Abstract
Culture filtrate from Mycobacterium tuberculosis contains protective antigens of relevance for the generation of a new antituberculosis vaccine. We have identified two previously uncharacterized M. tuberculosis proteins (TB7.3 and TB10.4) from the highly active low-mass fraction of culture filtrate. The molecules were characterized, mapped in a two-dimensional electrophoresis reference map of short-term culture filtrate, and compared with another recently identified low-mass protein, CFP10 (F. X. Berthet, P. B. Rasmussen, I. Rosenkrands, P. Andersen, and B. Gicquel. Microbiology 144:3195–3203, 1998), and the well-described ESAT-6 antigen. Genetic analyses demonstrated that TB10.4 as well as CFP10 belongs to the ESAT-6 family of low-mass proteins, whereas TB7.3 is a low-molecular-mass protein outside this family. The proteins were expressed in Escherichia coli, and their immunogenicity was tested in cultures of peripheral blood mononuclear cells from human tuberculosis (TB) patients, Mycobacterium bovis BCG-vaccinated donors, and nonvaccinated donors. The two ESAT-6 family members, TB10.4 and CFP10, were very strongly recognized and induced gamma interferon release at the same level (CFP10) as or at an even higher level (TB10.4) than ESAT-6. The non-ESAT-6 family member, TB7.3, for comparison, was recognized at a much lower level. CFP10 was found to distinguish TB patients from BCG-vaccinated donors and is, together with ESAT-6, an interesting candidate for the diagnosis of TB. The striking immunodominance of antigens within the ESAT-6 family is discussed, and hypotheses are presented to explain this targeting of the immune response during TB infection.
For a number of years, a major effort has been put into the development of a new vaccine against tuberculosis (TB) and better methods for the diagnosis of the disease. The search for candidate molecules has in the recent years focused on proteins released from dividing bacteria based on the reasoning that live bacteria generally induce higher levels of protection than killed preparations (4, 23).
For a number of years, the components of culture filtrate have been investigated by using narrow-molecular-mass fractions as a guide to identify immunologically active single molecules (2, 3, 5). Low-molecular-mass proteins between 6 and 12 kDa were, in this way, demonstrated to be strongly recognized by T cells isolated from human TB patients (10), as well as mice and cattle experimentally infected with TB (2, 6, 25).
Until recently, only a few small Mycobacterium tuberculosis proteins were known. The 10-kDa GroES molecule was the first antigen to be identified in this region and was found to be present in both M. tuberculosis (8) and Mycobacterium leprae (19). This antigen is abundant and constitutes a major component in culture filtrate as well as in cell wall preparations (17). This molecule, in the native form, was strongly recognized by TB patients and infected mice (8). However, several studies have tested recombinant GroES and reported only modest T-cell responses by TB patients and TB-infected mice (10, 20, 27a, 29); I. Rosenkrands and P. Andersen, unpublished data). The ESAT-6 antigen was identified in the low-molecular-mass fraction of culture filtrate due to a strong T-cell response with high levels of gamma interferon (IFN-γ) released (2). This antigen has now, in a number of studies, been demonstrated to have good stimulatory antigenic properties and is recognized strongly by a high percentage of TB patients (21, 26, 33), as well as different animal species infected with TB (11, 14, 25). Recently, a few other small proteins have been identified from various mycobacterial extracts and evaluated for their immunological relevance (13, 34). A recent development in this field was the identification of a 10-kDa molecule (CFP10) encoded in the same operon as ESAT-6 (9). The sequence of the cfp10 gene is homologous (approximately 40%) to esat-6, and both proteins are members of the ESAT-6 family of small proteins homologous to ESAT-6 and organized in operon-like structures on the mycobacterial genome (9, 12). However, so far no immunological data on this molecule have been presented.
The present study identifies two novel low-mass M. tuberculosis proteins, TB10.4 and TB7.3. One of these proteins, TB10.4, was found to be a member of the ESAT-6 family, whereas TB7.3 is a low-mass protein without the features characteristic for this family. Our data demonstrate that the three members of this family tested so far (TB10.4, CFP10, and ESAT-6) all share a striking immunodominance in the human immune response against M. tuberculosis and are more strongly recognized than TB7.3. CFP10 distinguished TB patients from Mycobacterium bovis BCG-vaccinated donors and is, together with ESAT-6, an interesting candidate for the diagnosis of TB.
MATERIALS AND METHODS
Bacterial strains.
The Escherichia coli strains used for cloning and expression were One Shot (Invitrogen, Leek, The Netherlands) and XL1 Blue (Stratagene, La Jolla, Calif.). Mycobacterial strains used for the interspecies study are listed in Table 1.
TABLE 1.
Mycobacterium interspecies distribution of tb7.3, tb10.4, cfp10, and esat-6
| Species | Sourcea | Distribution of the following geneb
|
|||
|---|---|---|---|---|---|
| tb7.3 | tb10.4 | cfp10 | esat-6c | ||
| M. tuberculosis H37Rv | ATCC 27294 | + | + | + | + |
| M. bovis MNC 27 | SSId | + | + | + | + |
| BCG Danish 1331 | SSIe | + | + | − | − |
| BCG Tokyo | WHO | + | + | − | − |
| M. avium | ATCC 15769 | + | + | − | − |
| M. intracellulare | ATCC 15985 | + | + | − | − |
| M. kansasii | ATCC 12478 | − | + | + | + |
| M. marinum | ATCC 927 | + | + | + | + |
| M. scrofulaceum | ATCC 19275 | − | − | − | − |
| M. fortuitum | ATCC 6841 | − | − | − | − |
| M. xenopi | Isolated from a Danish patient | + | − | − | − |
| M. szulgai | Isolated from a Danish patient | + | − | + | + |
ATCC, American Type Culture Collection, Manassas, Va.; SSI, Statens Serum Institut, Copenhagen Denmark; WHO, World Health Organization.
