Abstract
Previous studies on the regulation of polyadenylation of the immunoglobulin (Ig) heavy-chain pre-mRNA argued for trans-acting modifiers of the cleavage-polyadenylation reaction operating differentially during B-cell developmental stages. Using four complementary approaches, we demonstrate that a change in the level of hnRNP F is an important determinant in the regulated use of alternative polyadenylation sites between memory and plasma stage B cells. First, by Western analyses of cellular proteins, the ratio of hnRNP F to H or H′ was found to be higher in memory B cells than in plasma cells. In memory B cells the activity of CstF-64 binding to pre-mRNA, but not its amount, was reduced. Second, examination of the complexes formed on input pre-mRNA in nuclear extracts revealed large assemblages containing hnRNP H, H′, and F but deficient in CstF-64 in memory B-cell extracts but not in plasma cells. Formation of these large complexes is dependent on the region downstream of the AAUAAA in pre-mRNA, suggesting that CstF-64 and the hnRNPs compete for a similar region. Third, using a recombinant protein we showed that hnRNP F could bind to the region downstream of a poly(A) site, block CstF-64 association with RNA, and inhibit the cleavage reaction. Fourth, overexpression of recombinant hnRNP F in plasma cells resulted in a decrease in the endogenous Ig heavy-chain mRNA secretory form-to-membrane ratio. These results demonstrate that mammalian hnRNP F can act as a negative regulator in the pre-mRNA cleavage reaction and that increased expression of F in memory B cells contributes to the suppression of the Ig heavy-chain secretory poly(A) site.
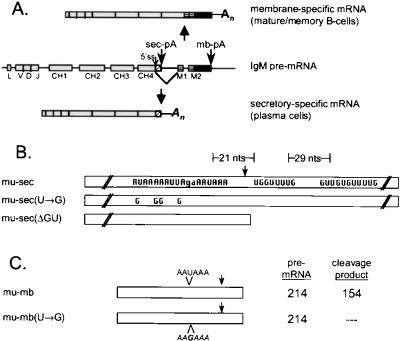
The immunoglobulin (Ig) heavy-chain μ transcription unit and the two heavy-chain mRNAs it encodes are shown in Fig. 1A (reviewed in reference 8). In mature and memory B cells the promoter-distal membrane-specific poly(A) site (mb-pA) is selected and splicing to the downstream M1 exon occurs via a 5′ splice site within the coding region of CH4. The secretory-species-specific poly(A) (sec-pA) and mb-pA sites are used with equal frequency in mature and memory B cells and their tumor analogs, lymphoma cells. Plasma cells are terminally differentiated B cells; myeloma cells are their tumor counterparts, which accurately reflect their pattern of Ig gene expression. In plasma and myeloma lines polyadenylation takes place preferentially at the weaker, promoter-proximal sec-pA site, precluding the splicing to membrane-specific exons; the sec-pA site is used up to 100-fold more often than the mb-pA site in plasma cells (23). Polyadenylation at the promoter-proximal secretory site and splicing of CH4 to M1 are mutually exclusive events; consequently, it is the balance between these two that determines the final ratio of secretory-form to membrane mRNA (sec-to-mb mRNA ratio) (26). Previous experiments demonstrated that regulation of Ig heavy-chain mRNA production occurs primarily at the level of polyadenylation, not message stability, transcription termination, or splicing efficiency (14–16, 19, 22, 27).
FIG. 1.
IgM transcription unit and substrates used for in vitro studies. (A) Schematic representation of the Ig μ heavy-chain transcription unit. Boxes, exons; lines, introns; boxes with diagonal lines, secretory-form-specific coding sequence; boxes with horizontal lines, membrane-specific exons; angled line, potential splicing of the CH4 exon to the M1 exon. (B and C) Ig μ secretory-form-specific constructs (B) and Ig μ membrane-specific constructs (C) used for in vitro studies. The mu-sec construct contains the wild-type Ig μ heavy-chain sec-pA site and surrounding elements in a pGEM vector. The mu-sec(U→G) mutations disrupt the upstream AU-rich region in four sites, while in mu-sec(ΔGU) the downstream GU-rich element is deleted. Arrows, cleavage sites for poly(A) addition. The mu-mb construct contains the wild-type Ig mu-mb-pA site and surrounding elements. The mu-mb(U→G) substrate has-a mutation in the single mu-mb AAUAAA hexanucleotide poly(A) signal. nts, nucleotides.
Several cis-acting elements are required for the cleavage and polyadenylation reactions that produce the poly(A) tail found on the 3′ end of most mature mRNAs (11, 43). These signals include the highly conserved AAUAAA consensus polyadenylation sequence located 10 to 30 nucleotides upstream of the cleavage site and a downstream element that is usually GU or U rich. Additional sequences influencing processing may be located either upstream or downstream of the polyadenylation site.
Six trans-acting protein factors necessary and sufficient for basal in vitro cleavage and polyadenylation have been identified (43). These include cleavage and polyadenylation specificity factor (CPSF), which recognizes the AAUAAA poly(A) signal, cleavage stimulation factor (CstF), cleavage factors Im and IIm, poly(A) polymerase, and poly(A)-binding protein II. CPSF and CstF form a stable complex with pre-mRNA and confer specificity to the cleavage-polyadenylation reaction. CstF comprises three unique subunits of 77, 64, and 50 kDa; the 77-kDa subunit of CstF bridges the 64- and 50-kDa subunits (35) and interacts directly with CPSF (34). The 64-kDa subunit of CstF (CstF-64) contains an amino-terminal ribonucleoprotein-type RNA-binding domain and, in the presence of CPSF and the other CstF subunits, interacts directly with the downstream GU- or U-rich element of transcripts containing poly(A) signals (3, 4, 17, 32, 36, 39).
The 10-fold overexpression of CstF-64 in chicken B-cell transfection experiments shifted the balance of the processing of the IgM heavy chain towards the secretory form (33). However, the analyses of regulation of polyadenylation during normal B-cell development argue against control influenced solely by changes in CstF-64 levels in human primary B cells (18), in mouse B-cell lines (7), and in chicken B cells, according to a recent study. In chicken B cells Ig μ secretory-form mRNA was induced by knocking out the histone deactylase 2 gene; CstF-64 levels remained unperturbed (37). In contrast, the RNA-binding activity of CstF-64 but not its amount increased in nuclear extracts prepared from plasma versus memory B-cell lines (7). Therefore, changes in CstF-64 levels alone cannot account for regulated polyadenylation site usage in B-cell stages, suggesting the need for other changes in the cells. Taken together the data describing the regulation of polyadenylation during B-cell development argue for the existence of differentially expressed, trans-acting modifiers of 3′-end processing (7, 18, 26, 29, 40).
The DSEF-1 protein interacts with a G-rich region downstream of the simian virus 40 (SV-40) late (SVL) poly(A) site to enhance cleavage at that site; DSEF-1 is identical in sequence to hnRNP H′, a member of the closely related hnRNP H/H′/F family, members of which represent strong candidates for trans-acting modifiers of 3′-end processing (1, 2). The hnRNP H, H′, and F proteins bind to G-rich sequences (21) and vary widely in their expression levels (13); H and F have been shown to be involved in regulation of neuronal splicing (5, 24). hnRNP F has also been shown to interact with the cap binding protein (10) and recently has been shown to interact with the carboxyl-terminal domain of RNA polymerase II (41).
We have identified unique protein–pre-mRNA complexes containing hnRNP H, H′, and F in nuclear extracts from memory B cells; these complexes are absent in plasma cell extracts in which sec-pA site use is differentially up-regulated in vivo. We show that the levels of these hnRNP proteins vary between plasma and B-cell lines and have characterized their in vitro interactions with poly(A) site-containing pre-mRNAs. We show that hnRNP F can block CstF-64 binding to pre-mRNA and inhibit RNA cleavage activity. Furthermore, overexpression of recombinant hnRNP F reduces the Ig sec-to-mb mRNA ratio in plasma cells. Results presented here establish hnRNP F as a regulator of poly(A) site choice during B-cell development.
MATERIALS AND METHODS
Cell culture and nuclear extract preparation.
Mouse lymphoid cell lines were grown by the Cell Culture Center (Cellex Biosciences, Inc., Minneapolis, Minn.) in “slosh” culture in Iscove's modified Dulbecco medium containing 5% horse serum, as described previously (7, 23). A20 lymphoma cells are representative of a memory B-cell line (IgG2a sec-to-mb ratio, 1:1), and the AxJ myeloma line behaves like a plasma cell line in its IgG2a (sec-to-mb ratio, >30) expression pattern (23).
Nuclear extracts were prepared as described previously (7) with the following modifications. The nuclei were extracted in buffer containing 300 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride, 100 μg of tosylsulfonyl phenylalanyl chloromethyl ketone/ml, and 1 μg of aprotinin, leupeptin, pepstatin A, and trypsin-chymotrypsin inhibitor/ml. The nuclear extracts were dialyzed against a buffer containing 20 mM HEPES (pH 7.9), 230 mM potassium glutamate, 1 mM MgCl2, 0.2 mM EDTA, 20% (vol/vol) glycerol, and protease inhibitors as described above. Protein concentrations were determined by a Bradford assay (Bio-Rad, Hercules, Calif.) and were typically between 4 and 6 mg/ml. Extracts prepared by this method are competent for in vitro cleavage, polyadenylation, and splicing (data not shown).
