Abstract
Achieving long-term retroviral expression in primary cells has been problematic. De novo DNA methylation of infecting proviruses has been proposed as a major cause of this transcriptional repression. Here we report the development of a mouse stem cell virus (MSCV) long terminal repeat-based retroviral vector that is expressed in both embryonic stem (ES) cells and hematopoietic stem (HS) cells. Infected HS cells and their differentiated descendants maintained long-term and stable retroviral expression after serial adoptive transfers. In addition, retrovirally infected ES cells showed detectable expression level of the green fluorescent protein (GFP). Moreover, GFP expression of integrated proviruses was maintained after in vitro differentiation of infected ES cells. Long-term passage of infected ES cells resulted in methylation-mediated silencing, while short-term expression was methylation independent. Tissues of transgenic animals, which we derived from ES cells carrying the MSCV-based provirus, did not express GFP. However, treatment with the demethylating agent 5-azadeoxycytidine reactivated the silent provirus, demonstrating that DNA methylation is involved in the maintenance of retroviral repression. Our results indicate that retroviral expression in ES cells is repressed by methylation-dependent as well as methylation-independent mechanisms.
Retroviral vectors are appealing vehicles for gene transfer. However, long-term expression mediated by integrated proviruses in primary cells has been difficult to achieve. Retroviral regulatory elements are repressed in numerous cell types, including embryonic stem (ES) cells and hematopoietic stem (HS) cells (1, 3). For example, vectors that are functional in mature hematopoietic cells are often not expressed in blood cells of animals transplanted with the infected stem cells (18, 19, 31). In particular, the lack of significant provirus transcription in ES cells and their differentiated descendants has hampered the use of retroviral vectors in transgenic experiments (5, 12, 32). Interestingly, this block in provirus expression is maintained upon differentiation of infected cells despite the fact that primary infection of cells after differentiation results in efficient expression (6, 7, 26).
Transcriptional repression is thought to be mediated by both cis-acting de novo methylation of the integrated proviruses and cell-type-specific trans-acting transcriptional repressors (5, 9, 23). The effect of trans-acting factors on retroviral expression through binding of specific sequences within the promotors of retroviruses has been examined in many studies (29, 30, 35). In fact, the mouse stem cell virus (MSCV) long terminal repeat (LTR) was generated by the modification of the sequences within the LTR to increase the affinity for positive factors and decrease the affinity for negative regulators (20).
In contrast, the role of methylation in silencing has been less clear. DNA methylation is thought to be a general mechanism used by cells to silence foreign DNA and may be involved in the cell defense against transposable elements (39). DNA methylation has also been associated with the repression of gene expression and the silencing of viral control elements (2, 14, 38). Exogenously introduced retroviruses silenced in vitro and in vivo can be reactivated by treatments that result in genomewide demethylation. In addition, transcriptionally silent endogenous retroviral elements are reactivated upon loss of genomic methylation in Dnmt1 knockout mice (38). Therefore, DNA methylation is thought to causally repress expression of retroviral promoters in a variety of cell types.
ES cells provide a good model to study the role of DNA methylation in retroviral silencing. First, it was demonstrated that ES cells have high de novo methylation activity, which leads to effective methylation of integrated retroviral vectors, while little or no de novo methylation activity was detected in differentiated cells (21). In addition, ES cells were genetically modified to alter the endogenous level of DNA methylation by the targeted disruption of the maintenance methyltransferase gene Dnmt1. ES cells homozygous for this mutation proliferate normally with their genomic DNA highly demethylated, while differentiated cells and mice die due to the loss of genomic methylation (21, 22). Therefore, these modified ES cells are useful to study the effect of DNA methylation on retroviral gene expression. In addition, ES cells can be induced to differentiate in vitro or in vivo, allowing the study of DNA methylation and its effect on long-term expression.
Both Moloney virus-based and MSCV-based retroviral vectors have been used for gene transduction in a variety of cells. The MSCV vector is different from the typical Moloney virus vector in that the mutations in the LTR have allowed expression in a larger host range (8, 20). To this end, we modified MSCV to express the green fluorescent protein (GFP) as a sensitive reporter for gene expression (37). Using this vector, we demonstrated efficient expression in both ES and HS cells. We also demonstrated that silencing of retroviruses involves two mechanisms: (i) trans-acting factors that affect the initial expression of Moloney virus-based vectors but not MSCV-based vectors and (ii) long-term DNA methylation-dependent silencing that directly restricts expression of MSCV in ES cells and during embryogenesis. Silencing of the MSCV vector in wild-type ES cells and in in vivo differentiated ES cells was reversed by 5-azadeoxycytidine (5-azadC) treatments that demethylated the retroviral sequences, demonstrating that DNA methylation directly controls the maintenance of retroviral repression.
MATERIALS AND METHODS
Tissue culture.
ES cells were cultured as described previously (21). To generate ES cell clones for injection into blastocysts, the ES cells were maintained on irradiated mouse embryonic fibroblasts (MEFs) with 500 U of leukemia inhibitory factor (LIF) per ml (22). For other experiments, the ES cells were cultured without MEFs in 1,000 U of LIF per ml. 293 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS), penicillin, streptomycin, and glutamine. Abelson virus-transformed B cells were maintained in RPMI 1640 supplemented with 10% defined FBS (HyClone), penicillin, streptomycin, glutamine, and 50 μM β-mercaptoethanol. ES cells with retroviral integrants were in vitro differentiated as follows: the cells were passaged without LIF in the absence of MEFs on bacterial plastic petri dishes for 4 days, trypsinized, and cultivated with or without retinoic acid for 2 weeks (25).
Plasmids.
The retroviral vectors MfgGFP, pMXGFP, and MSCViresGFP have been described elsewhere (27, 33, 37). The MSCViresGFP vector was modified by introducing either the Cre recombinase or the human Bcl-2 gene upstream of the internal ribosome entry site (IRES)-GFP cassette as described elsewhere (11, 37). The replication-incompetent helper plasmid pCL-eco was used (24).
Retroviral infections.
