Abstract
Several assays have been developed for detection of immunoglobulin G antibodies to Human herpesvirus 8 (HHV-8), including immunofluorescence assays (IFAs) and enzyme-linked immunosorbent assays (ELISAs). However, the specificity and sensitivity of these assays are not completely defined due to the lack of a “gold standard.” Although IFAs based on primary effusion lymphoma (PEL) cell lines are used widely, the assays can be confounded by nonspecific reactions against cellular components and potential cross-reaction with antibodies against other herpesviruses. To provide more reliable IFAs, we established recombinant Semliki Forest viruses (rSFVs) expressing the HHV-8-specific proteins ORF73 and K8.1 and used BHK-21 cells infected with these rSFVs for IFA (ORF73-IFA and K8.1-IFA). Expression of the HHV-8-specific proteins at very high levels by the rSFV system allowed easy scoring for IFA and thereby increased specificity. The rSFV system also allowed detection of antibodies against glycosylation-dependent epitopes of K8.1. Titers measured by rSFV-based IFAs and PEL-based IFAs correlated well (correlation coefficients of >0.9), and concordances of seroreactivities between rSFV-based and PEL-based IFAs were >97% (κ > 0.93). K8.1-IFA was more sensitive than either ORF73-IFA or peptide ELISAs. Using PEL-based lytic IFA as a reference assay, the sensitivity and specificity of K8.1-IFA were estimated to be 94 and 100%, respectively. HHV-8 prevalences determined by K8.1-IFA among the human immunodeficiency virus (HIV)-positive (HIV+) Kaposi's sarcoma (KS) patients, HIV+ KS− patients, and healthy controls were 100, 65, and 6.7%, respectively, which were consistent with prior reports. Therefore, our rSFV-based IFAs may provide a specific and sensitive method for use in epidemiology studies. In addition, they will provide a basis for further development of diagnostic tests for HHV-8 infection.
Human herpesvirus 8 (HHV-8) was identified in 1994 by Chang et al. (7). Subsequent studies have demonstrated that the virus has an etiologic role in Kaposi's sarcoma (KS), as well as a strong association with body-cavity-based lymphoma (BCBL)/primary effusion lymphoma (PEL) and certain forms of multicentric Castleman's disease (reviewed in references 24, 33, and 36). HHV-8 is not readily isolated in cell culture, and HHV-8 infection is usually diagnosed by detection of viral DNA or by serology. Therefore, reliable methods for antibody detection are crucial for fully understanding the epidemiology and biology of the virus, as well as for clinical diagnosis of infection.
The first generation of methods for HHV-8 antibody detection included indirect immunofluorescence assay (IFA) and immunoblotting using PEL cell lines that harbor HHV-8 genomes as antigens (13, 14, 25, 34, 35). PEL cell lines express the latency-associated nuclear antigen (LANA) under standard culture conditions. The HHV-8 orf73 gene product is the major component of LANA (17, 31) and is essential for maintenance of episomal HHV-8 genomes in latently infected cells (2). After treatment with 12-O-tetradecanoylphorbol-13-acetate (TPA), 10 to 30% of cells produce cytoplasmic antigens that correspond to structural proteins and replication enzymes associated with HHV-8 lytic infection. PEL-based assays can be confounded by nonspecific reactions against cellular components because matched HHV-8-negative PEL cell lines are not available for use as a negative control. In some experiments, B-cell lines that have a genetically different background from PEL cell lines have been used as controls or for adsorption (14). PEL-based assays, especially those using TPA-induced cells, also have potential for cross-reaction with antibodies against other herpesviruses. Nevertheless, PEL-based assays using TPA-induced cells were the most sensitive serological assays. In one study, bacterially expressed major viral capsid (ORF25) protein but not minor viral capsid (ORF26) protein cross-reacted with Epstein-Barr virus (EBV)-seropositive sera (1), although in other studies antibodies against EBV did not cross-react with HHV-8 lytic antigens in immunoblots (22, 35) or IFA (20). Modifications to reduce nonspecific reactions and increase sensitivity include isolation of nuclei from PEL cells for a latent assay (18) and use of a mouse monoclonal antibody against human immunoglobulin G (IgG) for IFA (20).
The current generation of assays includes enzyme-linked immunosorbent assays (ELISAs) using defined antigens such as purified whole virions, recombinant proteins, and oligopeptides (1, 9, 10, 26, 30, 37). However, the HHV-8 seropositivities in human immunodeficiency virus (HIV)-positive (HIV+) KS patients obtained by some of these assays were substantially lower than those obtained by other assays (1, 10, 30, 37). A recent blinded comparison of the two recombinant protein ELISAs, whole-virion ELISA, and four PEL-based IFAs demonstrated the need for both new assay development and standardization (30).
K8.1 is one of the most immunogenic HHV-8 proteins (6, 29). Because K8.1 has no homolog in other herpesviruses, assays based on the protein should provide high specificity. Very recently, such assays have been developed in several formats: an IFA using COS cells transfected with the K8.1 gene, immunoblotting with a K8.1–glutathione S-transferase fusion protein produced from a recombinant baculovirus, and ELISAs with bacterially expressed K8.1 and with an oligopeptide derived from K8.1 (19, 21, 39; T. J. Spira, L. Lam, S. C. Dollard, Y.-X. Meng, C.-P. Pau, J. B. Black, D. Burns, B. Cooper, M. Hamid, J. Huong, K. Kite-Powell, and P. E. Pellett, submitted for publication).
To develop reliable and sensitive serologic assays, it is critical to express HHV-8-specific antigens at a high level. Of the several systems developed for gene expression in mammalian cells, virus vectors based on alphaviruses have advantages in easy construction, wide host range, and transient high-level protein production (12, 15). Semliki Forest virus (SFV), an alphavirus, is a small enveloped virus with a single-stranded RNA genome of positive polarity. The 5′ two-thirds of the viral genome encodes nonstructural proteins (nsPs 1 to 4) and the 3′ one-third encodes structural proteins. Upon infection, the RNA genome functions as mRNA for translation of nsPs that replicate the viral RNA genome to >105 copies per cell. nsPs also promote transcription at the internal subgenomic promoter on the minus strand and produce large amounts of subgenomic RNA that encodes the capsid and envelope proteins. The capsid protein recognizes a packaging signal buried in the nsP2 open reading frame (ORF) and forms a nucleocapsid with genomic RNA. After acquisition of envelope glycoproteins, viral particles bud from cells. In the DNA-based SFV vector system (11), production of recombinant SFV (rSFV) is based on cotransfection of two plasmids. One is the vector plasmid, which contains the nsPs gene driven by the cytomegalovirus (CMV) immediate-early (IE) promoter, the subgenomic promoter followed by the heterologous gene, and segments derived from the 5′ and 3′ ends of the genome that are required for replication. The other is the helper plasmid, which contains the 5′ and 3′ replication signals and a subgenomic promoter followed by the structural gene but which lacks the nsP2 gene containing the packaging signal. The rSFV obtained in this way contains a defective genome that can replicate in infected cells but cannot generate progeny. The rSFV-based gene expression system has been used for various purposes, including serologic assays (3, 28).