Transfection of AxJ cells with recombinant hnRNP F.
Actively growing cells were transferred to Opti-MEM (Gibco/BRL), transfected with DNA containing hnRNP F cloned into the Flag tag vector (Sigma Chemical Co., St. Louis, Mo.) plus Superfect reagent (Qiagen), incubated for several hours at 37°C and 5% CO2, and then diluted with medium containing serum and antibiotics for an overnight incubation. The next day cells were diluted about twofold to approximately 5 × 105/ml and dispensed to a 24-well plate. On day 3, 1 ml of medium with serum, antibiotics, and 1 mg of G418 (Genticin; Gibco/BRL)/ml was added per well. Cells were fed for several more days with the same G418-containing medium. Individual clones arose about 10 to 14 days later and were analyzed as indicated below.
Plasmid constructs and in vitro transcription of RNA.
The IgM sec-pA site constructs were generously provided by Cathy Phillips (30). The construct H11G11 is labeled mu-sec, H00G11 is labeled mu-sec(U→G), and H11G00 is labeled mu-sec(ΔGU). The mu-sec construct contains the weak Ig mu-sec-pA site cloned into the pGEM-T vector. The mu-sec(U→G) construct mutates the two upstream AU-rich elements, including the AAUAAA polyadenylation signal, by replacing U residues with Gs (Fig. 1B). The mu-sec(ΔGU) construct deletes the two GU-rich elements downstream of the cleavage site. The clones used to produce T7 promoter-driven, 32P-labeled antisense RNA to probe the Northern blots of the transformed AxJ cells were derived from GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and a clone specific to the IgG2a CH3 exon (exon 347), both previously described (9). The mutation of the μ-mb-pA (mu-mb-pA) site was accomplished by oligonucleotide mutagenesis (38).
Plasmids were linearized and transcribed with either SP6 or T7 RNA polymerase in a 20- to 100-μl reaction mixture containing 500 μM cap analog [m7G(5′)ppp(5′)G], 250 μM recombinant GTP, 25 or 250 μM recombinant UTP, (depending on the specific activity of the RNA), 500 μM (each) recombinant ATP and CTP, 20 U of RNasin RNase inhibitor (Promega Corp., Madison, Wis.), and 20 to 50 μCi of [α-32P]UTP (New England Nuclear, Boston, Mass.). For biotinylated transcripts, 25 to 30 μM biotin-11-UTP (ENZO Diagnostics, Inc.; supplied through Sigma Chemical Co.) was included, resulting in the incorporation of from one to three biotins per molecule. Transcription reactions were carried out at 37 to 40°C for 60 to 90 min, the template was digested by DNase I for 15 min, and the transcribed RNA was extracted with phenol-chloroform, precipitated with ethanol, and purified either over a G-50 spin column or from a urea-polyacrylamide gel as previously described (7).
An hnRNP F cDNA clone in a pET vector (His tagged) was kindly provided by Doug Black (5). The BamH1 fragment containing the entire hnRNP coding portion plus a small segment of a multicloning site was removed from this clone and inserted into the unique BamH1 site of pFlag-tag (Sigma; E1775). The sequence of the resulting clone (clone 407) was determined, and the hnRNP F protein was verified as being in frame with the vector AUG from the Flag epitope.
Western analysis and antibodies.
Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electroblotted to a polyvinylidene difluoride (PVDF) membrane (PolyScreen PVDF transfer membrane; NEN Life Science Products, Inc., Boston, Mass.), and probed with antibodies. Antipeptide antibodies against CstF-64, CstF-77, and CPSF-160 were raised in rabbit hosts as previously described (18). The 3A7 monoclonal, anti-CstF-64 antibody was generously provided by Clint MacDonald.
Rabbit antisera variously recognizing hnRNP H and H′ and/or F were raised by coupling peptides corresponding to the predicted amino acid sequences of the human hnRNP proteins through a terminal cysteine to keyhole limpet hemocyanin with gluteraldehyde as described previously (18). Injections and collections of sera were performed at Charles River Pharmservices (Southbridge, Mass.). An antiserum (NH2) that reacts with both hnRNP H and H′ as well as F was generated for human hnRNP H′ (amino acids 4 to 21 with a carboxyl-terminal cysteine; STEGREGFVVKVRGLPWSC). An antiserum recognizing predominantly hnRNP H and H′ (C) was generated for CDQVLDENSSDYQSN, corresponding to amino acids 434 to 447 of H′. An Antiserum recognizing predominantly hnRNP F (I) was generated for CTARRYIGIVKQAGLER, corresponding to amino acids 215 to 230 of the F sequence. An equal mixture of C and I antibodies was used for most of the Western blots.
Antiserum to the whole CstF-64 molecule was raised by injecting chickens with a recombinant glutathione S-transferase–CstF-64 fusion protein; the IgY was prepared from eggs. Injections and collections of antibodies were performed at Charles River Pharmservices. Rabbit polyclonal antibodies against CPSF-100 and CPSF-30 were generously provided by Silvia Barabino, Ursula Rüegsegger, and Walter Keller. Chicken anti-rat polypyrimidine tract binding protein (PTB) was obtained from Paula Grabowski. Anti-GAPDH antibodies were purchased from Chemicon Corp. Peroxidase-coupled secondary antibodies were purchased from Sigma (mouse), Roche Molecular Biochemicals (rabbit), and Jackson Laboratory (chicken) and detected by chemiluminescence. Anti-Flag epitope antibodies were purchased from Sigma.
Polyadenylation complex assembly, gel filtration chromatography, and affinity purification.
Polyadenylation complex formation was permitted to occur for 20 min at 30°C in a reaction mixture containing 15 to 25 pmol of substrate RNA with biotin and 32P (38), 3.75 to 6.25 mg of nuclear extract, 40 g of yeast tRNA/ml, 1 mM ATP, 10 mM creatine phosphate, 0.7 mM MgCl2, 2 mM dithiothreitol (DTT), 80 U of RNasin RNase inhibitor/ml, and 20% (vol/vol) glycerol in 20 mM HEPES, pH 7.9.
After the assembly and incubation were complete, the mixture was loaded over a 1.5- by 50-cm Sephacryl S-500HR (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) gel filtration column at 4°C and separated using a 1× PCB-1 buffer system (150 mM KCl, 0.1% Triton X-100, 0.5 mM [each] DTT and phenylmethylsulfonyl fluoride in 20 mM Tris, pH 7.6). Prior to use, the columns were blocked with nuclear extract to minimize the nonspecific interaction of complex-associated proteins with the matrix. Columns were run at a flow rate of 0.35 ml · min−1. Fractions of 1.25 ml were collected until at least one bed volume was eluted from the column.
Cerenkov counts (20) were acquired for 100 μl of each collected 1.25-ml fraction to determine the elution profile of the RNA from the column. Peak fractions were pooled for subsequent purification. A portion of each pool was removed for analysis of total (unselected) protein content. The remaining samples were incubated with 15 to 20 μl of avidin-D–agarose (Vector Laboratories, Inc., Burlingame, Calif.)/ml with gentle rocking at 4°C for at least 6 h. The affinity resin was then washed at 4°C with gentle rocking once for 15 min plus three times for 10 min with 150 mM NaCl in 20 mM Tris, pH 7.6, and then once for 10 min with 50 mM NaCl in 20 mM Tris, pH 7.6. Cerenkov counts for the washed resin were measured to determine the fraction of input RNA associated with the resin. Approximately 60% of the RNA counts in pooled fractions containing biotinylated RNA bind to the avidin resin, versus less than 1% binding of nonbiotinylated RNA.
The RNA-protein complexes were released from the affinity resin by incubation with 0.6 μg of RNase A at 37°C for 20 min. The proteins were then eluted with 250 μl of elution buffer (2% SDS, 20 mM DTT in 20 mM Tris, pH 7.6) with gentle rocking at 4°C for 10 min. The resin was recounted to determine the efficiency of RNase incubation and elution; generally, 85 to 95% of bound counts were released. The eluents were heated to 65 to 70°C for 5 to 10 min, 40 μg of yeast tRNA or glycogen carrier was added, and 4 volumes of ice-cold acetone were added to precipitate the proteins. The samples were incubated on ice for 10 to 20 min and pelleted in a 4°C microcentrifuge for 20 min. The air-dried pellets were resuspended in 1.5× SDS sample buffer (0.15 M DTT, 3% SDS, 120 mM Tris [pH 6.8], 15% glycerol, 0.3% bromophenol blue) for subsequent analysis by SDS-PAGE and Western immunoblotting.
UV cross-linking assays of nuclear extract proteins, immunoprecipitations, and cleavage reactions.
