Abstract
Sp3 is a ubiquitous transcription factor closely related to Sp1. Previous analyses showed that, unlike Sp1, Sp3 fails to activate transcription in certain promoter settings. This is due to the presence of an inhibitory domain located between the second glutamine-rich activation domain and the DNA-binding domain. To further analyze the transcriptional properties of Sp3, we have expressed and purified recombinant Sp3 and Sp1 as epitope-tagged proteins from stable transfected insect cells. We found that Sp3 does act as a strong activator similar to Sp1 in an in vitro transcription assay using Sp1/Sp3-depleted HeLa nuclear extract. However, on the same promoter Sp3 is almost inactive when transfected into cells. Mutational studies demonstrate that a single lysine residue is responsible for the low transcriptional activity of Sp3 in vivo. We show that Sp3, but not a mutant of Sp3 that lacks this lysine residue, is highly acetylated in vivo. Our results strongly suggest that the transcriptional activity of Sp3 is regulated by acetylation. The consequences of acetylation for the activity of Sp3 are discussed.
INTRODUCTION
Sp3 is a ubiquitously expressed transcription factor closely related to Sp1. Both proteins contain a highly conserved DNA-binding domain close to the C-terminus and two glutamine-rich activation domains in the N-terminal moiety (1). Consistent with the high conservation of the DNA-binding zinc finger region, both Sp1 and Sp3 recognize the classical GC box present in many promoters with identical affinity (2,3). The high degree of structural similarity of Sp1 and Sp3 suggested at first that they exert similar functions. The physiological roles of Sp1 and Sp3, however, appear to be significantly different. Sp1 knockout mouse embryos are severely retarded in growth and die after day 10 of embryonic development (E10) (4). Sp3-deficient embryos develop until birth, but invariably die of respiratory failure immediately after birth (5). In addition, late tooth and bone developmental processes are impaired in Sp3–/– mice (5,6).
Functional analyses of the transcriptional properties of Sp1 and Sp3 also revealed significant differences between these two transcription factors. On many reporter constructs containing multiple Sp-binding sites Sp3 is, unlike Sp1, inactive or acts only as a weak activator (3). The molecular basis for the inactivity of Sp3 under these conditions has been mapped to an inhibitory domain located between the second glutamine-rich activation domain and the zinc finger region (7). The integrity of a charged amino acid triplet (KEE) within this domain is absolutely essential for inhibitor function. Mutation of these amino acids converted Sp3 to a strong activator (7) that is almost indistinguishable from Sp1. The presence of two strong activation domains that are silenced by an inhibitory domain suggested that Sp3 might be the target of regulatory events. Regulation of the activity of transcription factors is a common theme and can involve covalent modifications by phosphorylation and acetylation, as well as intermolecular interactions with coactivators or corepressors.
We have initiated a detailed analysis of the molecular mechanisms responsible for the transcriptional properties of Sp3. Here, we report the expression and purification of recombinant dual epitope-tagged Sp1 and Sp3. We show that highly purified recombinant Sp3 acts as a strong activator similar to Sp1 in a cell-free in vitro transcription system. This finding differs from results obtained in transfection assays, in which Sp3 is almost inactive in the same promoter context. We demonstrate that a single lysine residue is responsible for the low transcriptional activity of Sp3 in cell culture experiments. Most significantly, an arginine residue cannot functionally replace the lysine residue. We further show that wild-type Sp3, but not a mutant lacking this lysine residue, is acetylated in vivo, suggesting that acetylation regulates the transcriptional activity of Sp3.
MATERIALS AND METHODS
Plasmid constructions
Transient expression of Sp3 mutants in Drosophila Schneider SL2 cells (8) was driven by the Drosophila actin 5C 5′-flanking region. Construction of the parental wild-type Sp3 expression plasmids pPacUSp3 and pGEX-2TK-Sp3 has been described (7). Expression plasmids containing Sp3 point mutations were obtained by replacing the wild-type human Sp3 cDNA of pPacUSp3 by the mutated Sp3 cDNAs obtained from pGEX-2TK-Sp3 as BamHI–XhoI fragments. The appropriate point mutations were introduced into pGEX-2TK-Sp3 by PCR amplification of a Bst1107I–XhoI fragment using the appropriate oligonucleotides. Detailed information will be provided upon request. To generate pGEX-2TK-Sp3-B and pGEX-2TK-Sp3-BID (GST–Sp3B and GST–Sp3-BID expression vectors), a 0.8 kb Bst11071(blunt)–XbaI fragment or a 1 kb XbaI fragment of pSG-Gal4-Sp3 (dam– DNA) (7) was inserted into the SalI(blunt)–XbaI- or XbaI-restricted pGEX-2TK vector, respectively.