To generate retroviral supernatants, 293 cells were transiently transfected by calcium phosphate-mediated coprecipitation with 5 μg of the replication-incompetent helper vector pCL-eco and 10 μg of the reporter retroviral vector as stated elsewhere (28). The cells were fed at 24 h postinfection, and the retroviral supernatant was used at 48 h. The cells continued to produce high-titer retroviruses for another 2 days, and that supernatant was used if needed for additional experiments. The supernatant was collected, brought to 4 μg of Polybrene per ml–10 mM HEPES, and filtered (0.45-μm-pore-size filter) for use.
ES cells for infection were washed and trypsinized. They were then plated at 106 cells per well of a six-well dish and centrifuged. The ES cell medium was removed, and retroviral supernatant was added at 1 ml/106 cells. Next, the plate was centrifuged for 45 min at 2,500 rpm at room temperature. The retroviral supernatants were removed; the cells were resuspended in ES cell medium and plated onto gelatinized dishes. ES cells used to generate mice were plated onto irradiated MEFs.
Bone marrow was infected as follows (36). C57BL/6 mice were obtained from the Jackson Laboratory (Bar Harbor, Maine). Bone marrow cells were harvested from the tibias and femurs of C57BL mice 5 days after they received an intraperitoneal injection of 5 mg of 5-fluorouracil (Sigma) in Dulbecco's phosphate-buffered saline (Gibco/BRL). These cells were then cultured for 4 days at 2 × 106 cells/ml with recombinant mouse interleukin-3 (rmIL-3; 20 ng/ml), rmIL-6 (50 ng/ml), and (50 ng/ml; recombinant mouse stem cell factor; R&D Systems) in Dulbecco's modified Eagle medium containing 10% FBS. After 48 and 72 h, the bone marrow cells were spin infected with the retroviral supernatant generated as described above. Then the retroviral supernatant was removed and replaced with growth medium containing cytokines.
FACS (fluorescence-activated cell sorting) analysis and sorting.
Adherent cell lines were trypsinized, washed, and resuspended in complete medium to achieve a single-cell suspension at the time points indicated. Nonadherent cells were used directly for analysis. Organs were disrupted manually and passed through a 70-μm mesh to generate a single-cell suspension. The cells were analyzed for viability using scatter properties and the exclusion of propidium iodide. The level of GFP expression was monitored by fluorescence without compensation to detect cells with low levels of GFP expression. The ES cells were sorted into ES cell medium and plated immediately onto either gelatinized plates or MEFs for blastocyst injections. The survival of ES cells after sorting was approximately 50%, as measured by the number of colonies generated divided by the expected number of colonies.
5-AzadC treatments.
ES cells were treated with 0.15 μM 5-azadC (Sigma) at days 1 and 3 postplating. The cells were fed, allowed to recover, and then assayed 4 to 8 days later. The red blood cells in whole blood were lysed (5), and the remaining cells were stained with fluorescently labeled anti-H2-b, anti-H2-d, anti-B220, anti-TCRa (Pharmingen) at 1:200 as indicated. At day 0, splenocytes were treated with either anti-CD3 or anti-CD40 (Pharmingen); 0.15 μM 5-azadC was added at day 1, and the anti-CD3-treated cells were assayed at day 4. 5-AzadC was added again to the B-cell cultures with fresh anti-CD40 at day 4, and the cells were assayed at day 6.
Staurosporine-mediated cell death.
ES cells were infected with the stated retrovirus and treated with staurosporine at day 4 postinfection for 24 h with the indicated concentration of drug. The percentage of viable, GFP-positive cells was determined by flow cytometry (6). Data are presented as a percentage of GFP-positive cells before treatment. Results from one representative experiment of three performed are shown.
LacZ staining.
ES cells were infected with the stated retrovirus and sorted for GFP expression at day 3 postinfection. The ES cells were plated and cultured for an additional 5 days and stained for LacZ expression as described elsewhere (41).
Adoptive transfers.
Recipient mice (10) received a total of 1,200 rads of whole-body radiation in two doses (800 and 400 rads) 3 h apart and were then injected with 2 × 106 to 5 × 106 infected bone marrow cells. Irradiated mice were maintained on trimethaprim-sulfamethoxazole in sterile cages for 4 to 6 weeks to prevent opportunistic infections (34). Serial passages were performed by harvesting bone marrow from mice 6 to 8 weeks postreconstitution and transferring 2 × 106 to 5 × 106 cells into irradiated recipients. Mice were analyzed 8 to 12 weeks posttransfer to allow reconstitution of the T-cell compartment. These experiments were repeated multiple times with similar results.
Southern blot analysis.
The genomic DNA was isolated as described elsewhere (19). Ten micrograms of DNA was digested with the stated restriction enzyme overnight. The products were resolved on an agarose gel, transferred to a nylon membrane, and detected using a probe that spans the entire GFP coding sequence.
RESULTS
High-efficiency retroviral expression in ES cells.
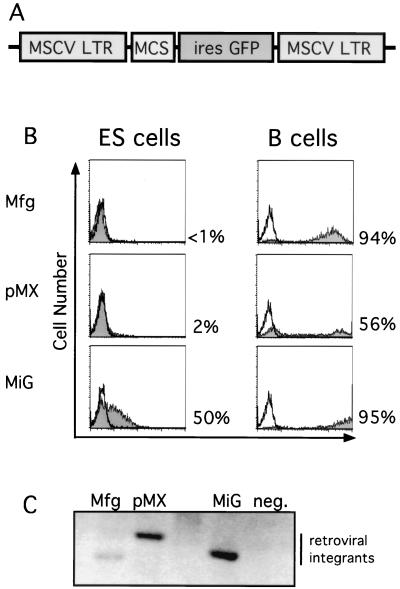
Retroviral vectors based on the MSCV LTR were constructed with a multiple cloning site followed by an IRES driving expression of the gene for GFP as schematically diagrammed in Fig. 1A (MiG) (37). We generated high-titer retroviruses by transient transfection and infected ES cells with an adapted spin infection protocol. Using this protocol, we reproducibly achieved high-efficiency (>50%) infection of ES cells as measured by flow cytometry; uninfected control cells were negative for GFP expression (Fig. 1B). The intracellular concentration of GFP is directly proportional to the fluorescence intensity measured by flow cytometry.
