Abstract
Splicing of the c-src N1 exon in neuronal cells depends in part on an intronic cluster of RNA regulatory elements called the downstream control sequence (DCS). Using site-specific cross-linking, RNA gel shift, and DCS RNA affinity chromatography assays, we characterized the binding of several proteins to specific sites along the DCS RNA. Heterogeneous nuclear ribonucleoprotein (hnRNP) H, polypyrimidine tract binding protein (PTB), and KH-type splicing-regulatory protein (KSRP) each bind to distinct elements within this sequence. We also identified a new 60-kDa tissue-specific protein that binds to the CUCUCU splicing repressor element of the DCS RNA. This protein was purified, partially sequenced, and cloned. The new protein (neurally enriched homolog of PTB [nPTB]) is highly homologous to PTB. Unlike PTB, nPTB is enriched in the brain and in some neural cell lines. Although similar in sequence, nPTB and PTB show significant differences in their properties. nPTB binds more stably to the DCS RNA than PTB does but is a weaker repressor of splicing in vitro. nPTB also greatly enhances the binding of two other proteins, hnRNP H and KSRP, to the DCS RNA. These experiments identify specific cooperative interactions between the proteins that assemble onto an intricate splicing-regulatory sequence and show how this hnRNP assembly is altered in different cell types by incorporating different but highly related proteins.
Alternative splicing is a common mechanism for regulating gene expression in eukaryotes, allowing the generation of diverse proteins from the same primary RNA transcript (46, 77, 78). The alteration of splice site choice is thought to be determined by regulatory proteins that bind to the pre-mRNA transcript and affect spliceosome assembly on particular exons or splice sites. The best characterized of these splicing-regulatory proteins are a set of polypeptides called SR proteins that, among many other properties, bind to exonic splicing enhancer sequences (7, 10, 35, 47, 75). The SR proteins bound to an exonic enhancer are thought to stimulate spliceosome assembly at the adjacent splice sites. Another group of pre-mRNA binding proteins are the heterogeneous nuclear ribonucleoproteins (hnRNPs) (19, 66). These are a diverse group of molecules that coat nascent pre-mRNAs, forming complex but little understood hnRNP structures (42, 52). The assembly of the spliceosome occurs after formation of these hnRNP complexes, and some hnRNPs have been implicated in splicing regulation. For example, hnRNP A1 is able to counteract the effect of SR proteins in some assays and can also apparently repress splicing through splicing silencer sequences (3, 7, 8, 11, 31). However, the assembly of a pre-mRNP complex is poorly understood. It is apparently highly cooperative, but the interactions between the different hnRNPs in these complexes are mostly unknown.
Although widely expressed, the SR proteins and hnRNPs do vary in concentration between different tissues (31, 39). Changes in splicing patterns are thought to be determined, in part, by subtle changes in the combinations of these proteins present in different cell types. Indeed, the ratio of hnRNP A1 to particular SR proteins can strongly affect the splicing pattern of some transcripts (50, 51). However, it is likely that more tissue-specific proteins also direct changes in splicing; how a cell achieves the precise tissue-specific control of a splicing pattern is still a mystery.
Polypyrimidine tract binding protein (PTB or hnRNP I) is a member of the hnRNP group of proteins (24, 26, 61, 74). PTB is implicated as a negative regulator of splicing for several alternative exons. In the β-tropomyosin (βTM) pre-mRNA, the skeletal muscle-specific exon 7 is apparently repressed by PTB in nonmuscle tissues (29, 57). This protein also represses the splicing of α-tropomyosin (αTM) exon 3 in smooth muscle (27, 62). Neuron-specific exons in the c-src, γ-aminobutyric acid A (GABAA) γ2 receptor, clathrin light chain B, and N-methyl-d-aspartate (NMDA) pre-mRNAs are also thought to be repressed by PTB (2, 13, 81). With the exception of αTM, these systems are similar in that PTB seems to repress a highly tissue-specific exon outside of that particular tissue. Although PTB is an apparent inhibitor of splicing, its mechanism of action is not clear. PTB often binds to pyrimidine-rich elements within or near the 3′ splice site of the repressed exon (74). In some cases, this PTB can block binding of the required splicing factor, U2AF, to the 3′ splice site (45, 65). However, sequence elements apart from the 3′ splice site are often required for splicing repression, indicating the need for more than the simple binding of PTB to the 3′ splice site (13, 27). It is also not clear how PTB-mediated splicing repression is relieved in particular cell types. Interestingly, an altered form of PTB has been found in extracts of neuronal cell lines and rat brain (2, 13). The electrophoretic mobility of this neural protein differs from that of the three known isoforms of HeLa cell PTB, indicating the existence of a tissue-specific form of the polypeptide. It has been speculated that this neural protein may prevent PTB-mediated repression of particular neuron-specific exons (28).
We are characterizing the N1 exon of the mouse c-src pre-mRNA as a model for understanding the neuron-specific regulation of splicing. The small (18-nucleotide [nt]) exon N1 is inserted into the src mRNA in neurons but skipped in other cells (71). This regulation can be observed in vitro using extracts of nonneural HeLa cells that skip the N1 exon and extracts of WERI-1 retinoblastoma cells where the N1 exon is spliced. The regulation of N1 splicing requires two regulatory regions in the src pre-mRNA (5, 12, 55, 56). The 3′ splice site upstream of the N1 exon is needed for the repression of N1 splicing in nonneural cells. We have shown that PTB binds to CUCUCU elements within this 3′ splice site and is required for splicing repression (13, 17). The second N1 regulatory region, encompassing nt 17 to 142 downstream of the N1 5′ splice site, is an intronic splicing enhancer and is also required for splicing repression by the upstream elements. Within this enhancer, nt 37 to 70 are highly conserved between mouse and human. This core sequence, called the downstream control sequence (DCS), contains the elements GGGGGCUG and UGCAUG that are needed for the proper function of the N1 enhancer in vivo and have been found in other intronic splicing enhancers. When present in more than one copy, the DCS alone can induce high levels of N1 exon splicing in vivo (56). However, the DCS normally requires adjacent elements to function properly (55). The DCS binds specific RNA binding proteins that can be detected in WERI-1 nuclear extract and are thought to be important for splicing regulation. Three of these proteins have been identified as hnRNP F, hnRNP H, and the KH-type splicing-regulatory protein (KSRP) (16, 53, 54). There are additional unidentified components of the DCS complexes. Moreover, none of the identified DCS binding proteins is specific to the WERI extract even though the larger of the observed DCS-RNP complexes is enriched in this extract over the HeLa extract.
The roles of the individual sequence elements within the DCS and the proteins that bind to them have been difficult to resolve. In vivo, the DCS sequence acts as a general splicing enhancer activating splicing to some extent in either cell type (56). Competition with a DCS RNA in an in vitro splicing reaction inhibits splicing, implying a positive role for some DCS RNA binding proteins (5, 53). Immunodepletion experiments and the direct addition of antibodies to splicing extract also support this role (16, 53, 54). However, CUCUCU elements within and around the DCS are also required for the repression of splicing in nonneural extract (13, 17). The DCS CUCUCU element binds to PTB in HeLa extract. Therefore, some DCS binding proteins are apparently also effectors of splicing repression or derepression.
The structure and assembly of the DCS-RNP complex is poorly understood. Here we characterize where on the DCS sequence the various DCS binding proteins contact the RNA and identify some of their protein-protein interactions. We also report the cloning and characterization of nPTB, a new protein binding to the CUCUCU element of the DCS RNA in WERI-1 nuclear extract. nPTB is very similar in sequence to PTB but is expressed primarily in the brain. The binding properties of nPTB and its reduced inhibitory activity on splicing imply roles in controlling the assembly of other splicing-regulatory proteins.
MATERIALS AND METHODS
DNA constructs and in vitro transcription.
The N70W, BS7, BS27, HβΔ6, and adenovirus pSPAd constructs are described elsewhere (13, 41, 53, 68). BS7 and BS27 templates were linearized with NotI, pSPAd was linearized with SmaI, and HβΔ6 was linearized with BamHI. The N70A1 template was obtained by placing DCS DNA under control of the T7A1 Escherichia coli RNA polymerase (RNAP) promoter using two-step PCR.
T7 RNA polymerase was used for transcription of N70W, BS7, and BS27. SP6 RNA polymerase was used for transcription of pSPAd and HβΔ6. Transcription reactions were carried out in the presence of cap analog and [α-32P]UTP by standard procedures (13).
Synthesis of 4-thiouridine-substituted RNA.
Site-specific introduction of 4-S-UTP (a gift of A. Mustaev, Public Health Research Institute, New York, N.Y.) into the DCS RNA was achieved through the “walking” transcription method using a His6-tagged E. coli RNAP bound to Ni-nitrilotriacetic acid agarose (Qiagen) (48, 58). The RNAP used in these experiments was purified, as described elsewhere, from the E. coli strain bearing the His-tagged β′ subunit of polymerase incorporated into the chromosome (a kind gift from R. Landick, University of Wisconsin, Madison, Wis.) RNAP (2 μg) and N70A1 template (2 μg) were preincubated for 5 min at 37°C in 20 μl of the transcription buffer TB (50 mM HEPES-HCl [pH 7.9], 100 mM KCl, 10 mM MgCl2), mixed with 10 μl of Ni-nitrilotriacetic acid beads preequilibrated with TB, and incubated for another 5 min. To this resin-bound complex, 2 μl of transcription starting mix (200 μM CpUpApC primer and 250 μM each ATP and GTP) was added. After a 5-min incubation, the beads were washed three times with 1.5 ml of TB. The elongation complex containing an RNA 11-mer was then walked to a desired position along the template by repeated alterations of washing and RNA chain extension with a subset of 10 μM deoxynucleoside triphosphates (NTPs) for 3 min at room temperature. The radiolabeled NTP and 4-S-UTP were then introduced into the system, and the reaction mix was incubated for 10 min at room temperature. After three washes with TB, all four NTPs at final concentrations of 200 μM each were added to generate a runoff RNA product. The modified RNA thus synthesized was purified on a 12% denaturing polyacrylamide gel.
Gel mobility shift and UV cross-linking.
UV cross-linking and gel shift experiments were carried out under the same conditions reported previously (53). A 4-μl volume (60 μg) of the WERI-1 or HeLa nuclear extract or the equivalent amount of 40% ammonium sulfate precipitate fraction (ASP40) was used in the site-specific cross-linking experiments. After incubation under splicing conditions in the presence of 13 μg of heparin per ml, the samples were irradiated with 360-nm UV light on ice for 15 min using a handheld UV lamp (UVP) and then treated with 1 μg of RNase A each for 15 min at 37°C. The cross-linked proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized on a PhosphorImager (Molecular Dynamics).
In the gel shift experiments, the amount of purified nPTB, PTB, and recombinant hnRNP H varied from 50 to 800 ng. These proteins were incubated with 100,000 cpm (15 fmol) of labeled DCS RNA for 10 min before being loaded onto a 6% native polyacrylamide gel (40:1 acrylamide-to-bisacrylamide ratio). The gel was run in 0.5× Tris-borate-EDTA (TBE) buffer and visualized as described previously.
Immunoprecipitation and Western blot analysis.
