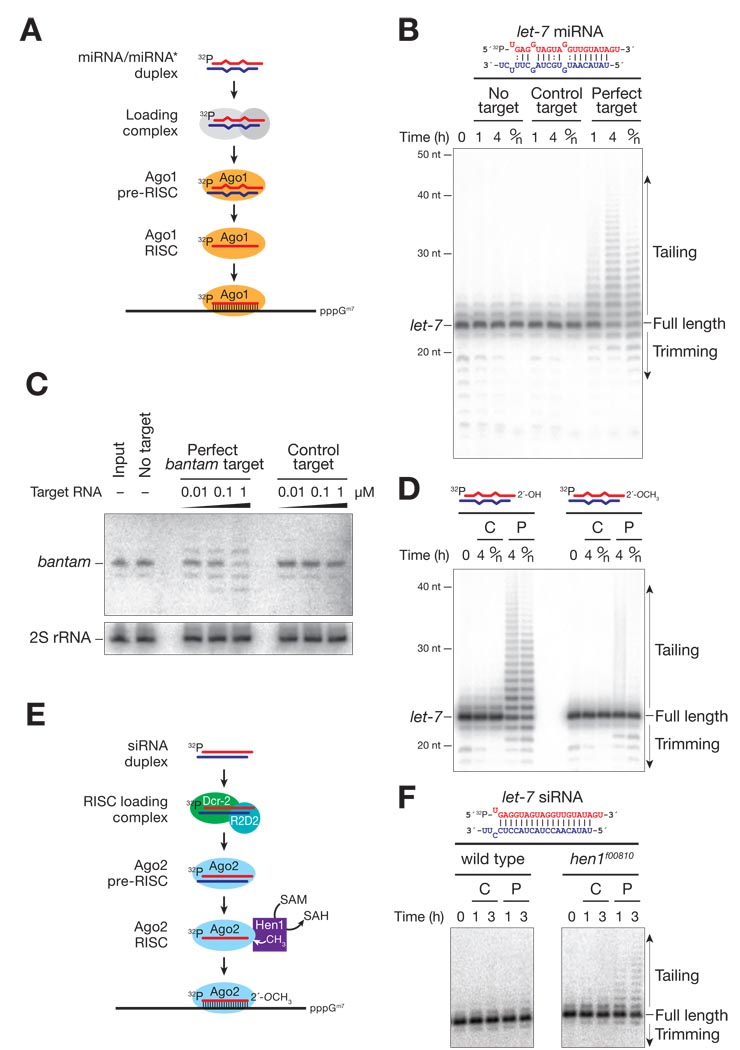
Target RNA-dependent remodeling of small RNAs in vitro
To decipher the mechanism by which a complementary target RNA triggers miRNA destabilization, we followed the fate of a miRNA in the presence of a perfectly complementary target RNA. A small RNA duplex comprising 5′ 32P-radiolabeled let-7 bound to let-7* was assembled into Ago1 in Drosophila embryo lysate (Fig. 3A). When a fully complementary, 7-methyl guanosine capped, target RNA was added to the lysate after loading let-7 into Ago1, the amount of intact let-7 declined and new longer (“tailed”) and shorter (“trimmed”) forms of let-7 appeared; let-7 was unaffected by addition of a control target (Fig. 3B and fig. S7). Similar results were obtained for a miRNA with different sequence (fig. S8). The added nucleotides in the tailed let-7 species were adenines (Table S1).
Fig. 3.
Terminal 2′-O-methyl modification and Argonaute protein identity control small RNA stability. (A) The in vitro miRNA stability assay used in (B) and (D). (B) In the presence of a fully complementary target RNA, but not an unrelated control RNA, let-7 was tailed and trimmed. (C) Endogenous bantam miRNA was tailed and trimmed when Drosophila embryo extract was incubated overnight with a fully complementary target RNA, but not a control target. bantam and 2S rRNA were detected by Northern blotting. (D) 2′-O-methyl modification of an Ago1-loaded miRNA blocked tailing and trimming. (E) The in vitro siRNA stability assay used in (F). (F) A fully complementary target RNA directed tailing and trimming of an Ago2-loaded siRNA in the absence of Hen1. C, control target RNA; P, perfectly complementary target RNA; o/n, overnight.
