Abstract
The p300/CREB-binding protein (CBP) family of proteins consists of coactivators that influence the activity of a wide variety of transcription factors. Although the mechanisms that allow p300/CBP proteins to achieve transcriptional control are not clear, it is believed that the regulation of chromatin is an important aspect of the process. Here, we describe a new level of p300-dependent control mediated through the functional interaction between p300/CBP and members of the family of nucleosome assembly proteins (NAP), which includes NAP1, NAP2, and TAF1. We find that NAP proteins, which have previously been implicated in the regulation of transcription factor binding to chromatin, augment the activity of different p300 targets, including p53 and E2F, through a process that is likely to involve the physical interaction between p300 and NAP. NAP proteins can form oligomers, and the results show that NAP proteins can bind to both core histones and p300 coactivator proteins, perhaps in a multicomponent ternary complex. We also provide data in support of the idea that histones can influence the interaction between p300 and NAP protein. These results argue that NAP is a functionally important component of the p300 coactivator complex and suggest that NAP may serve as a point of integration between transcriptional coactivators and chromatin.
Transcription of the eukaryotic genome is a dynamic process in which genes are constantly being switched on and off. Mammalian genomic DNA exists as chromatin, which is subject to the action of sequence-specific transcription factors and their associated coactivator complexes that regulate the transcription of target genes. The p300/CREB-binding protein (CBP) family is a group of coactivator proteins that act to nucleate the assembly of diverse cofactors into a coactivator complex (13, 14, 28, 48, 62). Furthermore, p300/CBP proteins have been implicated in regulating a variety of sequence-specific transcription factors, including the p53 tumor suppressor protein and the cell cycle-regulating transcription factor E2F (4, 21, 37, 38, 54). Although the molecular complexity of the p300/CBP coactivator complex has yet to be resolved, cofactors that have been found to associate with p300/CBP include P/CAF, P/CIP (ACTR or AIB1), SRC1, and JMY (2, 12, 29, 47, 53, 60, 61).
The gene-regulating properties of chromatin can be influenced by posttranslational modification, such as acetylation, and a role for transcriptional coactivator complexes in mediating and regulating the acetylation of chromatin has been suggested (7, 19, 52, 57, 58). For example, p300/CBP proteins together with P/CAF, P/CIP, and SRC1 possess an intrinsic histone acetyltransferase activity (HAT) that can acetylate histones (5, 12, 43, 50, 61). Moreover, p300/CBP and P/CAF are capable of acetylating nonhistone proteins, including p53, which may be required to augment p53 activity in vivo (6, 20, 24, 46). Taken together, these observations suggest that sequence-specific transcription factors recruit p300/CBP coactivator complexes to target genes and that the acetylation of the local chromatin environment facilitates its accessibility to transcription factors and other protein components required to stimulate transcription.
The most basic repeating unit of chromatin is the nucleosome, which is composed of an octamer of core histones (an H3-H4 tetramer bound to two H2A-H2B dimers) wrapped around about 146 bp of DNA. Chromatin is generally inhibitory to transcription and can impede the binding of certain transcription factors (32, 33, 51). In the cell, the need to regulate the influence of chromatin appears to be achieved in part through utilizing multicomponent remodeling activities, such as ACF, CHRAC, SWI-SNF, RSC, and NURF, which possess the common property of perturbing chromatin in an ATP-dependent fashion (3, 10, 27, 31). The transcriptional activation of a target gene in a chromatin environment is therefore most likely to involve the coordinated interplay between transcription factors, coactivator complexes, and multicomponent chromatin remodeling activities.
The nucleosome assembly protein/template activating factor (NAP/TAF) family (from now on referred to as NAP) is a group of histone chaperone-like proteins which have been credited with playing a variety of roles related to transcriptional control and possibly DNA replication. For example, the TAF1 member of the family was identified on the basis of its ability to stimulate adenovirus replication of a viral chromatin template (42). Furthermore, the TAF1 gene (also called set) is the subject of a chromosomal aberration in myeloid leukemogenesis (56), implying that deregulated NAP activity may play a role in oncogenesis. Another member of the family, yeast NAP1, was shown to possess chromatin assembly and histone binding capability (16), while similar activities have been established for the other NAP proteins (42, 45), and NAP1 can augment the binding of transcription factors to a nucleosome-containing binding site (59). The stimulation of transcription factor binding and nucleosome displacement occurs in a manner similar to the action of nucleoplasmin and through a process that requires nucleosome disassembly (59). Drosophila NAP1 was further shown to exist as a complex of H2A and H2B (9, 26), similar results having been observed in HeLa cells (11), suggesting that NAP1 may act as a histone chaperone. NAP proteins may also function in the assembly of regularly spaced nucleosomal arrays, since the ability of ACF to assemble nucleosomal arrays requires core histones, ATP, DNA, and NAP1 or CAF1 (27). In addition, NAP proteins have been shown to be associated not only with core histones in cytosolic extracts (11, 25) but also with cyclin B within an independent complex (30) and undergo nucleocytoplasmic shuttling during cell cycle progression (25). Overall, therefore, the NAP family comprises a group of multifunctional proteins that can participate in different aspects of chromatin-related activities.
Here, we report that p300/CBP proteins functionally interact with members of the NAP family of proteins. By studying the effect of NAP on p300/CBP-dependent transcription factors, such as p53 and E2F, we find that NAP augments p300/CBP-dependent transcription. Moreover, we find that NAP proteins can form both homomers and heteromers and that all members of the NAP family analyzed can bind directly to both core histones and p300 coactivator proteins. Most importantly, the data suggest that NAP proteins can form a ternary complex involving p300 and histones. These results argue that NAP proteins are likely to function as important components of p300/CBP-dependent effects and suggest that NAP proteins serve as a point of integration between coactivators and chromatin.
MATERIALS AND METHODS
Isolation of p300-interacting proteins.
For the isolation of p300-interacting proteins, the yeast strain CTY10.5 containing the LexA–β-galactosidase reporter vector pLex (HIS) and pLex-p300611–2283 were as described previously (8, 47). Screening a 10.5-day-postcoitum mouse embryo random-primed cDNA library fused to the VP16 trans activation domain (55) yielded two positive clones containing NAP2 and one containing the NAP1 sequence. Full-length NAP2 clones were isolated through screening cDNA libraries prepared from F9 EC (18).
Immunoprecipitation.
For immunoprecipitation, U2OS cells were transfected with pG4-p300611–2283 (30 μg), pG4 (30 μg), or pCMV-HA-NAP2 (30 μg) and after 48 h harvested in a solution containing 50 mM Tris-HCl (pH 7.4), 120 mM NaCl, 5 mM EDTA, 0.5% NP-40, 50 mM NaF, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.2 mM sodium orthovanadate, leupeptin (0.5 μg/ml), trypsin inhibitor (0.5 μg/ml), aprotinin (0.5 μg/ml), and bestatin (40 μg/ml) and incubated on ice for 30 min. The cell extract was precleared by incubation with protein A-agarose for 1 h at 4°C, and the supernatant was incubated with a mouse anti-Gal4 monoclonal antibody (Santa Cruz) for a further 1 h at 4°C. Samples were incubated with protein A agarose for another 1 h, collected, and washed three times in extraction buffer. Immunocomplexes were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and thereafter immunoblotted with a mouse antihemagglutinin (anti-HA) monoclonal antibody (Boehringer Mannheim).
