Abstract
Treatment of HIV-1-infected individuals with a combination of anti-retroviral agents results in sustained suppression of HIV-1 replication, as evidenced by a reduction in plasma viral RNA to levels below the limit of detection of available assays1,2. However, even in patients whose plasma viral RNA levels have been suppressed to below detectable levels for up to 30 months, replication-competent virus can routinely be recovered from patient peripheral blood mononuclear cells3,4 and from semen5. A reservoir of latently infected cells established early in infection6 may be involved in the maintenance of viral persistence despite highly active anti-retroviral therapy3-5. However, whether virus replication persists in such patients is unknown. HIV-1 cDNA episomes are labile products of virus infection and indicative of recent infection events. Using episome-specific PCR, we demonstrate here ongoing virus replication in a large percentage of infected individuals on highly active anti-retroviral therapy, despite sustained undetectable levels of plasma viral RNA. The presence of a reservoir of ‘covert’ virus replication in patients on highly active antiretroviral therapy has important implications for the clinical management of HIV-1-infected individuals and for the development of virus eradication strategies.
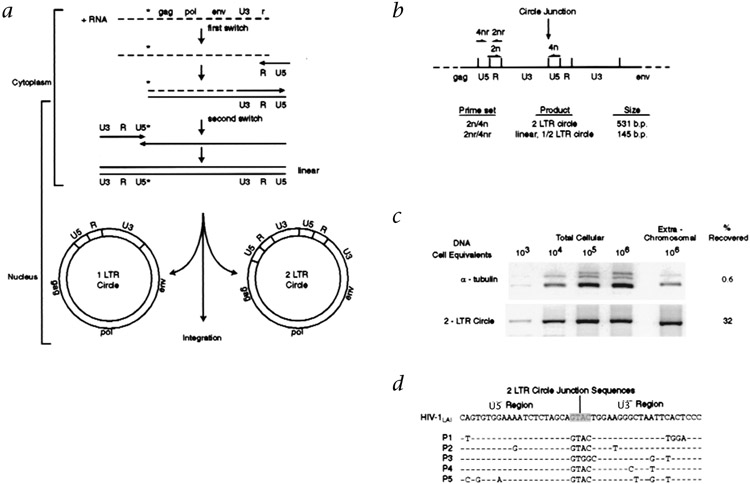
After HIV-1 infection of the cell, HIV-1 cDNA synthesis is initiated in the cytoplasm and proceeds at the same time as viral cDNA translocation to the host cell nucleus (Fig. 1a). Within the nucleus, some linear full-length cDNA genomes circularize to form episomes containing either one long-terminal repeat (1-LTR circle) or two long-terminal repeats (2-LTR circle) (Fig. 1a). As HIV-1 lacks necessary factors for the maintenance of episomal replication, these episomes may be labile and thus, indicative of recent infection events. To specifically detect 2-LTR circles, we used a PCR approach7 in which 5′ and 3′ primers direct the amplification of viral sequences across the junction that is formed after end-to-end 5′ and 3′ LTR ligation (Fig. 1b). To facilitate the detection of rare viral episomes in a background of a large proportion of uninfected peripheral blood mononuclear cells (PBMCs), we adapted a plasmid DNA isolation procedure that allowed for the selective isolation of viral episomes from genomic cellular DNA (Fig. 1c). Viral cDNA amplified from patient PBMC DNA by 2-LTR circle-specific primers represented genuine 2-LTR circle junctions (Fig. 1d).
Fig. 1.
Detection of 2-LTR replication intermediates of HIV-1. a, cDNA intermediates in HIV-1 reverse transcription. Dashed lines, viral RNA; solid lines, cDNA; *, primer binding site. Far left, subcellular location of the main stages in reverse transcription (the exact boundaries are not yet defined). After translocating to the nucleus, linear cDNA either undergoes integration to form the provirus or circularizes to form episomes containing 1-LTR or 2-LTRs. b, Strategy for 2-LTR-specific PCR amplification, with organization of the LTRs at the circle junction and binding sites for 2-LTR circle-specific primers. For amplification of all forms of viral cDNA, the 2-LTR circle-specific primers were reversed in orientation, such that amplification is internal of the LTR. c, Selective isolation of episomal forms of viral cDNA. Log dilutions of HIV-1-infected MT-4 cells (CD4+ T-cell line) were adjusted to 1 × 106 total cells by the addition of uninfected cells. Total cellular DNA and extrachromosomal DNA were isolated and amplified with α-tubulin-specific primers21 and with 2-LTR-specific primers. The recovery of cellular DNA and 2-LTR circles was 0.6 and 32%, respectively, indicating selective enrichment for 2-LTR circle forms of viral cDNA. d, 2-LTR circle junction sequences amplified from extrachromosomal DNA of five HIV-1-infected individuals (left margin, P1–P5) using 2-LTR circle-specific primers.
