Abstract
Cdc2 kinase is a master regulator of cell cycle progression in the fission yeast Schizosaccharomyces pombe. Our data indicate that Cdc2 phosphorylates replication factor Orp2, a subunit of the origin recognition complex (ORC). Cdc2 phosphorylation of Orp2 appears to be one of multiple mechanisms by which Cdc2 prevents DNA rereplication in a single cell cycle. Cdc2 phosphorylation of Orp2 is not required for Cdc2 to activate DNA replication initiation. Phosphorylation of Orp2 appears first in S phase and becomes maximal in G2 and M when Cdc2 kinase activity is required to prevent reinitiation of DNA replication. A mutant lacking Cdc2 phosphorylation sites in Orp2 (orp2-T4A) allowed greater rereplication of DNA than congenic orp2 wild-type strains when the limiting replication initiation factor Cdc18 was deregulated. Thus, Cdc2 phosphorylation of Orp2 may be redundant with regulation of Cdc18 for preventing reinitiation of DNA synthesis. Since Cdc2 phosphorylation sites are present in Orp2 (also known as Orc2) from yeasts to metazoans, we propose that cell cycle-regulated phosphorylation of the ORC provides a safety net to prevent DNA rereplication and resulting genetic instability.
Cyclin-dependent kinases (CDKs) are essential activators of replication initiation and are also required for once-per-cell-cycle control of DNA replication. Eukaryotic DNA replication initiates at many replication origins so that many replication forks can work simultaneously. This allows replication of large genomes in a short S phase. However, replication of each origin must be limited to once per cell cycle to maintain ploidy and genome stability.
In the yeasts Schizosaccharomyces pombe and Saccharomyces cerevisiae, initiation depends on DNA sequences called autonomously replicating sequences. To function as a replication origin, autonomously replicating sequence DNA must bind the origin recognition complex (ORC) (3). The ORC has six protein subunits named Orc1 through Orc6 and is an essential replication initiator in yeasts as well as metazoans (reviewed in reference 64). At least in some systems, the ORC stays bound to DNA throughout the cell cycle (2, 17, 45, 57). Beginning in late M or early G1 phase, the ORC recruits additional proteins so as to form a large preinitiation complex (reviewed in reference 54). One of the first of these additional proteins is Cdc18 (S. pombe name; also known as Cdc6 in S. cerevisiae) (11, 42, 63). Cdc18 is likely to be an ATPase, and is thought to function with the ORC to load the six MCM proteins (Mcm2 through Mcm7) onto the chromatin (12, 61). The MCMs may serve as the replicative helicase (reviewed in reference 72). Other essential initiator proteins that associate with the ORC include Cdc45, Mcm10, and Cdt1 (reviewed in references 39 and 70).
The preinitiation complex is apparently fully assembled in G1 phase, and yet initiation does not occur. Initiation is not triggered until two protein kinases become active. These kinases are Hsk1 (S. pombe name; also known as Cdc7) and Cdc2 (also known as Cdc28 in S. cerevisiae). Cdc2 is a cyclin-dependent kinase, and at different times of the cell cycle is activated by various different cyclins (reviewed in references 31, 48, and 67).
In S. pombe, much of the cell cycle is controlled by Cdc2 and its various cyclin partners. For instance, Cdc2-cyclin complexes control not only DNA replication, but also growth polarity, spindle pole body duplication, chromosome condensation, mitotic spindle functions, mitosis, and cytokinesis (56). Though much has been learned about how Cdc2 kinase itself is regulated, relatively little is understood about how this kinase in turn regulates the many events of the cell cycle, and it is not well-known what proteins are phosphorylated by Cdc2 to accomplish the many cell cycle transitions under Cdc2 control. In particular, while it is clear that initiation of replication depends on Cdc2 kinase activity, it is not known what proteins are phosphorylated by Cdc2 to trigger replication. In view of the global regulation of the cell cycle by Cdc2, regulation of replication by Cdc2 might be indirect. However, experiments in S. cerevisiae provide strong evidence that Cdc28 activity is required throughout S phase for the firing of individual replication origins, and this suggests a direct involvement (19).
Cdc2 kinase activity is low during G1 phase when the preinitiation complex is assembled, in part because of a lack of cyclin and in part because of high levels of the Cdc2 inhibitor Rum1. Later, after assembly of the preinitiation complex, Cdc2 kinase is activated when the cyclins Puc1 and Cig2 are synthesized and when Rum1 is degraded. This Cdc2 kinase activity (along with Hsk1 kinase activity) allows the preinitiation complex to fire, triggering S phase (24, 46, 51). Cyclins accumulate during S phase and G2 (15), but full Cdc2 activity is held in check by inhibitory phosphorylation by the Wee1 and Mik1 kinases. At the end of G2, Cdc2 binds the mitotic cyclin Cdc13 and is dephosphorylated by the Cdc25 phosphatase. This generates a high level of Cdc2 kinase activity sufficient for mitosis (reviewed in reference 4). At the end of mitosis, Cdc2 is inactivated by several mechanisms, including destruction of cyclin, expression of Rum1, and possibly removal of the activating T-loop phosphorylation (5, 13, 41, 52).
Strikingly, the low level of Cdc2 kinase activity in G1 is probably essential for assembly of the preinitiation complex; that is, when Cdc28 kinase is activated in G1 by some experimental manipulation, it prevents formation of the preinitiation complex (63). In addition, if Cdc2 or Cdc28 kinase activity is artificially lowered in G2, then preinitiation complexes re-form and can fire, leading to rereplication (8, 16, 27). Thus, the cycle in Cdc2 kinase activity (low in G1 phase, moderate in S and G2, high in M) is tightly linked to the cycle in replication. The low kinase activity in G1 allows preinitiation complexes to form, and the appearance of Cdc2 kinase activity in S allows them to fire. At the same time, this moderate or high Cdc2 kinase activity in S, G2, and M prevents preinitiation complexes from re-forming, and so rigorously prevents any rereplication.
In light of the links between replication and Cdc2 kinase, it is notable that several components of the preinitiation complex have conserved clusters of sites for Cdc2 phosphorylation. These components include Orp2 (also known as Orc2), Cdc18 (also known as Cdc6), Mcm4, and Mcm10. At present, there is no evidence that phosphorylation of any of these sites is involved in triggering initiation. However, there is evidence that some of these sites in MCMs and Cdc18 help prevent DNA rereplication by preventing assembly of new preinitiation complexes once S phase has begun. In particular, Cdc18 phosphorylated by Cdc2 is inactivated via ubiquitin-mediated degradation (21, 23, 30, 32, 37). Degradation is not the only way to inactivate Cdc18, and in human and Xenopus laevis cells, much of the Cdc6 is not degraded but is exported from the nucleus apparently as a result of CDK phosphorylation (60, 62, 66). In budding yeast, phosphorylation of Cdc6 and MCMs probably masks nuclear localization signals on these proteins, rendering them cytoplasmic (33, 38, 55). Cdc18 inactivation and inhibition of MCM function are two mechanisms for preventing assembly (or reassembly) of the preinitiation complex once Cdc2 kinase is active.
Although the Cdc2 phosphorylation sites in Cdc18 are involved in preventing reinitiation, they certainly are not the only such mechanism, since mutant proteins lacking the phosphorylation sites do not allow rereplication (at least when expressed at wild-type levels) (21, 44, 60, 71). Furthermore in S. pombe, unlike S. cerevisiae, the MCM proteins are constitutively nuclear (28, 55, 58, 59, 68). Thus, there must be additional, undiscovered controls preventing reinitiation. One such control in higher eukaryotes involves binding of an inhibitor, geminin, to the Cdt1 factor; however, no analogous pathway is known in yeasts (50, 69, 73). Existence of multiple pathways is not surprising, since even a small degree of reinitiation could have serious effects on genome stability.
We have found that S. pombe Cdc2 interacts with the S. pombe ORC protein Orp2 (the homolog of Orc2). Based on this observation, we proposed that Cdc2 regulates DNA replication directly at replication origins (40). Orp2 and its homologs have a conserved N-terminal cluster of consensus Cdc2 phosphorylation sites. Thus, since (i) Orp2 binds to Cdc2, (ii) Orp2 has appropriate sites for phosphorylation by Cdc2, and (iii) Cdc2 is known to trigger replication and to prevent rereplication, we asked whether phosphorylation of Orp2 by Cdc2 was involved in these processes. We found that Orp2 is phosphorylated by Cdc2, assessed the phosphorylation of Orp2 through the cell cycle, mutated the Cdc2 phosphorylation sites in Orp2, and investigated the biological impact of these mutations.
MATERIALS AND METHODS
Yeast methods, strains, and plasmids.