For immunoprecipitation of endogenous NAP2, extracts were prepared from A31 cells as described above. Immunoprecipitation was performed as described above with anti-NAP2 peptide antibody in the presence or absence of the competing homologous peptide (see below). After electrophoresis, immunoblotting with anti-NAP2, anti-p300 (NM11; Calbiochem), or anti-JMY (47) was performed.
Expression vectors.
The NAP2 1–123, 98–386, 1–279, 234–386, 98–386, Δ110–230, and 110–230 derivatives were prepared by PCR using the appropriate primers, and the resulting fragments were isolated and cloned into the pCDNA3 (Invitrogen) backbone at the BamHI/XhoI site. For the glutathione S-transferase (GST)–p300 constructs, the appropriate fragments were removed by a restriction enzyme digest from Gal4-p300 (37) and cloned directly into the pGEX-3KG vector (Pharmacia). pCBP HAT was as previously described (40), and expression vectors for p53 and E2F-1, together with pBax-luc, were as previously described (37). pG4-p30019–567 was prepared by subcloning the relevant fragment from Gal4-p300 into pCDNA3.
NAP and p300 binding assays.
For the in vitro binding assay, the indicated NAP2 proteins were in vitro translated by using a TNT-coupled reticulocyte lysate system (Promega) in the presence of [35S]methionine-cysteine (Amersham) and incubated with GST beads loaded with the fusion protein in TNN buffer (50 mm Tris [pH 8.0], 150 mM NaCl, 0.5% NP-40, 1 mM EDTA, and protease inhibitor cocktail [Calbiochem]). After 1 h of incubation at 4°C, the beads were washed three times with 1 ml of TNN buffer. Protein bound to the beads was eluted directly into SDS loading buffer. Flag epitope-tagged p300 protein (residues 1135 to 2414) and wild-type p300 protein (43) were expressed in Sf9 cells in the baculovirus system and bound to M2-anti-Flag antibody–agarose (Sigma). For the control, the M2 beads were treated with cell extract from uninfected Sf9 cells. NAP2 protein in pET28 (Novagen) was expressed and purified using nickel beads according to the manufacturer's instructions (Pharmacia). The eluted NAP2 protein was dialyzed against 50 mM Tris (pH 7.5)–100 mM KCl–20% glycerol–0.2 mM PMSF–0.1 mM DTT. NAP2 protein was incubated with either p300 coupled or control beads with or without histones at 4°C for 2 h in buffer containing 50 mM Tris (pH 7.5), 1.5 mM MgCl, 0.2 mM EDTA, 0.5% NP-40, 10% glycerol, protease inhibitor cocktail (Calbiochem), 0.1 mM DTT, 0.1 mM PMSF, and 0.5 mg of bovine serum albumin per ml. The protein complex on the beads was washed three times in TNN buffer and subjected to SDS-PAGE, followed by a Western blot with anti-NAP2 antiserum. Alternatively, in vitro-translated [35S]methionine-labeled NAP2 and mutant derivatives were used in the binding reaction, and binding efficiency was assessed after SDS-PAGE.
Mammalian two-hybrid assays.
For the mammalian two-hybrid assay, 0.5 μg of pG4-p300611–2283, -p300611–1257, -p3001302–1572, -p3001572–2283, or -p3001572–1906 (37) was transfected with 0.5 μg of pVP16 or pVP16-NAP2 into U2OS cells. The Gal4 reporters pG5-luc and pG4-AdML-luc have been described previously (37).
Transfection.
Transfection of SAOS2 and U2OS cells was carried out as previously described (47, 49). Immunoblotting of transfected cell extracts was performed as previously described using anti-p53 (DO1; Santa Cruz), anti-p21 (C19; Santa Cruz), and anti-NAP2 peptide antiserum.
Anti-NAP2 peptide antibody.
The anti-NAP2 peptide antibody was raised in rabbits against the peptide taken from the C-terminal region of NAP2 containing residues GDEEGEDEDDDDDDADVNPKK.
Core histone octamer preparation.
Core histone octamers were prepared from chicken blood as described previously (22). The composition was checked by SDS-PAGE and verified to contain H2A, H2B, H3, and H4.
Histone binding assays.
Glutathione beads containing Drosophila GST histone tails (17) derived from H2A, H3, H2B, and H4 (2 μg of each) and in vitro-translated NAP were incubated in the binding buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 150 mM NaCl, 0.1% Triton X-100, 0.5 mM PMSF, 1 mM DTT) for 1 h at 4°C. Beads were collected by centrifugation, washed twice with the same buffer, and resuspended in SDS sample buffer. [35S]methionine-radiolabeled NAP was resolved on a 7.5% gel.
Sucrose gradient analysis.
NAP2 and p300 (residues 1135 to 2414) proteins were purified as described above, and core histones were obtained from Sigma. For the in vitro analysis, protein complexes were formed in 50 mM Tris [pH 7.5], 1.5 mM MgCl2, 0.4 mM EDTA, 10% glycerol, and 0.25 mg of bovine serum albumin per ml at 4°C for 30 min. A 20 to 50% sucrose gradient was sedimented at 47,000 rpm for 17 h in an SW55 rotor (Beckman). Fractions were collected from the bottom of the gradient. Typically, about 25 fractions of about 200 μl each were collected and analyzed by immunoblotting with either the anti-NAP2 peptide antibody, anti-p300 (NM11; Calbiochem), or anti-histone H2A-H2B (Santa Cruz), as described above. Each gradient analysis was repeated several times. Standard molecular mass proteins (Amersham Pharmacia Biotech) included in the gradient analysis were aldolase (158 kDa), catalase (232 kDa), and ferritin (440 kDa).
RESULTS
NAP proteins bind to p300.
To investigate the mechanisms through which p300/CBP coactivators regulate transcription, we screened for proteins capable of interacting with p300 in the yeast two-hybrid assay. Using LexA-p300611–2283 as the bait, a 10.5-day-postcoitum mouse embryo activation domain-tagged library was screened from which we isolated a previously identified protein, known as NAP1, together with another less well characterized and more recently identified member of the family, referred to as NAP2 (23, 45). A comparison of NAP1 and NAP2 indicated that both proteins are of similar size (391 to 386 amino acid residues, respectively) and in certain regions possess striking similarity (Fig. 1a). The NAP family of proteins also includes TAF1, which is closely related to both NAP1 and NAP2 (Fig. 1a) and exists as two alternatively spliced variants, known as α and β (42). In the following study, we present data derived mostly from assays performed with NAP2, although it should be noted that unless otherwise stated, the properties of all three NAP proteins (NAP1, NAP2, and TAF1) were similar in the study reported here.