Expression and purification of epitope-tagged Sp proteins
The construction of plasmids designed for expression of inducible dual epitope-tagged Sp1, Sp3 and the Sp3SD mutant as well as the generation of stable SL2 cell lines have been described (9). The Sp3SD mutant lacks a highly charged region of 13 amino acids (DIRIKEEEPDPEE) (7). Stable transfected SL2 cells were grown in suspension in Schneider medium supplemented with 10% fetal calf serum, 100 IU/ml penicillin/streptomycin, 2 mM glutamine, 0.1% pluronic F-68 and 10 U/ml heparin. Expression of Sp protein was induced at a density of 12 × 106 cells/ml with 500 µM CuSO4 for 24 h. Nuclei were extracted according to Dignam et al. (10). Purification of the recombinant proteins included either DNA-binding affinity chromatography or immune affinity chromatography. For DNA-binding affinity chromatography nuclear extracts were precipitated with 53% (NH4)2SO4, resuspended in 2 ml of TM buffer (50 mM Tris–HCl pH 7.9, 12.5 mM MgCl2, 20% glycerol, 0.1% NP-40) and applied to a Sephacryl S-300 gel filtration column. The Sp activity of the collected fractions was analyzed by electrophoretic mobility shift assay (EMSA) as described (11). Sp protein-containing fractions were pooled and further purified by non-specific and specific DNA affinity chromatography essentially as described (12,13). Sp3 was alternatively purified by immune affinity chromatography. Nuclear extracts of Sp3-expressing SL2 cells were first pre-cleared by immune precipitation using Sp3 preimmune serum bound to protein A–Sepharose. HA/FLAG-tagged Sp3 was then immune precipitated with anti-FLAG M2 affinity gel (Sigma). The matrix was washed three times with BC150 buffer (20 mM Tris–HCl pH 7.9, 20% glycerol, 0.2 mM EDTA, 150 mM KCl). Sp3 was eluted with 100 ng/ml FLAG peptide (Sigma) in BC150 buffer. Sp3 activity was analyzed by EMSA. Prior to use, 1 mM DTT and 0.2 mM PMSF were freshly added to all buffers.
Transient transfection assays
SL2 cells were transfected as described (7,14). Every plate received 4 µg reporter plasmid (BCAT-2 or pGL3) and 2 µg β-galactosidase expression plasmid p97b as internal reference. Variable amounts of pPacUSp3 expression plasmids were compensated with the plasmid pPac. Twenty-four hours after addition of DNA, the medium was changed and 24 h later the cells were washed twice with PBS and harvested. Chloramphenicol acetyltransferase (CAT) was assayed by ELISA according to the manufacturer’s instructions (Boehringer Mannheim). Luciferase and β-galactosidase assays were carried out as described (15,16). Expression of CAT or luciferase enzymatic activity was normalized to β-galactosidase activity. Each transfection was repeated at least three times.
In vitro transcription
Supercoiled BCAT-2 (200 ng) or E(II)*CAT (200 ng) plasmid DNA was transcribed for 1 h at 30°C with HeLa nuclear extract (10) in a 30 µl reaction containing 57 mM KCl, 1 mM DTT, 2.5 mM MgCl2, 0.5 mM rNTPs. The conditions of transcription and primer extension were as described (17). HeLa nuclear extract was depleted of Sp1 and Sp3 by incubating with a one-twentieth volume of DNA affinity matrix for 30 min at 4°C. DNA affinity beads were removed by centrifugation and the depletion was repeated once.
In vivo acetylation assay
SL2 cells stably transfected with expression vectors for Sp3 or Sp3SD were plated on 60 mm plates. At 70% confluence, expression of Sp3 factors was induced with 500 µM CuSO4 for 3 h. Then 1 mCi/ml sodium [3H]acetate (8 Ci/mmol) was added to the medium. After 30 min, 0.5 µM trichostatin A (TSA) (ICN Pharmaceuticals) was added and incubation continued for another 30 min. Cells were washed with PBS and nuclear extracts were prepared as described (18). All buffers contained a protease inhibitor cocktail, 0.2 mM PMSF, 1 mM DTT and 0.5 µM TSA.
Immunoprecipitation and western blotting
For immunoprecipitation, 100 µl of nuclear extract obtained from a 60 mm plate were diluted with 180 µl of BC0 buffer (20 mM Tris–HCl pH 7.9, 20% glycerol, 0.2 mM EDTA) supplemented with protease inhibitors and 2 µM TSA. Extracts were incubated for 1.5 h at 4°C with 10 µl of slurry containing the Sp3 preimmune serum bound to protein A–Sepharose beads. After centrifugation, the supernatant was subjected to immunoprecipitation by incubation with 10 µl of a slurry containing an Sp3 antibody (3) bound to protein A–Sepharose beads for 2 h at 4°C using a head-over-tail rotator. Complexes were pelleted, washed three times with TBS and resolved by SDS–PAGE. Proteins were electrotransferred to PVDF membranes and probed with anti-HA antibodies (Santa Cruz) or anti-acetyl-lysine antibodies (Upstate). Visualization was performed by chemoluminescence detection (ECL, Amersham-Pharmacia).