In this study, we established rSFV expressing HHV-8-specific gene products ORF73 and K8.1 and evaluated IFAs that employed BHK-21 cells infected with these rSFVs as antigens.
MATERIALS AND METHODS
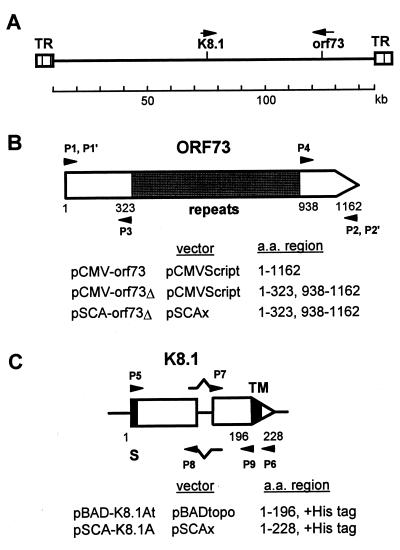
Plasmid construction.
Plasmids used in this study (Fig. 1) were constructed by standard techniques as follows. The HHV-8 orf73 gene was obtained from genomic DNA of BC-1 cells (5) (a gift of Y. Chang, Columbia University) by PCR amplification with primers P1 and P2 (see below). The PCR product was cloned between the EcoRI and HindIII sites of pCMVScript (Stratagene, La Jolla, Calif.), resulting in pCMV-orf73, which encodes the full-size of ORF73 protein. pCMV-orf73Δ consists of two segments of the orf73 gene obtained by PCR with primers P1 and P3 and with primers P2 and P4. pCMV-orf73Δ encodes a truncated ORF73 protein that lacks the region of repeated amino acid sequence (amino acids 324 to 937). pSCA-orf73Δ was constructed from pCMV-orf73Δ by PCR with primers P1′ and P2′, followed by cloning the PCR product into the BamHI site of pSCAx. pSCAx was obtained by self-ligation of a BamHI digest of pSCAβ (a gift of R. Bremner, University of Toronto) (11) to remove the lacZ gene. A DNA fragment containing the K8.1 gene was obtained by PCR from genomic BC-1 cell DNA with primers P5 and P6 and then cloned into pBADtopo (Invitrogen, Carlsbad, Calif.). The cDNA version of K8.1 (K8.1A) was obtained by PCR of the plasmid with four primers (P5, P6, P7, and P8). The PCR product was cloned into pBADtopo, resulting in pBAD-K8.1A, which encodes the K8.1A protein with a His6 tag. pBAD-K8.1At encoding a truncated form of K8.1A was obtained by PCR of pBAD-K8.1A with primers P5 and P9. The NotI-XbaI fragment of pBAD-K8.1A was repaired, ligated with 8-nucleotide BamHI linkers (New England Biolabs, Beverly, Mass.), and then cloned into the BamHI site of pSCAx, resulting in pSCA-K8.1A. The sequences of the primers are as follows (the epitope tag is underlined, non-HHV-8 sequences are italicized, and the start or stop codons are in boldface): P1, 5′-GGAATTCCACCATGGCGCCCCCGGGAATGCGCC; P1′, 5′-GGAAGATCTGGCGCCCCCGGGAATGC; P2, 5′-CCCAAGCTTATGTCATTTCCTGTGGAGAGTCCC; P2′, 5′-GGAAGATCTTATGTCATTTCCTGTGG; P3, 5′-CCGGATATCCTTGTCAACCTGACT; P4, 5′-CCGGATATCTTGCACGGGTCGTCA; P5, 5′-AGCGGCCGCGATCTGTACGACGATGACGATAAGATGAGTTCCACACAGATTCGCACAGAAATC; P6, 5′-TCTAGATTAATGATGATGATGATGATGCACTATGTAGGGTTTCTTACG; P7, 5′-TGGTTCCCCAGATGAATATGATCCTGAAAAGGCTGATATTAAGGCATC; P8, 5′-GATGCCTTAATATCAGCCTTTTCAGGATCATATTCATCTGGGGAACCA; and P9, 5′-ATGATGATGATGATGATGATGGGTCCGTATTTCTGCATTGTAGTG.
FIG. 1.
(A) Diagram of HHV-8 genome structure and the locations of orf73 and K8.1 genes. (B) Construction of plasmids for orf73 gene expression in mammalian cells. (C) Construction of plasmids for K8.1 gene expression in E. coli (pBAD-K8.1At) and in mammalian cells (pSCA-K8.1A).
Purification of K8.1 protein expressed in Escherichia coli.
E. coli BL21 (Stratagene) containing pBAD-K8.1At was grown to an optical density at 600 nm (OD600) of 0.5 to 0.7 and then incubated for 3 h in the presence of 0.05% l-(+) arabinose to induce gene expression from the araBAD promoter. After harvest, cells were suspended in buffer A (50 mM Tris-HCl, pH 8.0; 0.5 M NaCl; 5 mM dithiothreitol [DTT]; 0.1% Tween 20; 10 mM imidazole-HCl; 100 μg of lysozyme per ml; 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) and then sonicated. Cell extracts were obtained by centrifugation at 27,000 × g for 1 h. The K8.1At protein was purified on Talon resin (cobalt-nitriloacetic acid; Clontech, Palo Alto, Calif.) under the following conditions: binding of cell extracts prepared from 1 liter of the original culture with 1.5 ml of resin at 4°C for 2 h, washing with 50 bed volumes of buffer B (50 mM Tris-HCl, pH 8.0; 0.5 M NaCl; 8 M urea; 20 mM imidazole), and stepwise elution with 3 ml each of buffer B containing 0.1, 0.2, 0.3, or 0.5 M imidazole. Eluates were dialyzed against buffer C (50 mM Tris-HCl, pH 8.0; 5 mM EDTA; 10 mM DTT; 0.4 mM PMSF; 6 M urea) and then applied to a 5-ml Econo-Pac High-Q anion-exchange column (Bio-Rad Laboratories, Hercules, Calif.). The column was washed with buffer C containing 0.1 M NaCl and then with a linear gradient of 0.1 M and 0.7 M NaCl in buffer B. Purified protein was dialyzed against 0.1 M NaHCO3-Na2CO3 (pH 9.4). The purity of the protein was more than 90%, based on silver staining (Protein Silver Stain Kit; Bexel, Union City, Calif.). Approximately 40 to 60 μg of the purified protein was obtained from 1 liter of the bacterial culture.
ELISAs.
ELISAs with oligopeptides containing antigenic epitopes of K8.1 and ORF65 as antigens (K8.1-ELISA and ORF65-ELISA) were as described previously (26; Spira et al., submitted). A serum dilution of 1:100 was used. The cutoff was set at the mean plus 5 standard deviations of OD values of sera from healthy controls.
Transfection and rSFV production.