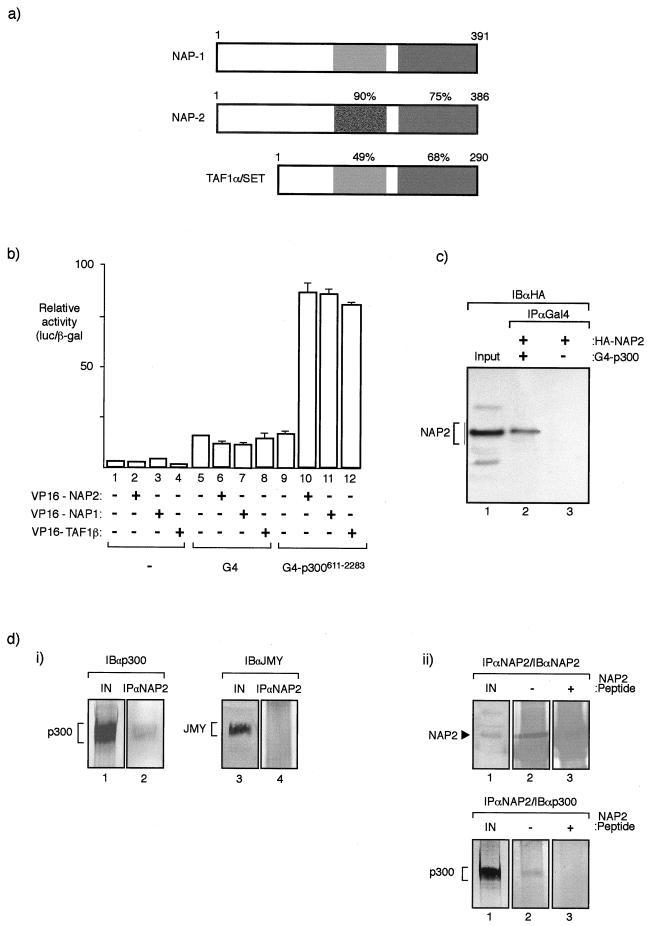
FIG. 1.
NAP proteins interact with p300. (a) Diagrammatic summary of members of the NAP family (NAP1, NAP2, and TAF1α) and the level of similarity between each human protein. The protein regions were compared with NAP2 from residue 159 to 270 and residue 337 to 382. The corresponding regions in NAP1 were 167 to 278 and 344 to 387, and in TAF they were 91 to 189 and 243 to 288. It should be noted that TAF1α and TAF1β are proteins that arise through alternative splicing and differ in a region in the N-terminal domain (42). (b) Mammalian two-hybrid assay performed in U2OS cells where the indicated VP16-NAP or TAF1β expression vectors (0.5 μg) were transfected together with expression vectors encoding G4 or pG4-p300611–2283 (0.5 μg). The values shown represent the average of three separate readings and the ratio of luciferase derived from the reporter pG4-luc and the internal control pCMV-βgal. (c) Coimmunoprecipitation of NAP2 and p300 hybrid proteins from U2OS cells transfected with pG4-p300611–2283 (lane 2) or pG4 (lane 3) together with pCMV-HA-NAP2 (lanes 1, 2, and 3). After extraction, immunoprecipitation was performed with anti-Gal4 antibody (IPαGal4) followed by immunoblotting with anti-HA (12CA5) (IBαHA). An extract from cells transfected with HA-NAP2 is shown in lane 1. (d) Panel i, coimmunoprecipitation of endogenous NAP2 and p300 from murine A31 cell extracts was performed using anti-NAP2 peptide antiserum followed by immunoblotting with either anti-p300 (lane 2) or, as a control, anti-JMY (lane 4). Lanes 1 and 3 show the input extract. Note that p300, but not JMY (47), is present in the NAP2 immunoprecipitate and further that A31 cell extracts were used because the anti-NAP2 antibody recognizes murine NAP2 only. Panel ii, coimmunoprecipitation of endogenous NAP2 and p300 from A31 cells was performed using anti-NAP2 peptide antiserum in the presence (+) (lane 3) or absence (−) (lane 2) of homologous peptide followed by immunoblotting with either anti-NAP2 (top) or anti-p300 (bottom). Lane 1 shows the input extract, and NAP2 and p300 are indicated. Note that the NAP2 polypeptide is absent in the presence of the competing NAP2 peptide (+) (top) and that p300 in the immunoprecipitate correlates with the presence of NAP2 (bottom).
We addressed whether NAP family proteins can interact with p300 in mammalian cells, using both two-hybrid cell-based assays and immunoprecipitation, which we performed on both exogenous and endogenous proteins. In a two-hybrid assay performed with U2OS cells with the bait G4-p300611–2283, clear and specific stimulation of reporter gene activity occurred when VP16 activation domain-tagged NAP1, NAP2, or TAF1β was coexpressed (Fig. 1b), supporting the idea that each of these three NAP family proteins can interact with p300. The stimulation in activity was specific for G4-p300611–2283, since VP16-NAP had little effect on the Gal4 DNA binding domain (Fig. 1b). Further evidence for an interaction between NAP and p300 was provided by the introduction of expression vectors for G4-p300611–2283 and HA-NAP2 into U2OS cells, followed by immunoprecipitation with anti-Gal4 and immunoblotting with anti-HA, where NAP2 was found to be specifically coimmunoprecipitated with p300 (Fig. 1c). Importantly, similar observations were made when the immunoprecipitation was performed on endogenous proteins from nontransfected murine A31 cells, where p300 was found to coimmunoprecipitate with endogenous NAP2. Thus, by using an anti-NAP2 peptide antibody that recognized murine NAP2, we found that when the immunoprecipitation was performed with anti-NAP2 in the presence or absence of a competing NAP2 peptide, p300 was detected only in the presence of the control peptide (Fig. 1d, panel ii). In summary, therefore, these results provide considerable support for the idea that p300 can physically interact with members of the NAP family of proteins and further that this interaction occurs under physiological conditions.
Binding domains in p300 and NAP2.
To resolve which region in p300 is responsible for the interaction with NAP2, we used the mammalian two-hybrid assay together with a panel of G4-p300 hybrids which were studied in the presence of coexpressed VP16-NAP2. An interaction was apparent between VP16-NAP2 and the C-terminal region of p300, encompassed within residues 1572 to 2283, which was further resolved to the region between amino acid residue 1572 and 1906 (Fig. 2a). These results were further confirmed through a biochemical binding assay in which different regions of p300 were expressed as GST fusion proteins and incubated with in vitro-translated NAP2. In agreement with the two-hybrid results, we found that GST-p3001572–1906 bound efficiently to NAP2, whereas the overlapping fusion proteins GST-p3001302–1737 and GST-p3001818–2080 failed to do so (Fig. 2b and d). Moreover, using in vitro-translated luciferase as a negative control, we could not detect any binding between luciferase and p300 (Fig. 2c). Overall, these results indicate that p300 and NAP2 are likely to directly interact in mammalian cells, and the results identify a C-terminal region in p300, encompassing the CH3 region, as a likely binding domain that can interact with NAP2.
