Abstract
BUR1, which was previously identified by a selection for mutations that have general effects on transcription in Saccharomyces cerevisiae, encodes a cyclin-dependent kinase that is essential for viability, but none of its substrates have been identified to date. Using an unbiased biochemical approach, we have identified the carboxy-terminal domain (CTD) of Rpb1, the largest subunit of RNA polymerase II, as a Bur1 substrate. Phosphorylation of Rpb1 by Bur1 is likely to be physiologically relevant, since bur1 mutations interact genetically with rpb1 CTD truncations and with mutations in other genes involved in CTD function. Several genetic interactions are presented, implying a role for Bur1 during transcriptional elongation. These results identify Bur1 as a fourth S. cerevisiae CTD kinase and provide striking functional similarities between Bur1 and metazoan P-TEFb.
The largest subunit of RNA polymerase II (Pol II), Rpb1, contains a highly conserved carboxy-terminal domain (CTD) that has a central role in transcriptional regulation in vivo (3, 11). The Rpb1 CTD consists of multiple repeats of the consensus heptapeptide sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser, which is repeated 26 times in Saccharomyces cerevisiae, 42 times in Drosophila melanogaster, and 52 times in humans and mice (9). Although the CTD is not required for RNA polymerase activity in promoter-independent assays, it is essential in vivo; deletion of the entire CTD in Drosophila and S. cerevisiae results in lethality, while truncation to 11 repeats in yeast confers conditional growth and promoter-specific transcriptional defects (36).
Phosphorylation of the CTD is important for regulation of Pol II activity during the transcription cycle: unphosphorylated Pol II is preferentially recruited into the preinitiation complex (PIC) (33) and then becomes phosphorylated during the transition from initiation to elongation (28). CTD phosphorylation thus has both stimulatory and inhibitory roles; phosphorylation prior to PIC assembly inhibits initiation, while phosphorylation after PIC assembly stimulates promoter escape and elongation. Phosphorylation occurs primarily on serine 2 and serine 5 of the consensus CTD repeat, with serine 2-phosphorylated Rpb1 being enriched distally from the promoter and serine 5-phosphorylated Rpb1 being enriched at promoter-proximal regions (26). Hyperphosphorylation of the CTD is also linked to other essential events during mRNA synthesis, including recruitment of mRNA modification enzymes and pre-mRNA splicing factors (reviewed in reference 50).
The importance of CTD phosphorylation for Pol II regulation has prompted efforts to identify the kinases and phosphatases that determine the CTD phosphorylation state. Several kinases capable of phosphorylating the CTD in vitro have been identified in Drosophila, human, and rodent cell extracts (reviewed in reference 11), but it is not clear whether they all function as CTD kinases in vivo. In S. cerevisiae, where sophisticated genetic analysis can be readily combined with biochemistry to determine their biological roles, three CTD kinases have been identified to date: Kin28, the kinase subunit of TFIIH (8, 12); Srb10, which is a component of the Pol II holoenzyme (31); and Ctk1, the catalytic subunit of the CTDK1 kinase complex (29). These three CTD kinases clearly have different functions in vivo: KIN28 is essential for viability (49), whereas srb10Δ and ctk1Δ strains are each viable yet display distinct mutant phenotypes (3, 27, 29, 51, 56). A recent hypothesis proposes that the functional differences between the kinases are not due to different catalytic activities or substrate preferences within the CTD repeats but instead are due to temporal differences in activity during the transcription cycle (18). Kin28 phosphorylates the CTD after PIC formation, thereby releasing Pol II from the promoter and positively regulating Pol II activity. Srb10, by contrast, phosphorylates Pol II prior to PIC formation, inhibiting recruitment of Pol II to the promoter. Less is known about the specific role of Ctk1, but a ctk1 deletion results in altered CTD phosphorylation, indicating that it is relevant to Pol II function (29, 40), and Ctk1 increases the transcription elongation rate in vitro (30).
An additional CTD kinase with a role in transcription has been identified in human and Drosophila cell extracts. Cdk9 is the catalytic subunit of P-TEFb, a CTD kinase that associates with cyclin T or cyclin K and is required for normal transcription elongation (44). Interestingly, P-TEFb associates with human immunodeficiency virus Tat and is required for Tat-dependent elongation across the human immunodeficiency virus type 1 genome (34, 59). Although sequence and functional homologs of Kin28 and Srb10 have been identified in other eukaryotes, including humans (47, 48, 52), it is not clear whether P-TEFb function is conserved in yeast. The identification of a yeast P-TEFb homolog would permit complementary analysis of P-TEFb function in vivo, including its genetic interactions with other CTD kinases and elongation factors.
We have previously selected for mutations that increase transcription from an upstream activation sequence-less promoter, with the expectation that these mutations would additionally cause general defects in transcription and thereby identify regulators of the basal transcription machinery (43). Two of the genes identified by this selection include BUR1 (also called SGV1) and BUR2, which cause a similar spectrum of pleiotropic mutant phenotypes and encode a cyclin-dependent kinase and its cyclin subunit, respectively (22, 43, 58). The Bur1-Bur2 complex is important for normal growth; BUR1 is essential for viability (22), while a bur2Δ mutation causes extremely slow growth (58). Despite its importance, no substrates have been identified to date for the Bur1-Bur2 complex. Here we provide evidence that the Rpb1 CTD is a substrate for the Bur1-Bur2 cyclin-dependent kinase. Rpb1 coimmunoprecipitates with, and is phosphorylated by, Bur1 on serine 5 within the CTD repeats in a coimmunoprecipitation-kinase assay, and Bur1 phosphorylates a CTD fusion protein. Several genetic interactions that extend the connections between Bur1 and the CTD and strongly imply a role for Bur1 during transcription elongation are described.
MATERIALS AND METHODS
Strains and growth conditions.
The S. cerevisiae strains used in this study are listed in Table 1. All media, including rich yeast-peptone-dextrose (YPD) synthetic complete (SC) drop-out medium (e.g., SC-Ura) and minimal and sporulation media, were made as described previously (46). 6-Azauracil (6AU) plates contained SC-Ura drop-out mix and 50 μM 6AU. Standard genetic methods for mating, sporulation, and tetrad analysis (46) were used throughout.
TABLE 1.