In vitro reaction mixtures for UV cross-linking of nuclear extract proteins to RNA contained 60 fmol (at least 6 × 105 cpm) of high-specific-activity 32P-labeled substrate RNA, 5 μg of nuclear extract, 40 μg of yeast tRNA/ml, 1 mM ATP, 0.7 mM MgCl2, 0.2 mM EDTA, 10 to 20% glycerol, and 230 mM potassium glutamate in 20 mM HEPES, pH 7.9. Reaction mixtures were incubated at 30°C for 5 min, placed on ice, and irradiated with 1.8 J of UV light/cm2. Unprotected RNA was digested with 50 g of RNase A/ml at 37°C for 15 to 20 min; proteins tagged with radiolabeled RNA were subjected to preclearing and then to immunoprecipitation with anti-CstF-64 antibodies coupled to protein A-Sepharose. Antigen-antibody-resin was washed with 50 mM Tris, pH 7.4–50 mM NaCl–0.5% NP-40 and resuspended in 20 μl of SDS-gel loading buffer, boiled for 5 min, and resolved by SDS–7.5% PAGE. RNA-protein complexes were visualized by autoradiography.
Cleavage assays with AxJ nuclear extracts were conducted as previously described using a 32P-labeled pSVL runoff transcript (38). A 32P-labeled runoff transcript of pSVL cut with HincII, 6 nucleotides shorter than the cleavage product, was included in the 5% acrylamide–8 M urea gel as a marker.
Band shift assays.
Band shift assays with SV40 late pre-mRNA substrates were performed as previously described (1, 2). SVL-Gem, which contained a 22-base substitution at positions 202 to 224 (from the transcription start site) that removes sequences downstream of the CstF binding site on SVL pre-mRNA, was prepared by a PCR approach. PCR mixtures containing pSVL, the SP6 promoter-specific primer (5′ CATACGATTTAGGTGACACT), and a mutagenic primer (5′-ACCGAGCTCGAATTCCGTGTATTCTGAACCTGAAACAT) yielded a 247-bp DNA fragment. The substituted region contains sequences from the pGEM4 polylinker. Transcription of this template yielded a 224-base RNA.
Purification of recombinant hnRNP F.
His-tagged hnRNP F was purified as previously described (5). The bacterial cell pellet was incubated in 50 mM NaPO4, pH 8.0–300 mM NaCl–200 μg of lysozyme/ml and then sonicated and brought to 6 M urea at room temperature. The denatured proteins were loaded onto a Ni-nitrilotriacetic acid column and refolded on the column by gradually decreasing the urea concentration to 1 M. The protein was eluted with buffer containing 0.25 M imidazole, and imidazole was removed by extensive dialysis.
RESULTS
GU- or U-rich and AAUAAA element-dependent binding of CstF-64 to pre-mRNA is increased in plasma cells in the absence of an increase in the amount of protein.
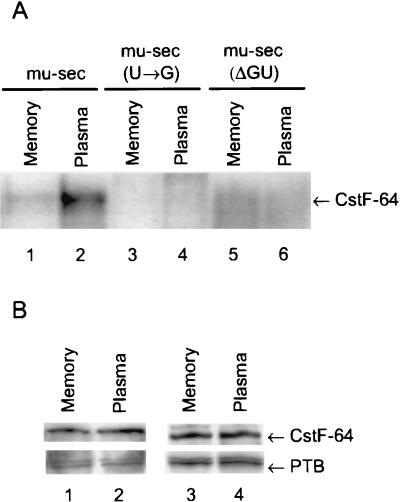
We analyzed the ability of CstF-64 to bind to 32P-labeled pre-mRNAs made from Ig mu-sec constructs (Fig. 1B) by immunoprecipitation of CstF-64 from nuclear extracts prepared from A20 memory B or lymphoma cells or AxJ plasma or myeloma cells using either polyclonal or COOH-specific anti-CstF-64 antibodies. Both extracts were competent for polyadenylation and cleavage (7). The results shown in Fig. 2A for the polyclonal anti-CstF-64 antiserum confirm that there is increased UV cross-linking of the Ig mu-sec poly(A) site to CstF-64 in the plasma extract relative to that in the memory cell extract. The results further demonstrate that CstF-64 binding in the plasma cell extract is disrupted upon mutation of either the AAUAAA binding sites recognized by CPSF [mu-sec(U→G) construct] or the GU- or U-rich downstream element seen by CstF [mu-sec(ΔGU) constructs]. Levels of protein CstF-64 in the extracts representing different B-cell stages and endogenous Ig sec-to-mb mRNA ratios are identical as shown by the Western analyses using two different antibodies to CstF-64, either a serum prepared in chickens containing a polyclonal antibody to the whole molecule (Fig. 2B, lane 1 versus 2) or a rabbit antiserum to the COOH-terminal 25 amino acids (Fig. 2B lane 3 versus 4). As a loading control the blot was probed with an antibody against PTB also located in the nucleus (42); the results in Fig. 2B show that there was an equal amount of protein in the memory and plasma cell nuclear extracts. Antibodies to the proteins B, D, and D′ of the small nuclear RNPs were also used to verify equal protein loads (data not shown).
FIG. 2.
CstF-64 cross-linking differs while the amount of protein remains the same in different B-cell stages. (A) UV cross-linking pattern. The A20 (memory or lymphoma) and AxJ (plasma or myeloma) nuclear extracts were incubated with the indicated 32P-labeled constructs under conditions permitting polyadenylation complex formation. The proteins were UV cross-linked to the RNAs, immunoprecipitated with the polyclonal chicken anti-CstF-64 antibody, and separated by SDS–8% PAGE as described in Materials and Methods. (B) Western blot of CstF-64 showing equal amounts in the two extracts. Proteins from the indicated nuclear extracts (5 μg) were separated by SDS–10% PAGE and electroblotted to PVDF. The membrane was probed with either the polyclonal chicken antibody to recombinant CstF-64 (lanes 1 and 2) or the rabbit antibody raised to the COOH terminus of human CstF-64 (lanes 3 and 4). As a loading control, the blot was then probed with an antibody against PTB.
hnRNP F level versus H or H′ level is decreased in plasma cells.
The hnRNP H/H′/F subfamily consists of three highly homologous but distinct RNA-binding proteins encoded by genes mapping to separate chromosomes (12, 21). The hnRNP H′ (DSEF-1) was identified because it bound to the G-rich element downstream of the SVL poly(A) site and was subsequently shown to bind to the downstream elements of several poly(A) sites with only GU-rich elements, such as the Ig mu-mb-pA site (1, 2, 31). Given their affinity for G- and GU-rich sequences (5, 24), one or more members of the hnRNP H/H′/F subfamily, we hypothesized, might influence CstF-64 binding to the sequences found downstream of polyadenylation sites.
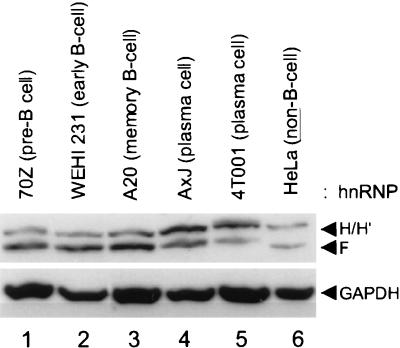
Using antipeptide antibodies that distinguish between the H and F members of the hnRNP H/H′/F family (see Materials and Methods) we examined the protein levels in whole-cell lysates prepared from various plasma and B-cell lines. Figure 3 shows the results of a representative Western analysis of hnRNP H, H′, and F proteins, demonstrating at least a twofold decrease in hnRNP F protein levels and at least a twofold increase in the H and H′ species in plasma cells relative to immature (70Z), mature (WEHI231), and memory (A20) B-cell lines. We conclude that the F-to-H or -H′ ratio is decreased appreciably in plasma cells.
FIG. 3.
The hnRNP F-to-H or -H′ ratio is reduced in plasma cells. The indicated whole-cell lysates (5 μg per lane) were separated by SDS–10% PAGE and electroblotted to PVDF, and the blot was probed with a mixture of antibodies recognizing hnRNP H and H′ and hnRNP F as described in Materials and Methods. As a loading control, the blot was then probed with an antibody against GAPDH. The cell lines in lanes 1 to 5 represent various stages of B-cell development: lane 1, pre-B; lane 2, early B; lane 3, memory B; lane 4, a fusion of memory and plasma cells with the phenotype of a plasma cell; lane 5, plasma cell. HeLa cell lysate (5 μg) was run in lane 6 as a size marker.
Large complexes containing hnRNPs H and H′ and hnRNP F are associated with the mu-sec-pA site in memory B-cell extracts.