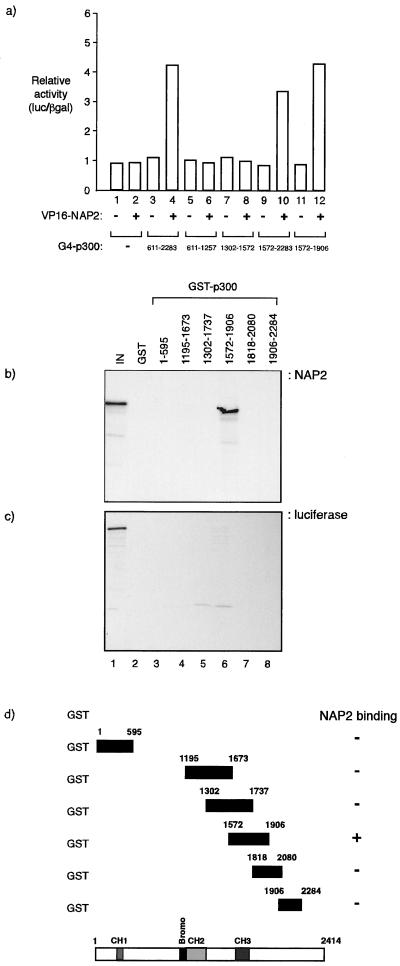
FIG. 2.
NAP proteins bind to p300. (a) A mammalian two-hybrid assay was performed with U2OS cells transfected with the indicated expression vectors, VP16 or VP16-NAP2 (0.5 μg), together with pG4, pG4-p300611–2283, pG4- p300611–1257, pG4-p3001302–1572, pG4-p3001572–2283, or pG41572–1906 (0.5 μg). The values shown are derived from triplicate readings (luciferase/β-galactosidase ratio [luc/βgal]) and represent the fold increase in the presence of VP16-NAP2 relative to VP16 alone. (b, c, and d) Binding assay between the indicated GST-p300 fusion proteins and in vitro-translated NAP2 (b) or luciferase (c) in which about 5.0 μg of GST fusion protein was incubated with the in vitro translate. Lane 1 shows the input (10%) NAP2 (b) or luciferase (c). Panel d shows a summary of the data, together with the location of the relevant domains in p300.
Using similar approaches, we investigated the region in NAP2 that is required for the interaction with p300 and present data derived from a biochemical binding assay, although similar conclusions were drawn from a mammalian two-hybrid analysis (data not shown). A comparison of the activity of different in vitro-translated NAP2 mutant derivatives in an assay that assessed binding to GST-p3001572–1906 indicated that there are two broad regions that can bind to p300. Although the N-terminal region of NAP2 from residue 1 to 123 possessed marginal binding activity, an internal domain from residue 110 to 230 and the C-terminal region from 234 to 386 both bound with greater efficiency to GST-p3001572–1906 (Fig. 3a, b, and c). These results therefore suggest that NAP2 contains at least two autonomous and separable interaction domains for p300 (Fig. 3g).
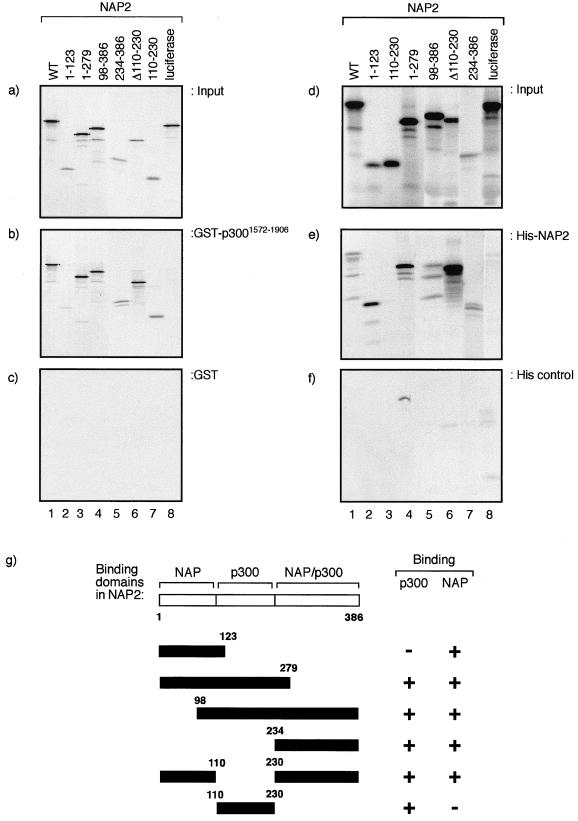
FIG. 3.
Domains in NAP2 that interact with p300 and NAP proteins form dimers. (a, b, and c) The indicated regions of NAP2 were in vitro translated (a) and assessed for binding to GST-p3001572–1906 (about 5 μg) (b) or GST alone (about 5 μg) (c). (d, e, and f) The indicated regions of NAP2 were in vitro translated (d) and assessed for binding to His-NAP2 (about 5 μg) (e) or His control beads (f). Note that the His control beads (f) were treated with bacterial extract without NAP2 induction. (g) Summary of the binding domains in NAP2 for p300 and NAP proteins.
We also tested whether NAP proteins could form homo- or heteromeric protein complexes with other members of the NAP family. By assessing biochemical binding activity, we found that His-NAP2 could bind to in vitro-translated NAP2 (Fig. 3d, e, and f); similar results were obtained for NAP1 and TAF1, which formed complexes with NAP1, TAF1, and NAP2 (data not shown). An analysis of the panel of NAP2 mutant derivatives indicated that there were at least two distinct domains capable of allowing complex formation, one encompassed within the N-terminal region up to amino acid residue 123 and the other in the C-terminal region from residue 234 to 386; an internal region from residue 110 to 230 failed to bind to NAP2 (Fig. 3e). The data derived from these binding studies indicate that NAP family proteins can form both homo- and heteromeric protein complexes and that at least two binding domains are involved in facilitating these interactions (Fig. 3g).
NAP proteins augment p300-dependent transcription.
To investigate the functional consequence of NAP2 on the activity of p300, we studied the effect of NAP2 on two p300-dependent transcription factors, p53 and E2F-1 (4, 21, 37, 38, 54). The transcriptional activity of p53 was assayed on different p53-responsive promoters, and we present data derived from the bax promoter, which is a p53-responsive gene that encodes a protein involved with inducing apoptosis (41). As expected, the bax promoter was efficiently activated in the presence of p53, and a titratable increase in p53-dependent transcription was apparent as the level of NAP2 was increased, with a marginal but significant enhancement evident upon the coexpression of p300 (Fig. 4a). The stimulation of transcription by NAP2 and p300 was dependent upon the integrity of the N-terminal activation domain, since a p53 derivative containing an inactive trans activation domain, p5322/23 (39), failed to respond to NAP2 and p300 (Fig. 4a). We also found that both NAP1 and TAF1β could similarly induce p53 activity (data not shown).