FIG. 1.
Efficient retroviral infection of ES cells. (A) Schematic diagram of MiG vector containing the MSCV LTR followed by a multiple cloning site (MCS) and an IRES-GFP cassette. (B) MSCV-based (MiG) but not Moloney virus-based (Mfg and pMX) retroviruses express in ES cells. B cells or ES cells were infected by the indicated retroviruses and assayed by flow cytometry 2 days postinfection. Uninfected cells (unshaded) and infected cells (shaded area) were electronically gated for live cells and subsequently analyzed for GFP fluorescence and for cell number. Percentages of GFP-positive cells are indicated. (C) Comparable levels of integration of different retroviruses into ES cells, determined by Southern blot analysis of genomic DNA purified from infected and uninfected ES cells 2 days postinfection, digested with KpnI, a restriction site present within the LTRs, and probed with the GFP coding sequence.
Next, we compared expression of the MSCV-based retrovirus and Moloney virus-based retroviral vectors in ES cells. GFP expression was detectable with the MSCV LTR-containing MiG vector but not with the two Moloney virus-based viruses pMX (27) and Mfg (33) (Fig. 1B). This was not due to inefficient genomic integration of the provirus or to a lower titer. Southern blot analysis of genomic DNA demonstrated that all three proviruses were integrated in the ES cells (Fig. 1C). Also, when parallel B-cell cultures were infected with the retroviral supernatant used to infect ES cells, all of the retroviral vectors were expressed in B cells at comparable efficiencies (Fig. 1B).
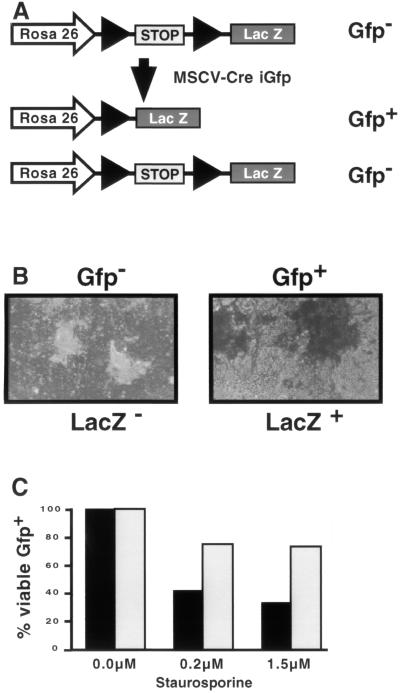
GFP expression driven by the MSCV LTR in ES cells was substantially lower than in other differentiated cell lines tested (Fig. 1B and data not shown) (20). To determine whether this low level of expression was sufficient to drive functional expression of other gene products, we cloned the Cre recombinase upstream of the IRES-GFP cassette to generate MSCVCreiresGFP. We tested for Cre activity by infecting ES cells that contain a translational stop sequence flanked by loxP sites located between the Rosa 26 promoter and a lacZ reporter schematically diagrammed in Fig. 2A (34). If Cre is expressed at functional levels in these ES cells, the protein will catalyze recombination of the loxP sites, leading to loss of the stop sequences and the expression of LacZ. Indeed, we found that >99% of GFP-positive ES cells that were infected with the Cre-expressing retrovirus were also LacZ positive (Fig. 2B). Uninfected cells were both GFP negative and LacZ negative (data not shown). This indicates that virus-mediated gene transfer resulted in functional Cre expression.
FIG. 2.
Expression from the MSCV LTR is sufficient to drive functional gene expression. (A) Schematic diagram of the Rosa 26 locus in Cre reporter ES cells. Before Cre-mediated recombination, LacZ expression is prevented by the presence of a stop fragment. Retroviral infection with a Cre-expressing retrovirus with a GFP reporter results in two populations of cells. Cells that are GFP+ become LacZ+ due to efficient Cre-mediated recombination of the stop fragment. In contrast, cells that are GFP− were not infected and thus remained LacZ−. (B) ES cells were infected with the MSCVCreiresGFP retrovirus and sorted for either Gfp− or Gfp+ as indicated. The cells were subsequently cultured and stained for LacZ expression. Gfp− cells are white (and therefore LacZ−) while Gfp+ cells are blue (and therefore LacZ+). More than 99% of the Gfp+ cells were LacZ+ in multiple experiments. (C) ES cells were infected with either MSCViresGFP (■) or MSCVBcl-2iresGFP (□) and treated with the indicated amounts of staurosporine. The percentage of viable, Gfp+ (infected) cells was determined by flow cytometry. The results are shown as a percentage of Gfp+ cells before treatment. The results are from one representative experiment of three performed.
Because Cre activity is required only transiently for LacZ expression, we tested a second gene product that must be stably expressed throughout the experiment. It has been demonstrated that Bcl-2 expression protects many cell types against staurosporine-mediated apoptosis (10). Therefore, we examined whether Bcl-2 could protect ES cells from apoptosis when expressed from the MSCV LTR. We cloned human Bcl-2 upstream of the IRES-GFP cassette to generate MSCV Bcl-2iresGFP. Wild-type ES cells were infected with either the Bcl-2-expressing retrovirus or the control virus lacking Bcl-2. Increasing concentrations of staurosporine were added to the cultures, and flow cytometry was used to assay for both viability and GFP expression. GFP-positive cells infected with the Bcl-2-containing virus were significantly protected from staurosporine-mediated cell death compared to the GFP-negative cells or GFP-positive cells infected with the control retrovirus (Fig. 2C). Therefore, the level of expression from the MSCV LTR is sufficient for stable functional gene expression in ES cells.
Short-term transcriptional silencing in ES cells is methylation independent.
