Abstract
A critical mediator of the cellular response to hypoxia is hypoxia-inducible factor 1 (HIF-1). Increased levels of HIF-1α are often associated with increased tumor metastasis, therapeutic resistance, and poorer prognosis. We recently identified a novel interaction between HIF-1α and the mammalian septin family member, SEPT9_v1. Septins are a highly conserved family of GTP-binding cytoskeletal proteins that are implicated in multiple cellular functions, including cell division and oncogenesis. SEPT9_v1 binds and stabilizes HIF-1α protein and stimulates HIF-1 transcriptional activity. SEPT9_v1-HIF-1 activation promotes tumor growth and angiogenesis. The structural and functional relationships between SEPT9_v1 and HIF-1α were analyzed. We found that SEPT9_v1 binds specifically with HIF-1α but not with HIF-2α. The GTPase domain of SEPT9_v1 was identified as essential for HIF-1α binding. A GTPase domain-derived polypeptide, corresponding to amino acids 252–379, was able to disrupt HIF-1α-SEPT9_v1 interaction and to inhibit HIF-1 transcriptional activity. SEPT9_v1 also protected HIF-1α from degradation induced by HSP90 inhibition by preventing the interaction of HIF-1α with the RACK1 protein, which promotes its oxygen-independent proteasomal degradation. In conclusion, a new mechanism of oxygen-independent activation of HIF-1 has been identified that is mediated by SEPT9_v1 blockade of RACK1 activity on HIF-1α degradation.
A key transcription factor for cells to respond and adapt to survive hypoxia is hypoxia-inducible factor-1 (HIF-1)3 (1). HIF-1 target genes encode proteins involved in anaerobic metabolism (e.g. glycolytic enzymes and glucose transporters), angiogenesis (e.g. vascular endothelial growth factor), and erythropoiesis (e.g. erythropoietin) (2). HIF-1 is a heterodimer and consists of a constitutively expressed subunit, HIF-1β, and an oxygen-regulated subunit, HIF-1α. HIF-1 transcriptional activity is directly affected by the abundance and stability of HIF-1α. HIF-1α protein expression is tightly regulated by O2 levels. Under normoxic conditions, human HIF-1α is hydroxylated by prolyl hydroxylases at two specific proline residues, Pro402 and Pro564 (3–5). The “proline” hydroxylated form of HIF-1α is recognized by von Hippel-Lindau tumor suppressor protein as part of a complex with E3 ubiquitin ligase activity that leads to ubiquitination and proteasome-dependent degradation of HIF-1α (6, 7). HIF-1 transcriptional activity is regulated by an additional mode of hydroxylation. Under normoxia, HIF-1α is also hydroxylated at asparagine residue 803 by factor inhibiting HIF-1α (FIH-1) leading to inhibition of CBP recruitment to the HIF-1α C-terminal transactivation domain (8–10). The hydroxylation reactions utilize O2 andα-ketoglutarate as substrates. Under hypoxia, HIF-1α is therefore not hydroxylated and becomes stabilized to heterodimerize with HIF-1β and to recruit co-factors such as CBP for transcriptional activation.
In tumor cells HIF-1 can also be regulated and activated by other genetic factors such as oncogenes (Ras and phosphoinositide 3-kinase) or loss of tumor suppressors (VHL or PTEN) even under aerobic conditions. On another level of post-translational regulation, Liu et al. (11) have recently identified RACK1 (receptor of activated protein kinase C 1) as a HIF-1α-interacting protein that competes with heat shock protein 90 (HSP90) for binding to HIF-1α and mediates O2- and prolyl hydroxylase/VHL-independent ubiquitination and proteasomal degradation (12).
We have recently described another novel activation pathway of HIF-1 under normoxia by a member of the mammalian septin family, SEPT9_v1 (septin 9 protein, variant 1) (13). We discovered that SEPT9_v1 interacts with HIF-1α to prevent its ubiquitination and degradation. SEPT9_v1 protein activates HIF-1 transcriptional activity and increases tumor growth and angiogenesis in prostate cancer xenografts (13). Similarly, Gonzalez et al. (14) have shown that high expression of SEPT9_v1 is also associated with accelerated growth kinetics and increased motility in human breast cancer cells. Therefore, we investigated the mechanisms of HIF-1 activation by SEPT9_v1. We found that SEPT9_v1 is involved in the regulation of the O2- and prolyl hydroxylase/VHL-independent degradation of HIF-1α, which is mediated by RACK1.
EXPERIMENTAL PROCEDURES
Cell Culture and Hypoxia Treatment—Human prostate cancer PC-3 cells were cultured in RPMI 1640 as recommended by the American Type Culture Collection. Stably transfected PC-3 cells with SEPT9_v1 (PC-3-SEPT9_v1) or with the empty vector (PC-3-Neo) were designed and characterized previously (13). PC-3 cells were infected with retroviral vector expressing either empty vector (PC-3-Puro) or shSEPT9_v1 (PC-3-shSEPT9_v1) as previously described (15). HEK293 cells were maintained in Dulbecco's modified Eagle's medium. All of the media were supplemented with 10% fetal calf serum and antibiotics. The cells were cultured at 37 °C in a humidified atmosphere and 5% CO2 in air. For hypoxic exposure, the cells were placed in a sealed modular incubator chamber (Billups-Rothenberg, Del Mar, CA) flushed with 1% O2, 5% CO2, and 94% N2.
Plasmids and Reagents—Human untagged-HIF-1α (GenBank™ accession number NM_001530), FLAG-HIF-1α, and FLAG-SEPT9_v1 (GenBank™ accession number AF189713) were previously constructed (13). FLAG-tagged mutated and truncated forms of HIF-1α were constructed into p3XFLAG-Myc-CMV-25 (Sigma-Aldrich), whereas SEPT9_v1-Myc truncated forms were constructed into pcDNA4/Myc-His (A) (Invitrogen). Deletion and point mutations were performed by multiple overlapping PCRs using the specific primers as listed in Table 1. Human RACK1 (GenBank™ accession number NM_006098) was subcloned at HindIII/XbaI sites into pcDNA4/Myc-His (A) to produce RACK1-Myc-tagged fusion protein (Table 1). Hemagglutinin (HA)-HIF-2α expression vector was a generous gift from Dr. W. G. Kaelin (Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA). All of the constructs were verified by DNA sequencing (Danyel Biotech, Rehovot, Israel).
TABLE 1.
