Abstract
Splicing of the K-SAM alternative exon of the fibroblast growth factor receptor 2 gene is heavily dependent on the U-rich sequence IAS1 lying immediately downstream from its 5′ splice site. We show that IAS1 can activate the use of several heterologous 5′ splice sites in vitro. Addition of the RNA-binding protein TIA-1 to splicing extracts preferentially enhances the use of 5′ splice sites linked to IAS1. TIA-1 can provoke a switch to use of such sites on pre-mRNAs with competing 5′ splice sites, only one of which is adjacent to IAS1. Using a combination of UV cross-linking and specific immunoprecipitation steps, we show that TIA-1 binds to IAS1 in cell extracts. This binding is stronger if IAS1 is adjacent to a 5′ splice site and is U1 snRNP dependent. Overexpression of TIA-1 in cultured cells activates K-SAM exon splicing in an IAS1-dependent manner. If IAS1 is replaced with a bacteriophage MS2 operator, splicing of the K-SAM exon can no longer be activated by TIA-1. Splicing can, however, be activated by a TIA-1–MS2 coat protein fusion, provided that the operator is close to the 5′ splice site. Our results identify TIA-1 as a novel splicing regulator, which acts by binding to intron sequences immediately downstream from a 5′ splice site in a U1 snRNP-dependent fashion. TIA-1 is distantly related to the yeast U1 snRNP protein Nam8p, and the functional similarities between the two proteins are discussed.
Many eucaryotic genes are made up of exons and introns (43). They are transcribed into pre-mRNAs, from which the intron sequences are removed by splicing. Exons to be included in mRNA must be identified as such. This involves interaction of short sequences at or close to the exon's 5′ and 3′ splice sites (5′ss and 3′ss, respectively) with spliceosome components such as snRNPs and associated proteins (for reviews, see references 4, 29, and 43). Exon splicing can be controlled, and several sequences which participate in the control of tissue-specific or developmentally controlled alternative splicing events have been described (for a review, see reference 32). These sequences are particularly interesting to study, as they may yield information on both splicing activation mechanisms and tissue-specific control mechanisms of gene expression. We have been studying fibroblast growth factor receptor 2 (FGFR-2) pre-mRNA splicing for this reason.
FGFR-2 alternative exons K-SAM and BEK are spliced in a tissue-specific, mutually exclusive manner, and the two types of FGFR-2 obtained bind different subsets of FGF family members (38). The K-SAM exon is under complex control. It has weak splice sites, and it contains an exon splicing silencer (ESS) which functions by recruiting hnRNP A1 (13). To overcome the activity of this silencer, at least three activating sequences in the downstream intron are required (6, 10, 12). One of these, IAS1, lies immediately downstream of the 5′ss and is a U-rich sequence (10). In the absence of IAS1 (10), or if IAS1 is moved further downstream from the 5′ss (F. Del Gatto-Konczak, unpublished data), the K-SAM exon is very poorly spliced, unless the ESS is inactivated also. IAS1 and the ESS are thus major determinants of K-SAM exon splicing. However, neither element may be directly responsible for the tissue-specific splicing of the K-SAM exon. Both elements can control splicing of heterologous exons in cells, and we have not detected any difference in their activities between cells which splice the K-SAM exon and cells which do not (reference 11 and our unpublished data).
The necessary proximity of IAS1 to the 5′ss suggests a model for activation in which a protein bound to IAS1 interacts with U1 snRNP bound itself to the 5′ss. Searching for the activator based on its ability to bind IAS1 has not proved easy, as many nuclear proteins bind U-rich sequences, including U2AF (52), polypyrimidine tract-binding protein (PTB) (18), or hnRNP C (44). Recent work on splicing in Saccharomyces cerevisiae has suggested a different approach, however. Yeast U1 snRNP is considerably more complex than mammalian U1 snRNP (21), and several yeast U1 snRNP proteins have no known metazoan counterpart. One such protein is Nam8p. Nam8p activity is necessary for efficient 5′ss recognition when U1 snRNP binding to the 5′ss is poor (39). In commitment complexes, Nam8p contacts nonconserved nucleotides in yeast pre-mRNA downstream of the 5′ss. Its activity is optimal when these sequences are U rich (39, 53). This led to the suggestion that a mammalian counterpart of Nam8p could be involved in activation of weak 5′ss followed by U-rich sequences, such as the 5′ss of the K-SAM exon (39).
The known mammalian proteins most closely related to Nam8p are a pair of very similar proteins called TIA-1 (47) and the related protein TIAR (27). TIA-1 was originally believed to be a precursor to a cytotoxic T-lymphocyte granule protein. However, it is now known that this is not the case (34). TIA-1 and TIAR are widely expressed RNA-binding proteins (3, 33). Both TIA-1 and TIAR are involved in stress-induced translational arrest, colocalizing after stress with poly(A)+ RNA in the cytoplasmic foci known as stress granules (28), and it has been reported previously (22) that TIAR binds to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA in macrophages and may be involved in translational repression. However, under normal conditions, the proteins are in general located mainly in the nucleus (28; C. Le Guiner, unpublished data). Predominantly nuclear localization has also been described previously for UBP1, a recently characterized TIA-1–TIAR relative in plants, which enhances splicing of suboptimal introns and also protects mRNAs from exonucleolytic degradation (30).