Yeast strains
| Strain | Genotype |
|---|---|
| FY886 | MATahis4-912δ lys2-128δ suc2ΔUAS(−1900/−390) ura3-52 leu2Δ1 bur1-2 |
| GHY296 | MATα his3Δ200 lys2-128δ ura3-52 leu2Δ1 ppr2Δ::HISG-URA3 |
| GY100 | MATα his4-912δ lys2-128δ suc2ΔUAS(−1900/−390) ura3-52 ade8 bur1-2 |
| GY169 | MATahis4-912δ lys2-128δ suc2ΔUAS(−1900/−390) ura3-52 trplΔ63 bur1-2 |
| GY170 | MATα his4-912δ lys2-128δ suc2ΔUAS(−1900/−390) ura3-52 trplΔ63 bur1-2 |
| GY320 | MATahis4-912δ lys2-128δ ura3-52 leu2Δ1 Δ(HTA1-HTB1)::LEU2 |
| GY386 | MATa his3Δ200 lys2-128δ suc2ΔUAS(−1900/−390) ura3-52 ade8 bur1-2 |
| GY458 | MATahis4-912δ lys2-128δ suc2ΔUAS(−1900/−390) ura3-52 trplΔ63 |
| GY460 | MATahis4-912δ lys2-128δ suc2ΔUAS(−1900/−390) ura3-52 leu2Δ1 |
| GY604 | MATα his3Δ200 lys2-128δ ura3-52 trplΔ63 leu2Δ1 bur1-2 |
| GY628 | MATahis3Δ200 lys2-128δ suc2ΔUAS(−1900/−390) ura3-52 ade8 leu2Δ1 bur1-2 |
| GY752 | MATα his3Δ200 lys2-128δ suc2ΔUAS(−1900/−390) ura3-52 leu2Δ1 ctk1Δ::HIS3 |
| GY755 | MATahis4-912δ lys2-128δ suc2ΔUAS(−1900/−390) ura3-52 trplΔ63 leu2Δ1 bur1-2 |
| GY777 | MATα his3Δ200 lys2-128δ ura3-52 trplΔ63 leu2Δ1 ppr2Δ::G-URA3 |
| GY778 | MATahis4-912δ lys2-128δ suc2ΔUAS(−1900/−390) ura3-52 trplΔ63 leu2Δ1 srb10Δ::TRP1 |
| OY2 | MATα his4-912δ lys2-128δ his3Δ200 suc2ΔUAS(−1900/−390) ura3-52 leu2 rpb1Δ::HIS3 (pC3 = LEU2 CEN rpb1Δ103) |
| OY52 | MATα his4-912δ lys2-128δ his3Δ200 suc2ΔUAS(−1900/−390) ura3-52 leu2 rpb1Δ::HIS3 (pRP114 = LEU2 CEN RPB1) |
| OY75 | MATα his3 lys2-128δ suc2ΔUAS(−1900/−390) leu2 trpl kin28-ts3 |
Plasmids.
pGP112 contains a 3.3-kb Sau3A-EcoRI Bur1 fragment in pRS426. pGP211 is identical to pGP112, except it contains bur1-3, a D213A mutant allele created by oligonucleotide-directed mutagenesis. pSM21 contains BUR1 FLAG tagged at the portion corresponding to the N terminus in a pRS426 plasmid background. pSM14 contains FLAG–bur1-3 in a pRS426 plasmid background. The RPB1+ and rpb1 truncation plasmid series has been described in Nonet et al. (36). pRU8 contains both FLAG-BUR1 and His6-BUR2 in pRS426, and pRU9 contains FLAG–bur1-3 and His6-BUR2 in pRS426.
Immunoprecipitation and Western blots.
Yeast transformants were grown to an A600 of 1.0 in selective SC drop-out media. Extracts were made by bead beating in lysis buffer containing 450 mM NaCl essentially as described previously (2). All protein manipulations were carried out at 4°C in the presence of protease inhibitors (aprotinin [100 μg/ml], pepstatin A [70 μg/ml], leupeptin [50 μg/ml], and phenylmethylsulfonyl fluoride [1 mM]). For some experiments, 10 mM NaF, 20 nM okadaic acid, and 1 mM EGTA were added to inhibit phosphatase activity. Immunoprecipitations were performed essentially as described previously (2), except 1 mg of protein extract in lysis buffer was incubated with 50 μl of an M2 anti-FLAG antibody-bead conjugate slurry (Sigma) in a final volume of 1 ml. We have also used a HEPES-acetate immunoprecipitate-/wash buffer (50 mM HEPES, 150 mM potassium acetate, 1 mM magnesium acetate, 1 mM EDTA, 0.05% Tween, 1 mM dithiothreitol) to eliminate background bands for some experiments as described in the figure legends. Following immunoprecipitation, beads were either resuspended in lysis buffer for subsequent kinase assays or in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Western analysis of immunoprecipitated samples was performed as described previously (42). Primary antibodies used were either M2 anti-FLAG (Sigma) or anti-Rpb1 (antibody E2, which was raised against the second exon of Drosophila Rpb1, was a gift of A. Greenleaf, Duke University).
Immunoprecipitation-kinase assay.
A small aliquot (10 μl) of immunoprecipitated sample was added to an equal volume of kinase buffer (25 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, 1 mM dithiothreitol, 10 mM MgCl2), and the reaction was initiated by the addition of 1 μCi of [γ-32P]ATP. The reaction mixtures were incubated at 30°C for 30 min, after which they were stopped by the addition of SDS-PAGE sample buffer. Products were resolved by SDS-PAGE and visualized by autoradiography. For the kinase assay using unlabeled ATP, ATP was added to a final concentration of 2.5 mM.
CTD kinase assay.
FLAG-Bur1 protein was immunoprecipitated as described above. A small aliquot (10 μl) of immunoprecipitate was added to 34 μl of kinase buffer (25 mM Tris-HCl [pH 7.8], 10 mM MgCl2, 0.1% Tween 20, 0.1% nonfat milk) containing 5 μl of β-galactosidase (Gal)–CTD substrate (kind gift of A. Greenleaf, Duke University); the reaction was initiated by the addition of 1 μCi of [γ-32P]ATP, and the mixture was incubated at 30°C for 30 min. Reactions were stopped by the addition of SDS-PAGE sample buffer. Products were resolved by SDS-PAGE and visualized by autoradiography.