Immunoprecipitation experiments used rabbit polyclonal antibodies raised against the PTB C-terminal peptide (CGAHHLRVSFSKSTI), the C2742 antiserum against residues 172 to 711 of KSRP, the 4606 antiserum against FUSE binding protein (FBP) (a gift of D. Levens), and the anti-Sm monoclonal antibody Y12 (a gift of J. Steitz). In a typical experiment, 10 μl of protein A or GammaBind Plus Sepharose (Pharmacia), preequilibrated with TETN150 buffer (25 mM Tris [pH 7.5], 5 mM EDTA 150 mM NaCl, 0.1% Triton X-100) and blocked with 2.5 mg of bovine serum albumin per ml in TETN150, was preincubated with 1 to 15 μl of antibody for 1 h at 4°C, washed with TETN150, and then incubated with the cross-linked sample for 1 h at 4°C. Western blot analysis also used the following: rabbit antibodies raised against the C-terminal peptides of KSRP (DB-KS) and U1-70k; full-length U2AF65 and hnRNP H proteins; mouse monoclonal antibodies 9H10 against hnRNP A1 (G. Dreyfuss), AK-96 against ASF/SF2 (A. Krainer), BB7 against PTB, and MAb104 recognizing phosphorylated SR proteins (M. Roth and K. Neugebauer); and human sera Hu134 against nucleolin (P. Bouvet) and LE against LA antigen (J. Steitz).
In vitro splicing.
Each splicing reaction, conducted as described previously (13), was performed in a mixture containing 8 μl of WERI-1 extract (approximately 100 μg of protein). This was supplemented with 40 to 400 ng of nPTB or PTB or with 0.3 to 3 μl of WERI- or HeLa-derived 0.3M KCl fractions from the DCS RNA affinity column, providing an equivalent amount of either protein. These reactions were terminated after a 4-h incubation for the BS7 and BS27 templates, a 90-min incubation for the adenovirus major late transcript (SPAd), and a 2-h incubation for the β-globin transcript (Hβ · 6).
nPTB purification.
nPTB was purified from 10 ml (150 mg of protein) of WERI-1 nuclear extract by using a method modified from that described previously (61). Chromatography over 20-ml DEAE-Sepharose Fast Flow, 10-ml heparin–Sepharose CL-6B, and 2-ml poly(U)-Sepharose 4B columns (Pharmacia) resulted in a fraction containing a mixture of PTB and nPTB proteins. A 1-ml HiTrap Blue Sepharose column (Pharmacia) was used next to purify nPTB from PTB. The poly(U) fraction containing these proteins was loaded on the column in buffer A (20 mM Tris [pH 8.0], 3 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 1 mM mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) containing 150 mM KCl. A 30-ml KCl gradient (150 to 500 mM) was then applied to the column, resulting in the elution of homogeneous nPTB at approximately 250 mM KCl. The bulk of the PTB eluted at 500 mM KCl. The fractions containing each protein were combined, dialyzed against buffer DG (20 mM HEPES [pH 7.9], 80 mM potassium glutamate, 1 mM dithiothreitol, 20% glycerol, 0.2 mM EDTA, 0.2 mM PMSF) overnight, and concentrated using Centricon-30 (Amicon) concentrators. HeLa PTB was purified in a similar fashion.
RNA affinity column purification.
RNA coupling to adipic acid hydrazide agarose (Pharmacia) was performed as specified by the manufacturer. A 40-nmol (470 μg) portion of a chemically synthesized 37-mer DCS RNA oligonucleotide (CUGAGGCUGGGGGCUGCUCUCUGCAUGUGCUUCCUGG) or the same amount of a 37-mer UR (unrelated) RNA oligonucleotide (CGAAUUGGGUACCGGGCCCAGCGCCGCCGUCGUGCCG) was dissolved in 980 μl of 20 mM Tris-HCl (pH 7.5). A 20-μl volume of freshly made 1 M sodium periodate solution was added, and the mixture was incubated on ice in the dark for 1 h. Oxidized RNA was ethanol precipitated and resuspended in 1 ml of 0.1 M sodium acetate (pH 5). This RNA was then incubated for 3 h at 4°C with 0.5 ml of adipic acid hydrazide agarose beads preequilibrated with the same buffer. The beads were then washed with 2 M NaCl and equilibrated with buffer DG. An 8-ml volume of either HeLa or WERI-1 nuclear extract supplemented with 0.1 mg of heparin per ml was added to the beads, which were then incubated for 4 h at 4°C with gentle mixing. The slurry was packed into disposable 2-ml columns and extensively washed either with buffer DG plus 20 mM KCl or with buffer D (20 mM HEPES [pH 7.9], 100 mM KCl, 1 mM dithiothreitol, 20% glycerol, 0.2 mM EDTA, 0.2 mM PMSF). The bound proteins were eluted with 4 to 6 ml of buffer A containing 0.3, 0.5, 1, or 2 M KCl or 6 M urea. The fractions were dialyzed against buffer DG and concentrated to approximately 200 μl using Centricon-10 (Amicon) concentrators.
Proteins of each fraction were resolved by SDS-PAGE (10% polyacrylamide) and analyzed by nanospray mass spectrometry (MS). Coomassie blue-stained proteins from the gel were subjected to an in-gel digestion with Endoproteinase Lys-C (Roche Diagnostics), and the extracted peptides were desalted over a 100-nl gel loader pipette tip column filled with POROS R2 resin (Perseptive Biosystems). The peptides were eluted off the resin in 1 μl of 50% methanol–5% formic acid into a nanospray glass capillary (PROTANA). The peptide solution was then infused into an LCQ mass spectrometer (FinniganMat), and individual peptide ions were subjected to tandem MS-MS analysis. The acquired MS-MS data were compared to the nonredundant protein database (National Center for Biotechnology Information) using the SEQUEST software (John Yates and Jimmy Eng, University of Washington). Identified proteins were confirmed by Western blot analysis.
Northern blot analysis.
nPTB cDNA tissue distribution was assessed using a commercial multiple-human-tissue Northern blot as specified by the manufacturer (Clontech). The membrane was probed with a 56-nt nPTB oligonucleotide or nPTB cDNA, a 1-kb PCR-generated fragment of PTB cDNA (nt 550 to 1448 of the PTB open reading frame), or a 2.0-kb human β-actin cDNA probe provided with the blot.
Total RNA from each cell line was isolated using the guanidine thiocyanate method (15). A 10-μg portion of total cellular RNA was loaded per lane, separated on a 1% formaldehyde–agarose gel, and transferred to a nylon membrane (Schleicher & Schuell). This blot was probed with the same β-actin cDNA or PTB cDNA fragments as the multiple-tissue Northern blot or with a full-length nPTB cDNA. The results were visualized by PhosphoImager (Molecular Dynamics) analysis.
nPTB cDNA cloning.
Purified nPTB was subjected to SDS-PAGE (10% polyacrylamide), and the nPTB band was excised and subjected to in-gel tryptic digestion. The eluted peptides were fractionated by high-pressure liquid chromatography on a Vydac C18 column and sequenced on an automated protein sequencer (Perkin-Elmer model 492). A search of the GenBank database with the peptides that differed from PTB identified no expressed sequence tag (EST) matches but resulted in the retrieval of two short bacterial artificial chromosome tags of human genomic clone CIT-HSP 2282L8 (DDBJ/EMBL/GenBank accession no. AQ000290 and AQ006967). The short identifier sequence from the end of this clone is in the nPTB exon encoding most of the NNQFQALLQYGDPVNAQQAK peptide. The 56-nt NNQ oligonucleotide (ATACTCAAGCTCTGCTCCAGTATGGTGATCCAGTAAATGCTCAACAAGCAAAACTA) encoding most of this peptide was used to probe a WERI-1 cDNA Lambda Zap library. Six positive clones were obtained. Random hexamer cDNA synthesis of WERI-1 total RNA followed by PCR using the NNQ oligonucleotide and a degenerate oligonucleotide corresponding to the FFQDHK peptide of nPTB resulted in an 821-bp PCR fragment containing approximately half of the nPTB open reading frame. This PCR product was then used to probe the same WERI-1 cDNA library. One positive clone identified by both probes was plaque purified and sequenced.
Nucleotide sequence accession number.
The human nPTB cDNA sequence has been submitted to GenBank and has been given accession no. AF176085.
RESULTS
Proteins that bind to individual sequence elements within the DCS RNA.
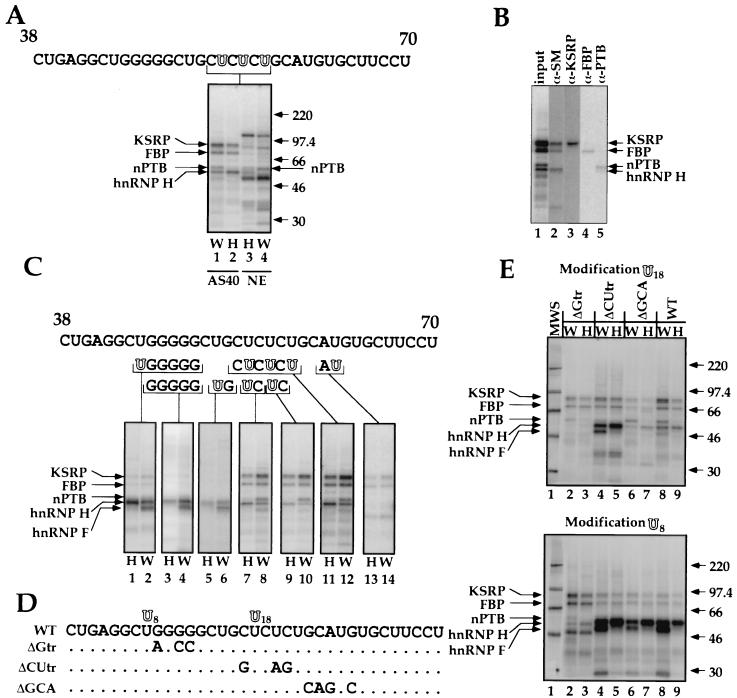
The DCS acts in vivo as part of an intronic splicing enhancer needed for inclusion of the N1 exon (56). The DCS also contains a CUCUCU element that mediates repression of N1 splicing in nonneural cells and that cross-links to PTB in a full-length spliceable src RNA (12, 13, 17). A short DCS RNA probe was previously shown to assemble into two RNA-protein complexes, “nonspecific” and “specific” (53). The nonspecific complex can be competed away by any RNA and is hence non-sequence specific. It is also present in both HeLa and WERI extracts. This complex contains a prominent 50- to 55-kDa protein observable by shortwave UV cross-linking. The specific complex is enriched by removing the 55-kDa protein by ammonium sulfate fractionation of the extract. This specific complex contains hnRNP H, hnRNP F, and KSRP as well as other factors. Although present in HeLa extract, the specific complex is more prominent in WERI extract (16, 53, 54). This complex is also sequence specific, since it is competed efficiently only with a DCS RNA. Although PTB binds to the CUCUCU element of the DCS in a full length pre-mRNA, we had not previously seen PTB binding to this element in the complexes formed on a short DCS RNA probe. To characterize proteins interacting specifically with the CUCUCU element of the DCS, we generated DCS RNAs where the U's in this element were replaced with the photoaffinity label 4-thiouridine (4-thioU). The 5′ phosphate of each cytosine in the CUCUCU element was further replaced with 32P. This modified DCS RNA was incubated with HeLa or WERI nuclear extract and irradiated with 366-nm UV light. The extract was treated with RNase A, and the cross-linked proteins were resolved by SDS-PAGE.
