Abstract
Background
The interferon-inducible transmembrane protein-3 (IFITM3) is a protein that restricts multiple pathogenic viruses such as influenza virus. The single nucleotide polymorphism rs12252-C, which is rare in Caucasian populations but much more common in Han Chinese, has been found in much higher homozygous frequency in patients with severe acute influenza. Until now there has been no study on the effect of this genetic variant on the clinical control of other viral infections.
Objectives
To investigate the impact of IFITM3-rs12252 genotypes on primary HIV-1 infection progression in an acute HIV-1 infected cohort in Beijing (PRIMO), China.
Design and methods
We identified IFITM3-rs12252 genotypes of 178 acute HIV-1 infected patients and 196 HIV negative candidates from the PRIMO cohort. HIV-1 viral load and CD4+T cell counts were monitored at multiple time points during the first year of infection and the association between IFITM3-rs12252 genotype and disease progression was evaluated.
Results
The current study shows that the IFITM3-rs12252 genetic variant affects the progression of HIV-1 infection but not the acquisition. A significantly higher frequency of the CC/CT genotypes was found in rapid progressors compared to non-progressors. Patients with CC/CT genotypes showed an elevated peak viraemia level and significantly lower CD4+T cell count at multiple time points during the first year of primary infection, and a significantly higher risk of rapid decline of the CD4+T cell count to below 350 cells/mm3.
Conclusion
A novel association between IFITM3 gene polymorphism and rapid disease progression is reported in an acute HIV-1 infected MSM cohort in China.
Introduction
The interferon-inducible transmembrane (IFITM) proteins are known to inhibit the replication of pathogenic viruses such as influenza A, HIV-1, dengue, filoviruses (Marburg virus and Ebola virus) and coronaviruses (e.g., severe acute respiratory syndrome (SARS)) [1, 2, 3, 4, 5, 6]. IFITM proteins interfere with viral replication preceding fusion of the viral and cellular membrane [7]. Further details of the anti-viral mechanism of IFITMs remain unknown.
In recent years, the crucial role of IFITM3 protein mostly has been studied in controlling influenza A virus infection. Everitt et al. (2012) [8], used a knock out mouse model, to show mice lacking IFITM3 gene display fulminant viral pneumonia on infection with a low-pathogenicity influenza virus. Likewise, in an in vitro study, an increase in viral replication was observed in absence of IFITM3, and reintroducing of IFITM3 restricted the replication of influenza A virus [8].
In humans, a rare single nucleotide polymorphism (SNP) rs12252-C allele of IFITM3, which, if homozygous, alters the outcome of influenza virus infection, has been identified [8, 9]. The allele frequency of this SNP is much higher in the Han Chinese population (minor allele frequency (MAF: 0.5)) compared to the Northern European population (MAF: 0.034) [9]. Previously, we studied the association of the rs12252-C allele with severity of the influenza virus infection in Chinese patients. We found that 69% of Chinese patients with severe pandemic influenza A H1N1/09 virus infection carry the CC genotype, compared with 25% in those with mild infection [9].
Two studies have shown a restrictive effect of IFITM3 on the progression of acute HIV-1 infection [10, 11]. However, the role of the rs12252 C allele in acute HIV-1 infection has not been explored. Jia et al. (2012) [10] showed that IFITM3 inhibits viruses that enter the cells via different routes. The presence of IFITM3 on late endosome blocks cytosolic access of pH-dependent viruses, such as influenza A. However, for pH-independent viruses such as HIV-1, IFITM3 appears to restrict viral cell entry at the cell periphery or early endosome. Furthermore, IFITM3 inhibits HIV-1 replication in vitro by reducing the expression of HIV-1 proteins such as Gag, Vif and Nef [11]. This raises the question: does the minor allele (of Caucasians) have the same effect on acute HIV-1 infection as it does on influenza virus? This is especially pertinent to growing concern about the spread of HIV in China, in particular, among men who have sex with men (MSM). To address to this question we screened 6000 individuals from MSM cohort (PRIMO cohort) from Beijing, China and compared the clinical outcome in rs12252 CC/CT and TT genotyped carriers.