The analysis was done by Southern blotting. +, positive reaction; −, no reaction.
Data obtained from reference 15.
Specifically from the Mycobacteria Department at SSI.
Specifically from the BCG Laboratory at SSI.
ST-CF and antibodies.
Short-term culture filtrate (ST-CF) of M. tuberculosis H37Rv was produced as described previously (3, 31).
The monoclonal antibody (MAb) PV-2 was described previously (2). Polyclonal antisera were raised against recombinant (r) TB7.3 and CFP10 as follows. Rabbits were immunized five times with 50 μg of recombinant antigen adjuvanted with incomplete Freund's adjuvant at 2-week intervals. The animals were bled, and sera were tested for reactivity against the recombinant protein and ST-CF by Western blotting.
Identification and characterization of low-mass proteins.
TB7.3 was identified from ST-CF as described by Rosenkrands et al. (27). In brief, ST-CF proteins were separated by thiophilic adsorption chromatography on an Affi-T gel column (Kem-En-Tec, Copenhagen, Denmark), and the TB7.3-containing fractions were further purified by anion-exchange chromatography (HR 5/5 Mono Q connected to a fast protein liquid chromatography system; Pharmacia, Uppsala, Sweden). Proteins were eluted by a 0 to 1 M NaCl gradient, and fractions enriched in the band representing TB7.3, when analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), were collected. For the identification of TB10.4 from ST-CF, chromatofocusing on a PBE 94 column equilibrated with 25 mM piperazine-HCl, pH 5.5, and elution with 10% PB74-HCl, pH 4.0, was applied.
The native protein preparations of TB7.3 and TB10.4 were separated on a 10 to 20% Tricine–SDS-PAGE gel and blotted onto Problott polyvinylidene difluoride membranes (Applied Biosystems, Foster City, Calif.), from which the protein bands of interest were excised and subjected to N-terminal amino acid analysis by automated Edman degradation with a Procise 494 sequencer (Applied Biosystems).
The gene encoding TB10.4 was identified by screening a λgt11 M. tuberculosis genome library constructed by R. Young et al. (36) with the MAb PV-2 as described previously (1, 2). A single positive clone (AA242) was found, and the EcoRI M. tuberculosis fragment was subcloned into the E. coli expression vector pBluescript II SK(+) (Stratagene) (AA251) for subsequent sequencing. The obtained nucleotide sequence was analyzed, and the open reading frame (ORF) (tb10.4) encoding the MAb PV-2 reactive protein (TB10.4) was identified.
Obtained sequences were used to search the SWISSPROT and the Sanger sequence databases with the BLASTP and the BLASTN algorithms. DNA star version 3.08b was used for molecular mass and pI calculations.
The proteins were mapped by two-dimensional electrophoresis (2-DE) as described previously (35).
Cloning, expression, and purification of recombinant proteins.
Primers were designed, based on the DNA sequences from the Sanger sequence database, and constructed to create unique restriction sites, up- and downstream of the start and stop codons, respectively, for use in the cloning procedure. The primers were synthesized at Statens Serum Institute by using a ABI-391 DNA synthesizer (Applied Biosystems). The primers used were as follows: tb7.3-sense, AAGAGTAGATCTATGATGGCCGAGGATGTTCGCG (creates a BglII site); tb7.3-antisense, CGGCGACGACGGATCCTACCGCGTCGG (creates a BamHI site); tb10.4-sense, GCAACACCCGGGATGTCGCAAATCATG (creates a SmaI site); and tb10.4-antisense, CTACTAAGCTTGGATCCCTAGCCGCCCCATTTGGCGG (creates a BamHI site). PCR was carried out in a thermal reactor (Rapid cycler; Idaho Technology, Salt Lake City, Utah) by using standard protocols (28). As template, M. tuberculosis H37Rv chromosomal DNA or plasmid DNA was used for the cloning of tb7.3 and tb10.4, respectively. The tb7.3 PCR product was cloned into the pCR2.1 cloning vector and transformed into One Shot cells (Invitrogen) as described by the manufacturer. Plasmid DNA (tb7.3) or PCR product (tb10.4) was digested with the appropriate restriction enzymes and cloned into either pMCT6 (16) (tb7.3) or pMST24 (32) (tb10.4) in frame with eight- or six-histidine residues, respectively. The correct insert was confirmed by sequencing of both DNA strands. DNA sequencing was performed at Statens Serum Institut by using the cycle sequencing system in combination with an automated gel reader (model 373A; Applied Biosystems).
The gene encoding CFP10 was cloned as described previously (9).
The histidine-tagged recombinant proteins (rTB7.3, rTB10.4, and rCFP10) were expressed and purified by metal affinity chromatography by using a Talon column (Clontech, Palo Alto, Calif.) in the presence of 8 M urea, essentially as described by the manufacturer. Purification of the proteins to homogeneity was done by anion chromatography (35) with 1-ml Hitrap columns (Pharmacia).