RESULTS
Rpb1 coimmunoprecipitates with, and is phosphorylated by, Bur1 in vitro.
BUR1 and BUR2 encode a Cdk-cyclin protein kinase complex proposed to have a general role in Pol II-dependent transcription. To understand the function of this complex and identify potential substrates in as unbiased a manner as possible, we established a Bur1- and Bur2-dependent immunoprecipitation-kinase assay. Briefly, Bur1 was tagged at its amino terminus with the FLAG epitope and expressed in yeast on a high-copy-number plasmid from its own promoter. Neither the FLAG epitope nor the overexpression interfered with BUR1 function, since FLAG-BUR1 complemented bur1 mutations, including bur1Δ, as efficiently as did BUR1+, and overexpression of BUR1+ or FLAG-BUR1+ did not cause any mutant phenotypes. Extracts were prepared, FLAG-Bur1 was immunoprecipitated with FLAG-specific M2 monoclonal antibody beads, and kinase activity towards coimmunoprecipitated proteins was assayed by incubation with [γ-32P]ATP and subsequent gel electrophoresis and autoradiography. Two controls were utilized to determine whether the phosphorylated products were Bur1 dependent: extracts were prepared from a strain that expressed untagged BUR1 and from a strain that expressed FLAG–bur1-3, an allele that is predicted to be catalytically inactive, based on analogous mutations that inactivate other protein kinases (14).
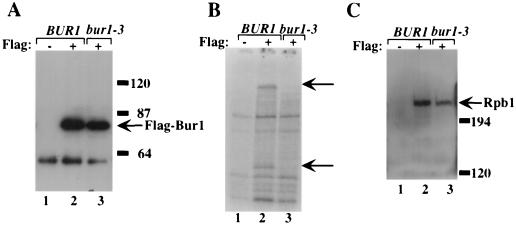
Using this assay, two Bur1-specific substrates were observed, with molecular masses of ∼80 and ∼210 kDa (Fig. 1B); generation of these phosphorylated products required the presence of both the FLAG epitope and the active Bur1. Western blots of the immunoprecipitated material demonstrated that FLAG-BUR1 and FLAG–bur1-3 were expressed and immunoprecipitated to equivalent levels (Fig. 1A); reduced phosphorylation of the ∼80- and ∼210-kDa proteins in the bur1-3 lane (Fig. 1B, compare lanes 2 and 3) is therefore due to loss of kinase activity and not simply to a physical absence of the kinase. Phosphorylation of both substrates was also dependent upon the Bur2 cyclin (58). Because Rpb1 migrates at ∼210 kDa in SDS-polyacrylamide gels, similar to our larger candidate substrate, we first determined whether the ∼210-kDa substrate was Rpb1. FLAG-Bur1 was immunoprecipitated with anti-FLAG agarose beads, and the immunoprecipitated material was probed in Western blots using an Rpb1-specific antibody. The result (Fig. 1C) demonstrated that Rpb1 coimmunoprecipitates with FLAG-Bur1 and that coimmunoprecipitation requires the FLAG epitope, indicating that it is not due to nonspecific binding of Rpb1 to the antibody beads. Coprecipitation of Bur1 and Rpb1 does not require Bur1 kinase activity, as Rpb1 coimmunoprecipitates with the catalytically inactive Bur1-3. Bur1 and Rpb1 coimmunoprecipitated even under the stringent conditions of 0.6 M NaCl (data not shown), indicating that this interaction was highly specific.
FIG. 1.
The largest subunit of RNA Pol II (Rpb1) coimmunoprecipitates with Bur1. (A) Extracts were prepared from yeast cells expressing untagged Bur1 (lane 1), FLAG-Bur1 (lane 2), or FLAG–Bur1-3 (lane 3). FLAG-Bur1 and FLAG–Bur1-3 were immunoprecipitated, and the beads were washed using a Tris-NaCl buffer containing 450 mM NaCl. Immunoprecipitates were resolved by SDS-PAGE on a 7.5% acrylamide gel. Epitope-tagged Bur1 was detected using the M2 anti-FLAG antibody. (B) Immunoprecipitates from panel A were assayed for in vitro kinase activity by incubation with [γ-32P]ATP. The reaction products were resolved by SDS-PAGE on a 7.5% acrylamide gel. Arrows indicate the major Bur1-dependent phosphorylated products. (C) Western analysis of immunoprecipitates from panel A using the anti-Rpb1 E2 antibody, which was raised against the second exon of Drosophila Rpb1. In all panels, the presence or absence of the epitope tag is denoted by a “+” or “−,” respectively. Molecular weight markers are indicated on the right.
The preceding experiment demonstrates that Rpb1 coimmunoprecipitates with Bur1. To determine whether the ∼210-kDa protein phosphorylated in the in vitro kinase assay was Rpb1, immunoprecipitation-kinase assays were performed using extracts prepared from a strain that contained an rpb1 allele (rpb1Δ103) in which the CTD is truncated from 26 to 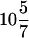 repeats (36). Strains carrying the rpb1Δ103 CTD truncation are viable, and the Rpb1Δ103 protein migrates faster than Rpb1+ in SDS-polyacrylamide gels. We therefore expressed Bur1 and FLAG-Bur1 in Rpb1+ and rpb1Δ103 strains. When the immunoprecipitation-kinase assay was performed using extracts prepared from an rpb1Δ103 strain, the ∼210-kDa phosphorylated protein was not observed; instead, a protein with slightly greater mobility that comigrates with Rpb1Δ103 was phosphorylated (Fig. 2A), demonstrating that the ∼210-kDa substrate is Rpb1. The mobility of the ∼80-kDa substrate was unchanged in the RPB1+ and rpb1Δ103 lanes (data not shown). Combined, these results demonstrate that Rpb1 coimmunoprecipitates with Bur1 and is a substrate for Bur1 in vitro.
repeats (36). Strains carrying the rpb1Δ103 CTD truncation are viable, and the Rpb1Δ103 protein migrates faster than Rpb1+ in SDS-polyacrylamide gels. We therefore expressed Bur1 and FLAG-Bur1 in Rpb1+ and rpb1Δ103 strains. When the immunoprecipitation-kinase assay was performed using extracts prepared from an rpb1Δ103 strain, the ∼210-kDa phosphorylated protein was not observed; instead, a protein with slightly greater mobility that comigrates with Rpb1Δ103 was phosphorylated (Fig. 2A), demonstrating that the ∼210-kDa substrate is Rpb1. The mobility of the ∼80-kDa substrate was unchanged in the RPB1+ and rpb1Δ103 lanes (data not shown). Combined, these results demonstrate that Rpb1 coimmunoprecipitates with Bur1 and is a substrate for Bur1 in vitro.