Lysates from 0–2 h old Drosophila embryos contain endogenous bantam miRNA loaded into Ago1 (4). Incubation of these lysates with an in vitro transcribed, 7-methyl guanosine capped, target RNA containing one site fully complementary to bantam triggered tailing and trimming of bantam, whereas a control target did not alter bantam length or abundance (Fig. 3C). Thus, fully complementary target RNAs triggered tailing and trimming in vitro for both an exogenous miRNA assembled into Ago1 in vitro (let-7 and bantam), and an endogenous miRNA (bantam) assembled in vivo, before the lysate was prepared.
To determine if 3′ terminal 2′-O-methylation—a modification found on Ago2- but not Ago1-associated small RNAs (8)—influences target-directed miRNA tailing or trimming, we assembled let-7 bearing a 2′-O-methyl modified 3′ end into Ago1 in embryo lysate using a let-7/let-7* duplex; the 2′-O-methyl modified let-7 was loaded normally into Ago1 (fig. S9). The presence of a methyl group at the 2′ position of the 3′ terminal nucleotide blocked tailing and trimming of Ago1-bound let-7 (Fig. 3D). A 3′ terminal, 3′ deoxy modification also inhibited target-directed effects (fig. S10). siRNAs, which load into Ago2 in embryo lysate, were not affected (Fig. 3E and F). However, in extracts prepared from embryos of homozygous mutant hen1f00810 flies (8, 9), the Ago2-bound siRNA became susceptible to target-dependent tailing and trimming (Fig. 3F). These data suggest that methylation of small RNAs by Hen1 renders them resistant to the small RNA modifying and trimming enzymes recruited by a small RNA-bound complementary target RNA.
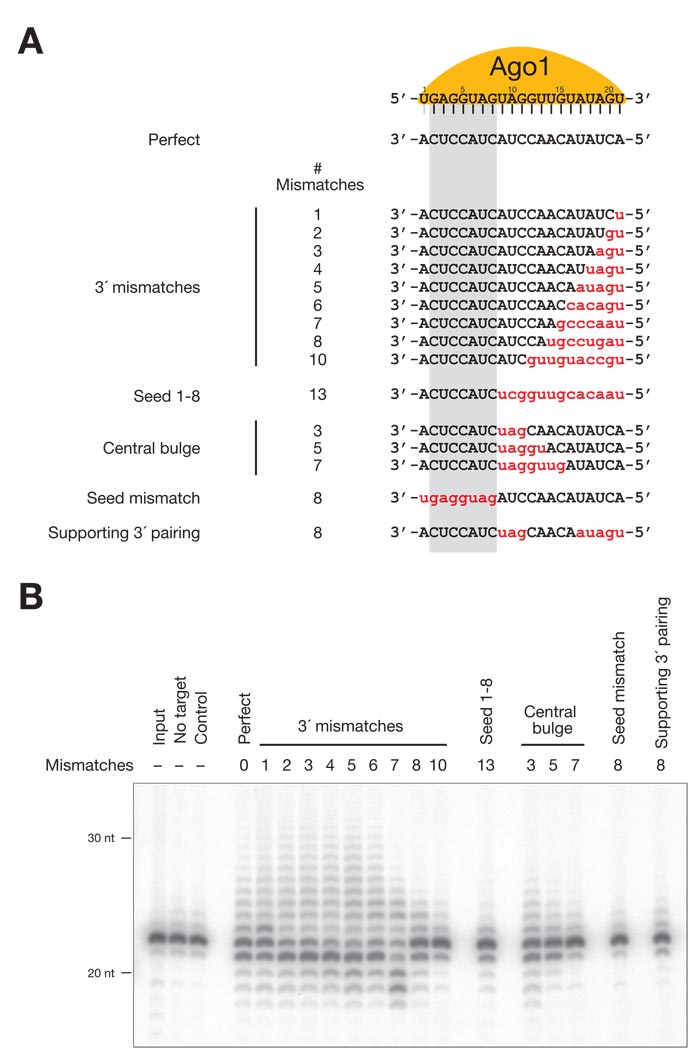
Sequence requirements for miRNA remodeling
Compared to plants, animal miRNAs are generally less complementary to their target sites (19). To determine the extent of complementarity required to elicit tailing and trimming, we analyzed a set of target RNAs with various different degrees of complementarity to a 32P-radiolabeled miRNA loaded into Ago1 in embryo lysate (Fig. 4A). Target sites that resembled the majority