Abstract
We report the characterization of the ccpA gene of Lactobacillus plantarum, coding for catabolite control protein A. The gene is linked to the pepQ gene, encoding a proline peptidase, in the order ccpA-pepQ, with the two genes transcribed in tandem from the same strand as distinct transcriptional units. Two ccpA transcription start sites corresponding to two functional promoters were found, expression from the upstream promoter being autogenously regulated through a catabolite-responsive element (cre) sequence overlapping the upstream +1 site. During growth on ribose, the upstream promoter showed maximal expression, while growth on glucose led to transcription from the downstream promoter. In a ccpA mutant strain, the gene was transcribed mainly from the upstream promoter in both repressing and non repressing conditions. Expression of two enzyme activities, β-glucosidase and β-galactosidase, was relieved from carbon catabolite repression in the ccpA mutant strain. In vivo footprinting analysis of the catabolite-controlled bglH gene regulatory region in the ccpA mutant strain showed loss of protection of the cre under repressing conditions.
Lactobacillus plantarum is a lactic acid bacterium with broad applications, being used for starter cultures in vegetable, meat, fodder, and beverage fermentation. It has also been selected for use in the development of functional and therapeutic foods and of potential live oral vaccines (1, 8). Despite its wide biotechnological applications, very little is known about the regulatory mechanisms of carbon metabolism in this organism. Genes coding for two L. plantarum β-glucosidases have recently been identified (22, 23). Preliminary studies showed that expression of these genes is under carbon catabolite control and suggested the involvement of a catabolite control protein A (CcpA)-mediated regulatory mechanism.
In gram-positive bacteria of low G+C content, carbon catabolite repression (CCR) involves negative regulation mediated by CcpA (10, 30). Genes and operons coding for enzymes involved in the catabolism of less favorable carbon sources are regulated by CcpA at the transcriptional level in the presence of rapidly metabolizable sugars like glucose or fructose. Null mutations in the ccpA gene partially or completely relieve expression from CCR. CcpA binds to DNA target sites termed catabolite-responsive elements (cre). The proposed consensus for cre is a 14-bp sequence containing a partial dyad symmetry (12), whose A+T-rich flanking regions mediate high-level CCR (36). Various effectors have been shown to stimulate the DNA-binding activity of CcpA. One of the most important CcpA effectors is a phosphorylated form of HPr, the phospho-carrier protein of the phosphoenolpyruvate-dependent phosphotransferase system (PTS), whose phosphorylation state reflects glycolytic activity. Being part of the PTS, HPr is phosphorylated by enzyme I at histidine 15 and transfers the phosphoryl group to the sugar-specific enzyme IIAs. In Bacillus subtilis, the gram-positive bacterium used most frequently in CCR studies, HPr is phosphorylated at serine 46 during growth on glucose. This ATP-dependent phosphorylation is carried out by HPr kinase, which appears to be the key component in signal transduction leading to CCR (28). The seryl-phosphorylated HPr enhances the binding of CcpA to cre sequences within regulatory and coding regions of catabolite-controlled genes, leading to repression of gene expression (5). In other systems CcpA-cre binding is enhanced by high concentrations of early glycolytic intermediates such as glucose-6-phosphate (9) or by a combination of seryl-phospharylated HPr and NADP (14). In addition to HPr, an HPr-like protein called Crh (catabolite repression HPr) was shown to participate in CCR (24).
CcpA is a master regulator which can function either as a repressor or as an activator of transcription. Activation was shown in the expression of genes involved in excretion of excess carbon, such as ackA of B. subtilis, and in the expression of the las operon of Lactococcus lactis (19, 33). This activating function of CcpA accounts for the fact that disruption of the ccpA gene in L. lactis and B. subtilis not only reduces catabolite repression of several target genes but also decreases the growth rate on both PTS and non-PTS sugars. Recent data show that independent mutations in the Bacillus megaterium ccpA gene separate growth effects from catabolite repression (15). Moreover, gene activation mediated by CcpA is responsible for adaptation of L. lactis to low temperature (35).