S. pombe methods were essentially as described by Moreno et al. (53): YES is rich medium, and EMM2 is defined medium and was supplemented with leucine (L), uracil (U), adenine (A), and histidine (H) as indicated. YSO is rich medium low in adenine (36). The vitamin B1 thiamine (B1) was added to 2.7 mg/liter as indicated. Genotypes are listed in Table 1.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| FWP162 | h+leu1-32 ura4-294 | C. Hoffman |
| JLP46 | h+/h−ade6-M216/ade6-M210 ura4-D18/ura4-D18 leu1-32/leu1-32 his7-366/his7-366 orp2+/orp2Δ::ura4+ | 40 |
| JLP52 | h+/h−ade6-M216/ade6-M210 ura4+/ura4-D18 leu1-32/leu1-32 his7-366/his7-366 orp2+/orp+ | 40 |
| JLP166 | h−ade6-M216 ura4-D18 leu1-32 his7-366 orp2Δ::ura4+porp2TKade | 36 |
| JLP230 | h−ade6-M216 ura4-D18 leu1-32 his7-366 orp2Δ::ura4+::orp2+ (leu1+) | This study |
| JLP238 | h−ade6-M210 ura4-D18 leu1-32 his7-366 orp2Δ::ura4+::orp2-T4A (leu1+) | This study |
| JLP306 | h−ade6-M216 leu1-32 his7-366 orp2Δ::orp2-T4A (leu1+) | This study |
| JLP308 | h−ade6-M210 leu1-32 his7-366 orp2Δ::orp2+ (leu1+) | This study |
| JLP515 | h+leu1-32 ura4-294::GST-cdc18-T4A (ura4+) | This study |
| JLP518 | ade6-M216 leu1-32 orp2Δ::orp2-T4A (leu1+) ura4-294::GST-cdc18-T4A (ura4+) | This study |
| JLP519 | ade6-M216 leu1-32 his7-366 orp2Δ::orp2-T4A (leu1+) ura4-294::GST-cdc18-T4A (ura4+) | This study |
| JLP523 | ade6-M210 leu1-32 orp2Δ::orp2+ (leu1+) ura4-294::GST-cdc18-T4A (ura4+) | This study |
| JLP524 | ade6-M210 leu1-32 his7-366 orp2Δ::orp2+ (leu1+) ura4-294::GST-cdc18-T4A (ura4+) | This study |
Potential phosphorylation sites [(S/T)P] encoded by orp2 were mutated by site-directed mutagenesis using PCR. To generate orp2-T4A, plasmid pJL207 (orp2+ cloned into pREP3 [40]) was used as a template. Primers P1 and P2 amplified the 5′ coding region of orp2+; P2 has nucleotide changes that altered Thr to Ala. Separately, primers P3 and P4 were used to amplify the region of orp2+ 3′ of the sequence amplified by P1-P2. The P3 primer has nucleotide changes to alter two Thr codons to Ala codons and introduces an XhoI site. The P1-P2 and P3-P4 products were gel isolated, phosphorylated, and ligated together, and the ligation product was amplified using P1 and P4. This product was cloned to replace the corresponding region of the orp2+, and the clone was sequenced. Resulting amino acid changes are shown in Fig. 1. The mutated region was subcloned into appropriate orp2+ vectors as follows: pJL211 (pRep1-GST-orp2 [40]) was changed to make pJL329 (pRep1-GST-orp2-T4A). pJL320 (genomic orp2+, leu1+ integrating vector [see below]) was changed to make pJL322 (genomic orp2-T4A, leu1+ integrating vector). Primer sequences were P1 (CTGAAGATACGTTTTCAACCATTTTTAAAG), P2 (CGCCTAAAAAAGGAGTTCTTGAAGAT), P3 (CAACAACTTCTGGTGCTAATGTAGAAGG), and P4 (CTCCTCGAGCACCAGGACATCGAA).
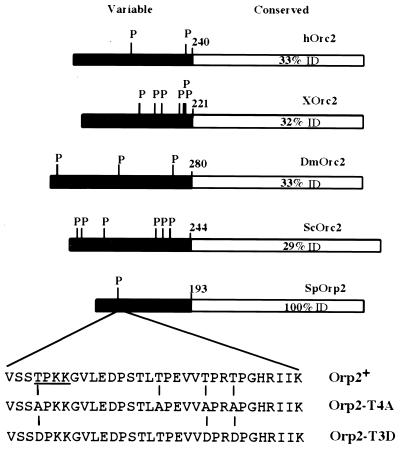
FIG. 1.
Cdc2 phosphorylation sites in Orc2 homologs. The N-terminal region shown in black is least conserved. The beginning of the conserved region is indicated (amino acid 193 in Orp2). Bars are drawn to scale. Percent amino acid identity (ID) of the various C-terminal domains compared with Orp2 is shown. P represents consensus Cdc2 phosphorylation sites [(S/T)PX(K/R)]; SP and TP sites that are not perfect matches to the consensus are not shown. The amino acid sequences of Orp2 wild type, Orp2-T4A, and Orp2-T3D show the mutations made, and the single consensus Cdc2 phosphorylation site in Orp2 is underlined. Abbreviations for homologs: h, human; X, Xenopus; Dm, D. melanogaster; Sc, S. cerevisiae; Sp, S. pombe.
To express alleles of orp2 from the orp2 promoter in the genome, an orp2+ leu1+ vector (pJL320) was constructed. pJL320 contains the orp2 promoter and coding sequence followed by the nmt1 terminator cloned between the PstI-SacI sites of integrating vector pJK148 (leu1+) (35). Steps in construction of pJL320 were as follows: The 1.7-kb PstI-NdeI fragment from pER17 (40) was cloned into PstI/NdeI-digested pJL207 (pREP3-orp2+) (40), thus replacing the nmt1 promoter with the orp2 promoter. From this, the PstI-SacI fragment containing the orp2 promoter and coding sequence and the nmt1 terminator was moved into PstI-SacI-digested pJK148 to create pJL320. pJL320 was cut with PflMI (in the orp2 5′ noncoding region) to target integration at orp2Δ::ura4+ to create orp2Δ::ura4+::orp2+ (leu1+).
Construction of JLP230 orp2+ and JLP238 orp2-T4A strains was as follows: plasmids pJL320 (orp2+ leu1+) and pJL322 (orp2-T4A leu1+) were integrated at the orp2Δ::ura4+ locus in diploid JLP46. Transformants were sporulated, and strains JLP230 and JLP238 were isolated and checked by Southern blot analysis. JLP306 and 308 are Ura− derivatives of JLP230 and JLP238 made by one-step gene replacement of the ura4+ within the orp2Δ::ura4+ by transformation with HindIII-digested pJL378 (pJL378 is an unmarked deletion of orp2 coding sequences cloned into the HindIII site of pBluescript). Ura− transformants were obtained using YES–0.1% 5-fluoroorotic acid to create orp2Δ::ura4+::orp2+ (leu1+).
Construction of nmt1GST-Cdc18* strains was as follows: pAL27-GST-cdc18-T4A has Thr codons at positions 26, 98, 104 and 134 mutated to Ala (44). pAL27-GST-cdc18-T4A was linearized with StuI and integrated at ura4-294 in strain FWP162 h+ leu1-32 ura4-294. GST-Cdc18-T4A is referred to as Cdc18*. The transformant JLP515 h+ leu1-32 ura4-294::GST-cdc18-T4A, ura4+was selected on plates lacking Ura, and stable integrants were identified as isolates that failed to grow on 0.1% 5-fluoroorotic acid–YES plates. JLP515 was mated to JLP306 (orp2Δ::orp2-T4A leu1+) to obtain strains JLP518, JLP519, and additional congenic isolates. JLP515 was mated to JLP308 (orp2Δ::orp2+ leu1+) to obtain JLP523, JLP524, and additional congenic isolates. All isolates were obtained by tetrad dissection.
Cdc18 induction and flow cytometry.
For induction of Cdc18* from the nmt1 promoter (49), cells were grown overnight in YES-thiamine. This preculture was used to inoculate EMM2-LAUH-B1 to an optical density at 600 nm (OD600) of 0.1 and grown for at least 10 h at 32°C to an OD600 of not more than 1.0. Cells were harvested and washed twice in water to remove thiamine. Cells were reinoculated into EMM2-LAUH (without thiamine) to a calculated OD600 of 0.01. These cultures were grown at 32°C and harvested for flow cytometry, Western blotting, and microscopy at the times indicated. Cells were processed for flow cytometry as described (14) except that cells were stained with 1 μM Sytox Green (Molecular Probes) instead of propidium iodide. Fluorescence was analyzed using a Becton Dickinson FACScan and CellQuest software.