Transfection of 293T cells (27) was done by the calcium phosphate method (32). For rSFV production, 293T cells were transfected with 16 μg of a helper plasmid pSCA-Helper and 4 μg of a vector plasmid per 10-cm-diameter dish. SFV-βgal, SFV-orf73Δ, and SFV-K8.1A were obtained with vector plasmids, pSCAβ, pSCA-orf73Δ, and pSCA-K8.1A, respectively. Culture supernatants were harvested 40 to 44 h after transfection. Cell debris in the supernatant was removed by passage through 0.45-μm filters. After 1 h of incubation of the filtered supernatant with 0.5 mg of α-chymotrypsin per ml, rSFV vector particles were precipitated by 3 h of incubation at 4°C in the presence of 10% polyethylene glycol (PEG) 8000 and 0.5 M NaCl and then resuspended in Dulbecco's modified Eagle's medium (DMEM). Titers of SFV-βgal stocks were determined by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside staining, and titers of SFV-orf73Δ and SFV-K8.1A stocks were determined by IFA.
Generation of rabbit anti-K8.1 peptide and anti-ORF73 peptide sera.
Oligopeptides P2700 (RIRVSPVAENGRNSGASNRV) and P2599 (KSSPDSLAPSTLRSLR), corresponding to K8.1A amino acid residues 154 to 173 and ORF73 amino acid residues 185 to 200, respectively, were conjugated to keyhole limpet hemocyanin using glutaraldehyde. The conjugate (0.5 ml of a 1-mg/ml concentration) was mixed in equal volume of Freund's incomplete adjuvant and used to immunize New Zealand White rabbits three to four times (Genelabs, Baton Rouge, La.). Rabbit anti-His6 tag antibody was purchased from a commercial source (Santa Cruz Biotechnology, Santa Cruz, Calif.).
Immunoblotting.
Cells were rinsed with phosphate-buffered saline (PBS), lysed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer containing 2-mercaptoethanol, and then sonicated four times for 30 s each time (power level 10, model W-375; Heat Systems-Ultrasonic, Inc., Farmingdale, N.Y.). Proteins were separated by SDS-PAGE and then transferred onto nylon membrane filters (Hybond-N, Amersham, Arlington, Ill.). The blots were incubated overnight in 5% skim milk in PBS containing 0.05% Tween 20 (PBST) and reacted with primary antibodies for 2 h. After three 10-min washes with PBST, the blots were reacted with horseradish peroxidase-conjugated goat anti-rabbit or anti-human immunoglobin (Amersham) for 1 h, washed, and then developed with a commercial detection kit (ECL Plus; Amersham) according to the vendor's protocol.
The relative expression levels of the antigens were quantified by two ways. One was by quantification of the intensity of bands after scanning the X-ray films. The other was by dilution of the sample to obtain similar intensity in a gel. The relative amount of antigens was also determined by coating cell lysate on ELISA plates, followed by detection with rabbit antibodies or with an HIV+ KS+ human serum specimen (Jose Corchero and N.I., unpublished).
IFA.
Mouse monoclonal antibody-enhanced IFAs (mIFAs) using untreated and TPA-induced BCBL-1 cells (latent-mIFA and lytic-mIFA) were as described previously (20). IFAs using rSFV-infected BHK-21 cells as antigens were performed as follows. First, 2 × 106 BHK-21 cells in one 10-cm dish were rinsed with PBS containing 10 mM MgCl2 and 10 mM CaCl2, incubated with rSFV diluted in DMEM without fetal bovine serum (FBS) at 37°C for 4 h, and then rinsed with PBS and incubated in DMEM supplemented with 10% of FBS for 20 h. rSFV infection was performed at a multiplicity of infection (MOI) of 0.3 so that approximately 30% of cells expressed the antigens. The cells were recovered by trypsinization. Approximately 104 cells were applied to each 5-mm-diameter well of Teflon-coated slides (12 wells per slide), dried, and then fixed in cold acetone for 10 min. Fixed cells on the slides were incubated for 1 h at 37°C with twofold dilutions of human sera beginning at 1:20. After three washes with PBS, the slides were incubated for 30 min with fluorescein isothiocyanate-conjugated goat anti-human IgG (Sanofi Diagnostics Pasteur, Chaska, Minn.). The slides were counterstained with Evans blue. In the upper row of each slide the wells contained cells infected with SFV-K8.1 or with SFV-orf73Δ (antigen wells), and in the bottom row the wells contained SFV-βgal-infected cells (control wells). Nonspecific reactions were evaluated by two ways. One was to use the 70% of uninfected cells in the same well as a negative control. The other was to compare reactions between the antigen well and the control well. The endpoint titer was defined as the highest serum dilution at which specimens reacted with >10% of cells in the antigen well.
Serum specimens were coded, and the IFAs were done blindly. Each specimen was examined by IFA at least twice; differences were within fourfold in all cases, and the mean of the repeated measurements was defined as the titer.
Human sera.
A total of 27 sera from HIV+ KS+ patients, 23 sera from HIV+ KS− patients, 2 sera from HIV− KS+ patients, and 10 sera from HIV− individuals with some risk factors (RF+) were obtained from a cross-sectional study in Atlanta (P.E. Pellett et al., manuscript in preparation) and from M. Offermann. Of 23 HIV+ KS− patients, 12 were men who have sex with men (MSM), and 2 were men with intravenous drug use (IVDU). Both of the HIV− KS+ sera were MSM. Of 10 RF+ individuals, 2 were MSM, 4 were men attending a sexually transmitted disease (STD) clinic, and 4 were women attending an STD clinic. Thirty serum specimens were obtained from the American Red Cross blood bank for use as healthy controls.
Statistical analyses.
Correlations between titers obtained by two different methods were evaluated by Pearson's correlation coefficient. The degree of concordance between the two assays was assessed by using the kappa statistic, which measures the excess of the observed agreement over that expected purely by chance. Kappa (κ) equals 1.0 for perfect agreement and 0 for no agreement beyond chance; values between 0.4 and 0.8 were considered to represent fair to good agreement beyond chance. The significance of the difference in seroreactivities between two groups was measured by the chi-square test.
RESULTS
We used the DNA-based rSFV system (11) for high-level expression of HHV-8-specific gene products and established rSFV-based IFAs for the detection of antibodies against HHV-8 in human sera. We compared antibody titers in each assay to demonstrate the specificity and then compared seroreactivities of all human specimen groups to characterize the sensitivity.
Optimization of rSFV production.
Based on the recommendation of the developer of the system, we used 293 cells to obtain a high-titer stock of rSFV (R. Bremner, personal communication). We then compared the production of rSFV in 293T cells with that in 293 cells, because expression from the CMV IE promoter in 293T cells is higher than in 293 cells (38). We found that use of 293T cells increased rSFV production by about 10-fold (data not shown). To simplify the purification, we tested precipitation of rSFV with PEG instead of sucrose density gradient centrifugation. rSFV particles were efficiently recovered after incubation in the presence of 10% PEG–0.5 M NaCl (data not shown).
Under optimized conditions, transfected 293T cells yielded >107 rSFV particles per ml of culture supernatant. Starting from five 10-cm dishes of 293T cells, >5 × 108 rSFV can be prepared easily within 4 days. This is sufficient for preparation of more than 5,000 12-well IFA slides.
rSFV expression of ORF73 and K8.1.