Like Nam8p, TIA-1 and TIAR are composed of an N-terminal domain containing three RNA recognition motifs, linked to a C-terminal domain (27, 47). The similarity between Nam8p and TIA-1–TIAR (approximately 26% sequence identity) is limited to their RNA recognition motif-containing domains. Both TIA-1 and TIAR bind to RNA, with the preferred binding sequence being U rich (14). These observations encouraged us to test if TIA-1 is able to activate splicing of exons linked to a U-rich sequence like IAS1. In this article, we show that TIA-1 activates 5′ss use and that activation depends on the intron sequence downstream from the 5′ss, with IAS1 being a preferred sequence. We show that TIA-1 can bind to IAS1 in cell extracts; this binding is optimal if a 5′ss is adjacent to IAS1, and the binding is U1 snRNP dependent. We discuss the functional similarities between TIA-1 and Nam8p.
MATERIALS AND METHODS
Plasmids. (i) Splicing constructs.
β-Tropomyosin constructs for in vitro synthesis of pre-mRNA substrates were derived from the previously described Tropo 6A-7 clone (1) using standard techniques. Constructs with two competing 5′ss were derived from a truncated adenovirus E1A gene (Sp1), in which the 13S 5′ss (D1 site) is replaced with a polylinker allowing insertion of a region containing two 5′ss (5).
(ii) Expression vectors.
A mouse TIA-1 cDNA clone was obtained as an I.M.A.G.E. consortium clone (identification no. 1261161) containing the TIA-1 coding sequence lacking alternative exon 5. It was used to make pTIA-1, an expression vector for an N-terminal FLAG-tagged TIA-1, and pTIA-coat, an expression vector for an N-terminal FLAG-tagged TIA-1–coat fusion. The coat expression vector pcoat (or pCI-MS2) and the hnRNP A1-coat fusion vector have been described elsewhere (13). The entire coding sequence of hnRNP C1 was introduced into the StuI site of pCI-MS2-NLS-FLAG (13) to make an expression vector for an N-terminal FLAG-tagged hnRNP C1-coat fusion (in which coat sequences are C terminal). The hnRNP C1 expression vector phnRNP C1 was obtained from it by eliminating coat sequences by BamHI digestion and religation.
(iii) Minigenes.
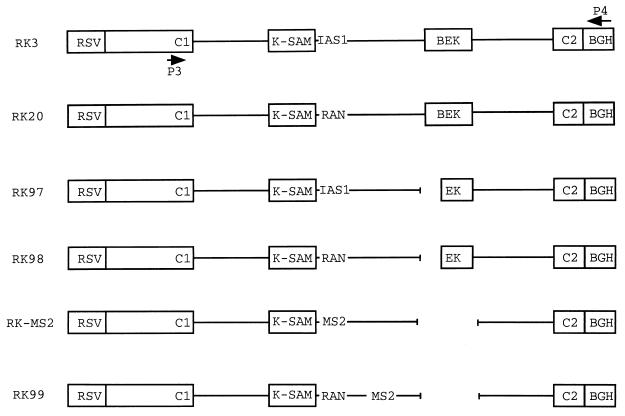
Rat preprotachykinin minigene pBPSVpA+2-7 (37) was a gift from P. J. Grabowski. The CD44 minigene was obtained by cloning a 6.9-kb ClaI-SmaI fragment of the human CD44 gene containing exons v8 to v10 and flanking introns (41) between the KpnI and HindIII sites of pRK3. pRK3 and pRK20 have been described elsewhere (10). RK-MS2 was made by inserting a 137-bp SpeI-EcoRI fragment of pIII/MS2-2 carrying MS2 coat protein binding sites (42) between nucleotides 15 and 505 of the 1,220-bp intron downstream of the K-SAM exon, in an RK3 derivative missing the BEK exon (deletion 1156–1412 of Fig. 1 in reference 12). pRK97 and pRK98 were made from pRK3 and pRK20, respectively, by deletion of the BEK exon's 3′ss and associated polypyrimidine sequence (deletion 1156–1233 of Fig. 1 in reference 12). pRK99 was made from pRK20 by deleting the BEK exon (deletion 1156–1412 of Fig. 1 in reference 12) and then replacing nucleotides 213 to 505 of the intron downstream of the K-SAM exon with the fragment carrying coat binding sites.
Extract preparations and in vitro splicing assays.
HeLa nuclear extract and cytoplasmic S100 extract were prepared as described previously (40). For preparation of whole-cell extracts (WCE) from 293-EBNA cells, cells resuspended in lysis buffer (20 mM Tris-HCl [pH 7.6], 400 mM KCl, 20% glycerol, 1 mM dithiothreitol, 0.2% NP-40, and a cocktail of protease inhibitors) were sonicated and then centrifuged at 10,000 rpm for 10 min in an SS-34 rotor. The supernatant (WCE) was dialyzed against buffer D for 5 h. For preparation of WCE from 293-EBNA cells overexpressing TIA-1 (WCE/TIA-1), cells were transfected with 6 μg of TIA-1-expressing vector and 14 μg of pBluescript SK(+) (Stratagene) and collected 48 h later, and WCE were prepared from them as described above.
Capped 32P-labeled pre-mRNA substrates were made by runoff in vitro transcription with SP6 RNA polymerase as described in reference 8. For the tropomyosin-derived transcripts, in vitro splicing was performed as described in reference 5 (25-μl final volume, using 12 μl of nuclear extract, in 60 mM KCl–1.3 mM MgCl2). Splicing of the E1A-derived transcripts was performed under a variety of conditions as indicated in the figure legends. Reaction mixtures were incubated at 30°C for 90 to 120 min, and splicing products were resolved on denaturing 5 to 6% polyacrylamide gels, followed by autoradiography.
