Abstract
tmRNA (also known as 10Sa RNA or SsrA) plays a central role in an unusual mode of translation, whereby a stalled ribosome switches from a problematic mRNA to a short reading frame within tmRNA during translation of a single polypeptide chain. Research on the mechanism, structure and biology of tmRNA is served by the tmRNA Website, a collection of sequences for tmRNA and the encoded proteolysis-inducing peptide tags, alignments, careful documentation and other information; the URL is http://www.indiana.edu/~tmrna. Four pseudoknots are usually present in each tmRNA, so the database is rich with information on pseudoknot variability. Since last year it has doubled (227 tmRNA sequences as of September 2001), a sequence alignment for the tmRNA cofactor SmpB has been included, and genomic data for Clostridium botulinum has revealed a group I (subgroup IA3) intron interrupting the tmRNA T-loop.
BACKGROUND
tmRNA is a bacterial RNA containing both tRNA-like and mRNA-like domains that is thought to solve problems arising from ribosomes stalled in translation. A ribosome might stall if the mRNA lacks an in-frame stop codon or if a particular charged tRNA species is lacking; stalling could result in sequestration of a substantial fraction of ribosomes and the release of an incomplete protein that may be deleterious. In the current model (1) for tmRNA usage, the alanine-charged form enters the stalled ribosome, the nascent peptide is transferred to its alanine moiety, the stalled mRNA is ejected and the ‘resume codon’ of the reading frame within tmRNA is positioned in the A-site. Standard translation proceeds on tmRNA to a stop codon and the protein is released, a product of reading frames from two separate RNAs; its tmRNA-directed peptide tag targets it for proteolysis. Thus, the stalled ribosome is freed and its incomplete product destroyed.
An example of the standard secondary structure of tmRNA (2,3) is shown in Figure 1A. The tRNA-like domain has a perfect analog of the acceptor branch of tRNAAla, with the appropriate modified bases (4). Analogy with the anticodon branch is strained: the anticodon stem–loop is replaced by the long P2 stem, the region corresponding to the D stem–loop has the conserved sequence 5′-N3–5GGN1–3UCGA with no conserved internal pairing and no modified bases, and the analog of the variable loop is uniformly 3 nt (usually 5′-GAC), shorter than for any tRNA. These features must be responsible for the unusually large (111–137°) angle that was recently measured between the acceptor and P2 stems in the Escherichia coli tRNA-like domain (5). This measurement was presented with a structural model of the tRNA-like domain, which, although welcome, may suffer from the assumption of a 2 bp D-stem that is not supported phylogenetically (e.g. Fig. 1A). The remainder of standard tmRNA is a large loop containing four pseudoknots and the tag reading frame. Variations on the standard structure are found in cyanobacteria, where the last pseudoknot is found as two short abutting pseudoknots, and in plastids, where the region corresponding to the last three pseudoknots appears largely unstructured (6). The most striking structural variation is found in α-proteobacteria and a small lineage in the cyanobacteria, where tmRNA is found in two pieces with the break in the large loop; this structure results from gene permutation, which has occurred independently in the two clades (7). Permuted tmRNA-like sequence was found in the mitochondrial genome of Reclinomonas americana, but lacking a tag reading frame.
Figure 1.
Intron in the C.botulinum tmRNA. (A) Standard secondary structure of the presumed post-splicing tmRNA; tag reading frame in lower case. The alanine charging tmRNA and the protein product of tmRNA translation are indicated. (B) Canonical features of group IA3 intron. Lower case, exon sequence; outline, bases highly conserved among group I introns; triangles, splice sites; dotted lines: proposed tetraloop–receptor interactions; rounded boxes, P10 partners. Sequence data from the Sanger Centre (http://www.sanger.ac.uk).
THE tmRNA WEBSITE
The tmRNA Website was established in 1997 to facilitate research on tmRNA biology, structure and mechanism of action (6). tmRNA sequences and alignments are presented with careful documentation. The many ongoing genome projects are diligently monitored to obtain new sequences, and the website is updated frequently. This year, an alignment of SmpB sequences has been included in the website; SmpB is thought to be a cofactor of tmRNA because the purified protein binds the RNA specifically and an smpB knockout has the same phenotype as an ssrA knockout (8).