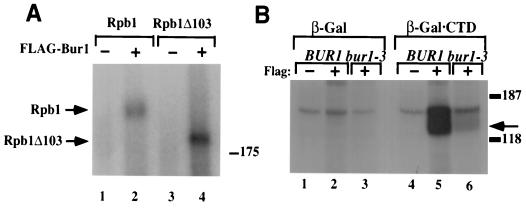
FIG. 2.
Bur1 phosphorylates the Rpb1 CTD in vitro. (A) Bur1+ or FLAG-Bur1 was expressed in strains containing either Rpb1+ or the Rpb1Δ103 CTD truncation as indicated at the top. FLAG-Bur1 was immunoprecipitated using a Tris-acetate buffer, the immunoprecipitates were assayed for kinase activity by the addition of [γ-32P]ATP, and the products were separated in an SDS-PAGE (7.5% acrylamide) gel. The positions of Rpb1 and Rpb1Δ103 are indicated. (B) FLAG antibodies were used to immunoprecipitate Bur1 (lanes 1 and 4), FLAG-Bur1 (lanes 2 and 5), or FLAG–Bur1-3 (lanes 3 and 6) from cellular lysates. Immunoprecipitates were incubated with purified recombinant β-Gal (lanes 1 through 3) or β-Gal–CTD, and reactions were initiated by the addition of [γ-32P]ATP. Reaction products were resolved by SDS-PAGE in a 7.5% acrylamide gel, and phosphorylated products were visualized by autoradiography. The arrow indicates the position of phosphorylated β-Gal–CTD. Phosphorylation of β-Gal–CTD by Bur1 results in a smear of phosphorylated products reminiscent of the hyperphosphorylated forms of Rpb1.
Bur1 phosphorylates a CTD fusion protein in vitro.
The immunoprecipitation-kinase assays described above demonstrate that Rpb1 is phosphorylated by Bur1 in vitro. To determine whether Bur1 phosphorylates Rpb1 within the CTD, we utilized a β-Gal–CTD fusion that has previously been used to investigate CTD phosphorylation (29). When FLAG-Bur1 and FLAG–Bur1-3 were immunoprecipitated and assayed for activity on this model CTD substrate, phosphorylation of β-Gal–CTD was observed using FLAG-Bur1 (Fig. 2B). Phosphorylation required the CTD (Fig. 2B, compare lanes 2 and 5) and epitope-tagged Bur1 (Fig. 2B, compare lanes 4 and 5) and was greatly reduced using the catalytically impaired FLAG–Bur1-3 (Fig. 2B, compare lanes 5 and 6), indicating that it was due to Bur1 activity and not coimmunoprecipitation of another CTD kinase.
Phosphorylation site specificity.
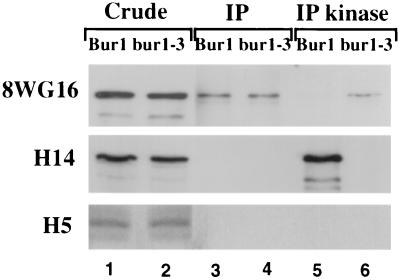
Rpb1 is differentially phosphorylated in vivo, primarily on serine 2 and serine 5 of the YSPTSPS CTD consensus repeat. To determine whether Bur1 associates with a specific phosphorylated subset of Rpb1, FLAG-Bur1 was immunoprecipitated with anti-FLAG beads and the immunoprecipitated material was probed with monoclonal antibodies that recognize unphosphorylated CTD repeats (antibody 8WG16), CTD phosphorylated on serine 2 (antibody H5), and CTD phosphorylated on serine 5 (antibody H14). The Rpb1 population that coimmunoprecipitated with FLAG-Bur1 and FLAG–Bur1-3 cross-reacted with 8WG16 but not with H14 or H5 (Fig. 3, lanes 3 and 4), suggesting that Bur1 associates primarily with Rpb1-containing unphosphorylated CTD repeats. The H5 and H14 antibodies readily recognized Rpb1 in the crude extract (Fig. 3, lanes 1 and 2), indicating that the lack of a signal in the immunoprecipitates was not simply due to the absence of serine 2- and serine 5-phosphorylated forms in the crude extract.
FIG. 3.
Serine specificity of Bur1 interactions. FLAG-Bur1 or FLAG–Bur1-3 was expressed in a BUR+ yeast strain (GY458) from pRU8 and pRU9 and immunoprecipitated (IP) using anti-FLAG beads. The immunoprecipitated material was then incubated in a kinase assay containing nonradioactive ATP. Samples were probed in Western blots using antibodies 8WG16 (specific for unphosphorylated CTD repeats), H14 (phosphoserine 5 specific), or H5 (phosphoserine 2 specific). Lanes 1 and 2 contain 100 μg of crude extract, while lanes 3 through 6 contain material immunoprecipitated from 1 mg of extract.
To determine whether Bur1 phosphorylates the CTD on serine 2 or serine 5 of the consensus repeat, FLAG-Bur1 and its associated proteins were immunoprecipitated, kinase assays were performed using nonradioactive ATP, and the reaction products were then probed with the phosphorylation state-specific antibodies. Although the coimmunoprecipitated Rpb1 was not recognized by H14, after the kinase assay, H14 reactivity was readily detected (Fig. 3, compare lanes 3 and 5). H14 reactivity was dependent upon Bur1 activity, since it was not observed in reaction mixtures containing the inactive FLAG–Bur1-3 (Fig. 3, compare lanes 5 and 6). Bur1-dependent phosphorylation in vitro reduced reactivity with 8WG16 (Fig. 3, compare lanes 5 and 6), as expected, since 8WG16 recognizes only the unphosphorylated repeat. No reactivity with H5 was observed either before or after the kinase reaction, indicating that Bur1 neither associates with nor phosphorylates serine 2 of the CTD repeat to any detectable level. Combined, these results suggest that Bur1 primarily associates with Rpb1 containing unphosphorylated CTD repeats and then phosphorylates Rpb1 on serine 5.