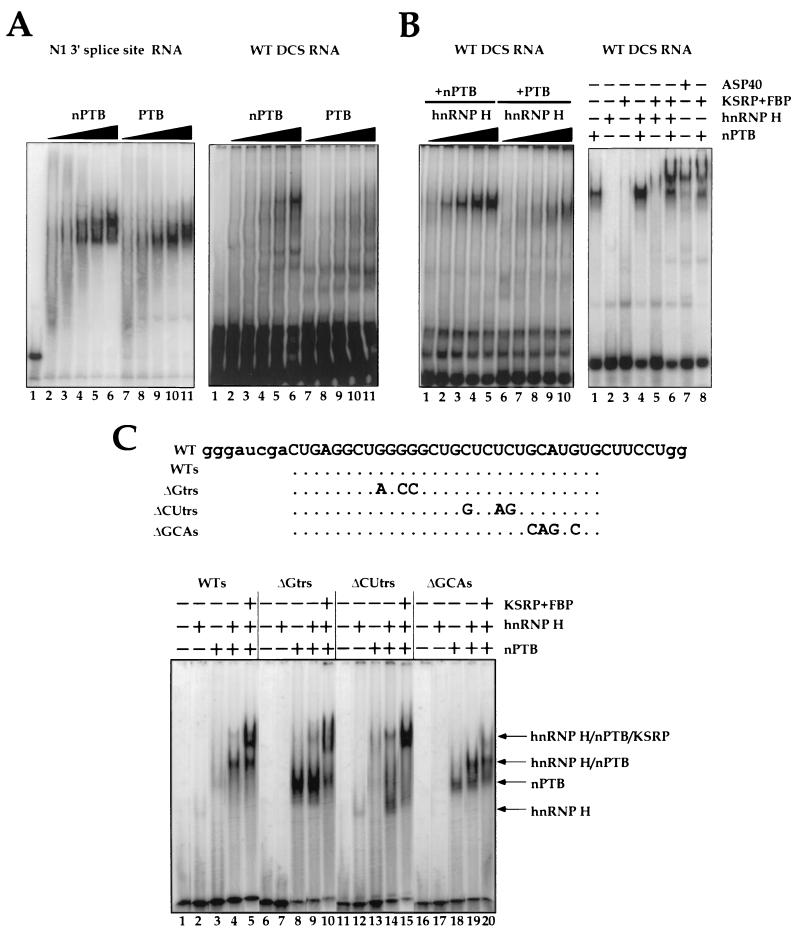
As with shortwave UV cross-linking, 4-thioU cross-linking to the DCS RNA in the unfractionated extract gave a prominent band of 50 to 55 kDa as well as a band of approximately 100 kDa (Fig. 1A lanes 3 and 4). These proteins are thought to be the La and nucleolin proteins nonspecifically bound to the RNA (see below). 4-ThioU cross-linking also gave a 60-kDa band (marked nPTB) that was not previously observed with shortwave UV cross-linking. This 60-kDa protein was more prominent in WERI extract. We showed previously that the RNP complex that is specific for the DCS sequence is enriched in the 40% ammonium sulfate pellet fraction (ASP40) of the extract (53). 4-ThioU cross-linking in this fraction again gave protein bands similar to those seen by shortwave UV (53) (Fig. 1A lanes 1 and 2). These proteins were identified by immunoprecipitation (Fig. 1B). The top band, migrating at 80-kDa, was identified as KSRP, and the 70-kDa band was identified as FBP, which is highly related to KSRP (Fig. 1B, lanes 3 and 4) (18, 20). The other prominent band common to both extracts runs at 55 kDa and was identified as hnRNP H, as indicated by immunoprecipitation with Y12 anti-SM antibodies (lane 2). We previously found that hnRNP H is recognized by this antibody (16). KSRP and hnRNP H were identified earlier as components of the specific DCS complex. The coimmunoprecipitation of KSRP and FBP with hnRNP H is presumably due to their presence in the same complex, since anti-SM serum has not been observed to react with KSRP and FBP. This coimmunoprecipitation varies with the reaction conditions and is not seen when precipitating with anti-KSRP or anti-FBP antibodies (lanes 3 and 4).
FIG. 1.
Structure and composition of the DCS complex. (A) nPTB cross-links to the CUCUCU element of the DCS RNA. The DCS RNA sequence is shown at the top. Modified bases are shown as hollow letters. Site-specific labeling of proteins binding to the DCS RNA is shown below the sequence. Cross-linking in the ASP40 fraction of WERI-1 (lane 1) or HeLa (lane 2) extract and in total HeLa (lane 3) or WERI-1 (lane 4) nuclear extracts is shown. Positions and sizes of the molecular weight standards (in thousands) are indicated on the right. (B) Immunoprecipitation of the proteins binding to the DCS RNA in the WERI ASP40 fraction. Antibodies used were anti-SM Y12 (lane 2), anti-KSRP (lane 3), anti-FBP (lane 4), and anti-PTB (lane 5). (C) Probing hnRNP H, hnRNP F, KSRP, FBP, and nPTB binding sites on the DCS RNA by site-specific labeling. Modified (hollow) and radiolabeled nucleotides are shown below the DCS RNA sequence. Cross-linking was done in the ASP40 fraction of WERI-1 (lanes W) or HeLa (lanes H) nuclear extract. (D) DCS RNA mutants used in the cross-linking experiments. Positions of 4-thioU modifications are indicated as U8 and U18 above the wild-type (WT) sequence, and the mutations are indicated below. (E) Effect of mutations in DCS RNA on the cross-linking of proteins to positions U18 (top) and U8 (bottom) of the DCS in the ASP40 fraction of WERI-1 extract (lanes W) or HeLa extract (lanes H).
Interestingly, the 60-kDa protein seen binding in the whole nuclear extract is also enriched in the ASP40 fraction and is immunoprecipitable with anti-PTB antibodies (Fig. 1B, lane 5). We reported earlier on a WERI-specific form of PTB that bound to the N1 3′ splice site (13). This may be the same protein seen here binding to the DCS sequence in WERI extract. As explained below, we call this protein nPTB (for neurally enriched homolog of PTB). However, since WERI extract also contains the normal HeLa form of PTB, these bands could be mixtures of the two proteins. Nevertheless, it is clear that a PTB-related protein binds the CUCUCU element of the DCS.
An equivalent DCS-cross-linked PTB band is present in HeLa extract but is less pronounced. It is not clear why PTB cross-linking in HeLa extract is inefficient. It could be that PTB actually binds less well to the DCS than nPTB does. Alternatively, the precise interaction of nPTB and PTB with the RNA may differ, leading to inefficient cross-linking of PTB despite its binding to the RNA. Given that the DCS CUCUCU element is needed for splicing repression in HeLa extract, we thought that the latter possibility was more likely (see below).
We next extended this cross-linking approach to more precisely localize the binding sites for each protein along the DCS RNA. By incorporating the 4-thioU and 32P at different positions, we could assess which nucleotides were in close contact with each protein (Fig. 1C). When the photoaffinity label was placed just 5′ or just 3′ of the G5 element in the DCS, the predominant cross-linked bands were hnRNP H and F (lanes 1 and 2 and lanes 5 and 6, respectively). This was also seen when the RNA was 32P labeled in the G tract without the 4-thioU and the cross-linking was by shortwave UV (lanes 3 and 4). Interestingly, hnRNP F, although present in both extracts, cross-linked well to the RNA only in the WERI extract. When the 4-thioU was moved downstream to the CU tract, cross-linking to the F protein was lost but cross-linking to hnRNP H, nPTB, KSRP, and FBP was seen (lanes 7 to 12). This contact with the H protein was primarily with the 5′-most U in the CU tract; when the label was only in the central U, the intensity of the H band was decreased (lanes 9 and 10). As before, the PTB band was enriched in the WERI extract over the HeLa extract. When the 4-thioU was placed in the element UGCAUG, just downstream of the CU tract, cross-linking was weaker but was predominantly to KSRP, FBP, and an unidentified protein of about 50 kDa. With the label in this position, cross-linking to PTB or nPTB and to hnRNP H and F was lost. The proteins that bind to this position are particularly interesting because this UGCAUG element is crucial to the activity of the DCS as an enhancer. Overall, these results give a clearly ordered arrangement of the proteins along the DCS sequence, going from 5′ to 3′, hnRNP H and F then nPTB, then KSRP and FBP.
The importance of each element in the binding of the individual proteins was assessed by repeating the cross-linking on RNAs where the small sequence elements were mutated (Fig. 1D and E). These experiments used labels in two positions along the RNA sequence, just 5′ of the G tract (modification U8 [Fig. 1D and E, bottom]) or within the CU tract (modification U18 [Fig. 1D and E, top]). Cross-linking at U8 adjacent to the G tract gave predominantly the hnRNP H and F proteins as before, unless the G tract was mutated, which caused a loss of hnRNP H and F cross-linking (Fig. 1E, bottom). This was also seen with the label at U18 and confirms the need for the G tract in hnRNP H and F protein binding (Fig. 1E, lanes 2 and 3). The effects of the other mutations were most easily observed with the label at U18 (Fig. 1E, top). Mutation of the CU tract reduced nPTB cross-linking and increased cross-linking to the hnRNP H and F proteins (lanes 4 and 5). It is not clear whether the increased signal for hnRNP H and F is due to increased binding of these proteins to the mutant RNA, perhaps due to changes in secondary structure, or to increased interaction with the label at this position because of the loss of PTB binding (see below). Mutation of the GCA sequence within the UGCAUG element does not affect nPTB binding but reduces KSRP and FBP binding (compare lanes 6 and 7 with lanes 8 and 9). In sum, the cross-linking to the mutant RNAs confirms the position of contact for each protein along the RNA.
The cross-linking experiments delineated the relative position of each protein along the RNA. However, these experiments did not give any information on the stoichiometry of the proteins binding to the DCS and did not distinguish how many complexes are formed by the various cross-linked proteins. For example, when we see both KSRP and nPTB cross-linking, we do not know whether these are binding simultaneously in one complex or independently in separate complexes. Previous gel shift and antibody supershift experiments indicated that hnRNP H and KSRP are in the same complex (16, 53, 54). As a first step in testing whether PTB is also in this complex with hnRNP H and KSRP, we performed coimmunoprecipitation experiments (data not shown). Using an anti-PTB monoclonal antibody, we immunoprecipitated PTB from HeLa extract in the presence or absence of the DCS RNA. KSRP was strongly coprecipitated with PTB only in the presence of the DCS RNA, indicating that these proteins do indeed interact with the DCS RNA in the same complex (data not shown). Since the majority of the KSRP is present in the DCS complex under these conditions, these results place PTB in a complex with both hnRNP H and KSRP rather than in a complex that is independent of the previously characterized DCS complex. Moreover, since these experiments were done with HeLa extract containing PTB rather than nPTB, they demonstrate that PTB is indeed binding to the DCS in spite of its weak cross-linking. The common binding of these proteins into a single complex is addressed further below.
Characterization of DCS RNA binding proteins by RNA affinity chromatography.
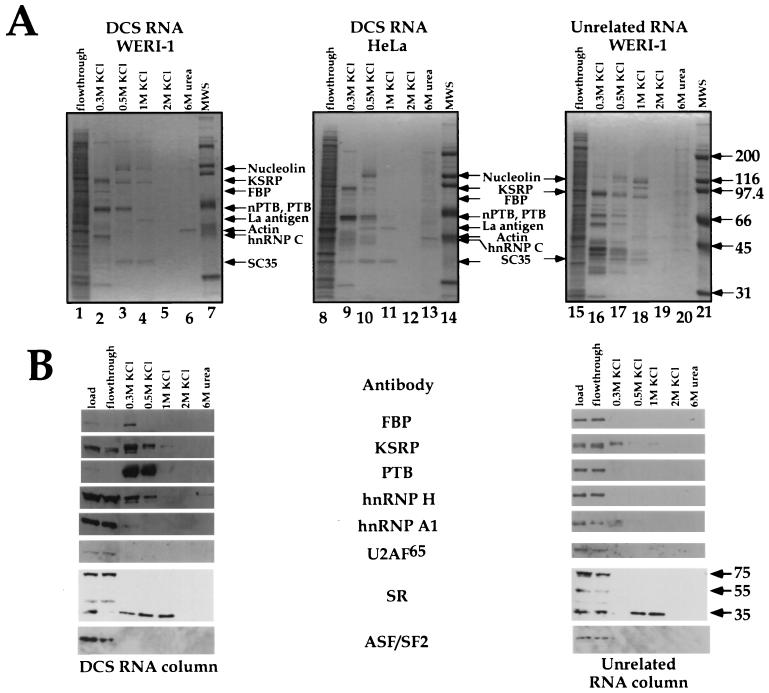
To further examine the proteins binding to the DCS RNA in the absence of cross-linking, we performed RNA affinity chromatography. After periodate treatment, the DCS RNA was chemically attached through its 3′ end to adipoyl hydrazido-Sepharose. We find that this resin shows lower nonspecific protein binding than do RNA affinity resins based on the avidin-biotin interaction. To further minimize nonspecific protein binding, nuclear extract was supplemented with 0.1 mg of heparin per ml before being loaded on the column. Based on gel shift data, this amount of heparin disrupts nonspecific RNA-protein complexes while leaving the specific DCS complex intact.