Competitive transformations.
JLP306 and JLP308 were grown separately overnight at 30°C in 5 ml of YES and then were mixed 1:1, spread on a YES plate, and grown overnight at 30°C. For each transformation, approximately 5 μl of cells from the plate was mixed with 150 μl of 0.1 M lithium acetate (pH 4.7) and incubated at 25°C for 1 h; 0.5 to 1 μg of plasmid DNA was added; 350 μl of 50% polyethylene glycol was added; and cells were incubated overnight at 25°C, harvested, washed gently with EMM2, and resuspended in 1 ml of EMM2. A 5- to 10-μl aliquot of this suspension was diluted and spread for single colonies on EMM2-LAUH (total viable), and the remainder was spread on four to six plates of EMM2-LAH (transformants). Colonies were picked from EMM2-LAH (transformants) or EMM2-LAUH (total) onto YSO. Red or pink colonies on YSO were counted after 2 days.
Antibodies and immunoblotting.
GST-Orp2 made in Escherichia coli was used to raise rabbit antibodies. The 1.3-kb XhoI fragment from pACT-166 (which contains orp2 cDNA cloned into the two-hybrid vector pAS1) (40) was subcloned into the SalI site of pGEX-KG (Novagen). The resulting glutathione S-transferase (GST)–Orp2 fusion protein was produced in E. coli (BL21), partially purified by binding to GSH-Sepharose (Pharmacia), and used to raise rabbit polyclonal antisera (26). Crude antisera readily detected Orp2 overexpressed in fission yeast. To detect wild-type levels of Orp2, antibody was affinity purified by binding to purified GST-Orp2 protein immobilized on a nylon membrane. Bound antibodies were eluted with 100 mM glycine (pH 2.0), neutralized with Tris (pH 8.5) and stored in 50% glycerol at −20°C, and used at a 1:100 dilution.
To distinguish which immunoreactive bands were due to Orp2 protein, extract from cells lacking the orp2+ gene were compared to extracts from wild-type cells and to extracts from cells overexpressing orp2+ (Fig. 2B and data not shown). Because orp2+ is essential, selective spore germination was used to obtain the population of cells lacking the gene (orp2Δ::ura4+), as described (40). Other experiments with epitope-tagged Orp2 have since confirmed the assignment of Orp2 specific bands. Strong cross-reacting bands were not detected when the primary antibody incubation included sarcosyl and sodium dodecyl sulfate (SDS) (Fig. 3A); however, antibody activity was rapidly lost in this solution. Standard Tris-buffered saline–0.3% Tween 20–5% powdered milk (26) was used in all other experiments. For analysis of GST-Cdc18*, whole-cell extracts were made by vortexing with glass beads as described (40), total protein was quantitated using Bradford's reagent (Bio-Rad), and 10 μg of protein was loaded per lane on an SDS–10% polyacrylamide gel. GST-Cdc18* was detected using rabbit anti-GST (gift of L. Hengst) at a 1:500 dilution.
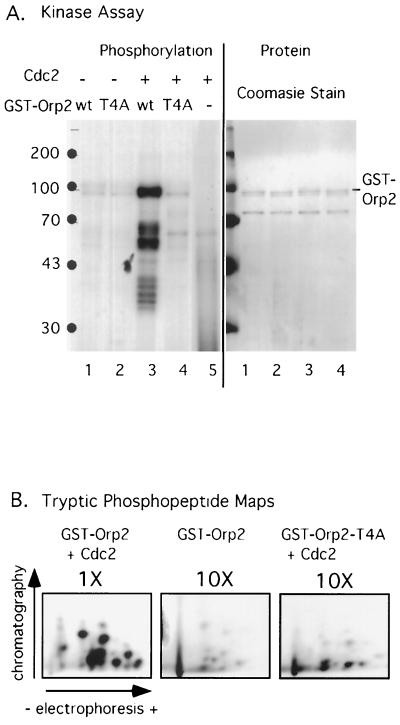
FIG. 2.
In vitro phosphorylation of Orp2 by Cdc2 kinase. (A) GST-Orp2+ (wt) and GST-Orp2-T4A (T4A) were incubated with S. pombe Cdc2 and [γ-32P]ATP and analyzed by SDS-PAGE. Incorporation of 32P into GST-Orp2 was visualized by autoradiography (left). Purity and relative amounts of GST-Orp2 and GST-Orp2-T4A added to the reactions were seen by Coomassie staining (right). (B) Phosphopeptide maps of proteins from kinase reactions analyzed in panel A. The GST-Orp2 or GST-Orp2-T4A bands were cut out of the gel, digested with trypsin, and analyzed. To visualize phosphopeptides, 10-fold more material (10×) was analyzed for the low-level reactions with GST-Orp2 (no kinase) and GST-Orp2-T4A (site mutant) than for GST-Orp2 plus Cdc2.
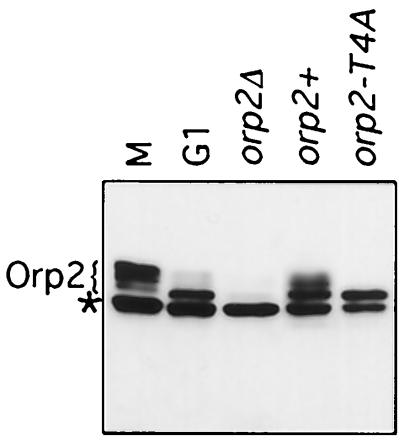
FIG. 3.
In vivo modification of Orp2 requires sites needed for Orp2 phosphorylation in vitro. Endogenous Orp2 protein in whole-cell extract was analyzed by SDS-PAGE and immunoblotting using Orp2 antibodies. Several mobility forms of Orp2 were observed. Orp2-specific bands are indicated. A cross-reacting band that is not Orp2 is present in all cells including those lacking Orp2 (orp2Δ) and is marked (*). Lanes are as follows: M, nuc2 mutant cells arrested in mitosis; G1, cdc10 mutant cells arrested in G1; orp2Δ, selective spore germination and growth of cells lacking orp2; orp2+, wild type; orp2-T4A, mutant in which orp2-T4A is the only orp2 gene in the cells, strain JLP238.
Kinase assays and analysis.
GST, GST-Orp2, and GST-Orp2-T4A were expressed in S. pombe from plasmids pJL205, pJL211 (40), and pJL329 (this study). GST proteins were isolated from yeast lysates by binding GSH-Sepharose (40), except that lysis and binding were in NETN.5 (50 mM Tris [pH 8], 500 mM NaCl, 5 mM EDTA, 10% glycerol, 1% NP-40) with protease and phosphatase inhibitors as previously described. Bound complexes were washed four times with NETN.5, and GST proteins were eluted with 40 mM glutathione (pH 8.0) and dialyzed against a solution containing 50 mM Tris (pH 8), 150 mM NaCl, 50% glycerol, and 1% NP-40.
Cdc2 kinase was obtained from S. pombe nuc2 mutant cells arrested at 36.5°C for 4 h. Anti-Cdc13 (gift of P. Russell) bound to protein A-Sepharose or p13suc1 beads (R. Aligue and P. Russell) were used to precipitate Cdc2 kinase. Assays were as described (52).
Phosphorylation of GST fusion proteins was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), and the 32P-labeled GST-Orp2 and GST-Orp2-T4A bands were excised after autoradiography. Tryptic peptide mapping was performed as described (7) using pH 1.9 buffer for the first dimension of electrophoresis and phosphochromatography buffer for the second dimension of ascending chromatography. Equal amounts of radioactivity were analyzed for each map, so it was necessary to use 1/10 as much of the Cdc2-GST-Orp2 sample as for the other samples. Phosphoamino acid analysis was performed as described (7), and results were quantitated using a phosphoimager (Molecular Dynamics).
RESULTS
Cdc2 phosphorylation of Orc2.
Consensus Cdc2 phosphorylation sites [(S/T)PX(K/R)] (29) are present in the Orc2 and Orp2 homolog proteins in fission yeast, budding yeast, Drosophila melanogaster, Xenopus, and mammals (Fig. 1). These sites occur exclusively within the N-terminal regions of the proteins. This region is poorly if at all conserved. Multiple sequence alignments and PSI-BLAST (1) were used to determine an N-terminal boundary for sequence similarity. The first Orc2 sequence motif conserved from yeast to humans, (F/V)(D/E)EYF, begins at amino acid 193 of S. pombe Orp2. This conserved motif is the boundary between the variable and conserved regions shown in Fig. 1. Even among mammals, there is a sharp change in sequence conservation between the N-terminal region (64% identical between human and mouse) and the C-terminal region (89% identical between human and mouse). The consensus Cdc2 phosphorylation sites occur exclusively within the variable N-terminal region.