Protein concentrations were determined by the bicinchoninic acid test (Micro BCA Protein Assay Reagent kit; Pierce, Oud-Beijerland, The Netherlands). Lipopolysaccharide content in these preparations, measured by the Limulus amoebocyte lysate test (7), was always below 0.05 ng of lipopolysaccharide/μg of protein.
Southern blotting.
Genomic DNA from the different mycobacterial species listed in Table 1 was prepared as described previously (3). The Southern blotting was carried out as described elsewhere (22) with the following modifications: 2 μg of purified chromosomal DNA was digested to completion with PvuII and run on a 0.8% agarose gel. The gel was blotted onto a Hybond-N+ membrane (Amersham, Life Science, Buckinghamshire, Little Chalfont, England) in a vacuum transfer device (Milliblot-V system; Millipore).
The probe holding the whole ORF of the selected protein was amplified by PCR from plasmid DNA with the primers described above, and the probes were purified by a QIAquick PCR Purification kit (Qiagen, Hilden, Germany) and labeled by using the ECL direct nucleic acid labeling system (Amersham). Hybridization and detection were performed according to the instructions provided by the manufacturer.
Human lymphocyte cultures.
Seventeen Danish TB patients diagnosed and treated at the Department of Pulmonary Medicine, University Hospital of Copenhagen, Copenhagen Denmark, were asked to participate in the study. Blood samples were drawn between 0 and 6 months after diagnosis. Seven BCG-vaccinated and 7 nonvaccinated healthy individuals with no known history of exposure to patients with TB or laboratory exposure were recruited as controls. Blood samples were drawn 2 months to 40 years after BCG vaccination.
Separation, culture of peripheral blood mononuclear cells (PBMC), and measurement of IFN-γ in the supernatants were done as described previously by Ravn et al. (26). A dose-response study of the three recombinant proteins (rTB7.3, rTB10.4, and rCFP10) was carried out by using 0.3 to 10 μg of antigen/ml of culture. ST-CF was used at 5 μg/ml. The detection limit of the IFN-γ assay was 50 pg/ml. Positive responses were defined as delta values (IFN-γ release in the antigen-stimulated well minus IFN-γ release in the unstimulated well) above 200 pg/ml. IFN-γ release in unstimulated wells was generally below 100 pg/ml. All IFN-γ analyses were done in duplicates on supernatants pooled from three wells and were given as means. The variation on duplicate wells was always less than 10% of the mean. This part of the study was approved by the Local Ethical Committee for Copenhagen and Frederiksberg (RH 01-282/96 and KF 01-369/98).
RESULTS
Identification and recombinant expression of low-mass M. tuberculosis proteins.
Despite the high biological activity of the low-mass fraction of culture filtrate, the components in this fraction have mostly remained elusive, as no distinct spots indicating significant quantities of protein can be detected in this region by 2-DE of culture filtrate preparations (30, 35).
We employed two different purification strategies to obtain fractions enriched in low-mass culture filtrate proteins. A 7-kDa protein was isolated by thiophilic adsorption chromatography, followed by anion-exchange chromatography. The 15 N-terminal amino acids of the purified protein were determined (AEDVRAEIVASVLEV) and used to search the Sanger sequence database (http://www.sanger.ac.uk). The search identified an ORF of 216 bp which encoded a protein (TB7.3) with a theoretical molecular mass of 7.3 kDa and a pI of 3.8 (Table 2). TB7.3 was similar to the C terminus of oxaloacetate decarboxylases and biotin carboxyl carrier proteins, and in agreement with this observation, TB7.3 was found to be biotinylated (I. Rosenkrands and P. Andersen, unpublished data).
TABLE 2.
Characteristics for two novel low-mass proteins from M. tuberculosis
| Protein | Molecular mass (kDa) | pI | Sanger identification nomenclature | Antibody | Homology to other M. tuberculosis proteins | Notes |
|---|---|---|---|---|---|---|
| TB7.3 | 7.3 | 3.8 | Rv3221c | PAb α-TB7.3 | Biotin binding sitea | |
| TB10.4 | 10.4 | 4.5 | Rv0288 | MAb PV-2 | 84% identity to Rv3019c | Ala-rich protein |
| AJ002067b | 65% identity in a 69-amino-acid overlap with Rv3017c | Member of mycobacterial protein family containing ESAT-6 |
Rosenkrands, unpublished results.
Accession number from the EMBL nucleotide sequence database.