The RNA resin was incubated with either HeLa or WERI nuclear extract, packed into a column, and washed extensively in 100 mM salt buffer. DCS RNA binding proteins were then eluted in a step gradient of KCl. In a control experiment, a column carrying RNA unrelated to the DCS sequence was used. The protein composition of each fraction was analyzed after gel separation by Coomassie blue staining and Western blot analysis (Fig. 2). The elution profiles for the HeLa and WERI nuclear extracts on the DCS RNA column were very similar. The most prominent WERI protein, eluting between 0.3 and 0.5 M salt, was nPTB, as confirmed by Western blot using an anti-PTB serum that reacts with both proteins. In HeLa extract, PTB eluted in the same fractions but ran as a doublet on the gel. Note that due to differences in the running of the two gels, they cannot be perfectly aligned. The PTB in the HeLa panel is slightly below that in the WERI panel. The binding of the nPTB and PTB proteins was very efficient; these proteins were almost completely depleted from the WERI flowthrough fraction (Fig. 2B).
FIG. 2.
Purification of proteins bound to the DCS RNA or an unrelated RNA in the WERI-1 or HeLa extracts using RNA affinity chromatography. (A) Coomassie blue-stained gels displaying fractions from the DCS RNA affinity column (lanes 1 to 6 and lanes 8 to 13) or an unrelated RNA affinity column (lanes 15 to 20). Proteins identified by MS are indicated by arrows. Lanes 7, 14, and 21 contain protein markers. (B) Western blot analysis of the same fractions. MAb104 monoclonal antibody was used for identification of the SR proteins. The positions of the SC35 and ASF/SF2 (SRP 35), SRp55, and SRp75 proteins are indicated by arrows.
Western blot analysis of the column fractions identified other proteins binding to the DCS RNA. Known components of the DCS gel shift complex, KSRP, FBP, and hnRNP H, all bound the DCS RNA column and eluted in 0.3 to 0.5 M salt (Fig. 2B). By Coomassie staining, hnRNP H appeared to bind to the column less efficiently than nPTB or KSRP did. The control RNA binding proteins hnRNP A1 and U2AF65 both failed to bind to the column and eluted in the flowthrough fraction. None of these proteins bound well to the nonspecific RNA column, although some KSRP and hnRNP A1 were present in the 0.3 M fraction.
Another protein eluting from the DCS RNA column was identified as SC35. This 35-kDa protein was reactive with MAb104 anti-SR and anti-SC35 monoclonal antibodies but not with ASF/SF2 antibodies. This was the only SR protein seen binding to the DCS column. However, it also bound to the unrelated RNA column. Given the known effects of SR proteins on splicing, we are investigating whether SC35 plays a role as a DCS binding protein, even though its binding specificity is somewhat in doubt.
Several other proteins also bound to the DCS RNA column. These were identified by MS analysis of their tryptic peptides as nucleolin, La antigen, hnRNP C, and actin. The nucleolin, SC35, and La antigen proteins bound tightly to the DCS RNA, eluting mostly at 0.5 to 1 M salt. Both La and nucleolin recognize short stem-loop RNA structures containing pyrimidine-rich sequences on their termini, and the DCS RNA can potentially fold into such a structure (22, 25, 70). Thus, the binding of these proteins to the column is probably due to similarities between the DCS RNA and natural RNA substrates for these proteins. La is presumably the ∼50-kDa protein observed in the nonspecific gel shift complex with the DCS RNA (53). hnRNP C eluted from the column at 0.3 M salt. hnRNP C has affinity for pyrimidine-rich RNAs, and such elements are present in the DCS RNA (69, 72, 73). Some actin protein was also observed to stick very tightly to the column, eluting only in 6 M urea. This may result from polymerized actin filaments, trapped at the top of the column, eluting from the column when denatured into monomers.
Most of the proteins binding to the DCS RNA, including nPTB and PTB, were not retained on the unrelated RNA column, which showed a different spectrum of bands. The exceptions to this were SC35 and nucleolin, which bound tightly to both RNA columns. The nucleolin elution profile varied from batch to batch of the extract. Note that there is a prominent nucleolin breakdown product migrating just below KSRP in some of the WERI and HeLa extract fractions.
The RNA affinity chromatography was repeated with RNAs carrying mutations in each of the various subelements of the DCS RNA (data not shown). This confirmed that PTB and nPTB required the CU tract for stable binding, hnRNP H binding needed the G tract and the UGCAUG element, and KSRP binding needed the CU tract and the UGCAUG element.
Finally, the affinity chromatography results indicate that HeLa PTB binds to the DCS RNA even though it does not cross-link efficiently. Moreover, as seen previously, the eluted HeLa PTB migrates as a doublet with slightly different mobility from the WERI nPTB-PTB mixture.
Purification and cloning of nPTB.
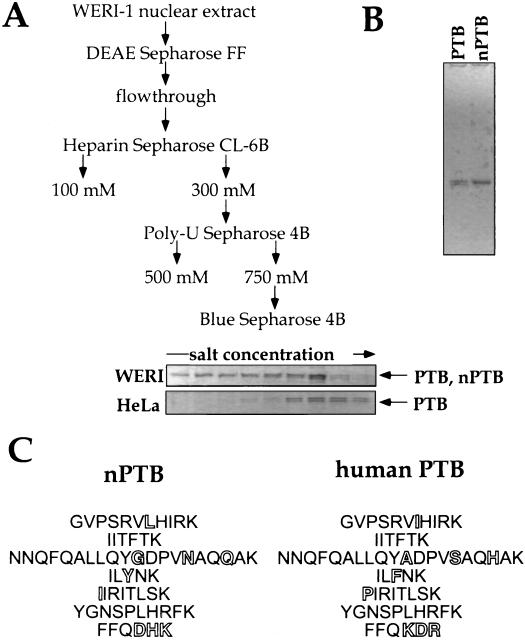
The different cross-linking patterns of PTB in the two extracts indicated a difference in the protein from these two sources. To allow a detailed comparison of the PTBs in the two extracts, we purified the two proteins using a modification of the published PTB purification protocol (61). The HeLa and WERI PTBs copurified over the first three columns: DEAE, heparin, and poly(U)-Sepharose. When the WERI extract fractions from the poly(U) column were loaded onto a HiTrap Cibacron Blue Sepharose column and eluted with a shallow gradient of KCl, the two forms of PTB were resolved. Under these conditions, nPTB came off the column earlier than the PTB isoforms seen in HeLa extract (Fig. 3A). As a result, a nearly homogeneous sample of the nPTB protein, free of PTB, was obtained (Fig. 3B).
FIG. 3.
Purification of human nPTB. (A) Purification scheme. Coomassie blue-stained gels of PTB- and nPTB-containing fractions from the HiTrap Blue column are shown below the scheme. (B) Coomassie blue staining of an SDS–10% polyacrylamide gel containing purified WERI-1 nPTB and HeLa PTB. (C) nPTB peptide sequences identified by Edman degradation (left) and corresponding human PTB-1 peptides (right). Differences between two proteins are shown by hollow letters.
nPTB was subjected to tryptic digestion followed by microsequencing of seven peptides. Two of the analyzed peptides matched the PTB sequence exactly, whereas the others contained one or more changes in amino acid sequence from the corresponding PTB peptides (Fig. 3C). The unique nPTB peptides were distributed along the PTB sequence, indicating that nPTB is a new protein rather than a new PTB splice variant.
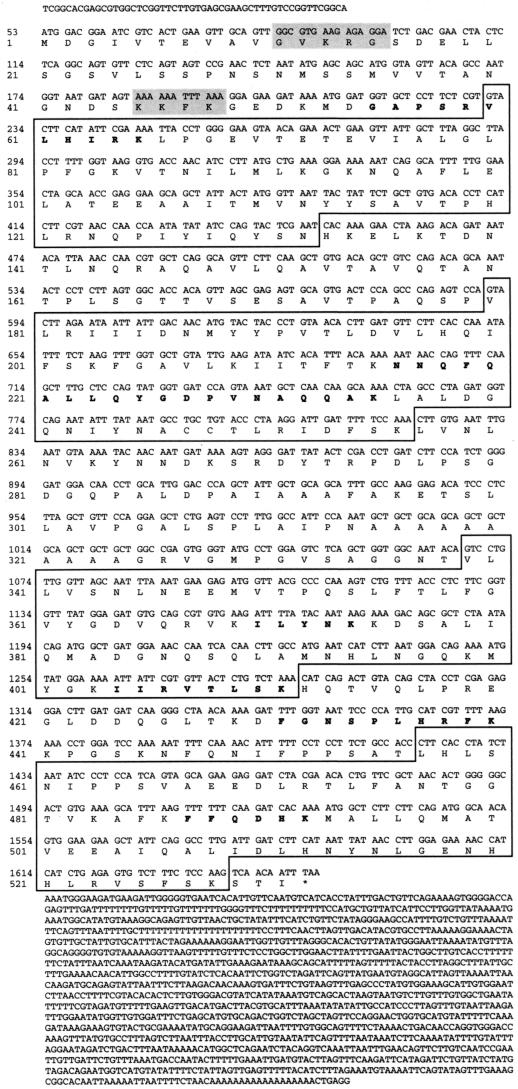
Searching GenBank databases for the nPTB peptide sequences resulted in only one positive match. There were no matches to ESTs. However, the longest identified peptide, NNQFQALLQYGDPVNAQQAK, matched the sequence from the end of the human genomic BAC clone AQ006967. This clone contained 56 nt of an apparent nPTB exon encoding most of the peptide. Reverse transcription-PCR (RT-PCR) of WERI cell total RNA, using the 56-nt exon oligonucleotide as a sense-oriented primer and an 18-nt degenerate antisense primer corresponding to the FFQDHK peptide, produced a partial nPTB cDNA, 821 nt in length. Both the 56-nt exon sequence and the RT-PCR product were used to screen a WERI cell λZap cDNA library. A clone containing the apparent full-length cDNA of nPTB was identified and sequenced (Fig. 4). This 3,060-nt cDNA contains a 1,593-nt open reading frame encoding a 531-amino-acid 57,454-Da protein. All seven nPTB peptides are present in the amino acid sequence of the encoded protein (Fig. 4).
FIG. 4.
Human nPTB cDNA sequence. The NLS sequences are shaded. The four RRM domains are boxed. Sequenced peptides are in bold. The asterisk indicates the stop codon.