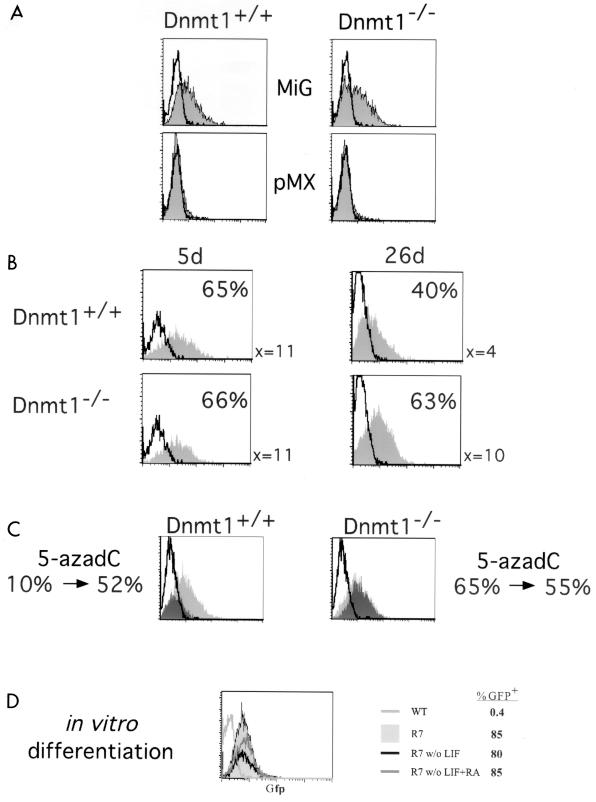
It long has been hypothesized that retroviruses are transcriptionally silenced in embryonic cells by DNA methylation (12, 14, 21). Therefore, it was possible that DNA methylation of the MSCV LTR was responsible for the decreased level of expression in ES cells compared to other cell types (Fig. 1B). In addition, we sought to test whether DNA methylation of the Moloney virus-based vectors in the wild-type ES cells was the mechanism by which the Moloney virus-based LTRs were silenced (9, 13). To this end, we infected ES cells deficient for the maintenance DNA methyltransferase gene, Dnmt1, the loss of which results in genomewide hypomethylation (21, 22). Dnmt1−/− ES cells are demethylated, and proviral sequences remain unmethylated. The Moloney virus-based retroviruses such as pMX remained silent even when introduced into Dnmt1−/− ES cells, whereas MSCV expressed similar levels of GFP in both Dnmt1+/+ and Dnmt1−/− ES cells (Fig. 3A). Therefore, the initial block in transcription directed by Moloney virus LTRs in ES cells is independent of DNA methylation and is presumably due to the binding of trans-acting factors. In addition, the mean fluorescence intensities of GFP were comparable between the Dnmt1+/+ and Dnmt1−/− ES cells, indicating that the basal level of expression of the MSCV LTR is independent of DNA methylation.
FIG. 3.
Long-term expression of retroviruses is repressed by methylation. (A) MSCV-based (MiG) but not Moloney virus-based (pMX) retroviruses express in ES cells independent of the methylation status of the cells. Dnmt1+/+ or Dnmt1−/− ES cells were not infected (unshaded) or infected by the indicated retroviruses (shaded) and assayed by flow cytometry 2 days postinfection as for Fig. 1B. (B) Long-term expression of GFP in ES cells is decreased in Dnmt1+/+ cells but not Dnmt1−/− cells. The ES cells were infected with MiG, passaged for 5 or 26 days postinfection, and assayed by flow cytometry as above. The mean fluorescent intensity for the population and the percentage of GFP-positive cells are indicated. (C) Treatment with 5-azadC rescues the expression of retroviruses silenced by long-term passage. Dnmt1+/+ or Dnmt1−/− ES cells were infected by MiG and passaged for >40 days. The cells were divided, and half were treated with 5-azadC. Then uninfected ES cells (unshaded), MiG-infected untreated ES cells (dark shading), and MiG-infected 5-azadC-treated ES cells (light shading) were assayed by FACS analysis. Numbers below the FACS plots are percentages of GFP-positive cells before and after 5-azadC treatment. (D) In vitro differentiation of ES cells does not affect retroviral expression. A clonal ES cell line (R7) infected with MiG or an uninfected wild-type (WT) ES cell control was in vitro differentiated by passage without feeders and LIF, with or without retinoic acid (RA) as indicated. The cells were assayed by flow cytometry, and the percentage of GFP-positive cells is indicated.
DNA methylation constrains long-term retroviral expression.
MiG-infected GFP-expressing ES cells were continually passaged to test the effect of DNA methylation on long-term expression. Though GFP expression was high in both Dnmt1−/− and Dnmt1+/+ ES cells at 5 days postinfection, a substantial fraction of the infected wild-type ES were GFP negative at 26 days postinfection. This was apparent by both a loss in the percentage of GFP-positive cells as well as a decrease in the mean fluorescence intensity of the bulk population of wild-type ES cells and was observed in both bulk cultures and individual cloned lines containing single integrants (Fig. 3B and data not shown). The fraction of GFP-positive cells continues to decrease with additional passages, as shown in Fig. 3C. These results suggest that long-term expression was suppressed by DNA methylation. To directly test whether retroviral repression was due to de novo methylation of the newly integrated retroviruses, we treated the long-term cultures with 5-azadC, a drug that leads to hypomethylation of genomic DNA (16). If DNA methylation was preventing expression of the MSCV LTR, treatment with the drug should activate retroviral expression. Indeed, we found that 5-azadC treatment of ES cells that had lost expression of GFP through long-term passage reactivated the provirus (Fig. 3C). In contrast, Dnmt1−/− ES cells infected with the retrovirus did not lose expression of GFP; thus, treatment with 5-azadC did not significantly affect retroviral expression (Fig. 3C). We also analyzed clonal lines containing single proviral integrants in which GFP expression was progressively silenced and found that treatment with 5-azadC resulted in the reactivation of gene expression in all cases (data not shown). This demonstrates that DNA methylation controls long-term but not short-term expression of retroviruses in ES cells.
Expression is maintained after in vitro differentiation.