The database has doubled in the last year. To accommodate the increased size, phylogenetically limited alignments and species lists are now available in addition to full versions. Some growth came from continued monitoring of ongoing genome projects. A second major source was tmRNA gene sequencing projects that took advantage of the ease with which tmRNA sequences can be obtained through PCR (3), and that produced eight new β-proteobacterial, 18 Gram-positive and 38 γ-proteobacterial tmRNA sequences (9–11). These have led to the first proposals for tertiary interactions outside of the tRNA-like domain, and also show the utility of tmRNA sequence data for classifying bacteria, the value of environmental samples as a source of new tmRNA sequences, and interesting idiosyncratic elaborations on the basic secondary structure. A stem–tetraloop that had earlier been proposed within the second pseudoknot in β-proteobacterial tmRNA (2) was found to have been elaborated in some species into a pair of stem–tetraloops or even a double-branching structure (9). Another clear example of a small idiosyncratic change is seen in Yersinia; the Yersinia enterocolitica tmRNA sequence is most similar to the Yersinia pestis sequence, which in turn is much like that of other enterobacteria. However, Y.enterocolitica seems to have recently undergone one or two small insertions that elongate the P5 stem–loop and the corresponding portion of the tag reading frame.
Another interesting finding this year was an interaction with a micromolar dissociation constant between tmRNA and the first tRNA that it specifies, tRNAAla (12). Preloading of the incoming tRNA onto tmRNA outside of the ribosome had been proposed earlier from the behavior of mutants affecting resume codon utilization (13).
GROUP I INTRON IN A tmRNA T-LOOP
Genome sequence data for Clostridium botulinum, released from the Sanger Centre, shows that a group I intron (Cb.ssrA) occurs before the last nucleotide of the tmRNA T-loop (Fig. 1). The intron is small (287 nt), contains no ORF and exhibits canonical primary and secondary structural features of group I, many of whose members are known to self-splice. Two tetraloop–receptor interactions can be proposed (dotted lines in Fig. 1B), based on identity or similarity to motifs demonstrated in the phage T4 td intron and in the Synechococcus tRNALeu intron (14,15). The presumed splicing product would yield a tmRNA like those of other clostridia, with no apparent defects; the pre-tmRNA is strongly expected to be spliced in vivo, although this remains to be demonstrated. This is the first report of an intron of any type in a T-loop, or in tmRNA.
The origin of the intron Cb.ssrA is unclear; its unelaborated P9, 5 bp P7, and the presence of P7.1–2 place it in the IA3 subgroup (16), all other members of which are found in organelles, usually in rRNAs (17). Thus, it does not appear particularly closely related to any known bacterial or bacteriophage intron (18–20). The other group I intron found in Gram-positive bacteria (not counting bacteriophages) resides in an mRNA of Clostridium difficile and can be generally classified as IA, but this transposase-associated intron bears no close affinity to Cb.ssrA or any other IA intron (18). The distribution of the tmRNA intron is very limited; tmRNA is uninterrupted in C.difficile, Clostridium perfringens and Clostridium acetobutylicum. However, the intron is suspected to occur in tmRNA of Clostridium tetani, because the BLAST server for that genome project (Göttingen Genomics Laboratory, which does not release sequences or alignments) gave high scores from the same contig for separate queries with the C.botulinum major exon or intron, that were additive for the united query.
How did Cb.ssrA transpose into the tmRNA gene? A possible first step in transposition would be reverse splicing into a new target site in an abundant cellular RNA (21,22); the intron encodes no protein, as would be expected for transposase-mediated transposition (18) or endonuclease-mediated homing (19). Reverse splicing may explain the sites of group I introns in tRNAs, all in anticodon loops. The currently known pattern of distribution of group I introns in tRNAs, putting aside cases of horizontal transfer of the same interrupted gene, requires as few as four or five original independent transposition events of closely related introns (subgroup IC3) into different bacterial tRNA genes (19,23) (Fig. 2). Why the anticodon loop and not, say, the T loop? One consideration is that group I intron 5′ splice sites are always preceded by a U, and three of the seven T-loop positions are never U. More importantly for a reverse splicing mechanism, the most exposed part of tRNA for interaction with other RNAs is its anticodon loop (24). Reverse splicing into a T-loop would seem less likely since it is expected to be occupied in tertiary interactions. tmRNA has no equivalent to the anticodon loop, and was invaded by an intron of a different subgroup (IA3) from those in tRNAs. Interestingly, the internal guide sequence of Cb.ssrA, by its adaptation to the tmRNA T-loop, would fortuitously match many tRNAs; in the completely sequenced C.acetobutylicum, 15 of 73 tRNA genes produce T-loops that would also be perfectly targeted for reverse splicing. Occurrence only in tmRNA, through a reverse splicing hypothesis, would suggest either that tmRNA expression levels exceed the aggregate for such tRNA species, or that the T-loop in tmRNA is more exposed than those of tRNAs. To complete transposition, reverse splicing should be followed by reverse transcription and homologous recombination. tRNA and tmRNA genes may favor the latter stage of transposition. tRNA genes are known to be hot spots for genetic rearrangements (25). When new site-specifity emerges among the integrases of prokaryotic genetic elements, in the majority of cases it is for tRNA and tmRNA genes; for many integrase subfamilies, the only sublocations used within tRNA and tmRNA genes are the anticodon or T-loops (26). It may be that tRNAs and tmRNAs somehow mark their genes for all these genetic activities, perhaps by hybridizing to their genes at low frequency, either as mature RNAs or nascent transcripts.