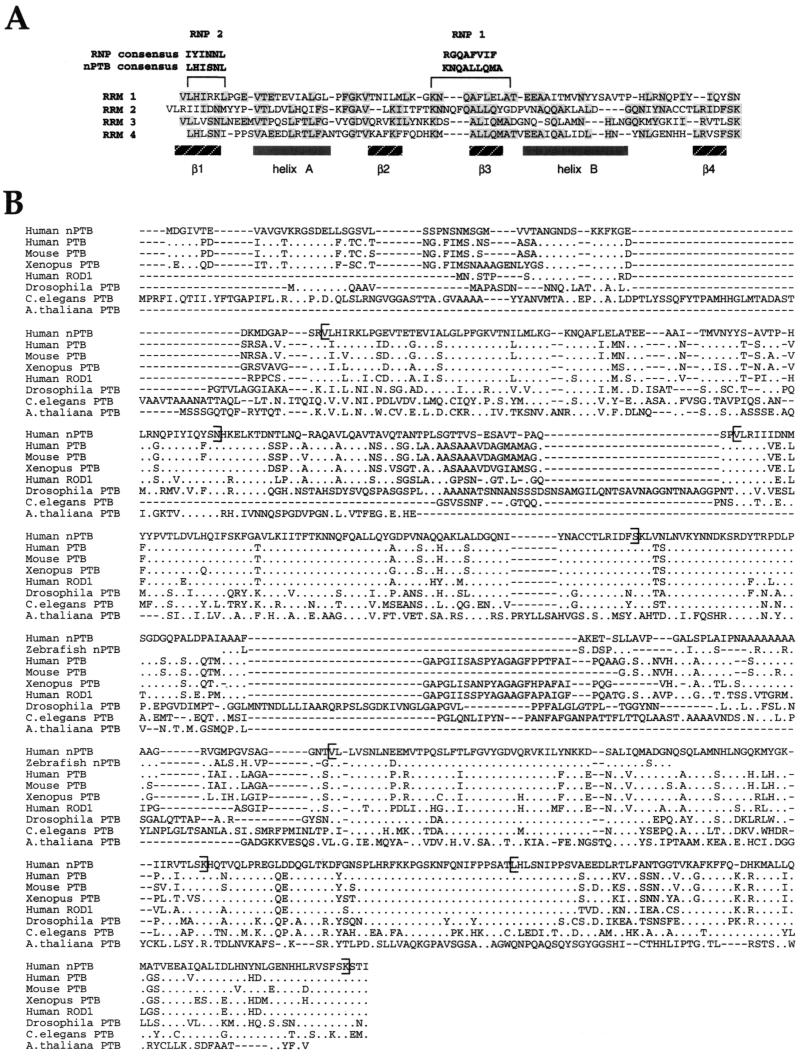
As expected, nPTB is very similar to PTB (∼74% identical, depending on how the gaps are weighted), containing four unusual RNA recognition motif (RRM) domains and a putative bipartite nuclear localization signal (NLS) near the N terminus. This NLS domain consists of two short sequences, GVKRG and KKFK, separated by a 29-amino-acid gap, and matches the consensus nucleoplasmin NLS better than the corresponding PTB domain does (63) (Fig. 4). The four 80-amino-acid RRM domains of nPTB can be aligned with each other and are similar to the PTB RRM domains. A typical RRM domain contains two consensus elements, RNP1 and RNP2, that comprise β-strands 3 and 1 of the domain (32, 60, 76, 79). Like PTB, the RNP elements of nPTB diverge from those in the consensus RRM (Fig. 5A). Alignment of the four nPTB RRM domains also shows common residues within α-helix A, within β-strand 2 and following β-strand 4. In the known structures of RRMs complexed with RNA, the β-sheets both form the hydrophobic core of the domain and make extensive contact with the RNA on the interaction surface (1, 32, 60, 79). The conservation of the nPTB sequence in these regions may reflect folding constraints or could result from similarities in RNA recognition.
FIG. 5.
Comparative analysis of the nPTB amino acid sequence. (A) Structural organization of the nPTB RRMs. RNP1 and RNP2 consensus sequences for the typical RRM and for the nPTB RRMs are shown at the top. The domain secondary structure is schematically shown below the RRM sequences. Residues identical in at least two RRMs are shaded. (B) Comparison of human nPTB with related PTB sequences. The human nPTB sequence is aligned with PTB sequences from human (accession no. X62006), mouse (accession no. X52101), Xenopus laevis (accession no. AAF00041), D. melanogaster (accession no. AAF22979), C. elegans (accession no. CAA85411), and Arabidopsis thaliana (accession no. AF076924), with a partial sequence of nPTB from Danio rerio (accession no. AA566427) and with the sequence of human ROD1 (accession no. NM005156) using the MegAlign program (DNAStar). Residues identical to those of nPTB are shown as dots. Brackets indicate the borders of RRM domains. (C) A phylogenetic tree of human nPTB and mammalian, plant, and C. elegans PTBs generated by MegAlign based on the protein alignment. The length of the branch on the x axis indicates the percent sequence divergence.
The variation in sequence between human nPTB and PTB far exceeds that between PTBs from different mammals. Human nPTB is presumably the homolog of the mouse brain PTB protein recently isolated by the Darnell laboratory, since they are nearly identical (accession no. AF095718). An EST database search with the nPTB sequence also identified the partial cDNA sequence of an apparent zebrafish nPTB (accession no. AA566427). Phylogenetic comparison of the known PTB sequences places nPTB on a distinct branch, roughly equally related to all the mammalian PTBs (Fig. 5C). nPTB is also 67% identical to ROD1, another mammalian PTB homolog identified as a regulator of differentiation in Schizosaccharomyces pombe (80). Searches of the complete Caenorhabditis elegans and Drosophila melanogaster genomes each uncovered a single gene with similarity to PTB. These proteins were divergent from both mammalian proteins, but at several positions they were more similar to PTB than nPTB. These results are consistent with the idea that nPTB is relatively recently evolved and is perhaps specific to the vertebrates.
Tissue distribution of nPTB.
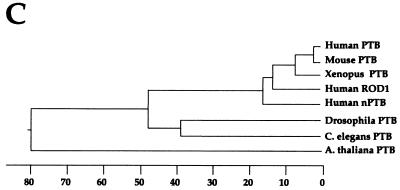
The biochemical assays for nPTB indicated its presence in WERI but not HeLa extract. We next examined the expression of nPTB and PTB on a multiple-tissue Northern blot (Fig. 6A). The full-length nPTB cDNA and a 1-kb fragment of the PTB coding region were used as probes. A human β-actin cDNA was used as a control. nPTB mRNA appeared as a single band approximately 3.4 kb in length, while PTB gave rise to a doublet of 3.6- and 4.7-kb bands. The upper band may be a cross-hybridizing mRNA or possibly an immature form of PTB mRNA. PTB mRNA was present in a wide range of tissues, with the lowest levels observed in the brain. In contrast, nPTB mRNA was most abundantly expressed in the brain, although there was detectable expression in other tissues, notably the heart and skeletal muscle.
FIG. 6.
Tissue and cell line distribution of nPTB. (A) Northern blot analysis of poly(A)+ RNA from the indicated human tissues using an nPTB oligonucleotide probe (top) or the PTB cDNA probe (middle) probe. Loading was verified by hybridization to a β-actin control cDNA probe (bottom). (B) Northern blot analysis of total RNA from the indicated neural (lanes 1 and 2) and nonneural (lanes 3 and 4) cell lines. Probes included the full-length nPTB cDNA (top), a 1-kb fragment of PTB (middle), and the β-actin control cDNA probe (bottom). (C) Western blot analysis of proteins from neural (lanes 1 to 3) and nonneural (lanes 4 to 6) cell lines using rabbit polyclonal anti-PTB antibodies recognizing both the PTB and nPTB proteins (top). The positions of the PTB isoforms and of nPTB are indicated by arrows. The same blot was probed with anti-U170K antibody as a protein-loading control (bottom).
The level of nPTB mRNA was also examined in tissue culture cell lines (Fig. 6B). Abundant expression was observed in the WERI-1 neural cell line, as expected. Only minimal mRNA was observed in the nonneural cell lines, HeLa and HEK293. Significantly, only low levels of nPTB mRNA were seen in the human neuroblastoma cell line, LA-N-5. This was confirmed by RT-PCR experiments, where LA-N-5 cells showed higher expression of nPTB mRNA than did HeLa or HEK cells but significantly lower expression than did WERI-1 cells (data not shown). Since LA-N-5 cells show strong inclusion of the src N1 exon, the level of nPTB is not the only factor that determines N1 splicing. These cell lines all expressed abundant PTB mRNA, confirming that WERI cells express both PTB and nPTB (Fig. 6B). This was further confirmed by Western blot analysis (Fig. 6C). nPTB could be observed in WERI cells as a reactive band running between the two major isoforms of PTB.
nPTB is a weaker repressor of N1 splicing than is PTB.
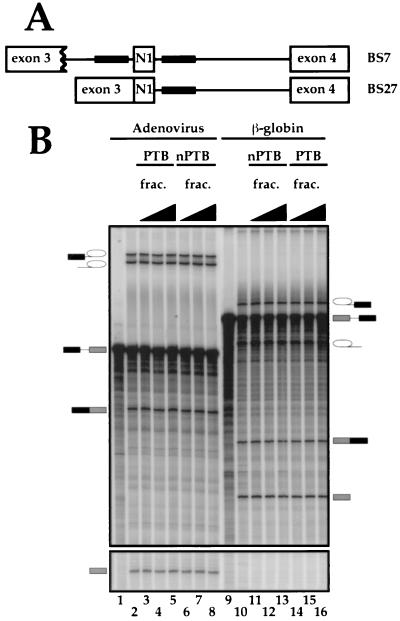
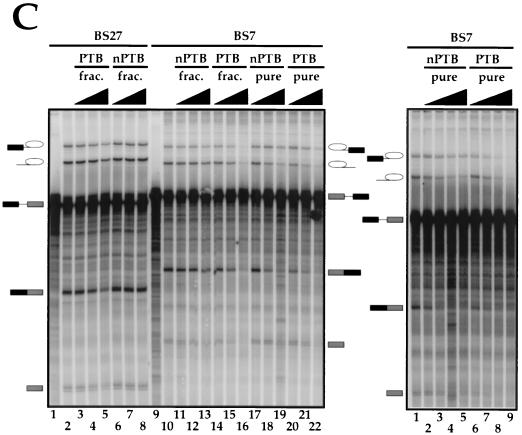
To assess the functional differences between nPTB and PTB, we analyzed the behavior of each protein in the in vitro splicing assay by using two src minigene transcripts. BS7 contains both the upstream and downstream regulatory sequences, each containing a pair of CUCUCU elements (Fig. 7A). BS27, obtained by precise deletion of the intron upstream of the N1 exon, contains only the two downstream CUCUCU elements. We previously showed that BS27 is spliced in the HeLa nuclear extract, where BS7 is repressed. Both transcripts splice equally well in WERI extract (12).
FIG. 7.
Effect of PTB and nPTB on N1 exon splicing in vitro. (A) Maps of the src splicing substrates. Black boxes indicate splicing-regulatory elements. (B) Splicing of adenovirus major late (lanes 1 to 8) and β-globin (lanes 9 to 16) transcripts in WERI-1 nuclear extract in the absence (lanes 2 and 10) or presence of 0.3, 1, or 3 μl of HeLa (lanes 3 to 5 and 14 to 16) or WERI-1 (lanes 6 to 8 and 11 to 13) 0.3 M KCl RNA affinity fractions. (C) The left panel shows splicing of the BS27 (lanes 1 to 8) or BS7 (lanes 9 to 22) transcripts in the WERI-1 nuclear extract. Lanes 1 and 9 contain no extract. Lanes 2 and 10 contain WERI extract without any additional factors. Lanes 3 to 5 and 14 to 16 contain WERI extract plus 0.3, 1, or 3 μl of HeLa 0.3 M KCl RNA affinity fraction. Lanes 6 to 8 and 11 to 13 contain WERI extract plus 0.3, 1, or 3 μl of WERI 0.3 M KCl RNA affinity fraction. Lanes 17 to 19 contain 40, 120, or 400 ng of nPTB. Lanes 20 to 22 contain 40, 120, or 400 ng of PTB. The right panel shows splicing of BS7 transcript in the WERI-1 nuclear extract using different preparations of nPTB, PTB, and nuclear extract from the left panel. Lane 1 contains WERI extract without any additional factors. Lanes 2 to 5 contain 50, 100, 200, or 400 ng of nPTB. Lanes 6 to 9 contain 50, 100, 200, or 400 ng of PTB.