Genetic links between BUR1 and CTD function.
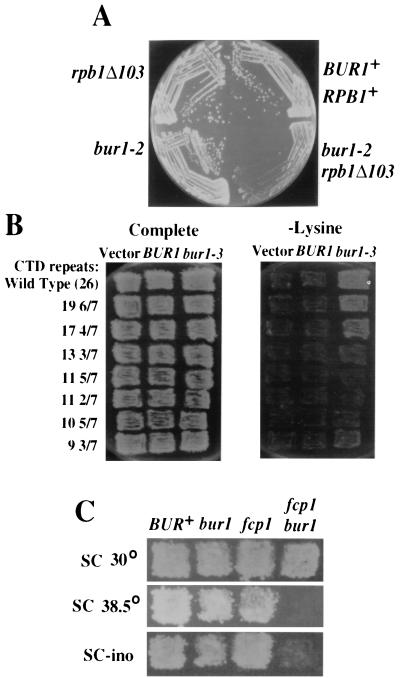
The results described above demonstrate that Rpb1 coimmunoprecipitates with, and is a substrate for, Bur1 in vitro. To determine whether these activities are physiologically relevant, we performed the following genetic tests examining whether the functions of Bur1 and the Rpb1 CTD overlap in vivo. First, bur1 mutations were crossed with rpb1 CTD truncation mutants to allow examination of the double-mutant phenotype. The bur1-2 allele was chosen for the following tests because it causes several clear phenotypes yet does not cause a severe growth defect by itself that might complicate interpretation of double-mutant phenotypes. The bur1-2 rpb1Δ103 double mutants grew extremely poorly compared to the individual mutants (Fig. 4A), suggesting that BUR1 and the Rpb1 CTD participate in the same process. Second, when the catalytically impaired bur1-3 allele is overexpressed from a high-copy-number plasmid, it causes increased transcription from the lys2-128δ promoter insertion allele, resulting in lysine-independent growth (Fig. 4B). This dominant negative Spt− phenotype (57) is suppressed by rpb1 CTD truncations (Fig. 4B), indicating that the bur1-3 dominant negative Spt− phenotype requires an intact CTD. Third, if Bur1 is a physiological CTD kinase, then we would predict genetic interactions between bur1 mutations and mutations in genes that encode other CTD kinases or phosphatases. We therefore crossed bur1-2 with deletions of SRB10 and CTK1, two temperature-sensitive alleles of KIN28, and an fcp1 allele (10). FCP1 encodes an essential conserved CTD phosphatase (1, 5) that is broadly required for transcription in vivo (25) and stimulates both initiation and elongation by Pol II under specific assay conditions in vitro (7). Interestingly, bur1-2 was lethal in combination with ctk1Δ, but no effects were observed in combination with the two kin28 temperature-sensitive alleles or with srb10Δ (Table 2). These results link the functions of BUR1 and CTK1 and distinguish these two kinases functionally from KIN28 and SRB10. We also detected a genetic interaction between bur1 and fcp1; the bur1-2 fcp1-110 double mutants grew extremely slowly and were very tightly Ts− and Ino−, unlike either of the single mutants (Fig. 4C). To determine the extent of functional overlap with BUR1, we also tested whether kin28, srb10Δ or ctk1Δ mutations cause a Bur− phenotype. In agreement with the synthetic lethality results that suggest partial redundancy between CTK1 and BUR1, ctk1Δ is weakly Bur−, while srb10Δ and kin28 temperature-sensitive alleles are Bur+ (data not shown). In summary, we have documented genetic interactions between Bur1, the Rpb1 CTD, another CTD kinase, and a CTD phosphatase that strongly link Bur1 and CTD functions in vivo.
FIG. 4.
RPB1-BUR1 genetic interactions. (A) Yeast strains with the indicated genotypes were streaked onto YPD media and grown at 30°C for 3 days. The bur1-2 rpb1Δ103 double mutants grew extremely poorly relative to the single mutants. (B) Yeast strains containing RPB1 or rpb1 CTD truncation alleles were transformed with either a 2μm vector (pRS426), a 2μm BUR1 plasmid (pGP112), or a 2μm bur1-3 plasmid (pGP211). Transformants were replica plated to an SC complete plate (left) and an SC plate lacking lysine (right) and were grown at 30°C for 3 days. The CTD truncations suppressed the bur1-3 Spt− phenotype. (C) Yeast strains with the indicated genotypes were replica plated to complete plates at 30 and 38.5°C and to complete plates lacking inositol.
TABLE 2.
bur1 genetic interactionsa
| Allele | Function | Growth phenotype
|
|
|---|---|---|---|
| BUR+ | bur1-2 | ||
| None | ++++ | +++ | |
| ctk1Δ::HIS3 | CTD kinase | +++ | − |
| spt4Δ | DSIF subunit | ++++ | − |
| spt5-4 | DSIF subunit | ++++ | − |
| rpb1Δ103 | Pol II subunit | +++ | + |
| rpb2-7 | Pol II subunit | +++ | ++ |
| bur2-1 | Cyclin | +++ | + |
| spt6-14 | Binds histones | ++++ | + |
| ppr2Δ | TFIIS | ++++ | ++ |
| fcp1-110 | CTD phosphatase | +++ | + |
| kin28-ts4 | CTD kinase | +++ | +++ |
| srb10Δ::TRP1 | CTD kinase | +++ | +++ |
| srb4-138 | Mediator component | ++++ | +++ |
| Δ(hta1-htb1)::LEU2 | Histones H2A and H2B | ++++ | +++ |
| spt16-197 | FACT subunit | ++++ | ++++ |
Double mutants were created between bur1-2 and the mutations in the leftmost column. The relative growth phenotype of the double mutants is shown in the rightmost column, next to the growth rate of the single mutant. Wild-type growth is indicated by ++++, synthetic lethality is indicated by −, and intermediate growth rates are estimated by the number of plus signs.
Genetic evidence implicating Ctk1 and Bur1 in elongation.