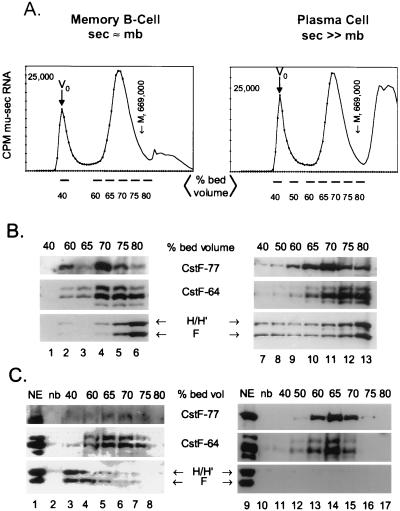
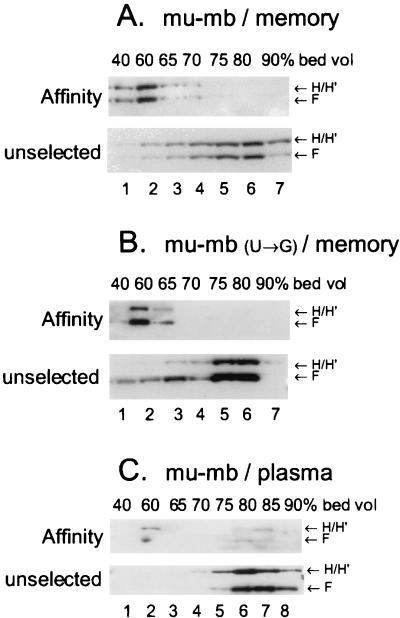
We wanted to determine if hnRNPs H and H′ and hnRNP F were differentially associated with exogenously added pre-mRNA in nuclear extracts prepared from memory B versus plasma cells. We used a two-step purification procedure to isolate and characterize RNA-protein complexes assembled in vitro (38). Nuclear extract was incubated with biotinylated mu-sec pre-mRNA (Fig. 1B) with a 32P-labeled tracer; the resultant complexes were fractionated by gel filtration on Sephacryl S-500HR. The bulk of the labeled RNA eluted at between 60 and 75% of the bed volume of the column, as shown in Fig. 4A, in both the memory B and the plasma cells. A portion of the RNA elutes in the void volume (40% of the bed volume) in very large complexes. The RNA eluting at >80% of the bed volume contained free nucleotides, short RNAs, and nucleolytic degradation products; the amounts of these varied from RNA preparation to preparation. Figure 4A indicates the fractions between 40 and 80% of the bed volume that were pooled for affinity purification and Western analysis. Thyroglobulin, used as a protein molecular mass marker (669,000 Da), eluted from the Sephacryl column at approximately 75 to 80% of the bed volume. We analyzed the total protein levels in the 40- to 80%-bed-volume samples by removing a portion of the fractions eluted from Sephacryl before the avidin affinity step; the immunoblot of those unselected fractions is shown in Fig. 4B. Coincident elution of CstF subunits CstF-64 and -77 occurred in a broad range of fractions from 65 to 85% of the total column volume in both cell types (Fig. 4B, lanes 2 to 6 and 9 to 13). Factors CPSF-160, -100, and -30 coeluted with the CstF subunits in the same broad range of fractions (not shown). The bulk of the polyadenylation and cleavage factors must remain associated with endogenous RNA to form these complexes. The elution of hnRNP H and H′ and hnRNP F from the column without the affinity selection was somewhat variable; representative Western blots are shown in Fig. 4B and 6. While the elution of these hnRNPs was broad, the bulk of the hnRNPs eluted predominantly at 70 to 80% of the bed volume in both cell types.
FIG. 4.
Large complexes containing hnRNPs H and H′ and F form on the weak Ig mu-sec-pA site in memory B-cell but not plasma cell nuclear extracts. Biotinylated and 32P-labeled mu-sec pre-mRNA was incubated with A20 memory B-cell or AxJ plasma cell nuclear extract under conditions permitting poly(A) complex formation. The reaction mixture was fractionated by gel filtration on Sephacryl S500 as described in Materials and Methods. (A) Elution profiles of mu-sec RNA from the column. The fractions pooled for subsequent analysis are indicated. The radioactivity in 10% of each fraction was determined for the RNA profile. Column void and total included volumes (Vo and Vtot, respectively) and the elution volume of protein size marker thyroglobulin (Mr = 669,000), whose profile was monitored by measuring optical density at 280 nm in a parallel column, are also shown. Lines below the x axis, fractions pooled for subsequent analyses. (B) Column elution profile before affinity selection of complexes. An aliquot representing 10% of the total proteins from each indicated pool was removed prior to the affinity selection step and analyzed by SDS–8% PAGE and Western immunoblotting with the indicated antibodies. (C) Column elution profile of affinity-purified proteins. The indicated fractions were pooled, and the proteins bound to the biotinylated RNA in each were affinity purified by avidin-agarose and eluted by RNAse A and SDS treatment. Proteins from each pool were analyzed by SDS–8% PAGE and Western immunoblotting with the indicated antibodies. Lanes NE, 2 μg of nuclear extract; lanes nb, nonbiotinylated RNA incubated with 300 μg of nuclear extract and run through the affinity purification steps as a control for background binding to the affinity resin.
FIG. 6.
Large complexes containing hnRNPs H, H′, and F form on the Ig mu-mb-pA site in memory B-cell but not plasma cell extracts. Biotinylated and 32p-labeled mu-mb or mu-mb(U→G) mutated pre-mRNA was incubated with A20 memory B-cell or AxJ plasma cell nuclear extract under poly(A) complex-forming conditions, and the reaction mixture was fractionated by gel filtration and affinity purified as described in Materials and Methods. Both affinity-purified fractions (affinity) and 10% of the column fractions prior to affinity selection (unselected) from each pool were analyzed by SDS–8% PAGE and Western immunoblotting with an antibody against hnRNP H and H′ and then hnRNP F in two steps. The position of the affinity-purified CstF-64 in the samples (A and C) was identical to that seen in Fig. 4C. (A) mu-mb pre-mRNA construct with memory (A20) nuclear extract. (B) mu-mb pre-mRNA site with U-to-G mutation incubated with memory B-cell (A20) nuclear extract. (C) mu-mb pre-mRNA site with plasma cell (AxJ) nuclear extract.
As a second step of purification of the RNA-protein complexes, pooled fractions from the column, as indicated in Fig. 4A, were incubated with avidin-agarose overnight at 4°C as previously described (38). The proteins in the resin-bound, biotinylated RNA-protein complexes were released by treatment with RNase and SDS and analyzed by SDS–8% PAGE and Western immunoblotting. The CstF-64 and -77 proteins that were affinity purified with the biotinylated RNA eluted from the Sephacryl column predominantly between 60 and 70% of the bed volume (Fig. 4C). Copurifying with CstF-RNA complexes were the CPSF subunits of 160, 100, and 30 kDa (data not shown). The affinity-purified proteins elute at a position in the column commensurate with their forming a complex on the input RNA, with an apparent molecular mass on the order of 2 × 106 to 4 × 106 Da, in both cell types. This is slightly larger than the average size of the endogenous complexes. The amounts of the CstF-64-, CstF-77-, and CstF-50-containing complexes were significantly reduced in the memory B-cell extracts compared with plasma cells; note the relative intensities of the CstF-77 lanes in Fig. 4C. In order to show detail, the CstF-64 signal from memory cells in Fig. 4C was exposed for a much longer time than the CstF-64 signal from plasma cells.
The striking observation was that, in the memory B cells, large complexes containing hnRNP H, H′, and F proteins were formed with mu-sec pre-mRNA (Fig. 4C, lanes 3 to 5), while in plasma cells these complexes were not detectable (Fig. 4C, lanes 11 to 17) even after prolonged exposure of the blot. The hnRNP proteins were in fact present in the total fractions in each cell type (Fig. 4B, lanes 2 to 6 and 8 to 13). The affinity-purified complexes containing hnRNP H, H′, and F in the memory B cells were on average larger than those formed with the CstF subunits and eluted at about 60% of the bed volume. The absence of these complexes in nuclear extracts prepared from plasma cells might explain the observed higher efficiency with which CstF-64 binds to pre-mRNAs in those extracts (Fig. 2).
Deletion of downstream sequences, but not mutation of the AAUAAA poly(A) signal, disrupts hnRNP H, H′, and F association with mu-sec RNA.
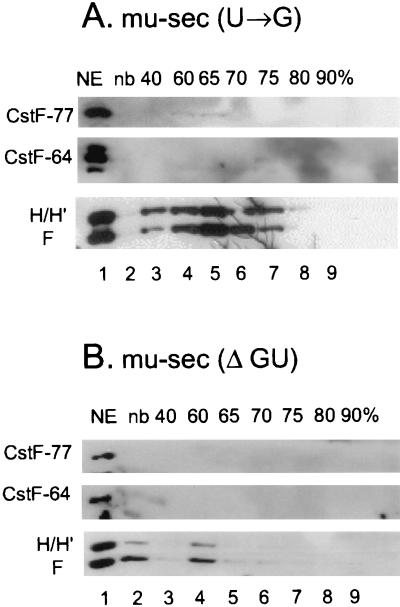
We hypothesized that a competition between the hnRNP H, H′, and F proteins and CstF-64 for binding to the same region of the RNA could occur. We therefore determined if hnRNPs H and H′ and/or F associate with mu-sec pre-mRNA via downstream sequences. Using the Sephacryl column and an affinity purification technique we analyzed the RNA-protein complexes formed in A20 memory B-cell extracts on two mu-sec mutants (constructs are shown in Fig. 1B). In the mu-sec(U→G) construct, four U residues have been converted to Gs in the AAUAAA polyadenylation signal and adjacent upstream AU-rich element; in mu-sec(ΔGU) the two downstream GU-rich elements where CstF binds were deleted along with the adjacent downstream sequences. Disruption of either the AAUAAA polyadenylation signal or downstream sequences, respectively resulted in a loss of CstF binding to the RNA as shown in a representative experiment in Fig. 5. Association of CPSF subunits with both of these mu-sec mutants was also blocked (data not shown). However, binding of hnRNPs H, H′, and F to the RNA was retained in the mu-sec(U→G) construct (Fig. 5A), where the GU- or U-rich element and adjacent downstream sequences are undisturbed. In contrast, deletion of these downstream sequences results in a dramatic reduction of hnRNP H, H′, and F binding to the mu-sec(ΔGU) RNA (Fig. 5B). Thus, downstream sequences are required for hnRNP H, H′, and F to associate with the RNA in the memory B-cell extracts. Curiously, the region downstream of the mu-sec poly(A) site contains only limited regions of G richness. The hnRNP H, H′, and F proteins might interact with the GU-rich region either directly or through protein-protein interactions.