CcpA and homologues have been identified in various gram-positive bacteria, including B. subtilis (11), B. megaterium (13), Lactobacillus casei (27), Lactococcus lactis (19), Lactobacillus pentosus (20), Streptococcus mutans (32), Enterococcus faecalis (16), and Streptococcus thermophilus (34). In all of these examples except S. mutans, glucose repression was relieved in ccpA mutant strains.
We report here the identification of the ccpA gene of L. plantarum. Expression of the gene is driven from two promoters, with the distant promoter autogenously regulated through a cre sequence overlapping the upstream +1 site. A ccpA null mutation negatively affected growth on glucose and relieved from CCR the expression of β-glucosidase and β-galactosidase activities.
MATERIALS AND METHODS
Bacterial strains.
L. plantarum LM3 (K. Thompson, K. McConville, L. McNeilly, C. Nicholson, and M. Collins, Abstr. 6th Symp. Lactic Acid Bacteria Genet. Metab. Appl., p. E5, 1999) was used throughout this study. L. plantarum was grown in MRS medium (prepared without carbon source) supplemented with 2% glucose, 1% ribose, 1% lactose, or 0.4% salicin. When needed, erythromycin (5 μg ml−1) or chloramphenicol (10 μg ml−1) was added to the MRS medium. The Escherichia coli TG1 was used for plasmid cloning.
DNA amplification, cloning, and sequencing.
Total DNA from L. plantarum LM3 was prepared as described elsewhere (17) and used as the template in PCR with primers A1 (5′-GGAATTCGTGTCGATGGCAACGGTTTCT-3′) and A2 (5′-CGTCTAGACGCATCGCTACTGCACCAAT-3′) to amplify the ccpA internal fragment. The two primers were designed on the basis of the B. megaterium ccpA sequence; primer A1 was the coding sequence for the central part of the helix-turn-helix domain, and primer A2 was the coding sequence for the N-terminal conserved domain of the protein. PCR was carried out with 35 amplification cycles of 1 min at 94°C, 1 min at 40°C, and 2 min at 72°C. The PCR amplification product, an 891-bp fragment, was cloned into the EcoRI-XbaI sites of pJDC9 (4) to yield plasmid pLM1. The 891-bp fragment was sequenced and used to probe an enriched EcoRI-SalI L. plantarum chromosomal DNA library constructed with pUC19 as the recipient vector. A positive recombinant clone, yielding plasmid pLM10, was used to complete sequencing of the ccpA 3′ end and its flanking region. The 5′ end of the gene was sequenced directly on chromosomal DNA as follows. An enriched 6-kb SalI fragment, hybridizing with the ccpA probe, was purified from an agarose gel and precipitated with 12% polyethylene glycol 6000–1.5 M NaCl; 500 ng of this DNA fraction was used for direct sequencing with a Thermo Sequenase radiolabeled terminator cycle sequencing kit (U.S. Biochemicals). PCR was carried out with 60 amplification cycles of 30 s at 95°C, 30 s at 42°C, and 1 min at 72°C.
Primer extension and Northern blot analysis.
Total RNA from L. plantarum cells grown to mid-exponential phase on MRS medium supplemented with 2% glucose or 1% ribose was isolated as described by Leong-Morgenthaler and coworkers (18). Primer extension products of bglH and ccpA transcripts were obtained using oligonucleotides SP1 (5′-GCCACCTGGTAACCGATCCGC-3′) and A3 (5′-GCGGGTTTAACGTTGGGATTACC-3′), respectively. The experiment was performed as described previously (23). Northern blotting was performed on total RNA extracted from cells grown on ribose, using the 891-bp internal fragment of the ccpA gene as the probe. The experiment was performed as described elsewhere (31).
Plasmid construction.
Plasmid pLM2 was constructed as follows. The C1′ (399-bp) and the C2′ (433-bp) regions of the ccpA gene were individually amplified from L. plantarum chromosomal DNA by PCR and cloned in the SacI-SmaI and SalI-HindIII sites of plasmid pJDC9 (4), respectively, in the same transcriptional orientation. C1′ corresponds to the 5′ portion of the gene lacking initial 36 bp of the coding sequence; C2′ corresponds to the 3′ portion lacking the last 91 bp. The cat cassette was inserted between the C1′ and C2′ fragments in the SmaI-SalI sites to yield plasmid pML2. Electrotransformation of L. plantarum with the integration plasmid pLM2 was performed as described elsewhere (17).