Methods and Materials
Study population
Every two months 6000 volunteers from PRIMO cohort in Beijing were checked for early signs of acute HIV-1 infection, as described previously [12]. In addition, their plasmas were collected and tested for HIV-1 antibodies, and HIV-1 RNA levels. Amongst them, 178 individuals were diagnosed with acute HIV-1 infection, and another 196 volunteers, who were HIV-1 negative, were enrolled in the control group. Every three months, thereafter from detection of seronversion, whole-blood specimens were collected. Following blood collection plasma and peripheral blood mononuclear cells (PBMCs) were separated for further studies. This study was approved by the Institutional Review Board of Beijing You’An Hospital, and written informed consent was obtained from all patients.
Clinical definitions
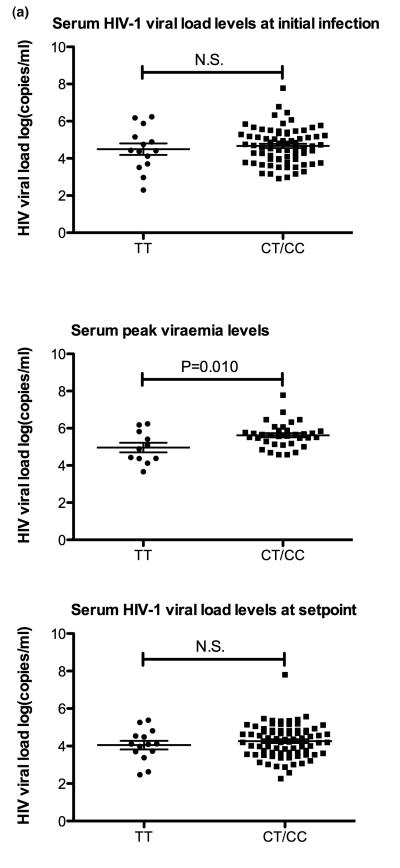
Acute HIV-1 infection was defined by a positive HIV-1 RNA test and negative/indeterminate HIV-1 ELISA and Western blot tests [12]. Rapid clinical progression was defined as when the time to treatment initiation (CD4<350 cells/mm3) or time to an estimated CD4+ lymphocyte count lower than 350 cells/mm3 was within one year of seroconversion. Those who did not show a significant CD4 T cell decline were defined as non-progressors [13]. Initial and peak viraemia levels were defined according to the dynamics of HIV-1 viraemia in acute HIV-1 infected patients [14]: initial viral load indicates HIV-1 viral load detected at Fiebig stage II-III [14], and peak viraemia level refers to the highest viral load detected during the acute HIV-1 infection. For setpoint definition, the established criteria described by Fellay et al. [13] were strictly followed, defining the 3-phase evolution of HIV-1 viraemia to calculate the average viral load setpoint (Table 1).
Table 1.
a. Clinical and laboratory characteristics.
| HIV negative MSM (196) | Acute HIV infection (178) |
P value | |||
|---|---|---|---|---|---|
| Total (178) | Rapid progressors (74) | Non progressors (104) | |||
| Age (years) | 33.03±8.79 | 32.05±8.97 | 32.40±8.12 | 31.81±9.55 | 0.675 |
| Gender (F/M) | 0/196 | 0/178 | 0/74 | 0/104 | N.A. |
| CD4+T cell count (cell/μl) | |||||
| seroconversion | N.A. | 501.08±170.16 | 384.45±124.17 | 570.53±155.60 | <0.001 |
| 3rd month | N.A. | 509.20±173.46 | 379.61±112.05 | 602.14±148.42 | <0.001 |
| 6th month | N.A. | 477.73±187.99 | 335.79±89.91 | 566.79±78.65 | <0.001 |
| 12th month | N.A. | 432.55±172.71 | 278.01±78.77 | 515.76±150.87 | <0.001 |
| HIV-1 VL (log/ml) | |||||
| Initial VL | N.A. | 4.64±0.99 | 4.85±0.89 | 4.54±1.03 | 0.165 |
| Peak VL | N.A. | 5.46±0.74 | 5.59±0.81 | 5.41±0.72 | 0.487 |
| Setpoint VL | N.A. | 4.22±0.87 | 4.61±0.68 | 4.02±0.89 | 0.002 |
| 1b. Allele and genotype frequencies of IFITM3 rs12252 in Chinese population, MSM HIV-1 negative control group and HIV-1 acute cohort. | ||||
|---|---|---|---|---|
| Genotype | HIV-1 (−) MSM (196) | Acute HIV infection |
||
| Total (178) | Rapid progressors (74) | Non progressors (104) | ||
| CC | 69 (35.20%) | 57 (32.02%) | 22 (29.73%) | 35 (33.65%) |
| CT | 86 (43.88%) | 89 (50%) | 46 (62.16%) | 43 (41.35%) |
| TT | 41 (20.92%) | 32 (17.98%) | 6 (8.11%) | 26 (25%) |
| Allele C | 224 (52.78%) | 203 (57.02%) | 90 (60.81%) | 113 (54.33%) |
| Allele T | 168 (47.22%) | 153 (42.98%) | 58 (39.19%) | 95 (45.67%) |
| Statistical analysis* | ||||
| Parameter | ||||
| Genotype# | x2 | 1.441 | 9.215 | 0.655 |
| P-value | 0.487 | 0.010 | 0.721 | |
| Allele# | x2 | 0.001 | 0.594 | 0.438 |
| P-value | 0.973 | 0.441 | 0.508 |
Compared with HIV-1 (−) MSM.