FIG. 5.
The region downstream of the poly(A) site facilitates binding of hnRNP H, H′, and F to the Ig mu-sec-pA site. The indicated biotinylated and 32p-labeled pre-mRNAs were incubated with A20 (memory B-cell) nuclear extracts under conditions permitting poly(A) complex formation. The reaction mixtures were fractionated by gel filtration and affinity purified as described for Fig. 4C and in Materials and Methods. Affinity-purified proteins from pooled fractions were analyzed by SDS–8% PAGE and Western immunoblotted with the indicated antibodies. (A) Proteins bound to mu-sec pre-mRNA with multiple U-to-G mutations in the AU-rich poly(A) signal region. (B) Proteins bound to mu-sec pre-mRNA truncated to remove the GU-rich downstream region. NE, nuclear extract; nb, no-biotin control. NE and nonbiotinylated RNA were prepared as described for Fig. 4.
CPSF and CstF also failed to associate with constructs lacking downstream or AAUAA regions in nuclear extracts prepared from plasma cells (data not shown). Consistent with our observations with the wild-type mu-sec construct in plasma extracts (Fig. 4C), hnRNP H, H′, and F family proteins did not associate with either of the mu-sec poly(A) region mutants in plasma nuclear extracts (data not shown). Therefore the association of hnRNP H, H′, and F with the downstream regions of the mu-sec polyadenylation signal appears to be developmentally regulated.
Large complexes containing hnRNPs H, H′, and F are associated with the mu-mb-pA site in memory B-cell extracts.
hnRNP H, H′, and F complex formation with pre-mRNA in memory but not plasma B cells was investigated with the mu-mb pre-mRNA. Figure 6 shows the results of experiments performed using the strong mu-mb-pA site and a point mutation of the mu-mb AAUAAA hexanucleotide to AAGAAA; for simplicity, only the hnRNP profile is shown. Affinity-purified CstF eluted from the columns with mu-mb RNA at 60 to 70% of the bed volume as had been seen with mu-sec RNA in extracts from both cell types (data not shown). However, complexes of hnRNP H, H′, and F eluting at 40 to 60% of the bed volume, a position in the column larger than those of the CstF-containing fractions, were seen only in the memory B-cell extracts (compare affinity fractions in Fig. 6A and C). Mutation of the poly(A) signal hexanucleotide to AAGAAA resulted in the predicted loss of CstF-CPSF complex formation but still supported the assembly of large hnRNP H, H′, and F-containing complexes in the memory B-cell extract (Fig. 6B, affinity fraction). This observation suggests that association of the hnRNP H/H′/F family proteins with the RNA requires neither an intact hexanucleotide poly(A) signal nor assembly of a polyadenylation complex and occurs on both the mu-mbpA and mu-sec-pA sites.
In the nuclear extracts from plasma cells, the mu-mb pre-mRNA supported the formation of a polyadenylation complex containing CstF subunits and CPSF subunits eluting from the column at 65 to 75% of the column bed volume (data not shown). However, only a small amount of hnRNP H, H′, or F was capable of being affinity purified with the mu-mb RNA (Fig. 6C, affinity fraction). A similar observation was also made with the SVL poly(A) site and the Ig γ heavy-chain sites in plasma cells (data not shown). These results confirm the conclusion reached for the mu-sec site, namely, that in plasma nuclear extracts hnRNP H, H′, and F do not efficiently form complexes with input pre-mRNA. Similar results were obtained with pre-mRNA containing SVL sequences (data not shown).
Competition between hnRNP F and H′ for binding to RNA.
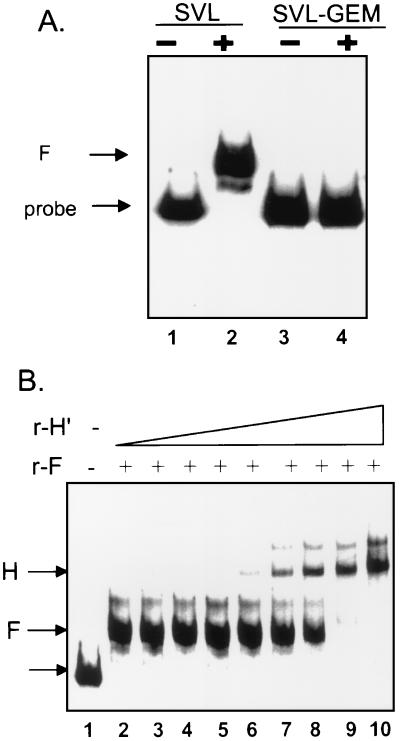
The data described above demonstrate that, in memory B cells, hnRNP F protein, in a large complex, can interact with the three polyadenylation signals tested. To determine if hnRNP F was capable of direct interaction with the pre-mRNA, we allowed recombinant hnRNP F to bind to 32P-labeled SVL pre-mRNA and performed a gel retardation assay (Fig. 7A). The binding of hnRNP F to SVL pre-mRNA seen with the intact SVL pre-mRNA (lane 2) is abolished by the replacement of the G-rich region downstream of the CstF binding site with polylinker sequences (Fig. 7A, lane 4). This indicates that the binding of hnRNP F to SVL pre-mRNA is direct and in close proximity to the GU- or U-rich downstream region of the SVL pre-mRNA. Purified recombinant hnRNP F also binds to Ig μRNA and Ig γ pre-mRNAs, both at the membrane and secretory form-encoding sites, as determined by UV-cross-linking experiments (data not shown).
FIG. 7.
Recombinant hnRNP F binding to pre-mRNA is influenced by the region downstream of the poly(A) site and by hnRNP H′. (A) RNA substrates containing SVL pre-mRNA or SVL pre-mRNA with nearly the entire region downstream of the CstF binding site replaced with polylinker sequences (SVL-Gem) were incubated with 200 ng of recombinant hnRNP F (lanes +), and protein-RNA complexes were analyzed on a nondenaturing 5% acrylamide gel. Lanes −, input RNA only. (B) RNA substrate containing SVL pre-mRNA (lane 1) was incubated with 200 ng of recombinant hnRNP F (lane 2) and increasing concentrations of hnRNP H′ (lane 3, 10 ng; lane 4, 12 ng; lane 5, 15 ng; lane 6, 17 ng; lane 7, 20 ng; lane 8, 22 ng; lane 9, 25 ng; lane 10, 30 ng). Complexes were analyzed for panel A. Arrows, migration of the markers: bottom arrow, RNA probe alone; middle arrow (F), hnRNP F plus probe; top arrow (H), hnRNP H′ plus probe.
Since hnRNP H and H′ and hnRNP F have a preference for G-rich sequences, they may be competing for a shared binding site on the pre-mRNA. Addition of recombinant hnRNP H′ to a mixture containing hnRNP F and intact SVL pre-mRNA resulted in a shift from an F-specific complex to one that is the same size previously observed with H′ alone (1) (Fig. 7B). The ratio of recombinant H′ to F necessary to achieve this shift in complex size is approximately the same as the F/H protein ratio seen by Western analyses in memory B-cell extracts, leading us to conclude that the hnRNP F-to-H′ ratio influences the binding characteristics of F to pre-mRNA.
The amount of recombinant F protein needed for maximal binding in Fig. 7 (approximately 200 ng) is much larger than that previously measured with hnRNP H′ (1). This could be due to a low affinity of hnRNP F for polyadenylation substrates or alternatively to the relative activity of the recombinant protein preparations. Purification of His-tagged hnRNP F on a nickel column requires a denaturation-and-renaturation step (5), which may reduce its specific activity. Finally, F may be modified in eucaryotic cells, and those modifications would be lacking in the bacterially produced protein, necessitating the large amounts necessary for an effect here.
HnRNP F reduces CstF-64 RNA-binding activity and the cleavage reaction.
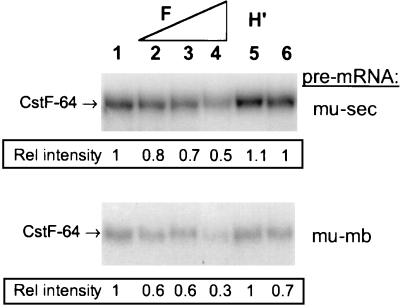
We wanted to test the hypothesis that excess hnRNP F, as seen in the memory cells, could interfere with CstF-64 binding to pre-mRNA containing a poly(A) site and GU- or U-rich downstream sequence. We added increasing concentrations of purified recombinant hnRNP F protein to plasma nuclear extract and then asked how efficiently CstF-64 could bind to 32P-labeled pre-mRNA. The reaction mixture was treated with anti-CstF-64 antiserum and the amount of 32P label UV cross-linked to CstF-64 that was immunoprecipitated was determined by autoradiography (Fig. 8). The ability of CstF-64 to bind to both the mu-sec and mu-mb pre-mRNAs was diminished when increasing amounts of recombinant hnRNP F protein were added (compare lanes 4 and 1).