List of designed constructs and primes used in this study
The underlining indicates the additional restriction enzyme sites, and italics indicate mutations. F, forward primer; R, reverse primer; DPM, double proline mutation (P402A and P564A).
| Gene | Primer sequences | |
|---|---|---|
| FLAG-HIF-1constructs | ||
| ΔHLH | F: CCGCGCCCGCGGCCGCGATGGTTCTCACAGATGATGGTG | |
| R: CAGTTCTAGATTATCAGTTAACTTGATCCAAAGCTC | ||
| ΔHLH/PAS | F: ACGTGCGGCCGCGTTTTCTACTCAGGACACAGATTTAGACTTG | |
| R: CAGTTCTAGATTATGAGTTAACTTGATCCAAAGCTCTGAG | ||
| ΔPAS | Amino acids 1-85 | F: ACGTGCGGCCGCGATGGAGGGCGCCGGCGGCGCGAACG |
| R: CCTGTACTGTCCTGTGGTGACTTGCATGTCATCTTCAATATCCAAATC | ||
| Amino acids 298-826 | F: GATTTGGATATTGAAGATGACATGCAAGTCACCACAGGACAGTACAGG | |
| R: CAGTTCTAGATTATGAGTTAACTTGATCCAAAGCTCTGAG | ||
| HLH/PAS | F: ACGTGCGGCCGCGATGGAGGGCGCCGGCGGCGCGAACG | |
| R: CAGTTCTAGATTAAGAATTCTTGGTGTTATATATGACAGTTG | ||
| ΔODDD | Amino acids 1-343 | F: ACGTGCGGCCGCGATGGAGGGCGCCGGCGGCGCGAACG |
| R: GTTCTTCCTCAGGAACTGTAGTACTCACAACGTAATTCACACATAC | ||
| Amino acids 698-826 | F: GTATGTGTGAATTACGTTGTGAGTACTACAGTTCCTGAGGAAGAAC | |
| R: CAGTTCTAGATTATGAGTTAACTTGATCCAAAGCTCTGAG | ||
| DPM | Amino acids 1-402 with P402A | F: ACGTGCGGCCGCGATGGAGGGCGCCGGCGGCGCGAACG |
| R: GTCTCCAGCGGCTGCGGCCAGCAAAG | ||
| Amino acids 402-826 | F: CTTTGCTGGCCGCAGCCGCTGGAGAC | |
| R: CAGTTCTAGATTATGAGTTAACTTGATCCAAAGCTCTGAG | ||
| Amino acids 1-564 with P564A | F: ACGTGCGGCCGCGATGGAGGGCGCCGGCGGCGCGAACG | |
| R: CATTGGGATATAGGCAGCTAACATCTC | ||
| Amino acids 564-826 with P564A | F: GAGATGTTAGCTGCCTATATCCCAATG | |
| R: CAGTTCTAGATTATGAGTTAACTTGATCCAAAGCTCTGAG | ||
| SEPT9_v1-Myc constructs | ||
| Full length | F: ACTAAGCTTATGAAGAAGTCTTACTCAGGAGGCACGCGGACC | |
| R: ACGTTCTAGACATCTCTGGGGCTTCTGGCTCCTTCTCCTCC | ||
| 1-251 | F: ACTAAGCTTATGAAGAAGTCTTACTCAGGAGGCACGCGGACC | |
| R: GTTCTAGAGTCGCCAACGCAGCTGGGAGC | ||
| 252-586 | F: ACTAAGCTTATGGCCGACACCCCCAGAGATG | |
| R: ACGTTCTAGACATCTCTGGGGCTTCTGGCTCCTTCTCCTCC | ||
| 252-379 | F: ACTAAGCTTATGGCCGACACCCCCAGAGATG | |
| R: ACGTTCTAGAGATGGGCTGCCAGCAGTTCTCG | ||
| 380-586 | F: ACTAAGCTTATGAAGTTCATCAATGACCAGTACGAG | |
| R: ACGTTCTAGACATCTCTGGGGCTTCTGGCTCCTTCTCCTCC | ||
| RACK1-Myc (full length) | F: CCGCGCCCAAGCTTATGACTGAGCAGATGACCCTTC | |
| R: CCGCGCCCTCTAGAGCGTGTGCCAATGGTCA |
17-Allylamino-17-demethoxy-geldanamycin (17-AAG) was purchased from Sigma-Aldrich. The following primary antibodies were used: mouse monoclonal antibodies to HIF-1α and to RACK1 (BD BioSciences, San Diego, CA; BD610959 and 610177, respectively), mouse monoclonal anti-HIF-1β (Novus Biologicals, Littleton, CO; ab2771–100), mouse monoclonal antibodies to FLAG and to tubulin (Sigma-Aldrich; F3165 and T9026, respectively), mouse monoclonal anti-HA.11 (Covance, Berkeley, CA; MMS-101R), goat polyclonal anti-TOPO-I (Santa Cruz Biotechnology, CA; sc-5342), and rabbit affinity-purified anti-Myc (Bethyl Laboratories, Montgomery, TX; A190-105A). Rabbit polyclonal antibodies to SEPT9_v1 were previously prepared (13). Secondary antibodies were horseradish peroxidase-conjugated (Jackson ImmunoResearch, West Grove, PA).
Protein Extraction and Western Blot—Whole cell and nuclear extracts were prepared as previously described (16). Protein concentration was determined using a bicinchoninic acid protein assay kit (Pierce). The proteins were analyzed by SDS-PAGE and immunoblotting with antibodies as displayed in the figures as described (13, 16). Sixty μg of protein were loaded in each lane.
Transfections and Reporter Gene Assays—The cells were seeded in 10-cm or 6-well plates and were transfected with a total of 5 μg DNA or 1 μg of DNA/well, respectively, using GenePorter transfection reagent (Gene Therapy Systems, Inc., San Diego, CA), according to the manufacturer's instructions. HRE-dependent luciferase activity was performed using the pBI-GL construct (pBI-GL V6L) containing six tandem copies of the VEGF HRE as previously described (17, 18). Briefly, duplicate sets of transfected cell-culture dishes were then separated and incubated under normoxic and hypoxic conditions for 16 h. Luciferase enzymatic activity was measured with commercial kit TROPIX (Bedford, MA) using a BMG Labtechnologies LUMIstar Galaxy luminometer following the manufacturer's instructions. Arbitrary luciferase activity units were normalized to the amount of protein in each assay point.
Immunoprecipitation (IP) Assays—Cells grown in 10-cm plates were lysed in 20 mm HEPES (pH 7.5), 0.1 m NaCl, 2 mm EDTA, 0.5% Nonidet P-40, and 10% glycerol 48 h after they had been transfected and treated accordingly. Protease and phosphatase inhibitors were added to the lysis buffer. The lysates were centrifuged at 14,000 rpm for 15 min at 4 °C. Ten percent of the extract was kept, and the rest was incubated with the appropriate antibodies for 3 h at 4 °C. The samples were next rotated with protein G-Sepharose (Sigma-Aldrich) for 2 h at 4 °C. The beads were washed six times with lysis buffer, and immunoprecipitates were eluted in 2× Laemmli sample buffer for SDS-PAGE analysis.
17-AAG Treatment—Cells, either transfected or nontransfected, grown in culture dishes until 70% confluence, were treated with vehicle (0.1% Me2SO) or 17-AAG at the concentrations indicated in the figures at 37 °C under normoxic or hypoxic conditions. After 5 or 16 h, the cells were washed with ice-cold phosphate-buffered saline, harvested, and subjected to IP or HRE-luciferase assays, respectively.