x2 and P-value were calculated by x2
Laboratory detection
For detection of specific antibodies to HIV-1, standard HIV type 1 (HIV-1) enzyme-linked immunosorbent assay (Abbott, IL), and western blot analysis (Genelabs, CA) kits were used. The copies of plasma HIV-1 RNA were quantified in serum with Roche Amplicor kit (Roche). The absolute counts of CD3+/CD4+/CD8-T cells (CD4+T) and CD3+/CD4-/CD8+T cells (CD8+T) were measured, using TriTEST Three-Color reagents (BD Company, USA), and MultiSET software in the FACSCalibur Flow Cytometry System.
To determine the HIV-1 subtypes, HIV-1 gag genes from 174 infected individuals were amplified and sequenced (Beijing Institute of Genomics (BGI), Shenzhen, China). The nucleotide sequences were analyzed, using Recombinant Identification Program: RIP 3.0 software (Alamos HIV sequence database).
Sequencing and genotyping of rs12252
Genomic DNA was extracted from PBMCs using the PureGene DNA Isolation kit (Gentra Systems, USA). The region encompassing the human IFITM3 rs12252 sequences was amplified, sequenced and analyzed as described previously [9].
Statistical analysis
Statistical analysis of genetic data was performed using the χ2-test. For association testing under different genetic models Fisher’s exact test was used since the χ2 approximation might not hold for small sample size. Student’s t-test was used to compare values between IFITM3-TT and CT/CC groups where data were normally distributed (evaluated with Kolmogorov-Smirnov test), and non-parametric t-test (Mann-Whitney test) was used where data were not normally distributed. For survival analysis, Log-rank (Mantel-Cox) test was employed to identify the difference of progression to CD4 cells below 350 between patients with TT and CT/CC genotype. Statistical test differences were considered significant if the P-values were <0.05. Analyses were performed with the GraphPad Prism v 5 (GraphPad Software, LaJolla, CA).
Results
Clinical and laboratory characteristics of study subjects
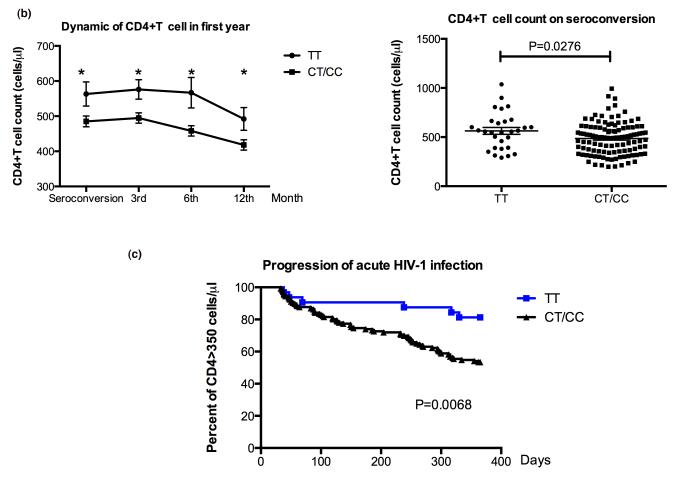
From 178 HIV-1 positive infected individuals, 74 were identified as rapid progressors (as described in Methods), and the other 104 patients with no significant CD4 decline were grouped as non-progressors. A significant difference in CD4+T cell count between rapid progressors and non-progressors was observed at seroconversion (384.45±124.17 vs. 570.53±155.60, P-value < 0.001), three (379.61±112.05 vs. 602.14±148.42, P-value < 0.001), six (335.79±89.91 vs. 566.79±78.65, P-value < 0.001), and twelve (278.01±78.77 vs. 515.76±150.87, P-value < 0.001) months after the seroconversion. In addition, progressors showed to have a significantly higher viral load at set point (4.61±0.68) compared to the non-progressors (4.02±0.89; P-value = 0.002; Table 1A).
