Abstract
Respiratory infections remain a major global health concern. Tuberculosis is one of the top 10 causes of death worldwide, while infections with Non‐Tuberculous Mycobacteria are rising globally. Recent advances in human tissue modeling offer a unique opportunity to grow different human “organs” in vitro, including the human airway, that faithfully recapitulates lung architecture and function. Here, we have explored the potential of human airway organoids (AOs) as a novel system in which to assess the very early steps of mycobacterial infection. We reveal that Mycobacterium tuberculosis (Mtb) and Mycobacterium abscessus (Mabs) mainly reside as extracellular bacteria and infect epithelial cells with very low efficiency. While the AO microenvironment was able to control, but not eliminate Mtb, Mabs thrives. We demonstrate that AOs responded to infection by modulating cytokine, antimicrobial peptide, and mucin gene expression. Given the importance of myeloid cells in mycobacterial infection, we co‐cultured infected AOs with human monocyte‐derived macrophages and found that these cells interact with the organoid epithelium. We conclude that adult stem cell (ASC)‐derived AOs can be used to decipher very early events of mycobacteria infection in human settings thus offering new avenues for fundamental and therapeutic research.
Keywords: airways, infection, mycobacteria, organoids, tuberculosis
Flowchart of host–pathogen interactions in microinjected human bronchiolar airway organoids. Organoids were injected using a micromanipulator coupled to a microinjection system (left). The capillary was pushed into the lumen of airway organoids (AOs) to deliver mycobacteria (middle). The readouts used to study host–pathogen interactions within the organoids are summarized (right). Created with Biorender.com.
![]()
1. INTRODUCTION
Pulmonary diseases due to mycobacteria cause significant morbidity and mortality to human health. Although the global burden of tuberculosis (TB) has declined over the last decades, Mycobacterium tuberculosis is still one of the deadliest infectious agent worldwide with an estimated 10 million disease cases and 1.4 million deaths in 2019 (World Health Organization, 2020). By contrast, Nontuberculous Mycobacteria (NTM) infections are rising globally, especially in developed countries, causing opportunistic pulmonary infections affecting individuals who are immunocompromised and who have underlying health conditions (Ratnatunga et al., 2020; To et al., 2020). Among them, Mycobacterium abscessus is considered one of the most antibiotic‐resistant mycobacteria and is associated with pulmonary disease such as cystic fibrosis (Johansen et al., 2020).
The lung is the main entry port for mycobacteria and the main site of disease. Bacteria‐containing droplets or aerosols first navigate through the respiratory tract interacting with airway epithelium functions in order for mycobacteria to reach the alveolar space and thrive in alveolar macrophages (Corleis & Dorhoi, 2020; Leiva‐Juárez et al., 2018; Torrelles & Schlesinger, 2017). However, how mycobacteria interact with the airway to establish a successful primary infection remains poorly understood. One of the challenges to address remains the complexity of host–pathogen interactions, particularly within human multicellular tissue.
Conventional 2D cell cultures of epithelial and immune cells, in vitro human‐based 3D granuloma, and animal models have contributed to deciphering key host–pathogen mechanisms at play during mycobacterial infection (Bermudez et al., 2002; Bernut et al., 2014, 2015; Bielecka et al., 2017; Cohen et al., 2018; Elkington et al., 2019; Fonseca et al., 2017; Parasa et al., 2014; Puissegur et al., 2007; Rampacci et al., 2020; Silva‐Miranda et al., 2015). More recently, in vitro lung models have been developed based on organ‐on‐chip and organoid technologies to mimic the structural and biological properties of the in vivo human lung environment to better replicate human disease and drug profiles (Cidem et al., 2020; Dutta & Clevers, 2017; Rossi et al., 2018). Lung‐on‐chip technology reconstructs the alveolar epithelium/endothelium interface and has deciphered the crucial role of surfactant in controlling Mtb replication (Thacker et al., 2020). We, and others, have applied organoid technology for respiratory infection modeling, including Cryptosporidium (Heo et al., 2018), RSV (Sachs et al., 2019), influenza viruses (Hui et al., 2018; Zhou et al., 2018), SARS‐CoV‐2 (Lamers et al., 2020, 2021), and Pseudomonas aeruginosa (Bagayoko et al., 2021) to decipher pathogen fitness, cell tropism, and host responses.
While the ongoing COVID‐19 pandemic has very recently fueled the development of new human lung organoid models (Salahudeen et al., 2020; Youk et al., 2020), the first human lung organoid model described was derived from lung adult stem cells (ASC’s) and composed of a polarized, pseudostratified airway epithelium containing basal, secretory, and multi‐ciliated cells, thus reproducing the bronchiolar part of the airway (Sachs et al., 2019). AOs display functional mucus secretion and ciliate beating (Sachs et al., 2019), therefore, constituting a human 3D system in which to model host–mycobacteria interactions (Iakobachvili & Peters, 2017).
Here, we have set out to use AOs as a model in which to study mycobacteria interaction with airway epithelial cells. Our data demonstrate that mycobacteria can be readily found in the lumen of AOs with some internalization by airway epithelial cells. While Mtb growth was overall controlled, Mabs readily replicated in the lumen of AOs. In response to Mtb infection, we showed that AOs induced the secretion of cytokines and antimicrobial peptides. Interestingly, both Mabs and Mtb inhibited the expression of mucins which function in pathogen clearance. The option to model innate cell recruitment by co‐culturing human macrophages with bacteria‐injected AOs was demonstrated in this work.
2. RESULTS AND DISCUSSION
First of all, we evaluated the morphology and cell composition of AOs. As shown in Figure S1, AOs displayed a cystic structure (Figure S1a) and are composed of basal cells lined with ciliated and goblet cells on the lumen facing side (Figure S1b and Movie S1). AOs presented functional ciliate beating (Movies S2 and S3) and evident from the live‐cell imaging is the functional mucociliary system where cilia beat secreted mucus and cell debris around the lumen (Movie S4).
Due to the innate cystic structure of AOs, where the pathogen‐sensing apical side faces the lumen, DsRed‐expressing H37Rv Mtb or dTomato‐expressing Mabs were microinjected via a BSL‐3‐ or a BSL‐2‐approved custom‐made micro‐injection system (Figure 1a). Mycobacteria could be found in the lumen of AOs and occasionally making contact with epithelial cells but without causing obvious alterations to organoid architecture and ultrastructure (Figures 1b,c, S2 and Movie [Link], [Link], [Link]). We evaluated whether mycobacteria infection could trigger cell death and found neither Mtb nor Mabs infection triggered additional cell death compared to uninfected AOs, which undergo regular cell turnover (Figure 1d).
FIGURE 1.