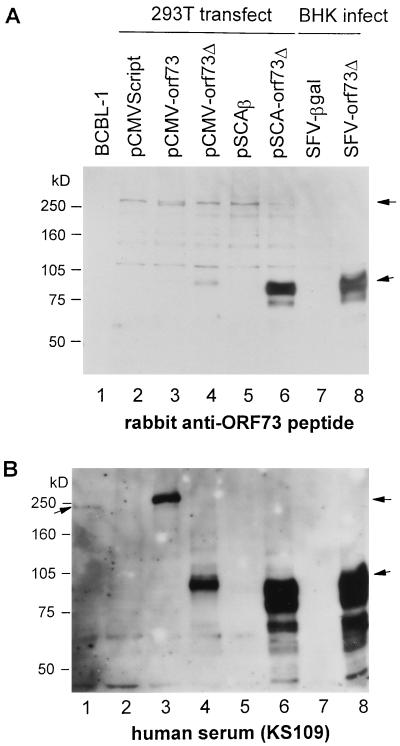
ORF73 protein expressed in BCBL-1 cells was detected as a band with an apparent molecular mass of 240 kDa in immunoblots immunoblots with an HIV+ KS+ human serum, as reported previously (17, 31) (Fig. 2B, lane 1). The same-sized band was barely detected with rabbit anti-ORF73 antibody in the short exposure shown (Fig. 2A, lane 1) but was faintly detectable after a longer exposure (data not shown). We cloned the orf73 gene from BC-1 cells and constructed plasmids pCMV-orf73 and pCMV-orf73Δ for expression of the full-length and truncated versions of ORF73 under control of the CMV IE promoter (Fig. 1). Both versions of the ORF73 protein expressed from these plasmids were detected in immunoblotting with anti-ORF73 peptide antibody and with an HIV+ KS+ patient serum (Fig. 2, lanes 3 and 6). The difference in size of the full-length version of ORF73 proteins (lanes 1 and 3) is due to the difference in the length of the repeated region between ORF73 from BC-1 and BCBL-1 cells (31). Use of the SFV vector plasmid pSCA-orf73Δ markedly increased the production of the truncated version of ORF73 (lane 6) due to amplification of the vector and subgenomic RNAs by SFV nsPs expressed from the plasmid after transfection (11). A further slight increase was observed after transduction of BHK-21 cells with SFV-orf73Δ (lane 8).
FIG. 2.
Expression of ORF73 in mammalian cells. Cell lysates were prepared from BCBL-1 cells (lane 1); 293T cells transfected with pCMVScript (lane 2), pCMV-orf73 (lane 3), pCMV-orf73Δ (lane 4), pSCAβ (lane 5), or pSCA-orf73Δ (lane 6); and BHK-21 cells infected with SFV-βgal (lane 7) or SFV-orf73Δ (lane 8) at an MOI of 3. Lysates were prepared from an equal number of cells (4 × 104 cells/lane) except for BCBL-1 (8 × 104 cells/lane) and then separated by SDS–7.5% PAGE. ORF73 proteins were detected by immunoblotting with rabbit anti-ORF73 peptide antibodies (A) and HIV+ KS+ patient serum KS109 (B). Arrows indicate the full-size and truncated ORF73 proteins.
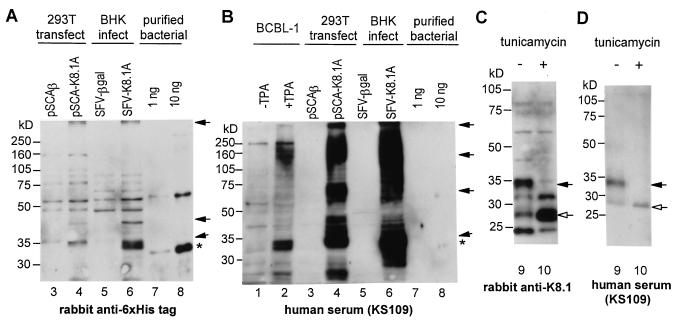
K8.1A proteins with apparent molecular masses of 35 to 37 kDa, as well as antigenically related species of higher molecular weight, were detected in TPA-treated BCBL-1 cells in immunoblots with an HIV+ KS+ human serum (Fig. 3, lane 2) and with rabbit anti-K8.1 peptide antibody (data not shown). We cloned K8.1 ORF from BC-1 cells (K8.1A) and constructed the SFV vector plasmid pSCA-K8.1A which encodes K8.1A with His6 tag at the carboxyl end (Fig. 1). Large amounts of K8.1A proteins were produced in 293T cells transfected with the plasmid and in BHK-21 cells infected with SFV-K8.1A (Fig. 3, lanes 4, 6, and 9). As inferred by comparison with purified K8.1At, approximately 1 ng of K8.1A was expressed per 104 cells (Fig. 3A, lanes 6 to 8); this result indicated that >106 molecules of K8.1A were produced in each cell.
FIG. 3.
Expression of K8.1 in mammalian cells. Cell lysates were prepared from TPA-uninduced (lane 1) and -induced (lane 2) BCBL-1 cells, 293T cells transfected with pSCAβ (lane 3) or pSCA-K8.1A (lane 4), and BHK-21 cells infected with SFV-βgal (lane 5) or SFV-K8.1A (lanes 6, 9, 10) at an MOI of 3. Lysates were prepared from the equal number of cells (4 × 104 cells/lane) except for BCBL-1 (8 × 104 cells/lane) and then separated on 10% (A and B) or 12% (C and D) SDS-PAGE gels. K8.1 proteins were detected by immunoblotting with anti-His6 tag antibody (A), HIV+ KS+ patient serum KS109 (B and D), and anti-K8.1 peptide antibody (C). K8.1At protein expressed in E. coli and purified (∗) was run in parallel for quantitation (lanes 7 and 8). BHK-21 cells were treated with tunicamycin (0.2 μg/ml) during SFV-K8.1A infection (lane 10). Closed arrows indicate the K8.1 proteins, and open arrows indicate unglycosylated K8.1 protein.
Glycosylation-dependent epitopes of K8.1.
Tunicamycin treatment demonstrated that the expressed K8.1A proteins were glycosylated as reported previously (6, 21, 29) (Fig. 3C, lane 10). Although antibodies against a K8.1 peptide (Fig. 3C) and the His6 tag (data not shown) reacted similarly with glycosylated and unglycosylated forms of the K8.1A protein, human serum KS109 (Fig. 3D) reacted with the glycosylated form much more strongly than with the unglycosylated form. Similar results were obtained with two other human sera (data not shown). These observations suggest that human sera contain antibodies against epitopes that depend on glycosylation of the protein. This observation is supported by differences in antibody reactivities between the K8.1 proteins expressed in E. coli and in BHK-21 cells. Rabbit antibodies against the His6 tag (Fig. 3A, lanes 6 and 8) and against a K8.1 peptide (data not shown) detected the K8.1A protein expressed in 4 × 104 BHK-21 cells with an intensity similar to the reaction with 10 ng of the bacterially expressed K8.1At protein. However, human serum KS109 detected the K8.1A protein expressed in 4 × 104 BHK-21 cells more strongly than 10 ng of the bacterially expressed K8.1At protein (Fig. 3B, lanes 6 and 8).