Data Analysis—The experiments presented in the figures are representative of three or more independent repetitions. Using TINA software version 2.0, quantification of band densities was performed. The data are expressed as means ± S.D. Student's t test was used to compare differences between particular conditions. Pearson Chi-Square was calculated using SPSS software version 15. Statistical significance was set at p < 0.05.
RESULTS
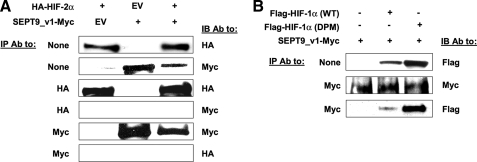
SEPT9_v1 Binds HIF-1α but Not HIF-2α Regardless of Proline Hydroxylation Status—We previously reported that SEPT9_v1 interacts with HIF-1α protein to activate HIF-1 transcriptional activity (13). To further characterize the specificity of HIF-1α-SEPT9_v1 interactions, we studied whether SEPT9_v1 would or would not also associate with HIF-2α protein. HEK293 cells were transiently co-transfected with expression vectors encoding SEPT9_v1-Myc and HA-HIF-2α. The IP experiment was performed in parallel using both antibodies to Myc and HA. The immunoprecipitated proteins from each lysate were analyzed by Western blotting using their counterpart tag antibodies (Fig. 1A). Although both HIF-2α and SEPT9_v1 were immunoprecipitated, each by their corresponding tag antibody, no interaction between the two proteins was observed (Fig. 1A). These findings demonstrate that SEPT9_v1 does not interact with HIF-2α.
FIGURE 1.
Specificity of SEPT9_v1/HIF-α interaction. A, HEK293 cells were transiently co-transfected with expression vectors encoding HA-HIF-2α, SEPT9_v1-Myc or their corresponding empty vector (EV) as indicated. After 48 h the cells were lysed, subjected to immunoprecipitation (IP) using antibodies (Ab) to HA or Myc, and then immunoblotted (IB) with HA and Myc antibodies. None, no immunoprecipitation (whole cell extract samples). B, HEK293 cells were transiently co-transfected with expression vectors encoding, either full-length wild type (WT) FLAG-HIF-1α or full-length double proline mutant (DPM) together with SEPT9_v1-Myc and their corresponding empty vector (–) as indicated. After 48 h the cells were lysed, subjected to IP using antibodies to Myc, and then analyzed by immunoblotting with FLAG and Myc antibodies.
Because the interaction between endogenous HIF-1α and SEPT9_v1 proteins was more prominent under normoxia rather than under hypoxia (13), we hypothesized that hydroxylation of the proline residues 402 and 564 within the oxygen-dependent degradation domain (ODDD) of HIF-1α may modulate the interaction. We therefore designed and inserted alanine point mutations in these two proline residues (P402A/P564A) and tested the interaction with SEPT9_v1 (Fig. 1B). As expected, the protein expression level of HIF-1α P402A/P564A was much higher than the wild type HIF-1α polypeptide (Fig. 1B). Regardless of the amino acid substitution, the double proline mutant species interacted with SEPT9_v1 protein in a similar fashion as the wild type HIF-1α (Fig. 1B). The results show that neither proline 402 nor proline 564 is required for HIF-1α/SEPT9_v1 interaction.
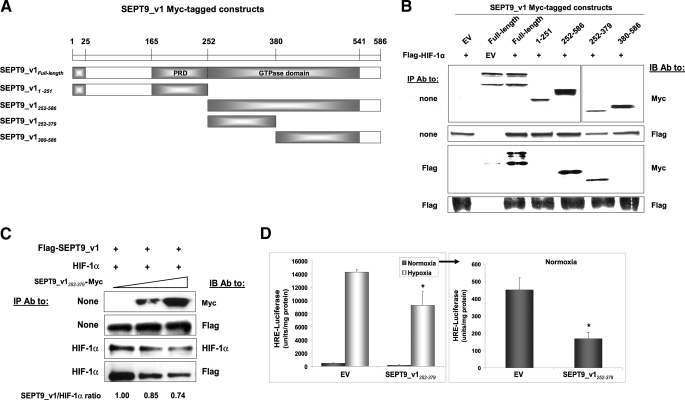
Amino Acids 252–379 of SEPT9_v1 Are Sufficient to Bind HIF-1α and to Inhibit HIF-1 Transcriptional Activity—Our initial studies indicated that activation of HIF-1 by SEPT9_v1 is dependent on the N-terminal part (amino acids 1–25) and the GTP-binding site (amino acids 305–312) of SEPT9_v1, although none was required for the interaction itself (13). As schematically outlined in Fig. 2A, a series of Myc-tagged SEPT9_v1 constructs were designed to further identify the domain of SEPT9_v1 that binds to HIF-1α protein. These constructs were co-expressed with HIF-1α in HEK293 cells and subjected to an IP assay. The C-terminal part of SEPT9_v1 (amino acids 252–586) consisting of the conserved GTPase domain bound to full-length HIF-1α protein, whereas the N-terminal part (amino acids 1–251) did not interact with HIF-1α (Fig. 2B). Further narrowing of the C-terminal part revealed that amino acids 252–379 (SEPT9_v1252–379) were sufficient to bind to HIF-1α (Fig. 2B). Consequently, we tested whether SEPT9_v1252–379 would be able to interfere with HIF-1α/SEPT9_v1 interaction and to affect HIF-1 activity. HEK293 cells were transfected with SEPT9_v1252–379 in the presence of HIF-1α and SEPT9_v1. Increasing amounts of SEPT9_ v1252–379 reduced HIF-1α protein expression and caused a dose-dependent inhibition of HIF-1α association with SEPT9_v1 (Fig. 2C). Expression of SEPT9_v1252–379 also led to a significant inhibition, respectively, of HIF-1 transcriptional activity measured by HRE-dependent reporter gene assay (60% under normoxia and 35% under hypoxia) (Fig. 2D). Similar results were also obtained in PC-3 cells (data not shown). Altogether, the minimal interacting domain identified of SEPT9_v1 (SEPT9_v1252–379) is able to disrupt the HIF-1α/SEPT9_v1 complex and is capable of inhibiting HIF-1 transcriptional activity.
FIGURE 2.