FIG. 8.
Alteration of the hnRNP H′-to-F ratio influences CstF-64 RNA-binding activity. Recombinant hnRNP F or hnRNP H′ proteins were added to either 32P-labeled mu-sec or mu-mb pre-RNA prior to the addition of the AxJ nuclear extract. Reaction mixtures were incubated at 30°C for 5 min, placed on ice, and irradiated with UV light as described in Materials and Methods. Unprotected RNA was digested with RNase A, the proteins were immunoprecipitated with anti-CstF-64 antibody, and the 32p-tagged CstF-64 was electrophoresed on SDS–7.5% PAGE gel and quantified by densitometry. An anti-COOH-terminal CstF-64 antibody was used for the immunoprecipitations. Lanes 1 and 6, no recombinant protein; lanes 2 to 4, recombinant hnRNP F (0.25, 0.5, and 1 μg respectively), added; lane 5, 1 μg of recombinant H′ added.
While the amounts of protein added are large, addition of similar amounts of control proteins such as DSEF-1 (Fig. 8, lane 5), glutathione S-transferase, or bovine serum albumin did not interfere with the ability of CstF-64 to bind to pre-mRNA (data not shown) (1).
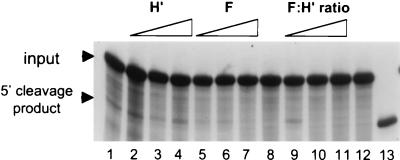
Since recombinant hnRNP F had a negative effect on CstF-64 binding, we wondered if it would have an influence on the cleavage reaction on pre-mRNA. Using nuclear extracts from plasma cells and SVL pre-mRNA we found that addition of increasing amounts of hnRNP F blocked production of the cleavage product (Fig. 9, lanes 5 to 7). When equivalent amounts of recombinant hnRNP H′ (lanes 2 to 4) or a His-tagged recombinant sucrose nonfermenting protein (not shown) were added to the plasma cell extract, neither enhancement nor diminution of activity was seen. When the hnRNP F-to-H′ ratio was increased, the cleavage reaction was also blocked (Fig. 9, lanes 9 to 11). We therefore conclude that high levels of hnRNP F are inhibitory for the cleavage reaction. The regulatory function of hnRNP F may extend beyond blocking CstF-64 from RNA binding since F seems to act in vitro at a lower level to block cleavage than is required to inhibit CstF-64 binding. It is known that hnRNP F blocks other cleavage and/or splicing factors (see Discussion).
FIG. 9.
Recombinant hnRNP F inhibits the nuclear cleavage reaction. The 32p-labeled SVL pre-mRNA was incubated in the in vitro cleavage system from plasma cells for 30 min at 30°C. Products were analyzed on a 5% acrylamide gel containing 8 M urea. The positions of the input pre-mRNA and the 5′ cleavage product are indicated. Lanes 1 to 7 and 9 to 11 contain nuclear extract, while in lanes 8 and 12 no nuclear extract was added. Lane 1, no recombinant protein; lane 2, 100 ng of recombinant H′ (rH′); lane 3, 300 ng of rH′; lane 4, 600 ng of rH′; lane 5, 100 ng of recombinant F (rF); lane 6, 300 ng of rF; lane 7, 600 ng of rF; lane 9, 100 ng of rF plus 500 ng of rH′; lane 10, 300 ng of rF plus 300 ng of rH′; lane 11, 500 ng of rF plus 100 ng of rH′; lane 13, 32P-labeled runoff transcript of an SVL template cut with HincII that is 6 nucleotides shorter than the cleavage product. Recombinant hnRNP F and H′ proteins were purified as described in Materials and Methods; recombinant proteins were preincubated with the RNA for 5 min at room temperature prior to the addition of the nuclear extract.
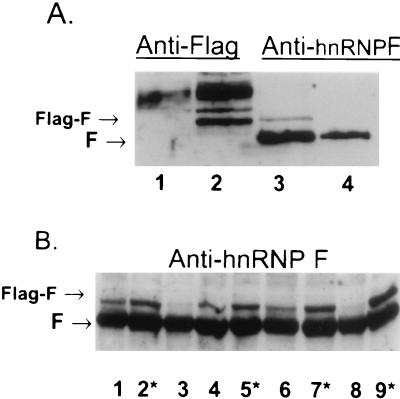
Plasma cells stably transfected with hnRNP F show reduced Ig sec-to-mb mRNA ratios.
AxJ myeloma and plasma cells expressing Ig heavy-chain mRNA with a 36:1 ratio of secretory form-to membrane-encoding species were stably transfected with recombinant Flag-tagged hnRNP F driven by the cytomegalovirus promoter. A Western blot of the protein expressed in individual G418-resistant transfectants expressing various levels of the Flag-tagged hnRNP F are shown in Fig. 10. In Fig. 10A, there is a protein in the transfected cells (lanes 2 and 3) which reacts with both anti-Flag and anti-F antibodies. This Flag-F protein is missing in the untransfected AxJ cells. There is a protein that cross-reacts with the anti-Flag in the untransformed and transfected AxJ cells (lanes 1 and 2), but this protein is not related to hnRNP F (lanes 3 and 4). Several clones of AxJ transfected with F were isolated; in some but not all of the transfected cells the overall levels of Flag-F increased significantly (Fig. 10B). The increase in F levels in the transfectants is not as high as the increase in authentic F levels in memory and plasma cells (Fig. 3, compare hnRNP F in lane 3 with that in lane 4 or 5).
FIG. 10.
Western analysis of the hnRNP F transfectants. Proteins from AxJ plasma cells or transfectants of AxJ with hnRNP F linked to the Flag-tagged epitope were separated on an SDS–10% PAGE gel and blotted to membranes. The positions of the authentic hnRNP F and the Flag-tagged F protein are indicated. (A) One gel was run and cut in half vertically. Lanes 1 and 2 were probed with antibodies to the Flag epitope, while lanes 3 and 4 were probed with antibodies to hnRNP F. Lanes 1 and 4 contain AxJ cell lysates, while lanes 2 and 3 contain lysate from AxJ transfectant B4. (B) Nine individual G418-resistant clones were isolated and analyzed. Asterisks indicate transformants chosen for further study. Lane 1, transformant B6; lane 2, B5; lane 3, B4; lane 4, C4; lane 5, C5; lane 6, C6; lane 7, D4; lane 8, D5; lane 9, D6.
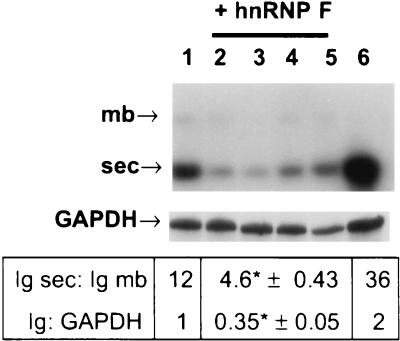
To determine if the increased expression of F influenced the poly(A) site choice in the resident Ig heavy-chain gene, the poly(A)+ selected RNA from the indicated (Fig. 11) Flag-F transfectants was obtained and probed on a Northern blot for both the Ig secretory form-encoding (1.8-kb) and membrane-encoding (3.6-kb) species (23). A sample of one of the four determinations is shown and all the data obtained are summarized in Fig. 11. In cells expressing endogenous as well as recombinant hnRNP F, the ratio of secretory form- to membrane-encoding species decreased dramatically, while transfection with the Flag vector alone had a more modest effect. In addition, the overall amount of Ig heavy-chain mRNA, relative to that of GAPDH control mRNA, was decreased in the Flag-F transfectants, consistent with what we previously observed in memory B cells (23). We conclude that overexpression of the Flag-tagged hnRNP F significantly alters the regulation of the endogenous Ig heavy-chain gene in plasma cells.
FIG. 11.
Northern analysis of the hnRNP F transfectants. Poly(A)+ RNA was isolated from AxJ cells or the transfected AxJ cells, run on an formamide-formaldehyde-containing 0.8% agarose gel, blotted, and then probed sequentially with 32P-labeled antisense RNA derived from the IgG2a CH3 exon and then GAPDH. The locations of the 1.8- and 3.6-kb messages bearing, respectively, the sec-pA site (sec) and the mb-pA site (mb) are indicated. The amount of each Ig heavy-chain species and the amount of total Ig mRNA relative to that of GAPDH were quantified in the phosphorimager analysis of four separate Northern blots. The mean values (± the standard errors) are beneath the corresponding lanes of the blot. Lane 1, values obtained with AxJ cells transfected with the empty Flag vector alone; lanes 2 to 5, values obtained from four separate Northern blots of the transfectants receiving the hnRNP F-Flag recombinant, transfectants A5, B5, D4, and D6, respectively; lane 6, RNA from untransfected AxJ. Asterisks indicate results that were considered to be extremely statistically different (P < 0.0002) from the other values by sequential, two-tailed, unpaired t tests.