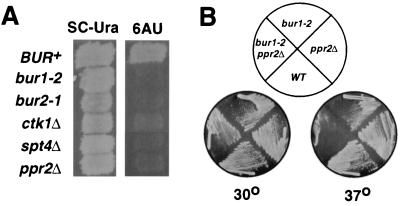
The synthetic lethality observed between bur1-2 and ctk1Δ mutations suggested that Bur1 and Ctk1 have overlapping functions. Although the role of CTK1 in vivo is not completely understood, Ctk1 has been shown to stimulate transcription elongation in vitro (30). To determine whether Ctk1 and Bur1 might function during elongation in vivo, we examined their response to 6AU. Sensitivity to 6AU is a strong indicator of a role during elongation, as mutations in the genes that encode the elongation factors TFIIS, the Spt4-Spt5 complex (DSIF), and the elongation-defective forms of Pol II all are 6AU sensitive (45). The bur1-2, bur2-1, and ctk1Δ alleles each conferred sensitivity to 6AU (Fig. 5A), similar to the deletion alleles of PPR2 (TFIIS) or SPT4 (DSIF subunit), suggesting that they participate in elongation in vivo. In contrast, mutations in KIN28 or SRB10, which each encode CTD kinases, are 6AU resistant (data not shown).
FIG. 5.
Genetic evidence for a Bur1 role during elongation. (A) 6AU sensitivity of CTD kinase mutants. Strains containing mutations shown on the left were replica plated to SC-Ura and SC-Ura plus 6AU plates. The photographs were taken after 2 days of growth at 30°C. (B) Temperature sensitivity of bur1-2 ppr2Δ double mutants. Strains GHY296 and FY886 were crossed, and representative progeny with the indicated genotypes, derived from a single tetrad, were streaked onto YPD plates and grown at 30 or 37°C for 2 days.
To further test whether Bur1 has a role during elongation in vivo, bur1-2 mutants were crossed with strains containing mutations in genes encoding other known elongation factors. Several combinatorial effects were observed. In particular, bur1-2 was lethal in combination with spt4Δ and spt5-4 (Table 2), indicating that in the presence of a defective Spt4-Spt5 complex (DSIF), normal BUR1 function is essential for viability. Synthetic growth defects were also observed in combination with spt6 and ppr2 mutations, and bur1-2 ppr2Δ mutants were strongly temperature sensitive, unlike either of the single mutants (Fig. 5B). The bur1 synthetic double-mutant phenotypes were specific for elongation factors, since no combinatorial defects were observed when bur1-2 was combined with mutations in the genes encoding the Srb4 subunit of the Pol II holoenzyme, the Srb10 and Kin28 CTD kinases, or deletion of one of the histone H2A-H2B loci (Table 2). An enhancement of the bur1-2 growth defect and other bur1 mutant phenotypes was also observed in combination with the elongation-deficient rpb2-7 and rpb2-10 alleles. These effects were not very strong, however, and might simply be due to additive effects between these bur1 and rpb2 alleles. Interestingly, bur1-2 also showed no synthetic phenotype when combined with a mutation in the gene encoding the Spt16 subunit of the FACT complex, which is also involved in elongation. Although this result stands in contrast to the interactions of bur1-2 with the SPT4, SPT5, SPT6, and PPR2 genes, it is consistent with recent observations that FACT and DSIF likely have different, and perhaps even antagonistic, modes of action (37, 54).
DISCUSSION
Phosphorylation of the Rpb1 carboxy-terminal domain plays an important role in regulating Pol II elongation (11), Rpb1 stability (6, 21, 35), and interactions with both transcription factors (4, 38, 53) and enzymes involved in RNA processing (19). It is thus not altogether surprising that multiple protein kinases target the CTD. Three cyclin-dependent kinases (Srb10, Kin28, and Ctk1) were previously proposed to phosphorylate the Rpb1 CTD during transcription in yeast. Here we present evidence that Bur1 is a fourth physiological CTD kinase that affects transcription. First, Rpb1 coimmunoprecipitates with Bur1, even under highly stringent conditions. Second, Bur1 phosphorylates Rpb1 in an immunoprecipitation-kinase assay; phosphorylation in this assay requires functional Bur1, as it does not occur using the catalytically impaired Bur1-3 or in the absence of the Bur2 cyclin. Third, Bur1 phosphorylates a CTD fusion protein previously used to assay CTD kinase activity. Fourth, several genetic tests establish strong connections between BUR1 and CTD function: rpb1 and bur1 mutations cause overlapping Spt−, Ino−, and 6AU− mutant phenot ypes; bur1 mutations cause poor growth in combination with CTD truncations; and a bur1 mutation causes lethality and synthetic phenotypes in combination with mutations in CTK1 and FCP1, which encode a CTD kinase and a CTD phosphatase, respectively. These combined biochemical and genetic results provide a compelling argument that Bur1 affects transcription through phosphorylation of the Rpb1 CTD.
Although the four yeast CTD kinases each affect transcription, genetic analysis indicates that they nonetheless have different specific roles in vivo: KIN28 and BUR1 are essential for viability (22, 49), whereas srb10 and ctk1 deletion strains are relatively healthy, and viable mutations in each of these genes confer distinct phenotypes (3, 27, 29, 51, 56). The functions of Kin28, Srb10, and Ctk1 have been well documented; as a subunit of TFIIH, Kin28 plays a positive role in transcription of most genes, Srb10 represses 3% of the yeast genome as a subunit of the Pol II holoenzyme (20), and Ctk1 stimulates the elongation efficiency of Pol II in vitro (30). By contrast, little was known about Bur1 beyond the recent finding that its kinase activity was dependent upon the Bur2 cyclin (58). In addition to identifying Bur1 as a physiological CTD kinase, our results further indicate that Bur1 and Ctk1 functions are more related to each other than they are to Kin28 and Srb10. We found that bur1 mutations are lethal in combination with ctk1Δ but are unaffected by srb10Δ or kin28 temperature-sensitive mutations (Table 2). Because synthetic lethality is considered a genetic hallmark of overlapping or redundant function (16), the simplest interpretation of these results is that BUR1 and CTK1 functions are partially redundant and distinct from those of KIN28 and SRB10. This conclusion is supported by our finding that bur1 and ctk1Δ mutations each cause a Bur− phenotype, while srb10Δ and kin28 mutations are Bur+ (G. Prelich, unpublished observations). The presence of phenotypic distinctions between the four CTD kinases likely reflects their requirement during successive stages of the transcription cycle. The bur1 and ctk1Δ 6AU sensitivity and the genetic interactions that we detected between bur1-2 and spt4 and spt5 (DSIF subunits) and ppr2 (TFIIS) mutations strongly link Bur1 and Ctk1 with elongation, whereas Srb10 and Kin28 are involved in PIC formation and promoter escape.