The fact that consensus (and usually additional nonconsensus SP or TP) Cdc2 phosphorylation sites are always present in the N-terminal region of Orc2 homologs, coupled with the fact that Cdc2 kinase is important for regulating DNA synthesis, suggests that these Cdc2 sites could be targets of Cdc2 kinase. The first test of this idea is to find out whether Orc2 is a substrate of Cdc2 in vitro. Therefore, a GST-Orp2 fusion protein was mixed with active Cdc2 kinase from fission yeast, and it was found that the GST-Orp2 became phosphorylated (Fig. 2A). GST-Orp2 was a better substrate than was histone H1, while GST did not become phosphorylated at all (data not shown). Identical results were obtained using Cdc2 isolated from fission yeast lysates by binding to p13suc1 beads or by immunoprecipitation of the cyclin Cdc13. These results show that Cdc2 and Cdc13 complexes can phosphorylate Orp2 in vitro.
Phosphorylation of Orp2 in vitro depends on the consensus Cdc2 phosphorylation sites.
To see if the phosphorylation of Orp2 in vitro depends on the Cdc2 phosphorylation sites, various (S/T)P sequences in Orp2 were mutated to nonphosphorylatable AP sequences by site-directed mutagenesis. Changing the four TP sequences between amino acids 43 and 66 (Fig. 1) yielded the mutant orp2-T4A, which is the mutant characterized in this study. GST-Orp2 and the mutant GST-Orp2-T4A were purified from fission yeast and compared as Cdc2 substrates in vitro. GST-Orp2 was readily phosphorylated, whereas GST-Orp2-T4A was not (Fig. 2A). Quantitation showed that 10-fold more radioactive phosphate was incorporated into GST-Orp2 than into GST-Orp2-T4A. That equal amounts of substrate proteins were added to each reaction was determined by Coomassie staining of the total proteins in the reactions after separation by SDS-PAGE (Fig. 2A). Hereafter, we will refer to Orp2-T4A as the “nonphosphorylatable” Orp2, even though it might be a substrate of other kinases and could even have weak residual or alternative sites for Cdc2 phosphorylation.
Phosphopeptide mapping showed that the major Cdc2 phosphorylation sites in Orp2 were eliminated by the mutation of four threonines in Orp2-T4A (Fig. 2B). Consistent with the idea that threonines are the phosphorylation targets of Cdc2 in Orp2, phosphothreonine was the main (>90%) phosphoamino acid detected in GST-Orp2 phosphorylated by Cdc2 (datum not shown). Some phosphoserine was also detected in this reaction. However, most of this phosphoserine was independent of the addition of Cdc2 and came from a low level of kinase associated with the GST-Orp2 (datum not shown). This other kinase is not Cdc2, because the phosphopeptides differ from those generated by Cdc2 (Fig. 2B), because no Cdc2 was detected by Western blotting in preparations of GST-Orp2 (data not shown), and because this other kinase phosphorylates GST-Orp2 and GST-Orp2-T4A equally well (Fig. 2A).
Phosphorylation of Orp2 in vivo requires the same sites that are phosphorylated by Cdc2 in vitro
Phosphorylation of Orp2 in vivo causes a shift to a slower-migrating species in SDS-PAGE analysis (45). If the slowly migrating Orp2 results from phosphorylation by Cdc2, then the same Cdc2 sites required for Orp2 phosphorylation in vitro should also be required for phosphorylation and mobility shift of Orp2 in vivo. To test this, isogenic strains were constructed having either wild-type orp2+ or orp2-T4A. The orp2-T4A strains were viable with no obvious phenotype (see below). Immunoblot analysis showed that asynchronous wild-type cells had several mobility forms of Orp2, whereas the mutant orp2-T4A had a single, rapidly migrating band (Fig. 3). This single form of Orp2-T4A comigrated with the single, rapidly migrating form of wild-type Orp2 observed in G1 cells (Fig. 3). Mutation of the single consensus Cdc2 phosphorylation site was not sufficient to abolish the cell cycle-regulated phosphorylation (data not shown). That the same Cdc2 sites required for Orp2 phosphorylation in vitro are also required for generation of slowly migrating forms of Orp2 in vivo suggests that Orp2 is an in vivo substrate of Cdc2.
Cell cycle regulation of Orp2 phosphorylation.
To find out when in the cell cycle Orp2 is phosphorylated, we synchronized cells in G2 using block and release of a cdc25-22 mutant and followed Orp2 protein through two cell cycles (Fig. 4A). Orp2 shifted from slowly migrating forms at the G2 block to faster migrating forms at the end of mitosis. The fastest migrating (unphosphorylated) form of Orp2 appeared simultaneously with the peak in septation. Typically, fission yeast cells are in the S phase of the next cell cycle by the end of septation; thus, the peak in septation and in abundance of fast-migrating Orp2 occurs near the G1/S boundary.
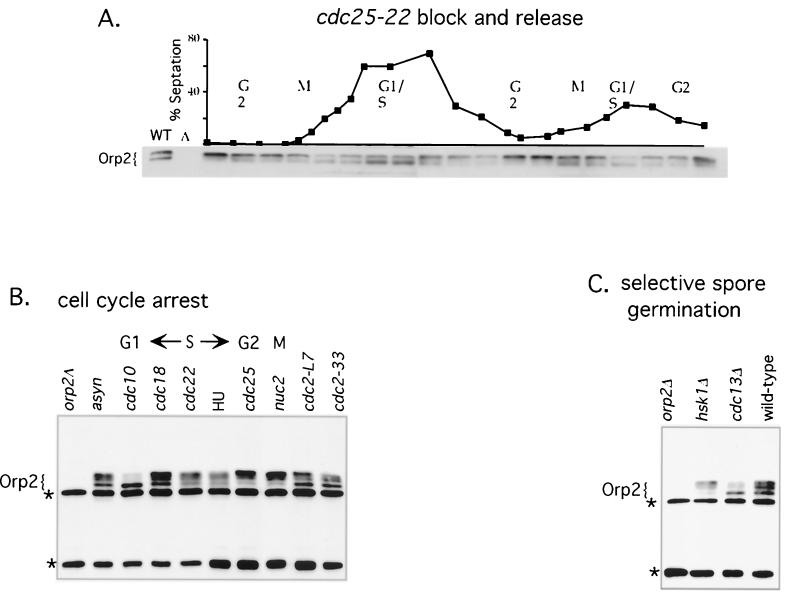
FIG. 4.
Cell cycle-regulated phosphorylation of Orp2. (A) Orp2 protein throughout the cell cycle. Cells were synchronized by block and release of a cdc25-22 mutant. Samples were harvested for analysis every 20 min through two consecutive cell cycles. Whole-cell extracts were analyzed by SDS-PAGE and immunoblotting using affinity-purified Orp2 antibodies. Synchrony and cell cycle progression were determined by septation index (above). Asynchronous cells (WT) and an orp2 deletion control (Δ) on the left show which bands are Orp2 specific; the orp2Δ was made by selective spore germination (see Materials and Methods). (B) Orp2 protein during cell cycle arrest. Mutants were arrested by shift to 36.5°C for 4 h, the mutant is indicated above each lane, and the stage of cell cycle arrest is shown at the top. Orp2 bands are indicated. Cross-reacting bands that are not Orp2 are marked (*). Abbreviations: asyn, asynchronous wild type shifted to 36.5°C for 4 h; HU, wild type incubated with 12 mM HU for 4 h at 32°C; orp2Δ, cells with orp2 deletions from selective spore germination of an orp2Δ/orp2+ heterozygote lack Orp2-specific bands. (C) Orp2 protein in cells lacking essential genes. Immunoblot analysis of Orp2 from cells obtained by selective germination of spores with deletions of essential genes orp2, cdc13, and hsk1 as indicated. wild-type, selective spore germination of a wild-type diploid heterozygous at ura4 (ura4-D18/ura4+, JLP52).
As a second method for examining cell cycle regulation of phosphorylation of Orp2, cells were blocked in various stages of the cell cycle using cell cycle mutants or the replication inhibitor hydroxyurea (HU). Orp2 isolated from the arrested cells was analyzed by Western blotting (Fig. 4B). Orp2 was in the fastest migrating form at the cdc10 arrest (G1 phase). More slowly migrating (phosphorylated) forms first appeared in the S phase (cdc22 and HU arrests) and when the replication factor Cdc18 was inactivated (cdc18). In G2 and early M (cdc25 and nuc2), Orp2 accumulated in the slowest-migrating forms. It is notable that Orp2 is fully modified at the cdc25 mutant arrest (G2). Cdc25 is the tyrosine phosphatase required for activating Cdc2 to the high levels required for mitosis. If Cdc2 is the kinase modifying Orp2, then the pool of Cdc2 that phosphorylates Orp2 may not require activation by Cdc25.