The GTPase domain of SEPT9_v1 is required for the interaction with HIF-1α. A, Myc-tagged constructs of SEPT9_v1 were designed as illustrated. Amino acids 1–25 are unique for SEPT9_v1 variant. PRD, proline-rich domain. B, Myc-tagged SEPT9_v1 constructs were co-expressed with FLAG-HIF-1α or corresponding empty vectors (EV) in HEK293 cells for immunoprecipitation (IP) with antibodies (Ab) to FLAG and then immunoblotted (IB) with FLAG and Myc antibodies. None, no immunoprecipitation (whole cell extract samples). C, HEK293 cells were transiently co-transfected with expression vectors encoding untagged-HIF-1α, FLAG-SEPT9_v1, and increasing amounts (0, 1.2, and 2.4 μg/well) of SEPT9_v1252–379-Myc. After 48 h the cell lysates were subjected to IP using antibodies to HIF-1α and then IB with FLAG, Myc, and HIF-1α antibodies. The ratio of immunoprecipitated SEPT9_v1 by HIF-1α was calculated using densitometric quantification and outlined below the figure where 1.0 stands for IP without SEPT9_v1252–379. D, PC-3 cells were co-transfected with the plasmid pBI-GL V6L (0.2 μg) expressing luciferase under the control of HRE and with SEPT9_v1252–379-Myc (0.9 μg) or empty vector (EV) (0.9 μg). After 24 h of transfection, the cells were grown for overnight under normoxia or hypoxia and then analyzed for luciferase activity. Relative luciferase activity represents units/mg of protein at each assay point. Columns, means; bars, S.D. (n = 3). *, p < 0.01 between SEPT9_v1252–379 and EV in normoxia and hypoxia, respectively.
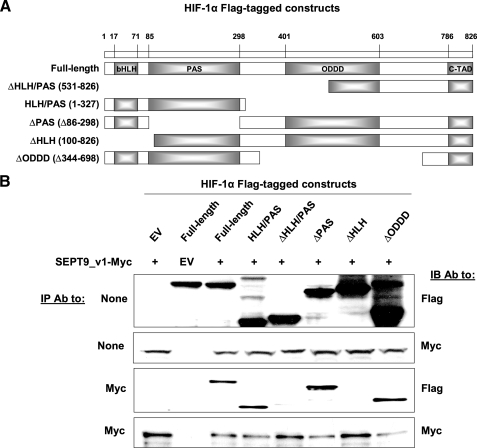
The Helix-Loop-Helix Domain of HIF-1α Is Required for the Interaction with SEPT9_v1—To map the domain of HIF-1α that interacts with SEPT9_v1, a number of FLAG-tagged HIF-1α constructs were designed as illustrated in Fig. 3A. Myc-tagged full-length SEPT9_v1 and HIF-1α constructs were expressed in HEK293 cells and subjected to an IP assay (Fig. 3B). Consistent with the HIF-1α double proline mutant results, HIF-1α ODDD was not required for the interaction with SEPT9_v1 (Fig. 3B). Only the species that included HIF-1α HLH domain interacted with SEPT9_v1, whereas all other HIF-1α species lacking the HLH domain did not interact with SEPT9_v1 (Fig. 3B). Importantly, our results demonstrated that the HLH domain, which is one of the essential domains required for heterodimerization with HIF-1β, is also required for the interaction with SEPT9_v1.
FIGURE 3.
The HLH of HIF-1α is required for binding to SEPT9_v1. A, FLAG-tagged constructs of HIF-1α were designed as illustrated. bHLH, basic helix-loop-helix; ODDD, oxygen-dependent degradation domain; c-TAD, C-terminal transactication domain. B, FLAG-HIF-1α constructs were co-expressed with SEPT9_v1-Myc or corresponding empty vectors (EV) in HEK293 cells for immunoprecipitation (IP) with antibodies (Ab) to Myc and then immunoblotted (IB) with FLAG and Myc antibodies. None, no immunoprecipitation (whole cell extract samples).
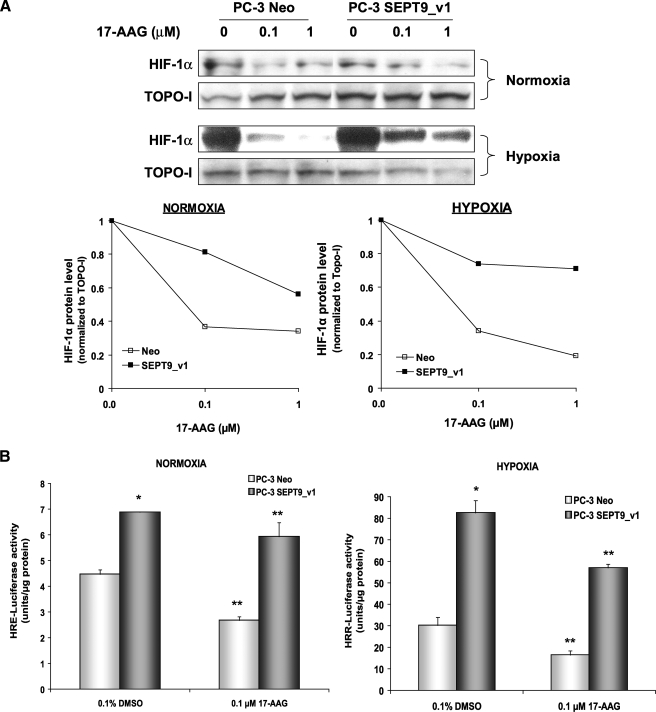
SEPT9_v1 Functions to Protect HIF-1α from RACK1-mediated Degradation—We have shown previously that SEPT9_v1 interacts with HIF-1α and prevents its ubiquitination (13). We investigated the mechanism of HIF-1α stabilization or shielding by SEPT9_v1. Given that SEPT9_v1 does not interact with the ODDD of HIF-1α and that the interaction is not influenced by proline modification, we sought for alternative regulation pathways other than the well known VHL-dependent degradation pathway. The anchoring protein, RACK1 interacts with HIF-1α and competes with HSP90 to promote HIF-1α ubiquitination and degradation that is independent of VHL (11). Because the N-terminal half of HIF-1α comprising the HLH/PAS domain is involved with HSP90 and RACK1 interaction and regulation (11, 19), we postulated that stabilizing the HIF-1α protein by SEPT9_v1 could be a result of a competition between RACK1 and SEPT9_v1 for HIF-1α binding. To test this hypothesis, we used PC-3 stably expressing SEPT9_v1 protein (PC-3-SEPT9_v1) and its control counterpart expressing the empty vector (PC-3-Neo). Both PC-3 cells subtypes were treated with increasing concentrations of the HSP90 inhibitor 17-AAG, a less toxic analog of geldanamycin. The IC50 of HIF-1α protein down-regulation by 17-AAG was 10-fold higher in PC-3-SEPT9_v1 cells compared with PC-3-Neo cells (>1 μm versus 0.1 μm) under normoxic and hypoxic conditions (Fig. 4A). On the functional level, 0.1 μm 17-AAG induced 40% versus 15% (p < 0.001, 2-sided Parson Chi-Square) inhibition of HIF-1 transcriptional activity in PC-3-Neo cells compared with PC-3-SEPT9_v1 cells under normoxic conditions (Fig. 4B). Under hypoxia, 0.1 μm 17-AAG also inhibited HIF-1 transcriptional activity by 45% versus 30% (p = 0.028, 2-sided Parson Chi-Square) in PC-3-Neo cells compared with PC-3-SEPT9_v1 cells (Fig. 4B). These results suggest that SEPT9_v1 protects HIF-1α from a degradation mediated by the 17-AAG-RACK1 pathway.