Previously, in vitro differentiation of ES cells had been demonstrated to silence expression of retroviral sequences (12, 20). Thus, we tested whether GFP expression from the MiG retrovirus in ES cells was affected by in vitro differentiation. We cultured MiG-infected wild-type ES cells in the absence of embryonic feeder cells and LIF in suspension to generate embryoid bodies. Disaggregated embryoid bodies were replated either with or without retinoic acid. We found no change in GFP expression in MiG-infected bulk cultures or individual subclones containing one to several integrants upon in vitro differentiation with either method, as shown for one clonal line containing multiple integrants in Fig. 3D. GFP expression was unchanged in all in vitro-differentiated ES cell lines, regardless of whether the subclones contained only a single or multiple integrants. This indicates that the MSCV-based MiG retrovirus is not silenced by in vitro differentiation.
Generation of mice from GFP-expressing MiG-infected ES cells.
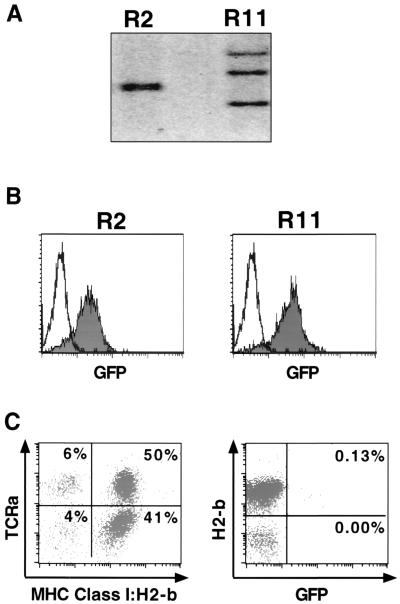
We next determined whether expression of the MSCV-based MiG vector was affected by in vivo differentiation of the infected ES cells. Cells from the chimeric animals were derived by injection of MiG-infected wild-type ES cells (derived from 129-Sv/Jae mice) into BALB/c blastocysts. MiG-infected Dnmt1−/− ES cells cannot be used for injection into blastocysts, because Dnmt1−/− ES cells die upon differention and therefore do not contribute significantly to adult mice (22). MiG-infected wild-type ES cells were sorted for GFP expression by flow cytometry prior to injection, and two GFP-expressing clones, R2 and R11, were isolated (Fig. 4B). Southern blot analysis demonstrated that R2 contained two integrants that comigrate on an agarose gel, and R11 contained three proviral integrants (Fig. 4A). High-contribution chimeras (>80% by coat color) were generated from the R2 and R11 ES cells, which transmitted the proviruses to their offspring (data not shown).
FIG. 4.
Retrovirally infected, GFP-expressing ES cells generated nonexpressing mice. (A) Two retrovirally infected clones sorted for GFP expression were analyzed for proviral integrants by Southern blot analysis. R2 contained two integrants, while R11 contained three. Uninfected cells are negative. (B) The clones were passaged after sorting for GFP-expressing cells by flow cytometry and reanalyzed for GFP expression. Both R2 and R11 express GFP (shaded) compared to uninfected controls (unshaded). (C) PBMCs from the R2 chimera (more than 50% contribution by coat color) were analyzed by flow cytometry. ES cell contribution to the chimera was determined by phycoerythrin-H2-b staining and cyc-TCRa staining and demonstrated contribution to the T-cell compartment. The percentage of cells in each quadrant is listed. The cells were also monitored for GFP expression. The percentage of GFP+ cells that are either major histocompatibility complex (MHC) class I H2-b+ (ES cell derived) or H2-b− (blastocyst derived) is listed in the quadrant.
To test whether the chimeras expressed the integrated retroviruses, we isolated peripheral blood mononuclear cells (PBMCs) from both the R2- and R11-derived chimeras. To distinguish whether the PBMCs were derived from the ES cell donor or the host blastocyst, we stained the cells with antibodies that recognized specific major histocompatibility complex class I haplotypes (Pharmingen). The donor ES cells (129 derived) are H2-b, and the blastocysts (BALB/C derived) are H2-d (Fig. 4C and data not shown). In addition, we stained the PBMCs with a pan-T-cell (TCRa) (Fig. 4C) or pan-B-cell (B220) antibody (data not shown) to determine the ES cell contribution to these lineages. Using this strategy, we found that approximately 90% of the PBMCs from either the R2 or R11 chimera were ES cell derived as measured by H2-b staining (Fig. 4C, data not shown). However, the majority of the cells did not express GFP in either chimera (Fig. 4C and data not shown). On the order of 0.1% of the PBMCs that were ES cell derived were GFP positive, compared to less than 0.01% that were blastocyst derived (Fig. 4C). Similar results were also obtained with cells from the R11 chimera (data not shown). The results indicate that the MSCV LTR is repressed during in vivo differentiation to lymphocytes. Nevertheless, a small number of cells escaped silencing and expressed GFP. This transcriptional repression of the MiG provirus in the chimeras is in contrast to the GFP expression both in the donor ES and after in vitro differentiation (Fig. 4B, data not shown).
To determine if other somatic cells expressed the retroviral integrants, we analyzed the progeny of the chimeras. We isolated spleen, thymus, kidney, and liver cells from an animal carrying the two proviral integrants present in the R2 chimera and a littermate control containing no retroviral integrants. We analyzed these cells for GFP expression by FACS analysis and found no detectable expression of GFP in the splenocytes, thymocytes, renal cells, or hepatocyes (Fig. 5A and data not shown).
FIG. 5.
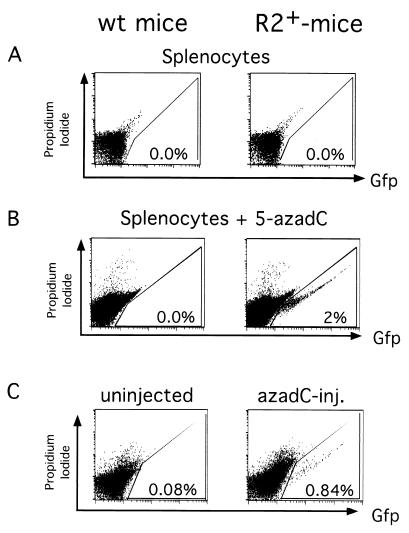
Silenced retroviruses can be reactivated with 5-azadC. (A) Splenocytes from an R2+ or R2− littermate do not express GFP by flow cytometry. Cells were stained with propidium iodide to exclude dead cells, and the percentage of GFP+ cells is indicated. (B) The splenocytes from panel B were induced to proliferate with anti-CD3 and treated with 5-azadC. Cells were stained with propidium iodide to exclude dead cells and analyzed by flow cytometry. The percentage of GFP+ cells is indicated. (C) Flow cytometric analysis of the splenocytes of littermates that were either uninjected or injected with 5-azadC at passage 5 and analyzed at passage 14 for GFP expression. The percentage of GFP+ splenocytes is indicated.