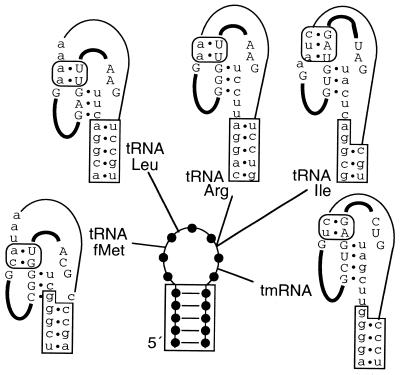
Figure 2.
Effect of group I introns interrupting tRNA and tmRNA loops on the flanking stems. A generic 7 nt loop shows intron positions in tRNAs (subgroup IC3 in anticodon loops) or tmRNA (subgroup IA3 in T-loop). RNAs are shown at a stage between the two splicing steps, with intron in upper case and thick lines, and exons in lower case and thin line. Satisfying proposed P10 (rounded box) and P1 (unboxed) pairings comes at the expense of tRNA stem pairing (boxed) in some cases. Scytonema hofmanni tRNAfMet (28); Anabaena PCC7120 tRNALeu (29); Agrobacterium tumefaciens tRNAArg (29); Azoarcus sp. tRNAIle (29).
For a group I intron in tRNALeu it was demonstrated that splicing depends strongly on pairing of the anticodon stem in the pre-tRNA (27); this exon–exon pairing, otherwise uncommon among group I introns, was proposed to compensate for the unusually short (3 bp) intron–exon pairing in P1 that is characteristic of all the tRNA group I introns. It can be noted that exon–exon pairing is essential for the protein-based splicing of tRNAs in archaea and eukaryotic nuclei. Striking as is the limitation of tRNA introns to anticodon loops, they nevertheless occur at different points within the loops (Fig. 2). Exon–exon pairing has not been investigated experimentally for other tRNA group I introns, but satisfying proposed P1 and P10 stems would come at the expense of some anticodon stems and the tmRNA T-stem. Partial forms of the anticodon stem may occur in pre-spliced RNAs; a complete anticodon stem for the pre-tRNAfMet is especially unlikely. However, the other two conflicts would be resolved if P10 consists of a single base pair, which is all that can form for natural variants of some of these introns.
Acknowledgments
ACKNOWLEDGEMENTS
I thank the many genome projects that keep their data freely available, for the tremendous service they are providing to the scientific community, and Brice Felden and Scott Kelley for sending sequences prior to publication. This work was supported by NIH grant GM59881.