Purified HeLa PTB efficiently repressed BS7 splicing when titrated into the WERI nuclear extract (Fig. 7C, left, lanes 20 to 22). The levels of both the splicing intermediates and products were decreased 3- to 10-fold by the added PTB. Similarly, the 0.3 M fraction of the HeLa extract from the DCS RNA column (Fig. 2) also repressed splicing (Fig. 7C, left, lanes 14 to 16). The splicing repression required the same amount of PTB whether it was introduced as 0.3 M fraction or as a purified protein. At the highest point of the titration, the total PTB concentration was about twofold higher than that of the endogenous protein in HeLa extract. Bacterially expressed recombinant PTB also repressed splicing but was about 10 times less active than the purified protein (data not shown). This may be due to aberrant folding of the recombinant protein, to a lack of posttranscriptional modification, or to other effects.
PTB was a much stronger repressor of splicing than was nPTB. At the highest concentration of nPTB, the levels of the second-step products of splicing were somewhat reduced but the levels of the intermediate products of the first step were unaffected (Fig. 7C, left, lanes 17 to 19). Most interestingly, when the 0.3 M RNA affinity column fraction of the WERI extract was used, virtually no repression of splicing was observed (lanes 11 to 13). Thus, the presence of other factors in the 0.3 M fraction may further reduce the repressor activity of nPTB.
Control introns from adenovirus and β-globin were unaffected by either nPTB or PTB or by the column fractions, indicating the specificity of nPTB and PTB for the src substrate (Fig. 7B and data not shown). Moreover, the inhibitory effect of PTB and nPTB required the CU element containing sequences upstream of the N1 exon, since the BS27 RNA was only weakly repressed by the PTB fraction and not at all repressed by nPTB (Fig. 7C, left, lanes 1 to 8).
The amounts of PTB and nPTB used in these experiments were carefully measured to ensure that equal amounts of protein were added in each of the titrations. These fractions also have equal binding activity for the N1 3′ splice site, as shown below. To further ensure that the observed difference in repression activity was not due to variations in our preparation of the proteins, we repeated these experiments with several independently isolated samples of PTB and nPTB and with two preparations of WERI extract. The results were always the same; PTB strongly repressed splicing, while nPTB only partially reduced the second-step products. Another titration of PTB and nPTB into a different sample of WERI extract is shown in Fig. 7C (right).
Assembly of the DCS complex from purified components.
Three sequence elements were defined previously in functional assays and are defined here as binding sites for particular proteins: the G tract, the CU tract, and the UGCAUG element. These three elements all reside within a 20-nt portion of the DCS sequence. The RNA contacts of an RRM or a KH domain can be between 4 and 7 nt, making these elements reasonable binding sites for individual RNA binding proteins (43, 76). However, one imagines that there must be extensive protein-protein contacts between the DCS complex proteins if they are simultaneously bound to this short sequence. To examine how the binding of each DCS protein affected the binding of the others, we performed gel shift experiments using recombinant or purified fractions of the individual proteins.
First, we tested whether nPTB and PTB were distinguishable in their binding properties. Purified nPTB and PTB were used in electrophoretic mobility shift assays with both the DCS RNA and the polypyrimidine tract of the N1 3′ splice site as probes (Fig. 8A, right and left panels, respectively). Both proteins bound the 3′ splice site RNA, with saturation reached at the same concentration of protein, although the mobilities of the PTB and nPTB complexes were slightly different. In contrast, with the DCS RNA probe, nPTB formed a larger and more abundant complex than PTB did. In the PTB binding reaction, the predominant RNA-protein complex was much faster migrating and only weakly present in the nPTB reaction. The overall binding of the two proteins to the DCS RNA sequence was much weaker than that to the 3′ splice site. Nevertheless, the affinity of nPTB for the DCS RNA probe was clearly higher than that of PTB.
FIG. 8.
Binding of purified nPTB and PTB to the src N1 exon splicing-regulatory elements in the presence and absence of other protein components of the DCS complex. (A) The left panel shows a gel mobility shift analysis of nPTB (lanes 2 to 6) or PTB (lanes 7 to 11) complexed with an N1 3′ splice site polypyrimidine tract RNA. Lane 1 contains free RNA probe (20 fmol, 105 cpm). Lanes 2 to 6 contain 50, 100, 200, 400, and 800 ng of purified nPTB. Lanes 7 to 11 contain equivalent amounts of PTB. The right panel shows a gel mobility shift analysis of nPTB (lanes 2 to 6) or PTB (lanes 7 to 11) complexed with an src DCS RNA. The amount of protein in each lane is equivalent to that in the left panel. (B) The left panel shows that hnRNP H enhances PTB and nPTB binding to the DCS RNA. Binding-reaction mixtures contained 100 ng of either nPTB (lanes 1 to 5) or PTB (lanes 6 to 10). This was supplemented with 25 ng (lanes 1 and 6), 50 ng (lanes 2 and 7), 100 ng (lanes 3 and 8), 200 ng (lanes 4 and 9), or 400 ng (lanes 5 and 10) of hnRNP H. The right panel shows assembly of the DCS-like complex from purified and recombinant factors. A total of 800 ng of purified nPTB, 300 ng of recombinant hnRNP H, and/or 400 ng of a KSRP-FBP fraction were used where indicated by +. (C) DCS RNA mutants used in the gel shift experiments. The sequence of the original DCS probe is shown at the top. The WTs probe sequence is truncated as indicated. The ΔGTrs, ΔCUTrs, and ΔGCA mutations are indicated. Gel mobility shift analysis of nPTB complexed with src DCS RNA mutants in the presence of hnRNP H, KSRP, and FBP is shown below the sequences. A total of 200 ng of purified nPTB, 300 ng of recombinant hnRNP H, and/or 400 ng of a KSRP-FBP fraction were used where indicated by +. The WTs and mutant probes are indicated above. The position of each complex is shown by an arrow.
Next, we examined the effect of recombinant hnRNP H on the binding of nPTB and PTB. hnRNP H interacted very weakly with the DCS RNA by itself (Fig. 8B, right, lane 2). However, the effect of combining the proteins was striking; binding of both nPTB and PTB was improved by hnRNP H (Fig. 8B, left). Complex formation in the presence of 300 ng of nPTB and 400 ng of hnRNP H was 4- to 10-fold higher than for either protein alone (Fig. 8B, right, lane 4). Oddly, the gel mobility of the complex formed with the combination of hnRNP H and nPTB was similar to that formed with nPTB alone (see below). hnRNP H also stimulated PTB binding, although equivalent reactions with PTB and hnRNP H exhibited at least fourfold less complex formation than with nPTB at all points in the titration (Fig. 8B, left). From these results, it is clear that hnRNP H strongly affects both nPTB and PTB binding to the DCS RNA.
A purified fraction containing almost exclusively KSRP and FBP did not give an observable complex with the DCS RNA in the gel shift assay (Fig. 8B, right, lane 3). Similarly, combining the KSRP fraction with hnRNP H gave only a weak new complex (lane 5). This KSRP fraction, when added to a complex containing both hnRNP H and nPTB, bound strongly to the DCS RNA (lane 6). The mobility of this hnRNP H-nPTB-KSRP complex was lower than that of the hnRNP H-nPTB complex and about equivalent to that of the complex formed in the crude ASP40 fraction (lane 7).
These gel shift experiments with purified proteins indicate that nPTB and/or PTB are essential for the cooperative assembly of hnRNP H and KSRP into the RNP complex. However, the comigration of the hnRNP H-nPTB complex with the complex formed with nPTB alone was confusing. A combination of cross-linking and antibody supershift experiments indicated that both proteins were indeed in the hnRNP H-nPTB complex (data not shown). The comigration was thus apparently due to a change in the stoichiometry or conformation of the nPTB complex upon addition of H. In addition to the CU tract, the DCS RNA used for these experiments has a second potential PTB binding site. This is a group of pyrimidine residues at the extreme 3′ end of the DCS RNA that were known to affect PTB binding in the RNA affinity chromatography assay (data not shown). To resolve the nPTB and hnRNP H-nPTB complexes, we shortened the DCS probe to remove these residues (Fig. 8C, top, probe WTs). With this WTs probe, nPTB gives a weak complex on its own (Fig. 8C, lane 3). hnRNP H alone gave a faster-migrating complex that was barely detectable (lane 2). The combination of these two proteins allowed the formation of a strong RNP complex band that was larger than that of either protein alone, as well as a weak band still higher in the gel (lane 4). Finally, the addition of KSRP and FBP shifted the nPTB-hnRNP H complex higher in the gel to make a three-protein DCS complex (lane 5).
The resolution of the nPTB and hnRNP H-nPTB complexes allowed us to examine the importance of the different sequence elements in the binding of each protein, using the gel shift assay. These results agreed well with the cross-linking and affinity chromatography results. Mutation of the G tract eliminated the weak binding of hnRNP H, as expected (Fig. 8C, lane 7). Interestingly, this mutation stimulated nPTB binding (compare lane 8 with lane 3). The DCS sequence can form a secondary structure that pairs the G tract with the CU tract. Mutation of the G tract presumably allows easier access of nPTB to the CU element by disrupting this RNA structure. This may be part of the stimulatory activity of hnRNP H for nPTB binding; by binding to the G tract, hnRNP H may make the nPTB site more accessible. The addition of hnRNP H to the DCS with a mutant G tract did not further stimulate nPTB binding, although a faint higher-molecular-weight complex was seen (lane 9). The addition of KSRP and FBP shifted the PTB complex further, although this complex was smeared and was apparently less stable than the hnRNP H-nPTB-KSRP complex seen on wild-type RNA (lane 10).
Mutation of the CU tract also disrupted the potential secondary structure between this element and the G tract. However, the binding of hnRNP H was only weakly stimulated by this change (lane 12). Presumably, hnRNP H needs nPTB to bind well, whether the secondary structure is there or not. As expected, the CU tract mutation nearly eliminated nPTB binding (lane 13). Interestingly, the combination of hnRNP H and nPTB gave some complexes with this RNA (lane 14). These ranged in mobility from the size of hnRNP H alone to the size of the full DCS complex. It is possible that the interaction between nPTB and hnRNP H stimulates binding even if not all the normal RNA contacts can be made to the mutant RNA. These hnRNP H-nPTB complexes coalesced into a strong DCS complex in the presence of KSRP (lane 15).
Mutation of the UGCAUG element also behaved as predicted. PTB formed a complex on its own (lane 18). This was shifted to an hnRNP H-nPTB complex when hnRNP H was added, although the stimulation of binding was not as strong as that seen with the wild-type RNA (lane 19). Most significantly, the UGCAUG is essential to KSRP binding, since the hnRNP H-nPTB complex did not show a substantial further shift in the presence of KSRP (lane 20).
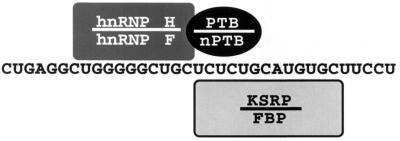
Taken together, the binding data provide a consistent picture of the DCS complex (Fig. 9). This complex is held together by both protein-RNA contacts and protein-protein contacts. The G tract is the binding site for hnRNP H and F, although it is not clear yet whether hnRNP H and F are in separate complexes or bind as a heterodimer (16). nPTB binds to the CU tract, and this interaction is critical in stimulating the subsequent assembly of the other proteins. Finally, KSRP and FBP bind to the UGCAUG element, although, again, these two proteins may bind separately or together.
FIG. 9.
Diagram of the DCS RNP complex. The position of each protein along the DCS RNA is indicated. These contacts are supported by cross-linking, affinity chromatography, and gel shift assays using wild-type and mutant DCS RNA sequences. Homologous pairs of proteins are indicated (hnRNPs H and F, PTB and nPTB, and KSRP and FBP). The stoichiometry of hnRNP F and FBP binding relative to their homologs is not yet known.