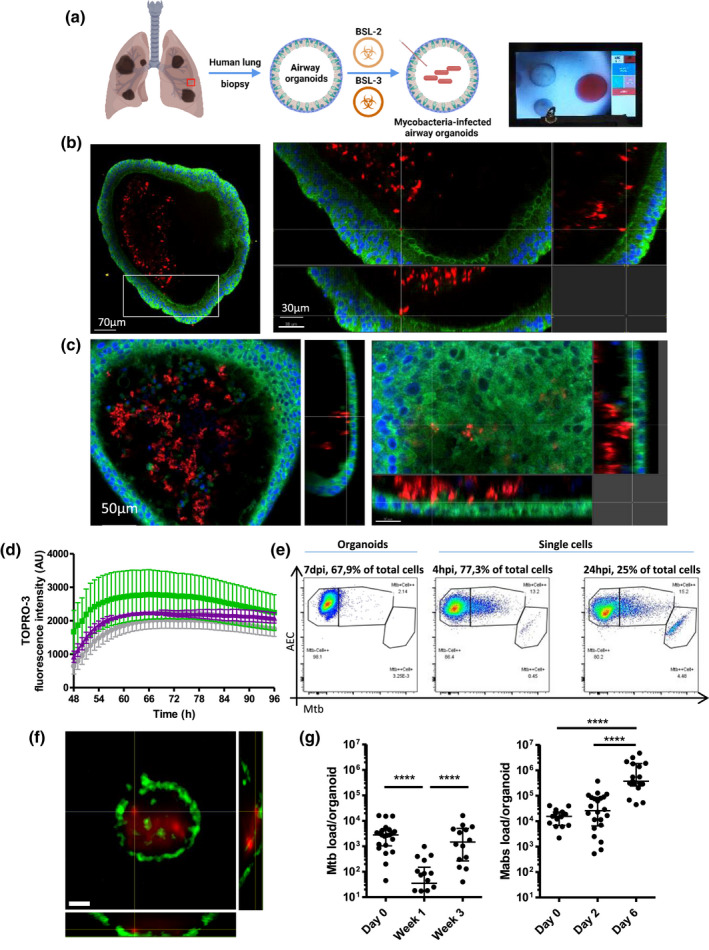
Human airway organoids (AOs) infected with Mtb and Mabs. (a) Experimental scheme and bright‐field image of the microinjection procedure. Created with Biorender.com. (b) Confocal microscopy of DsRed‐expressing H37Rv inside AOs, 4 days post‐infection. Nuclei are labeled with DAPI (blue); cellular membranes with CellMask green (green). (c) Confocal microscopy of dTomato‐expressing Mabs inside AOs, 4 days post‐infection. Nuclei are labeled with DAPI (blue); cellular membranes with CellMask green (green). (d) AOs were injected with PBS (grey), H37Rv Mtb (green), or Mycobacterium abscessus (purple), stained with ToPRO3 and imaged hourly for four consecutive days after embedding in matrigel. Fluorescence intensity from ToPRO3 incorporation, and therefore epithelial cell death, was quantified using Fiji and plotted for each condition. The experiment was performed three times independently. (e) Representative flow cytometry dot plots of cells associated with H37Rv after 4 (left) or 24 hr (middle) incubation with AOs‐derived single cells or 7 days incubation in whole organoids (right). (f) Representative image of a sorted epithelial cell with intracellular DsRed‐expressing Mtb, scale bars = 5 µm. (g) Colony‐forming unit (CFU) counts from individual organoids on the day of microinjection (day 0), 7 and 21 days post‐infection. Each dot represents one organoid. Lines indicate median CFU counts. The experiment was performed at least three times independently. *** p < .001 by a two‐tailed Mann‐Whitney test
Mtb is known to infect bronchial epithelial cells in 2D conditions (Reuschl et al., 2017), and pneumocytes in vitro (Ryndak et al., 2015) and in vivo (Cohen et al., 2018), but with low efficiency. To determine whether Mtb could infect airway epithelial cells, Mtb‐infected AOs were dissociated and analyzed by flow cytometry to quantify cells positive for Mtb in an unbiased manner. Only 2% of the cells composing the organoid were positive for Mtb (Figure 1e). As mucociliary movement could minimize bacteria interaction with epithelial cells, to favor optimal contact between epithelial cells and bacteria, AOs were dissociated, and single cells were infected with Mtb and analyzed by flow cytometry. Approximately 13% and 15% of epithelial cells were found associated with bacteria after 4 and 24 hr of infection, respectively (Figure 1e), confirming that Mtb has a low tropism for epithelial cells as already described with primary bronchial cells (Reuschl et al., 2017). To evaluate bacterial internalization by epithelial cells, double positive cells sorted after flow cytometry were imaged by confocal microscopy. As shown in Figure 1f, epithelial cells containing Mtb could be found, suggesting cell invasion by a yet unknown mechanism and confirming that the majority of mycobacteria are extracellular in the lumen of AOs. The functioning mucociliary system within AOs is likely responsible for preventing mycobacterial contact with, and internalization by, epithelial cells (Leiva‐Juárez et al., 2018; Whitsett & Alenghat, 2015).
We next investigated mycobacterial survival in AOs. Bacteria‐infected AOs were collected individually, lysed and analyzed for bacterial load by CFU assay. Mtb demonstrated a bi‐phasic curve, with a significant decrease of bacterial load after 7 days followed by an increase at 21 days post‐infection back to the initial load, without affecting AO viability (Figure 1d). This suggests an early stage of Mtb control by the AO microenvironment followed by bacterial adaptation and proliferation, with cell debris present in the lumen which could foster bacterial replication, as already shown in macrophages (Mahamed et al., 2017). By contrast, Mabs growth displayed a 2‐day lag phase followed by significant bacterial replication and an increase in bacterial burden over 6 days (Figure 1g). These results also confirm that the airway constitutes a hospitable microenvironment for Mabs to thrive, compared to Mtb that establishes its replicative niche in alveolar macrophages (Corleis & Dorhoi, 2020; Johansen et al., 2020).
Next, we determined whether AOs mounted an inflammatory/antimicrobial response to mycobacteria infection. Mtb‐ or Mabs‐infected AOs were collected and subjected to RT‐qPCR analysis to evaluate cytokine, antimicrobial peptide, and mucin gene expression (Figure 2a). Mtb induced significant gene expression of IL‐8 (Figure 2b), a immune cell chemoattractant (Baggiolini & Clark‐Lewis, 1992), while IL‐8 secretion did not reach significance (Figure 2c). Non‐significant expression of IP‐10, GM‐CSF, and CCL5, cytokines mediating the accumulation of neutrophils and macrophages for optimal granuloma formation and control of Mtb (Domingo‐Gonzalez et al., 2016) was observed. The antimicrobial peptides β‐defensin‐1 and 2, lactoferrin, hepcidin, and RNAse7 were enhanced upon Mtb infection, being significant for only β‐defensin‐1 (Figure 2b). β‐Defensin‐1 and RNAse7 may participate in Mtb restriction during early infection, promoting pore formation on the bacteria cell membrane (Arranz‐Trullén et al., 2017; Torres‐Juarez et al., 2018), while β‐defensin‐2 improves the chemoattraction of macrophages (Rivas‐Santiago et al., 2005; Semple & Dorin, 2012). Interestingly, β‐defensin‐3 and 4, which are related to long‐term control of mycobacterial infection (Dong et al., 2016) were downregulated. The upregulation of Hepcidin and Lactoferrin could contribute to decrease the transmembrane transport of iron (Sow et al., 2007, 2009) and modulate its extracellular pool by hijacking it (Schaible et al., 2002), making the airway microenvironment more hostile for Mtb survival (Boelaert et al., 2007).