DISCUSSION
The results presented in this paper demonstrate that changes in the levels of hnRNP F are an important determinant in the regulated use of alternative polyadenylation sites in memory and plasma cells. The ratio of hnRNP F to H or H′ is higher in memory B cells than in plasma cells. Large assemblages with pre-mRNA substrates containing hnRNP H, H′, and F but deficient in CstF-64 are found in memory B-cell extracts but not in plasma cells. Formation of these large complexes is dependent on the region downstream of AAUAAA in pre-mRNA, suggesting that CstF-64 and the hnRNPs bind to similar regions of the RNA in a mutually exclusive fashion. We showed that purified recombinant hnRNP F could bind to the region downstream of a poly(A) site and reduce the association of CstF-64 in a nuclear extract with pre-mRNA. Addition of recombinant hnRNP F to a nuclear extract from plasma cells was able to block the in vitro cleavage reaction even more efficiently. The cleavage inhibition occurred when the level of recombinant hnRNP F equaled or exceeded that of added recombinant H′. Finally, overexpression of hnRNP F in plasma cells, where the secretory poly(A) site is normally dominant, resulted in a significant decrease in the endogenous Ig heavy-chain sec-to-mb mRNA ratio. We therefore conclude that hnRNP F plays an important role in the regulated use of alternative polyadenylation sites by memory and plasma cells. Our experiments are the first to show a role for the mammalian hnRNP F in the cleavage and polyadenylation reaction. Previously yeast hnRNP Nab4p/Hrp1p, with homology to mammalian hnRNP A and B, was shown to prevent utilization of alternative cleavage sites (25).
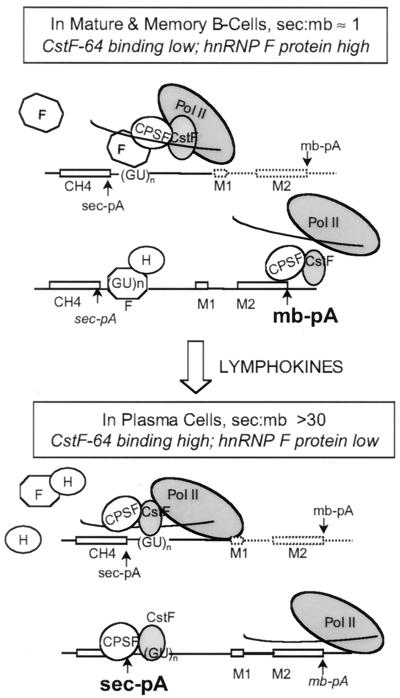
Based on the available data, we propose a model in which hnRNP F participates in the regulation of alternative poly(A) site selection during B-cell development (Fig. 12). Carried along by the carboxyl-terminal domain (CTD) of RNA Pol II (41), hnRNP F can bind to the pre-mRNA in the region downstream of the sec-pA site as it is synthesized. When levels of hnRNP F are high, as in mature and memory B cells, F may prevent the association of CstF-64 with the GU- or U-rich downstream element, thereby blocking formation of a stable polyadenylation complex at that site. The in vitro effect we have shown here, i.e., that of free hnRNP F blocking CstF-64 association with pre-mRNA, is consistent with this model. Following lymphokine stimulation of mature or memory B cells, the reduction of hnRNP F levels in authentic plasma cells could relieve the F-CTD association and therefore the competition with CstF-64 for the sec-pA GU-rich region would be considerably diminished, thus favoring use of the weak promoter-proximal sec-pA site. We have shown that both the sec- and the mb-pA sites are influenced by free hnRNP F in vitro. However, in understanding the regulation of this locus the positions of the sites relative to the promoter and their distance from each other are critical parameters (28). The weak sec-pA site is promoter proximal, and the stronger membrane-encoding site is over 2 kb away. Any increase in polyadenylation efficiency favors the first of the two linked sites, and use of the first site precludes use of the second. The regulation achieved by a balance between the F and H or H′ ratios satisfies a model we proposed several years ago. We proposed both an inhibitor (such as F), acting in early and memory cells, and an activator (such as H or H′), acting in plasma cells (7). Our previous data established hnRNP H′ as a stimulator of polyadenylation in HeLa cells (1). As seen in the data in Fig. 9, H′ does not have a large effect in plasma cells where the endogenous levels are already high. The negative influence of F on processing efficiency appears to be dominant over that of H′, suggesting that heterodimers between the two are likely to be inhibitory in cells. This suggests that the association of the hnRNP H, H′, and F proteins seen in the complexes purified from memory cell extracts may act in vivo to block the association of CstF-64 with the secretory-form-specific poly(A) site and/or the cleavage reaction at that site.
FIG. 12.
Model for the regulation of poly(A) site choice during B-cell development. In mature and memory B cells, hnRNP F protein levels are high while CstF-64 binding activity is low. The binding of hnRNP F and hnRNP H and H′ to the sec-pA site near the downstream GU- or U-rich element immediately after that portion of the RNA is synthesized may inhibit the association of CstF-64 with pre-mRNA and hinder the assembly of a stable polyadenylation complex at that site. Consequently, there is an increased chance that the strong downstream mb-pA site will be transcribed by a polymerase molecule still carrying the polyadenylation factors and serve as a cleavage or polyadenylation signal. Upon lymphokine stimulation and differentiation to an antibody-secreting plasma cell, hnRNP F protein expression is decreased, which leads to an increase in the apparent affinity of CstF-64 for the sec-pA site as it is transcribed. The cleavage-polyadenylation complex can form at the sec-pA site, even before the polymerase reaches the membrane site, thus favoring use of the promoter-proximal (sec) site. This plasma cell phenotype can be reversed by overexpression of recombinant hnRNP F.
Previously, we compared the expression of the endogenous Ig heavy-chain mRNA in memory B cells to that in plasma cells. We found that the final Ig heavy-chain sec/mb mRNA ratio was about 1:1 in memory cells versus 36:1 in plasma cells. In addition, compared with memory cells, the plasma cells had about a 10-fold higher production of total Ig heavy-chain mRNA than control messages such as GAPDH and cyclophilin (23). We attributed this increase in both the sec/mb ratio and overall amount of Ig heavy-chain mRNA in plasma cells to increased processing and polyadenylation of the primary transcript since a comparison of the two cell types revealed no significant differences in transcription rates of the Ig gene or in half-life between the Ig mRNA species. In the plasma cells that we transformed here with excess hnRNP F, the Ig sec/mb ratio fell from 36:1 to 4.6:1 and the level of Ig mRNA fell from 2 times to about 0.35 times that of GAPDH. Both of these observations indicate that there is an overall decrease in the processing and polyadenylation of the Ig heavy-chain primary transcript that is almost but not quite as large as that seen in memory cells. In keeping with our model, access to the Ig sec-pA site appears to be blocked by the overexpression of hnRNP F. Before reaching the mb-pA site (>2 kb downstream), the polyadenylation machinery might uncouple from CTD. This disengagement could lead to a transcript that would remain unprocessed and hence turn over rapidly in the nucleus, leading to lower levels of mature Ig heavy-chain mRNA. Since hnRNP F is an abundant protein, we were not able to increase its expression up to the level seen in memory cells with the cytomegalovirus promoter; this may explain why the effect of overexpression of hnRNP F on the Ig sec/mb ratios is not complete.
At the amino acid level, hnRNP H (molecular mass, 56 kDa) has 96% identity with hnRNP H′ (molecular mass, 56 kDa) and 78% identity with hnRNP F (molecular mass, 53 kDa); hnRNPs H′ and F are 75% identical. Each protein has three RNA-binding domains which include submotifs resembling RNP-1, RNP-2, CS-1, and CSR-3 consensus sequences (6, 12, 21). It remains to be determined if hnRNP H and H′ differ in function. HnRNPs H, H′, and F were purified via their affinity for poly(rG) (21) but were not analyzed rigorously for their favored RNA targets. Interestingly, the levels of these proteins and their ratios to each other vary widely in the many tissues examined (13). Both hnRNP H and F are components of a neural cell-specific complex which assembles on the G-rich control sequence of the src proto-oncogene pre-mRNA and directs the alternative splicing of the short N1 exon (5, 24).
In addition, hnRNP F protein has been reported to interact with the cap binding protein complex (CBC) and to associate more strongly with CBC-RNA than with naked RNA (10). The extent of these interactions in the context of other nuclear proteins is unclear, as the experiments examined only a single modified U1 RNA substrate containing no processing signals and two purified proteins. Based on the general role of the CBC in splicing, hnRNP F was hypothesized to be a general splicing factor. Interestingly, addition of low levels of recombinant hnRNP F to immunodepleted HeLa extracts partially restored in vitro splicing, but addition of high levels of hnRNP F led to increased inhibition of splicing. The mechanism of this inhibition is not understood. The potential exists for a complex competition between the binding of hnRNP F at the GU-rich downstream element of a pre-mRNA to block polyadenylation and at G- or GU-rich regions near splice sites to activate cryptic signals.