The second approach was to separate ST-CF by large-scale preparative SDS-PAGE into narrow fractions highly enriched in low-mass components. One of these fractions (4 to 8 kDa) was used to immunize mice, and a MAb (PV-2) (2) directed against a low-mass molecule was obtained. This MAb was used to screen a λgt11 M. tuberculosis genome library, and a phage clone with an insert containing an ORF of 291 bp was identified. The ORF encoded a protein of 96 amino acids (TB10.4) with a theoretical molecular mass of 10.4 kDa and a pI of 4.5. Searching the Sanger database, we identified two other deduced proteins (Rv3017c and Rv3019c) with homology to TB10.4 (Table 2). To confirm the correct identification of TB10.4, we purified the protein recognized by the MAb PV-2 from culture filtrate and obtained the N-terminal sequence (GHAGDMAGYAGTLQS). This sequence corresponds to residues 13 through 27 in TB10.4, and in addition to confirming our identification, it suggests an alternative start site (the Leu [encoded by ttg] in position 12 or the Met in position 11) or a partial cleavage of the protein in culture filtrate. The sequences of TB7.3 and TB10.4 were analyzed by using the Signal P database (http://www.cbs.dtu.dk/service/SignalP) and were not found to encode conventional signal sequences. Interestingly, the database searches identified both TB10.4 and the recently identified low-mass protein CFP10 (9) as members of the ESAT-6 family, which consists of small proteins homologous to ESAT-6 (Fig. 1) and organized in operon-like structures on the mycobacterial genome (9, 12). TB7.3, on the other hand, was found to be a low-molecular-mass antigen outside the ESAT-6 family.
FIG. 1.
Alignment of the protein sequence of ESAT-6 with the two ESAT-6 family proteins TB10.4 and CFP10. Identical residues are marked by vertical lines.
The genes encoding the two newly identified proteins as well as ESAT-6 and CFP10 were expressed and purified as histidine-tagged products.
Characterization of low-mass M. tuberculosis proteins.
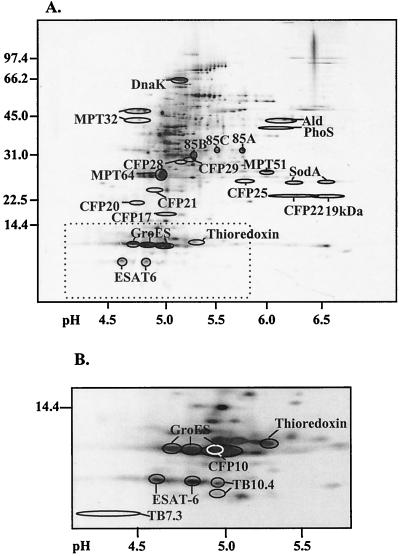
CFP10 has not been characterized in detail, and we therefore decided to investigate this molecule in comparison with TB7.3 and TB10.4. The molecules were mapped in a 2-DE reference map of M. tuberculosis ST-CF components (35). This was done by probing 2-DE immunoblots sequentially with MAb PV-2 and the polyclonal antisera directed against TB7.3 (polyclonal antibody [PAb] α-TB7.3) and CFP10 (PAb α-CFP10). The newly identified molecules are present in low quantities in ST-CF, and in particular TB7.3 is difficult to distinguish after silver staining. Western blotting allowed mapping of the molecules to distinct positions around ESAT-6 in the region below 10 kDa. These positions were subsequently transferred to silver-stained duplicate gels and compared to the already characterized proteins in this region (Fig. 2). No reaction was seen to higher molecular mass components in ST-CF, indicating that these molecules are mature proteins and not fragments of larger molecules.
FIG. 2.
2-DE map of novel and previously identified M. tuberculosis proteins in ST-CF. (A) Silver-stained two-dimensional SDS-PAGE of total ST-CF. Previously identified proteins have been indicated. (B) Silver-stained two-dimensional SDS-PAGE of the low-molecular-mass region of ST-CF. The novel molecules have all been mapped by Western blotting of duplicate gels, and their positions (indicated by rings) have been transferred to the silver-stained reference gels. The white ring indicates CFP10. TB7.3 is present in only very low concentrations in culture filtrate and is not detectable by silver staining. Previously characterized proteins are shown for comparison. Molecular masses are given to the left in kilodaltons. The pH scale is shown below.
The interspecies distribution of the molecules was evaluated by Southern blotting on genomic DNA from several different mycobacterial species (Table 1), using the coding regions of tb7.3, tb10.4, and cfp10 as probes. The data were compared to the previously published species distribution of esat-6 (15) (Table 1).
The three genes were all detected in M. tuberculosis and M. bovis. The close relationship between cfp10 and esat-6 was confirmed, as these genes were identically distributed. Both genes were absent in the two BCG strains tested and the majority of the environmental mycobacteria.
tb7.3 and tb10.4 were broadly distributed and could be detected in BCG, the Mycobacterium avium complex, and other environmental mycobacteria (Table 1).
Immunological recognition of low-mass M. tuberculosis proteins.