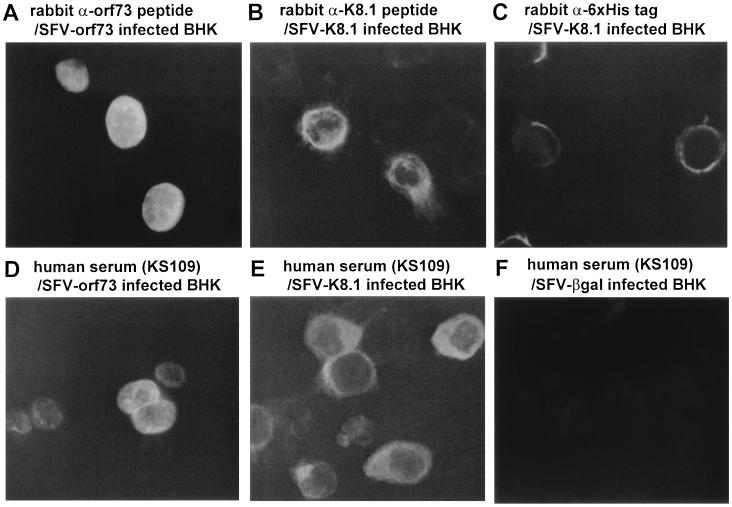
Cellular localization of ORF73 and K8.1.
Localizations of K8.1A and ORF73 proteins expressed in rSFV-infected cells were determined by IFA (Fig. 4). K8.1A was present on cell surfaces and in the cytoplasm, while the truncated ORF73 was distributed diffusely in nuclei. Localizations of the full-length and the truncated ORF73 proteins in 293T cells transfected with plasmids were identical to that in SFV-orf73Δ-infected BHK-21 (data not shown), indicating that the deleted region is not required for nuclear localization.
FIG. 4.
Localization of ORF73 and K8.1. ORF73 (A and D) and K8.1 (B, C, and E) proteins expressed in rSFV-infected BHK-21 cells were detected by IFA with anti-ORF73 peptide (A), anti-K8.1 peptide (B), and anti-His6 tag (C) antibodies and human serum KS109 (D to F). BHK-21 cells infected with SFV-βgal were used as a negative control (F).
Comparison of rSFV-based and PEL-based IFAs.
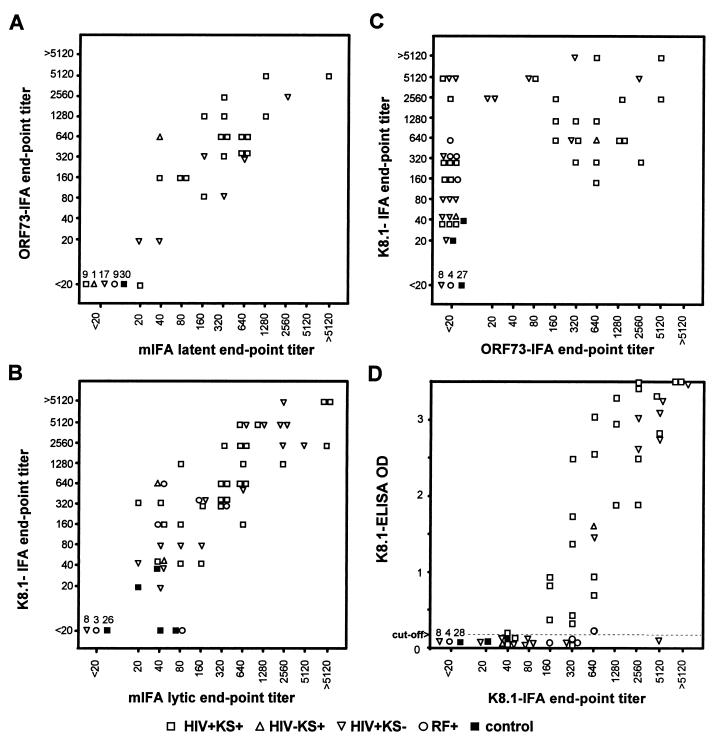
SFV-orf73Δ- and SFV-K8.1A-infected BHK-21 cells were used as antigens in IFAs (ORF73-IFA and K8.1-IFA). SFV-βgal-infected BHK-21 cells were used as negative controls to evaluate nonspecific reactions. We determined endpoint antibody titers against ORF73 and K8.1 by rSFV-based IFAs, as well as those against latent and lytic antigens by PEL-based mIFAs. A total of 92 serum specimens, including 27 HIV+ KS+ patients, 23 HIV+ KS− patients, 2 HIV− KS+ patients, 10 HIV− individuals with risk factors (RF+), and 30 healthy blood donors, were analyzed. The endpoint titers obtained by latent-mIFA and ORF73-IFA and those obtained by lytic-mIFA and K8.1-IFA correlated well, respectively (correlation coefficients of >0.92) (Fig. 5A and B). One serum specimen from an HIV+ KS+ patient was negative (titer of <20) in ORF73-IFA but weakly positive in latent-mIFA (titer of 20) (Fig. 5A). Three sera were negative in K8.1-IFA but positive in lytic-mIFA (Fig. 5B). Two of them were from healthy controls; these sera were negative in the other assays used in this study. The other serum was from an RF+ individual; this serum reacted in ORF65-ELISA but not in K8.1-ELISA or latent-mIFA (data not shown). The concordance of seroreactivities (titers of ≥20 are positive, ones that are <20 are negative) between latent-mIFA and ORF73-IFA was 99% (κ = 0.97), and 97% (κ = 0.93) between lytic-mIFA and K8.1-IFA, indicating that PEL-based IFAs and rSFV-based IFAs for latent and lytic antigens, respectively, provided nearly identical results. The sensitivity and specificity of K8.1-IFA using lytic-mIFA as a reference were 94 and 100%, respectively, and 45 and 100%, respectively, for ORF73-IFA.
FIG. 5.
(A) Comparison of ORF73-IFA titers with latent-mIFA titers. (B) Comparison of K8.1-IFA titers with lytic-mIFA titers. (C) Comparison of ORF73-IFA and K8.1-IFA titers. (D) Comparison of K8.1-IFA titers with OD measurements by K8.1-ELISA. A total of 92 sera from HIV+ KS+ (□), HIV− KS+ (▵), HIV+ KS− (▿), RF+ (○), and healthy control (■) individuals were examined. Due to nonspecific reactions, serum titers from two RF+ individuals were not available for IFAs.
Antibody titers measured by ORF73-IFA and K8.1-IFA had a statistically significant correlation (correlation coefficient of 0.65) (Fig. 5C), and seroreactivities determined by the two assays provide a fair concordance (71%, κ = 0.45). K8.1-IFA was more sensitive than ORF73-IFA. The degree of correlation between K8.1-IFA and ORF73-IFA was weaker than between K8.1-IFA and lytic-mIFA.
Comparison of rSFV-based IFAs with ELISAs.
We compared the titers obtained by K8.1-IFA with OD measurements obtained in two ELISAs that employ peptides as antigens: K8.1-ELISA (Fig. 5D) and ORF65-ELISA (data not shown). The K8.1-ELISA OD measurements correlated well with the K8.1-IFA endpoint titers (correlation coefficient of 0.84). Most serum specimens with K8.1-IFA titers of ≥640 were positive in these ELISAs, while few of those with titers between 20 and 80 in K8.1-IFA were positive in the ELISAs. Concordances of seroreactivities between K8.1-IFA and K8.1-ELISA and between ORF73-IFA and K8.1-ELISA were 80% (κ = 0.61) and 90% (κ = 0.77), respectively.