REFERENCES
- 1.Keiler K.C., Waller,P.R. and Sauer,R.T. (1996) Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science, 271, 990–993. [DOI] [PubMed] [Google Scholar]
- 2.Williams K.P. and Bartel,D.P. (1996) Phylogenetic analysis of tmRNA secondary structure. RNA, 2, 1306–1310. [PMC free article] [PubMed] [Google Scholar]
- 3.Felden B., Himeno,H., Muto,A., McCutcheon,J.P., Atkins,J.F. and Gesteland,R.F. (1997) Probing the structure of the Escherichia coli 10Sa RNA (tmRNA). RNA, 3, 89–103. [PMC free article] [PubMed] [Google Scholar]
- 4.Felden B., Hanawa,K., Atkins,J.F., Himeno,H., Muto,A., Gesteland,R.F., McCloskey,J.A. and Crain,P.F. (1998) Presence and location of modified nucleotides in Escherichia coli tmRNA: structural mimicry with tRNA acceptor branches. EMBO J., 17, 3188–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stagg S.M., Frazer-Abel,A.A., Hagerman,P.J. and Harvey,S.C. (2001) Structural studies of the tRNA domain of tmRNA. J. Mol. Biol., 309, 727–735. [DOI] [PubMed] [Google Scholar]
- 6.Williams K.P. and Bartel,D.P. (1998) The tmRNA Website. Nucleic Acids Res., 26, 163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keiler K.C., Shapiro,L. and Williams,K.P. (2000) tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: a two-piece tmRNA functions in Caulobacter. Proc. Natl Acad. Sci. USA, 97, 7778–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karzai A.W., Susskind,M.M. and Sauer,R.T. (1999) SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J., 18, 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felden B., Massire,C., Westhof,E., Atkins,J.F. and Gesteland,R.F. (2001) Phylogenetic analysis of tmRNA genes within a bacterial subgroup reveals a specific structural signature. Nucleic Acids Res., 29, 1602–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelley S., Harris,J.K. and Pace,N.R. (2001) Evaluation and refinement of tmRNA structure using gene sequences from natural microbial communities. RNA, 7, 1310–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schönhuber W., Le Bourhis,G., Tremblay,J., Amann,R. and Kulakauskas,S. (2001) Utilization of tmRNA sequences for bacterial identification. BMC Microbiol., 1, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillet R. and Felden,B. (2001) Transfer RNA(Ala) recognizes transfer-messenger RNA with specificity; a functional complex prior to entering the ribosome? EMBO J., 20, 2966–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams K.P., Martindale,K.A. and Bartel,D.P. (1999) Resuming translation on tmRNA: a unique mode of determining a reading frame. EMBO J., 18, 5423–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa M. and Michel,F. (1995) Frequent use of the same tertiary motif by self-folding RNAs. EMBO J., 14, 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikawa Y., Naito,D., Aono,N., Shiraishi,H. and Inoue,T. (1999) A conserved motif in group IC3 introns is a new class of GNRA receptor. Nucleic Acids Res., 27, 1859–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel F. and Westhof,E. (1990) Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol., 216, 585–610. [DOI] [PubMed] [Google Scholar]
- 17.Damberger S.H. and Gutell,R.R. (1994) A comparative database of group I intron structures. Nucleic Acids Res., 22, 3508–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun V., Mehlig,M., Moos,M., Rupnik,M., Kalt,B., Mahony,D.E. and von Eichel-Streiber,C. (2000) A chimeric ribozyme in Clostridium difficile combines features of group I introns and insertion elements. Mol. Microbiol., 36, 1447–1459. [DOI] [PubMed] [Google Scholar]
- 19.Edgell D.R., Belfort,M. and Shub,D.A. (2000) Barriers to intron promiscuity in bacteria. J. Bacteriol., 182, 5281–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett K.D., Kahane,S., Bush,R.M. and Friedman,M.G. (1999) An unspliced group I intron in 23S rRNA links Chlamydiales, chloroplasts, and mitochondria. J. Bacteriol., 181, 4734–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roman J., Rubin,M.N. and Woodson,S.A. (1999) Sequence specificity of in vivo reverse splicing of the Tetrahymena group I intron. RNA, 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodson S.A. and Cech,T.R. (1989) Reverse self-splicing of the tetrahymena group I intron: implication for the directionality of splicing and for intron transposition. Cell, 57, 335–345. [DOI] [PubMed] [Google Scholar]
- 23.Rudi K. and Jakobsen,K.S. (1999) Complex evolutionary patterns of tRNA Leu(UAA) group I introns in the cyanobacterial radiation. J. Bacteriol., 181, 3445–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scarabino D., Crisari,A., Lorenzini,S., Williams,K. and Tocchini-Valentini,G.P. (1999) tRNA prefers to kiss. EMBO J., 18, 4571–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giroux S. and Cedergren,R. (1989) Evolution of a tRNA operon in gamma purple bacteria. J. Bacteriol., 171, 6446–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams K.P. (2002) Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res., 30, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaug A.J., McEvoy,M.M. and Cech,T.R. (1993) Self-splicing of the group I intron from Anabaena pre-tRNA: requirement for base-pairing of the exons in the anticodon stem. Biochemistry, 32, 7946–7953. [DOI] [PubMed] [Google Scholar]
- 28.Biniszkiewicz D., Cesnaviciene,E. and Shub,D.A. (1994) Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J., 13, 4629–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinhold-Hurek B. and Shub,D.A. (1992) Self-splicing introns in tRNA genes of widely divergent bacteria. Nature, 357, 173–176. [DOI] [PubMed] [Google Scholar]