UV cross-linking and immunoprecipitation assays.
UV cross-linking was performed as described previously (7) with minor modifications. RNA probes were synthesized in vitro from pBluescript SK(+)-based plasmids containing appropriate sequences (5′ss-IAS1, 5′ss-RAN, IAS1, or RAN) downstream of the T7 promoter. They were uniformly labeled at high specific activity using [α-32P]UTP, and 50 fmol was used per assay. Cross-linking assay mixtures (15 μl) containing 3 μl of cell extracts, supplemented or not with 150 ng of TIA-1 or hnRNP C1, were incubated with RNA probes in 0.6× buffer D containing 2.6% polyvinyl alcohol and 0.5 μg of Escherichia coli tRNA. After a 20-min incubation at 30°C, reaction mixtures were exposed to UV light for 15 min at 4°C and then treated with a mixture of RNases A (750 ng) and T1 (250 U) for 30 min at 37°C. For direct analysis, samples were diluted with a 2× sodium dodecyl sulfate (SDS) protein loading buffer and resolved by SDS-polyacrylamide gel electrophoresis (PAGE) on 12% polyacrylamide gels. For UV cross-linking and immunoprecipitation assays, the RNase-treated samples were diluted to 60 μl with IPP buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% NP-40), and 5 μg of anti-TIA-1 polyclonal antibody (Santa Cruz Biotech) or 10 μg of anti-hnRNP C1 monoclonal antibody (a generous gift from G. Dreyfuss) was added. After incubation at 4°C for 3 h, 10 μl of protein G-Sepharose beads was added and incubation was continued overnight. After three washes of the beads with 200 μl of IPP buffer, bound proteins were eluted in 20 μl of SDS loading buffer at 100°C for 5 min and loaded on an SDS–12% polyacrylamide gel.
To analyze the role of U1 snRNP in TIA-1 binding to the 5′ss-IAS1 probe, 3-μl aliquots of extracts (WCE or WCE/TIA-1) were pretreated with 0.5 μg of a 14-nucleotide oligodeoxynucleotide complementary to the 5′ end of U1 snRNA, or with a nonrelated probe complementary to the T7 promoter, in the presence of 0.5 U of RNase H, for 30 min at 30°C before cross-linking to the 5′ss-IAS1 probe and immunoprecipitation as described above.
Recombinant proteins and total SR preparation.
Total SR proteins from HeLa cells were purified as described in reference 51. To produce recombinant TIA-1 and hnRNP C1 in E. coli, the corresponding coding sequences from the second (TIA-1) or first (hnRNP C1) codon up to the stop codon were inserted in frame between the BamHI and EcoRI sites of pET28-b (Novagen). Resulting plasmids were used for production of six-His-tagged proteins. They were expressed and purified under nondenaturing conditions as recommended by the manufacturer and dialyzed against buffer D for 1 h.
Transfections and RT-PCR.
Transfection of SVK14 (46) and 293-EBNA cells (Invitrogen) was performed as described previously (10, 13). For cotransfections, 2 μg of minigene was cotransfected with 18 μg of the appropriate expression vector. Forty-eight hours later, RNA was harvested and analyzed by reverse transcription-PCR (RT-PCR) using reporter-specific primers. Preprotachykinin primers P1 and P2 were as follows: P1, GGAAATCGGTGCCAACG; P2, GAGAGATCTGACCATGCC. CD44 and FGFR-2 primers were as follows: P3, ATCCAGTGGATCAAGCAC, and P4, GGCAACCTAGAAGGCACAG. Twenty cycles of amplification were used so as to remain in the range of exponential amplification. PCR products were caused to migrate on agarose gels, transferred to nylon filters (Hybond N+; Amersham), and hybridized with different probes. Experiments were carried out at least in triplicate, and representative results are shown here. 32P-labeled RNA probes used were obtained by in vitro transcription of DNA fragments corresponding to (i) nucleotides 3 to 134 of the 148-nucleotide K-SAM exon; (ii) linked C1 and C2 exon sequences; and (iii) linked exons 2, 3, and 5 of the preprotachykinin gene.
RESULTS
IAS1 activation of heterologous exons.
Efficient recognition of the chicken β-tropomyosin gene's exon 6A (1) normally requires a 33-nucleotide pyrimidine-rich sequence (S4) starting 37 nucleotides downstream from its 5′ss. Can IAS1 replace S4 in this system? Various pre-mRNA substrates (Fig. 1A) containing exons 6A and 7 were used for in vitro splicing assays in HeLa cell nuclear extract. As expected from previous work (17), splicing (Fig. 1B) of a pre-mRNA lacking S4 (6A-Δ4-7, lane 7) was inefficient compared to splicing of pre-mRNAs with S4 (Tropo 6A-7, lane 2) or a purine-rich sequence (6A-P3AS-7, lane 3). When S4 was replaced with IAS1 (IAS1 down, Fig. 1A), splicing dropped to levels below those observed in the absence of S4 (compare lanes 6 and 7, Fig. 1B). In the IAS1 down pre-mRNA, IAS1 lies 43 nucleotides downstream of the exon 6A-intron junction. In vivo, IAS1 activates splicing only if it lies immediately downstream from the K-SAM exon's 5′ss (F. Del Gatto-Konczak, unpublished data). A further pre-mRNA was made (IAS1 up, Fig. 1A) which lacked S4 but contained IAS1 positioned immediately downstream from the exon 6A 5′ss. Splicing of IAS1 up was at least as efficient as splicing of substrates containing S4 or the purine-rich sequence (compare lanes 2, 3, and 4, Fig. 1B). Pre-mRNA RAN (Fig. 1A) is a version of IAS1 up in which IAS1 has been replaced with a random sequence incapable of activating K-SAM exon splicing in vivo (10). Splicing of pre-mRNA RAN was significantly less efficient than splicing of substrate IAS1 up (Fig. 1B, compare lanes 4 and 5) while being slightly more efficient than splicing of pre-mRNA 6A-Δ4-7 (compare lanes 5 and 7). These results show that IAS1 can activate splicing of a heterologous exon in vitro, provided that it is positioned immediately downstream of the exon's 5′ss. They also show that the random sequence does not act as a repressor of an adjacent 5′ss in vitro.