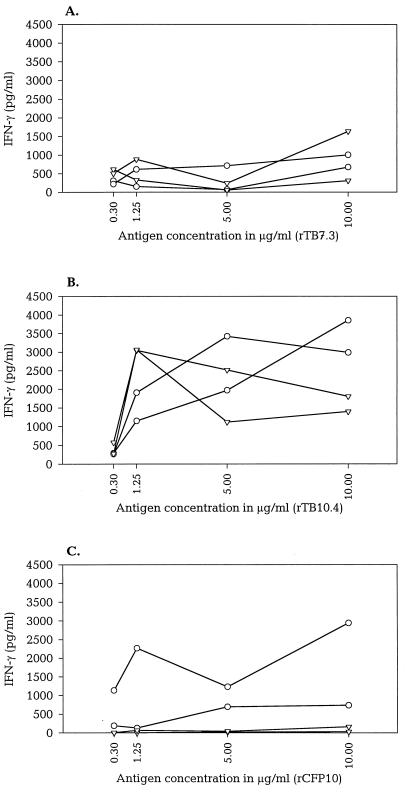
The immunological recognition of the purified low-mass recombinant proteins was evaluated by stimulating PBMC isolated from TB patients, BCG-vaccinated donors, and healthy nonvaccinated donors. A dose-response investigation was conducted for TB7.3, TB10.4, and CFP10 with concentrations ranging from 0.3 to 10 μg/ml (Fig. 3). Figure 3 shows the IFN-γ levels in lymphocyte cultures from two Danish TB patients and two healthy Danish BCG-vaccinated donors stimulated with the antigens. The lymphocyte response after stimulation with TB7.3 was moderate with IFN-γ releases generally below 1,000 pg/ml (Fig. 3A). Neither IFN-γ nor proliferative responses to this antigen (data not shown) reached more than 20% of the responses seen with ST-CF. For the two ESAT-6 family antigens (CFP10 and TB10.4), high levels of IFN-γ were induced with increasing antigen concentrations (Fig. 3B and C). Optimal concentrations of the antigens were between 1.25 and 10 μg/ml, and these concentrations gave responses in the range of 1,000 to 4,000 pg of IFN-γ/ml. The concentration of 5 μg of purified recombinant antigen per ml was chosen for the subsequent comparative evaluation of the three antigens and ESAT-6.
FIG. 3.
Human lymphocyte responses to rTB7.3, rTB10.4, and rCFP10. The IFN-γ response resulting from stimulation of PBMC from two human TB patients (circles) and two healthy BCG-vaccinated human donors (upside-down triangles) with increasing concentrations of rTB7.3 (A), rTB10.4 (B), and rCFP10 (C). The data depicted are the means of duplicate analysis.
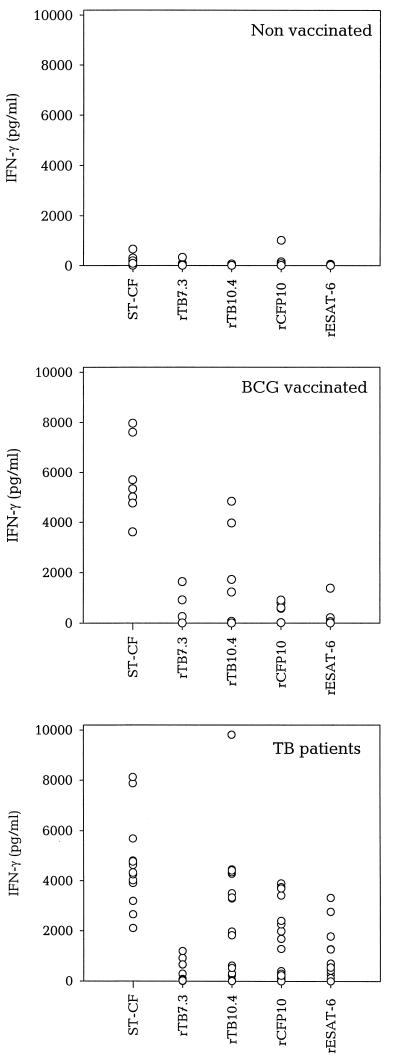
The four low-mass antigens were investigated in 13 to 17 TB patients, 4 to 7 BCG-vaccinated donors, and 7 nonvaccinated donors (Fig. 4). TB7.3 was recognized, but at low levels in both patients and BCG-vaccinated donors. Thirty-eight percent (5 of 13) of the TB patients recognized this molecule at a level significantly above background, and for these donors, the median response was 659 pg of IFN-γ/ml versus 4,024 pg of IFN-γ/ml in the same donors for ST-CF. The ESAT-6 family members were all recognized at a much higher level. TB10.4 was recognized by both BCG-vaccinated donors (71% responders; median IFN-γ = 3,968 pg/ml versus 5,335 pg/ml in the same donors for ST-CF) and TB patients (88% responders; median IFN-γ = 3,298 pg/ml versus 4,707 pg/ml in the same donors for ST-CF). In the TB patients, CFP10 induced a pronounced release of IFN-γ (median IFN-γ = 2,135 pg/ml versus 4,755 pg/ml in the same donors for ST-CF). As would be expected from the species distribution, reactivity to CFP10 and ESAT-6 was TB specific. These two antigens were recognized only by individuals infected with M. tuberculosis and not by BCG-vaccinated and unvaccinated healthy Danes (Fig. 4).
FIG. 4.
IFN-γ responses to low-mass antigens from M. tuberculosis in different groups of donors. Seven healthy nonvaccinated donors, 7 healthy BCG-vaccinated donors, and 17 TB patients were stimulated with 5 μg of ST-CF or recombinant antigens. Individual antigen-specific responses are shown as delta values (IFN-γ release in the antigen-stimulated well minus IFN-γ release in the unstimulated well). Positive responses are defined as delta values above 200 pg of IFN-γ/ml. IFN-γ release in unstimulated wells was generally below 100 pg of IFN-γ/ml. Three TB patients induced an IFN-γ release against ST-CF that was higher than 10,000 pg/ml. These results are not included in the figure.
Compared to ESAT-6, TB10.4 induced significantly higher levels of IFN-γ in TB patients (P = 0.0017, Wilcoxon signed-rank test), whereas T-cell responses to CFP10 and ESAT-6 were similar (P = 0.121).