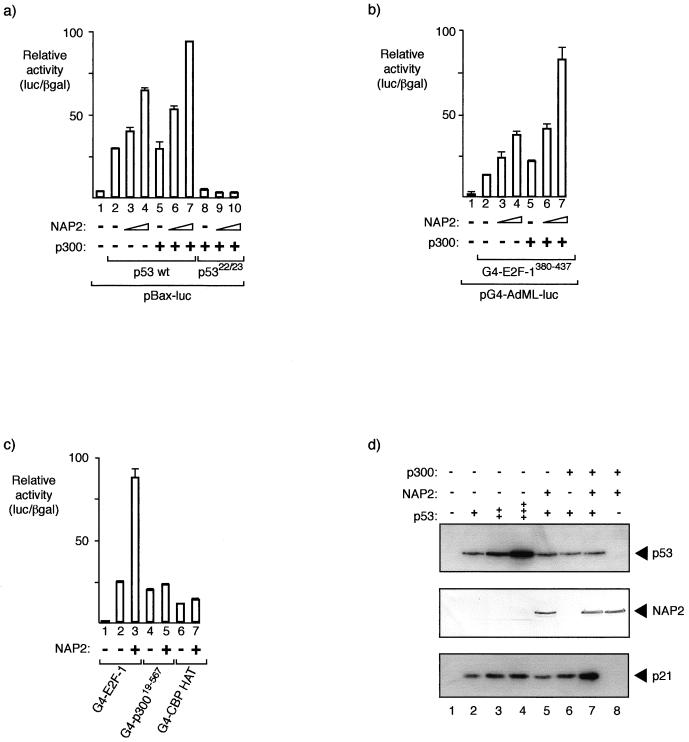
FIG. 4.
NAP proteins augment the p300-dependent transcription factors p53 and E2F-1. (a) The p53 reporter pBax-luc (0.5 μg) together with expression vectors for wild-type p53 (50 ng), p5322/23 (50 ng), p300 (3 μg), or NAP2 (2 or 5 μg) was introduced into SAOS2 cells as indicated. The values shown are the average of duplicate readings and represent the ratio of the level of luciferase to the β-galactosidase activity from the internal control. (b) The Gal4 reporter pG4-AdML-luc (0.5 μg) together with expression vectors for Gal4-E2F-1380–437 (50 ng), p300 (+ indicates 4 μg), or NAP2 (2 or 4 μg) was introduced into SAOS2 cells as indicated, and the luciferase/β-galactosidase ratio was calculated as described above. (c) The Gal4 reporter pG4-AdML-luc (0.5 μg) together with expression vectors for Gal4-E2F-1380–437 (50 ng), Gal4-p30019–567 (50 ng), or Gal4-CBP HAT (50 ng) and NAP2 (+ indicates 4 μg) was introduced into SAOS2 cells as indicated, and the luciferase/β-galactosidase ratio was calculated as described above. (d) Regulation of endogenous p21Waf1/Cip1 levels by NAP2 and p300. The indicated plasmids were transfected into SAOS2 cells (p53, 0.3, 1.0, and 2.0 μg; NAP2 and p300, 5 μg), and the transfected cells were harvested at 48 h. Cell extracts were immunoblotted with anti-p53 (DO-1), anti-NAP2 peptide antiserum, or anti-p21 (C19).
We studied the effect of NAP2 on another p300-responsive transcription factor, namely, E2F-1 (37, 54). The E2F-1 trans activation domain, which is located in the C-terminal region of E2F-1, is a Gal4 hybrid protein-activated transcription of a Gal4-responsive promoter (Fig. 4b). Coexpression of NAP2 increased the level of transcription of G4-E2F-1380–437, and there was a further increase in activity upon the coexpression of p300 (Fig. 4b). Overall, these results indicate that NAP proteins can enhance the activity of different p300-dependent transcription factors.
The specificity of NAP2 for transcriptional activation was assessed by employing derivatives of p300/CBP expressed as hybrid proteins with the Gal4 DNA binding domain. These p300/CBP hybrids were chosen because they are known to be transcriptionally active and thus could be used to test the specificity of the effect of NAP2 on transcription. Two hybrids were studied, G4-p30019–567, encompassing the CH1 (48) region, and G4-CBP HAT, which contains the HAT domain (residue 1099 to 1758) taken from CBP (40); both hybrids are transcriptionally active (40) (Fig. 4c). While G4-p30019–567 and G4-CBP HAT were transcriptionally active (at a level similar to that observed for G4-E2F-1380–437), there was little effect on their transcriptional activity upon coexpression of NAP2 (Fig. 4c), thus implying that NAP2 does not function as a general activator of transcription.
Although the previous results show that NAP2 can augment the transcriptional activity of p53 and E2F-1, the data were derived from experiments performed on transfected templates, and therefore the chromatin environment of the template may not necessarily reflect the situation encountered with endogenous cellular genes. To address this point, we assessed the effect of NAP and p300 upon endogenous genes by studying the activity of the p53-responsive gene that encodes p21Waf1/Cip1 (15). We introduced p53 into SAOS2 cells (which are p53−/− and Rb−/−) together with NAP2 and p300, either alone or together, and monitored the levels of endogenous p21. As expected, we found that p53 could stimulate p21 expression as the level of p53 was increased (Fig. 4d). At a subsaturating amount of p53, we introduced NAP2 and p300. Whereas neither NAP2 nor p300 had a profound effect on the p53-dependent induction of p21, when both NAP2 and p300 were introduced together a greater level of p21 was apparent, usually exhibiting about a threefold induction (Fig. 4d). These results therefore further strengthen the idea that NAP2 and p300 can functionally interact in the induction of genes within a chromatin environment.
NAP binds to core histone tails.
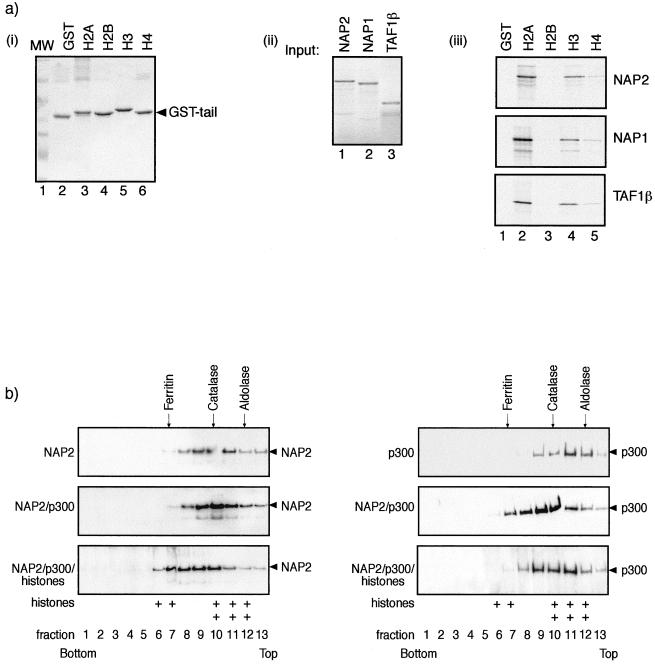
In the next series of experiments, we explored the biochemical properties of NAP proteins and thereafter established a likely relevance of the association between p300 and NAP. Previous studies have indicated that NAP1 copurifies with the core histones H2A and H2B, an interaction that may facilitate the assembly of nucleosomes onto a DNA template (9, 11, 16). We extended these observations by determining if NAP2 showed binding activity for N-terminal tails and thereafter whether there was any specificity for the N-terminal tail region of core histones. In an assay using GST tail proteins derived from Drosophila H2A, H2B, H3, and H4 (17), we found that NAP2 could bind to isolated histone tails and reproducibly exhibited stronger binding activity for the H2A and H3 tail regions, although it was able to bind specifically to H2B and H4 (Fig. 5a). Similar results were observed on the tail binding specificity of NAP1 and TAF1β, which also exhibited preferential binding activity for the H2A and H3 tail regions (Fig. 5a). It is important to note that this binding specificity did not appear to reflect the conservation of protein sequence between the Drosophila and murine tail regions, which were all greater than 90% identical in sequence across the N-terminal 50 residues. Thus, these results suggest that NAP proteins can directly interact with the tail region of core histones and further imply that a level of specificity may exist that allows NAP proteins to bind more efficiently to certain core histone tails.