FIGURE 4.
SEPT9_v1 reduces the sensitivity of HIF-1α to 17-AAG. A, PC-3 cells stably expressing SEPT9_v1 (PC-3-SEPT9_v1) or the empty vector (PC-3-Neo) were treated with increasing concentrations of 17-AAG (0–1 μm) for 4 h under normoxic and hypoxic conditions. Nuclear extracts were prepared, analyzed by SDS-PAGE, and immunoblotted with antibodies to HIF-1α and TOPO-I as loading control. Normalized HIF-1α levels to TOPO-I under normoxia and hypoxia were analyzed by densitometry and plotted over 17-AAG concentration (lower panels). B, PC-3-Neo and PC-3-SEPT9_v1 cells were transfected with pBI-GL V6L (1μg/well) expressing luciferase under the control of HRE. After 24 h of transfection, the cells were treated with 0.1% dimethyl sulfoxide (DMSO, vehicle control) or 0.1 μm 17-AAG for overnight under normoxia and hypoxia and then analyzed for luciferase luminescence assay. Relative luciferase activity represents units/μg of protein at each assay point. Columns, means; bars, S.D. (n = 3). *, p < 0.001 between PC-3-SEPT9_v1 and PC-3-Neo (induction); **, p < 0.05 between 17-AAG and EV (inhibition) in normoxia and hypoxia, respectively.
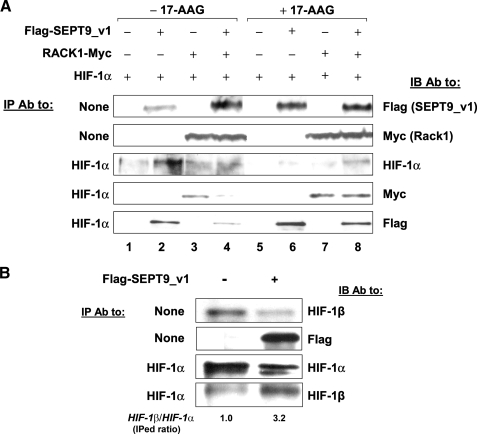
To confirm that SEPT9_v1 prevents RACK1 direct binding to HIF-1α, HEK293 cells were co-transfected with HIF-1α, SEPT9_v1, or RACK1 for IP experiment (Fig. 5A). The association of HIF-1α with RACK1 was weaker in the presence of SEPT9_v1 (compare lane 4 with 3). Furthermore, the association of HIF-1α with SEPT9_v1 was also weaker in the presence of RACK1 comparing lane 4 with lane 2. As predicted, HIF-1α protein expression was increased by SEPT9_v1 and decreased by RACK1 (compare lanes 3 and 4 with lane 1). The HSP90 inhibitor 17-AAG inhibited HIF-1α protein levels and to a greater extent did so in the presence of RACK1 (compare lanes 5 and 7 with lane 1). Similar to RACK1, SEPT9_v1 was associated more with HIF-1α after 17-AAG treatment (compare lane 7 with lane 3 and lane 6 with lane 2). SEPT9_v1 was able to protect HIF-1α from degradation induced by 17-AAG and RACK1 (compare lane 8 with lane 5). In addition, even in the presence of 17-AAG, the relative association of SEPT9_v1 and RACK1 with HIF-1α was affected, as manifested by comparing lane 8 with lane 6 and comparing lane 8 with lane 7. Taken together, these results indicate that, as an activator of function, SEPT9_v1 and, as an inhibitor of function, RACK1 compete with each other to directly bind HIF-1α. Although RACK1 binds to promote degradation, SEPT9_v1 binds HIF-1α to protect against degradation by RACK1 and confers stabilization of HIF-1α.
FIGURE 5.
SEPT9_v1 competes with RACK1 for binding HIF-1α. A, HEK293 cells were transfected with expressing vectors encoding untagged-HIF-1α, FLAG-SEPT9_v1, RACK1-Myc, and their corresponding empty vectors (–) as indicated. After 48 h the cells were treated with 0.1% Me2SO (–17-AAG) or with 5 μm 17-AAG (+17-AAG) for 4 h. Subsequently, the cells were lysed, and equal amounts of extracted proteins normalized to equal volumes were subjected to immunoprecipitation (IP) with antibodies (Ab) to HIF-1α. The whole amount of eluted immunoprecipitates from each condition was loaded on SDS-PAGE and then analyzed by immunoblotting (IB) with antibodies to HIF-1α, FLAG, and Myc. In parallel 10% (40–60 μg) of the whole cell extract (input) were also analyzed (None, no IP). B, PC-3 cells were transiently transfected with FLAG-SEPT9_v1 or empty vector (–) and subjected for IP assay with HIF-1α antibodies. Immunoprecipitates were analyzed by IB using antibodies to HIF-1α and HIF-1β. The ratio of immunoprecipitated (IPed) HIF-1β and IPed HIF-1α was calculated using densitometric quantification and outlined below the panel where 1.0 stands for the ratio without SEPT9_v1. None, no IP (whole cell extract samples).
SEPT9_v1 Promotes HIF-1α/HIF-1β Heterodimerization—Our previous results showed that SEPT9_v1 does not bind HIF-1β (13). A major role of septins is to scaffold, anchor, and/or stabilize other proteins (20, 21). Because SEPT9_v1 binds the HLH domain of HIF-1α and the HLH domain required for heterodimerization with HIF-1β in addition to the PAS domains, we examined whether SEPT9_v1 “modifies” HIF-1α in a way that recruits HIF-1β rather than competes with it. PC-3 cells were transfected with SEPT9_v1 and then were subjected to an IP experiment using HIF-1α antibodies (Fig. 5B). Analysis of the immunoprecipitates showed that the amount of endogenous HIF-1β protein associated with endogenous HIF-1α protein in the presence of excess SEPT9_v1 was increased in the control samples (Fig. 5B). Similar results were also obtained in HEK293 cells co-transfected with HIF-1α and SEPT9_v1 (data not shown). Because SEPT9_v1 does not compete with HIF-1β to bind HIF-1α, HIF-1β is more associated with HIF-1α in the presence of SEPT9_v1. There is no direct interaction between SEPT9_v1 and HIF-1β. These strongly support the model that SEPT9_v1 has an important cellular role in HIF-1α/HIF-1β heterodimerization.