FIGURE 2.
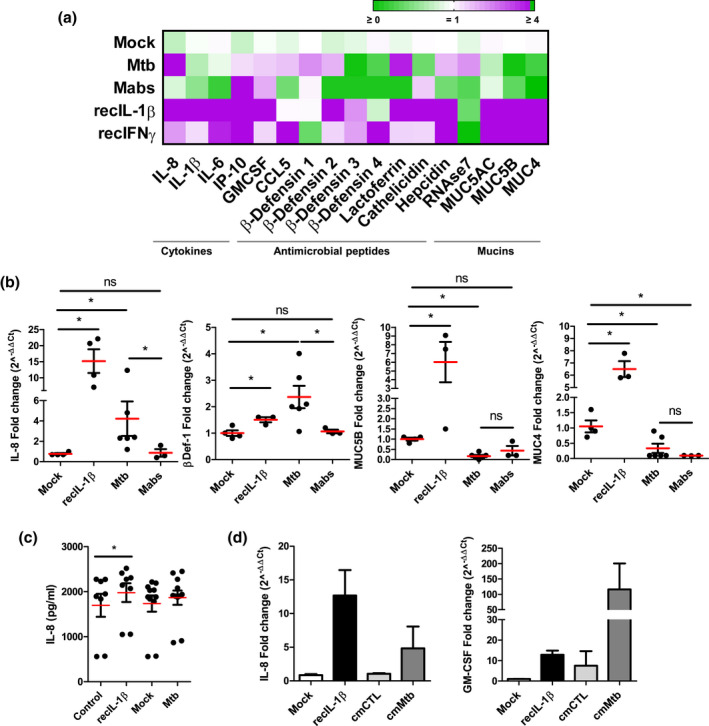
Mycobacteria‐induced host responses in AOs. (a) Heat map displaying modulation of cytokines, antimicrobial peptides, and mucins in AOs in response to Mtb H37Rv or Mabs injection compared to mock‐injected organoids. As positive controls, AOs were treated with recombinant IL‐1β and IFNβ. The experiment was performed four times independently. (b) Statistically significant expression changes of IL‐8, β‐defensin‐1, MUC5B, and MUC4 as determined by RT‐qPCR at 48 hr post‐infection. * p < .05 by a two‐tailed Mann‐Whitney test. (c) ELISA quantification of IL‐8 secretion by H37Rv‐infected AOs at 48 hr post‐infection. IL‐8 secretion in H37Rv‐infected AOs was almost significantly (p = .053 by two‐tailed Wilcoxon matched‐pairs signed‐rank test), recIL‐1β‐treated AOs (recIL‐1β) was used as positive control. The experiment was performed three times independently. (d) Expression changes of IL‐8 and GM‐CSF as determined by RT‐qPCR at 72 hr after conditioning with cmCTR and cmMTB, defined as conditioned media from non‐infected and Mtb‐infected macrophages, respectively. The experiments were performed two times independently with pooled conditioned medium from three independent donors
By contrast, Mabs infection did not enhance the expression of cytokines and antimicrobial peptides (Figure 2a). This could be due to the presence of glycopeptidolipids on the surface of its cell wall (Johansen et al., 2020), which mask other lipids and prevent the interaction of Mabs with pattern recognition receptors expressed by host cells (Rhoades et al., 2009), resulting in overall low responsiveness of lung epithelial cells to bacterial challenge (Matsuyama et al., 2018). Surprisingly, both Mabs and Mtb significantly downregulated the expression of mucins, including MUC5B and MUC4 (Figure 2b). Mucin expression and secretion are normally enhanced in response to inflammatory challenge and form part of an efficient clearance system for pathogen removal from the airway (Leiva‐Juárez et al., 2018; Ridley & Thornton, 2018; Whitsett & Alenghat, 2015). Our data also showed that mycobacterial infection is associated with downregulation of NFkB‐regulated genes such as IL‐1b and IL‐6 (Figure 2a), which could participate in mucin expression impairment as IL‐1b is a powerful inducer of mucin expression in the airway (Chen et al., 2019). Pathogens such as Helicobacter pylori have been shown to downregulate the expression of mucin genes to promote stomach tissue colonization (Cooke et al., 2009). How downregulation of mucin expression by mycobacteria might interfere with mucociliary function and infection outcome remains to be elucidated.
Upon Mtb infection, macrophages mount an inflammatory response that modulates the lung microenvironment (Fonseca et al., 2017; Lastrucci et al., 2015). AOs were stimulated with the supernatant of Mtb‐infected human macrophages (cmMTB) and analyzed for gene expression compared to stimulation with the supernatant of non‐infected macrophages (cmCTR). As shown in Figure 2d, the expression of IL‐8 and GM‐CSF, major cytokines for macrophage control of Mtb infection, were significantly enhanced in cmMTB‐stimulated AOs compared to those treated with cmCTR which mimics the paracrine macrophage‐epithelial signaling naturally occurring during lung Mtb infection (Reuschl et al., 2017).
Finally, due to the essentiality of macrophages in TB disease (Cadena et al., 2016; Corleis & Dorhoi, 2020), we co‐cultured human monocyte‐derived macrophages, alongside mycobacteria‐injected organoids, and observed hourly by confocal microscopy over the course of 4 days. Due to the complex nature of this experiment, it was optimized and set up under BSL‐2 conditions using Mycobacterium bovis BCG. Human macrophages were found to migrate within the collagen matrix and in some instances, moved towards organoids containing mycobacteria (Movie S8). While we found no evidence of macrophages being able to traverse the basal side and enter the organoid lumen to clear mycobacteria, we did observe macrophages migrating to and interacting with the basal edge of the AO, capturing and ingesting bacteria (Movie S8). This resembles the natural process of peri‐bronchiolar macrophage migration to the site of infection and phagocytosis of pathogenic material (Cambier et al., 2014).
Using AOs as a human 3D system to study the early steps of mycobacterial infection, we have shown that Mtb remains viable for up to 21 days within the lumen of AOs, while Mabs actively replicated over 7 days (Figure 1g). During this timeframe, while AO integrity remained uncompromised (Figures 1b,c, S2 and Movies S5 and S6), molecular interactions began as early as 48 hr after injection with the modulation of cytokines and antimicrobial peptides, and the inhibition of mucin expression (Figure 2b–d). Within 72 hr, innate immune cells can be recruited to the surface of infected AOs (Movie S8). Together, these data indicate that AOs offer a valuable 3D system to assess early interactions of mycobacteria with the human airway.