Exploring the details of our model and the novel role(s) of hnRNP F as a potential linker of polyadenylation and splicing may elucidate important new control mechanisms by which Ig gene expression is directed in a B-cell stage-specific manner. Since the levels of hnRNP F versus those of H or H′ are so different in various tissues, these mechanisms may extend to a variety of cell types and genes beyond those of lymphoid origin.
ACKNOWLEDGMENTS
We are grateful to Clinton MacDonald and Paula Grabowski for providing antisera, to Cathy Phillips for the mu-sec clones, and to Douglas L. Black for the hnRNP F clone. These colleagues also provided useful insights. Large-scale cell culture services were provided by the Cell Culture Center, sponsored by the National Center for Research Resources, NIH.
J.W. was supported by GM56434. C.M. was supported by CA86433 from NCI.
REFERENCES
- 1.Bagga P, Arhin G, Wilusz J. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 1998;26:5343–5350. doi: 10.1093/nar/26.23.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagga P S, Ford L P, Chen F, Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyer K, Dandekar T, Keller W. RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J Biol Chem. 1997;272:26769–26779. doi: 10.1074/jbc.272.42.26769. [DOI] [PubMed] [Google Scholar]
- 4.Chen F, MacDonald C, Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou M-Y, Rooke N, Turck C W, Black D L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol Cell Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreyfuss G, Swanson M, Pinol-Roma S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem Sci. 1988;13:86–91. doi: 10.1016/0968-0004(88)90046-1. [DOI] [PubMed] [Google Scholar]
- 7.Edwalds-Gilbert G, Milcarek C. Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol Cell Biol. 1995;15:6420–6429. doi: 10.1128/mcb.15.11.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwalds-Gilbert G, Veraldi K, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaspohler J A, Boczkowski D, Hall B L, Milcarek C. The 3′-untranslated region of membrane exon 2 from the gamma 2a immunoglobulin gene contributes to efficient transcription termination. J Biol Chem. 1995;270:11903–11911. doi: 10.1074/jbc.270.20.11903. [DOI] [PubMed] [Google Scholar]
- 10.Gamberi C, Izaurralde E, Beisel C, Mattaj I. Interaction between the human nuclear cap-binding protein complex and hnRNP F. Mol Cell Biol. 1997;17:2587–2597. doi: 10.1128/mcb.17.5.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graber J, Cantor C, Mohr S, Smith T. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc Natl Acad Sci USA. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honore B, Rasmussen H, Vorum H, Dejgaard K, Liu X, Gromov P, Madsen P, Gesser B, Tommerup N, Celis J. Heterogeneous nuclear ribonucleoproteins H, H′, and F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J Biol Chem. 1995;270:28780–28789. doi: 10.1074/jbc.270.48.28780. [DOI] [PubMed] [Google Scholar]
- 13.Kamma H, Portman D S, Dreyfuss G. Cell type-specific expression of hnRNP proteins. Exp Cell Res. 1995;221:187–196. doi: 10.1006/excr.1995.1366. [DOI] [PubMed] [Google Scholar]
- 14.Kobrin B J, Milcarek C, Morrison S L. Sequences near the 3′ secretion-specific polyadenylation site influence levels of secretion-specific and membrane-specific immunoglobulin G2b mRNA in myeloma cells. Mol Cell Biol. 1986;6:1687–1697. doi: 10.1128/mcb.6.5.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lassman C R, Matis S, Hall B L, Toppmeyer D L, Milcarek C. Plasma cell-regulated polyadenylation at the Ig gamma 2b secretion-specific poly(A) site. J Immunol. 1992;148:1251–1260. [PubMed] [Google Scholar]
- 16.Lassman C R, Milcarek C. Regulated expression of the mouse γ2b Ig H chain gene is influenced by polyA site order and strength. J Immunol. 1992;148:2578–2585. [PubMed] [Google Scholar]
- 17.MacDonald C, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martincic K, Campbell R, Edwalds-Gilbert G, Souan L, Lotze M, Milcarek C. Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the Go to S phase transition. Proc Natl Acad Sci USA. 1998;95:11095–11100. doi: 10.1073/pnas.95.19.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matis S A, Martincic K, Milcarek C. B-lineage regulated polyadenylation occurs on weak poly(A) sites regardless of sequence composition at the cleavage and downstream regions. Nucleic Acids Res. 1996;24:4684–4692. doi: 10.1093/nar/24.23.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews H R. The application of Cerenkov counting to column chromatography of 32P labelled substances. J Chromatogr. 1968;36:302–308. doi: 10.1016/s0021-9673(01)92946-2. [DOI] [PubMed] [Google Scholar]
- 21.Matunis M, Xing J, Dreyfuss G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22:1059–67. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milcarek C, Hall B. Cell-specific expression of secreted versus membrane forms of immunoglobulin gamma 2b mRNA involves selective use of alternate polyadenylation sites. Mol Cell Biol. 1985;5:2514–2520. doi: 10.1128/mcb.5.10.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milcarek C, Hartman M, Croll S. Changes in abundance of IgG 2a mRNA in the nucleus and cytoplasm of a murine B-lymphoma before and after fusion to a myeloma cell. Mol Immunol. 1996;33:691–701. doi: 10.1016/0161-5890(96)00009-0. [DOI] [PubMed] [Google Scholar]
- 24.Min H, Chan R, Black D. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 25.Minvielle-Sebastia L, Beyer K, Krecic A, Hector R, Swanson M, Keller W. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 1998;17:7454–7468. doi: 10.1093/emboj/17.24.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson M L. RNA processing and expression of immunoglobulin genes. In: Snow E C, editor. Handbook of B and T lymphocytes. San Diego, Calif: Academic Press; 1994. pp. 321–342. [Google Scholar]
- 27.Peterson M L, Gimmi E R, Perry R P. The developmentally regulated shift from membrane to secreted μ- mRNA production is accompanied by an increase in cleavage-polyadenylation efficiency but no measurable change in splicing efficiency. Mol Cell Biol. 1991;11:2324–2327. doi: 10.1128/mcb.11.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson M L, Perry R P. Regulated production of mu-m and mu-s mRNA requires linkage. Proc Natl Acad Sci USA. 1986;83:8883–8887. doi: 10.1073/pnas.83.23.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips C, Schimpl A, Dietrich-Goetz W, Clements J, Virtanen A. Inducible nuclear factors binding the IgM heavy chain pre-mRNA secretory poly(A) site. Eur J Immunol. 1996;26:3144–3152. doi: 10.1002/eji.1830261247. [DOI] [PubMed] [Google Scholar]
- 30.Phillips C, Virtanen A. The murine IgM secretory poly(A) site contains dual upstream and downstream elements which affect polyadenylation. Nucleic Acids Res. 1997;25:2344–2351. doi: 10.1093/nar/25.12.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian Z, Wilusz J. An RNA binding protein specifically interacts with a functionally important domain of the downstream element of the simian virus 40 late polyadenylation signal. Mol Cell Biol. 1991;11:5312–5320. doi: 10.1128/mcb.11.10.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takagaki Y, MacDonald C, Shenk T, Manley J. The human 64-kDa polyadenylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc Natl Acad Sci USA. 1992;89:1403–1407. doi: 10.1073/pnas.89.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takagaki Y, Manley J. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 34.Takagaki Y, Manley J. A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature. 1994;372:471–474. doi: 10.1038/372471a0. [DOI] [PubMed] [Google Scholar]
- 35.Takagaki Y, Manley J L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takagaki Y, Manley J L. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takami Y, Kikuchi H, Nakayama T. Chicken histone deacetylase-2 controls the amount of the IgM H-chain at the steps of both transcription of its gene and alternative processing of its pre-mRNA in the DT40 cell line. J Biol Chem. 1999;34:23977–23990. doi: 10.1074/jbc.274.34.23977. [DOI] [PubMed] [Google Scholar]
- 38.Veraldi K, Edwalds-Gilbert G, MacDonald C, Wallace A, Milcarek C. Isolation and characterization of polyadenylation complexes assembled in vitro. RNA. 2000;6:768–777. doi: 10.1017/s135583820099246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilusz J, Shenk T, Takagaki Y, Manley J. A multicomponent complex is required for the AAUAAA -dependent cross-linking of a 64-kilodalton protein to polyadenylation substrates. Mol Cell Biol. 1990;10:1244–1248. doi: 10.1128/mcb.10.3.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan D-H, Weiss E, Nevins J. Identification of an activity in B-cell extracts that selectively impairs the formation of an immunoglobulin μs poly(A) site processing complex. Mol Cell Biol. 1995;15:1901–1906. doi: 10.1128/mcb.15.4.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida T, Kokura K, Makino Y, Ossipow V, Tamura T. Heterogeneous nuclear RNA-ribonucleoprotein F binds to DNA via an oligo(dG)-motif and is associated with RNA polymerase II. Genes Cells. 1999;4:707–719. doi: 10.1046/j.1365-2443.1999.00295.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L, Liu W, Grabowski P J. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA. 1999;5:117–130. doi: 10.1017/s1355838299981530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]