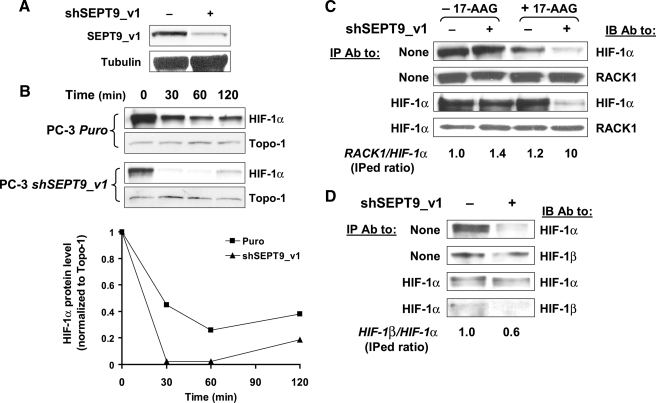
Silencing SEPT9_v1 Facilitates RACK1-mediated HIF-1α Degradation and Disrupts HIF-1α/HIF-1β Interaction—To study the effects of endogenous SEPT9_v1 on RACK1-mediated HIF-1α degradation and on HIF-1α/HIF-1β interaction, we utilized PC-3 cells stably expressing specific short hairpin RNA to knockdown SEPT9_v1 (PC-3-shSEPT9_v1) and their counterpart PC-3 cells expressing the empty vector (PC-3-Puro) (Fig. 6). SEPT9_v1 protein expression was down-regulated by 40–50% in PC-3-shSEPT9_v1 cells compared with control cells (Fig. 6A). In PC-3-shSEPT9_v1 cells, 17-AAG induced faster degradation (2-fold) of endogenous HIF-1α compared with control cells (Fig. 6B). In accordance with these results, knockdown SEPT9_v1 also increased endogenous RACK1 binding to endogenous HIF-1α, especially in the presence of 17-AAG (Fig. 6C). On the other hand, knockdown SEPT9_v1 caused a weaker interaction between endogenous HIF-1α and HIF-1β (Fig. 6D). Altogether, the data obtained from SEPT9_v1 silencing show and further confirm the preventive effects of SEPT9_v1 on HIF-1α degradation by RACK1 as well as the effects on HIF-1α/HIF-1β heterodimerization.
FIGURE 6.
Knocking down SEPT9_v1 increases RACK1-mediated HIF-1α degradation and affects HIF-1α/HIF-1β interaction. A, PC-3 cells were stably infected with either the pSUPER.retro empty vector or pSUPER.retro expressing shRNA directed against SEPT9_v1 (shSEPT9_v1). After selection with puromycin the cells were analyzed by Western blotting using antibodies to SEPT9_v1 and tubulin. B, PC-3-Puro and PC-3-shSEPT9_v1 cells were treated with 1 μm 17-AAG for various time points under normoxic conditions. Nuclear extracts were prepared, analyzed by SDS-PAGE, and immunoblotted with antibodies to HIF-1α and TOPO-I. Normalized HIF-1α levels to TOPO-I under normoxia were analyzed by densitometry and plotted over time (lower panel). C, PC-3-Puro and PC-3-shSEPT9_v1 cells were treated with 0.1% Me2SO (–17-AAG) or with 1 μm 17-AAG (+17-AAG). Subsequently, the cells were lysed and subjected to immunoprecipitation (IP) with antibodies (Ab) to HIF-1α and then immunoblotted (IB) with antibodies to HIF-1α and RACK1. The ratio of immunoprecipitated (IPed) RACK1 and IPed HIF-1α was calculated using densitometric quantification and outlined below the panel, where 1.0 stands for the ratio in PC-3-Puro cells in the absence of 17-AAG. D, PC-3-Puro and PC-3-shSEPT9_v1 cells were subjected for IP assay with HIF-1α antibodies. Immunoprecipitates were analyzed by IB using antibodies to HIF-1α and HIF-1β. The ratio of IPed HIF-1β and IPed HIF-1α was calculated using densitometric quantification and outlined below the panel where 1.0 stands for the ratio in PC-3-Puro cells. None, no immunoprecipitation (whole cell extract samples).
DISCUSSION
In these studies, we present mechanistic evidence of the involvement of the septin family member, SEPT9_v1 in HIF-1 O2-independent activation in prostate cancer cells. SEPT9_v1 acts as a HIF-1α “protective factor” that inhibits RACK1-mediated HIF-1α proteasomal degradation. As a consequence, SEPT9_v1 elevates intracellular HIF-1α levels. Furthermore, we show that SEPT9_v1 contributes to optimal heterodimerization of HIF-1α with HIF-1β to create a functional HIF-1 transcription complex. These findings shed light on one of the important epigenetic processes that could lead to the aerobic activation of HIF-1 in cancer cells.
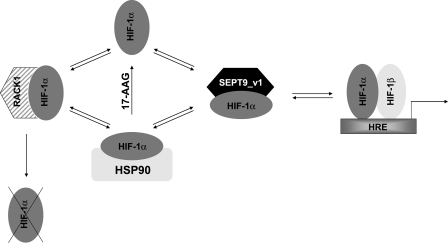
The action of SEPT9_v1 on HIF-1α appears to function like HSP90 in several respects. First, both proteins stabilize and protect HIF-1α from proteasomal degradation (13, 18, 19, 22). HSP90 interacts with HIF-1α, but it is not co-translocated with HIF-1α into the nucleus (23). In contrast, SEPT9_v1 was shown to bind with HIF-1α predominantly in the nucleus (13). We therefore propose that SEPT9_v1 has an important role in protecting HIF-1α from ubiquitination and proteasomal degradation that commences in the nucleus (24). Second, HLH and PAS domains of HIF-1α were critical for HSP90 binding (23, 25, 26). We discovered that the HLH domain but not the PAS domain is required for SEPT9_v1 binding (Fig. 3). Third, HSP90 (11, 27) and SEPT9_v1 (Fig. 5) compete with RACK1 for binding HIF-1α. In addition, in the presence of the HSP90 inhibitor, 17-AAG, where the interaction of HIF-1α with HSP90 is disrupted, the association of SEPT9_v1 with HIF-1α is favored, leading to increased HIF-1α stabilization and function (Figs. 4 and 5A). Moreover, when SEPT9_v1 protein levels were reduced in the presence of 17-AAG, the interaction between HIF-1α and RACK1 becomes dramatically more efficient, leading to increased HIF-1α degradation (Fig. 6, B and C). Taken together, these data indicate that there is a dynamic interplay between the three proteins, HSP90, SEPT9_v1, and RACK1 involved in the fine-tuning of the nonhypoxic transcriptional activation of HIF-1 as schematically illustrated in Fig. 7. On the other hand, unlike HSP90, which also binds HIF-2α (26), SEPT9_v1 interacts specifically with HIF-1α (Fig. 1A). Of interest is whether other members of the conserved septin family may be involved in the regulation of other HIF-α subunits.
FIGURE 7.
A proposed model of HIF-1 activation by SEPT9_v1. As long as HIF-1α protein is shielded by either HSP90 or SEPT9_v1, it is protected from RACK1-mediated degradation via the proteasome. In the presence of 17-AAG, HIF-1α-HSP90 association is disrupted, leading to a competition between RACK1, as an inhibitor of function, and SEPT9_v1, as an activator of function, for binding HIF-1α.