FIG. 1.
IAS1 activates splicing of a heterologous tropomyosin exon. (A) Schematic representations of pre-mRNAs used for in vitro splicing. In Tropo 6A-7 pre-mRNA, exons 6A and 7 are separated by a 284-nucleotide intron including the S4 activating sequence. In 6A-P3AS-7 and IAS1 down, S4 has been replaced with a purine-rich sequence and with IAS1, respectively. In 6A-Δ4-7, the S4 sequence is deleted. In IAS1 up and RAN, IAS1 and the random RAN sequence, respectively, have been inserted immediately downstream of the 5′ss. The last nucleotides of exon 6A are boxed, and the IAS1 and RAN sequences are shown in boxes. (B) In vitro splicing assays using pre-mRNAs shown in panel A in HeLa cell nuclear extract. mRNAs obtained by splicing exons 6A and 7 together are identified, as well as excised introns. The space of migration between the pre-mRNAs and mRNA has been reduced. The amounts of excised introns (I) and remaining pre-mRNA (P) were quantified using a Fuji phosphorimager, and the percentage of splicing was determined as I/(I + P) × 100%. The mean of three determinations is given below the lanes. nt, nucleotides.
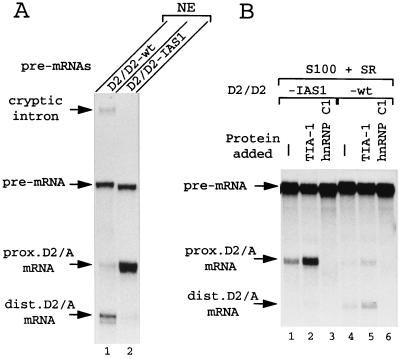
In the FGFR-2 pre-mRNA, IAS1 is involved in a competitive splicing choice. This encouraged us to test if IAS1 can function when two 5′ss are in competition. We used pre-mRNAs derived from the adenovirus E1A gene (Fig. 2), in which the unique 13S 5′ss (D1) has been replaced with different pairs of competing splice sites (5). Pre-mRNA D2/D2-wt contains two identical copies of the E1A 12S 5′ss D2. When this pre-mRNA is spliced in nuclear extract, the distal D2 5′ss is markedly preferred to the proximal D2 5′ss (Fig. 3A, lane 1). This preference for the distal 5′ss is probably due to involvement of the nuclear cap binding complex (CBC), which acts to favor use of the 5′ss closer to the pre-mRNA's cap (31). However, placing IAS1 immediately downstream from the proximal site (pre-mRNA D2/D2-IAS1, Fig. 2) induces a very strong shift of splicing toward use of this latter site (Fig. 3A, lane 2).
FIG. 2.
Schematic representations of pre-mRNAs with competing 5′ss used for in vitro splicing. Part of the E1A pre-mRNA is shown, with the natural competing D2 and D1 5′ss. In the other pre-mRNAs, the D1 5′ss has been replaced with a pair of competing 5′ss as shown, and the major splicing reactions observed are indicated.
FIG. 3.
Effect of IAS1 and TIA-1 on competing D2 5′ss in vitro. In vitro splicing assays were performed using pre-mRNAs shown in Fig. 2. For spliced mRNAs, the donor (D) and acceptor (A) (the E1A 3′ss) splice sites used are identified. (A) Splicing was carried out in HeLa cell nuclear extract (NE). Note that a cryptic splicing reaction occurs with the D2/D2-wt pre-mRNA using a cryptic 5′ss located 88 nucleotides upstream of the distal D2 site. The cryptic intron is visible on the photo, but the corresponding mRNA has not been retained. (B) Splicing was in cytoplasmic S100 extract (9 μl) with 0.5 μg of SR proteins added (S100+SR). Lanes 1 to 3, D2/D2-IAS1 pre-mRNA spliced in extract alone (lane 1) or in extract with 600 ng of TIA-1 (lane 2) or 600 ng of hnRNP C1 (lane 3). Lanes 4 to 6, D2/D2-wt pre-mRNA spliced in extract alone (lane 4) or with 600 ng of TIA-1 (lane 5) or 600 ng of hnRNP C1 (lane 6) added.
TIA-1 activation of 5′ss.