The ability to model these early timeframes in a responsive, multicellular and functionally similar system to the human airway, but without the complications, monetary and ethical restrictions of animal research, constitutes a major advance for the field. Organoid technology provides a complementary strategy to model in vitro human tissues, and infection by human‐related pathogens. Organoids based on lung ASC’s are advantageous as they generate AOs via a one‐step proliferation and differentiation protocol and naturally self‐organize in 3D to recapitulate the complexity of the lung epithelium. The resulting epithelium comprises all the different cell types and related secreted products (mucus, surfactant proteins), thus providing a reductionist system to assess host–pathogen interactions at the tissue level. Due to the self‐renewal of ASC’s, one of the most important advantages of organoids is that they are a genetically stable during long‐term culture, allowing for their cryopreservation in biobanks.
One of the main limitations of the organoid system comes from the absence of air‐liquid‐mucosa interface due to their closed, cystic structure. Very recent advances in bioprinting and microfluidic approaches allowed intestinal organoid‐based tissue engineering to create tube‐shaped epithelia and a continuous lumen (Nikolaev et al., 2020), with the potential of extending such an approach to airway models. Lung‐on‐a‐chip technology also constitutes an alternative approach to reproduce the air‐liquid‐mucosa interface and model mycobacterial infection (Huh et al., 2013; Thacker et al., 2020). However, lung‐on‐chips are generally developed using induced Pluripotent Stem Cells (iPSC) or epithelial cell lines, require complex expertise in both iPSC cultures based on long stepwise differentiation protocols, and in nano‐micro fabrication for chip devices. Therefore, organoids and on‐chip organs are complementary approaches that provide alternatives to animal experimentation thus complying with the “3R′s”—Replacement, Reduction, Refinement—which call for in vitro or a alternatives to model functional organs and pathologies (Gkatzis et al., 2018).
As mycobacteria, especially Mtb, establish their replicative niche in the alveolar part of the lung, another limitation comes from the absence of pneumocytes in the AO model. Recent advances in modeling the human alveolar part of the human lung as organoids (Salahudeen et al., 2020; Youk et al., 2020) will allow further modeling of the mycobacterial replicative niche. Finally, the ability to introduce human macrophages allows functional modeling of a key cell type and its cellular network, overcoming another limitation of organoid systems. Extension of the BALO technology, combining connected airway with alveoli and engraftment of macrophages (Vazquez‐Armendariz et al., 2020), to human stem cells could open new venues for mycobacterial infection modeling. We believe that this work forms a starting point for modeling a wide range of human respiratory pathogens using lung organoids, as exemplified with SARS‐CoV‐2, (Lamers et al., 2020, 2021; Salahudeen et al., 2020; Si et al., 2021; Thacker et al., 2021), but also respiratory infections driven by host disorders such as cystic fibrosis.
3. METHODS
3.1. Ethic statements
The collection of patient data and tissue for AO generation was performed according to the guidelines of the European Network of Research Ethics Committees following European and national law. In the Netherlands and France, the responsible accredited ethical committees reviewed and approved this study in accordance with the Medical Research Involving Human Subjects Act. Human lung tissue was provided by the Primary Lung Culture Facility (PLUC) at MUMC+, Maastricht, The Netherlands. Collection, storage, use of tissue and patient data were performed in agreement with the “Code for Proper Secondary Use of Human Tissue in the Netherlands” (http://www.fmwv.nl). The scientific board of the Maastricht Pathology Tissue Collection approved the use of materials for this study under MPTC2010‐019 and formal permission was obtained from the local Medical Ethical Committee (code 2017‐087). The CHU of Toulouse and CNRS approved protocol CHU 19 244 C and Ref CNRS 205782. All patients participating in this study consented to scientific use of their material; patients can withdraw their consent at any time, leading to the prompt disposal of their tissue and any derived material.
Human buffy coats were obtained from volunteers with informed consent via Sanquin (NVT0355.01) or établissement français du sang (Agreement 21PLER2017‐0035AV02).
3.2. Organoid culture
Healthy tissue from five independent donors receiving surgical treatment for lung cancer were used to derive organoids. AOs were characterized as described (Sachs et al., 2019).
3.3. Bacterial culture and microinjection
DsRed‐WT H37Rv Mtb strains were obtained by complementation with the pMRF plasmid containing a DsRed cassette, and were cultured in the continuous presence of 20 μg/ml of the selective antibiotic hygromycin under BSL‐3 conditions (Simeone et al., 2015). Mtb strains (BSL‐3) and GFP expressing M. bovis BCG (BSL‐2) were grown under the selection of 20 μg/ml Kanamycin in BSL‐2 conditions. M. abscessus sensu stricto (Mabs) strain CIP104536T (ATCC19977T) morphotype Smooth (S) carrying pASTA3 plasmid (Addgene, plasmid 24,657) that express red fluorescent protein (tdTomato), was cultured in presence of 500 μg/ml of the antibiotic hygromycin B as previously described (Bernut et al., 2014). All the bacterias were prepared for microinjection as described (Lastrucci et al., 2015). Bacterial density was adjusted to OD600 = 1, and phenol red was added at 0.005% to visualize successful microinjection (Bartfeld & Clevers, 2015). Pulled borosilicate capillaries (1.2 mm diameter, Harvard Apparatus Ltd.) were backfilled with the bacterial suspension using Microloader Tips (Eppendorf) and the tapered end broken off using tweezers until the smallest size droplet could be injected using a Femtojet micro‐injector (Eppendorf). We determined the CFU of these droplets that were destined for organoids by injecting them into 100 µl media and plating directly onto agar for enumeration. This was a reproducible way to quantify the initial bacterial load at the start of our experiments. An in‐house custom‐made Raspberry Pi‐controlled digital camera was used to observe organoids and the capillary. The capillary was attached to a TransferMan micro‐manipulator (Eppendorf) to facilitate semi‐automated injection. Injected organoids were allowed to recover for 2 hr at 37°C, individually collected, washed to remove extracellular bacteria and re‐seeded into fresh matrix for subsequent analysis.
3.4. Microscopy
For time‐lapse imaging, injected organoids were seeded in IBIDI 4 well chambers (IBIDI) and stained with CellMask™ Green Plasma Membrane Stain (1/1000, Molecular Probes) for 30 min at 37°C. Organoids were washed and fresh medium containing TOPRO‐3 Iodide (1/1000, Molecular Probes) was added. Organoids were imaged using a FEI CorrSight at Maastricht University (BSL‐2) or Andor/Olympus Spinning Disk Confocal CSU‐X1 (10× Air 0,4 NA, 3,1 mm WD) at IPBS (BSL‐3). Z‐stacks were acquired every hour for the duration of experiments and data analyzed using Fiji/Image J and IMARIS. The fluorescent labels used in this work are summarized in Table 2.
TABLE 2.