The phosphorylation state of Orp2 was assayed in cdc2 mutant cells. Phosphorylation of Orp2 was reduced but not abolished in cdc2-L7 and cdc2-33 mutants at restrictive temperature (Fig. 4B). Because Cdc2 is essential both for G1-to-S and G2-to-M transitions, these cdc2 mutants arrest as a mixture of 10 to 20% G1 cells and 80 to 90% G2 cells. These data show that inactivation of Cdc2 that is sufficient to arrest cell cycle progression does not result in fully dephosphorylated Orp2.
The phosphorylation state of Orp2 was also assayed in cells lacking cdc13. Cdc13 is a B-type cyclin essential for mitosis and for once-per-cell-cycle control of DNA replication. Selective spore germination was used to obtain a culture of cells lacking Cdc13. These cells cannot proceed past G2 into mitosis, and they rereplicate DNA. The Orp2 in these cells was in the fast-migrating (unphosphorylated) form (Fig. 4C). Since cells arrested in G2 by other mutations (Fig. 4B) accumulate phosphorylated forms of Orp2, the lack of Orp2 phosphorylation in the cdc13 mutant suggests that the Cdc13-Cdc2 complex may be directly required for phosphorylation of Orp2. Furthermore, this observation suggests that phosphorylation of Orp2 may not be needed for replication, since cdc13 deletion mutants replicate and rereplicate DNA (27).
Selective spore germination was also used to examine the role of another kinase, Hsk1, in generating phosphorylated Orp2. Hsk1 is essential for DNA replication and is the fission yeast homolog of Cdc7 (47). Cdc7 is bound to replication origins via the ORC and is required for replication initiation at individual origins throughout the S phase (6, 18, 20, 25). Orp2 is fully shifted to the phosphorylated forms even in the hsk1 mutant (Fig. 4C). Furthermore, Hsk1 failed to phosphorylate Orp2 in vitro (data not shown). These results indicate that Hsk1 does not phosphorylate Orp2. This is consistent with the idea that the relevant Hsk1 substrate is Mcm2 and possibly additional MCMs (reviewed in reference 70).
Phosphorylation of Orp2 is not required for DNA replication.
Cdc2 is essential for initiation of replication, and so presumably it has some substrate that is important for initiation. We and others expected that Orp2 could be such a substrate (40, 45). Our data suggest otherwise. Cells carrying orp2-T4A as their only allele of orp2 were viable and healthy, with no obvious phenotype. Likewise, cells with orp2-T4A and also mutated at two additional (S/T)P sites (T159→A and S162→A) were also viable and healthy. Finally, cells expressing GST-orp2-Δ1-127 as the only allele of orp2 were viable and healthy. This GST-Orp2-Δ1-127 fusion protein lacks the first 127 amino acids of Orp2 (approximately two-thirds of the variable region), including all the T-P sequences mutated in orp2-T4A. Except for S139, which has not been tested, all (S/T)P sequences in the variable region of Orp2 can be mutated or deleted without significantly altering replication initiation.
The orp2-T4A allele was integrated into the chromosome at the orp2Δ::ura4+ locus under control of the orp2 promoter. Several experiments were done to see if this mutant had a defect in initiation. Flow cytometry showed that the S phase progressed similarly in orp2-T4A and wild-type cells (data not shown). There was no detectable change in cell size at division, which would have indicated a cell cycle delay. There was no genetic interaction between orp2-T4A and cdc18, cdc19, or cdc21 temperature-sensitive alleles defective for replication factors.
To perform a possibly more sensitive assay, we compared the efficiency with which orp2-T4A and wild-type strains could be transformed with an ars plasmid as has been done to evaluate ars sequences in S. pombe (10, 22). The assays described above may be insensitive detectors of initiation defects, because each chromosome has many origins. Even if the efficiency of initiation declined to, say, 30% of wild-type values, this 30% may be adequate to give a wild-type phenotype by growth rate or flow cytometry assays, etc. Since plasmids have only a single origin, any defect in origin usage lowers transformation frequency. To control for other factors affecting transformation and cell viability, the mutant and wild-type strains were mixed and transformed in one tube (Materials and Methods). The orp2-T4A strain was marked with ade6-M210, and the orp2+ strain was marked with ade6-M216. Both alleles are Ade−, but on rich medium low in adenine (YSO), ade6-M216 yields pink colonies while ade6M-210 yields red colonies. The orp2+ ade6-M216 and orp2-T4A ade6-M210 strains were mixed 1:1, grown overnight, and then transformed with ura4+ tester plasmids containing wild-type and mutant derivatives of ars3002 (Table 2) (22). Transformants were selected on medium lacking uracil. In addition, cells were plated on medium containing uracil to determine the number of cells of each genotype surviving the transformation protocol. Colonies from both surviving cells and from transformed (i.e., Ura+) cells were picked to YSO to determine whether they were orp2+ ade6-M216 or orp2-T4A ade6-M210. The ratio of transformants of each genotype was divided by the ratio of surviving cells to normalize for survival and determine the relative transformation efficiency (Materials and Methods).
TABLE 2.
Transformation efficiencies of orp2-T4A and orp2-T3D strains relative to orp2+
| Strains compared | Plasmida | Relative ars functionb | Ratio ofc:
|
Relative transformationd | |
|---|---|---|---|---|---|
| Colonies | Transformants | ||||
| orp2-T4A/orp2+ | ars3002 | 100 | 125/176 | 383/189 | 2.9 |
| ars3002Δ5 | 40–50 | 131/139 | 388/144 | 2.9 | |
| ars3002Δ9 | 20–30 | 201/227 | 266/180 | 1.6 | |
| ars3002Δ11 | 10–20 | 302/277 | 303/266 | 1.4 | |
| ars3002Δ8 | <5 | 134/149 | 283/138 | 1.9 | |
| orp2-T3D/orp2+ | ars3002 | 100 | 92/91 | 80/120 | 0.7 |
| ars3002Δ5 | 40–50 | 84/103 | 79/120 | 0.8 | |
| ars3002Δ9 | 20–30 | 190/223 | 93/107 | 1.0 | |
| ars3002Δ11 | 10–20 | 92/115 | 93/107 | 1.1 | |
| ars3002Δ8 | <5 | 75/119 | 76/124 | 1.0 | |
As shown in Table 2, the orp2-T4A mutant had transformation efficiencies up to threefold higher than the wild-type strain. The plasmids used in this assay contained wild-type and mutant derivatives of ars3002. This test was performed with several different ars constructs. Deletion mutants of ars3002 were chosen because they have been extensively characterized (22). In fact, the wild-type ars showed the greatest difference between orp2-T4A and orp2+. Perhaps the differences detected in this assay are obscured by the defective function of the ars mutants. In any case, the results give no indication of any initiation defect in the mutant, but rather suggest that the Cdc2 phosphorylation sites in Orp2 could have an inhibitory function.
Introducing mutations that might mimic phosphorylation of Orp2 had little effect on Orp2 function. Threonines in three of the putative Cdc2 phosphorylation sites were changed to aspartate (Fig. 1). The resulting mutant, orp2-T3D, has no obvious defect in growth or replication (data not shown). This orp2-T3D mutant has a transformation efficiency 0.7- to 1.1-fold that of the wild type in the transformation assay (Table 2); the variations in relative efficiency depended upon the plasmid ars tested. As with orp2-T4A, the greatest difference in transformation efficiency was observed with the wild-type ars, although in this case, the difference is a decrease in transformation efficiency in the mutant; however, all experiments suggest the orp2-T3D mutant is wild type or nearly wild type for initiation.
The Cdc2 phosphorylation sites in Orp2 help prevent rereplication.
Cdc2 is essential for limiting DNA replication to once per cell cycle. Inactivation of Cdc18 is one mechanism used by Cdc2 to prevent reinitiation, but there must be redundant mechanisms, since loss of the phosphorylation sites on Cdc18 does not in itself allow reinitiation. In S. pombe, overexpression of Cdc18 does allow rereplication, probably by supplying functional Cdc18 and also by interfering with other pathways for control of rereplication. Phosphorylation of Orp2 might be one of these additional mechanisms. If so, then we expect no obvious phenotype of mutating Cdc2 phosphorylation sites in Orp2 in wild-type cells, but the role of these sites in regulating rereplication might be revealed if Cdc18 is de-regulated. Therefore, we compared rereplication in orp2-T4A and wild-type strains when cdc18* was overexpressed in each of them. cdc18* lacks four Cdc2 sites and induces reinitiation and rereplication when overexpressed (as does cdc18+ itself) (44).