There was no correlation between the responses to the individual antigens in responsive patients, and several patients recognized only one or two of the three ESAT-6 family members.
DISCUSSION
In the present study, three novel low-mass M. tuberculosis proteins were characterized and immune responses to these molecules were evaluated. Two of these antigens, CFP10 and TB10.4, were strongly recognized by >70% of the TB patients with levels of IFN-γ comparable to or higher than that of ESAT-6, whereas the third molecule, TB7.3, elicited only modest responses. Interestingly, the two strong antigens, CFP10 and TB10.4, both have several points in common with ESAT-6. They have almost identical size and pI (10 kDa and 4.5, respectively), and both coding genes have approximately 40% identity to esat-6. Together with a number of other putative ORFs, these molecules constitute what has been called the ESAT-6 family (9, 12). Our study clearly demonstrates that the molecules from this gene family evaluated so far also share a very striking immunological activity.
By searching different protein databases, it has not been possible from the primary structure of any of the proteins in the ESAT-6 family to provide clues as to the biological functions of these molecules. However, since there is no obvious homology to known proteins from other organisms, these proteins possibly have important mycobacterium-specific functions, which may be related to the intracellular habitat of the macrophage phagosome. In this regard, the apparent discrepancy between the low quantity of these molecules in vitro and the prominent role they play as targets in vivo may suggest that the expression of these molecules is highly upregulated during intracellular growth.
The homology of these molecules raises the question of whether identical epitopes are being recognized on these different molecules. The homology at the protein level is, however, relatively low (<20%), and we found neither cross-reactivity between the two MAbs nor DNA cross-hybridization between the three ESAT-6 family proteins (data not shown). Furthermore, an alignment of the protein sequences (Fig. 1) illustrates that the identical residues in these proteins are scattered throughout the sequence with no stretches of epitope size. The strong recognition of TB10.4 by BCG-vaccinated donors, which do not respond to ESAT-6 and CFP10, and the fact that there is no correlation between the responses to the individual antigens in responsive patients further confirm that different epitopes are recognized on these different molecules. Therefore, the explanation for the immunodominance of the molecules within the ESAT-6 family should be sought elsewhere than in their sequence homology. As alluded to above, we speculate that these molecules may have a role in bacterial virulence, and their synchronous upregulation during a particular phase of the infection may be part of the explanation. Another common denominator is, of course, the small size of these molecules, which may render them more susceptible to proteolytic degradation, processing, and intracellular traffic. However, the small size is on its own not enough, as illustrated by the low activity found with TB7.3, which is not an ESAT-6 family member. Of relevance in this regard, the immunological activity observed for TB7.3 is in agreement with the general picture which emerges from studies of human T-cell recognition of mycobacterial antigens (10, 21, 33, 34). In a recent study by Mustafa and colleagues (21), the majority of eight mycobacterial antigens were recognized by no more than 25 to 50% of the TB patients tested and, in general, had IFN-γ levels much below the responses to complex antigens, such as ST-CF and purified protein derivative. Along the same line, a study by Ulrichs et al. (33) recently reported that less than 20% of the TB patients recognized the two mycobacterial antigens MPT64 and MPT63. However, both of these studies, as well as other recent studies (24, 26), have identified ESAT-6 as the target for an extraordinary strong T-cell recognition and IFN-γ release in 65 to 95% of patients with TB from various geographical regions (21, 24, 26, 33).
The interspecies analysis demonstrated an identical distribution pattern of cfp10 and esat-6. The two genes are located in the same operon and are regulated by the same promoter (9). The cfp10–esat-6 operon is located in the RD-1 region, deleted in BCG (18), and in agreement with this, the genes were not found in any of the BCG strains tested.
The three ESAT-6 family proteins were all found in low concentrations in culture filtrate, although none of the proteins were found to have a conventional leader sequence for protein secretion (9, 31). In this regard, the increasing sensitivity of our identification and purification methods now allows the definition of molecules in ST-CF, which are present in very low quantities. This is exemplified by the difficulties in detecting the ESAT-6 family members in 2-DE gels by highly sensitive silver staining. Therefore, either a specific and yet undefined secretion mechanism may lead to the release of the ESAT-6 family members or these proteins represent cytoplasmic proteins which escape to the filtrate in trace amounts. With the lack of information on alternative translocation mechanisms in mycobacteria, this distinction is at present impossible.
Interestingly, CFP10 was found to induce strong IFN-γ responses in PBMC from human TB patients, whereas low responses (<1,000 pg/ml) were seen with PBMC from BCG-vaccinated healthy donors. This is in agreement with recent data from the investigation of ESAT-6 responses in these two groups of donors (26) and suggests that a combination of CFP10 and ESAT-6 would have major potential as a diagnostic reagent.
The data from this study, taken together with other recent investigations of T-cell responses to ESAT-6, indicate a striking focusing of the host immune response toward members of the ESAT-6 family. Although further studies are needed to explain and fully understand the host pathogen interactions leading to this target selection, it is clear that the ESAT-6 family contains a number of immunodominant molecules of relevance for future TB vaccines and diagnostics.
ACKNOWLEDGMENTS
This investigation received financial support from the Danish National Association against Lung Diseases and The European Community (project no. BMH4-97-2134 and BMH4-97-2167).