The C allele at SNP 12252 was associated with rapid progression
Three hundred base pairs of the IFITM3 locus encompassing SNP rs12252 were sequenced in all acute HIV-1 infected patients and in the control group. 91.89% of rapid progressors carried CC/CT genotypes, a higher frequency than in HIV-1 negative individuals (79.08%, 155/196, P-value = 0.01), while the genotype frequency of the non-progressors was not significantly different from HIV-1 negative group (P-value = 0.721). The association of rs12252 with the progression of the infection was tested in other genetic models, as described previously [9]. A significantly higher frequency of the CC/CT genotypes was found in rapid progressors (91.89%, 68/74), comparing with non-progressors (75%, 78/104, P-value = 0.004). In particular, under the dominant model, the carriers of the C allele showed a threefold increase in the risk of rapid infection progression compared with carriers of the T allele (P-value = 0.004, odds ratio=3.778, 95% confidence interval (CI) 1.468-9.723)
Elevated peak viraemia level and degressive CD4+T cell count in acute HIV-1 infected patients with CC/CT genotypes
At peak viraemia the viral load was significantly higher in CC/CT carriers, compared to TT genotype carriers (5.624±0.114 vs. 4.962±0.260, P-value = 0.010; figure 1a). However, there was no significant difference in viral load levels between CC/CT and TT genotypes carriers at initial infection and at viral set point. Both heterozygotes and homozygotes for C allele showed significantly lower CD4+ T cell counts than homozygotes for the T allele, at the seroconversion time point (485.2±15.33 vs. 563.0±34.54, P-value=0.0276), to the 3rd (494.9±14.78 vs. 576.0±27.78, P-value = 0.0197), 6th (458.1±14.58 vs. 566.7±43.37, P-value = 0.0232), and at 12 months (418.3±14.80 vs. 492.0±32.64, P-value = 0.0324) after acute HIV-1 infection (Fig 1b). To identify whether there is a difference in disease progression in individuals with CD4+T cell counts below 350, who are CC/CT genotypes carries versus the TT genotype carriers, a Log-rank (Mantel-Cox) test was performed. The result of this test verified that CC/CT genotypes carries are more likely to become rapid progressors (P-value = 0.0068; Fig. 1c).
The effect of rs12252-C allele on rapid progression is independent of HIV-1 subtypes
Four different HIV-1 subtypes were identified among 125 acute HIV-1 infected patients in this cohort and their distribution were described: subtype B 34.4% (43/125), subtype BC 11.2% (14/125), subtype C 1.6% (2/125) and subtype AE 52.8% (66/125). We did not find any difference in the distribution of HIV-1 subtypes between rapid and non-progressors (data not shown). However, a higher percentage of rapid progressors were CC/CT genotypes carriers, regardless of the infecting HIV-1 subtype (data not shown).
Discussion
In the current study we show that the IFITM3-rs12252 C genetic variant is associated with more rapid progression of HIV-1 infection. A significantly higher frequency of the CC/CT genotypes was observed in rapid progressors compared to non-progressors. We further found that CC/CT genotype carriers had a higher peak viraemia level and lower CD4+T cell count. More strikingly, those with the CC/TC genotype had a significantly higher chance of their CD4+T cell counts falling to less than 350 cells/mm3 during the first year of infection. There was however no genetic association with acquisition of infection since there were no significant difference in the distribution of IFITM3 genotypes between acute HIV-1 infected patients and HIV-1 negative contros.
The current results could partly explain why there is more rapid progression of HIV-1 infection observed in the Beijing PRIMO cohort, a Chinese cohort, when compared to the Concerted Action on SeroConversion to AIDS and Death in Europe (CASCADE) cohort [15]. In the PRIMO study, CD4+ T cell decline and viral load increase were faster in this Chinese MSM cohort, 75% CT/CC genotype carriers compared to CASCADE, a population with only 2-8% CT/CC genotypes carriers [9]. However, many other factors, both in the host and virus, are likely to contribute to these differences.