In vitro acetylation assays
The histone acetyltransferase (HAT) domains of PCAF (amino acids 1–832), CBP (amino acids 1098–1758) and p300 (amino acids 1071–1715) were expressed as GST fusion proteins (PCAF and CBP) or as 6×His-tagged protein (p300) (19) in Escherichia coli strain BL21 or M15 (pREP 4), and allowed to bind to glutathione–Sepharose 4B (Amersham-Pharmacia) or Ni–NTA matrix (Qiagen) according to the manufacturer’s instructions. GST–Sp3 fusion proteins used as substrates for HAT activity were bound to glutathione–Sepharose 4B and either eluted with 10 mM glutathione or cleaved with thrombin. In vitro acetylation was performed essentially as described (20), in a 30 µl reaction containing 1 µl of [14C]acetyl-CoA (1.92 GBq/mmol). The reaction products were resolved by SDS–PAGE and visualized by Coomassie blue staining and autoradiography.
RESULTS
Expression and purification of recombinant Sp1 and Sp3
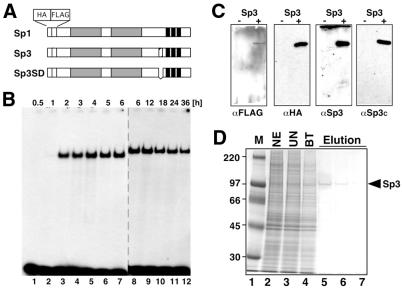
Our previous functional analyses of Sp3 in direct comparison with Sp1 revealed that, unlike Sp1, Sp3 did not exert an activation function on a promoter setting containing two Sp1/3 binding sites (3,7). To further dissect the mechanisms responsible for the inactivity or low activity of Sp3 compared to Sp1, we sought to establish an in vitro transcription system using purified Sp1 and Sp3. As a source for recombinant Sp proteins we have generated stable transfected SL2 cell lines that express Sp1, Sp3 or a mutant of Sp3 (SpSD) as dual epitope (FLAG and HA)-tagged proteins in an inducible manner (Fig. 1; 9). Sp3SD lacks 13 amino acids of the inhibitory domain of Sp3 (7). All three proteins were purified by immunoaffinity chromatography via the FLAG epitope or, alternatively, by DNA-binding affinity chromatography (13), yielding highly purified proteins (Fig. 1D and data not shown).
Figure 1.
Expression and purification of epitope-tagged Sp3 and Sp3SD in stable transfected SL2 cells. (A) Schematic representation of Sp1, Sp3 and the Sp3 mutant Sp3SD expressed in SL2 cells. Gray boxes indicate the glutamine-rich activation domains, three black bars the three zinc fingers of the Sp factors. The box with hatched stripes N-terminal to the zinc fingers in Sp3 depicts the inhibitory domain. All three proteins contain a hemagglutinin (HA) and a FLAG epitope at their N-termini. (B) Induction of epitope-tagged Sp3 in stable transfected SL2 cells. Expression of Sp3 was induced with 500 µM copper sulfate. At various times after induction (0.5–36 h) nuclear extracts were prepared and subjected to electrophoretic mobility shift analysis. In lanes 1–7 0.75 µg and in lanes 8–12 1.2 µg extracted protein were used for the GC box-binding reaction. (C) Detection of epitope-tagged Sp3 by western blotting. Extracts of epitope-tagged Sp3-expressing SL2 cells were subjected to western blot analyses using antibodies against the FLAG epitope (αFLAG), the HA epitope (αHA) and two different anti-Sp3 antibodies [αSp3 (3) and αSp3c (Santa Cruz)]. (D) Purification of epitope-tagged Sp3 from stable transfected SL2 cells using immune affinity chromatography. Aliquots of the various purification steps were analyzed by SDS–PAGE and stained with Coomassie blue. M, marker lane; NE, crude nuclear extract; UN, breakthrough of a non-specific immune affinity chromatography (preimmune αSp3 serum coupled to protein A–Sepharose); BT, breakthrough of a specific immune affinity chromatography (αFLAG matrix); Elution, consecutive 300 µl fractions upon elution with FLAG peptide.