Having established that IAS1 can activate splicing of an adjacent 5′ss in vitro, we searched for possible effects of TIA-1 on IAS1 activity. To stand a chance of observing an effect on splicing in vitro of an increase in TIA-1 levels, it is necessary to start with splicing extracts in which the concentration of TIA-1 is suboptimal. For this reason, we used S100 extract (with added SR proteins) for further experiments, as TIA-1 is less abundant in cytoplasmic S100 extracts than in nuclear extracts (data not shown). In the absence of added exogenous TIA-1, only use of the proximal D2 site linked to IAS1 was detected in S100 extract (Fig. 3B, lane 1). Splicing was less efficient than that in nuclear extract (compare with Fig. 3A, lane 2), most probably because several factors, including TIA-1, are limiting in the S100 extract. Addition of recombinant TIA-1 (600 ng) led to a strong stimulation of splicing using the D2-IAS1 site (Fig. 3B, lane 2). In contrast, addition of hnRNP C1, a protein which, like TIA-1, binds to U-rich sequences (20), actually decreased use of the D2-IAS1 site (lane 3). While no comparable stimulation by TIA-1 of use of a D2 site linked to the random sequence was observed (compare lanes 4 and 5), addition of 600 ng of TIA-1 did weakly stimulate use of both copies of the D2 5′ss in the D2/D2-wt substrate (compare lanes 4 and 5 in Fig. 3B). This suggests that TIA-1 can also activate 5′ss linked to sequences other than IAS1, at least in S100 extract supplemented with SR proteins, which, compared to nuclear extract, is suboptimal for splicing. Note that both D2 5′ss copies are stimulated to approximately the same extent, and so TIA-1 is not preferentially activating either the proximal or the distal 5′ss here. Importantly, activation by TIA-1 of the D2 5′ss not linked to IAS1 cannot be detected on the pre-mRNA containing a competing D2 5′ss adjacent to IAS1 (compare lanes 1 and 2, dist. D2/A mRNA). TIA-1 is thus showing a preference for the 5′ss adjacent to IAS1.
Can this effect of TIA-1 be reproduced using other pairs of splice sites, particularly when one of them is the K-SAM exon DSAM 5′ss? We have analyzed splicing of pre-mRNA from other constructs, which contain as competing splice sites the strong E1A 13S D1 5′ss and the weaker DSAM 5′ss. In pre-mRNA D1/DSAM-IAS1 (Fig. 2), the DSAM site is followed by IAS1. In pre-mRNA D1/DSAM-RAN, IAS1 has been replaced with the random sequence described above. When the D1/DSAM-IAS1 pre-mRNA was spliced in S100 extract (with added SR proteins), the D1 site was scarcely used for splicing (Fig. 4A, lane 2), while the DSAM 5′ss was preferred (Fig. 4A, lane 2). However, addition of TIA-1 (300 or 600 ng) strongly activated use of the DSAM 5′ss (approximately fourfold), without affecting the very weak use of the D1 5′ss (lanes 3 and 4). As observed previously (Fig. 3B), addition of hnRNP C1 decreased use of the IAS1-linked (DSAM) 5′ss (Fig. 4A, lane 5). No comparable TIA-1-induced activation was observed when the DSAM 5′ss was linked to the random sequence (compare lanes 6 and 7). However, addition of 600 ng of TIA-1 did stimulate use of the D1 5′ss in the D1/DSAM-RAN substrate (compare lanes 6 and 7 in Fig. 4A). Yet despite this, no activation of the D1 5′ss by TIA-1 could be observed when it was in competition with an IAS1-linked 5′ss (the DSAM-IAS1 5′ss) on D1/DSAM-IAS1 pre-mRNA (compare lane 2 to lanes 3 and 4). TIA-1 is once again showing a preference for the 5′ss adjacent to IAS1.
FIG. 4.
Effects of IAS1 and TIA-1 on competing D1 and DSAM 5′ss in vitro. In vitro splicing assays were performed using pre-mRNAs shown in Fig. 2. For spliced mRNAs, the donor (D) and acceptor (A) (the E1A 3′ss) splice sites used are identified. (A) Splicing was carried out in cytoplasmic S100 extract (9 μl) with 0.5 μg of SR proteins added (S100+SR). Lane 1, D1/DSAM-IAS1 pre-mRNA starting material. Lanes 2 to 5, D1/DSAM-IAS1 pre-mRNA spliced in extract alone (lane 2), in extract with 300 or 600 ng of TIA-1 added (lanes 3 and 4, respectively), or in extract with 600 ng of hnRNP C1 added (lane 5). Lanes 6 to 8, D1/DSAM-RAN pre-mRNA spliced in extract alone (lane 6), extract with 600 ng of TIA-1 added (lane 7), or extract with 600 ng of hnRNP C1 added (lane 8). (B) Splicing was in a 6:4 mixture of nuclear extract and S100 extract (NE/S100). Lane 1, D1/DSAM-IAS1 pre-mRNA starting material. Lanes 2 to 6, D1/DSAM-IAS1 pre-mRNA spliced in extract alone (lane 2); in extract with 200, 400, or 600 ng of TIA-1 added (lanes 3 to 5, respectively); or in extract with 400 ng of hnRNP C1 added (lane 6). Lanes 7 to 9, D1/DSAM-RAN pre-mRNA spliced in extract alone (lane 7) or in extract with 400 ng of TIA-1 (lane 8) or 400 ng of hnRNP C1 (lane 9) added. The radioactivities present in the mRNAs were determined by a phosphorimager, corrected for their content in C residues, and used to calculate the ratio of use of D1 versus that of DSAM.