FIG. 5.
NAP2 binds to core histones. (a) Panel i, about 1 μg of GST (lane 2), GST-H2A (lane 3), H2B (lane 4), H3 (lane 5), or H6 (lane 6) N-terminal tail fusion proteins was stained by Coomassie blue. Lane 1 shows the molecular weight standards. Panel ii, in vitro-translated NAP2 (lane 1), NAP1 (lane 2), and TAF1β (lane 3) showing 10% of the input used for panel iii. Panel iii, binding between in vitro-translated NAP2 (top), NAP1 (middle), or TAF1β (bottom) and the indicated GST proteins. Note that upon longer exposure, specific binding was apparent between the NAP proteins and H2B and H4 tails. (b) Sucrose gradient analysis was performed as described previously on either NAP2 alone, NAP2 and p300, or NAP2, p300, and core histones, using 2 μg of each pure protein preparation or 4 μg of core histones. Samples were subjected to 20 to 50% sucrose gradient centrifugation, and fractions were analyzed by SDS-PAGE and immunoblotted with anti-NAP2, anti-p300, or anti-H2A-H2B. Note that when NAP2, p300, and core histones were analyzed together, the extent of sedimentation increased. The distribution of core histones H2A-H2B in fractions 6, 7, and 8 in the NAP2-p300-histone gradient (bottom) is indicated by +; in the absence of all three proteins, histones appeared predominantly in fractions 10, 11, and 12. The positions of the standard molecular masses of aldolase (158 kDa), catalase (232 kDa), and ferritin (440 kDa) are shown. Wild-type His-NAP2 and Flag-p3001135–2414 were used in the analysis.
So far, the data indicate that NAP2 can interact with two types of protein, p300/CBP coactivators and core histones. It was therefore of interest to assess the likelihood that histones could form a ternary complex with NAP and p300. To gain evidence for this idea, we employed sedimentation analysis by centrifugation through a sucrose gradient with purified NAP2, p300, and core histones, which was performed either on isolated NAP2 or p300, on NAP2 and p300 together, or on all three proteins. By monitoring the distribution of NAP2, p300, or core histones, each analysis gave rise to a characteristic sedimentation profile, which reflected the properties of the protein complex in the gradient (Fig. 5b). Only when all three proteins were incubated together and thereafter analyzed on the gradient was there a significant and substantial shift in the sedimentation profile of NAP2 towards a protein complex with a greater mass (Fig. 5b), a result that is consistent with an interaction between all three types of proteins. Furthermore, the fractions containing the greater-mass NAP2 complex also contained p300 and core histones (Fig. 5b). The profiles of NAP2 alone and of NAP2 and p300 were similar (Fig. 5b), a result that may reflect the ability of NAP2 to form oligomers. Nevertheless, based on these results, the most likely explanation for the increased sedimentation properties observed with NAP2, p300, and histones is that they result from the formation of a ternary complex that involves all three proteins.
p300 efficiently binds to NAP in the presence of core histones.
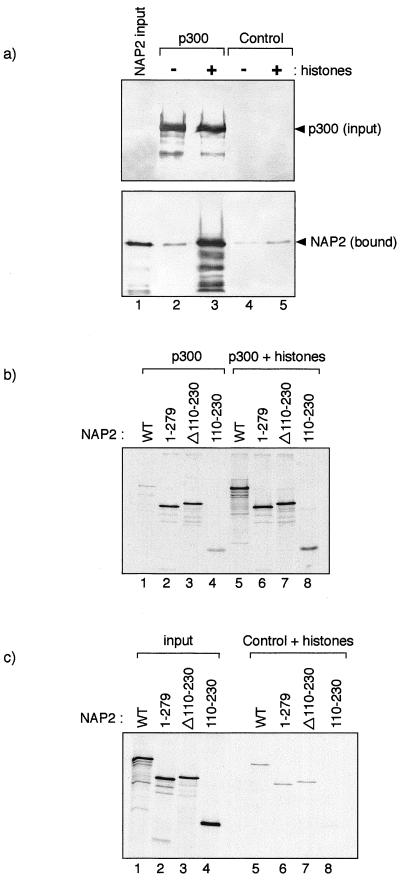
Having gained evidence that a ternary complex between p300, NAP2, and core histones may occur, we investigated the effect that core histones had on the interaction between p300 and NAP2. To pursue this question, we identified binding conditions in which subsaturating levels of NAP2 bound specifically to p3001135–2414 (Fig. 6a, compare lanes 2 and 4). We reasoned that under these conditions, histones may play a role in facilitating the interaction between NAP2 and p3001135–2414. We therefore assessed the effect of core histones and found that the addition of core histones caused a notable and specific increase in the efficiency of the interaction between p3001135–2414 and NAP2 (Fig. 6a, compare lanes 2 and 3), suggesting that the presence of core histones could act to enhance the interaction between p300 and NAP2. It should be noted that this effect was unlikely to be caused by the charged nature of histones, since polyanions, such as spermidine, failed to cause a similar effect (data not shown).
FIG. 6.
Binding of NAP to p300 is augmented in the presence of core histones. (a) Flag-tagged p300 (1135 to 2414; about 1 μg) was incubated with His-NAP2 (about 1 μg) in the presence (about 3 μg; indicated by +) (lane 3) or absence (−) (lane 2) of chicken core histones. Control Flag-tagged beads were treated in a similar fashion (lanes 4 and 5), and lane 1 represents the input (10%) NAP2 protein. Note that in the presence of core histones, the level of NAP2 associated with p300 is substantially increased and that the effect seen on the control Flag-tagged beads is far less dramatic. (b and c) The indicated NAP2 mutant derivatives were in vitro translated, and the binding to wild-type Flag-tagged p300 (about 1 μg) was assessed in the absence (b) (lanes 1 to 4) or presence (b) (lanes 5 to 8) of core histones (about 5 μg). The effect of histones on the binding to the NAP2 derivatives to control Flag beads is shown (c) (lanes 5 to 8); the input protein (10%) is shown in lanes 1 to 4. Note that the data shown in panels b and c were derived from the same experiment and represent similar exposure times and that the specific effect of histones on the interaction between NAP2 and p300 can be seen by comparing lanes 5 in panels b and c.