Summary of fluorescent markers used for flow cytometry and microscopy
| Dye | Concentration/Dilution | Marker of | Flow cytometry or microscopy |
|---|---|---|---|
| DAPI | 1 µg/ml | Nuclei | Microscopy |
| CellMask deep red | 1/30000 | Cell membranes | Microscopy and Flow Cytometry |
| CellMask Green | 1/1000 | Cell membranes | Microscopy |
| TOPRO‐3 iodide | 1/1000 | Dead cells | Microscopy |
| CellTracker CMAC blue | 20 µM | Cell cytosol | Microscopy |
For transmission electron microscopy (TEM), injected AOs were fixed in 4% PFA for a minimum of 3 hr at RT prior to removal from the containment lab and embedding in epon blocks as described in (Bartfeld & Clevers, 2015; Lamers et al., 2020). TEM data was collected autonomously as virtual nanoscopy slides on a 120 kV FEI Tecnai Spirit T12 Electron Microscope equipped with an Eagle CCD camera.
3.5. Colony‐forming unit (CFU) assay
Four to six Mtb or Mabs‐injected organoids were collected manually using a 200 µl pipette, washed in PBS, seeded into 24‐well plates, and cultured in complete AO medium for 2–21 days. At the relevant timepoint, organoids were lysed in 100 µl of 10% Triton X100 in water, serial dilutions were plated on 7H11 agar plates for Mtb or LB agar plates for Mabs and cultured for 3 weeks or 6 days respectively at 37°C.
3.6. RT‐qPCR
Uninfected control and Mtb‐ or Mabs‐infected AOs (15 per condition) were collected at 48 hr post‐infection, lysed in 1 ml of TRIzol Reagent (Invitrogen) and stored at −80°C for 2 days. As positive controls, AOs were stimulated with 0.02 µg/ml of human IL‐1β (Invivogen) or 0.1 µg/ml of IFNγ (PeproTech) for 48 hr. Total RNA was extracted using the RNeasy mini kit (Qiagen) and retrotranscribed (150 ng) using the Verso cDNA Synthesis Kit (Thermo Scientific). mRNA expression was assessed with an ABI 7,500 real‐time PCR system (Applied Biosystems) and the SYBRTM Select Master Mix (ThermoScientific). Relative quantification was determined using the 2−ΔΔCt method and normalized to GAPDH. Primer sequences are provided in Table 1.
TABLE 1.
List of primers used for RT‐qPCR experiments on airway organoids
| Gene | Primers 5′−3′ | Reference |
|---|---|---|
| Cytokines and chemokines | ||
| CCL5 (NM_002985) | F‐ CCTCATTGCTACTGCCCTCT | In‐house |
| R‐ CGGGTGACAAAGACGACTGC | ||
| GM‐CSF (NM_000758) | F‐ CCTGAACCTGAGTAGAGACACT | In‐house |
| R‐ CCTTGAGCTTGGTGAGGCTG | ||
| IL‐1β (NM_000576) | F‐ AGCTACGAATCTCCGACCAC | In‐house |
| R‐ GGGAAAGAAGGTGCTCAGGTC | ||
| IL‐6 (NM_000600.5) | F: ACTCACCTCTTCAGAACGAATTG | PrimerBank |
| R: CCATCTTTGGAAGGTTCAGGTTG | ||
| IL‐8 (NM_000584) | F‐ TACTCCAAACCTTTCCACCCC | In‐house |
| R‐ CTTCTCCACAACCCTCTGCA | ||
| IP‐10 (NM_001565) | F‐ GTGGCATTCAAGGAGTACCTC | In‐house |
| R‐ GATTCAGACATCTCTTCTCACCC | ||
| Antimicrobial peptides | ||
| β‐Defensin 1 (NM_005218) | F‐ ATGGCCTCAGGTGGTAACTTTC | In‐house |
| R‐ GGTCACTCCCAGCTCACTTG | ||
| β‐Defensin 2 (NM_004942) | F‐ATAGGCGATCCTGTTACCTGC | In‐house |
| R‐CCTCCTCATGGCTTTTTGCAG | ||
| β‐Defensin 3 (NM_018661) | F‐ TGGGGTGAAGCCTAGCAG | In‐house |
| R‐ ACTTGCCGATCTGTTCCTCC | ||
| β‐Defensin 4 | ||
| (NM_080389.3) | F: TGCCGGAAGAAATGTCGCA | In‐house |
| R: CGACTCTAGGGACCAGCAC | ||
| Cathelicidin LL37 (NM_004345) | F‐ ATGCTAACCTCTACCGCCTCC | In‐house |
| R‐ TCACCAGCCCGTCCTTCTTG | ||
| Hepcidin (NM_021175) | F‐ GTTTTCCCACAACAGACGGG | In‐house |
| R‐ AGATGGGGAAGTGGGTGTC | ||
| Lactoferrin (NM_002343) | F‐ CCCCTACAAACTGCGACCTG | In‐house |
| R‐ CAGACCTTGCAGTTCGTTCAG | ||
| RNAse 7 (NM_032572) | F‐ GGAGTCACAGCACGAAGACCA | In‐house |
| R‐ GGCTTGGCACTGACTGGGATC | ||
| Mucins | ||
| MUC4 (NM_018406.7) | F: CTCAGTACCGCTCCAGCAG | In‐house |
| R: CCGCCGTCTTCATGGTCAG | ||
| MUC5AC (NM_001304359.2) | F: CCAGTCCTGCCTTTGTACGG | In‐house |
| R: GACCCTCCTCTCAATGGTGC | ||
| MUC5B (NM_002458.3) | F: GCCCACATCTCCACCTATGAT | PrimerBank |
| R: GCAGTTCTCGTTGTCCGTCA | ||
| Housekeeping | ||
| GAPDH (NM_002046) | F‐CTCCAAAATCAAGTGGGGCGATG | In‐house |
| R‐GGCATTGCTGATGATCTTGAGGC | ||
3.7. Enzyme‐linked immunosorbent assay
Between 20 and 30 organoids were embedded in fresh BME Cultrex and cultured with 800 µl complete media. After 48 hr, the supernatant was collected, sterilized through double 0.22 µm filters and stored at −80°C until analysis. IL‐8 ELISA was performed according to manufacturer instructions (Qiagen).
3.8. Flow cytometry and cell sorting
Organoids were washed out of Matrigel and dissociated into single cells using TrypLE for 5 min at 37°C. A minimum of 5 × 105 cells/ml were incubated with dsRED expressing Mtb at an MOI of 10 in complete organoid media. After 4 or 24 hr for single cells, or 7 days for whole intact organoids, samples were washed with PBS, stained with CellMask Deep Red (1:30.000) to highlight all cells composing the organoid and fixed in 4% paraformaldehyde overnight at 4°C. Cells were pelleted and resuspended in PBS supplemented with 2% FCS. Samples were filtered just before analysis and sorted using a BD FACS ARIA Fusion. Cells were selected on their FSC‐A/SSC‐A profile, doublets were excluded on FSC‐W/FSC‐H and SSC‐W/SSC‐H parameters. Finally, double positive cells were gated based on Cell Mask Deep Red staining and DsRED‐expressing Mtb. The population of cells from P5 (positive for GFP/bacteria and APC/ organoid cells) was pooled and imaged by confocal microscopy to determine if bacteria was intracellular. The fluorescent dyes used are summarized in Table 2.