We are grateful to Iben Nielsen and Vita Skov for excellent technical assistance and thank Laurens van Pinxteren for critical reading of the manuscript.
REFERENCES
- 1.Andersen A B, Worsaae A, Chaparas S D. Isolation and characterization of recombinant lambda gt11 bacteriophages expressing eight different mycobacterial antigens of potential immunological relevance. Infect Immun. 1988;56:1344–1351. doi: 10.1128/iai.56.5.1344-1351.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen P, Andersen A B, Sorensen A L, Nagai S. Recall of long-lived immunity to Mycobacterium tuberculosis infection in mice. J Immunol. 1995;154:3359–3372. [PubMed] [Google Scholar]
- 3.Andersen P, Askgaard D, Gottschau A, Bennedsen J, Nagai S, Heron I. Identification of immunodominant antigens during infection with Mycobacterium tuberculosis. Scand J Immunol. 1992;36:823–831. doi: 10.1111/j.1365-3083.1992.tb03144.x. [DOI] [PubMed] [Google Scholar]
- 4.Andersen P, Askgaard D, Ljungqvist L, Bennedsen J, Heron I. Proteins released from Mycobacterium tuberculosis during growth. Infect Immun. 1991;59:1905–1910. doi: 10.1128/iai.59.6.1905-1910.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen P, Heron I. Simultaneous electroelution of whole SDS-polyacrylamide gels for the direct cellular analysis of complex protein mixtures. J Immunol Methods. 1993;161:29–39. doi: 10.1016/0022-1759(93)90195-d. [DOI] [PubMed] [Google Scholar]
- 6.Andersen P, Heron I. Specificity of a protective memory immune response against Mycobacterium tuberculosis. Infect Immun. 1993;61:844–851. doi: 10.1128/iai.61.3.844-851.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baek L. New, sensitive rocket immunoelectrophoretic assay for measurement of the reaction between endotoxin and Limulus amoebocyte lysate. J Clin Microbiol. 1983;17:1013–1020. doi: 10.1128/jcm.17.6.1013-1020.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes P F, Mehra V, Rivoire B, Fong S J, Brennan P J, Voegtline M S, Minden P, Houghten R A, Bloom B R, Modlin R L. Immunoreactivity of a 10-kDa antigen of Mycobacterium tuberculosis. J Immunol. 1992;148:1835–1840. [PubMed] [Google Scholar]
- 9.Berthet F X, Rasmussen P B, Rosenkrands I, Andersen P, Gicquel B. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10) Microbiology. 1998;144:3195–3203. doi: 10.1099/00221287-144-11-3195. [DOI] [PubMed] [Google Scholar]
- 10.Boesen H, Jensen B N, Wilcke T, Andersen P. Human T-cell responses to secreted antigen fractions of Mycobacterium tuberculosis. Infect Immun. 1995;63:1491–1497. doi: 10.1128/iai.63.4.1491-1497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandt L, Oettinger T, Holm A, Andersen P. Key epitopes on the ESAT-6 antigen recognized in mice during the recall of protective immunity to Mycobacterium tuberculosis. J Immunol. 1996;157:3527–3533. [PubMed] [Google Scholar]
- 12.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 13.Coler R N, Skeiky Y A, Vedvick T S, Bement T, Ovendale P J, Campos-Neto A, Alderson M R, Reed S G. Molecular cloning and immunologic reactivity of a novel low molecular mass antigen of Mycobacterium tuberculosis. J Immunol. 1998;161:2356–2364. [PubMed] [Google Scholar]
- 14.Elhay M J, Oettinger T, Andersen P. Delayed-type hypersensitivity responses to ESAT-6 and MPT64 from Mycobacterium tuberculosis in the guinea pig. Infect Immun. 1998;66:3454–3456. doi: 10.1128/iai.66.7.3454-3456.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harboe M, Oettinger T, Wiker H G, Rosenkrands I, Andersen P. Evidence for occurrence of the ESAT-6 protein in Mycobacterium tuberculosis and virulent Mycobacterium bovis and for its absence in Mycobacterium bovis BCG. Infect Immun. 1996;64:16–22. doi: 10.1128/iai.64.1.16-22.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harboe M, Wiker H G, Ulvund G, Malin A S, Dockrell H, Holm A, Jørgensen M C, Andersen P. B-cell epitopes and quantification of the ESAT-6 protein of Mycobacterium tuberculosis. Infect Immun. 1998;66:717–723. doi: 10.1128/iai.66.2.717-723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter S W, McNeil M, Modlin R L, Mehra V, Bloom B R, Brennan P J. Isolation and characterization of the highly immunogenic cell wall-associated protein of Mycobacterium leprae. J Immunol. 1989;142:2864–2872. [PubMed] [Google Scholar]
- 18.Mahairas G G, Sabo P J, Hickey M J, Singh D C, Stover C K. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehra V, Bloom B R, Bajardi A C, Grisso C L, Sieling P A, Alland D, Convit J, Fan X D, Hunter S W, Brennan P J. A major T cell antigen of Mycobacterium leprae is a 10-kD heat-shock cognate protein. J Exp Med. 1992;175:275–284. doi: 10.1084/jem.175.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehra V, Gong J H, Iyer D, Lin Y, Boylen C T, Bloom B R, Barnes P F. Immune response to recombinant mycobacterial proteins in patients with tuberculosis infection and disease. J Infect Dis. 1996;174:431–434. doi: 10.1093/infdis/174.2.431. [DOI] [PubMed] [Google Scholar]