We also purified K8.1 protein expressed in E. coli and used it as an antigen in an ELISA. However, this assay was less sensitive than either K8.1-ELISA or K8.1-IFA (data not shown).
Seroreactivities of human specimens.
The seropositivities of each human specimen group obtained with four IFAs and two ELISAs are shown in Table 1. Antibodies against K8.1 were detected in all of the HIV+ KS+ patient sera by K8.1-IFA. A total of 94% (50 of 53) of sera positive in the lytic-mIFA were positive in the K8.1-IFA. K8.1-IFA was more sensitive than the other four assays. All sera containing antibodies against ORF73 were positive by K8.1-IFA, and all sera positive by K8.1-IFA were positive by lytic-mIFA. With one exception, every serum found to be positive in either of the ELISAs was positive by K8.1-IFA.
TABLE 1.
Seroreactivities in each assay
| Assay | No. of specimens seroreactive/total no. (%)a
|
||||
|---|---|---|---|---|---|
| HIV+ KS+ (n = 27) | HIV− KS+ (n = 2) | HIV+ KS− (n = 23) | RF+ (n = 10)b | Control (n = 30) | |
| Lytic-mIFA | 27/27 (100) | 2/2 (100) | 15/23 (65) | 5/10 (50) | 4/30 (13) |
| K8.1-IFA | 27/27 (100) | 2/2 (100) | 15/23 (65) | 4/8 (50) | 2/30 (6.7) |
| K8.1-ELISA | 23/27 (85) | 1/2 (50) | 7/23 (30) | 3/10 (30) | 0/30 (0) |
| ORF65-ELISA | 23/27 (85) | 0/2 (0) | 7/23 (30) | 2/10 (20) | 0/30 (0) |
| Latent-mIFA | 18/27 (67) | 1/2 (50) | 6/23 (26) | 0/8 (0) | 0/30 (0) |
| ORF73-IFA | 17/27 (63) | 1/2 (50) | 6/23 (26) | 0/9 (0) | 0/30 (0) |
| ORF73-IFA/K8.1-IFAc | 17/27 (63) | 1/2 (50) | 6/15 (40) | 0/4 (0) | 0/2 (0) |
n, number of serum specimens in each group. Data are the number of serum specimens positive/number of specimens tested.
Due to nonspecific reactions against cellular components at a serum dilution 1:20, seroreactivities of two specimens in the RF+ group are not available for some of the IFAs. These sera were negative at a serum dilution of 1:40 in IFAs.
The data in the last row indicate the number of serum specimens positive by ORF73-IFA/number of K8.1-IFA positive specimens (%).
Because all sera positive in the ORF73-IFA were positive by K8.1-IFA, we analyzed whether individuals containing antibodies against both ORF73 and K8.1 and those containing antibodies against K8.1 but not against ORF73 belong to different population groups. Among sera positive by K8.1-IFA, seropositivity in ORF73-IFA was more frequent in KS+ patient sera (18 of 29, 62%) than in KS− sera (6 of 21, 29%) (χ2 test, P < 0.02) (Table 1).
DISCUSSION
Advantages of rSFV-based IFAs.
PEL-based IFAs are widely used and have provided a large volume of insightful serological data (reviewed in references 8, 24, 33, and 36). However, these conventional IFAs have two potential drawbacks: nonspecific reactions against cellular components and cross-reaction with antibodies against other herpesviruses. In PEL-based latent IFAs, a characteristic punctate distribution of LANA is used to distinguish specific from nonspecific reactions, since authentic negative control cells are not available. Because expression of HHV-8-specific genes at very high levels allows these drawbacks to be circumvented while increasing assay sensitivity, we expressed HHV-8-specific antigens ORF73 and K8.1 using the rSFV system. One of the advantages of rSFV-based IFAs is that they provide uninfected cells as a negative control in the same field with rSFV-infected cells and also negative control wells coated with SFV-βgal-infected cells. High-level expression of ORF73 and K8.1 also allowed easy IFA reading, thereby increasing sensitivity and specificity.
Sensitivity and specificity of rSFV-based IFAs.
Titers measured by rSFV-based IFAs and PEL-based mIFAs showed a good linear correlation, and OD measurements by K8.1-ELISA correlated well with titers measured by K8.1-IFA, indicating a high specificity of rSFV-based IFAs. To determine the sensitivity and specificity of the newly developed assays, it is essential to have a “gold standard” assay that is standardized among researchers. Unfortunately, in the case of HHV-8 infection, there is no gold standard assay. One way to determine the specificity and sensitivity is to use the most sensitive available method as a reference. Lytic-mIFA is currently the most sensitive assay, although its specificity is undefined. Using this assay as a reference, the overall sensitivity and specificity of K8.1-IFA were 94 and 100%, respectively.
Another way to determine the specificity and sensitivity in the absence of a gold standard assay is to use populations that are assumed to be definitely seropositive or seronegative as gold standards. KS patients can be employed as presumed seropositives; young children or virginal women can be employed as presumed seronegatives based on the assumption that HHV-8 transmission is mainly by sexual routes in the United States. However, in the KS patient cases, it is possible that decreased immune response due to HIV-1 infection prevents development of antibodies against some viral infections. In addition, it is reported that HHV-8 primary infection frequently occurs during childhood in Africa, probably by mother-to-child transmission (4, 16, 33), and we do not know whether that type of transmission occurs in the United States. It is possible that individuals with low-titer HHV-8-specific antibodies in low-risk populations were not detected previously due to low assay sensitivity. In this study, antibodies against K8.1 were detected in all KS patients, indicating a sensitivity 100% relative to this standard. Although we did not have a definite seronegative population, for the purpose of calculating specificity we could assume that the 6.7% of blood donors who reacted against K8.1 were “false-positives,” indicating that the “true” specificity of K8.1-IFA is >93.3%. Therefore, based on the two ways of calculation, the sensitivity and specificity of K8.1-IFA were estimated to be 94 to 100% and 93 to 100%, respectively. A further comparison of K8.1-IFA with different assays done in different laboratories will be essential to more precisely define its specificity and sensitivity.
Although in this study all sera positive in ORF73-IFA were positive in K8.1-IFA, a recent study identified two cases containing anti-ORF73 but not anti-lytic antigens antibodies among 265 specimens (39). Further studies with a larger number of sera will be required to determine the frequency of such cases. Because two of the three sera negative in K8.1-IFA but positive in lytic-mIFA were negative in the other four assays, there is no evidence to corroborate positive reactions in lytic-mIFA as being HHV-8 specific. Use of ORF65 as an antigen in addition to K8.1A in the immunoblot assay increased sensitivity slightly (39). Addition of other HHV-8-specific antigens in rSFV-based IFAs may enable K8.1-IFA false negatives to be distinguished from lytic-mIFA false positives.