3.9. CmMTB preparation and macrophage co‐cultures
Peripheral blood mononuclear cells were enriched from buffy coat using RosetteSep human monocyte enrichment cocktail according to the manufacturer’s instructions (Stem Cell Technologies). Monocytes were purified by density gradient centrifugation using Lymphoprep (Stem Cell Technologies) according to the manufacturer’s protocol. Monocytes were differentiated into macrophages by addition of 5 ng/ml macrophage colony‐stimulating factor (Sigma Aldrich) for 6 days. cmCTR and cmMTB were prepared and used as previously described (Lastrucci et al., 2015). Organoids were stained with CellMask Deep Red plasma membrane dye as previously described, and macrophages stained with 20 µM CellTracker Blue CMAC dye (ThermoFisher Scientific) for 1 hr in serum‐free media. Microinjected organoids and macrophages were resuspended in freshly prepared Rat Tail Collagen type 1 (Thermofisher, 1 mg/ml) and polymerized in a 4‐well, glass‐bottom µ‐slide (Ibidi) at 37°C for 30 min and imaged for 96 hr under a FEI CorrSight microscope.
CONFLICT OF INTEREST
Hans Clevers and Norman Sachs are inventors on patents related to organoid technology. Other authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
NI, CC, and PJP designed the experiments with the help of CLI, ON, EM, and GLV. NI, SALI, and CC performed the experiments with the contribution of KK, RBGR, SM, AP, CCh, KC, and CG. SR generated the fluorescent Mtb strains. MME and JM provided the lung biopsies. HC and NS developed the lung organoid technology. NI, CC, SALI, and PJP wrote the manuscript.
Supporting information
Fig S1
Fig S2
Movie S1
Movie S2
Movie S3
Movie S4
Movie S5
Movie S6
Movie S7
Movie S8
ACKNOWLEDGMENTS
Authors acknowledge C. Kuo (Stanford University, USA) for the stable expressing Rspo1‐Fc cell line, and the Hubrecht Institute for the stable expressing Noggin cell line; Genotoul TRI‐IPBS core facility for flow cytometry and imaging, in particular E. Näser and E. Vega; IPBS BSL‐3 facilities, in particular C. Verollet for technical support. Authors also acknowledge the Microscopy CORE lab and PLUC facility at Maastricht University. This work was supported by grants from Campus France PHC Van Gogh (40577ZE to GL‐V), the Agence Nationale de la Recherche (ANR‐15‐CE15‐0012 (MMI‐TB)) to GL‐V, FRM “Amorçage Jeunes Equipes” (AJE20151034460 to EM), ERC StG 693 (INFLAME 804249 to EM), ATIP to EM, ZonMW 3R’s (114021005) to PJP, the Nuffic Van Gogh Programme (VGP.17/10 to NI), and by the LINK program from the Province of Limburg, the Netherlands.
Iakobachvili, N. , Leon‐Icaza, S.A. , Knoops, K. , Sachs, N. , Mazères, S. , Simeone, R. , et al (2022) Mycobacteria–host interactions in human bronchiolar airway organoids. Molecular Microbiology, 117, 682–692. 10.1111/mmi.14824
Nino Iakobachvili and Stephen Adonai Leon‐Icaza are shared the first autorship.
Céline Cougoule and Peter J. Peters are shared the last autorship.
Contributor Information
Céline Cougoule, Email: Celine.Cougoule@ipbs.fr.
Peter J. Peters, Email: pj.peters@maastrichtuniversity.nl.
REFERENCES
- Arranz‐Trullén, J. , Lu, L. , Pulido, D. , Bhakta, S. & Boix, E. (2017) Host antimicrobial peptides: the promise of new treatment strategies against tuberculosis. Frontiers in Immunology, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagayoko, S. , Leon‐Icaza, S.A. , Pinilla, M. , Hessel, A. , Santoni, K. , Péricat, D. et al. (2021, February 17) Host phospholipid peroxidation fuels ExoU‐dependent cell necrosis and supports Pseudomonas aeruginosa‐driven pathology. bioRxiv, 17(9), e1009927. 10.1371/journal.ppat.1009927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggiolini, M. & Clark‐Lewis, I. (1992) Interleukin‐8, a chemotactic and inflammatory cytokine. FEBS Letters, 307, 97–101. [DOI] [PubMed] [Google Scholar]
- Bartfeld, S. & Clevers, H. (2015) Organoids as model for infectious diseases: Culture of human and murine stomach organoids and microinjection of Helicobacter pylori . Journal of Visualized Experiments, 2015, 53359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez, L.E. , Sangari, F.J. , Kolonoski, P. , Petrofsky, M. & Goodman, J. (2002) The efficiency of the translocation of Mycobacterium tuberculosis across a bilayer of epithelial and endothelial cells as a model of the alveolar wall is a consequence of transport within mononuclear phagocytes and invasion of alveolar epithelial cells. Infection and Immunity, 70, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernut, A. , Dupont, C. , Sahuquet, A. , Herrmann, J.‐L. , Lutfalla, G. & Kremer, L. (2015) Deciphering and imaging pathogenesis and cording of Mycobacterium abscessus in zebrafish embryos. Journal of Visualized Experiments, 2015, 53130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernut, A. , Herrmann, J.‐L. , Kissa, K. , Dubremetz, J.‐F. , Gaillard, J.‐L. , Lutfalla, G. et al. (2014) Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proceedings of the National Academy of Sciences, 111, E943–E952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielecka, M.K. , Tezera, L.B. , Zmijan, R. , Drobniewski, F. , Zhang, X. , Jayasinghe, S. et al. (2017) A Bioengineered three‐dimensional cell culture platform integrated with microfluidics to address antimicrobial resistance in tuberculosis. mBio, 8(1), e02073‐16. 10.1128/mBio.02073-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelaert, J.R. , Vandecasteele, S.J. , Appelberg, R. & Gordeuk, V.R. (2007) The effect of the host’s iron status on tuberculosis. Journal of Infectious Diseases, 195, 1745–1753. [DOI] [PubMed] [Google Scholar]
- Cadena, A.M. , Flynn, J.L. & Fortune, S.M. (2016) The importance of first impressions: Early events in Mycobacterium tuberculosis infection influence outcome. mbio, 7(2), e00342‐16. 10.1128/mBio.00342-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier, C.J. , Takaki, K.K. , Larson, R.P. , Hernandez, R.E. , Tobin, D.M. , Urdahl, K.B. et al. (2014) Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature, 505, 218–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G. , Sun, L. , Kato, T. , Okuda, K. , Martino, M.B. , Abzhanova, A. et al. (2019) IL‐1β dominates the promucin secretory cytokine profile in cystic fibrosis. Journal of Clinical Investigation, 129, 4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cidem, A. , Bradbury, P. , Traini, D. & Ong, H.X. (2020) Modifying and integrating in vitro and ex vivo respiratory models for inhalation drug screening. Front Bioeng Biotechnol, 8, 581995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S.B. , Gern, B.H. , Delahaye, J.L. , Adams, K.N. , Plumlee, C.R. , Winkler, J. et al. (2018) Alveolar macrophages provide an early Mycobacterium tuberculosis niche and initiate dissemination. Cell Host & Microbe, 24, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke, C.L. , An, H.J. , Kim, J. , Canfield, D.R. , Torres, J. , Lebrilla, C.B. et al. (2009) Modification of gastric mucin oligosaccharide expression in rhesus macaques after infection with Helicobacter pylori . Gastroenterology, 137, 1061–1071.e8. [DOI] [PubMed] [Google Scholar]