- 21.Mustafa A S, Amoudy H A, Wiker H G, Abal A T, Ravn P, Oftung F, Andersen P. Comparison of antigen specific T cell responses of tuberculosis patients using complex or single antigens of Mycobacterium tuberculosis. Scand J Immunol. 1998;48:535–543. doi: 10.1046/j.1365-3083.1998.00419.x. [DOI] [PubMed] [Google Scholar]
- 22.Oettinger T, Andersen A B. Cloning and B-cell-epitope mapping of MPT64 from Mycobacterium tuberculosis H37Rv. Infect Immun. 1994;62:2058–2064. doi: 10.1128/iai.62.5.2058-2064.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orme I M. Characteristics and specificity of acquired immunologic memory to Mycobacterium tuberculosis infection. J Immunol. 1988;140:3589–3593. [PubMed] [Google Scholar]
- 24.Pathan A, Brookes R, Pritchard H, Wilkinson R, Pasvol G, Hill A, Lalvani A. Human T cell responses to the antigen ESAT-6 characterize a vaccine candidate and potential diagnostic test for tuberculosis. Immun Infect. 1998;95:90. [Google Scholar]
- 25.Pollock J M, Andersen P. The potential of the ESAT-6 antigen secreted by virulent mycobacteria for specific diagnosis of tuberculosis. J Infect Dis. 1997;175:1251–1254. doi: 10.1086/593686. [DOI] [PubMed] [Google Scholar]
- 26.Ravn P, Demissie A, Eguale T, Wondwosson H, Lein D, Amoudy H, Mustafa A S, Jensen A K, Holm A, Rosenkrands I, Oftung F, Olobo J, von-Reyn C F, Andersen P. Human T cell responses to the ESAT-6 antigen from Mycobacterium tuberculosis. J Infect Dis. 1999;179:637–645. doi: 10.1086/314640. [DOI] [PubMed] [Google Scholar]
- 27.Rosenkrands I, Rasmussen P B, Carnio M, Jacobsen S, Theisen M, Andersen P. Identification and characterization of a 29-kDa culture filtrate protein from Mycobacterium tuberculosis recognized in mice during the recall of immunity. Infect Immun. 1998;66:2728–2735. doi: 10.1128/iai.66.6.2728-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Rosenkrands I, Weldingh K, Ravn P, Brandt L, Hojrup P, Rasmussen P B, Coates A R, Singh M, Mascagni P, Andersen P. Differential T-cell recognition of native and recombinant Mycobacterium tuberculosis GroES. Infect Immun. 1999;67:5552–5558. doi: 10.1128/iai.67.11.5552-5558.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Silveira H, Ordway D, Dockrell H, Jackson M, Ventura F. Cell-mediated immune responses to mycobacterial antigens in patients with pulmonary tuberculosis and HIV infection. Clin Exp Immunol. 1997;110:26–34. doi: 10.1046/j.1365-2249.1997.5091407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonnenberg M G, Belisle J T. Definition of Mycobacterium tuberculosis culture filtrate proteins by two-dimensional polyacrylamide gel electrophoresis, N-terminal amino acid sequencing, and electrospray mass spectrometry. Infect Immun. 1997;65:4515–4524. doi: 10.1128/iai.65.11.4515-4524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorensen A L, Nagai S, Houen G, Andersen P, Andersen A B. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect Immun. 1995;63:1710–1717. doi: 10.1128/iai.63.5.1710-1717.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Theisen M, Vuust J, Gottschau A, Jepsen S, Hogh B. Antigenicity and immunogenicity of recombinant glutamate-rich protein of Plasmodium falciparum expressed in Escherichia coli. Clin Diagn Lab Immunol. 1995;2:30–34. doi: 10.1128/cdli.2.1.30-34.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ulrichs T, Munk M E, Mollenkopf H, Behr-Perst S, Colangeli R, Gennaro M L, Kaufmann S H. Differential T cell responses to Mycobacterium tuberculosis ESAT-6 in tuberculosis patients and healthy donors. Eur J Immunol. 1998;28:3949–3958. doi: 10.1002/(SICI)1521-4141(199812)28:12<3949::AID-IMMU3949>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Webb J R, Vedvick T S, Aldersen M R, Guderian J A, Jen S S, Ovendale P J, Johnson S M, Reed S G, Skeiky Y A. Molecular cloning, expression, and immunogenicity of MTB12, a novel low-molecular-weight antigen secreted by Mycobacterium tuberculosis. Infect Immun. 1998;66:4208–4214. doi: 10.1128/iai.66.9.4208-4214.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weldingh K, Rosenkrands I, Jacobsen S, Rasmussen P B, Elhay M J, Andersen P. Two-dimensional electrophoresis for analysis of Mycobacterium tuberculosis culture filtrate and purification and characterization of six novel proteins. Infect Immun. 1998;66:3492–3500. doi: 10.1128/iai.66.8.3492-3500.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young R A, Bloom B R, Grosskinsky C M, Ivanyi J, Thomas D, Davis R W. Dissection of Mycobacterium tuberculosis antigens using recombinant DNA. Proc Natl Acad Sci USA. 1985;82:2583–2587. doi: 10.1073/pnas.82.9.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]