DISCUSSION
Cooperative assembly of an hnRNP complex.
The regulation of spliceosome assembly is generally thought to occur within the hnRNP complexes that assemble onto nascent pre-mRNAs as they are synthesized by RNAP II. The structures of these pre-mRNPs are extremely complex, with dozens of different hnRNPs binding to each RNA transcript (4, 42, 52). However, the interactions of these proteins with the RNA and each other are generally unknown. In this study, we have characterized the assembly of a small subset of these RNPs that bind to an important splicing regulatory region in the src pre-mRNA, the DCS. The DCS is interesting because it can have both enhancing and repressing activity on the splicing of an upstream exon, depending on the cellular environment and its adjacent sequences (55, 56).
The c-src N1 exon is controlled by an intricate combination of positive and negative regulatory elements that bind to an array of RNA binding proteins (13, 16, 53–56). The role of each DCS binding protein in splicing regulation is not yet understood. Individual proteins could be involved in splicing repression and derepression as well as in true splicing enhancement. Better functional data on all the DCS binding proteins are needed to resolve their activities. As we build up our picture of the RNP structure of the enhancer region and examine how it changes upon splicing activation, the roles of the individual proteins should become clearer. With this in mind, we examined how the DCS complex assembles from individual pre-mRNA binding proteins and identified the RNA binding sites for several of its components.
The highly related hnRNP H and F both bind to a GGGGGCUG element within the DCS. This element is important for src splicing enhancer function in vivo, and similar elements have been identified in other splicing enhancers (9, 23, 56, 67). hnRNP H has also been implicated as a negative regulator of tropomyosin splicing (14). The binding of hnRNP H and F to this element is consistent with the known high affinity of these proteins for poly(G) resin (49). Interestingly, although it is present in both extracts, hnRNP F is seen cross-linking to the DCS RNA only in WERI extract. It is tempting to speculate that this is due to a preferred interaction of hnRNP F with nPTB over PTB, but this has not yet been shown.
KSRP and FBP interact with the UGCAUG element in the DCS. This is the most important element in the function of the c-src enhancer in vivo and has been found in other splicing enhancers (34, 36, 37, 40, 44). KSRP apparently requires adjacent nPTB binding for stable binding to the UGCAUG. In other systems, there are UGCAUG elements that function as enhancers but do not have apparent adjacent PTB binding sites. Thus, either KSRP can cooperate with other adjacent proteins or it may not be the protein that mediates the enhancement effect of this element. KSRP and FBP also bind extremely tightly to poly(U) resin (54) and also bind to the single-stranded form of a DNA sequence element in the c-myc promoter (18, 20, 21). In this last capacity, these proteins have been implicated in the regulation of transcription, although this is probably a separate function from that studied here (33). There are additional copies of the hnRNP H and KSRP binding elements in the enhancer region surrounding the DCS. Thus, the larger enhancer RNP complexes are likely to have multiple copies of these proteins engaging in numerous cooperative interactions.
PTB and nPTB both bind to the CUCUCU element of the DCS, although with different apparent affinities. Of all the DCS binding proteins, PTB is the factor most strongly implicated in splicing regulation; PTB is required to repress splicing in nonneural extracts (13, 17). In HeLa extract, PTB binds to sites upstream and downstream of the exon, including the site in the DCS (13, 17). Examination of PTB binding to the full-length transcript in the HeLa extract indicates that cross-linking of PTB to the upstream CUCUCU elements requires intact downstream elements. This implies cooperation between the PTBs bound at the different elements (17). Here we see binding of PTB and nPTB to the upstream sites alone and weak binding to the DCS alone. This may reflect a more sensitive binding assay or differences between the short RNAs used here and the full-length transcript. In WERI extract, where splicing is derepressed, PTB and nPTB bind to the pre-mRNA but are removed from the upstream sites in an ATP-dependent process (17). The downstream protein remains bound under these conditions. We show here that this downstream protein is nPTB rather than PTB and that nPTB is a key protein in the assembly of the DCS complex in WERI extract. The adjacent hnRNP H and KSRP binding sites may stabilize the nPTB and prevent its removal from the DCS when it is lost from the 3′ splice site.
A new tissue-specific RNA binding protein.
We identified and cloned nPTB as a component of the DCS complex. nPTB is highly related to PTB/hnRNP I, but where PTB is broadly expressed, nPTB is enriched in the brain. In WERI-1 cell extracts, both nPTB and PTB are present.
Although PTB and nPTB both bind to the CUCUCU repressor elements, there are differences in their binding. Both proteins bind well to the upstream polypyrimidine tract containing two CUCUCU elements. The DCS portion of the downstream splicing enhancer contains one repressor element surrounded by the GGGGG and UGCAUG elements implicated in the positive control of splicing. Both PTB and nPTB bind to this sequence on an RNA affinity column. However, by gel shift analysis, nPTB shows significantly better binding to the DCS sequence than PTB does. This difference is enhanced by the presence of a second DCS binding protein, hnRNP H. These results resolve a mystery from earlier studies of the DCS binding proteins: what causes their tissue-specific assembly? The DCS RNA assembles into a complex that is enriched in the WERI extract. However, the previously identified components of the DCS complex are present in both extracts (16, 53, 54). The extract specificity of nPTB and its greater affinity for the DCS are the likely source of the observed neuron-specific assembly of the DCS complex. However, since PTB does bind to the DCS in other assays, the HeLa extract is likely to form a PTB-DCS complex that is just not stable in the gel shift assay.
The sequence differences between PTB and nPTB should provide clues to how they differ in activity. PTB and nPTB are most different in the long alanine-rich linkers separating RRMs 1 and 2 and RRMs 2 and 3. Interestingly, the linker between RRMs 2 and 3 is also where the PTB splice variants are altered (26). The third RRM domain of PTB plays a pivotal role in RNA recognition (63). nPTB has a number of unique residues in this region, including changes in the loop between β-strand 1 and α-helix A and the sequence from helix B through β-strand 4. In the β-strand 1-helix A loop, changes from Glu (nPTB) to Pro (PTB) and Met (nPTB) to Arg (PTB) could affect RNA binding specificity, since the corresponding residues in the U1A RRM are involved in RNA base recognition (1, 60). The residues unique to nPTB within β-strand 4 may also contribute to the RNA binding specificity, since their counterparts in both the Sxl and U1A proteins make contacts with the RNA (32, 60). RRM 2 (amino acids 169 to 264) has been implicated in PTB homodimer formation and heterodimer formation with hnRNP L (30, 59, 63). Most of the changes in RRM 2 cluster in helix B and between β-strand 1 and helix A. According to the solved RRM structures, helix B is on the opposite face from the RNA binding surface (32, 79). If nPTB does not dimerize with PTB, it will be interesting to determine whether these residues affect this specificity.
Models for splicing derepression.
Because of its extract specificity and binding properties, nPTB is a prime candidate for mediating the loss of splicing repression in WERI-1 cells. However, significant further work is required to prove this. Since nPTB is only weakly expressed in at least one cell line (LA-N-5) where the N1 exon is efficiently spliced, nPTB cannot be the sole determinant of N1 splicing. This is not entirely surprising, since previous work has shown that N1 splicing is controlled by a complex mixture of positive and negative elements across the region of the exon (55, 56). The inclusion of the exon can be increased by removing one of its repression mechanisms or by increasing its activation, and different cells seem to make different use of these mechanisms (55). The LA-N-5 cells may overcome the repression by PTB through increased enhancer activity. Indeed, the downstream enhancer is particularly strong and the repressor is relatively weak in LA-N-5 cells (55).
Without knowing more about how PTB represses splicing, it is difficult to speculate on whether or how this could be counteracted by nPTB. One possibility is that differences in the oligomerization properties of PTB and nPTB affect their repression activity. PTB can exist as a homodimer in solution (59, 63). Since the bulk of nPTB can be cleanly separated from PTB, it is not clear whether nPTB is forming heterodimers with PTB or homodimers with itself. The ability to form oligomers may allow PTB to assemble a higher-order complex on the four repressor elements surrounding the N1 exon (17). The resulting RNA loop could prevent the assembly of the spliceosome on the N1 exon. In neuronal extract, nPTB bound to the DCS may not stably interact with either PTB or nPTB bound to the other elements. This disruption of bridging between the CUCUCU elements could prevent the repression of splicing.
It is also likely that PTB and nPTB will interact with different sets of auxiliary factors that modify their behavior. One indication of this is that factors in the 0.3 M fraction from the DCS RNA affinity column apparently reduce the ability of nPTB to repress splicing. The importance of such protein-protein interactions is also illustrated by the stimulation of nPTB binding to the DCS RNA by hnRNP H.
The Grabowski laboratory also reported a strong correlation between the binding of a brain-specific form of PTB and the neuron-specific splicing of exons in the GABAA receptor γ2 subunit, the NMDA receptor NR1, and the clathrin light-chain pre-mRNAs (2, 28, 81). It seems likely that this rat brain protein is nPTB. In all three of these transcripts, a recombinant glutathione S-transferase–PTB fusion protein could repress the splicing of the exons in favor of the nonneural pathway. nPTB has also been cloned in a yeast two-hybrid screen as interacting with the Nova protein, a putative splicing regulator that is highly neuron specific (6, 38, 64). Nova is present in WERI extract but does not seem to affect N1 splicing (38) (V. Markovtsov, unpublished data). Nevertheless, it will be interesting to determine the relative affinities of PTB and nPTB for Nova, since it is a good example of the type of protein that may cooperate with nPTB in its function. Thus, nPTB is a potential player in the regulation of many different neural splicing systems. An important problem for the future will be understanding what happens after regulatory proteins bind to the pre-mRNA and filling the current gap between proteins in the DCS complex or other hnRNP complexes and the general splicing machinery.
ACKNOWLEDGMENTS
We thank Adrian Krainer, Philippe Bouvet, David Levens, Gideon Dreyfuss, Joan Steitz, Mark Roth, and Karla Neugebauer for providing antibodies and Hosung Min for providing the WERI-1 cDNA library. We thank Chris Smith, Charles Query, Jane Wu, Frederic Allain, and members of the Black laboratory for critical evaluation of the manuscript. We are particularly grateful to Robert Darnell and Valery Bliskovsky for communicating unpublished results and to Ray Chan for advice on protein purification.
This work was supported by NIH grant R01 GM49662 to D.L.B. V.M. is supported by a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. D.L.B. is an associate investigator of the Howard Hughes Medical Institute and a David and Lucile Packard Foundation Fellow.