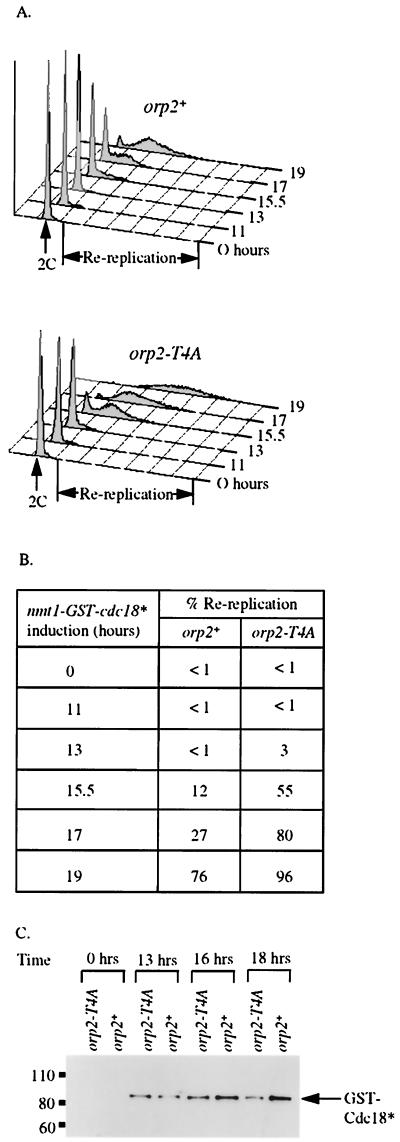
The orp2-T4A mutation enhanced rereplication induced by overexpression of cdc18*. A strain with a stable integrated copy of cdc18* controlled by the inducible nmt promoter was crossed with isogenic orp2-T4A and wild-type orp2+ strains that have wild-type levels of Orp2 expressed from its endogenous promoter. nmt-cdc18* orp2-T4A and nmt-cdc18* orp2+ strains were isolated by tetrad analysis. Cdc18* was induced, and rereplication was quantitated by flow cytometry (fluorescence-activated cell sorter analysis). The nmt promoter is repressed for at least 10 h after removal of thiamine and is not fully induced until 16 h after removal of thiamine (49). Western blot analysis showed that Cdc18* protein was detectable 13 h after induction in both genotypes, and expression was the same in both genotypes (Fig. 5C). Data shown in Fig. 5A and B are representative of quantitative analysis of four independent isolates of each genotype. An increase in rereplication in orp2-T4A compared with orp2+ was first observed when the inducible Cdc18 was wild type for Cdc2 phosphorylation sites (data not shown). However, the difference between orp2-T4A and orp2+ was magnified when Cdc2 sites in Cdc18 were mutated.
FIG. 5.
Sensitivity of orp2-T4A mutants to rereplication. Haploid isogenic strains JLP524 orp2+ and JLP519 orp2-T4A were compared upon induction of GST-Cdc18* from the nmt1 promoter. (A) DNA content of cells after removal of thiamine to induce the nmt1-driven GST-cdc18*. 2C DNA content is indicated. “Re-replication” marks DNA content greater than approximately 4C. (Top) orp2+ GST-Cdc18* (JLP524); (bottom) orp2-T4A GST-Cdc18* (JLP519). (B)
Overexpression of Cdc18 is still required to drive rereplication even when Cdc2 phosphorylation sites in both Cdc18 and Orp2 are mutated as in the nmt-cdc18* orp2-T4A strain. One explanation for this could be that nmt-cdc18* is not sufficient to supply replication initiation function when the nmt promoter is repressed; note that these cells also have a wild-type cdc18+ gene and so are not dependent on nmt-cdc18* for viability. We confirmed the published results that repressed levels of cdc18* can complement the essential replication initiation function of Cdc18 in the cdc18-K9 mutant at restrictive temperature. This suggests that that Orp2 phosphorylation and Cdc18 phosphorylation are redundant with yet more mechanisms for preventing DNA rereplication and that high level expression of Cdc18 is needed to override these mechanisms.
DISCUSSION
Cdc2 phosphorylation sites are present in the amino termini of Orc2 proteins from yeasts to humans. We have shown that the S. pombe Orc2, Orp2, is a substrate for Cdc2 in vitro and appears to be phosphorylated by Cdc2 in vivo. Mutation of Cdc2 phosphorylation sites in Orp2 results in a phenotype of enhanced rereplication when other controls preventing DNA rereplication are inactivated. We propose that phosphorylation of Orp2 is redundant with phosphorylation of Cdc18 to maintain once-per-cell-cycle control of DNA replication. Finally, our data suggest that Cdc2 phosphorylation of Orp2 is dispensable for Cdc2 activation of DNA replication.
There is evidence for phosphorylation of Orc2 proteins from budding yeast, fission yeast, and Xenopus to humans. As predicted from the Orp2 sequence, Orp2 can be phosphorylated in vitro by Cdc2. This Orp2 phosphorylation depends on putative Cdc2 sites. Lygerou and Nurse have shown that Orp2 is also phosphorylated in vivo, producing a change in electrophoretic mobility (45). We find that this in vivo phosphorylation likewise depends on the Cdc2 sites. The in vivo phosphorylation state of Orp2 varies according to cell cycle position, with maximal phosphorylation in the M phase and minimal phosphorylation in G1. This corresponds to the pattern of Cdc2 activation through the cell cycle. These observations suggest that Orp2 may be an in vivo substrate of Cdc2.
The failure to phosphorylate Orp2 in the cdc13 cyclin deletion mutant suggests that a Cdc2-Cdc13 complex is important to phosphorylate Orp2 in vivo. Failure to phosphorylate Orp2 in cells lacking Cdc13 is not easily explained as a failure of these cells to enter mitosis because other blocks to mitosis did not block Orp2 phosphorylation. That loss of the one cyclin Cdc13 nearly abolishes Orp2 phosphorylation suggests that other cyclins, such as the S-phase cyclin Cig2, are poor effectors of Orp2 phosphorylation (24, 51). Phosphorylation of Orp2 by a CDK complex primarily active in G2/M rather than an S-phase-specific complex is consistent with a model in which Orp2 phosphorylation helps prevent rereplication during G2/M rather than having a primary role in activating replication initiation in the S phase.
Orp2 phosphorylation was reduced but not abolished in Cdc2 mutants arrested at restrictive temperature. There are three likely causes for Orp2 remaining phosphorylated when Cdc2 is inactivated. (i) There may be a kinase other than Cdc2 that carries out the phosphorylation. This is possible but seems unlikely given the importance of the Cdc13 cyclin for Orp2 phosphorylation. (ii) The mutant Cdc2 may retain some residual activity. Indeed, these temperature-sensitive cdc2 alleles almost certainly do have residual function at restrictive temperature because complete inactivation of Cdc2 in G2 followed by reactivation of Cdc2 leads to rereplication and diploidization, which is not seen in these cdc2 mutants (27). (iii) Orp2 phosphorylation may be particularly stable. For instance, Cdc2 substrates might be rapidly dephosphorylated at the end of mitosis by a phosphatase analogous to Cdc14 of budding yeast. In this case, phosphates on Cdc2 substrates such as Orp2 might be quite stable until the end of mitosis. Notably, budding yeast orc2-1 is synthetically lethal with cdc14-1, which could be explained if Cdc14 removes inhibitory phosphates from Orc2 (43).
Since both Cdc2 and Orp2 are essential for replication initiation, we looked for defects in the initiation of replication in the nonphosphorylatable orp2-T4A mutant, but no defects were seen; growth rate was normal; S phase was normal as assayed by flow cytometry; cell size at division was normal; there was no genetic interaction with the cdc18-K9 mutant, and the transformation frequency of orp2-T4A mutants was, if anything, higher than that of the wild type. Finally, deletion of the first 127 amino acids of Orp2 did not obviously affect growth, suggesting that the N-terminal domain of Orp2 is not needed for initiation. Since each chromosome may have hundreds of origins of replication, even a severe reduction in the efficiency of initiation in these mutants (orp2-T4A and GST-Orp2-Δ127) would still allow many origins to fire, perhaps leading to normal growth rate, DNA content, and cell division kinetics. However, it is harder to explain how transformation frequency could be normal if the orp2-T4A mutant suffered from a significant initiation defect.