K8.1-IFA was more sensitive than the peptide-based ELISAs. Although combination of the results obtained from two ELISAs reduced the frequency of false negatives (Spira et al., submitted), it is still difficult to access the true status of specimens that give OD values that are near the cutoff value (data not shown). It is possible that the insensitivity of the ELISAs might be due to the use of a high serum dilution (1:100). We expressed K8.1A protein in E. coli and tried to use the purified protein in an ELISA. However, the low yield of the protein made it difficult to completely remove residual bacterial proteins that cross-reacted with human sera when large amounts of the proteins were used for plate coating and when lower serum dilutions were used (data not shown). In addition, we found that human sera contain antibodies against K8.1 that reacted more strongly with the glycosylated form of the protein. This might explain why a recently reported ELISA using bacterially expressed K8.1 protein as an antigen showed much lower seropositivities (19) compared to other studies that used K8.1 expressed in eukaryotic cells as antigen (21, 39). For high-throughput analyses such as the determination of prevalence in large populations, we are developing an ELISA with the K8.1 proteins expressed in the rSFV-infected cells.
Seroprevalence in different groups.
The six assays tested in this study exhibited a consistent pattern of decreasing positivity, with KS+ patients having the highest prevalence and blood donors having the lowest prevalence, although the assays based on latent antigens were less sensitive than the assays based on lytic antigens. Seroprevalences of HHV-8 based on K8.1-IFA in the HIV+ KS+ and HIV+ KS− groups were 100 and 65%, respectively, which were consistent with prior reports for lytic antigen-based assays (9, 20, 26, 30, 34, 39; Spira et al., submitted). K8.1-IFA detected 6.7% seropositivity in the U.S. blood donor group, while ORF73-IFA did not detect any seropositive individuals in that group. HHV-8 prevalence in the U.S. blood donors has been estimated to be 0 to 29%, depending on the assays (8, 9, 14, 20, 30, 34, 39). Prior studies by others also demonstrated that PEL-based lytic IFAs were more sensitive than the PEL-based latent IFAs (20, 30). Although the small amount of LANA in PEL cell lines might be the reason for the insensitivity of the latent IFAs, the nearly identical antibody titers between latent-mIFA and ORF73-IFA indicate that the amount of the antigen is not the limiting factor. It is likely that differences in sensitivity between latent and lytic assays reflect differences in immune responses against latent and lytic antigens in HHV-8-infected individuals. Assays based on latent antigens may underestimate the true prevalence of the virus.
Antibody profiles in different groups.
Among sera containing antibodies against K8.1, antibodies against ORF73 were significantly more prevalent in KS+ sera than in KS− sera. Although we do not have enough evidence at this point to draw a conclusion, it is possible that the appearance of antibodies against lytic antigens such as K8.1 precedes the increase of antibodies against LANA during KS progression. During primary EBV infection, antibodies against EBNA develop more slowly than those against viral capsid antigens (VCA). Anti-LANA positivity among KS patient sera containing anti-ORF65 antibodies was significantly higher than that among KS− sera containing anti-ORF65 antibodies (34). Comparison of seropositivities in lytic-mIFA and in latent-mIFA between the KS+ and KS− groups also supports the hypothesis (20). Previous studies demonstrated that antibodies against LANA or those against ORF73 are good markers for KS progression (13, 14, 23). Therefore, it will be interesting to monitor the difference in immune response against latent and lytic antigens such as K8.1 and ORF73 separately in longitudinal studies.
In conclusion, our rSFV-based IFA can provide a useful and reliable method for epidemiology studies and the basis for further development of serodiagnosis for HHV-8 infection. Because it is less ambiguous to interpret, K8.1-IFA may prove a useful substitute for lytic-mIFA, which is currently the most sensitive assay.
ACKNOWLEDGMENTS
We thank Y. Chang and D. Ganem for BC-1 and BCBL-1 cells and R. Bremner for pSCAβ and pSCA-Helper plasmids. We also thank F. R. Stamey for sequencing plasmid clones, T. J. Shepherd for performing part of the IFA reactions, Y.-X. Meng for critical reading of the manuscript, and the Atlanta HHV-8 Working Group and M. Offermann for the collection of the KS patient sera used in this study.
REFERENCES
- 1.André S, Schatz O, Bogner J R, Zeichhardt H, Stöffler-Meilicke M, Jahn H-U, Ullrich R, Sonntag A K, Kehm R, Haas J. Detection of antibodies against viral capsid proteins of human herpesvirus 8 in AIDS-associated Kaposi's sarcoma. J Mol Med. 1997;75:145–152. doi: 10.1007/s001090050099. [DOI] [PubMed] [Google Scholar]
- 2.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 3.Bouche F, Ammerlaan W, Berthet F, Houard S, Schneider F, Muller C P. Immunosorbent assay based on recombinant hemagglutinin protein produced in a high-efficiency mammalian expression system for surveillance of measles immunity. J Clin Microbiol. 1998;36:721–726. doi: 10.1128/jcm.36.3.721-726.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourboulia D, Whitby D, Boshoff C, Newton R, Beral V, Carrara H, Lane A, Sitas F. Serologic evidence for mother-to-child transmission of Kaposi sarcoma-associated herpesvirus infection. JAMA. 1998;280:31–32. doi: 10.1001/jama.280.1.31-a. [DOI] [PubMed] [Google Scholar]
- 5.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 6.Chandran B, Bloomer C, Chan S R, Zhu L, Goldstein E, Horvat R. Human herpesvirus-8 ORF K8.1 gene encodes immunogenic glycoproteins generated by spliced transcripts. Virology. 1998;249:140–149. doi: 10.1006/viro.1998.9316. [DOI] [PubMed] [Google Scholar]
- 7.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 8.Chatlynne L G, Ablashi D V. Seroepidemiology of Kaposi's sarcoma-associated herpesvirus (KSHV) Semin Cancer Biol. 1999;9:175–185. doi: 10.1006/scbi.1998.0089. [DOI] [PubMed] [Google Scholar]
- 9.Chatlynne L G, Lapps W, Handy M, Huang Y Q, Masood R, Hamilton A S, Said J W, Koeffler H P, Kaplan M H, Friedman-Kien A, Gill P S, Whitman J E, Ablashi D V. Detection and titration of human herpesvirus-8-specific antibodies in sera from blood donors, acquired immunodeficiency syndrome patients, and Kaposi's sarcoma patients using a whole virus enzyme-linked immunosorbent assay. Blood. 1998;92:53–58. [PubMed] [Google Scholar]
- 10.Davis D A, Humphrey R W, Newcomb F M, O'Brien T R, Goedert J J, Straus S E, Yarchoan R. Detection of serum antibodies to a Kaposi's sarcoma-associated herpesvirus-specific peptide. J Infect Dis. 1997;175:1071–1079. doi: 10.1086/516444. [DOI] [PubMed] [Google Scholar]