REFERENCES
- 1.Allain F H, Gubser C C, Howe P W, Nagai K, Neuhaus D, Varani G. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature. 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- 2.Ashiya M, Grabowski P J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 3.Bai Y, Lee D, Yu T, Chasin L A. Control of 3′ splice site choice in vivo by ASF/SF2 and hnRNP A1. Nucleic Acids Res. 1999;27:1126–1134. doi: 10.1093/nar/27.4.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett M, Piñol-Roma S, Staknis D, Dreyfuss G, Reed R. Differential binding of heterogeneous nuclear ribonucleoproteins to mRNA precursors prior to spliceosome assembly in vitro. Mol Cell Biol. 1992;12:3165–3175. doi: 10.1128/mcb.12.7.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black D L. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992;69:795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- 6.Buckanovich R J, Posner J B, Darnell R B. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron. 1993;11:657–672. doi: 10.1016/0896-6273(93)90077-5. [DOI] [PubMed] [Google Scholar]
- 7.Caceres J F, Krainer A R. Mammalian pre-mRNA splicing factors. In: Krainer A R, editor. Eukaryotic mRNA processing. Oxford, United Kingdom: Oxford University Press; 1997. pp. 174–217. [Google Scholar]
- 8.Caputi M, Mayeda A, Krainer A R, Zahler A M. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlo T, Sterner D A, Berget S M. An intron splicing enhancer containing a G-rich repeat facilitates inclusion of a vertebrate micro-exon. RNA. 1996;2:342–353. [PMC free article] [PubMed] [Google Scholar]
- 10.Chabot B. Directing alternative splicing: cast and scenarios. Trends Genet. 1996;12:472–478. doi: 10.1016/0168-9525(96)10037-8. [DOI] [PubMed] [Google Scholar]
- 11.Chabot B, Blanchette M, Lapierre I, La Branche H. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol Cell Biol. 1997;17:1776–1786. doi: 10.1128/mcb.17.4.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan R C, Black D L. Conserved intron elements repress splicing of a neuron-specific c-srcexon in vitro. Mol Cell Biol. 1995;15:6377–6385. doi: 10.1128/mcb.15.11.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan R C, Black D L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol Cell Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C D, Kobayashi R, Helfman D M. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomczynsky P, Sacchi N. Single-step method of RNA isolation by acid guanidine thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.Chou M Y, Rooke N, Turck C W, Black D L. hnRNP H is a component of a splicing enhancer complex that activates a c-srcalternative exon in neuronal cells. Mol Cell Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou M Y, Underwood J U, Nikolic J, Luu M H T, Black D L. Multisite binding and release of polypyrimidine tract binding protein during the regulation of c-src neural specific splicing. Mol Cell. 2000;5:949–957. doi: 10.1016/s1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- 18.Davis-Smyth T, Duncan R C, Zheng T, Michelotti G, Levens D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J Biol Chem. 1996;271:31679–31687. doi: 10.1074/jbc.271.49.31679. [DOI] [PubMed] [Google Scholar]
- 19.Dreyfuss G, Matunis M J, Piñol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 20.Duncan R, Bazar L, Michelotti G, Tomonaga T, Krutzsch H, Avigan M, Levens D. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–480. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- 21.Duncan R, Collins I, Tomonaga T, Zhang T, Levens D. A unique transactivation sequence motif is found in the carboxyl-terminal domain of the single-strand-binding protein FBP. Mol Cell Biol. 1996;16:2274–2282. doi: 10.1128/mcb.16.5.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan H, Goodier J L, Chamberlain J R, Engelke D R, Maraia R J. 5′ processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol Cell Biol. 1998;18:3201–3211. doi: 10.1128/mcb.18.6.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallego M E, Sirand-Pugnet P, Durosay P, Clouet d'Orval B, d'Aubenton-Carafa Y, Brody E, Expert-Bezancon A, Marie J. Tissue-specific splicing of two mutually exclusive exons of the chicken beta-tropomyosin pre-mRNA: positive and negative regulations. Biochimie. 1996;78:457–465. doi: 10.1016/0300-9084(96)84752-3. [DOI] [PubMed] [Google Scholar]
- 24.Ghetti A, Pinol-Roma S, Michael W M, Morandi C, Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghisolfi-Nieto L, Joseph G, Puvion-Dutilleul F, Amalric F, Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J Mol Biol. 1996;260:34–53. doi: 10.1006/jmbi.1996.0380. [DOI] [PubMed] [Google Scholar]
- 26.Gil A, Sharp P A, Jamison S F, Garcia-Blanco M A. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 1991;5:1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- 27.Gooding C, Roberts G C, Smith C W. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated alpha-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- 28.Grabowski P J. Splicing regulation in neurons: tinkering with cell-specific control. Cell. 1998;92:709–712. doi: 10.1016/s0092-8674(00)81399-9. [DOI] [PubMed] [Google Scholar]
- 29.Grossman J S, Meyer M I, Wang Y C, Mulligan G J, Kobayashi R, Helfman D M. The use of antibodies to the polypyrimidine tract binding protein (PTB) to analyze the protein components that assemble on alternatively spliced pre-mRNAs that use distant branch points. RNA. 1998;4:613–625. doi: 10.1017/s1355838298971448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahm B, Cho O H, Kim J E, Kim Y K, Kim J H, Oh Y L, Jang S K. Polypyrimidine tract-binding protein interacts with HnRNP L. FEBS Lett. 1998;425:401–406. doi: 10.1016/s0014-5793(98)00269-5. [DOI] [PubMed] [Google Scholar]
- 31.Hanamura A, Caceres J F, Mayeda A, Franza B R, Jr, Krainer A R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 32.Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 33.He L, Liu J, Collins I, Sanford S, O'Connell B, Benham C J, Levens D. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 2000;19:1034–1044. doi: 10.1093/emboj/19.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedjran F, Yeakley J M, Huh G S, Hynes R O, Rosenfeld M G. Control of alternative pre-mRNA splicing by distributed pentameric repeats. Proc Natl Acad Sci USA. 1997;94:12343–12347. doi: 10.1073/pnas.94.23.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hertel K J, Maniatis T. The function of multisite splicing enhancers. Mol Cell. 1998;1:449–455. doi: 10.1016/s1097-2765(00)80045-3. [DOI] [PubMed] [Google Scholar]
- 36.Huh G S, Hynes R O. Elements regulating an alternatively spliced exon of the rat fibronectin gene. Mol Cell Biol. 1993;13:5301–5314. doi: 10.1128/mcb.13.9.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huh G S, Hynes R O. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 1994;8:1561–1574. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- 38.Jensen K B, Dredge B K, Stefani G, Zhong R, Buckanovich R J, Okano H J, Yang Y Y, Darnell R B. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 39.Kamma H, Portman D S, Dreyfuss G. Cell type-specific expression of hnRNP proteins. Exp Cell Res. 1995;221:187–196. doi: 10.1006/excr.1995.1366. [DOI] [PubMed] [Google Scholar]
- 40.Kawamoto S. Neuron-specific alternative splicing of nonmuscle myosin II heavy chain-B pre-mRNA requires a cis-acting intron sequence. J Biol Chem. 1996;271:17613–17616. [PubMed] [Google Scholar]
- 41.Krainer A R, Maniatis T, Ruskin B, Green M R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 42.Krecic A M, Swanson M S. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 43.Lewis H A, Musunuru K, Jensen K B, Edo C, Chen H, Darnell R B, Burley S K. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 44.Lim L P, Sharp P A. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol Cell Biol. 1998;18:3900–3906. doi: 10.1128/mcb.18.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin C H, Patton J G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 46.Lopez A J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 47.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 48.Markovtsov V, Mustaev A, Goldfarb A. Protein-RNA interactions in the active center of transcription elongation complex. Proc Natl Acad Sci USA. 1996;93:3221–3226. doi: 10.1073/pnas.93.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matunis M J, Xing J, Dreyfuss G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22:1059–1067. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayeda A, Helfman D M, Krainer A R. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol. 1993;13:2993–3001. doi: 10.1128/mcb.13.5.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mayeda A, Krainer A R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 52.McAffee J G, Huang M, Soltaninassab S, Rech J E, Iyengar S, Lestourgeon W M. The packaging of pre-mRNA. In: Krainer A, editor. Eukaryotic mRNA processing. Oxford, United Kingdom: Oxford University Press; 1997. pp. 68–102. [Google Scholar]
- 53.Min H, Chan R C, Black D L. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 54.Min H, Turck C W, Nikolic J M, Black D L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 55.Modafferi E F, Black D L. Combinatorial control of a neuron-specific exon. RNA. 1999;5:687–706. doi: 10.1017/s1355838299990155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Modafferi E F, Black D L. A complex intronic splicing enhancer from the c-srcpre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17:6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mulligan G J, Guo W, Wormsley S, Helfman D M. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of beta-tropomyosin pre-mRNA. J Biol Chem. 1992;267:25480–25487. [PubMed] [Google Scholar]
- 58.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Transcription processivity: protein-DNA interactions holding together the elongation complex. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 59.Oh Y L, Hahm B, Kim Y K, Lee H K, Lee J W, Song O, Tsukiyama-Kohara K, Kohara M, Nomoto A, Jang S K. Determination of functional domains in polypyrimidine-tract-binding protein. Biochem J. 1998;331:169–175. doi: 10.1042/bj3310169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oubridge C, Ito N, Evans P R, Teo C H, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 61.Patton J G, Mayer S A, Tempst P, Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991;5:1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- 62.Perez I, Lin C H, McAfee J G, Patton J G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 63.Pérez I, McAfee J G, Patton J G. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry. 1997;36:11881–11890. doi: 10.1021/bi9711745. [DOI] [PubMed] [Google Scholar]
- 64.Polydorides A D, Okana H J, Yang Y Y L, Stefani G, Darnell R B. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Natl Acad Sci USA. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh R, Valcarcel J, Green M R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 66.Siomi H, Dreyfuss G. RNA-binding proteins as regulators of gene expression. Curr Opin Genet Dev. 1997;7:345–353. doi: 10.1016/s0959-437x(97)80148-7. [DOI] [PubMed] [Google Scholar]
- 67.Sirand-Pugnet P, Durosay P, Brody E, Marie J. An intronic (A/U)GGG repeat enhances the splicing of an alternative intron of the chicken beta-tropomyosin pre-mRNA. Nucleic Acids Res. 1995;23:3501–3507. doi: 10.1093/nar/23.17.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Solnick D. Trans splicing of mRNA precursors. Cell. 1985;42:157–164. doi: 10.1016/s0092-8674(85)80111-2. [DOI] [PubMed] [Google Scholar]
- 69.Soltaninassab S R, McAfee J G, Shahied-Milam L, LeStourgeon W M. Oligonucleotide binding specificities of the hnRNP C protein tetramer. Nucleic Acids Res. 1998;26:3410–3417. doi: 10.1093/nar/26.14.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stefano J E. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- 71.Sugrue M M, Brugge J S, Marshak D R, Greengard P, Gustafson E L. Immunocytochemical localization of the neuron-specific form of the c-src gene product, pp60c-src(+), in rat brain. J Neurosci. 1990;10:2513–2527. doi: 10.1523/JNEUROSCI.10-08-02513.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swanson M S, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Swanson M S, Dreyfuss G. RNA binding specificity of hnRNP proteins: a subset bind to the 3′ end of introns. EMBO J. 1988;7:3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valcarcel J, Gebauer F. Post-transcriptional regulation: the dawn of PTB. Curr Biol. 1997;7:R705–R708. doi: 10.1016/s0960-9822(06)00361-7. [DOI] [PubMed] [Google Scholar]
- 75.Valcarcel J, Green M R. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- 76.Varani G, Nagai K. RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct. 1998;27:407–445. doi: 10.1146/annurev.biophys.27.1.407. [DOI] [PubMed] [Google Scholar]
- 77.Wang J, Manley J L. Regulation of pre-mRNA splicing in metazoa. Curr Opin Genet Dev. 1997;7:205–211. doi: 10.1016/s0959-437x(97)80130-x. [DOI] [PubMed] [Google Scholar]
- 78.Wang Y-C, Selvakumar M, Helfman D. Alternative pre-mRNA splicing. In: Krainer A R, editor. Eukaryotic mRNA processing. Oxford, United Kingdom: Oxford University Press; 1997. pp. 242–279. [Google Scholar]
- 79.Xu R M, Jokhan L, Cheng X, Mayeda A, Krainer A R. Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure. 1997;5:559–570. doi: 10.1016/s0969-2126(97)00211-6. [DOI] [PubMed] [Google Scholar]
- 80.Yamamoto H, Tsukahara K, Kanaoka Y, Jinno S, Okayama H. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol Cell Biol. 1999;19:3829–3841. doi: 10.1128/mcb.19.5.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang L, Liu W, Grabowski P J. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA. 1999;5:117–130. doi: 10.1017/s1355838299981530. [DOI] [PMC free article] [PubMed] [Google Scholar]