Indeed the orp2-T4A mutant functioned up to threefold better than wild-type orp2 in transformation efficiency. This apparent gain of function could be explained if the Orp2 phosphorylation can inhibit initiation at least partially, especially at functional origins. For example, although Orp2 is not phosphorylated when the S phase begins, the presence of phosphorylated Orp2 at the end of mitosis might have an effect on the following S phase by delaying establishment of prereplicative complexes. The Orp2-T4A protein would suffer no such interference and so might have a higher efficiency of firing individual origins and a resulting higher transformation efficiency.
It is possible that phosphorylation of the N-terminal domain of Orp2 is important for initiation, but is completely redundant with phosphorylation of some other replication proteins. It is well known that other replication proteins (e.g., Cdc18, Mcm4, and Cdc23 [also known as Mcm10]) have clusters of Cdc2 phosphorylation sites. Thus, an initiation defect might appear only when multiple phosphorylation sites on multiple proteins are mutated simultaneously. The orp2 mutants used in this study may be useful to investigate this possibility in the future.
In this study, we find evidence suggesting that Orp2 phosphorylation is redundant with phosphorylation of other replication proteins to prevent reinitiation of replication. We were more successful in finding a phenotype when we looked for rereplication because we had some knowledge of other pathways that prevent rereplication and could deregulate those pathways. This deregulation was achieved by overexpression of Cdc18* from the nmt promoter, a condition in which both the transcription and phosphorylation of Cdc18 are deregulated and cells are driven to rereplicate DNA. Both the kinetics and magnitude of rereplication were affected by mutation of Cdc2 phosphorylation sites in Orp2. Rereplication occurred 4 to 6 h after the appearance of Cdc18* in the wild-type strains but only 2 h after the appearance of Cdc18* in the orp2-T4A strains. Thus, the orp2-T4A mutant is sensitized to rereplication, showing that the phosphorylation sites in Orp2 help to prevent rereplication.
The idea that phosphorylation of Orp2 is more important for inhibiting reinitiation of replication than for triggering initiation is consistent with the timing of Orp2 phosphorylation and with its dependence on cdc13+. Orp2 phosphorylation is first detected at about the time of the S phase, but is maximal in G2 and M when initiation is no longer occurring and when reinitiation must be prevented. Orp2 phosphorylation is not seen in the cdc13 mutant; this mutant replicates (suggesting phosphorylation of Orp2 is not essential for replication) and rereplicates (consistent with the idea that phosphorylation of Orp2 helps prevent rereplication).
The idea that phosphorylation of Orp2 helps prevent reinitiation is also consistent with Orc2 phosphorylation and ORC-CDK interactions in other systems. In Xenopus extracts, XOrc2 is unphosphorylated during interphase and becomes highly phosphorylated during mitosis (9). Xenopus ORC fails to bind the Cdk2-cyclin A or Cdk2-cyclin E complexes known to be important for activating initiation of replication but does bind the Cdc2-cyclin A complexes that might be important for preventing reinitiation (65). Finally, mutation of the consensus CDK phosphorylation sites in budding yeast Orc2 together with mutation of sites in Orc6 also sensitizes cells to rereplication (J. J. Li, personal communication).
In current models for replication initiation, the Orc proteins form a seemingly static platform onto which other initiation factors are assembled. How then might Orp2 phosphorylation inhibit reinitiation of DNA replication? Since binding of the ORC to DNA is not regulated and Orp2 is not targeted for destruction (34, 36, 45, 57), phosphorylation of Orp2 probably alters binding to other initiation factors (e.g., Cdc18) or promotes changes within the ORC complex. Our favored model is that the N-terminal domain of Orp2 is activated by phosphorylation to inhibit initiation, perhaps by binding a negative regulator analogous to geminin, or by interfering with the binding of other initiator proteins to the ORC (50, 69, 73). Another model is that the N-terminal domain of Orp2 has a positive function that is then inactivated by phosphorylation. This second model is hard to reconcile with the fact that most of the N-terminal domain of Orp2, including the phosphorylation sites, can be deleted without abolishing Orp2 function. One prediction of either model is that phosphorylated Orp2 would not initiate DNA replication. However, the orp2-T3D allele created to try to mimic Orp2 phosphorylation shows only a very weak initiation defect. Our simple models may be incorrect or the orp2-T3D mutant may fail to mimic the phosphorylated state of Orp2. Identifying what factors interact with the N terminus of Orp2 would help clarify exactly how phosphorylation modulates the ORC function.
In Xenopus and humans, the homologue of Cdc18, Cdc6, is not fully degraded during the S phase (reviewed in reference70). In these organisms, mechanisms for preventing reinitiation that are independent of Cdc6, such as phosphorylation of Orc2, may be even more important than they are in yeasts. Since CDK phosphorylation sites in the amino terminus of Orp2 are a conserved feature of Orc2 and its homologs, the phosphorylation of Orc2 may be a conserved mechanism for maintaining once-per-cell-cycle control of DNA replication. Of course, there must also be additional controls (since, for instance, the orp2-T4A mutant does not rereplicate unless Cdc18* is overexpressed). Multiple overlapping controls provide extra assurance that cells will not reinitiate replication. Any one control may be sufficient to prevent bulk rereplication of DNA but might allow occasional rereplication events from some origins. Such rare events would not be readily detectable in the laboratory except by specially designed genetic tests, but the genomic instability caused by even very rare reinitiation might reduce fitness in simple eukaryotes such as yeast and possibly lead to genetic diseases and cancer in humans.
ACKNOWLEDGMENTS
We thank Paul Russell and Antonia Lopez-Girona for intellectual inspiration and reagents. Thanks go to Beth Baber, Clare McGowan, and Kazuhiro Shiozaki for friendship and helpful advice critical to the success of the in vitro phosphorylation experiments. Thanks go to Joan Kiely and Anya Bogomez, whose experiments contributed to this work. Thanks go to Joachim Li for communicating results prior to publication. Special thanks go to Bruce Futcher for extensive help with the manuscript and to Nancy Reich for helpful comments.
This work was supported by a Beckman Scholars Award to W.M., Leukemia Society Special Fellow grant and Kimmel Scholar Award to J.L., and NIH grant R01 GM61532.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 3.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 4.Berry L D, Gould K L. Regulation of Cdc2 activity by phosphorylation at T14/Y15. Prog Cell Cycle Res. 1996;2:99–105. doi: 10.1007/978-1-4615-5873-6_10. [DOI] [PubMed] [Google Scholar]
- 5.Booher R N, Alfa C E, Hyams J S, Beach D H. The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell. 1989;58:485–497. doi: 10.1016/0092-8674(89)90429-7. [DOI] [PubMed] [Google Scholar]
- 6.Bousset K, Diffley J F. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle W J, Van Der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin layer cellulose plates. Methods Enzymol. 1991;210:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 8.Broek D, Bartlett R, Crawford K, Nurse P. Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature. 1991;349:388–393. doi: 10.1038/349388a0. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter P B, Mueller P R, Dunphy W G. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- 10.Clyne R K, Kelly T J. Genetic analysis of an ARS element from the fission yeast Schizosaccharomyces pombe. EMBO J. 1995;14:6348–6357. doi: 10.1002/j.1460-2075.1995.tb00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocker J H, Piatti S, Santocanale C, Nasmyth K, Diffley J F. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 12.Coleman T R, Carpenter P B, Dunphy W G. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 13.Correa-Bordes J, Nurse P. p25rum1 orders S phase and mitosis by acting as an inhibitor of the p34cdc2 mitotic kinase. Cell. 1995;83:1001–1009. doi: 10.1016/0092-8674(95)90215-5. [DOI] [PubMed] [Google Scholar]
- 14.Costello G, Rodgers L, Beach D. Fission yeast enters the stationary phase G0 state from either mitotic G1 or G2. Curr Genet. 1986;11:119–125. [Google Scholar]
- 15.Creanor J, Mitchison J M. The kinetics of the B cyclin p56cdc13 and the phosphatase p80cdc25 during the cell cycle of the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1996;109:1647–1653. doi: 10.1242/jcs.109.6.1647. [DOI] [PubMed] [Google Scholar]