of natural miRNA binding sites in animals—including sites paired only to the miRNA seed region or to both the seed and a small patch of 3′ sequence identified as a miRNA-binding determinant (19)—did not trigger tailing and trimming (Fig. 4B and fig. S11). Targets with eight or fewer mismatches with the 3′ end of the miRNA triggered tailing and trimming (Fig. 4A, B and fig. S11). Moreover, a small central bulge of three nucleotides, flanked by fully complementary sequence, triggered significant tailing and trimming, but five- or seven-nucleotide central bulges did not. We conclude that in vitro, target RNA-directed tailing and trimming of a miRNA requires extensive but not complete complementarity, whereas the canonical animal miRNA binding sites described previously do not alter miRNA stability.
Fig. 4.
Target sequence requirements for miRNA tailing and trimming. (A) Target RNAs assayed for their ability to direct tailing and trimming in vitro. Mismatches are indicated with red, lower case letters. The miRNA seed sequence is highlighted in gray. (B) Seed pairing plus extensive central and 3′ pairing between the miRNA and the target was required for efficient tailing and trimming. The miRNA duplex was assembled into Ago1 by incubation in Drosophila embryo lysate, then the target RNA was added and the reaction incubated overnight.
Hen1 protects Ago2-bound small RNAs
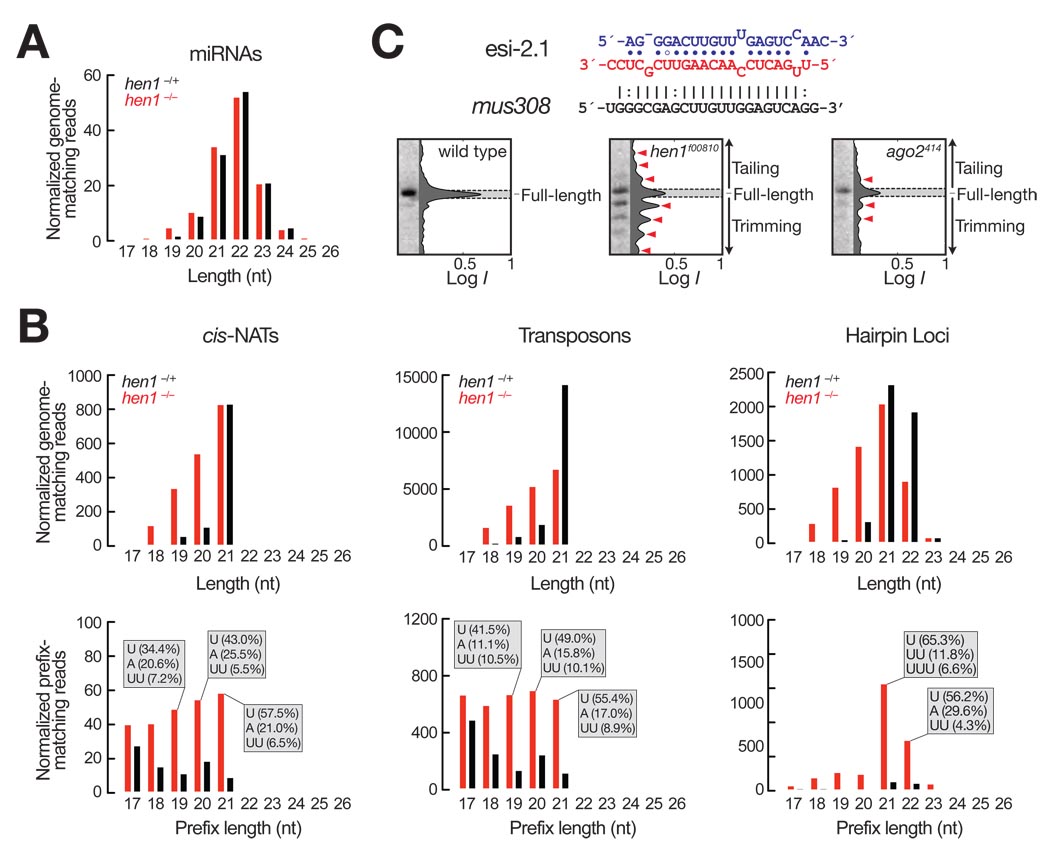
We sequenced 18–29 nt small RNAs from heterozygous or homozygous mutant hen1f00810 heads (Tables S2 and S3). Consistent with the idea that Hen1 does not act on Ago1-associated small RNAs, the absence of Hen1 in the mutant flies altered neither the abundance nor the length of most miRNAs (Fig. 5A). The most abundant class of Ago2-bound small RNAs are endogenous siRNAs (endo-siRNAs), and they are fully or extensively complementary to cellular or transposon-derived mRNAs (20–23). The length heterogeneity of perfectly genome-matching endo-siRNA reads increased in hen1f00810 mutant heads, compared to the heads of their heterozygous siblings (Fig. 5B, upper