In vitro reactivation of retroviral expression.
One possible explanation for transcriptional repression during in vivo differentiation was de novo methylation of the integrated retroviral LTR during embryonic development. To test this hypothesis, we cultured splenocytes from a mouse containing the R2 proviruses and from a littermate control, by treating the cells with either anti-CD3 or anti-CD40 to activate and induce proliferation of the T cells or B cells, respectively (4). We then assayed for GFP expression by flow cytometry and found that proliferation of the splenocytes did not activate expression of the retrovirus (data not shown). Next, we added 5-azadC to the splenocyte cultures to induce demethylation of the retroviruses. Indeed, treatment with 5-azadC activated expression in approximately 2% of the T cells (anti-CD3) (Fig. 5B) and 2% of the B cells (anti-CD40) (data not shown). In addition, when in vivo-differentiated cells, which had been isolated from the kidney of a transgenic mouse and transformed with simian virus 40 large T antigen (15), were treated with 5-azadC, activation of the silent provirus was observed in a similar fraction of the cells (data not shown). The extent of reactivation of expression of the provirus in in vivo-differentiated cells by 5-azadC was lower than in ES cells, where the reactivation of the provirus with 5-azadC was almost complete.
We next determined whether demethylation of the retrovirus in vivo would activate expression of the integrated retroviruses (13). Newborn mice were subcutaneously injected with 5-azadC at postnatal day 5 and subsequently analyzed at postnatal day 14 for GFP expression by flow cytometry. We found that 5-azadC-injected animals but not the uninjected controls had activated GFP expression of the proviruses in the spleen, thymus, and kidney (Fig. 5C and data not shown). When we injected higher concentration of 5-azadC in an effort to further demethylate the newborn mice, all injected animals died. This result demonstrated that repression by DNA methylation is, at least in part, responsible for silencing expression of the retroviral LTR in vivo.
Retroviral expression in HS cells after serial adoptive transfers.
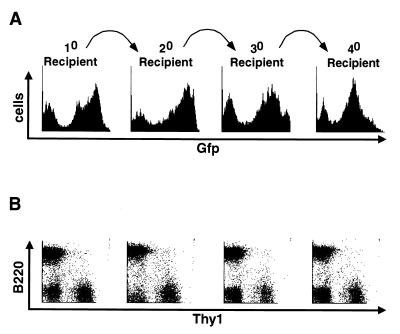
Bone marrow contains the HS cells that can stably repopulate the hematopoietic system after transfer to lethally irradiated mice. To determine whether HS cells can be effectively transduced and express the MiG retrovirus, we used infected bone marrow cells to reconstitute lethally irradiated mice (Fig. 6). We found that between 30 and 80% of the splenocytes from these primary recipients expressed the retrovirus, as measured by FACS analysis for GFP expression and shown for one representative experiment (Fig. 6A). The MiG virus was expressed in the B-cell, T-cell, and granulocyte compartments, as measured by a pan-B cell (B220), pan-T-cell (Thy-1), and pan-granulocyte (Gr-1) marker electronically gated on GFP-positive cells (Fig. 6B and data not shown). Because a large fraction of the splenocytes in the primary recipients are derived from relatively differentiated, lineage-committed progenitors, serial adoptive transfers are required to test for retroviral expression in the true HS cells (17). Therefore, we used bone marrow from these primary recipients to serially reconstitute lethally irradiated mice. This protocol requires substantial expansion from the stem cells and tests for long-term expression of the retrovirus. We observed no change in the percentage GFP-positive HS cells, and the level of GFP expression from the adoptive transfers into multiple recipients was stable over three additional passages (4° recipient). In addition, the infected cells gave rise to both B- and T-cell lineages at the expected ratios (Fig. 6B), demonstrating not only that the MiG retrovirus transduced the long-term repopulating HS cells but also that the MiG-mediated GFP expression was stable during in vivo hematopoietic differentiation. However, our results do not exclude the possibility that in addition to the transcriptionally active proviruses present within these cells, there are also copies of the virus that were transcriptionally silenced.
FIG. 6.
Serial adoptive transfers maintain expression of the MSCV-based retrovirus. (A) Bone marrow was infected with MiG and used to reconstitute multiple lethally irradiated mice to generate the 10 recipient. The spleen of the 10 recipient was analyzed for GFP expression by flow cytometry. The bone marrow of the 10 recipient was used to reconstitute lethally irradiated 20 recipients. The spleen of a 20 recipient was analyzed for GFP expression, and the bone marrow was used to reconstitute lethally irradiated 30 recipients. The spleen of a 30 recipient was analyzed for GFP expression, and the bone marrow was used to reconstitute lethally irradiated 40 recipients. A representative analysis is shown. (B) splenocytes from panel A, stained with pan-B-cell (B220) and pan-T-cell (Thy-1) antibodies and electronically gated for GFP+ cells, are shown below the GFP histogram they are derived from. The FACS diagrams are shown for these serially reconstituted spleens, demonstrating that the transferred cells contribute to both B- and T-cell lineages in the appropriate ratios.
DISCUSSION
We have investigated the role of DNA methylation in retroviral silencing. Retrovirus-based studies of stem cells have been hampered by the lack of expression. We have overcome the transcriptional repression in ES cells by using an MSCV-based vector in combination with a sensitive GFP reporter gene (MiG vector). The analysis of expression of the MiG vector and other Moloney virus-based vectors in Dnmt1−/− and Dnmt1+/+ ES cells has allowed us to determine whether DNA methylation directly controls retroviral gene expression in these cells. We found that both methylation-dependent and methylation-independent mechanisms exist to control retroviral gene expression.