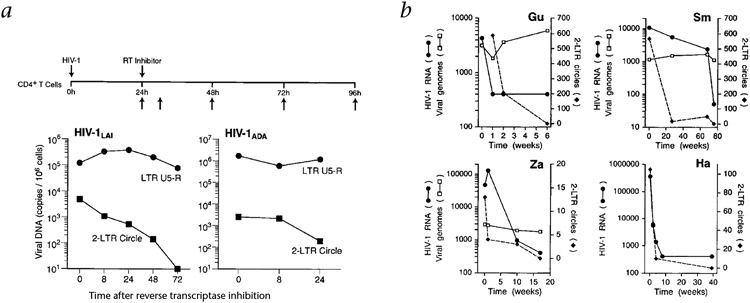
We initially assessed the stability of 2-LTR circle forms of viral DNA in acutely infected cells in vitro. We infected CD4+ MT-4 T cells and Jurkat-CCR5 cells with the X4 variant HIV-1LAI and the R5 variant HIV-1ADA, respectively. We allowed synthesis of viral cDNA to proceed for 24 hours and then restricted further rounds of virus infection and cDNA synthesis by the addition of reverse transcriptase inhibitors (Fig. 1). Within 24–48 hours after the addition of the reverse transcriptase inhibitors, 2-LTR circle numbers decreased by more than 90% in both HIV-1LAI-and HIV-1ADA-infected cells (Fig. 2a). The copy numbers of other viral DNA forms identified by the internal LTR primers (mostly linear and integrated viral genomes) remained relatively constant over the same interval (Fig. 2a). Thus, 2-LTR circles seem to be labile intermediates in the virus lifecycle.
Fig. 2.
Stability of 2-LTR circles in vitro and in vivo. a, Instability of 2-LTR circle forms of viral cDNA in vitro. CD4+ MT-4 T cells or CD4+ Jurkat-CCR5 T cells were infected with the X4 variant, HIV-1LAI or the R5 variant HIV-1ADA and viral infection and cDNA synthesis were allowed to proceed for 24 h, then further rounds of infection were then prevented by the addition of 5 μM ZDV or 1 μM Nevirapine, respectively; cells were then maintained in the presence of these reverse transcriptase (RT) inhibitors. Upward arrows, sampling intervals. Total cellular DNA was isolated (horizontal axis, time in hours) and amplified with 2-LTR circle-specific primers and with internal (LTR U5–R) primers (Fig. 1b). b, Instability of 2-LTR circle forms of viral cDNA in vivo. PBMCs were obtained from four HIV-1-infected individuals (patients Gu, Sm, Za and Ha) who, after adjustment of their anti-retroviral regimens to more potent combinations, showed rapid decreases in plasma viral RNA levels. The frequency of viral genomes was determined relative to the amount of genomic cellular DNA (measured directly by absorbance at 260 nm); 100 ng cellular DNA represents approximately 1 × 106 PBMCs.
We next evaluated whether 2-LTR circles were labile in vivo. We obtained PBMC samples from four HIV-1-infected individuals who, after adjustment of their anti-retroviral regimens to more potent combinations, showed steady decreases in plasma viral RNA levels. There were considerable decreases in 2-LTR circle copy numbers over the interval in which there was a rapid decrease in levels of plasma viral RNA (Fig. 2b). In contrast, when samples were analyzed in parallel with internal LTR primers (Fig. 1b), HIV-1 cDNA levels fluctuated by no more than 300% (Fig. 2b). These results indicate that 2-LTR circles are labile, both in vitro and in vivo, relative to integrated viral genomes.