panel and fig. S15). In contrast, endo-siRNA reads that contain 3′non-templated nucleotide additions (prefix reads) increased in the hen1 mutant heads (Fig. 5B lower panel). The sequences of prefix reads correspond to the reference fly genome for their first 15 or more nucleotides, then contain a short tail of 3′ nucleotides not found in genomic sequence. The majority of prefix reads contained a single 3′ uridine tail; the second most abundant nucleotide added was adenine. Longer tails comprising more than one non-genome matching addition were rare, but nearly always corresponded to homopolymeric stretches of uridines (Fig. 5B, lower panel). Such uridine tails are found on small RNAs in plants lacking Hen1 and are believed to tag siRNAs and miRNAs for destruction (12).
Fig. 5.
Small RNA tailing and trimming in vivo. (A) Length distribution of miRNAs from heterozygous (black) or homozygous (red) hen1f00810 fly heads. For each annotated pre-miRNA, reads were normalized to sequencing depth, then normalized miRNA reads for each distinct pre-miRNA were weighted equally to eliminate the influence of differences in transcriptional rates. (B) Length distribution of endo-siRNA sequence reads perfectly matching the fly genome (top panels) or matching only within a 5′ prefix (bottom panels) for heterozygous (black) or homozygous (red) hen1f00810 heads. The three classes of endo-siRNAs were analyzed separately: siRNAs derived from natural antisense transcripts (cis-NATs), from transposons, or from hairpin loci (esiRNAs). The most frequent non-genome matching nucleotide additions are indicated in the gray boxes as the percentage of all non-templated additions for specific prefix lengths. Reads are in parts per million (ppm). (C) The sequence of the esi-2.1 duplex and its cellular mRNA target, mus308. Northern analysis was used to detect esi-2.1 in total RNA from whole Oregon R (wild type), hen1f00810 or ago2414 mutant flies, and the absolute signal intensities (I, log10 scale) determined for each lane. Tailing and trimming products are marked with red arrowheads.