- 11.DiCiommo D P, Bremner R. Rapid, high-level protein production using DNA-based Semliki Forest virus vectors. J Biol Chem. 1998;273:18060–18066. doi: 10.1074/jbc.273.29.18060. [DOI] [PubMed] [Google Scholar]
- 12.Frolov I, Hoffman T A, Prágai B M, Dryga S A, Huang H V, Schlesinger S, Rice C M. Alphavirus-based expression vectors: Strategies and applications. Proc Natl Acad Sci USA. 1996;93:11371–11377. doi: 10.1073/pnas.93.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao S-J, Kingsley L, Hoover D R, Spira T J, Rinaldo C R, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore P S. Seroconversion to antibodies against Kaposi's sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi's sarcoma. N Engl J Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 14.Gao S-J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 15.Garoff H, Li K-J. Recent advances in gene expression using alphavirus vectors. Curr Opin Biotechnol. 1998;0:464–469. doi: 10.1016/s0958-1669(98)80030-x. [DOI] [PubMed] [Google Scholar]
- 16.He J, Bhat G, Kankasa C, Chintu C, Mitchell C, Duan W, Wood C. Seroprevalence of human herpesvirus 8 among Zambian women of childbearing age without Kaposi's sarcoma (KS) and mother-child pairs with KS. J Infect Dis. 1998;178:1787–1790. doi: 10.1086/314512. [DOI] [PubMed] [Google Scholar]
- 17.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kedes D H, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 19.Lang D, Hinderer W, Rothe M, Sonneborn H-H, Neipel F, Raab M, Rabenau H, Masquelier B, Fleury H. Comparison of the immunoglobulin-G-specific seroreactivity of different recombinant antigens of the human herpesvirus 8. Virology. 1999;260:47–54. doi: 10.1006/viro.1999.9804. [DOI] [PubMed] [Google Scholar]
- 20.Lennette E T, Blackbourn D J, Levy J A. Antibodies to human herpesvirus type 8 in the general population and in Kaposi's sarcoma patients. Lancet. 1996;348:858–861. doi: 10.1016/S0140-6736(96)03240-0. [DOI] [PubMed] [Google Scholar]
- 21.Li M, MacKey J, Czajak S C, Desrosiers R C, Lackner A A, Jung J U. Identification and characterization of Kaposi's sarcoma-associated herpesvirus K8.1 virion glycoprotein. J Virol. 1999;73:1341–1349. doi: 10.1128/jvi.73.2.1341-1349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin S F, Sun R, Heston L, Gradoville L, Shedd D, Haglund K, Rigsby M, Miller G. Identification, expression, and immunogenicity of Kaposi's sarcoma-associated herpesvirus-encoded small viral capsid antigen. J Virol. 1997;71:3069–3076. doi: 10.1128/jvi.71.4.3069-3076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin J N, Ganem D E, Osmond D H, Page-Shafer K A, Macrae D, Kedes D H. Sexual transmission and the natural history of human herpesvirus 8 infection. N Engl J Med. 1998;338:948–954. doi: 10.1056/NEJM199804023381403. [DOI] [PubMed] [Google Scholar]
- 24.Meng Y-X, Pellett P E. Herpesviruses 6, 7, and 8. In: Ahmed R, Chen I, editors. Persistent viral infections. Sussex, England: John Wiley & Sons, Ltd.; 1999. pp. 269–296. [Google Scholar]
- 25.Miller G, Rigsby M O, Heston L, Grogan E, Sun R, Metroka C, Levy J A, Gao S-J, Chang Y, Moore P S. Antibodies to butyrate-inducible antigens of Kaposi's sarcoma-associated herpesvirus in patients with HIV-1 infection. N Engl J Med. 1996;334:1292–1297. doi: 10.1056/NEJM199605163342003. [DOI] [PubMed] [Google Scholar]
- 26.Pau C-P, Lam L L, Spira T J, Black J B, Stewart J A, Pellett P E, Respess R A. Mapping and serodiagnostic application of a dominant epitope within the human herpesvirus 8 ORF65-encoded protein. J Clin Microbiol. 1998;36:1574–1577. doi: 10.1128/jcm.36.6.1574-1577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pear W S, Nolan G, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilot-Matias T J, Carrick R J, Coleman P F, Leary T P, Surowy T K, Simons J N, Muerhoff A S, Buijk S L, Chalmers M L, Dawson G J, Desai S M, Mushahwar I K. Expression of the GB virus C E2 glycoprotein using Semliki Forest virus vector system and its utility as a serologic marker. Virology. 1996;225:282–292. doi: 10.1006/viro.1996.0602. [DOI] [PubMed] [Google Scholar]
- 29.Raab M-S, Albrecht J-C, Birkmann A, Yaǧuboǧlu S, Lang D, Fleckenstein B, Neipel F. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J Virol. 1998;72:6725–6731. doi: 10.1128/jvi.72.8.6725-6731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabkin C S, Schulz T F, Whitby D, Lennette E T, Magpantay L I, Chatlynne L, Biggar R J. Interassay correlation of human herpesvirus 8 serologic tests. HHV-8 Interlaboratory Collaborative Group. J Infect Dis. 1998;178:304–309. doi: 10.1086/515649. [DOI] [PubMed] [Google Scholar]
- 31.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S-J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Laboratory Press; 1989. [Google Scholar]
- 33.Schulz T F. Epidemiology of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. Adv Cancer Res. 1999;76:121–160. doi: 10.1016/s0065-230x(08)60775-7. [DOI] [PubMed] [Google Scholar]
- 34.Simpson G R, Schulz T F, Whitby D, Cook P M, Boshoff C, Rainbow L, Howard M R, Gao S-J, Bohenzky R A, Simmonds P, Lee C, de Ruiter A, Hatzakis A, Tedder R S, Weller I V D, Weiss R A, Moore P S. Prevalence of Kaposi's sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet. 1996;348:1133–1138. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- 35.Smith M S, Bloomer C, Horvat R, Goldstein E, Casparian J M, Chandran B. Detection of human herpesvirus 8 DNA in Kaposi's sarcoma lesions and peripheral blood of human immunodeficiency virus-positive patients and correlation with serologic measurements. J Infect Dis. 1997;176:84–93. doi: 10.1086/514043. [DOI] [PubMed] [Google Scholar]
- 36.Spira T J, Jaffe H W. Human herpesvirus 8 and Kaposi's sarcoma. In: Scheld W M, Craig W A, Hughes J M, editors. Emerging infections 2. Washington, D.C.: ASM Press; 1998. pp. 81–104. [Google Scholar]
- 37.Tedeschi R, De Paoli P, Schulz T F, Dillner J. Human serum antibodies to a major defined epitope of human herpesvirus 8 small viral capsid antigen. J Infect Dis. 1999;179:1016–1020. doi: 10.1086/314657. [DOI] [PubMed] [Google Scholar]
- 38.Trobridge G D, Russell D W. Helper-free foamy virus vectors. Hum Gene Ther. 1998;9:2517–2525. doi: 10.1089/hum.1998.9.17-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu L, Wang R, Sweat A, Goldstein E, Horvat R, Chandran B. Comparison of human sera reactivities in immunoblots with recombinant human herpesvirus (HHV)-8 proteins associated with the latent (ORF73) and lytic (ORFs 65, K8.1A, and K8.1B) replicative cycles and in immunofluorescence assays with HHV-8-infected BCBL-1 cells. Virology. 1999;256:381–392. doi: 10.1006/viro.1999.9674. [DOI] [PubMed] [Google Scholar]