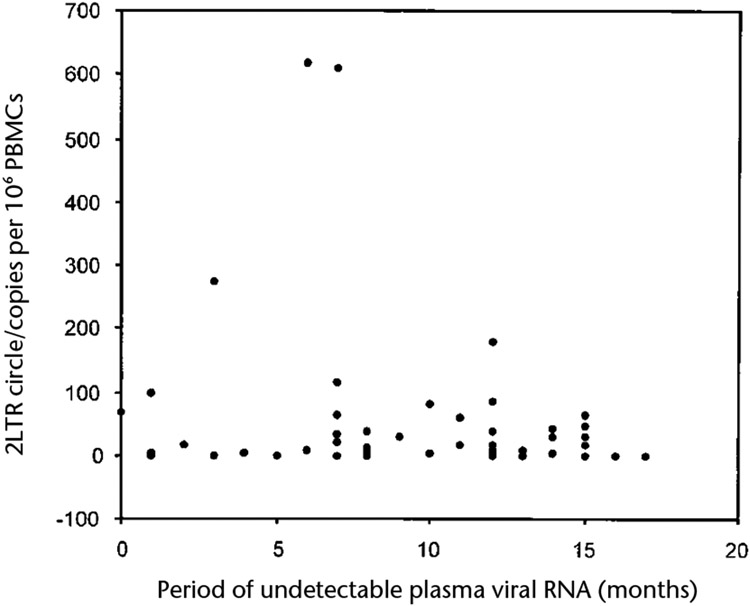
We next determined whether 2-LTR HIV-1 episomes were present in 63 patients who, throughout treatment with highly active anti-retroviral therapy (HAART), had undetectable levels of plasma viral RNA for sustained periods of time (Tables 1 and 2). Of these 63 patients, 50 (80%) had undetectable levels of plasma viral RNA (assay limit of sensitivity, 400 copies/ml) for 12 months or longer (Table 2). Of these 50 patients, 24 (48%) had undetectable levels of plasma viral RNA for 12 months or more using an assay with sensitivity as low as 50 copies/ml. In 48 of the 63 patients (76%), 2-LTR circles were detected in their PBMCs (Table 2). 2-LTR circle copy numbers ranged from less than 1 copy per 1 x 106 PBMCs to 620 copies per 1 × 106 PBMCs. There did not seem to be any substantial relationship between the frequency of 2-LTR circles in patient PBMCs and the time during which plasma viral RNA was undetectable (Fig. 3). These data indicate that labile replication intermediates are present in a substantial proportion of HIV-1 infected individuals who show sustained suppression of plasma viral RNA while on HAART. 2-LTR circles were not detectable in PBMCs from 15 (24%) patients (Table 2). However, this may not indicate an arrest of ongoing virus replication, because our assay does not account for extrachromosomal linear and 1-LTR circle forms of viral cDNA.
Table 1.
Characteristics of study subjects
| Patient number |
Drug regimen | CD4 T Cells (cells/mm3) |
Patient number |
Drug regimen | CD4 T cells (cells/mm3) |
|---|---|---|---|---|---|
| W1 | RTV,ZDV,3TC | 475 | L2 | 3TC,D4T,RTV | 852 |
| W2 | NFV,ZDV,3TC | 827 | L3 | ZDV,3TC,IDV | 448 |
| W3 | IDV,D4T,3TC | 436 | L4 | ZDV,3TC,RTV | 978 |
| W4 | IDV,D4T,3TC | 505 | L6 | D4T,RTV,SQV | 577 |
| W5 | IDV,D4T,3TC | 248 | L7 | D4T,ddI,NVP | 394 |
| W7 | SQV,D4T,3TC | 443 | L8 | ZDV,3TC,NFV | 173 |
| W8 | ddI,D4T | 870 | L9 | 3TC,D4T,EFV | 482 |
| W9 | NFV,D4T,3TC | 641 | L11 | ZDV,3TC,RTV | 615 |
| W10 | IDV,ZDV,3TC | 656 | L12 | 3TC,D4T,RTV | 389 |
| W11 | IDV,ZDV,3TC | 344 | L13 | D4T,SQV,NFV | 312 |
| W12 | ZDV,3TC,DLV | 626 | L14 | 3TC,D4T,IDV | 375 |
| W13 | NFV,ZDV,3TC | 699 | L15 | 3TC,RTV, SQV,ABV | 91 |
| W14 | NFV,D4T,3TC | 685 | L16 | 3TC,D4T,SQV,RTV | 575 |
| W15 | NFV,3TC,NVP | 866 | L17 | 3TC,D4T,SQV | 196 |
| W16 | RTV,D4T,3TC | 572 | L18 | ZDV,3TC,IDV | 175 |
| W17 | IDV,ZDV,3TC | 364 | L19 | 3TC,D4T,RTV,SQV | 499 |
| W18 | IDV,ZDV,3TC | 119 | L22 | ZDV,D4T,IDV | 223 |
| W19 | SQV,ZDV,3TC | 153 | L23 | 3TC,ddC,IDV | 534 |
| W20 | IDV,ZDV,3TC | 360 | L26 | 3TC,D4T,SQV,NFV | 911 |
| W21 | NFV,D4T,3TC | 208 | L27 | ZDV,3TC,IDV | 185 |
| W22 | D4T,3TC | 495 | L28 | D4T,ABV,EFV | 80 |