In an extension to this analysis, we assessed whether domains exist in NAP2 which can be influenced by the presence of core histones and therefore are likely to play a role in regulating the interaction of NAP2 with p300. Relative to the interaction between wild-type NAP2 and p300, we found in the absence of core histones that a C-terminal deletion in NAP2 up to residue 279 bound to p300 at about a 10-fold-greater efficiency than wild-type NAP2, a similar enhancement in binding efficiency being apparent for the internal deletion mutant Δ110–230 (Fig. 6b, compare lanes 1, 2, and 3). For both of these NAP2 mutants, we assessed the effect on the interaction with p300 of adding core histones, which we found enhanced the interaction to a far lesser extent than that observed for wild-type NAP2 binding to wild-type p300, which was, as previously noted, significantly enhanced by core histones (Fig. 6b, compare lanes 5, 6, and 7). The effects were specific to the interaction between NAP2 and p300, since similar effects were not observed on the binding of NAP2 to Flag-tagged control beads (Fig. 6b and c, compare lanes 5, 6, and 7). In contrast to results for the other two NAP2 mutants, the binding of the internal region of NAP2, 110 to 230, to p300 was enhanced by the presence of core histones (Fig. 6b), suggesting that this region of NAP2 possesses a domain that is influenced by the presence of core histones. Overall, these results suggest that the presence of core histones may influence the interaction between p300 and NAP.
DISCUSSION
NAP proteins functionally interact with p300/CBP coactivators.
The NAP proteins are an evolutionarily conserved family of chaperone-like proteins, to which the functions of both transcription and DNA replication-related mechanisms have been ascribed (1, 25, 26, 27, 42). For example, NAP proteins can act to facilitate the binding of sequence-specific transcription factors to a nucleosomal template (59) and further may play a role in nucleosome assembly by interacting with ACF (27). Other reports are consistent with NAP proteins taking part in the intracellular transport of histones (26), and a separate interaction with cyclin B has also been documented for yeast (30).
In this study, we have identified a hitherto unexpected role for NAP proteins in contributing to the activity of p300/CBP coactivators. The results strongly suggest that NAP proteins can physically associate with p300 and imply that this interaction allows NAP to augment p300 activity and thus p300-responsive transcription factors, such as p53 and E2F-1. Moreover, although NAP can form a complex with p300, we also found that NAP possesses the intrinsic capacity for binding to histone tails. Most interestingly, we acquired some evidence to suggest that NAP, p300, and core histones may interact to form a ternary complex and further that core histones may influence the interaction between NAP and p300. It seems likely, therefore, that certain aspects of p300 activity will entail a functional interaction with NAP proteins.
NAP proteins augment p300 activity.
Although it has previously been shown that NAP proteins can augment the binding of sequence-specific transcription factors to a nucleosomal template (59), the data described here define a new level of control through which NAP can regulate transcription. In this respect, it is interesting that we gained evidence to support the idea that the ability of NAP to bind to p300 may be influenced by the presence of core histones (Fig. 6), thus providing a plausible mechanism that could account for certain aspects of transcriptional control through NAP. Perhaps such a mechanism is important in maintaining the association of coactivator complexes with areas of chromatin that are destined to become, or already have become, transcriptionally active. In addition, another relevant consideration relates to the ability of NAP to influence nucleosome assembly in conjunction with ACF (27). It is possible that such an activity may favor the recruitment and stable association of p300 complexes with newly replicated DNA, which could be an important step in regulating the transcriptional activity of newly replicated genes. A role for the NAP-p300 interaction in a DNA replication-related context cannot therefore be excluded.
Transcriptional control by NAP.
Although the results reported here generally support the NAP-p300 interaction in transcriptional activation, the stage or stages at which this interaction is required in the activation process have yet to be determined. However, it has become clear that chromatin can provide a substantial barrier to transcription, mostly through the inability of sequence-specific transcription factors to penetrate to DNA in a chromatin environment, and that a variety of ATP-dependent chromatin-remodeling activities can act to overcome this barrier (3, 10, 31). One potential model suggests that chromatin-remodeling activities create a fluid chromatin environment by regulating the rate of interconversion between different chromatin states and that activating transcription complexes lock chromatin in an active state, whereas repressing complexes fix chromatin in an inactive state. In the context of this model, it is likely that the interaction between NAP and p300 functions as an activating complex which, in collaboration with a remodeling activity, locks transcription into the on state. Thus, another role for the interaction between p300 and NAP may be relevant to a later stage of a multistep activation process, once chromatin-remodeling activities have been targeted and chromatin has been modified. In this respect, it is noteworthy that our results show that NAP proteins can bind to histone tail regions. While the copurification of NAP1 with H2A and H2B has been previously noted to occur (9, 11, 26), the data presented here suggest in addition that NAP proteins can bind to the tail region of core histones. Furthermore, within this tail binding activity we obtained evidence that specificity resides for certain core histones. It will be most interesting to establish the relevance of this tail binding activity to NAP function in vivo.
The results presented in this study raise a number of important questions. For example, while we find that the activities of p53 and E2F-1 are likely to be influenced by NAP and p300, we have yet to determine whether this process represents a general feature of p300-dependent transcriptional activation. Furthermore, the p300 HAT activity appears to be important in allowing p300 to activate some transcription factors (34, 35, 36, 40, 44), although the results presented here suggest that NAP does not directly influence the isolated HAT domain (Fig. 4d). It is possible, though it has yet to be proven, that other HATs in p300 complexes are responsible for augmenting transcription when NAP assembles with p300 or, alternatively, that NAP augments p300 activity through a process that does not involve HAT.
A model for NAP-dependent transcriptional activation through p300 coactivators.
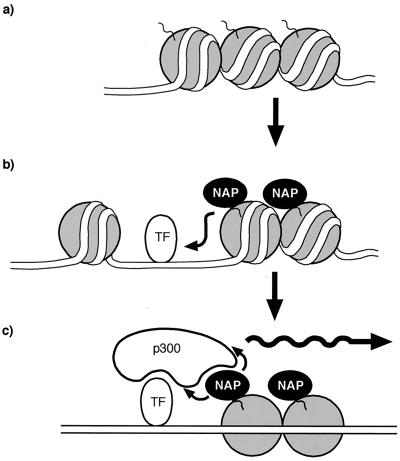
The ability of NAP to interact with both p300 and core histones, combined with the fact that the presence of core histones appears to regulate the interaction between p300 and NAP, implies a plausible model that may account for the capacity of NAP to stimulate p300-dependent transcription (Fig. 7). We suggest that the stable association of p300 coactivators with transcriptional activation domains and the subsequent p300 binding to NAP may result in an overall stabilizing influence which acts to strengthen the association of coactivator complexes with chromatin. According to the results presented here, the interaction between p300 and NAP may be augmented by the histone, perhaps nucleosomal, environment of a chromatin template. We suggest that this is a useful mechanistic feature that is a desirable property of transcriptional coactivators, since it provides a process that is destined to aid transcriptional activation. Moreover, we would also anticipate, based on the results in previous reports (59), that the presence of NAP in close proximity to a transcription factor binding site positioned within a nucleosome will favor DNA binding activity, which we imagine is an equally important event that will contribute to enhanced transcriptional activity.
FIG. 7.
Hypothetical model for NAP-dependent stimulation of p300 transcription. (a) Nucleosomal DNA is shown, which upon interacting with NAP (indicated by the arrow) facilitates the binding of transcription factors to their DNA binding site in a nucleosomal template (b). The recruitment and binding of p300 coactivators by the activation domains of sequence-specific transcription factors are strengthened through the interaction of p300 with NAP (c), which is suggested to augment transcriptional activation.