- Corleis, B. & Dorhoi, A. (2020) Early dynamics of innate immunity during pulmonary tuberculosis. Immunology Letters, 221, 56–60. [DOI] [PubMed] [Google Scholar]
- Domingo‐Gonzalez, R. , Prince, O. , Cooper, A. & Khader, S.A. (2016) Cytokines and chemokines in Mycobacterium tuberculosis infection. Microbiology Spectrum, 4(5). 10.1128/microbiolspec.TBTB2-0018-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H. , Lv, Y. , Zhao, D. , Barrow, P. & Zhou, X. (2016) Defensins: the case for their use against mycobacterial infections. Journal of Immunology Research, 2016, 7515687. 10.1155/2016/7515687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, D. & Clevers, H. (2017) Organoid culture systems to study host–pathogen interactions. Current Opinion in Immunology, 48, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkington, P. , Lerm, M. , Kapoor, N. , Mahon, R. , Pienaar, E. , Huh, D. et al. (2019) In vitro granuloma models of tuberculosis: potential and challenges. Journal of Infectious Diseases, 219, 1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, K.L. , Rodrigues, P.N.S. , Olsson, I.A.S. & Saraiva, M. (2017) Experimental study of tuberculosis: from animal models to complex cell systems and organoids. PLoS Pathogens, 13(8), e1006421. 10.1371/journal.ppat.1006421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkatzis, K. , Taghizadeh, S. , Huh, D. , Stainier, D.Y.R. & Bellusci, S. (2018) Use of three‐dimensional organoids and lung‐on‐a‐chip methods to study lung development, regeneration and disease. European Respiratory Journal, 52, 1800876. [DOI] [PubMed] [Google Scholar]
- Heo, I. , Dutta, D. , Schaefer, D.A. , Iakobachvili, N. , Artegiani, B. , Sachs, N. et al. (2018) Modeling cryptosporidium infection in human small intestinal and lung organoids. Nature Microbiology, 3, 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, D. , Kim, H.J. , Fraser, J.P. , Shea, D.E. , Khan, M. , Bahinski, A. et al. (2013) Microfabrication of human organs‐on‐chips. Nature Protocols, 811(8), 2135–2157. [DOI] [PubMed] [Google Scholar]
- Hui, K.P.Y. , Ching, R.H.H. , Chan, S.K.H. , Nicholls, J.M. , Sachs, N. , Clevers, H. et al. (2018) Tropism, replication competence, and innate immune responses of influenza virus: an analysis of human airway organoids and ex‐vivo bronchus cultures. The Lancet Respiratory Medicine, 6, 846–854. [DOI] [PubMed] [Google Scholar]
- Iakobachvili, N. & Peters, P.J. (2017) Humans in a dish: the potential of organoids in modeling immunity and infectious diseases. Frontiers in Microbiology, 8, 2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, M.D. , Herrmann, J.‐L. & Kremer, L. (2020) (2020) Non‐tuberculous mycobacteria and the rise of Mycobacterium abscessus . Nature Reviews Microbiology, 187(18), 392–407. [DOI] [PubMed] [Google Scholar]
- Lamers, M.M. , Beumer, J. , van der Vaart, J. , Knoops, K. , Puschhof, J. , Breugem, T.I. et al. (2020) SARS‐CoV‐2 productively infects human gut enterocytes. Science (80‐), 369, 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers, M.M. , van der Vaart, J. , Knoops, K. , Riesebosch, S. , Breugem, T.I. , Mykytyn, A.Z. et al. (2021) An organoid‐derived bronchioalveolar model for SARS‐CoV‐2 infection of human alveolar type II‐like cells. EMBO Journal, 40(5), e105912. 10.15252/embj.2020105912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastrucci, C. , Bénard, A. , Balboa, L. , Pingris, K. , Souriant, S. , Poincloux, R. et al. (2015) Tuberculosis is associated with expansion of a motile, permissive and immunomodulatory CD16+ monocyte population via the IL‐10/STAT3 axis. Cell Research, 25, 1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiva‐Juárez, M.M. , Kolls, J.K. & Evans, S.E. (2018) Lung epithelial cells: therapeutically inducible effectors of antimicrobial defense. Mucosal Immunology, 11, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamed, D. , Boulle, M. , Ganga, Y. , Arthur, C.M. , Skroch, S. , Oom, L. et al. (2017) Intracellular growth of Mycobacterium tuberculosis after macrophage cell death leads to serial killing of host cells. Elife, 6, e22028. 10.7554/eLife.22028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama, M. , Martins, A.J. , Shallom, S. , Kamenyeva, O. , Kashyap, A. , Sampaio, E.P. et al. (2018) Transcriptional response of respiratory epithelium to nontuberculous mycobacteria. American Journal of Respiratory Cell and Molecular Biology, 58, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev, M. , Mitrofanova, O. , Broguiere, N. , Geraldo, S. , Dutta, D. , Tabata, Y. et al. (2020) (2020) Homeostatic mini‐intestines through scaffold‐guided organoid morphogenesis. Nature, 585(7826), 574–578. [DOI] [PubMed] [Google Scholar]
- Parasa, V.R. , Rahman, M.J. , Hoang, A.T.N. , Svensson, M. , Brighenti, S. & Lerm, M. (2014) Modeling Mycobacterium tuberculosis early granuloma formation in experimental human lung tissue. Disease Models & Mechanisms, 7, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissegur, M.‐P. , Lay, G. , Gilleron, M. , Botella, L. , Nigou, J. , Marrakchi, H. et al. (2007) Mycobacterial lipomannan induces granuloma macrophage fusion via a TLR2‐dependent, ADAM9‐ and β1 integrin‐mediated pathway. The Journal of Immunology, 178, 3161–3169. [DOI] [PubMed] [Google Scholar]
- Rampacci, E. , Stefanetti, V. , Passamonti, F. & Henao‐Tamayo, M. (2020) Preclinical models of nontuberculous mycobacteria infection for early drug discovery and vaccine research. Pathogens, 9, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnatunga, C.N. , Lutzky, V.P. , Kupz, A. , Doolan, D.L. , Reid, D.W. , Field, M. et al. (2020) The rise of non‐tuberculosis mycobacterial lung disease. Frontiers in Immunology, 11, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuschl, A.‐K. , Edwards, M.R. , Parker, R. , Connell, D.W. , Hoang, L. , Halliday, A. et al. (2017) Innate activation of human primary epithelial cells broadens the host response to Mycobacterium tuberculosis in the airways. PLoS Pathogens, 13(9), e1006577. 10.1371/journal.ppat.1006577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, E.R. , Archambault, A.S. , Greendyke, R. , Hsu, F.‐F. , Streeter, C. & Byrd, T.F. (2009) Mycobacterium abscessus glycopeptidolipids mask underlying cell wall phosphatidyl‐myo‐Inositol mannosides blocking induction of human macrophage TNF‐α by preventing interaction with TLR2. The Journal of Immunology, 183, 1997–2007. [DOI] [PubMed] [Google Scholar]