Sp3 acts as a strong transcriptional activator in vitro
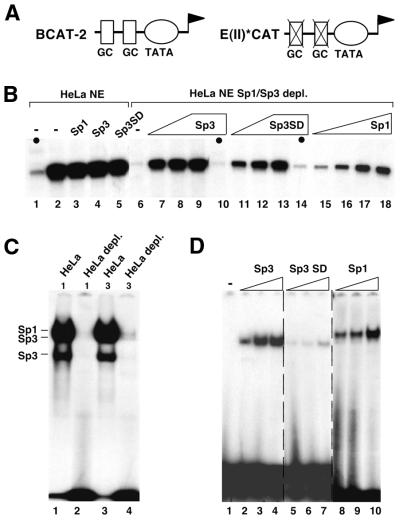
We questioned whether the purified Sp1 and Sp3 proteins could act as transcriptional regulators in a cell-free HeLa transcription assay system in the same way as their counterparts in vivo. As a reporter construct we chose BCAT-2 (21), which contains two Sp1/Sp3-binding sites fused to the E1B TATA box (Fig. 2A). As a control, we included the plasmid E(II)*CAT (22), which contains the same TATA box region but has mutated Sp-binding sites. Expression from the BCAT-2 promoter was already very high in HeLa nuclear extracts (Fig. 2B, lane 2) and addition of DNA-binding-competent purified recombinant Sp1, Sp3 or Sp3SD (see Fig. 2D) did not further increase transcription (Fig. 2B, lanes 3–5). EMSA showed that our HeLa nuclear extract contained high amounts of Sp1 and Sp3 (Fig. 2C, lanes 1 and 3). Therefore, we depleted the HeLa nuclear extract of GC box-binding proteins using a GC box affinity column (Fig. 2C, lanes 2 and 4). Sp-depleted nuclear extracts transcribed the CAT gene in BCAT-2 at a very low level, similar to the control promoter lacking the Sp-binding sites. Addition of recombinant Sp1, Sp3 and Sp3SD strongly enhanced transcription from the BCAT-2 but not the E(II)*CAT plasmid (Fig. 2B). This result shows that purified recombinant Sp1, Sp3 and Sp3SD expressed in insect cells act as strong activators in vitro. The transcriptional activity of Sp3 is similar to the activity of Sp1 as well as to the Sp3 mutant Sp3SD lacking the inhibitory domain. Thus, the inhibitory domain of Sp3 that is responsible for the low activity of Sp3 in vivo (7) does not exert its function in vitro.
Figure 2.
In vitro transcription with purified recombinant Sp1, Sp3 and Sp3SD. (A) Schematic representation of the reporter plasmids BCAT-2 and E(II)*CAT used as templates. BCAT-2 contains two adjacent GC boxes fused to the E1B TATA-box. Both GC boxes were mutated in E(II)*CAT. (B) Primer extension analyses of in vitro transcribed RNA. In vitro transcription reactions were performed with HeLa nuclear extract programmed with BCAT-2 or E(II)*CAT (the latter being indicated by black circles, lanes 1, 10 and 14) as templates. Increasing amounts of purified recombinant Sp3 (2, 4 and 8 µl, lanes 7–10), Sp3SD (2, 4 and 8 µl, lanes 11–14) or Sp1 (1, 2, 4 and 8 µl, lanes 15–18) were added to whole HeLa nuclear extract (lanes 1–5) or HeLa nuclear extract depleted of GC box-binding proteins (lanes 6–18). Reaction products were subjected to denaturing PAGE and autoradiography. (C) GC box-binding activity in HeLa nuclear extracts used for in vitro transcription assays. HeLa nuclear extract (1 and 3 µl) prior to (lanes 1 and 3) and after (lanes 2 and 4) incubation with a GC box affinity matrix was incubated with 0.1 ng 32P-labeled GC box oligonucleotide and subjected to EMSA. (D) Purified recombinant Sp3, Sp3SD and Sp1 fractions used for in vitro transcription experiments were analyzed by EMSA. Binding reactions contained 0.05 µl (lanes 2, 5 and 8), 0.075 µl (lanes 3, 6 and 9) or 0.1 µl (lanes 4, 7 and 10) of the protein extracts used for the transcription reactions shown in (B).
A single lysine residue is essential for inhibitory function
Differences between the in vivo and in vitro results may concern (i) covalent modifications that were eliminated during the purification procedure, (ii) potential corepressors that were not present in the HeLa nuclear extract or (iii) the chromatin structure of the template. Possibly, two or even all three of these parameters might determine the activity of the inhibitory domain of Sp3.