A major remaining challenge is to identify why phosphorylation of the CTD by these kinases has different biological effects. Several simple models can be envisioned: the CTD kinases may phosphorylate different residues within the repeats, they may be spatially restricted to different specific promoters, they may phosphorylate the CTD during different points within the transcription cycle, or they may have additional targets beyond the CTD. The distinct roles of Kin28 and Srb10 were previously proposed to be due to their acting during successive temporal stages of the transcription cycle, with Srb10 inhibiting Pol II recruitment to the PIC and Kin28 stimulating promoter escape (18). This temporal model for regulation of Rpb1 by the successive actions of CTD kinases can easily accommodate the inclusion of Bur1 and Ctk1 acting subsequently to Kin28 and Srb10 to facilitate transcript elongation. As described above, Bur1 and Ctk1 are both proposed to be involved in elongation yet their mutant phenotypes suggest that their specific roles are different. This difference may be accounted for by their target site specificity within the CTD repeats. We found that Bur1 primarily phosphorylates serine 5 in vitro (Fig. 3), while a previous result suggests that Ctk1 phosphorylates serine 2 (40). In light of a recent study demonstrating that Rpb1 phosphorylated on serine 5 preferentially cross-links to the promoter region and that Rpb1 phosphorylated on serine 2 is associated with the distal coding region (26), this implicates Bur1 in an early aspect of elongation, with Ctk1 functioning further downstream. Alternatively, like P-TEFb (13, 23, 24), Bur1 might also phosphorylate Spt5. Consistent with this idea, bur1 and spt5 mutations cause similar mutant phenotypes and interact genetically (Table 2) and SPT5 shows genetic interactions with the CTD and FCP1 similar to those demonstrated here for BUR1 (G. Hartzog, unpublished data). Thus, Bur1 might phosphorylate and regulate both Spt5 and Rpb1.
Is Bur1 or Ctk1 a functional homolog of P-TEFb?
P-TEFb is a cyclin-dependent protein kinase that stimulates elongation in Drosophila and mammalian cell extracts (44). Bur1 and Ctk1 were the best candidates for yeast P-TEFb catalytic subunits, as they were originally cited as being equally similar to Cdk9, the P-TEFb catalytic subunit in metazoans (59). A more recent phylogenetic sequence comparison (32) using the recently completed Drosophila, Caenorhabditis elegans, and available human genomic sequences indicates that Bur1 is the likely yeast ortholog of Cdk9, whereas Ctk1 is more closely related to a human kinase, CHED (15). Four results described here now provide experimental support for this idea, extending the similarities between Bur1 and P-TEFb beyond sequence alignments, into the functional level. First, like P-TEFb, Bur1 is a cyclin-dependent kinase that phosphorylates the Rpb1 CTD. Second, the 6AU sensitivity of bur1 mutations and synthetic lethality with ppr2Δ (TFIIS) mutations suggest that, like P-TEFb, Bur1 is also involved in elongation. Third, P-TEFb interacts biochemically with DSIF (17, 55), composed of Spt4 and Spt5 subunits, while bur1 mutations interact genetically with spt4 and spt5 mutations (Table 2) (G. Hartzog, unpublished observations). Fourth, like P-TEFb (39, 41), Bur1 physically associates with Pol II. Several of these characteristics are also shared with Ctk1. Thus, the functional similarities between Bur1, Ctk1, and P-TEFb, combined with the identification of CHED in humans, force us to consider the intriguing possibility that multiple P-TEFb-like CTD kinases might be involved in elongation in yeast and in other organisms.
ACKNOWLEDGMENTS
We thank Fred Winston for critical reading of the manuscript and Gerard Faye, Arno Greenleaf, and David Bregman for reagents and strains.
This work was supported by research grants GM60479 to G.H. and GM52486 to G.P. from the National Institutes of Health.
REFERENCES
- 1.Archambault J, Chambers R S, Kobor M S, Ho Y, Cartier M, Bolotin D, Andrews B, Kane C M, Greenblatt J. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:14300–14305. doi: 10.1073/pnas.94.26.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cang Y, Auble D T, Prelich G. A new regulatory domain on the TATA-binding protein. EMBO J. 1999;18:6662–6671. doi: 10.1093/emboj/18.23.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Carty S M, Goldstrohm A C, Sune C, Garcia-Blanco M A, Greenleaf A L. Protein-interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc Natl Acad Sci USA. 2000;97:9015–9020. doi: 10.1073/pnas.160266597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers R S, Kane C M. Purification and characterization of an RNA polymerase II phosphatase from yeast. J Biol Chem. 1996;271:24498–24504. doi: 10.1074/jbc.271.40.24498. [DOI] [PubMed] [Google Scholar]
- 6.Chang A, Cheang S, Espanel X, Sudol M. Rsp5 WW domains interact directly with the carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2000;275:20562–20571. doi: 10.1074/jbc.M002479200. [DOI] [PubMed] [Google Scholar]
- 7.Cho H, Kim T K, Mancebo H, Lane W S, Flores O, Reinberg D. A protein phosphatase functions to recycle RNA polymerase II. Genes Dev. 1999;13:1540–1552. doi: 10.1101/gad.13.12.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cismowski M J, Laff G M, Solomon M J, Reed S I. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corden J L. Tails of RNA polymerase II. Trends Biochem Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 10.Costa P J, Arndt K M. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics. 2000;156:535–547. doi: 10.1093/genetics/156.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 12.Feaver W J, Svejstrup J Q, Henry N L, Kornberg R D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 13.Garber M E, Mayall T P, Suess E M, Meisenhelder J, Thompson N E, Jones K A. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 Tat–P-TEFb complex to TAR RNA. Mol Cell Biol. 2000;20:6958–6969. doi: 10.1128/mcb.20.18.6958-6969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbs C S, Zoller M J. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266:8923–8931. [PubMed] [Google Scholar]
- 15.Grana X, De Luca A, Sang N, Fu Y, Claudio P P, Rosenblatt J, Morgan D O, Giordano A. PITALRE, a nuclear CDC2-related protein kinase that phosphorylates the retinoblastoma protein in vitro. Proc Natl Acad Sci USA. 1994;91:3834–3838. doi: 10.1073/pnas.91.9.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guarente L. Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet. 1993;9:362–366. doi: 10.1016/0168-9525(93)90042-g. [DOI] [PubMed] [Google Scholar]