In vivo, endo-siRNAs from long hairpin transcripts (esiRNAs) associate predominantly with Ago2, but a minor fraction can be detected bound to Ago1 (21, 22, 24). In the absence of Hen1, esiRNAs generally became shorter (Fig. 5B). In particular, esi-2.1 (also termed hp-CG4068B and esiRNA-sl-1)—the most abundant hairpin-derived endo-siRNA—was less abundant, tailed, and trimmed in both hen1f00810 and ago2414 whole mutant flies, compared to wild-type (Fig. 5C and fig. S12). We speculate that loss of Hen1 and Ago2 have distinct consequences: In a hen1 mutant, Ago2-bound esi-2.1 becomes tailed and trimmed, because it no longer possesses a protective, 3′ terminal, 2′-O-methyl modification. Thus, for small RNAs such as esi-2.1, tailed and shortened species comprise both Ago1- and Ago2-loaded RNAs. In contrast, in an ago2 mutant, the normally Ago2-bound small RNAs no longer exist, so the only remaining tailed and shortened species must derive from Ago1-bound esiRNAs. High throughput sequencing data suggest that trimming of esi-2.1 occurs almost exclusively in the 3′ to 5′ direction (Table S4).
A model for small RNA tailing and trimming in Drosophila
Our data suggest a model for the influence of target RNA complementarity on small RNA abundance in Drosophila (fig. S13). miRNAs typically direct Ago1 to bind target RNAs and repress their translation and decrease their stability (25). Such binding is nearly always mediated by complementarity between the miRNA seed sequence and the target, with few additional base pairs tethering the two RNAs together. The presence of transcripts with extensive complementarity to Ago1-bound small RNAs results in small RNA remodeling, which our data suggest involves a terminal nucleotide transferase and a 3′-to-5′ exonuclease. In contrast, Ago2-associated small RNAs are 2′-O-methyl modified by Hen1 as a final step of Ago2 loading. The methyl group blocks tailing and trimming; in hen1 mutants, the unmethylated but Ago2-bound small RNAs are subject to target-directed tailing and trimming.