Historically, retroviral expression of Moloney virus-based vectors in ES cells has been negligible. In contrast, the MSCV LTR not only transduces GFP expression in ES cells but also expresses other exogenous gene products such as the Cre recombinase and the antiapoptotic factor Bcl-2 at detectable level in ES cells. Therefore, the MSCV LTR can be used to express various transgenes in ES cells and their differentiated descendant cells.
It had been proposed that DNA methylation has evolved as a cellular mechanism to silence retroviral elements, preventing the spread of transposable elements through the genome (39). Indeed, de novo methylation of integrated proviral sequences has been observed in wild-type ES cells, which was correlated with the transcriptional silencing of the retrovirus (14). Our findings are the first demonstration that inhibition of the Dnmt1 methyltransferase gene prevents silencing of the retroviruses in ES cells. This result provides direct evidence that DNA methylation is causally involved in long-term retroviral repression. Consistent with this conclusion is the demonstration that the transcriptionally silenced proviruses present in long-term Dnmt1+/+ ES cell cultures can be reversed by treatments with 5-azadC.
In contrast, methylation-independent mechanisms determine initial retroviral expression in ES cells. Wild-type or Dnmt1−/− ES cells infected with Moloney virus-based vectors were transcriptionally silent, and therefore this silencing was independent of the DNA methylation status of the cells. Moreover, the basal level of expression from the MSCV-based vector was unaffected by the methylation status of the cells. This formally demonstrates that DNA methylation-independent mechanisms control initial retroviral gene expression in ES cells. Because the basal level of expression of the MSCV LTR in ES cells is lower than in differentiated cell types and not affected by the methylation status of the ES cells, trans-acting factors must regulate the initial level of expression.
Previous studies found that retroviruses, including the MSCV LTR, are silenced by the in vitro differentiation process (20). In contrast, we found for the first time that expression of this MSCV-based retrovirus in ES cells was maintained after in vitro differentiation with and without retinoic acid. We were also able to show long-term, stable GFP expression from the MiG vector in HS cells and their differentiated derivatives. MiG-mediated GFP expression from HS cells was stable through serial adoptive transfers, and the HS cells gave rise to GFP-expressing B- and T-cell lineages. Therefore, this MSCV-based retroviral transduction system should allow for a molecular analysis of stem cell biology and differentiation programs by forced expression of exogenous gene products.
It has been postulated that methylation-dependent mechanisms repress retroviral gene expression upon in vivo differentiation (13, 20). To test this, we injected GFP-expressing undifferentiated ES cells into recipient blastocysts and generated chimeric mice. Differentiated tissues derived from these in vivo-differentiated ES cells, such as PBMCs, lacked significant GFP expression. Treatment of ES cell-derived differentiated cells with 5-azadC in vitro or in vivo led to partial reactivation of expression of the silenced retroviruses in lymphoid and nonlymphoid tissues. We conclude from these results that the maintenance of retroviral silencing in vivo involves DNA methylation. However, only a small fraction of the 5-azadC-treated cells reactivated GFP expression, unlike the long-term ES cell cultures, in which every cell reactivated GFP expression. This suggests that methylation-independent mechanisms exist to suppress retroviral expression. Alternatively, 5-azadC treatment of differentiated cells, in contrast to ES cells, may not lead to a level of genomic demethylation sufficient for complete retroviral reactivation. The transgenic animals carrying the silenced MiG proviruses will be a valuable indicator for in vivo activation of GFP expression under different conditions.
ACKNOWLEDGMENTS
The first two authors, S. R. Cherry and D. Biniszkiewicz, contributed equally to this work.
We thank members of the Baltimore and Jaenisch labs for advice and discussions. We are grateful to Brian Bates for discussions and comments on the manuscript, George Daly for discussions, Jessie Dausman for blastocyst injections, Ruth Curry for mouse work, and Glen Paradis for FACS sorting.
D. Biniszkiewicz was supported by the Deutsche Akademische Austauschdienst. D. Baltimore and R. Jaenisch (NIH/NCI 5-R35-CA44339) are supported by NIH grants. This work was supported in part by the ERC program of the National Science Foundation under award EEC-9843342.
REFERENCES
- 1.Asche W, Colletta G, Warnecke G, Nobis P, Pennie S, King R M, Ostertag W. Lack of retrovirus gene expression in somatic cell hybrids of Friend cells and teratocarcinoma cells with a teratocarcinoma phenotype. Mol Cell Biol. 1984;4:923–930. doi: 10.1128/mcb.4.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cedar H. DNA methylation and gene activity. Cell. 1988;53:3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- 3.Challita P M, Kohn D B. Lack of expression from a retroviral vector after transduction of murine hematopoietic stem cells is associated with methylation in vivo. Proc Natl Acad Sci USA. 1994;91:2567–2571. doi: 10.1073/pnas.91.7.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coligan, J. E., et al. (ed.). Current protocols in immunology. J. Wiley & Sons, New York, N.Y.