If TIA-1 does really have a preference for the IAS1-linked 5′ss, it should be able to provoke a significant switch in splice site use under appropriate circumstances. To attempt to visualize such a switch, we set out to increase the use of the D1 site for splicing of the D1/DSAM-IAS1 pre-mRNA. On this pre-mRNA, the D1 site is distal, and use of a distal 5′ss close to a cap site is known to be facilitated by the CBC (31). Cap binding protein is less abundant in S100 extract than in nuclear extract, and its concentration in S100 extract may be suboptimal (25). We therefore performed splicing assays using the D1/DSAM-IAS1 pre-mRNA in the presence of a mixture of S100 extract and nuclear extract, trying in this way to limit starting concentrations of TIA-1 while benefiting from significantly higher levels of CBC. Under these conditions, the distal D1 site was indeed efficiently used (Fig. 4B, lane 2). The competing (proximal) DSAM 5′ss was also used for splicing, though at a lower level (Fig. 4B, lane 2). However, addition of TIA-1 (200, 400, or 600 ng) strongly activated use of the DSAM 5′ss (approximately fourfold), whereas splicing using the D1 5′ss concomitantly decreased (lanes 3 to 5). These results show that TIA-1 can effectively provoke a significant switch in splice site use, from predominant use of the D1 site (a strong 5′ss) to approximately equal use of the D1 and the IAS1-linked DSAM 5′ss. Taken together, our results show that TIA-1 activates 5′ss use in a manner dependent on the downstream intron sequence, with a preference for a downstream U-rich sequence.
Preferential binding of TIA-1 to IAS1 adjacent to a 5′ss.
We used UV cross-linking analysis to test whether TIA-1 can bind to IAS1. The first RNA probe used (5′ss-IAS1) contained a strong 5′ss sequence (AG/GUAAGU) linked to IAS1. The major protein cross-linked to this probe in HeLa cell nuclear extract was an approximately 60-kDa protein (which might be PTB or U2AF, both known to bind to pyrimidine-rich sequences); several smaller proteins were also detected (Fig. 5A, lane 1). A number of proteins cross-linked weakly to the 5′ss-IAS1 probe in the S100 extract, which contains only low amounts of TIA-1 and hnRNP C (Fig. 5A, lane 2). When the latter extract was enriched with recombinant TIA-1 or hnRNP C1, the probe cross-linked to major new proteins with net molecular masses of ≈45 (corresponding in size to recombinant TIA-1, lane 3) and ≈40 (corresponding in size to recombinant hnRNP C1, lane 4) kDa, respectively. We also tested S100 extract enriched with recombinant ASF/SF2 lacking its RS domain. This protein, which binds with high affinity to purine-rich sequences, was not detected with the 5′ss-IAS1 probe (data not shown).
FIG. 5.
Interaction between TIA-1 and IAS1. (A and B) RNA probes as shown were incubated with various extracts either alone (−) or with 150 ng of added recombinant TIA-1 (+ TIA-1) or hnRNP C1 (+ C1) as indicated before UV cross-linking and SDS-PAGE analysis. NE, HeLa nuclear extract; S100, HeLa S100 extract. WCE, WCE from 293-EBNA cells. WCE/TIA-1, WCE from cells transfected with pTIA-1. After UV cross-linking and RNase treatment, equivalent aliquots were resolved directly on an SDS-polyacrylamide gel. The positions of prestained protein standards (NOVEX) are indicated. Note that the apparent molecular mass of the adduct proteins is usually 3 to 5 kDa higher than that of the corresponding protein. (C and D) RNA probes were incubated with various extracts as in panels A and B. After UV cross-linking and RNase treatment, samples were divided into three parts. One was analyzed directly (total); the others were analyzed after immunoprecipitation with antibodies against either TIA-1 (αTIA-1) or hnRNP C1 (αC1). Analysis was performed by SDS-PAGE. The aliquots loaded on the gel for the samples analyzed directly (total) were one-third of the amount used for those analyzed after immunoprecipitation.
To test if TIA-1 expressed in mammalian cells can also bind to IAS1, we performed similar experiments using WCE prepared from human 293-EBNA cells, either transfected or not with the mouse TIA-1 expression vector pTIA-1. Transfection led to an approximately 10-fold increase in TIA-1 levels (data not shown). The 5′ss-IAS1 probe cross-linked to several proteins in the untransfected, control WCE (Fig. 5A, lane 5). In WCE from cells transfected with pTIA-1, a major new protein corresponding in size to TIA-1 was detected with the 5′ss-IAS1 probe (lane 6). However, when the 5′ss-RAN (the random sequence linked to a 5′ss) probe was used, no difference between WCE from untransfected (lane 8) and transfected (lane 9) cells could be observed. Thus, these results show that both TIA-1 produced bacterially and TIA-1 produced in a mammalian cell can bind specifically and efficiently to IAS1.
Somewhat different results were obtained when the probe used was IAS1 without an adjacent 5′ss (Fig. 5B). This probe detects a major ≈40-kDa protein in HeLa cell nuclear extract (lane 1) which is hnRNP C (Fig. 5D), and a major ≈80-kDa protein in S100 extract (lane 2). While we detected cross-linking of the IAS1 probe with recombinant TIA-1 (lane 3) or hnRNP C1 (lane 4) in S100 extract supplemented with these proteins, interaction of TIA-1 with the IAS1 probe appeared weaker than that with the 5′ss-IAS1 probe (compare lane 3, Fig. 5B, with lane 3, Fig. 5A). This difference between the two probes was also observed using WCE from cells overexpressing TIA-1 following transfection (compare Fig. 5A and B, lane 6). As expected, we did not detect any cross-linking of TIA-1 to the random sequence probe RAN (Fig. 5B, compare lanes 8 and 9).