Homologs of the Hen1 methyltransferase are found in plants, fungi and animals (8–10, 26, 27). In plants, miRNAs and siRNAs alike are methylated by Hen1 and the absence of Hen1 causes small RNAs to be 3′ uridylated and destroyed. In mammals, Hen1 methylates germ line-specific PIWI-interacting RNAs (piRNAs), but not miRNAs (8, 9, 26, 28).
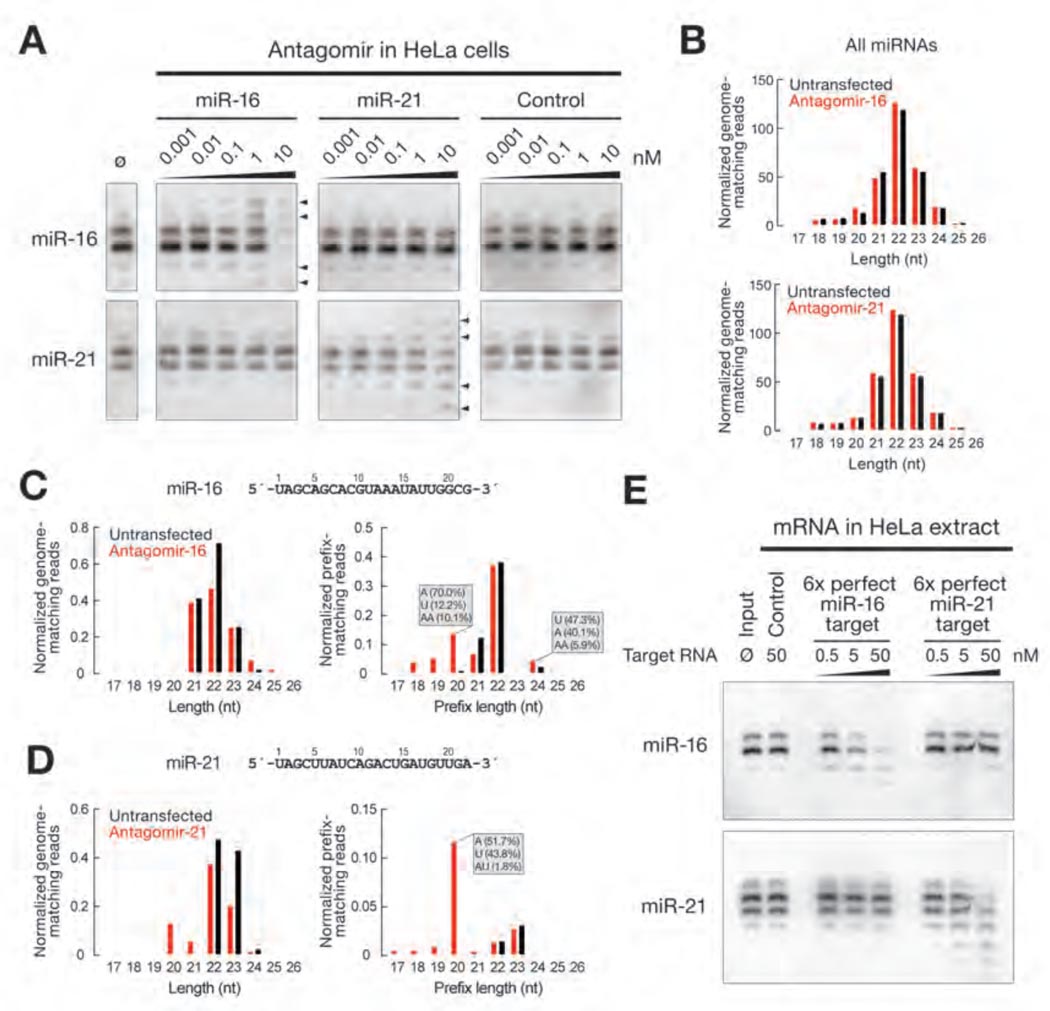
Stoffel and colleagues have previously described the sequence-specific degradation of miRNAs in response to synthetic, chemically modified RNA-analogs, “antagomirs”, designed to block miRNA function in vivo (29). In cultured HeLa cells, transfection of antagomirs to miR-16 or miR-21 triggered not only dose-dependent miRNA shortening, but also tailing (Fig. 6A and fig. S14). We sequenced 18–30 nt small RNAs from HeLa cells transfected with antagomirs against miR-16 or miR-21 and compared it to the small RNA profile of mock transfected HeLa cells (Tables S2 and S3). The profile of miRNA-matching reads in general did not change after transfection of either antagomir (Fig. 6B). However, we observed a sequence-specific decrease in full length miR-16 (Fig. 6C, left panel) and miR-21 (Fig. 6D, left panel) in response to treatment with the corresponding antagomir. Like tailing in flies (Fig. 5), decrease in genome-matching reads triggered by the antagomir was accompanied by an increased number of prefix reads for the antagomir-targeted, but not the control, miRNA (Fig. 6C and D, right panels and fig. S15). The non-templated nucleotides added to the 3′ end of the targeted miRNA were predominantly adenosine and uridine. As in Drosophila, target RNA-dependent trimming of small RNAs in HeLa cells was nearly always 3′to-5′ (Table S4). Moreover, we could recapitulate trimming of endogenous human miRNAs in HeLa cell cytoplasmic extract: incubation of the extract with in vitro transcribed, 7-methyl guanosine capped and poly-adenylated target mRNAs containing six sites fully complementary to miR-16 or miR-21 triggered shortening and loss of the corresponding miRNA (Fig. 6E). A control target did not alter miRNA length or abundance. These results suggest that target-dependent small RNA tailing and trimming is conserved between flies and mammals.