β-Glucosidase assay.
L. plantarum strains were grown to mid-exponential phase in MRS medium (50 ml) supplemented with either 2% glucose or 0.4% salicin, washed twice with 150 mM NaCl, and resuspended in 1 ml of 50 mM phosphate buffer, pH 6.2. Appropriate aliquots of cell suspensions were added to 800 μl of 30 mM salicin in phosphate buffer. After 20 min of incubation at 30°C, the reaction was stopped by addition of 500 μl of 1 M Na2CO3. The production of saligenin from salicin was detected as described elsewhere (21).
β-Galactosidase assay.
L. plantarum strains were grown to mid-exponential phase in MRS medium supplemented with 1% lactose, glucose, ribose, or lactose plus glucose. β-Galactosidase activity was detected on whole cells permeabilized with chloroform and sodium dodecyl sulfate as described by Miller (25), with o-nitrophenyl-β-d-galactopyranoside as a substrate.
In vivo footprinting.
L. plantarum cells were grown overnight in MRS medium supplemented with 0.4% salicin or 2% glucose. Cells were then diluted 1:100 and grown to mid-exponential phase. Methylation was performed by adding freshly diluted dimethyl sulfate (DMS; Aldrich) to a final concentration of 0.1% for 3 min at 30°C with shaking. The methylation reaction was stopped by adding an equal volume of ice-cold saline phosphate buffer (150 mM NaCl, 40 mM K2HPO4, 22 mM KH2PO4 [pH 7.2]). Cells were harvested by centrifugation at 10,000 × g for 10 min and washed twice with saline phosphate buffer. Chromosomal DNA was purified as described previously (17). Contaminating RNA was removed by treatment with RNases A and T1 followed by precipitation with polyethylene glycol 6000 (31).
Breakage points of the modified DNAs were revealed by a primer extension method adapted from Brewer and coauthors (2) as follows. A linear PCR using Taq polymerase was performed on chromosomal DNAs. Primer SP2 (5′-GCGGTATGGCTTCATCTATGTCG-3′) was used to probe the bottom strand of the bglH gene (22). End labeling was performed with (γ-32P)ATP and T4 polynucleotide kinase as described elsewhere (31). The primer extension reaction was carried out in a volume of 20 μl containing 150 ng of chromosomal DNA, 0.5 pmol of 32P-end-labeled oligonucleotide, 2 μl of 10× Taq polymerase reaction buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 20 mM MgCl2, 0.2% [wt/vol] gelatin), and a final concentration of 200 μM each deoxynucleoside triphosphate. The linear PCR and analysis of the products were performed as described previously (22).
Nucleotide sequence accession number.
The sequence of a 1,691-bp fragment, containing the ccpA gene and the 5′ end of the pepQ gene, was deposited in the EMBL databases under accession no. AJ310777.
RESULTS AND DISCUSSION
The ccpA locus of L. plantarum.
We isolated an 891-bp internal fragment of the L. plantarum ccpA gene by amplification of chromosomal DNA with two oligonucleotides designed on the basis of the B. megaterium ccpA sequence (see Materials and Methods). The 891-bp ccpA fragment was used to probe an enriched EcoRI-SalI L. plantarum chromosomal DNA library. Among the positive clones, one containing plasmid pLM10 carrying a 1.5-kb chromosomal fragment was isolated. The sequence analysis of the 1.5-kb fragment revealed the complete 3′ end of the ccpA gene. The nucleotide sequence of the ccpA 5′ end was completed by direct sequencing of an enriched 6-kb SalI fragment of chromosomal DNA. The deduced amino acid sequence (336 amino acids) showed 95% identity with CcpA of L. pentosus (20) and 63% identity with CcpA of L. casei (27). Downstream of the ccpA gene, an open reading frame whose putative product showed 69% identity with the proline peptidase coded by the L. pentosus pepQ gene was found. The ccpA and pepQ genes were found to be transcribed in the same orientation but on two separate transcriptional units, as determined by Northern blotting, by nucleotide sequence, and by primer extension analysis of the ccpA (see below) and pepQ (not shown) transcripts. Primer extension of the pepQ transcript allowed the determination of the promoter sequence and of the +1 transcriptional start, with the −35 sequence occurring 26 bases downstream of the ccpA stop codon (not shown). Northern blot analysis using the 891-bp internal fragment of the ccpA gene as probe showed the appearance of a single band of about 1.1 kb, indicating that ccpA is transcribed on a monocistronic unit (not shown). The finding that the gene order in L. plantarum is ccpA-pepQ, with the two genes transcribed in tandem, is quite surprising, since in all lactic acid bacteria so far analyzed, including the closely related L. pentosus, the gene order is pepQ-ccpA, with the two genes divergently transcribed (20, 34).