Further experiments were carried out to demonstrate that the ≈45-kDa protein detected in extracts with the 5′ss-IAS1 probe really was TIA-1. A variety of extracts were incubated with either the 5′ss-IAS1 probe or the IAS1 probe before UV cross-linking. Aliquots were analyzed either before immunoprecipitation (total) or after immunoprecipitation with antibodies recognizing either TIA-1 or hnRNP C1. Both TIA-1 and hnRNP C1 cross-linked to the 5′ss-IAS1 probe in HeLa cell nuclear extract (Fig. 5C, lanes 2 and 3), albeit weakly. This cross-linking appears to be weak because of competition for the probe by the major 60-kDa cross-linking protein (lane 1). (Note that this competition does not necessarily take place during in vitro splicing assays where bona fide splicing substrates are used.) Thus, when the IAS1 probe is used (Fig. 5D), cross-linking to the 60-kDa protein is much less marked (lane 1), while cross-linking to hnRNP C1 concomitantly increases greatly (lane 3). However, despite the apparently greater probe availability, no cross-linking of the IAS1 sequence to TIA-1 is seen (lane 2). This result is in agreement with those shown in Fig. 5A and B, which demonstrated that TIA-1 binds very weakly to IAS1 which is not adjacent to a 5′ss.
Using WCE from untransfected 293-EBNA cells (Fig. 5C, lanes 4 to 6), we detected efficient cross-linking of both TIA-1 (lane 5) and hnRNP C1 (lane 6) to the 5′ss-IAS1 probe. Furthermore, a strong increase of TIA-1 cross-linking was observed with WCE from cells transfected with pTIA-1 (compare lanes 5 and 8). Interestingly, when the IAS1 probe was used, TIA-1 was detected, and only weakly, solely in WCE from the transfected cells (Fig. 5D, compare lanes 5 and 8), while hnRNP C1 was detected in WCE from both untransfected (lane 6) and transfected (lane 9) cells. Taken together, the results in Fig. 5 demonstrate that TIA-1 interacts preferentially with the IAS1 motif when adjacent to a 5′ss. This conclusion was confirmed by competition experiments using WCE from transfected cells. Interaction of TIA-1 with the labeled 5′ss-IAS1 probe was strongly reduced (four- to fivefold) in the presence of an 80-fold excess of unlabeled 5′ss-IAS1 probe, while the presence of a 320-fold excess of unlabeled IAS1 probe had no significant effect (data not shown).
TIA-1 binding is U1 snRNP dependent.
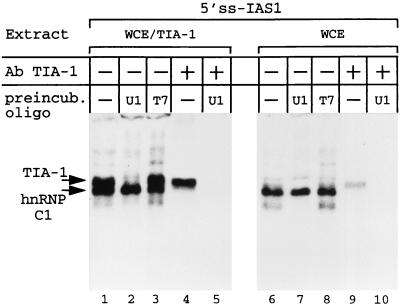
The preferred binding site for TIA-1 has been identified as a U-rich sequence by experiments analyzing TIA-1–RNA interaction in the absence of any other cellular proteins (14). Under these conditions, there was no indication that the U-rich sequence need be adjacent to a sequence resembling a 5′ss. It seemed possible to us that the preferred binding of TIA-1 to IAS1 adjacent to a 5′ss in cell extracts might reflect the presence, in these extracts, of U1 snRNP and its binding to the 5′ss. Is the binding of TIA-1 to IAS1 adjacent to a 5′ss in fact dependent on U1 snRNP binding to the 5′ss? To address this question, we incubated WCE from TIA-1-transfected cells with either an oligonucleotide complementary to the 5′ 14 nucleotides of U1 snRNA or an “irrelevant” oligonucleotide (corresponding to one strand of the T7 promoter), in the presence of RNase H. The former oligonucleotide provokes degradation of the 5′ nucleotides of U1 snRNA necessary for U1 snRNP binding to the 5′ss (and so abolishes this binding). The latter oligonucleotide has no such effect. When WCE from TIA-1-transfected cells were subjected to either a mock preincubation or preincubation with the T7 oligonucleotide before cross-linking to the 5′ss-IAS1 probe, probe cross-linking to both TIA-1 and hnRNP C1 was readily observed (Fig. 6, lanes 1 and 3). However, when preincubation was done with the antisense U1 snRNA oligonucleotide, cross-linking to TIA-1 was selectively eliminated (lane 2). This result was confirmed when aliquots of cross-linked material were subjected to immunoprecipitation using anti-TIA-1 antibodies (compare lanes 4 and 5). Similar results were observed when WCE from untransfected cells were analyzed (lanes 6 to 10), although the amount of cross-linked TIA-1 was, as expected, lower. The U1 snRNP-dependent cross-linking of TIA-1 to the probe could nevertheless be visualized after immunoprecipitation of samples with anti-TIA-1 antibodies (compare lanes 9 and 10). We conclude that TIA-1 binding to IAS1 in cell extracts is dramatically enhanced by U1 snRNP binding to an adjacent 5′ss.
FIG. 6.
U1 snRNP is involved in TIA-1 binding to IAS1. WCE from 293-EBNA cells (WCE) or from transfected 293-EBNA cells (WCE/TIA-1) were mock preincubated (−) or preincubated with oligonucleotides complementary to the 5′ end of U1 snRNA (U1) or complementary to the T7 promoter (T7) before cross-linking to the 5′ss-IAS1 probe. For lanes 1 to 3 and 6 to 8, each assay mixture was analyzed directly. In addition, immunoprecipitation with anti-TIA-1 antibodies was performed on the mock-preincubated samples (lanes 4 and 9) and the samples preincubated with the oligonucleotide complementary to the 5′ end of U1 snRNA (lanes 5 and 10). The aliquots loaded on the gels for the samples analyzed directly represent one-third of the amount used for those analyzed after immunopurification.