| W28 | NFV,ddI,D4T | 527 | L29 | ZDV,ddC,SQV,NFV | 121 |
| W30 | D4T,3TC | 575 | L32 | 3TC,D4T,EFV | 219 |
| M1 | NFV,D4T,NVP | 287 | L33 | 3TC, D4T,IDV | 610 |
| M3 | IDV,ddI,NVP | 440 | L36 | ddI, D4T, NFV | 172 |
| M4 | IDV,ZDV,3TC | 586 | L37 | ZDV,ddC,3TC,IDV | 279 |
| M6 | NFV,ZDV,3TC | 317 | L41 | ZDV,3TC,RTV | 390 |
| M7 | NFV,3TC,NVP | 175 | L42 | 3TC,D4T,SQV | 117 |
| M8 | IDV,ZDV,3TC,NVP | 357 | L46 | 3TC,D4T,NFV | 180 |
| M12 | NFV,D4T,3TC | 749 | |||
| M13 | ZDV,3TC,EFV | 670 | |||
| M14 | IDV,ZDV,3TC | 728 | |||
| M15 | IDV,ZDV,3TC | 565 | |||
| M16 | NFV,3TC,NVP | 403 |
CD4+ T-cell counts were determined at the time of 2-LTR circle measurement (Table 2). ZDV, zidovudine; 3TC, lamivudine; D4T, Stavudine; ddI, Didanosine; NVP, Nevirapine; RTV, Ritonavir; EFV, Efavirenz; SQV, Saquinavir; IDV, Indinavir, NFV, Nelfinavir; ddC, Zalcitabine; ABV, Abacavir.
Table 2.
Analysis of 2-LTR circles in 63 aviremic patients
| Patient number |
Period of undetectable viral RNAa (months) |
2-LTR Circles (copies/106 PBMC) |
PBMCs analyzedb (millions) |
Patient number |
Period of undetectable viral RNAa (months) |
2-LTR Circles (copies/106 PBMC) |
PBMCs analyzedb (millions) |
||
|---|---|---|---|---|---|---|---|---|---|
| W1 | 23 | (14) | 3 | L2 | 8 | (8) | 5 | ||
| W2 | 13 | (13) | <1 | 5.5 | L3 | 12 | (8) | 10 | |
| W3 | 23 | (14) | 27 | L4 | 21 | (12) | 180 | ||
| W4 | 23 | (12) | 37 | L6 | 10 | (7) | 0 | 4.0 | |
| W5 | 19 | (11) | 15 | L7 | 11 | (7) | 610 | ||
| W7 | 19 | (13) | 8 | L8 | 17 | (8) | 0 | ||
| W8 | 18 | (15) | <1 | 4.0 | L9 | 8 | (5) | <1 | 2.2 |
| W9 | 22 | (11) | 59 | L11 | 19 | (12) | 84 | ||
| W10 | 22 | (15) | <1 | 4.0 | L12 | 19 | (6) | 7 | |
| W11 | 22 | (15) | 65 | L13 | 15 | (3) | <1 | 7.8 | |
| W12 | 26 | (16) | 0 | 5.5 | L14 | 14 | (7) | 116 | |
| W13 | 13 | (13) | 0 | 5.5 | L15 | 30 | (17) | 0 | 1.5 |
| W14 | 21 | (15) | 47 | L16 | 12 | (12) | 4 | 8.1 | |
| W15 | 25 | (12) | 17 | L17 | 15 | (15) | 14 | ||
| W16 | 22 | (14) | 2 | 5.5 | L18 | 16 | (13) | 0 | 10.2 |
| W17 | 26 | (15) | 31 | L19 | 15 | (6) | 620 | ||
| W18 | 21 | (16) | 0 | 2.0 | L22 | 14 | (12) | 6 | |
| W19 | 16 | (10) | 4 | 4.0 | L23 | 14 | (12) | 0 | 4.8 |
| W20 | 27 | (15) | 0 | 4.0 | L26 | 17 | (8) | 36 | |
| W21 | 13 | (13) | 0 | 2.0 | L27 | 17 | (17) | 0 | 3.2 |
| W22 | 23 | (15) | 0 | 4.0 | L28 | 8 | (3) | 275 | |
| W28 | 22 | (8) | 9 | L29 | 21 | (1) | 3 | 2.0 | |
| W30 | 22 | (17) | 0 | 4.0 | L32 | 7 | (1) | <1 | 10.0 |
| M1 | 14 | (9) | 31 | L33 | 16 | (1) | <1 | 14.4 | |
| M3 | 16 | (7) | 22 | L36 | 14 | (4) | 2 | 5.6 | |
| M4 | 13 | (ND) | 264 | L37 | 13 | (7) | 0 | 5.6 | |
| M6 | 24 | (7) | 63 | L41 | 22 | (1) | 100 | ||
| M7 | 11 | (ND) | 4 | 5.5 | L42 | 18 | (1) | 0 | 2.0 |