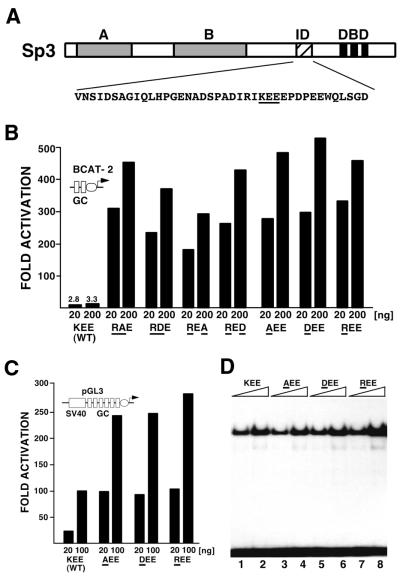
Given that the transcriptional properties of Sp3 differ significantly in vitro and in vivo, we sought to further determine the primary sequence requirements that are essential for the low activity of Sp3 in vivo. From previous studies we knew that the amino acid sequence KEE within the inhibitory domain of Sp3 is essential for silencing of Sp3 transcriptional capacity in vivo. Mutation of the KEE triplet to three alanine residues strongly enhanced the transcriptional activation potential of Sp3 (7). At first, we substituted the KEE wild-type sequence by the amino acids RAE, RDE, REA or RED (mutant amino acids underlined) in the context of full-length Sp3 (Fig. 3). The activity of the mutants was analyzed by transient transfection into the Drosophila Schneider SL2 cell line along with the BCAT-2 reporter. All four Sp3 mutants activated the BCAT-2 reporter up to 500-fold, whereas the wild-type Sp3 protein remained almost inactive (Fig. 3B). Since all four Sp3 mutants contained a substitution of the lysine residue, we asked next whether the lysine residue on its own might be essential for silencing. The lysine was mutated to an alanine, an aspartic acid or an arginine residue. All three lysine mutants activated the BCAT-2 promoter to a similar extent as the double mutants (Fig. 3B). EMSAs showed that all three proteins were expressed at equal levels after transfection (Fig. 3D). We also used the SV40 promoter as a target for the Sp3 mutants. The special setting of the SV40 promoter is activated by wild-type Sp3 in a dose-dependent manner (7). Nevertheless, the Sp3 lysine mutants further enhanced transcription 3- to 5-fold (Fig. 3C).
Figure 3.
Point mutations in the inhibitory domain strongly enhance Sp3 activation capacity. (A) Schematic drawing of Sp3. A and B indicate the glutamine-rich activation domains, ID the inhibitory domain and DBD the zinc finger DNA-binding domain. Double and single point mutations were introduced into the intact Sp3 molecule within the KEE sequence of the inhibitory domain. (B) SL2 cells were transfected with 4 µg BCAT-2 plasmid along with 20 and 200 ng expression plasmids for wild-type Sp3 or Sp3 mutants, respectively, as indicated. Amino acids that differ from the wild-type KEE sequence are underlined. The CAT values are expressed relative to the CAT activity obtained with the vector (pPac), which has been given the arbitrary value of 1. The values represent mean values of at least two independent transfections. (C) SL2 cells were transfected with 20 and 100 ng expression plasmids for the Sp3 lysine mutants along with 4 µg SV40 promoter-driven luciferase reporter plasmid pGL3. (D) Transient expression of Sp3 mutant proteins in SL2 cells. Gel retardation assays were performed with crude nuclear extracts from SL2 cells transfected with 10 µg expression plasmids for wild-type Sp3 (KEE, lanes 1 and 2) or the lysine mutants (AEE, lanes 3 and 4; DEE, lanes 5 and 6; REE, lanes 7 and 8) as indicated. All reactions contained 0.2 ng 32P-labeled GC box oligonucleotide and 1 µg (lanes 1, 3, 5 and 7) or 3 µg (lanes 2, 4, 6 and 8) protein extract.
These results show that the integrity of the lysine residue within the inhibitory domain of Sp3 is absolutely essential to silence the activation potential of Sp3. Most significantly, the silencing function of the lysine cannot be substituted by an arginine residue, indicating that it is not the positive charge of the lysine that is essential for inhibition but rather the specific amino acid structure.
Sp3 is acetylated in vivo
The crucial role of the lysine residue prompted us to ask whether Sp3 might be covalently modified in vivo by acetylation. To address this question, we performed pulse–chase experiments with [3H]acetate in the presence of the histone deacetylase inhibitor TSA. We employed our SL2 cell lines that either express epitope-tagged wild-type Sp3 or the mutant Sp3SD protein (Fig. 4A) in an inducible manner (see above). Wild-type Sp3 and the Sp3SD mutant were expressed at similar levels, as judged by αHA detection (Fig. 4B). However, the acetylation signal obtained with wild-type Sp3 was much stronger than the signal obtained with the Sp3SD mutant protein (Fig. 4C). This result suggests that the lysine residue essential for silencing of Sp3 activity is a substrate for acetylation in vivo.
Figure 4.