- 17.Hartzog G A, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hengartner C J, Myer V E, Liao S M, Wilson C J, Koh S S, Young R A. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 19.Hirose Y, Manley J L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 20.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 21.Huibregtse J M, Yang J C, Beaudenon S L. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irie K, Nomoto S, Miyajima I, Matsumoto K. SGV1 encodes a CDC28/cdc2-related kinase required for a G alpha subunit-mediated adaptive response to pheromone in S. cerevisiae. Cell. 1991;65:785–795. doi: 10.1016/0092-8674(91)90386-d. [DOI] [PubMed] [Google Scholar]
- 23.Ivanov D, Kwak Y T, Guo J, Gaynor R B. Domains in the Spt5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J B, Sharp P A. Positive transcription elongation factor b phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J Biol Chem. 2001;276:12317–12323. doi: 10.1074/jbc.M010908200. [DOI] [PubMed] [Google Scholar]
- 25.Kobor M S, Archambault J, Lester W, Holstege F C, Gileadi O, Jansma D B, Jennings E G, Kouyoumdjian F, Davidson A R, Young R A, Greenblatt J. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol Cell. 1999;4:55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- 26.Komarnitsky P, Cho E J, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laybourn P J, Dahmus M E. Phosphorylation of RNA polymerase IIA occurs subsequent to interaction with the promoter and before initiation of transcription. J Biol Chem. 1990;265:13165–13173. [PubMed] [Google Scholar]
- 29.Lee J M, Greenleaf A L. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1991;1:149–167. [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J M, Greenleaf A L. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J Biol Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- 31.Liao S M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Kipreos E T. Evolution of cyclin-dependent kinases (CDKs) and CDK-activating kinases (CAKs): differential conservation of CAKs in yeast and metazoa. Mol Biol Evol. 2000;17:1061–1074. doi: 10.1093/oxfordjournals.molbev.a026387. [DOI] [PubMed] [Google Scholar]
- 33.Lu H, Flores O, Weinmann R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marshall N F, Peng J, Xie Z, Price D H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 35.Morris D P, Greenleaf A L. The splicing factor, prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2000;275:39935–39943. doi: 10.1074/jbc.M004118200. [DOI] [PubMed] [Google Scholar]
- 36.Nonet M, Sweetser D, Young R A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987;50:909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- 37.Orphanides G, Wu W H, Lane W S, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 38.Otero G, Fellows J, Li Y, de Bizemont T, Dirac A M, Gustafsson C M, Erdjument-Bromage H, Tempst P, Svejstrup J Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 39.Parada C A, Roeder R G. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 1999;18:3688–3701. doi: 10.1093/emboj/18.13.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patturajan M, Conrad N K, Bregman D B, Corden J L. Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J Biol Chem. 1999;274:27823–27828. doi: 10.1074/jbc.274.39.27823. [DOI] [PubMed] [Google Scholar]
- 41.Ping Y H, Rana T M. Tat-associated kinase (P-TEFb): a component of transcription preinitiation and elongation complexes. J Biol Chem. 1999;274:7399–7404. doi: 10.1074/jbc.274.11.7399. [DOI] [PubMed] [Google Scholar]
- 42.Prelich G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2α homolog that has both positive and negative roles in transcription in vivo. Mol Cell Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prelich G, Winston F. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics. 1993;135:665–676. doi: 10.1093/genetics/135.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price D H. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reines D, Conaway R C, Conaway J W. Mechanism and regulation of transcriptional elongation by RNA polymerase II. Curr Opin Cell Biol. 1999;11:342–346. doi: 10.1016/S0955-0674(99)80047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 47.Serizawa H, Makela T P, Conaway J W, Conaway R C, Weinberg R A, Young R A. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature. 1995;374:280–282. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- 48.Shiekhattar R, Mermelstein F, Fisher R P, Drapkin R, Dynlacht B, Wessling H C, Morgan D O, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 49.Simon M, Seraphin B, Faye G. KIN28, a yeast split gene coding for a putative protein kinase homologous to CDC28. EMBO J. 1986;5:2697–2701. doi: 10.1002/j.1460-2075.1986.tb04553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinmetz E J. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 51.Surosky R T, Strich R, Esposito R E. The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose. Mol Cell Biol. 1994;14:3446–3458. doi: 10.1128/mcb.14.5.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tassan J P, Jaquenoud M, Leopold P, Schultz S J, Nigg E A. Identification of human cyclin-dependent kinase 8, a putative protein kinase partner for cyclin C. Proc Natl Acad Sci USA. 1995;92:8871–8875. doi: 10.1073/pnas.92.19.8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Usheva A, Maldonado E, Goldring A, Lu H, Houbavi C, Reinberg D, Aloni Y. Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell. 1992;69:871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- 54.Wada T, Orphanides G, Hasegawa J, Kim D K, Shima D, Yamaguchi Y, Fukuda A, Hisatake K, Oh S, Reinberg D, Handa H. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol Cell. 2000;5:1067–1072. doi: 10.1016/s1097-2765(00)80272-5. [DOI] [PubMed] [Google Scholar]
- 55.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wahi M, Johnson A D. Identification of genes required for alpha 2 repression in Saccharomyces cerevisiae. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winston F, Sudarsanam P. The SAGA of Spt proteins and transcriptional analysis in yeast: past, present, and future. Cold Spring Harbor Symp Quant Biol. 1998;63:553–561. doi: 10.1101/sqb.1998.63.553. [DOI] [PubMed] [Google Scholar]
- 58.Yao S, Neiman A, Prelich G. BUR1 and BUR2 encode a divergent cyclin-dependent kinase–cyclin complex important for transcription in vivo. Mol Cell Biol. 2000;20:7080–7087. doi: 10.1128/mcb.20.19.7080-7087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]