| M8 | 13 | (2) | 15 | L46 | 7 | (1) | 4 | 20.0 | |
| M12 | 12 | (7) | 35 | ||||||
| M13 | 10 | (0) | 67 | ||||||
| M14 | 14 | (14) | 41 | ||||||
| M15 | 10 | (10) | 82 | ||||||
| M16 | 12 | (8) | 3 | 4.0 | |||||
In all cases, maintenance of suppression of virus replication was confirmed by measurement of plasma viral RNA loads every 90–120 d.
Period for which plasma viral RNA was below the level of detection using an assay with a sensitivity as low as 400 copies/ml. Numbers in parenthesis, period for which viral RNA was below the level of detection using an assay with a sensitivity as low as 50 copies/ml. 2-LTR circle copy numbers in most cases were determined in duplicate on independent PBMC samples. Values less than 1 indicate that more than 1 × 106 PBMCs were required for detection of 2-LTR circles.
Total number of PBMCs from which extrachromosomal DNA was isolated and analyzed for the presence of 2-LTR circles. Except where indicated otherwise, quantitation was done on extrachromosomal DNA from 1 × 106 PBMCs. In all cases, 2-LTR circles were quantitated by fluorescence-based PCR. Similar 2-LTR circle numbers were obtained when samples were quantified by comparison of PCR. band intensity to a standard dilution series of a 2-LTR circle containing plasmid.
Fig. 3.
Relationship between 2-LTR circle frequency and interval during which plasma viral RNA was undetectable in plasma (fewer than 50 copies/ml). Data represent all 63 patients. Statistical analysis (rs = −0.143; P = 0.26; Spearman’s rank correlation) indicates that within the time frame of the analysis, there was no correlation between 2-LTR circle frequency and duration of undetectable plasma viral RNA.
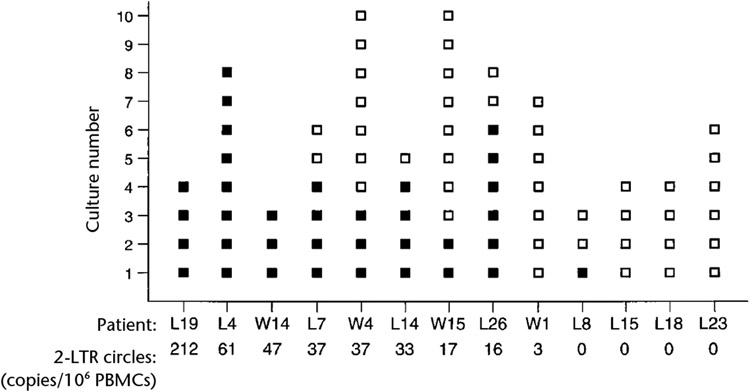
To determine whether patients with 2-LTR circles also had cells with replication-competent virus, we did high-input viral co-culture assays on PBMCs from nine patients with 2-LTR circles and four patients without 2-LTR circles (Fig. 4). Replication-competent virus could readily be isolated from eight of the nine patients with 2-LTR circles. Virus could not be isolated from patient W1, who had a very low circle copy number. Infectious virus could not be isolated from three patients who lacked 2-LTR circles, even though co-culture was done on 4 × 107–6 × 107 CD8-depleted patient PBMCs. For patient L8, who also lacked 2-LTR circles, only one of three cultures yielded infectious virus (Fig. 4). These results indicate a correlation between the presence of 2-LTR circles and cells with replication-competent virus. Plasma-based viral RNA assays therefore fail to demonstrate the full extent of viral activity in infected individuals who are being treated with HAART.