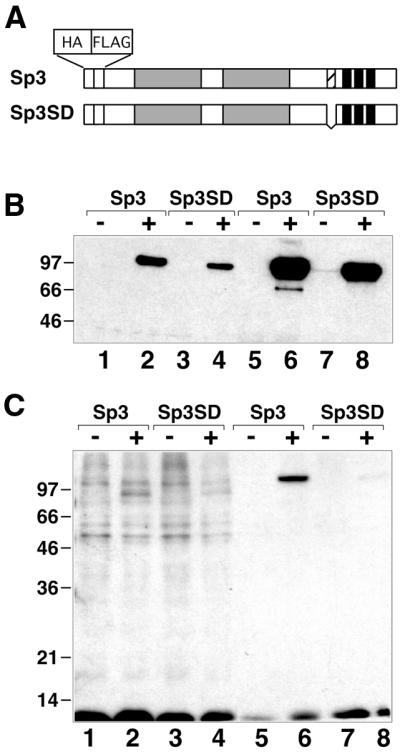
Acetylation of Sp3 in vivo. Stable SL2 transfectants containing expression constructs for epitope-tagged Sp3 or Sp3SD were incubated with [3H]acetate (1 mCi/ml) for 1 h. Nuclear extracts were prepared and subjected to immunoprecipitation with an αFLAG antibody. (A) Schematic representation of Sp3 and the mutant Sp3SD lacking a 13 amino acid stretch. (B) Nuclear extracts prepared from SL2 cells prior to (lanes 1–4) and after (lanes 5–8) immunoprecipitation with an αFLAG antibody were subjected to western blot analyses with an αHA antiserum and visualized by chemoluminescence detection. (–) and (+) indicate the absence and presence of copper sulfate (induction of Sp3 protein). (C) Autoradiogram showing [3H]acetate-labeled proteins of the same fractions as in (B).
We also performed western blot analyses using a commercially available antibody that specifically recognizes acetylated lysine residues. This anti-acetyl-lysine antibody recognized wild-type Sp3 obtained by immunoprecipitation of Sp3-expressing SL2 cells but not the Sp3SD mutant or bacterially expressed Sp3 (data not shown). These results support the notion that Sp3 is an acetylated transcription factor in vivo and that acetylation might occur predominantly at a single lysine residue within the inhibitory domain.
CBP and p300 but not PCAF can acetylate Sp3 in vitro
We asked whether enzymes that are known to catalyze the acetylation of histones and various other transcription factors are able to acetylate fragments of Sp3 containing the lysine residue of the inhibitory domain in vitro. Thus far, we have used the recombinant catalytic HAT domains of PCAF, CBP and p300 with GST–Sp3 fusion proteins as substrates to address this question. These experiments revealed that under conditions where we obtained strong acetylation of histones PCAF did not acetylate GST–Sp3 substrates (data not shown). In contrast, GST–Sp3 fusion proteins were found to be acetylated by the HAT domains of p300 and CBP (Fig. 5). However, to our surprise, the GST domain on its own was acetylated by these HATs (Fig. 5C, lane 3, and data not shown). Upon removal of the GST portion of GST–Sp3B and GST–Sp3BID by thrombin cleavage, the Sp3BID but not the Sp3B mutant protein lacking the inhibitory domain of Sp3 became acetylated (Fig. 5C, lanes 6 and 7). It should be noted that the acetylation signal that we obtained was rather weak. Thus it remains to be established whether CBP is the natural acetyltransferase responsible for acetylation of Sp3 in vivo.
Figure 5.
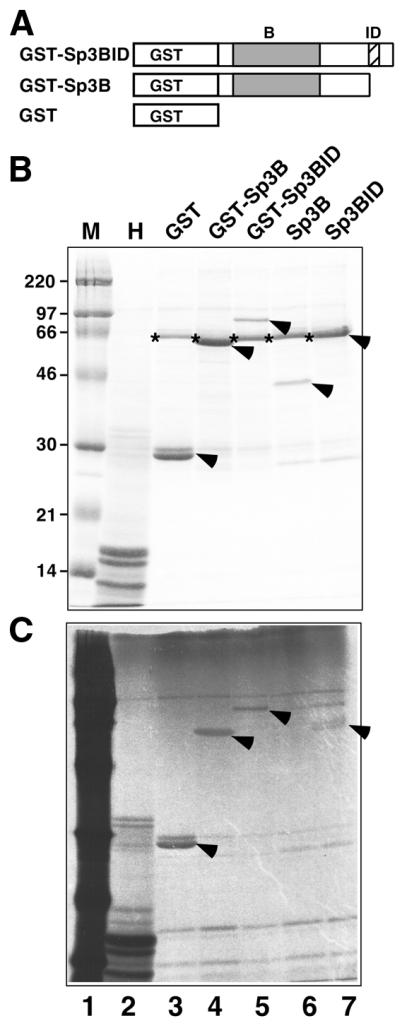
In vitro acetylation of recombinant Sp3 fusion proteins. (A) Schematic representation of GST fusion proteins used as substrates. Sp3B and Sp3BID were obtained by thrombin cleavage of the appropriate GST fusion proteins. GST, GST–Sp3BID, GST–Sp3B, Sp3B, Sp3BID and histones were incubated with GST–CBP-HAT in the presence of [14C]acetate and analyzed by SDS–PAGE and autoradiography. (B) Coomassie stained SDS–PAGE gel of fractionated proteins subjected to the in vitro acetylation reaction. Lane M is the marker lane and lane H contains histones. The stars indicate BSA that was included in the acetylation reactions. The arrows point to the GST, GST–Sp3BID, GST–Sp3B, Sp3B and Sp3BID proteins. (C) Autoradiogram of the gel shown in (B). The marker lane (M) contains 14C-labeled proteins. The arrows point to the acetylated GST, GST–Sp3BID, GST–Sp3B and Sp3BID proteins.