Disruption of the chromosomal ccpA gene.
To assess the role of CcpA in L. plantarum, we isolated a strain carrying a null mutation in the ccpA gene. Due to instability of mutants carrying sequence duplications derived from one-step chromosomal integration of suicide vectors (R. Marasco, unpublished data), we used a two-step homologous recombination process (7) leading to stable chromosomal disruption of ccpA. A copy of the ccpA gene truncated both at 5′ and 3′ ends and interrupted by the chloramphenicol resistance gene (cat) was cloned in the pJDC9 integrating vector, yielding plasmid pLM2. Clones in which the integration event had occurred were selected as chloramphenicol-erythromycin-resistant strains after electroporation of L. plantarum cells with plasmid pLM2 and were confirmed by PCR. Among these we chose clone LM3-1, in which the integration had occurred between the 3′ end of the ccpA fragment of pLM2 and the homologous chromosomal sequence. L. plantarum LM3-1 cells were grown in the presence of chloramphenicol for 40 generations, and the second homologous recombination event leading to excision of the plasmid was screened as loss of erythromycin resistance. We chose for further analysis one such strain, LM3-2, after confirming the excision event by PCR.
Effects of the ccpA null mutation.
To study the effects of the ccpA mutation on the regulation of catabolic pathways, the growth rate and two enzyme activities, β-glucosidase and β-galactosidase, were monitored under various growth conditions in the wild type and mutant strain.
The doubling time of the mutant strain was higher than that of the wild type with all sugars tested (glucose, ribose, and lactose), with a major effect found with glucose (data not shown). These growth defects found in the ccpA mutant might suggest the involvement of CcpA in gene activation as occurs for other lactic acid bacteria (19, 34), but this will need further investigations.
β-Glucosidase activity was analyzed by assaying the hydrolysis of salicin in whole cells of L. plantarum LM3 (wild type) and LM3-2 (ccpA::cat) grown in MRS medium supplemented with 2% glucose or 0.4% salicin. In agreement with our previous findings (22), salicin utilization in the wild-type strain, grown on glucose as a sole carbon source, was about 20-fold less than in cells grown on salicin. In contrast, a less than twofold decrease was detected in the ccpA mutant strain grown on glucose compared to salicin (Table 1).
TABLE 1.
β-Glucosidase activity in wild-type and mutant strains of L. plantarum
| Strain | Growth condition | Sp act (nmol of product formed/min/cell [1010])a |
|---|---|---|
| LM3 (wild type) | Salicin | 456 |
| Glucose | 25 | |
| LM3-2 (ccpA::cat) | Salicin | 428 |
| Glucose | 283 |
Means of three independent experiment for which standard deviations did not exceed 5%. A saturating substrate concentration (30 mM salicin) was used.
Table 2 shows that while a basal β-galactosidase activity in ribose-grown cells was induced by lactose in both the wild type and the ccpA mutant, and growth on glucose repressed to near zero both basal and induced wild-type activity, a marginal repression of only the lactose-induced activity was observed in the mutant.
TABLE 2.