- Ridley, C. & Thornton, D.J. (2018) Mucins: the frontline defence of the lung. Biochemical Society Transactions, 46, 1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas‐Santiago, B. , Schwander, S.K. , Sarabia, C. , Diamond, G. , Klein‐Patel, M.E. , Hernandez‐Pando, R. et al. (2005) Human β‐defensin 2 is expressed and associated with Mycobacterium tuberculosis during infection of human alveolar epithelial cells. Infection and Immunity, 73, 4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, G. , Manfrin, A. & Lutolf, M.P. (2018) Progress and potential in organoid research. Nature Reviews Genetics, 1911(19), 671–687. [DOI] [PubMed] [Google Scholar]
- Ryndak, M.B. , Singh, K.K. , Peng, Z. & Laal, S. (2015) Transcriptional profile of Mycobacterium tuberculosis replicating in type II alveolar epithelial cells. PLoS One, 5, 112–114. 10.1016/j.gdata.2015.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, N. , Papaspyropoulos, A. , Ommen, D.D.Z. , Heo, I. , Böttinger, L. , Klay, D. et al. (2019) Long‐term expanding human airway organoids for disease modeling. EMBO Journal, 38(4), e100300. 10.15252/embj.2018100300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahudeen, A.A. , Choi, S.S. , Rustagi, A. , Zhu, J. , van Unen, V. & Sean, M. et al. (2020) Progenitor identification and SARS‐CoV‐2 infection in human distal lung organoids. Nature 588(7839), 670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible, U.E. , Collins, H.L. , Priem, F. & Kaufmann, S.H.E. (2002) Correction of the iron overload defect in β‐2‐microglobulin knockout mice by lactoferrin abolishes their increased susceptibility to tuberculosis. Journal of Experimental Medicine, 196, 1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple, F. & Dorin, J.R. (2012) β‐Defensins: multifunctional modulators of infection, inflammation and more? Journal of Innate Immunity, 4, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si, L. , Bai, H. , Rodas, M. , Cao, W. , Oh, C.Y. , Jiang, A. et al. (2021) A human‐airway‐on‐a‐chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nature Biomedical Engineering, 2021, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva‐Miranda, M. , Ekaza, E. , Breiman, A. , Asehnoune, K. , Barros‐Aguirre, D. , Pethe, K. et al. (2015) High‐content screening technology combined with a human granuloma model as a new approach to evaluate the activities of drugs against Mycobacterium tuberculosis . Antimicrobial Agents and Chemotherapy, 59, 693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone, R. , Sayes, F. , Song, O. , Gröschel, M.I. , Brodin, P. , Brosch, R. et al. (2015) Cytosolic access of Mycobacterium tuberculosis: criticalimpact of phagosomal acidification control and demonstration of occurrence invivo. PLoS Pathogens, 11(2), e1004650. 10.1371/journal.ppat.1004650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sow, F.B. , Alvarez, G.R. , Gross, R.P. , Satoskar, A.R. , Schlesinger, L.S. , Zwilling, B.S. et al. (2009) Role of STAT1, NF‐κB, and C/EBPβ in the macrophage transcriptional regulation of hepcidin by mycobacterial infection and IFN‐γ. Journal of Leukocyte Biology, 86, 1247–1258. [DOI] [PubMed] [Google Scholar]
- Sow, F.B. , Florence, W.C. , Satoskar, A.R. , Schlesinger, L.S. , Zwilling, B.S. & Lafuse, W.P. (2007) Expression and localization of hepcidin in macrophages: a role in host defense against tuberculosis. Journal of Leukocyte Biology, 82, 934–945. [DOI] [PubMed] [Google Scholar]
- Thacker, V.V. , Dhar, N. , Sharma, K. , Barrile, R. , Karalis, K. & McKinney, J.D. (2020) A lung‐on‐chip model of early Mycobacterium tuberculosis infection reveals an essential role for alveolar epithelial cells in controlling bacterial growth. Elife, 9, 1–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker, V.V. , Sharma, K. , Dhar, N. , Mancini, G. , Sordet‐Dessimoz, J. , & McKinney, J.D. (2021) Rapid endotheliitis and vascular damage characterize SARS‐CoV‐2 infection in a human lung‐on‐chip model. EMBO Reports, 22(6), e52744. 10.15252/embr.202152744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To, K. , Cao, R. , Yegiazaryan, A. , Owens, J. & Venketaraman, V. (2020) General overview of nontuberculous mycobacteria opportunistic pathogens: Mycobacterium avium and Mycobacterium abscessus . Journal of Clinical Medicine, 9, 2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrelles, J.B. & Schlesinger, L.S. (2017) Integrating lung physiology, immunology and tuberculosis. Trends in Microbiology, 25, 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres‐Juarez, F. , Touqui, L. , Leon‐Contreras, J. , Rivas‐Santiago, C. , Enciso‐Moreno, J.A. , Hernández‐Pando, R. et al. (2018) RNase 7 but not psoriasin nor sPLA2‐IIA associates with Mycobacterium tuberculosis during airway epithelial cell infection. Pathogens and Disease, 76(2). 10.1093/femspd/fty005 [DOI] [PubMed] [Google Scholar]
- Vazquez‐Armendariz, A.I. , Heiner, M. , Agha, E.E. , Salwig, I. , Hoek, A. , Hessler, M.C. et al. (2020) Multilineage murine stem cells generate complex organoids to model distal lung development and disease. EMBO Journal, 39(21), e103476. 10.15252/embj.2019103476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsett, J.A. & Alenghat, T. (2015) Respiratory epithelial cells orchestrate pulmonary innate immunity. Nature Immunology, 16(1), 27–35. 10.1038/ni.3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . (2020) Tuberculosis. Accessed August 5, 2021, https://www.who.int/teams/global‐tuberculosis‐programme/data [Google Scholar]
- Youk, J. , Kim, T. , Evans, K.V. , Jeong, Y.‐I. , Hur, Y. , Hong, S.P. et al. (2020) Three‐dimensional human alveolar stem cell culture models reveal infection response to SARS‐CoV‐2. Cell Stem Cell, 27, 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Li, C. , Sachs, N. , Chiu, M.C. , Wong, B.‐H.‐Y. , Chu, H. et al. (2018) Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proceedings of the National Academy of Sciences of the United States of America, 115, 6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Movie S1
Movie S2
Movie S3
Movie S4
Movie S5
Movie S6
Movie S7
Movie S8