We also demonstrate that intact HIF-1α interaction with SEPT9_v1 is essential for HIF-1 activation. Overexpression of the interacting domain of SEPT9_v1, SEPT9_v1252–379, which comprises part of the GTPase domain, was capable of disrupting HIF-1α-SEPT9_v1 complex and inhibiting HIF-1 transcriptional activity (Fig. 2). These data strengthen the rationale for discovering small molecules with high throughput screening that interfere with or displace the binding HIF-1α-SEPT9_v1 interaction, making these small molecules a new class of HIF-1 inhibitors as HSP90 inhibitors have been previously classified (22). Moreover, it would be worthy to test whether the combination of HSP90 inhibitors with disruption of HIF-1α-SEPT9_v1 interaction will allow more efficient total down-regulation of HIF-1α.
Although the ODDD of HIF-1α was not required for the interaction with SEPT9_v1 (Fig. 3), still the affinity of HIF-1α to SEPT9_v1 under hypoxia was lowered (13). Our explanation for this weaker interaction is that SEPT9_v1 is displaced by HIF-1β under hypoxia. It is important to emphasize that both proteins, HIF-1β and SEPT9_v1, bind to HLH domain of HIF-1α without evidence of direct competition. In the presence of SEPT9_v1 it would be expected that the amount of HIF-1β associated with HIF-1α would be reduced. In contrast, the same amount of HIF-1α binds more molecules to HIF-1β in the presence of SEPT9_v1 (Fig. 5B) and vice versa when SEPT9_v1 levels are reduced (Fig. 6D). These results are consistent with findings that septins function as “stabilizing” proteins for critical cellular processes (20, 21). The data indicate that, in addition to stabilizing HIF-1α, SEPT9_v1 also functions to facilitate the interaction with HIF-1β for optimal heterodimerization and creation of transcriptionally functional HIF-1.
We find it provocative that SEPT9_v1 is involved in up-regulating HIF-1α by counter-regulation of the role of RACK1 in driving degradation of HIF-1α levels. SEPT9 was originally discovered as a fusion partner gene of myeloid/lymphoid or mixed lineage leukemia in acute myeloid leukemias and was initially named MSF (myeloid/lymphoid or mixed lineage leukemia septin-like fusion) (28, 29). One would predict that SEPT9_v1 should therefore be overexpressed in transformed cells and tumors. Indeed, Scott et al. (30) found consistent overexpression of SEPT9 mRNA in a wide range of human tumors (e.g. breast, central nervous, endometrium, kidney, liver, lung, and ovary) when they studied 7287 fresh frozen human tissue samples and 292 human cell lines. Later on, we described overexpression of SEPT9_v1 protein in multiple human cancer cells (e.g. prostate, breast, colon, and ovary), which was analyzed by Western blotting and was correlated significantly with resistance to microtubule-disrupting agents (15). Interestingly, Gonzalez et al. (14) recently have also found that SEPT9_v1 is highly expressed in a majority of primary human breast cancers by immunohistochemistry. Their functional studies in human breast cells showed that SEPT9_v1 accelerates growth kinetics, promotes cell invasion, increases genomic instability in culture manifested by development of aneuploidy, and stimulates morphologic changes associated with de-differentiation in human breast cells (14). These phenotypic increases in tumor invasiveness and metastatic potential could all be mechanistically attributed to the SEPT9_v1 downstream up-regulation of HIF-1. We and others have described up-regulation of HIF-1α in human breast cancer in vitro and in vivo (31–33). HIF-1 up-regulation is also associated with lymph node metastasis and micrometastatic implantation in bone metastases (34, 35). It will be of interest to assess whether SEPT9_v1 is contributing to the up-regulation of HIF-1α observed in clinical specimens.
HIF-1α-SEPT9_v1 binding was first discovered in human prostate cancer cells (13). Very recently, Setlur et al. (36) reported on the mRNA expression signature for prostate cancers with the most common chromosomal fusion between the transmembrane protease serine 2 (TMPRSS2) and ERG, a member of erythroblast transformation-specific transcription factor family. They identified an 87-gene expression signature that distinguishes TMPRSS2-ERG fusion prostate cancer as a discrete molecular entity. SEPT9 had a significantly higher expression in tumors with the TMPRSS2-ERG chromosomal fusions (36). Prostate cancers with the TMPRSS2-ERG fusion appear to have a more aggressive natural clinical history than other prostate cancers (37). It is now testable in prostate cancer that part of the downstream effect of TMRSS-ERG chromosomal fusions is SEPT9_v1 activation of HIF-1α in prostate tumorigenesis.
To our knowledge, the protective effects of SEPT9_v1 against RACK1 on HIF-1α have not been previously described. Future mechanistic studies to evaluate how SEPT9_v1 affects the fate of other HSP90 clients should provide additional novel insights to the functional roles of SEPT9_v1. Evaluation of SEPT9_v1 protein levels in vivo in prostate cancer and other human cancers appears to be of great interest in evaluating the contribution of SEPT9_v1 in the up-regulation of HIF-1 in human cancers.
Acknowledgments
We appreciate the critical editorial contribution of Rebecca Levine.
This work was supported by the M. K. Humanitarian Foundation, Prostate Cancer Foundation, and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation.
Footnotes
The abbreviations used are: HIF-1, hypoxia-inducible factor 1; HSP90, heat shock protein 90; 17-AAG, 17-Allylamino-17-demethoxy-geldanamycin; HRE, hypoxia-response elements; ODDD, oxygen-dependent degradation domain; HLH, helix-loop-helix; PAS, Per-aryl hydrocarbon receptor nuclear translocator-Sim; HA, hemagglutinin; IP, immunoprecipitation; VHL, von Hippel-Lindau.