- 5.Gautsch J W. Embryonal carcinoma stem cells lack a function required for virus replication. Nature. 1980;285:110–112. doi: 10.1038/285110a0. [DOI] [PubMed] [Google Scholar]
- 6.Gautsch J W, Wilson M C. Delayed de novo methylation in teratocarcinoma suggests additional tissue-specific mechanisms for controlling gene expression. Nature. 1983;301:32–37. doi: 10.1038/301032a0. [DOI] [PubMed] [Google Scholar]
- 7.Greiser-Wilke I, Ostertag W, Goldfarb P, Lang A, Furusawa M, Conscience J F. Inducibility of spleen focus-forming virus by BrdUrd is controlled by the differentiated state of the cell. Proc Natl Acad Sci USA. 1981;78:2995–2999. doi: 10.1073/pnas.78.5.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grez M, Akgun E, Hilberg F, Ostertag W. Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc Natl Acad Sci USA. 1990;87:9202–9206. doi: 10.1073/pnas.87.23.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoeben R C, Migchielsen A A, van der Jagt R C, van Ormondt H, van der Eb A J. Inactivation of the Moloney murine leukemia virus long terminal repeat in murine fibroblast cell lines is associated with methylation and dependent on its chromosomal position. J Virol. 1991;65:904–912. doi: 10.1128/jvi.65.2.904-912.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang D C, Cory S, Strasser A. Bcl-2, Bcl-XL and adenovirus protein E1B19kD are functionally equivalent in their ability to inhibit cell death. Oncogene. 1997;14:405–414. doi: 10.1038/sj.onc.1200848. [DOI] [PubMed] [Google Scholar]
- 11.Jacob J, Baltimore D. Modelling T-cell memory by genetic marking of memory T cells in vivo. Nature. 1999;399:593–597. doi: 10.1038/21208. [DOI] [PubMed] [Google Scholar]
- 12.Jaenisch R, Fan H, Croker B. Infection of preimplantation mouse embryos and of newborn mice with leukemia virus: tissue distribution of viral DNA and RNA and leukemogenesis in the adult animal. Proc Natl Acad Sci USA. 1975;72:4008–4012. doi: 10.1073/pnas.72.10.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaenisch R, Schnieke A, Harbers K. Treatment of mice with 5-azacytidine efficiently activates silent retroviral genomes in different tissues. Proc Natl Acad Sci USA. 1985;82:1451–1455. doi: 10.1073/pnas.82.5.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahner D, Jaenisch R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature. 1985;315:594–597. doi: 10.1038/315594a0. [DOI] [PubMed] [Google Scholar]
- 15.Jat P S, Cepko C L, Mulligan R C, Sharp P A. Recombinant retroviruses encoding simian virus 40 large T antigen and polyomavirus large and middle T antigens. Mol Cell Biol. 1986;6:1204–1217. doi: 10.1128/mcb.6.4.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones P A. Altering gene expression with 5-azacytidine. Cell. 1985;40:485–486. doi: 10.1016/0092-8674(85)90192-8. [DOI] [PubMed] [Google Scholar]
- 17.Jones R J, Wagner J E, Celano P, Zicha M S, Sharkis S J. Separation of pluripotent haematopoietic stem cells from spleen colony-forming cells. Nature. 1990;347:188–189. doi: 10.1038/347188a0. [DOI] [PubMed] [Google Scholar]
- 18.Kohn D B. The current status of gene therapy using hematopoietic stem cells. Curr Opin Pediatr. 1995;7:56–63. doi: 10.1097/00008480-199502000-00011. [DOI] [PubMed] [Google Scholar]
- 19.Krall W, Kohn D B. Expression levels by retroviral vectors based upon the N2 and the MFG backbones. Gene Ther. 1996;3:365. [PubMed] [Google Scholar]
- 20.Laker C, Meyer J, Schopen A, Friel J, Heberlein C, Ostertag W, Stocking C. Host cis-mediated extinction of a retrovirus permissive for expression in embryonal stem cells during differentiation. J Virol. 1998;72:339–348. doi: 10.1128/jvi.72.1.339-348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei H, Oh S P, Okano M, Juttermann R, Goss K A, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 22.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 23.Loh T P, Sievert L L, Scott R W. Evidence for a stem cell-specific repressor of Moloney murine leukemia virus expression in embryonal carcinoma cells. Mol Cell Biol. 1990;10:4045–4057. doi: 10.1128/mcb.10.8.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naviaux R K, Costanzi E, Haas M, Verma I M. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichols J, Evans E P, Smith A G. Establishment of germ-line-competent embryonic stem (ES) cells using differentiation inhibiting activity. Development. 1990;110:1341–1348. doi: 10.1242/dev.110.4.1341. [DOI] [PubMed] [Google Scholar]
- 26.Niwa O, Yokota Y, Ishida H, Sugahara T. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983;32:1105–1113. doi: 10.1016/0092-8674(83)90294-5. [DOI] [PubMed] [Google Scholar]
- 27.Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier L L, Gorman D M, Nolan G P, Miyajima A, Kitamura T. Applications of retrovirus-mediated expression cloning. Exp Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- 28.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen R, Kempler G, Barklis E. A stem cell-specific silencer in the primer-binding site of a retrovirus. Mol Cell Biol. 1991;11:1214–1221. doi: 10.1128/mcb.11.3.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prince V E, Rigby P W. Derivatives of Moloney murine sarcoma virus capable of being transcribed in embryonal carcinoma stem cells have gained a functional Sp1 binding site. J Virol. 1991;65:1803–1811. doi: 10.1128/jvi.65.4.1803-1811.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robbins P B, Skelton D C, Yu X J, Halene S, Leonard E H, Kohn D B. Consistent, persistent expression from modified retroviral vectors in murine hematopoietic stem cells. Proc Natl Acad Sci USA. 1998;95:10182–10187. doi: 10.1073/pnas.95.17.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson E, Bradley A, Kuehn M, Evans M. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature. 1986;323:445–448. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- 33.Silver D P, Spanopoulou E, Mulligan R C, Baltimore D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc Natl Acad Sci USA. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 35.Tsukiyama T, Ueda H, Hirose S, Niwa O. Embryonal long terminal repeat-binding protein is a murine homolog of FTZ-F1, a member of the steroid receptor superfamily. Mol Cell Biol. 1992;12:1286–1291. doi: 10.1128/mcb.12.3.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Parijs L, Refaeli Y, Abbas A K, Baltimore D. Autoimmunity as a consequence of retrovirus-mediated expression of C-FLIP in lymphocytes. Immunity. 1999;11:763–770. doi: 10.1016/s1074-7613(00)80150-8. [DOI] [PubMed] [Google Scholar]
- 37.Van Parijs L, Refaeli Y, Lord J D, Nelson B H, Abbas A K, Baltimore D. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity. 1999;11:281–288. doi: 10.1016/s1074-7613(00)80103-x. [DOI] [PubMed] [Google Scholar]
- 38.Walsh C P, Chaillet J R, Bestor T H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 39.Yoder J A, Walsh C P, Bestor T H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]