DISCUSSION
Purified recombinant Sp3 is a strong activator in vitro
We have expressed and purified recombinant Sp1 and Sp3 as dual epitope-tagged proteins from insect cells. Both proteins act as strong activators in a cell-free transcription system that has been depleted of endogenous GC box-binding activities. This result shows that Sp3 has a similar intrinsic potential to activate transcription as Sp1.
The finding that Sp3 is in vitro transcriptionally active on a promoter containing two adjacent Sp-binding sites was to some extent unexpected, because in transfection assays using the same reporter plasmid it is almost inactive. The molecular basis for the inactivity of Sp3 in cell culture assays had been mapped to an inhibitory domain located between the second glutamine-rich activation domain and the first zinc finger (7). Thus far, two principally different mechanisms underlying the function of this domain had seemed possible. One possibility would rely on an intramolecular masking of the activation domains; the other would involve intermolecular interactions. Our finding that the inhibitory domain is inactive in an in vitro transcription assay renders a simple intramolecular masking mechanism unlikely.
Sp3 is acetylated in vivo
The finding that the integrity of a single lysine residue is essential for silencing of the transcriptional activity of Sp3 in cell culture experiments prompted us to analyze whether Sp3 is acetylated in vivo. Our results showed that Sp3 is an acetylated transcription factor. It remains to be established whether Sp1 or other members of the Sp family of transcription factors are also targets of post-translational modification by acetylation. Wild-type Sp3 is acetylated at a significantly higher level than a Sp3 mutant lacking the lysine of the inhibitory domain. Nevertheless, the Sp3SD mutant is also acetylated to a certain degree, suggesting that other lysine residues might also be targets of acetylation. Alternatively, the conformation of the Sp3SD mutant might be slightly altered as compared to wild-type Sp3 such that a lysine becomes acetylated that is not accessible in the wild-type protein.
What might be the consequences of acetylation?
It has been known for some time that lysine residues in the N-terminal tails of histones can be covalently modified by acetylation, which leads to transcriptionally active chromatin (23). More recently it became evident that in addition to histones, other nuclear proteins can also be targets of acetylation events. Nuclear substrates, other than chromatin components, include retroviral gene products, transcriptional co-regulators and transcription factors (24). Acetylation of lysine residues in transcription factors has been described for p53 (25), GATA-1 (26,27), EKLF (28), MyoD (29), TCF (30), E2F1 (19), GATA-3 (31) and TAL1/SCL (32). The functional consequences of post-translational modification by acetylation of these transcription factors appear to be quite different. In the case of p53, E2F1, MyoD and Tal1 acetylation occurs close to the DNA-binding domain and results in stimulation of DNA binding (19,29,32). When EKLF becomes acetylated its transcriptional potency is enhanced (28). Other biochemical functions regulated by acetylation are protein–protein interactions and protein stability, as in the cases of dTCF (30) and E2F1 (19), respectively. In some cases, the physiological consequences of acetylation have also been analyzed. In the case of GATA-1, two acetylated lysine residues are essential to trigger terminal differentiation of erythroid cells (27). Acetylation of GATA-3 appears to affect T cell survival and homing to secondary lymphoid organs in vivo (31).
Our finding that the critical lysine residue, which is essential for silencing of Sp3 activity, is acetylated in vivo suggests that acetylation is involved in the process of silencing. However, one can also imagine alternative possibilities. In the course of our analyses of the inhibitory domain of Sp3 we realized that the amino acids surrounding the lysine residue within the inhibitory domain of Sp3 resemble sites that are known to be modified by the small ubiquitin-like modifier (SUMO) (33). Thus, it seems not unlikely that the same lysine residue could be modified not only by acetylation but also by other modifications. If this were the case, one could imagine an intriguing scenario in which acetylation would compete for sumoylation. Further analyses should clarify the functional consequences of acetylation of Sp3.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Iris Rohner for excellent technical assistance. Martha Kalff-Suske is gratefully acknowledged for critically reading the manuscript. Tony Kouzarides, Andrew Banister and Aurora Sanchez Pacheco generously provided us with the plasmids for CBP-HAT, p300-HAT and PCAF-HAT. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to G.S.