It is still not clear how SNP rs12252 affects the expression of IFITM3. It has been predicted that this mutation may result in expression of truncated IFITM3, which lacks the first 21 amino acids at the N-terminus [8]. Previous mechanistic studies have all been based on this assumption and the conclusions have been rather controversial [8, 16]). However, the existence of a truncated IFITM3 protein in cells with the SNP rs12252 C allele has not yet been shown. In cells that were transfected with a synthetic truncated version lacking the first 21 amino acids, IFITM3 was relocated from the late endosome to the cell periphery [10]. If IFITM3 is relocated to the cell surface it might affect HIV-1 viral entry [10], Moreover, Tarotour et al. have suggested that IFITM proteins are incorporated into HIV-1 virion particles and impair their infectivity [17]. These findings could partly explain our results.
Nonetheless, more studies are required to answer the following questions: does the presence of SNP rs12252 C allele results in expression of truncated IFITM3 protein, which localizes differently from the full length IFITM3, or does it simply affect the level of expression of IFITM3? We are currently investigating the answers to these questions.
In conclusion, our data clearly showed that CC/CT genotypes are associated with rapid progression of acute HIV-1 infection, regardless of HIV-1 subtype. However, it is not associates with the disease acquisition.
Acknowledgements
This work was supported by: National Natural Science Foundation of China (81271842, 81228020 and 81320108017), Beijing Natural Science Foundation (7132098), National S&T Major Project for Infectious Diseases Control (2012ZX10001006-001-008, 2013ZX10001004-001-002, 2012ZX10001003, 2012ZX10001006 and 2012ZX10004904-002-002), Beijing Municipal Science & Technology Commission (D131100005313004, D131100005313005, D141100000314005 and D141100000314002), Capital Health Development (2011-2018-06, 2011-1011-02), Medical Research Council, UK and Wellcome Trust, Institutional Strategic Support Fund.
References
- 1.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feeley EM, Sims JS, John SP, Chin CR, Pertel T, Chen LM, et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, et al. Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J Virol. 2010;84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weidner JM, Jiang D, Pan XB, Chang J, Block TM, Guo JT. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J Virol. 2010;84:12646–12657. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raychoudhuri A, Shrivastava S, Steele R, Kim H, Ray R, Ray RB. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J Virol. 2011;85:12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang YH, Zhao Y, Li N, Peng YC, Giannoulatou E, Jin RH, et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun. 2013;4:1418. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia R, Pan Q, Ding S, Rong L, Liu SL, Geng Y, et al. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J Virol. 2012;86:13697–13707. doi: 10.1128/JVI.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chutiwitoonchai N, Hiyoshi M, Hiyoshi-Yoshidomi Y, Hashimoto M, Tokunaga K, Suzu S. Characteristics of IFITM, the newly identified IFN-inducible anti-HIV-1 family proteins. Microbes Infect. 2013;15:280–290. doi: 10.1016/j.micinf.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X, Chen H, Li W, Li H, Jin X, Perelson AS, et al. Precise determination of time to reach viral load set point after acute HIV-1 infection. J Acquir Immune Defic Syndr. 2012;61:448–454. doi: 10.1097/QAI.0b013e31827146e0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol. 2010;10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X, Lodi S, Fox Z, Li W, Phillips A, Porter K, et al. Rate of CD4 decline and HIV-RNA change following HIV seroconversion in men who have sex with men: a comparison between the Beijing PRIMO and CASCADE cohorts. J Acquir Immune Defic Syndr. 2013;62:441–446. doi: 10.1097/QAI.0b013e31827f5c9a. [DOI] [PubMed] [Google Scholar]
- 16.Williams DE, Wu WL, Grotefend CR, Radic V, Chung C, Chung YH, et al. IFITM3 polymorphism rs12252-C restricts influenza A viruses. PLoS One. 2014;9:e110096. doi: 10.1371/journal.pone.0110096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tartour K, Fau - Appourchaux R, Appourchaux R, Fau - Gaillard J, Gaillard J, Fau - Nguyen X-N, Nguyen Xn, Fau - Durand S, Durand S, Fau - Turpin J, Turpin J, Fau - Beaumont E, et al. IFITM proteins are incorporated onto HIV-1 virion particles and negatively imprint their infectivity. Retrovirology. 2014;11:103. doi: 10.1186/s12977-014-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]