β-Galactosidase activity in wild-type and mutant strains of L. plantarum
| Strain | Growth condition | Sp act (nmol of o-nitrophenol released/min/OD600 unit of cell culture)a |
|---|---|---|
| LM3 (wild type) | Lactose | 1,240 |
| Glucose | 1.6 | |
| Ribose | 56 | |
| Lactose + glucose | 2.4 | |
| LM3-2 (ccpA::cat) | Lactose | 533 |
| Glucose | 60 | |
| Ribose | 58 | |
| Lactose + glucose | 235 |
Means of three independent experiment for which standard deviations did not exceed 5%. OD600, optical density at 600 nm.
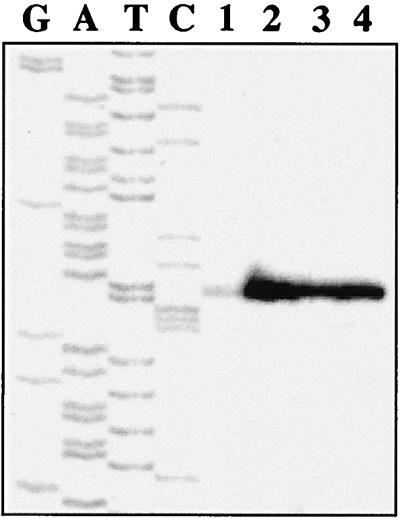
To verify the effect of the ccpA null mutation on transcription of the bglH gene, coding the β-glucosidase BglH, a primer extension was performed on mRNA from wild-type and mutant strains grown in repressing and nonrepressing conditions. Figure 1 shows that bglH transcription was repressed about 10-fold in wild-type cells grown on glucose versus ribose, while the same level of bglH transcription was found in the mutant strain grown on both sugars.
FIG. 1.
Transcriptional regulation of L. plantarum bglH. Primer extension products were obtained by using oligonucleotide SP1 and total RNA extracted from LM3 cells grown on glucose (lane 1) or ribose (lane 2) and from LM3-2 cells grown on glucose (lane 3) or ribose (lane 4). As a reference, sequencing reactions were performed with the same primer.
These results demonstrate that expression of the genes coding the two enzymes is regulated by CcpA-mediated CCR and that for bglH, this control occurs at the transcriptional level. The residual glucose effect on the two tested enzyme activities found in mutant strain LM3-2 may be due to other CcpA-independent regulatory mechanisms such as inducer exclusion or expulsion (29, 30). Although in principle this effect may also be due to an additional transcription factor (3), results of the primer extension experiment suggest that this hypothesis is excluded, at least for bglH transcription (Fig. 1).
Autogenous regulation of the ccpA gene.
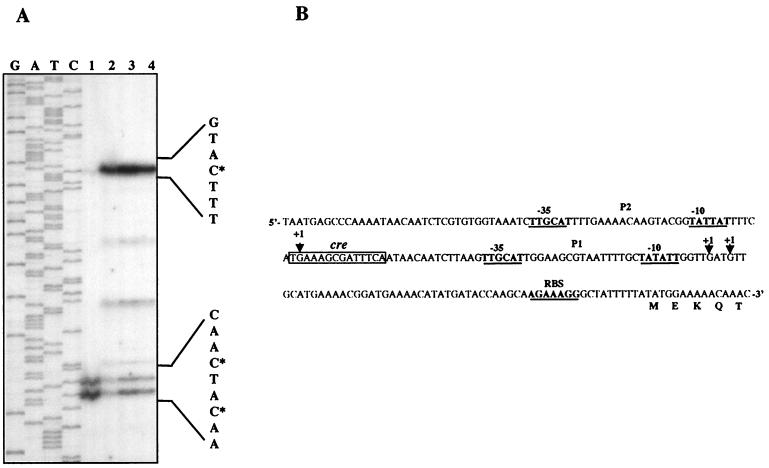
To determine the promoter sequence and study ccpA expression in various growth conditions, a primer extension analysis was performed on total RNA from L. plantarum cells grown in MRS medium supplemented with glucose (Fig. 2A, lane 1) or ribose (Fig. 2A, lane 2). The results indicate that ccpA is transcribed from three major transcriptional start points, depending on the carbon source used. In the presence of glucose, a double band revealed two adjacent start points driven by the same promoter, P1 (Fig. 2). Growth on ribose favored the use of an alternative upstream transcriptional start point, which allowed us to define promoter P2 (Fig. 2). The upstream transcriptional start point overlaps with the second base of a cre site which fully matches the consensus sequence (Fig. 2B). This finding may suggest that the activity of the upstream promoter P2 is down-regulated in the presence of an activated CcpA protein which occurs during growth on glucose, leading to autogenous regulation of the gene. This was further investigated by testing ccpA transcription in mutant strain LM3-2. In the absence of CcpA, P2 was the main functional promoter driving transcription from the upstream start point during growth on either glucose (Fig. 2A, lane 3) or ribose (Fig. 2A, lane 4), which confirms autogenous regulation of the gene. Autogenous regulation of ccpA has been described for the closely related L. pentosus (20), sharing 95% identity with L. plantarum CcpA, and for Staphylococcus xylosus (6), while constitutive expression of ccpA has been found in B. subtilis (26), B. megaterium (13), L. casei (27), and Streptococcus thermophilus (34).