Fig. 4.
High-input virus co-culture assays for replication-competent HIV-1. Virus co-culture results for replicate cultures (each containing 1 × 107 CD8-depleted PBMCs): ■, positive; □, negative. Treatment regimens, CD4+ T-cell status and time of undetectable plasma viral RNA, Table 1. There is significant correlation between the frequency of positive virus co-cultures and levels of 2-LTR circles (rs = 0.892, Spearman’s rank correlation; P < 0.01; P < 0.02, paired t-test).
One important variable with a big effect on the decay characteristics of viral compartments in patients on HAART is the possible contribution of a reservoir of ongoing virus replication. Such a reservoir would be predicted to provide a continuous source of infectious virus, which could sustain the pool of productively and latently infected cells. We have used a new approach to demonstrate ongoing virus replication in a substantial percentage of patients who have sustained undetectable levels of plasma viral RNA. Our observations support recent studies that presented evidence for ongoing virus replication in aviremic patients on the basis of viral sequence evolution8,9 and the presence of HIV-1-specific transcripts10.
Several characteristics of the reservoir in which virus replication continues in patients on HAART can be inferred from our analyses. 2-LTR circles are formed only after completion of viral cDNA synthesis and translocation of the viral cDNA to the host cell nucleus11,12, and both processes occur only after infection of cycling but not resting T cells13,14 or peripheral blood monocytes15. These extrachromosomal episomes in the host cell nucleus are distinct from the extrachromosomal cytoplasmic cDNA intermediates that result from the restricted infection of resting CD4+ T cells12-14. CD8+ T cells contain proviral DNA in AIDS patients16. We have made a preliminary analysis of purified cells in two patients with readily detectable 2-LTR circles (patients L7 and L19). 2-LTR circles were detected only in purified CD4+ lymphocytes and not in CD4− cells. Furthermore, 2-LTR circles were detected in both CD4+/RO+ and CD4+/RO− subpopulations (data not shown), which is in agreement with studies indicating that both naive and memory T cells are susceptible to HIV-1 infection in vitro17-19. One question that emerges from our analysis is the nature of the reservoir in which virus replication is ongoing. Although 2-LTR circles were detected in patient PBMCs, it is unlikely that this compartment was the source of the infectious virus. Rather, CD4+ T cells may have been infected during their passage through extravascular tissues. At present, we can only speculate on the tissue source of infectious virus. For example, ‘sanctuary sites’ may exist in some or all tissues where virus replication can occur in an environment that is relatively inaccessible to anti-retroviral agents.
Our study has implications for the development of strategies to eradicate virus replication in HIV-1-infected individuals. Although complete elimination of HIV-1 replication may be difficult with current anti-retroviral regimens, our study, along with other published observations9,20, indicates that there do seem to be examples in which even the most sensitive assays fail to demonstrate ongoing replication in some well-suppressed patients9,20. It is also likely that, as more potent anti-retroviral therapeutics enter the clinic, ongoing or ‘covert’ virus replication may be arrested in more patients. A better understanding of the nature of the reservoir that sustains virus replication in aviremic patients on HAART may lead to the development of more effective strategies for the arrest of virus replication.
Methods
Study Populations.