- 16.Dahmann C, Diffley J F, Nasmyth K A. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 17.Diffley J F, Cocker J H, Dowell S J, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 18.Donaldson A D, Fangman W L, Brewer B J. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 1998;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donaldson A D, Raghuraman M K, Friedman K L, Cross F R, Brewer B J, Fangman W L. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell. 1998;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- 20.Dowell S J, Romanowski P, Diffley J F. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- 21.Drury L S, Perkins G, Diffley J F. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubey D D, Kim S M, Todorov I T, Huberman J A. Large, complex modular structure of a fission yeast DNA replication origin. Curr Biol. 1996;6:467–473. doi: 10.1016/s0960-9822(02)00514-6. [DOI] [PubMed] [Google Scholar]
- 23.Elsasser S, Chi Y, Yang P, Campbell J L. Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol Biol Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisher D L, Nurse P. A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 1996;15:850–860. [PMC free article] [PubMed] [Google Scholar]
- 25.Hardy C F. Characterization of an essential Orc2p-associated factor that plays a role in DNA replication. Mol Cell Biol. 1996;16:1832–1841. doi: 10.1128/mcb.16.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harlow E, Lane D. Antibodies. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 27.Hayles J, Fisher D, Woollard A, Nurse P. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 28.Hennessy K M, Clark C D, Botstein D. Subcellular localization of yeast CDC46 varies with the cell cycle. Genes Dev. 1990;4:2252–2263. doi: 10.1101/gad.4.12b.2252. [DOI] [PubMed] [Google Scholar]
- 29.Holmes J K, Solomon M J. A predictive scale for evaluating cyclin-dependent kinase substrates. A comparison of p34cdc2 and p33cdk2. J Biol Chem. 1996;271:25240–25246. doi: 10.1074/jbc.271.41.25240. [DOI] [PubMed] [Google Scholar]
- 30.Jallepalli P V, Brown G W, Muzi-Falconi M, Tien D, Kelly T J. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev. 1997;11:2767–2779. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jallepalli P V, Kelly T J. Cyclin-dependent kinase and initiation at eukaryotic origins: a replication switch? Curr Opin Cell Biol. 1997;9:358–363. doi: 10.1016/s0955-0674(97)80008-7. [DOI] [PubMed] [Google Scholar]
- 32.Jallepalli P V, Tien D, Kelly T J. sud1(+) targets cyclin-dependent kinase-phosphorylated Cdc18 and Rum1 proteins for degradation and stops unwanted diploidization in fission yeast. Proc Natl Acad Sci USA. 1998;95:8159–8164. doi: 10.1073/pnas.95.14.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jong A, Young M, Chen G C, Zhang S Q, Chan C. Intracellular location of the Saccharomyces cerevisiae Cdc6 gene product. DNA Cell Biol. 1996;15:883–895. doi: 10.1089/dna.1996.15.883. [DOI] [PubMed] [Google Scholar]
- 34.Kearsey S E, Montgomery S, Labib K, Lindner K. Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 2000;19:1681–1690. doi: 10.1093/emboj/19.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keeney J B, Boeke J D. Efficient targeted integration at leu1–32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiely J, Haase S B, Russell P, Leatherwood J. Functions of fission yeast orp2 in DNA replication and checkpoint control. Genetics. 2000;154:599–607. doi: 10.1093/genetics/154.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kominami K, Toda T. Fission yeast WD-repeat protein pop1 regulates genome ploidy through ubiquitin-proteasome-mediated degradation of the CDK inhibitor Rum1 and the S-phase initiator Cdc18. Genes Dev. 1997;11:1548–1560. doi: 10.1101/gad.11.12.1548. [DOI] [PubMed] [Google Scholar]
- 38.Labib K, Diffley J F, Kearsey S E. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat Cell Biol. 1999;1:415–422. doi: 10.1038/15649. [DOI] [PubMed] [Google Scholar]
- 39.Leatherwood J. Emerging mechanisms of eukaryotic DNA replication initiation. Curr Opin Cell Biol. 1998;10:742–748. doi: 10.1016/s0955-0674(98)80117-8. [DOI] [PubMed] [Google Scholar]
- 40.Leatherwood J, Lopez-Girona A, Russell P. Interaction of Cdc2 and Cdc18 with a fission yeast ORC2-like protein. Nature. 1996;379:360–363. doi: 10.1038/379360a0. [DOI] [PubMed] [Google Scholar]
- 41.Lee K M, Saiz J E, Barton W A, Fisher R P. Cdc2 activation in fission yeast depends on Mcs6 and Csk1, two partially redundant Cdk-activating kinases (CAKs) Curr Biol. 1999;9:441–444. doi: 10.1016/s0960-9822(99)80194-8. [DOI] [PubMed] [Google Scholar]
- 42.Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 43.Loo S, Fox C A, Rine J, Kobayashi R, Stillman B, Bell S. The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol Biol Cell. 1995;6:741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez-Girona A, Mondesert O, Leatherwood J, Russell P. Negative regulation of Cdc18 DNA replication protein by Cdc2. Mol Biol Cell. 1998;9:63–73. doi: 10.1091/mbc.9.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lygerou Z, Nurse P. The fission yeast origin recognition complex is constitutively associated with chromatin and is differentially modified through the cell cycle. J Cell Sci. 1999;112:3703–3712. doi: 10.1242/jcs.112.21.3703. [DOI] [PubMed] [Google Scholar]
- 46.Martin-Castellanos C, Blanco M A, de Prada J M, Moreno S. The puc1 cyclin regulates the G1 phase of the fission yeast cell cycle in response to cell size. Mol Biol Cell. 2000;11:543–554. doi: 10.1091/mbc.11.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masai H, Miyake T, Arai K-i. hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 1995;14:3094–3104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masai H, Sato N, Takeda T, Arai K. CDC7 kinase complex as a molecular switch for DNA replication. Front Biosci. 1999;4:D834–D840. doi: 10.2741/masai. [DOI] [PubMed] [Google Scholar]
- 49.Maundrell K. nmt1 of fission yeast. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 50.McGarry T J, Kirschner M W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 51.Mondesert O, McGowan C H, Russell P. Cig2, a B-type cyclin, promotes the onset of S in Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:1527–1533. doi: 10.1128/mcb.16.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moreno S, Hayles J, Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell. 1989;58:361–372. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- 53.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 54.Newlon C S. Putting it all together: building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- 55.Nguyen V Q, Co C, Irie K, Li J J. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr Biol. 2000;10:195–205. doi: 10.1016/s0960-9822(00)00337-7. [DOI] [PubMed] [Google Scholar]
- 56.Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 57.Ogawa Y, Takahashi T, Masukata H. Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol Cell Biol. 1999;19:7228–7236. doi: 10.1128/mcb.19.10.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okishio N, Adachi Y, Yanagida M. Fission yeast Nda1 and Nda4, MCM homologs required for DNA replication, are constitutive nuclear proteins. J Cell Sci. 1996;109:319–326. doi: 10.1242/jcs.109.2.319. [DOI] [PubMed] [Google Scholar]
- 59.Pasion S G, Forsburg S L. Nuclear localization of Schizosaccharomyces pombe Mcm2/Cdc19p requires MCM complex assembly. Mol Biol Cell. 1999;10:4043–4057. doi: 10.1091/mbc.10.12.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pelizon C, Madine M A, Romanowski P, Laskey R A. Unphosphorylatable mutants of cdc6 disrupt its nuclear export but still support DNA replication once per cell cycle. Genes Dev. 2000;14:2526–2533. doi: 10.1101/gad.176300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perkins G, Diffley J F. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- 62.Petersen B O, Lukas J, Sorensen C S, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Piatti S, Bohm T, Cocker J H, Diffley J F, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 64.Quintana D G, Dutta A. The metazoan origin recognition complex. Front Biosci. 1999;4:D805–D815. doi: 10.2741/quintana. [DOI] [PubMed] [Google Scholar]
- 65.Romanowski P, Marr J, Madine M A, Rowles A, Blow J J, Gautier J, Laskey R A. Interaction of Xenopus Cdc2 · cyclin A1 with the origin recognition complex. J Biol Chem. 2000;275:4239–4243. doi: 10.1074/jbc.275.6.4239. [DOI] [PubMed] [Google Scholar]
- 66.Saha P, Chen J, Thome K C, Lawlis S J, Hou Z H, Hendricks M, Parvin J D, Dutta A. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol Cell Biol. 1998;18:2758–2767. doi: 10.1128/mcb.18.5.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sclafani R A. Cdc7p-Dbf4p becomes famous in the cell cycle. J Cell Sci. 2000;113:2111–2117. doi: 10.1242/jcs.113.12.2111. [DOI] [PubMed] [Google Scholar]
- 68.Sherman D A, Forsburg S L. Schizosaccharomyces pombe Mcm3p, an essential nuclear protein, associates tightly with Nda4p (Mcm5p) Nucleic Acids Res. 1998;26:3955–3961. doi: 10.1093/nar/26.17.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tada S, Li A, Maiorano D, Mechali M, Blow J J. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takisawa H, Mimura S, Kubota Y. Eukaryotic DNA replication: from pre-replication complex to initiation complex. Curr Opin Cell Biol. 2000;12:690–696. doi: 10.1016/s0955-0674(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 71.Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 72.Tye B K. MCM proteins in DNA replication. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 73.Wohlschlegel J A, Dwyer B T, Dhar S K, Cvetic C, Walter J C, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]