FIG. 2.
Transcriptional regulation of L. plantarum ccpA. (A) Primer extension analysis of ccpA mRNA. Primer extension products were obtained by using oligonucleotide A3 and total RNA extracted from LM3 cells grown on glucose (lane 1) or ribose (lane 2) and from LM3-2 cells grown on glucose (lane 3) or ribose (lane 4). Start points of transcription are indicated with asterisks. As a reference, sequencing reactions were performed with the same primer. (B) Nucleotide sequence of the ccpA promoter region. Putative ribosome-binding site (RBS) and promoter (P1 and P2) sequences are labeled. A cre motif is boxed. Triangles indicate transcriptional start points. The N-terminal CcpA sequence is shown.
The presence of two promoters, one of which active during CCR, seems to ensure constant levels of ccpA transcripts under different growth conditions.
Analysis of CcpA-cre binding.
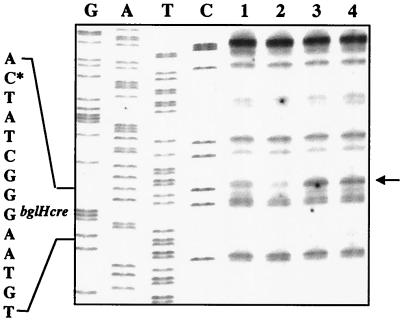
To assess the function of CcpA in DNA binding, we performed an in vivo footprinting analysis of a cre sequence in wild-type and ccpA mutant cells. Our previous in vivo footprinting experiments on the cre-containing regulatory region of the L. plantarum bglH gene strongly suggested the occurrence of CcpA-cre binding (22). We extended this analysis to both the wild-type and ccpA mutant. Chromosomal DNAs of LM3 and LM3-2 cells grown on glucose or on salicin were methylated with DMS during exponential growth. The analysis was focused on the G residue in position 13 (bottom strand) of the bglH cre protected from DMS attack during growth in the presence of glucose (22). The experiment in Fig. 3 shows that the analyzed G residue was protected in wild-type cells grown on glucose (lane 2) but not in those grown on salicin (lane 1), while no protection was observed in the mutant cells grown on either salicin (lane 3) or glucose (lane 4). This result demonstrates the involvement of CcpA in transcriptional regulation of the bglH gene.
FIG. 3.
In vivo footprinting analysis of the bglH regulatory region. The analysis was performed on methylated DNA extracted from LM3 cells grown on salicin (lane 1) or glucose (lane 2) and from LM3-2 cells grown on salicin (lane 3) or glucose (lane 4). Lanes G, A, T, and C indicate the nucleotide sequencing reactions for the bottom strand (G's in the methylated strand correspond to C's in the sequencing lane). The arrow points to the G residue protected from DMS attack, and the asterisk indicates its relative position in the cre sequence.
To our knowledge, this is the first demonstration of the occurrence of CcpA-cre binding in vivo.
ACKNOWLEDGMENTS
We thank M. Valenzi for computer assistance and C. Sole for technical assistance.