Fig. 6.
Target-dependent small RNA remodeling and destabilization in cultured human HeLa cells. (A) Northern analysis of total RNA from HeLa cells transfected with increasing amounts of 21 nt target RNA analogs (antagomirs) fully complementary to miR-16 or miR-21. Arrowheads mark tailed or trimmed miRNAs. (B) Total miRNA length distribution in mock transfected HeLa cells and in cells transfected with 1 nM antagomir targeting miR-16 (top) or 10 nM antagomir targeting miR-21 (bottom). (C) and (D) Small RNA profile from HeLa cells mock transfected (black) or transfected (red) with 1 nM antagomir targeting miR-16 (C) or 10 nM antagomir targeting miR-21 (D). Left panels: reads perfectly matching the miR-16-1 and miR-16-2 loci (C) or the miR-21 locus (D). Right panels: reads with prefixes matching miR-16-1 and miR-16-2 (C) or miR-21 (D). Non-genome matching nucleotides added to the 3′ end of the prefixes that most increased in response to antagomir transfection are shown (grey box). Reads are in ppm. (D) Endogenous miR-16 and miR-21 were destabilized when HeLa cytoplasmic extract was incubated with a fully complementary target mRNA, but not a control target. miR-16 and miR-21 were detected by Northern blotting.
Discussion
In flies, RNA methylation by Hen1 enables Ago2 to bind and cleave highly complementary target RNAs; the exclusion of Hen1 from the Ago1-loading pathway restricts Ago1-bound small RNAs to regulate only partially complementary targets. Thus, the methyl group deposited by Hen1 allows specialization of Drosophila Argonaute proteins. The finding that Ago1-associated small RNAs are sensitive to target-directed tailing and trimming is consistent with how Drosophila miRNAs bind their targets. In flies as in virtually all other animals, most predicted miRNA binding sites lack substantial pairing to the small RNA 3′ end (3, 19).
Even in hen1f00810 flies, small RNAs bound to Ago1 are more prone to target RNA-dependent tailing and trimming than those bound to Ago2 (Fig. 3). Ago1 is an inefficient ribonuclease whose catalytic rate is limited by the dissociation of its reaction products (18), whereas Ago2 is an efficient multiple-turnover enzyme (30). The ability of Ago2 to rapidly cleave its RNA targets may limit its susceptibility. In contrast, Ago1 likely resides on its target RNAs for longer than Ago2, making Ago1-bound small RNAs good substrates for target-directed tailing and trimming.
Uridylation of small RNAs as well as Ago2-cleaved, 5′ target RNA fragments have been linked to RNA turnover (12, 31–33). The molecular basis for the tailing and trimming of Argonaute-bound small RNAs is unknown. The ends of small RNAs are anchored to Argonaute proteins through binding of the small RNA 5′ phosphate to a pocket composed mainly of residues from the Mid domain and binding of the small RNA 3′ end to the PAZ domain (34–40). Access to the 3′ end of the small RNA likely requires dislodging it from the PAZ domain. Recent crystal structures of a eubacterial Argonaute protein confirm earlier suggestions that extensive pairing of the 3′ half of an siRNA with its target releases its 3′ end from the PAZ domain (41, 42). Our data are consistent with the idea that extensive 3′ pairing to a target RNA exposes a small RNA to nucleotidyl transferases and 3′-to-5′ exonuclease enzymes.