Patient samples were obtained from three centers: the Division of Infectious Diseases and Immunology at UMass/Memorial Health Care (Worcester, Massachusetts), the Chelsea and Westminster Hospital (London, UK) and the UMass/Memorial pediatric HIV program (Worcester, Massachusetts). All participants gave informed consent for these studies. For evaluation of 2-LTR circle stability in vivo, PBMCs were obtained from four HIV-1-infected individuals (patients Gu, Sm, Za and Ha) who showed steady decreases in plasma viral RNA after adjustment of anti-retroviral regimens to more potent combinations. Patient Gu, who had been maintained on a two-drug reverse transcriptase inhibitor combination, was subsequently changed (week 0) to a three-drug regimen (ZDV/3TC/NFV). Patient Sm, who had been on a two-drug regimen (ZDV/3TC) was changed at week 68 to ddI/EFV/NFV. Patient Za, who had been on the four-drug regimen 3TC/D4T/ddI/NFV was adjusted (week 1) to ZDV/ddC/NFV/RTV. Patient Ha, previously on the three-drug regimen ZDV/ddI/NVP was subsequently adjusted (week 0) to D4T/3TC/NVP. Drug name abbreviations: ZDV, zidovudine; 3TC, lamivudine; D4T, stavudine; ddI, didanosine; NVP, nevirapine; RTV, ritonavir; EFV, efavirenz; NFV, nelfinavir; ddC, zalcitabine.
Statistical Methods.
The relationship between 2-LTR circle frequency and either the duration of undetectable plasma viral RNA or the frequency of positive virus co-cultures was examined using Spearman’s correlation coefficient. The mean frequency of positive co-cultures in individuals with and without 2-LTR circles (Fig. 4) was further compared with a paired t-test.
Nucleic acid purification.
Ficoll-purified PBMCs (2 × 106–40 × 106) were collected by centrifugation at 1,300g for 2 min. Cell pellets were resuspended in buffer P1 and extrachromosomal DNA was purified by a QIAprep™ spin miniprep kit (Qiagen, Valencia, California) using the modification for the isolation of low-copy-number plasmids as recommended by the manufacturer. Chromosomal DNA was recovered from the sodium acetate–SDS precipitate using DNAzol™ reagent (Life Technologies) according to the manufacturer’s protocol. Total cellular DNA was purified using an Isoquick™ nucleic acid extraction kit (ORCA Research, Bothell, Washington).
Characterization of 2-LTR circle junctions.
2-LTR circle junctions were amplified from 10–30 μl of extrachromosomal DNA in a 50-μl reaction containing 1x HotStarTaq™ buffer, 200 nM nNTPs, 400 nM primers and 1.5 U HotStarTaq™ (Qiagen, Valencia, California). The reverse primer (2n) was 5′–CAGATCTGGTCTAACCAGAGA–3′ and the forward primer (4n) was 5′–GTAACTAGAGATCCCTCAGAC–3′, which annealed to nucleotides 9,157–9,137 (HIV-1 LTR R region) and nucleotides 130–150 (HIV-1 LTR U5 region) of HIV-1LAI, respectively (GenBank accession number K02013). After an initial denaturation step (95 °C for 10 min), PCR amplification proceeded for 45 cycles (95 °C for 30 s; 60 °C for 30 s; 72 °C for 60 s) followed by a final extension (72 °C for 5 min). To control for the effect of sequence polymorphisms at primer binding sites, amplification was done with internal primers (2nr and 4nr) that were reversed in orientation to those described above. Amplification with the internal LTR primers proceeded for 35 cycles using conditions described above. Polymorphisms in the region of the LTR that is recognized by the fluorogenic probe can affect annealing of the probe and potentially result in ‘false negatives’. To accommodate this, Taqman reaction products were subsequently analyzed on agarose–TBE gels and stained with ethidium bromide to ensure that those reactions did not contain episome-specific PCR products. For quantification of 2-LTR circle frequency in patient PBMCs, PCR reactions were done using an ABI prism 7700 sequence detection system with the addition of 200 nM fluorogenic probe to the reaction (5′–AGTGGCGAGCCCTCAGATGCTGC–3′), which anneals to nucleotides 9,081–9,103 of HIV-1LAI. The oligonucleotide probe was modified with 6-FAM (6-carboxyfluorescein) reporter dye on the 5′ end and 6-TAMRA (6-carboxytetramethylrhodamine) quencher dye on the 3′ end. Copy number estimates of 2-LTR circles were determined either by extrapolation from a plot of standards versus band intensity or by using the ABI prism 7700 quantification software. For sequencing, 2-LTR circle junctions were cloned into a TA cloning vector (Invitrogen, San Diego, California) and analyzed on an ABI 377 DNA sequencer according to the manufacturer’s protocol.