This work was supported by MIRAAF, “Piano Nazionale Biotecnologie Vegetali.” Partial support was also obtained from CNR, Progetto Finalizzato Biotecnologie, MURST-PRIN 1999, and MURST-PRIN 2000.
REFERENCES
- 1.Bengmark S. Colonic food: pre- and probiotics. Am J Gastroenterol. 2000;95:5–7. doi: 10.1016/s0002-9270(99)00807-2. [DOI] [PubMed] [Google Scholar]
- 2.Brewer A C, March P J, Patient R K. A simplified method for in vivo footprinting using DMS. Nucleic Acids Res. 1990;18:5574. doi: 10.1093/nar/18.18.5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chauvaux S, Paulsen I T, Saier M H., Jr CcpB, a novel transcription factor implicated in catabolite repression in Bacillus subtilis. J Bacteriol. 1998;180:491–497. doi: 10.1128/jb.180.3.491-497.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J D, Morrison D A. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Strepcoccus pneumoniae. Gene. 1988;64:155–164. doi: 10.1016/0378-1119(88)90489-1. [DOI] [PubMed] [Google Scholar]
- 5.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 6.Egeter O, Brückner R. Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol Microbiol. 1996;21:739–749. doi: 10.1046/j.1365-2958.1996.301398.x. [DOI] [PubMed] [Google Scholar]
- 7.Ferain T, Hobbs J N, Richardson J, Bernard N, Garmyn D, Hols P, Allen N E, Delcour J. Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum. J Bacteriol. 1996;178:5431–5437. doi: 10.1128/jb.178.18.5431-5437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geoffroy M C, Guyard C, Quatannens B, Pavan S, Lange M, Mercenier A. Use of green fluorescent protein to tag lactic acid bacterium strains under development as live vaccine vectors. Appl Environ Microbiol. 2000;66:383–391. doi: 10.1128/aem.66.1.383-391.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gösseringer R, Küster E, Galinier A, Deutscher J, Hillen W. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium results from sensing two different signals. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 10.Henkin T M. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1996;135:9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 11.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 12.Hueck C J, Hillen W, Saier M H., Jr Analysis of a cis-active sequence mediating catabolite repression in Gram-positive bacteria. Res Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 13.Hueck C J, Kraus A, Schmiedel D, Hillen W. Cloning expression and functional analysis of the catabolite control protein CcpA from Bacillus megaterium. Mol Microbiol. 1995;16:855–864. doi: 10.1111/j.1365-2958.1995.tb02313.x. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Voskuil M I, Chambliss G H. NADP, corepressor for the Bacillus catabolite control protein CcpA. Proc Natl Acad Sci USA. 1998;95:9590–9595. doi: 10.1073/pnas.95.16.9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Küster E, Hilbich T, Dahl M K, Hillen W. Mutations in catabolite control protein CcpA separating growth effects from catabolite repression. J Bacteriol. 1999;181:4125–4128. doi: 10.1128/jb.181.13.4125-4128.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leboeuf C, Leblanc L, Auffray Y, Hartke A. Characterization of the ccpA gene of Enterococcus faecalis: identification of starvation-inducible proteins regulated by CcpA. J Bacteriol. 2000;182:5799–5806. doi: 10.1128/jb.182.20.5799-5806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leer R J, Christiaens H, Verstraete W, Peters L, Posno M, Pouwels P H. Gene disruption in Lactobacillus plantarum strain 80 by site-specific recombination: isolation of a mutant strain deficient in conjugated bile salt hydrolase activity. Mol Gen Genet. 1993;239:269–272. doi: 10.1007/BF00281627. [DOI] [PubMed] [Google Scholar]
- 18.Leong-Morgenthaler P, Zwahlen M C, Hottinger H. Lactose metabolism in Lactobacillus bulgaricus: analysis of the primary structure and expression of the genes involved. J Bacteriol. 1991;173:1951–1957. doi: 10.1128/jb.173.6.1951-1957.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luesink E, van Herpen R, Grossiord B, Kuipers O P, de Vos W. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol Microbiol. 1998;30:789–798. doi: 10.1046/j.1365-2958.1998.01111.x. [DOI] [PubMed] [Google Scholar]