Overall, we suggest that NAP's ability to recruit and stabilize the interaction of p300 with chromatin, together with its capacity to facilitate stable transcription factor binding to a nucleosomal template, places it in a central position of control in regulating p300-dependent transcription.
ACKNOWLEDGMENTS
We thank Marie Caldwell for help in preparing the manuscript, T. Owen-Hughes and C. Peterson for discussion and technical advice, Y. Nakatani for p300 constructs, K. Nagata for TAF plasmids, and C. Wu for the GST-tail constructs.
We thank the Medical Research Council, the European Molecular Biology Organisation, and the Cancer Research Campaign for supporting this research. H.M.C. was supported by the Wellcome Trust.
REFERENCES
- 1.Adams C R, Kamakaka R T. Chromatin assembly: biochemical identities and genetic redundancy. Curr Opin Genet Dev. 1999;9:185–190. doi: 10.1016/S0959-437X(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 2.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong J A, Emerson B M. Transcription of chromatin: these are complex times. Curr Opin Genet Dev. 1998;8:165–172. doi: 10.1016/s0959-437x(98)80137-8. [DOI] [PubMed] [Google Scholar]
- 4.Avantaggiati M L, Ogryzko V V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 6.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 7.Brownell J E, Allis C D. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 8.Buck V, Allen K E, Sørensen T, Bybee A, Hijmans E M, Voorhoeve P M, Bernards R, La Thangue N B. Molecular and functional characterisation of E2F-5, a new member of the E2F family. Oncogene. 1995;11:31–38. [PubMed] [Google Scholar]
- 9.Bulger M, Ito T, Kamakaka R T, Kadonaga J T. Assembly of regularly spaced nucleosome arrays by Drosophila chromatin assembly factor 1 and a 56-kDa histone-binding protein. Proc Natl Acad Sci USA. 1995;92:11726–11730. doi: 10.1073/pnas.92.25.11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns B R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- 11.Chang L, Loranger S S, Mizzen C, Ernst S G, Allis C D, Annunziato A T. Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry. 1997;36:469–480. doi: 10.1021/bi962069i. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor co-activator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 13.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 14.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 15.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 16.Fujii-Nakata T, Ishimi Y, Okuda A, Kikuchi A. Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. J Biol Chem. 1992;267:20980–20986. [PubMed] [Google Scholar]
- 17.Georgel P T, Tsukiyama T, Wu C. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girling R, Partridge J F, Bandara L R, Burden N, Totty N F, Hsuan J J, La Thangue N B. A new component of the transcription factor DRTF1/E2F. Nature. 1993;362:83–87. doi: 10.1038/362083a0. [DOI] [PubMed] [Google Scholar]
- 19.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 20.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 21.Gu W, Shi X-L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–822. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 22.Hansen J C, Ausio J, Stanik V H, van Holde K E. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- 23.Hu R J, Lee M P, Johnson L A, Feinberg A P. A novel human homologue of yeast nucleosome assembly protein, 65 kb centromeric to the p57KIP2 gene, is biallelically expressed in fetal and adult tissues. Hum Mol Genet. 1996;5:1743–1748. doi: 10.1093/hmg/5.11.1743. [DOI] [PubMed] [Google Scholar]
- 24.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 25.Ito T, Tyler J K, Bulger M, Kobayashi R, Kadonaga J T. ATP-facilitated chromatin assembly with a nucleoplasmin-like protein from Drosophila melanogaster. J Biol Chem. 1996;271:25041–25048. doi: 10.1074/jbc.271.40.25041. [DOI] [PubMed] [Google Scholar]
- 26.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilising chromatin assembly and remodelling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 27.Ito T, Bulger M, Kobayashi R, Kadonaga J T. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol Cell Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janknecht R, Hunter T. Transcription. A growing co-activator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 29.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 30.Kellogg D R, Kikuchi A, Fujii-Nakata T, Turck C W, Murray A W. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J Cell Biol. 1995;130:661–673. doi: 10.1083/jcb.130.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 32.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multi-protein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 33.Kornberg R D, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 34.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific recruitment of co-activators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 35.Kraus W L, Manning E T, Kadonaga J T. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin template. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurokawa R, Kalafus D, Ogliastro M H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 37.Lee C W, Sørensen T S, Shikama N, La Thangue N B. Functional interplay between p53 and E2F through co-activator p300. Oncogene. 1998;16:2695–2710. doi: 10.1038/sj.onc.1201818. [DOI] [PubMed] [Google Scholar]
- 38.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP co-activators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 39.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domains are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 42.Nagata K, Kawase H, Handa H, Yano K, Yamasaki M, Ishimi Y, Okuda A, Kikuchi A, Matsumoto K. Replication factor encoded by a putative oncogene set, associated with myeloid leukemogenesis. Proc Natl Acad Sci USA. 1995;92:4279–4283. doi: 10.1073/pnas.92.10.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 44.Puri P L, Sartorelli V, Yang X-J, Hamaori Y, Ogryzko V V, Howard B H, Kedes L, Wang J Y J, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and pCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez P, Munroe D, Prawitt D, Chu L L, Bric E, Kim J, Reid L H, Davies C, Nakagama H, Loebbert R, Winterpacht A, Petruzzi M J, Higgins M J, Nowak N, Evans G, Shows T, Weissman B E, Zabel B, Housman D E, Pelletier J. Functional characterization of human nucleosome assembly protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics. 1997;44:253–265. doi: 10.1006/geno.1997.4868. [DOI] [PubMed] [Google Scholar]
- 46.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shikama N, Lee C W, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue N B. A novel cofactor for p300 that regulates the p53 response. Mol Cell. 1999;4:365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- 48.Shikama N, Lyon J, La Thangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 49.Sørensen T S, Girling R, Lee C-W, Gannon J, Bandara L R, La Thangue N B. Functional interaction between DP-1 and p53. Mol Cell Biol. 1996;16:5888–5895. doi: 10.1128/mcb.16.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spencer T E, Jenster G, Burcin M M, Assis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor co-activator-1 is a histone acetyltransferase. Nature. 1997;289:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 51.Svaren J, Horz W. Regulation of gene expression by nucleosomes. Curr Opin Genet Dev. 1996;6:164–170. doi: 10.1016/s0959-437x(96)80046-3. [DOI] [PubMed] [Google Scholar]
- 52.Torchia J, Glass C K, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 53.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 54.Trouche D, Cook A, Kouzarides T. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 1996;24:4139–4145. doi: 10.1093/nar/24.21.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 56.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol Cell Biol. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wade P A, Wolffe A P. Histone acetyltransferases in control. Curr Biol. 1997;7:R82–R84. doi: 10.1016/s0960-9822(06)00042-x. [DOI] [PubMed] [Google Scholar]
- 58.Wade P A, Pruss D, Wolffe A P. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 59.Walter P P, Owen-Hughes T A, Cote J, Workman J L. Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol Cell Biol. 1995;15:6178–6187. doi: 10.1128/mcb.15.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Westin S, Kurokowa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;305:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 61.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 62.Xu L, Glass C K, Rosenfeld M G. Co-activator and co-repressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]