References
- 1.Semenza, G. L. (2007) Sci. STKE 2007, cm8. [DOI] [PubMed] [Google Scholar]
- 2.Semenza, G. L., Shimoda, L. A., and Prabhakar, N. R. (2006) Novartis Found. Symp. 272, 2–16 [PubMed] [Google Scholar]
- 3.Bruick, R. K., and McKnight, S. L. (2001) Science 294, 1337–1340 [DOI] [PubMed] [Google Scholar]
- 4.Epstein, A. C., Gleadle, J. M., McNeill, L. A., Hewitson, K. S., O'Rourke, J., Mole, D. R., Mukherji, M., Metzen, E., Wilson, M. I., Dhanda, A., Tian, Y. M., Masson, N., Hamilton, D. L., Jaakkola, P., Barstead, R., Hodgkin, J., Maxwell, P. H., Pugh, C. W., Schofield, C. J., and Ratcliffe, P. J. (2001) Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 5.Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J. M., Lane, W. S., and Kaelin, W. G., Jr. (2001) Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 6.Kallio, P. J., Wilson, W. J., O'Brien, S., Makino, Y., and Poellinger, L. (1999) J. Biol. Chem. 274, 6519–6525 [DOI] [PubMed] [Google Scholar]
- 7.Cockman, M. E., Masson, N., Mole, D. R., Jaakkola, P., Chang, G. W., Clifford, S. C., Maher, E. R., Pugh, C. W., Ratcliffe, P. J., and Maxwell, P. H. (2000) J. Biol. Chem. 275, 25733–25741 [DOI] [PubMed] [Google Scholar]
- 8.Mahon, P. C., Hirota, K., and Semenza, G. L. (2001) Genes Dev. 15, 2675–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lando, D., Peet, D. J., Gorman, J. J., Whelan, D. A., Whitelaw, M. L., and Bruick, R. K. (2002) Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNeill, L. A., Hewitson, K. S., Claridge, T. D., Seibel, J. F., Horsfall, L. E., and Schofield, C. J. (2002) Biochem. J. 367, 571–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, Y. V., Baek, J. H., Zhang, H., Diez, R., Cole, R. N., and Semenza, G. L. (2007) Mol. Cell 25, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertout, J. A., Patel, S. A., and Simon, M. C. (2008) Nat. Rev. Cancer 8, 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amir, S., Wang, R., Matzkin, H., Simons, J. W., and Mabjeesh, N. J. (2006) Cancer Res. 66, 856–866 [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez, M. E., Peterson, E. A., Privette, L. M., Loffreda-Wren, J. L., Kalikin, L. M., and Petty, E. M. (2007) Cancer Res. 67, 8554–8564 [DOI] [PubMed] [Google Scholar]
- 15.Amir, S., and Mabjeesh, N. J. (2007) Cancer Biol. Ther. 6, 1926–1931 [DOI] [PubMed] [Google Scholar]
- 16.Mabjeesh, N. J., Escuin, D., LaVallee, T. M., Pribluda, V. S., Swartz, G. M., Johnson, M. S., Willard, M. T., Zhong, H., Simons, J. W., and Giannakakou, P. (2003) Cancer Cell 3, 363–375 [DOI] [PubMed] [Google Scholar]
- 17.Post, D. E., and Van Meir, E. G. (2001) Gene Ther. 8, 1801–1807 [DOI] [PubMed] [Google Scholar]
- 18.Mabjeesh, N. J., Post, D. E., Willard, M. T., Kaur, B., Van Meir, E. G., Simons, J. W., and Zhong, H. (2002) Cancer Res. 62, 2478–2482 [PubMed] [Google Scholar]
- 19.Isaacs, J. S., Jung, Y. J., Mimnaugh, E. G., Martinez, A., Cuttitta, F., and Neckers, L. M. (2002) J. Biol. Chem. 277, 29936–29944 [DOI] [PubMed] [Google Scholar]
- 20.Spiliotis, E. T., and Nelson, W. J. (2006) J. Cell Sci. 119, 4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinoshita, M. (2006) Curr. Opin. Cell Biol. 18, 54–60 [DOI] [PubMed] [Google Scholar]
- 22.Neckers, L. (2003) Curr. Med. Chem. 10, 733–739 [DOI] [PubMed] [Google Scholar]
- 23.Minet, E., Mottet, D., Michel, G., Roland, I., Raes, M., Remacle, J., and Michiels, C. (1999) FEBS Lett. 460, 251–256 [DOI] [PubMed] [Google Scholar]
- 24.Zheng, X., Ruas, J. L., Cao, R., Salomons, F. A., Cao, Y., Poellinger, L., and Pereira, T. (2006) Mol. Cell. Biol. 26, 4628–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isaacs, J. S., Jung, Y. J., and Neckers, L. (2004) J. Biol. Chem. 279, 16128–16135 [DOI] [PubMed] [Google Scholar]
- 26.Katschinski, D. M., Le, L., Schindler, S. G., Thomas, T., Voss, A. K., and Wenger, R. H. (2004) Cell Physiol. Biochem. 14, 351–360 [DOI] [PubMed] [Google Scholar]
- 27.Liu, Y. V., and Semenza, G. L. (2007) Cell Cycle 6, 656–659 [DOI] [PubMed] [Google Scholar]
- 28.Osaka, M., Rowley, J. D., and Zeleznik-Le, N. J. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 6428–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taki, T., Ohnishi, H., Shinohara, K., Sako, M., Bessho, F., Yanagisawa, M., and Hayashi, Y. (1999) Cancer Res. 59, 4261–4265 [PubMed] [Google Scholar]
- 30.Scott, M., Hyland, P. L., McGregor, G., Hillan, K. J., Russell, S. E., and Hall, P. A. (2005) Oncogene 24, 4688–4700 [DOI] [PubMed] [Google Scholar]
- 31.Zhong, H., De Marzo, A. M., Laughner, E., Lim, M., Hilton, D. A., Zagzag, D., Buechler, P., Isaacs, W. B., Semenza, G. L., and Simons, J. W. (1999) Cancer Res. 59, 5830–5835 [PubMed] [Google Scholar]
- 32.Laughner, E., Taghavi, P., Chiles, K., Mahon, P. C., and Semenza, G. L. (2001) Mol. Cell. Biol. 21, 3995–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bos, R., Zhong, H., Hanrahan, C. F., Mommers, E. C., Semenza, G. L., Pinedo, H. M., Abeloff, M. D., Simons, J. W., van Diest, P. J., and van der Wall, E. (2001) J. Natl. Cancer Inst. 93, 309–314 [DOI] [PubMed] [Google Scholar]
- 34.Woelfle, U., Cloos, J., Sauter, G., Riethdorf, L., Janicke, F., van Diest, P., Brakenhoff, R., and Pantel, K. (2003) Cancer Res. 63, 5679–5684 [PubMed] [Google Scholar]
- 35.Hiraga, T., Kizaka-Kondoh, S., Hirota, K., Hiraoka, M., and Yoneda, T. (2007) Cancer Res. 67, 4157–4163 [DOI] [PubMed] [Google Scholar]
- 36.Setlur, S. R., Mertz, K. D., Hoshida, Y., Demichelis, F., Lupien, M., Perner, S., Sboner, A., Pawitan, Y., Andren, O., Johnson, L. A., Tang, J., Adami, H. O., Calza, S., Chinnaiyan, A. M., Rhodes, D., Tomlins, S., Fall, K., Mucci, L. A., Kantoff, P. W., Stampfer, M. J., Andersson, S. O., Varenhorst, E., Johansson, J. E., Brown, M., Golub, T. R., and Rubin, M. A. (2008) J. Natl. Cancer Inst. 100, 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demichelis, F., Fall, K., Perner, S., Andren, O., Schmidt, F., Setlur, S. R., Hoshida, Y., Mosquera, J. M., Pawitan, Y., Lee, C., Adami, H. O., Mucci, L. A., Kantoff, P. W., Andersson, S. O., Chinnaiyan, A. M., Johansson, J. E., and Rubin, M. A. (2007) Oncogene 26, 4596–4599 [DOI] [PubMed] [Google Scholar]