Supplementary Material
Footnotes
Supporting Online Material
Supporting Text and Discussion
Materials and Methods
Figure Legends S1 to S15
figs. S1 to S15
Tables S1 to S5
References
References and Notes
- 1.Ghildiyal M, Zamore PD. Nat Rev Genet. 2009;10:94. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Science. 2001;293:1146. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 3.Brennecke J, Stark A, Russell RB, Cohen SM. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okamura K, Ishizuka A, Siomi H, Siomi MC. Genes Dev. 2004;18:1655. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwasaki S, Kawamata T, Tomari Y. Mol Cell. 2009;34:58. doi: 10.1016/j.molcel.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 6.Rhoades MW, et al. Cell. 2002;110:513. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 7.Pelisson A, Sarot E, Payen-Groschene G, Bucheton A. J Virol. 2007;81:1951. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horwich MD, et al. Curr Biol. 2007;17:1265. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 9.Saito K, et al. Genes Dev. 2007;21:1603. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu B, et al. Science. 2005;307:932. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park W, Li J, Song R, Messing J, Chen X. Current Biology. 2002;12:1484. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Yang Z, Yu B, Liu J, Chen X. Curr Biol. 2005;15:1501. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V. Developmental Biology. 2002;244:170. doi: 10.1006/dbio.2002.0594. [DOI] [PubMed] [Google Scholar]
- 14.Tomari Y, Du T, Zamore PD. Cell. 2007;130:299. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Czech B, et al. Mol Cell. 2009;36:445. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okamura K, Liu N, Lai EC. Mol Cell. 2009;36:431. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. RNA. 2010;16:43. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Cell. 2007;130:287. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. Cell. 2009;136:215. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghildiyal M, et al. Science. 2008;320:1077. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Czech B, et al. Nature. 2008;453:798. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamura K, et al. Nature. 2008;453:803. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamura K, Balla S, Martin R, Liu N, Lai EC. Nat Struct Mol Biol. 2008;15:581. doi: 10.1038/nsmb.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawamura Y, et al. Nature. 2008;453:793. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 25.Brodersen P, Voinnet O. Nat Rev Mol Cell Biol. 2009;10:141. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 26.Kirino Y, Mourelatos Z. Nat Struct Mol Biol. 2007;14:347. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 27.Kurth HM, Mochizuki K. RNA. 2009;15:675. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohara T, et al. Nat Struct Mol Biol. 2007;14:349. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 29.Krutzfeldt J, et al. Nature. 2005;438:685. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 30.Haley B, Zamore PD. Nat Struct Mol Biol. 2004;11:599. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 31.Shen B, Goodman HM. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 32.Mullen TE, Marzluff WF. Genes Dev. 2008;22:50. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Wolfswinkel JC, et al. Cell. 2009;139:135. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 34.Song JJ, et al. Nat Struct Biol. 2003;10:1026. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 35.Yan KS, et al. Nature. 2003;426:468. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 36.Lingel A, Simon B, Izaurralde E, Sattler M. Nature. 2003;426:465. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 37.Ma JB, Ye K, Patel DJ. Nature. 2004;429:318. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lingel A, Simon B, Izaurralde E, Sattler M. Nat Struct Mol Biol. 2004;11:576. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 39.Parker JS, Roe SM, Barford D. Nature. 2005;434:663. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma JB, et al. Nature. 2005;434:666. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomari Y, Zamore PD. Genes Dev. 2005;19:517. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, et al. Nature. 2009;461:754. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.We thank S. Ma and A. Boucher for help with fly husbandry, G. Farley for technical assistance, E. Pfister for cloning of miR-16 and miR-21 reporter constructs, and members of the Zamore lab for advice, suggestions, and critical comments on the manuscript. Sequence data are available through the NCBI Short Read Archive (www. ncbi.nlm.nih.gov/sites/sra) as GSM278704 (ago2414), SRA010045 (hen1f00810 and HeLa cell data), and GSE18806 (Oregon R). This work was supported in part by an EMBO long term fellowship (ALTF 522-2008) and an Erwin Schrödinger-Auslandsstipendium (Austrian Science Fund FWF, J2832-B09) to S.L.A. and by grants from the National Institutes of Health (GM62862 and GM65236) to P.D.Z.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.