REFERENCES
- 1.Suske G. (1999) The Sp-family of transcription factors. Gene, 238, 291–300. [DOI] [PubMed] [Google Scholar]
- 2.Hagen G., Müller,S., Beato,M. and Suske,G. (1992) Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res., 20, 5519–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagen G., Müller,S., Beato,M. and Suske,G. (1994) Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J., 13, 3843–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marin M., Karis,A., Visser,P., Grosveld,F. and Philipsen,S. (1997) Transcription factor Sp1 is essential for early development but dispensable for cell growth and differentiation. Cell, 89, 619–628. [DOI] [PubMed] [Google Scholar]
- 5.Bouwman P., Göllner,H., Elsässer,H.P., Eckhoff,G., Karis,A., Grosveld,F., Philipsen,S. and Suske,G. (2000) Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J., 19, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Göllner H., Dani,C., Phillips,B., Philipsen,S. and Suske,G. (2001) Impaired ossification in mice lacking the transcription factor Sp3. Mech. Dev., 106, 77–83. [DOI] [PubMed] [Google Scholar]
- 7.Dennig J., Beato,M. and Suske,G. (1996) An inhibitor domain in Sp3 regulates its glutamine-rich activation domains. EMBO J., 15, 5659–5667. [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider I. (1972) Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol., 27, 353–365. [PubMed] [Google Scholar]
- 9.Braun H. and Suske,G. (1999) Vectors for inducible expression of dual epitope-tagged proteins in insect cells. Biotechniques, 26, 1038–1042. [DOI] [PubMed] [Google Scholar]
- 10.Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun H. and Suske,G. (1998) Combinatorial action of HNF3 and Sp family transcription factors in the activation of the rabbit uteroglobin/CC10 promoter. J. Biol. Chem., 273, 9821–9828. [DOI] [PubMed] [Google Scholar]
- 12.Kadonaga J.T., Carner,K.R., Masiarz,F.R. and Tjian,R. (1987) Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell, 51, 1079–1090. [DOI] [PubMed] [Google Scholar]
- 13.Kadonaga J.T. (1991) Purification of sequence-specific binding proteins by DNA affinity chromatography. Methods Enzymol., 208, 10–23. [DOI] [PubMed] [Google Scholar]
- 14.Suske G. (2000) Transient transfection of Schneider cells in the study of transcription factors. Methods Mol. Biol., 130, 175–187. [DOI] [PubMed] [Google Scholar]
- 15.Brasier A.R. and Fortin,J.J. (1997) Nonisotopic assays for reporter gene activity. In Ausubel,F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds), Current Protocols in Molecular Biology. Wiley Interscience, Chichester, UK, Vol. 1 (suppl. 29), pp. 9.7.12–9.7.21.
- 16.Hall C.V., Jacob,P.E., Ringold,G.M. and Lee,F. (1983) Expression and regulation of E. coli lacZ gene fusions in mammalian cells. J. Mol. Appl. Genet., 2, 101–109. [PubMed] [Google Scholar]
- 17.Schmidt M.C., Zhou,Q. and Berk,A.J. (1989) Sp1 activates transcription without enhancing DNA-binding activity of the TATA box factor. Mol. Cell. Biol., 9, 3299–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrew N.C. and Faller,D.V. (1991) A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res., 19, 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Balbas M.A., Bauer,U.M., Nielsen,S.J., Brehm,A. and Kouzarides,T. (2000) Regulation of E2F1 activity by acetylation. EMBO J., 19, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- 21.Pascal E. and Tjian,R. (1991) Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes Dev., 5, 1646–1656. [DOI] [PubMed] [Google Scholar]
- 22.Dennig J., Hagen,G., Beato,M. and Suske,G. (1995) Members of the Sp transcription factor family control transcription from the uteroglobin promoter. J. Biol. Chem., 270, 12737–12744. [DOI] [PubMed] [Google Scholar]
- 23.Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- 24.Kouzarides T. (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J., 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu W. and Roeder,R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- 26.Boyes J., Byfield,P., Nakatani,Y. and Ogryzko,V. (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature, 396, 594–598. [DOI] [PubMed] [Google Scholar]
- 27.Hung H.L., Lau,J., Kim,A.Y., Weiss,M.J. and Blobel,G.A. (1999) CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol., 19, 3496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W. and Bieker,J.J. (1998) Acetylation and modulation of erythroid Krüppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl Acad. Sci. USA, 95, 9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sartorelli V., Puri,P.L., Hamamori,Y., Ogryzko,V., Chung,G., Nakatani,Y., Wang,J.Y. and Kedes,L. (1999) Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell, 4, 725–734. [DOI] [PubMed] [Google Scholar]
- 30.Waltzer L. and Bienz,M. (1998) Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature, 395, 521–525. [DOI] [PubMed] [Google Scholar]
- 31.Yamagata T., Mitani,K., Oda,H., Suzuki,T., Honda,H., Asai,T., Maki,K., Nakamoto,T. and Hirai,H. (2000) Acetylation of GATA-3 affects T-cell survival and homing to secondary lymphoid organs. EMBO J., 19, 4676–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang S., Qiu,Y., Shi,Y., Xu,Z. and Brandt,S.J. (2000) P/CAF-mediated acetylation regulates the function of the basic helix–loop–helix transcription factor TAL1/SCL. EMBO J., 19, 6792–6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melchior F. (2000) Sumo nonclassical ubiquitin. Annu. Rev. Cell. Dev. Biol., 16, 591–626. [DOI] [PubMed] [Google Scholar]