Virus co-culture assays.
Patient PBMCs were separated by Ficoll–Paque (Amersham–Pharmacia) and depleted of CD8+ T lymphocytes using antibody-coated beads (Dynal, Oslo, Norway). Cells were seeded in flasks in aliquots of 1 × 107 cells in RPMI 1640 medium supplemented with 10% fetal calf serum and were activated by addition of 5 μg/ml phytohemagglutinin for 12 h. CD8+-depleted PBMCs from HIV-1-seronegative individuals were activated for 12 h with phytohemagglutinin and added to flasks of patient PBMCs, in equal number, together with 20 IU/ml of interleukin 2 (Genzyme, Cambridge, Massachusetts). At weekly intervals, half of the culture supernatant was replaced with fresh medium containing 20 IU/ml IL-2 and 1 × 107 freshly isolated, CD8-depleted and phytohemagglutinin-activated donor PBMCs from HIV-1-seronegative individuals. HIV-1 Gag p24 antigen in culture supernatants was evaluated by enzyme-linked immunosorbent assay after 4 weeks (Beckman Coulter, Fullerton, California).
Acknowledgments
We thank P. Himlan, L. Mangini, C. Jaffarian, B. Cullen and L. O’Reilly for patient recruitment and scheduling; M. McManus for data management; C. Waterworth and G. Sontag of Imperial College and research associates in the laboratory of pediatric immunology at UMass Medical School for technical assistance; N. Bakker for manuscript preparation; and B. Mellor for assistance in preparation of the figures. Jurkat-CCR5 cells were obtained from M. Emerman. This work was supported in part by National Institutes of Health grants RR11589, HL57880 (M.S.) AI 32391 (K.L.), and AI32907 (J.L.S.). M. Sharkey is the recipient of a National Institutes of Health training grant (T32 AI07272). K. Luzuriaga is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation.
References
- 1.Perelson AS, Neumann AU, Markowtiz M, Leonard JM & Ho DD HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271, 1582–1586 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Gulick RM et al. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med 337, 734–739 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Wong JK et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1227 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Finzi D et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Zhang H et al. Human immunodeficiency virus type 1 in the semen of men receiving highly active antiretroviral therapy. N. Engl. J. Med 339, 1803–1809 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Chun TW et al. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 95, 8869–8873 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevenson M et al. Integration is not necessary for expression of human immunodeficiency virus type 1 protein products. J. Virol 64, 2421–2425 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L et al. Rapid clearance of simian immunodeficiency virus particles from plasma of rhesus macaques. J. Virol 73, 855–860 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L et al. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med 340, 1605–1613 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Furtado MR et al. Persistence of HIV-1 transcription in peripheral blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med 340, 1614–1622 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Brown PO, Bowerman B, Varmus HE & Bishop JM Correct integration of retroviral DNA in vitro. Cell 49, 347–356 (1987). [DOI] [PubMed] [Google Scholar]
- 12.Bukrinsky MI, Stanwick TL, Dempsey MP & Stevenson M Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254, 423–427 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zack JA et al. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61, 213–222 (1990). [DOI] [PubMed] [Google Scholar]
- 14.Stevenson M, Stanwick TL, Dempsey MP & Lamonica CA HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 9, 1551–1560 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonza S et al. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J. Virol 70, 3863–3869 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livingstone WJ, Moore M, Innes D, Bell JE & Simmonds P Frequent infection of peripheral blood CD8-positive T-lymphocytes with HIV-1. Edinburgh Heterosexual Transmission Study Group. Lancet 348, 649–654 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Chun TW et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387, 183–188 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Spina CA, Prince HE & Richman DD Preferential replication of HIV-1 in the CD45RO memory cell subset of primary CD4 lymphocytes in vitro. J. Clin. Invest 99, 1774–1785 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riley JL et al. Naive and memory CD4 T cells differ in their susceptibilities to human immunodeficiency virus type 1 infection following CD28 costimulation: implications for transmission and pathogenesis. J. Virol 72, 8273–8280 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finzi D et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nature Med. 5, 512–517 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Cowan NJ, Dobner PR, Fuchs EV & Cleveland DW Expression of human alpha-tubulin genes: interspecies conservation of 3′ untranslated regions. Mol. Cell. Biol 3, 1738–1745 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]