- 20.Mahr K, Hillen W, Titgemeyer F. Carbon catabolite repression in Lactobacillus pentosus: analysis of the ccpA region. Appl Environ Microbiol. 2000;66:277–283. doi: 10.1128/aem.66.1.277-283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marasco R, Lago C T, De Felice M. Utilization of cellobiose and other β-d-glucosides in Agrobacterium tumefaciens. Res Microbiol. 1995;146:485–492. doi: 10.1016/0923-2508(96)80294-4. [DOI] [PubMed] [Google Scholar]
- 22.Marasco R, Muscariello L, Varcamonti M, De Felice M, Sacco M. Expression of the bglH gene of Lactobacillus plantarum is controlled by carbon catabolite repression. J Bacteriol. 1998;180:3400–3404. doi: 10.1128/jb.180.13.3400-3404.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marasco R, Salatiello I, De Felice M, Sacco M. A physical and functional analysis of the newly identified bglGPT operon of Lactobacillus plantarum. FEMS Microbiol Lett. 2000;186:269–273. doi: 10.1111/j.1574-6968.2000.tb09116.x. [DOI] [PubMed] [Google Scholar]
- 24.Martin-Verstraete I, Deutscher J, Galinier A. Phosphorylation of HPr and Crh by HPrK, early steps in the catabolite repression signalling pathway for the Bacillus subtilis levanase operon. J Bacteriol. 1999;181:2966–2969. doi: 10.1128/jb.181.9.2966-2969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26.Miwa Y, Saikawa M, Fujita Y. Possible function and some properties of the CcpA protein of Bacillus subtilis. Microbiology. 1994;140:2567–2575. doi: 10.1099/00221287-140-10-2567. [DOI] [PubMed] [Google Scholar]
- 27.Monedero V, Gosalbes J M, Perez-Martinez G. Catabolite repression in Lactobacillus casei ATCC393 is mediated by CcpA. J Bacteriol. 1997;179:6657–6664. doi: 10.1128/jb.179.21.6657-6664.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reizer J, Hoischen C, Titgemeyer F, Rivolta C, Rabus R, Stülke J, Karamata D, Saier M H, Jr, Hillen W. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol Microbiol. 1998;27:1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 29.Saier M H., Jr Regulation of carbon metabolism in bacteria, a Forum in Microbiology. Res Microbiol. 1996;147:439–587. [PubMed] [Google Scholar]
- 30.Saier M H., Jr Multiple mechanisms controlling carbon metabolism in bacteria. Biotechnol Bioeng. 1998;58:170–174. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<170::aid-bit9>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Simpson C L, Russel R R. Identification of a homolog of CcpA catabolite repressor protein in Streptococcus mutans. Infect Immun. 1998;66:2085–2092. doi: 10.1128/iai.66.5.2085-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turinsky A J, Grundy F J, Kim J, Chamblis G H, Henkin T M. Transcriptional activation of the Bacillus subtilis ackA gene requires sequences upstream of the promoter. J Bacteriol. 1998;180:5961–5967. doi: 10.1128/jb.180.22.5961-5967.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Bogaard P, Kleerebezem M, Kuipers O P, de Vos W M. Control of lactose transport, β-galactosidase activity, and glycolysis by CcpA in Streptococcus thermophilus: evidence for carbon catabolite repression by a non-phosphoenolpyruvate-dependent phosphotransferase system sugar. J Bacteriol. 2000;182:5982–5989. doi: 10.1128/jb.182.21.5982-5989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wouters J A, Kamphuis H H, Hugenholts J, Kuipers O P, de Vos W M, Abee T. Changes in glycolytic activity of Lactococcus lactis induced by low temperature. Appl Environ Microbiol. 2000;66:3686–3691. doi: 10.1128/aem.66.9.3686-3691.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zelieckas J M, Wray L V, Jr, Fisher S H. Expression of the Bacillus subtilis acsA gene: position and sequence context affect cre-mediated carbon catabolite repression. J Bacteriol. 1998;180:6649–6654. doi: 10.1128/jb.180.24.6649-6654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]