TIA-1 enhances K-SAM exon splicing in vivo.
As described previously (10), the RK3 minigene (Fig. 7) contains a wild-type FGFR-2 gene fragment carrying the very similarly sized alternative K-SAM and BEK exons, together with flanking intron sequences and the upstream and downstream constitutive exons C1 and C2. Transfection of epithelial SVK14 cells with RK3 leads to preferential splicing of the K-SAM exon, while transfection of 293-EBNA cells leads to preferential splicing of the BEK exon. Skipping of both exons is also observed at a low level. Cotransfection of 293-EBNA cells with RK3 and pcoat (a bacteriophage MS2 coat protein expression vector) resulted in BEK exon splicing (Fig. 8A and B, lanes 1), reflected by major RT-PCR product C1BC2 (Fig. 8E). For RK3 and derivatives, two probes were used to identify RT-PCR products. The C1C2 probe detects all products, while the K-SAM probe detects only products containing the K-SAM exon. Cotransfection of RK3 with pTIA-1 resulted in splicing of the K-SAM exon to the BEK exon (Fig. 8A and B, lanes 2), reflected by the major product C1SBC2, though some C1SC2 product is also detected (Fig. 8E). Importantly, cotransfection of RK3 with the hnRNP C1 expression vector did not detectably activate K-SAM exon splicing (lanes 3), suggesting that the TIA-1 effect on the K-SAM exon cannot be reproduced by just any U-rich sequence binding protein. RK20 (Fig. 7) is a version of RK3 in which IAS1 has been replaced with the random sequence (10) unable to bind TIA-1. Cotransfection of RK20 with pTIA-1 did not lead to K-SAM exon splicing, as RT-PCR products of type C1BC2 (Fig. 8E) were obtained regardless of whether RK20 was cotransfected with pcoat (Fig. 7A and B, lanes 4), pTIA-1 (lanes 5), or phnRNP C1 (lanes 6). TIA-1 activation of K-SAM exon splicing is thus IAS1 dependent. Note that, in the absence of IAS1, TIA-1 activates splicing of C1 to C2 somewhat (Fig. 8A, lane 5) but that this effect cannot be detected when IAS1 is present (Fig. 8A, lane 2). This is in agreement with our in vitro splicing results, which show that, while TIA-1 can activate a variety of 5′ss, when a 5′ss linked to IAS1 is in competition with one not so linked, it is use of the former which is favored by TIA-1.
FIG. 7.
Schematic representations of FGFR-2 minigenes. The parent minigene RK3 is shown, with the Rous sarcoma virus long terminal repeat promoter (RSV), the alternative exons K-SAM and BEK, the upstream and downstream constitutive exons C1 and C2, and the bovine growth hormone polyadenylation sequence (BGH). Locations of primers P3 and P4 used for RT-PCR are marked. The U-rich intron-activating sequence IAS1 is identified. RK20 is similar to RK3, except for the replacement of IAS1 with a random sequence. In RK97, the BEK exon's polypyrimidine sequence and 3′ss have been deleted. RK98 was derived from RK97 by replacing IAS1 with the random sequence described in the text. RK-MS2 is derived from RK3 by replacing IAS1 and some downstream sequences with bacteriophage MS2 coat binding sites (MS2). In addition, the BEK exon is deleted. RK99 is similar to RK20, except that the BEK exon is deleted and bacteriophage MS2 coat binding sites (MS2) have been placed well downstream of the K-SAM exon's 5′ss.
FIG. 8.
TIA-1 activation of the K-SAM exon requires IAS1. Cells were cotransfected with minigenes and expression vectors for bacteriophage MS2 coat protein, TIA-1, or hnRNP C1 as shown. RT-PCR was carried out on transfected cell RNA using primers P3 and P4 shown in Fig. 7, and products were subjected to Southern analysis. Hybridization was performed first to a probe corresponding to the K-SAM exon (B and D), and then the same blot was dehybridized and rehybridized to a probe made up of exons C1 and C2 (A and C). RT-PCR products are identified using names corresponding to structures shown in panel E.
TIA-1 can activate K-SAM exon splicing not only to the BEK exon but also to the C2 exon. In RK97 (Fig. 7), the BEK exon's 3′ss and associated polypyrimidine sequence have been deleted, blocking its splicing. Cotransfection of RK97 with pcoat in 293-EBNA cells results mainly in skipping of the K-SAM exon. The major RT-PCR product obtained (Fig. 8C and D, lanes 1) is C1C2 (Fig. 8E); only a little splicing of the K-SAM exon is observed (product C1SC2). Cotransfection of RK97 with pTIA-1 leads to a marked increase of K-SAM exon splicing, as evidenced by an increase in the levels of the C1SC2 product (Fig. 8C and D, lanes 2). Once again, this activation of K-SAM splicing is IAS1 dependent, as cotransfection of pTIA-1 with RK98, a version of RK97 in which IAS1 has been replaced with the random sequence (Fig. 7), has no similar effect (compare lanes 3 and 4 of Fig. 8C and D).
Artificial recruitment of TIA-1 obviates the IAS1 requirement.
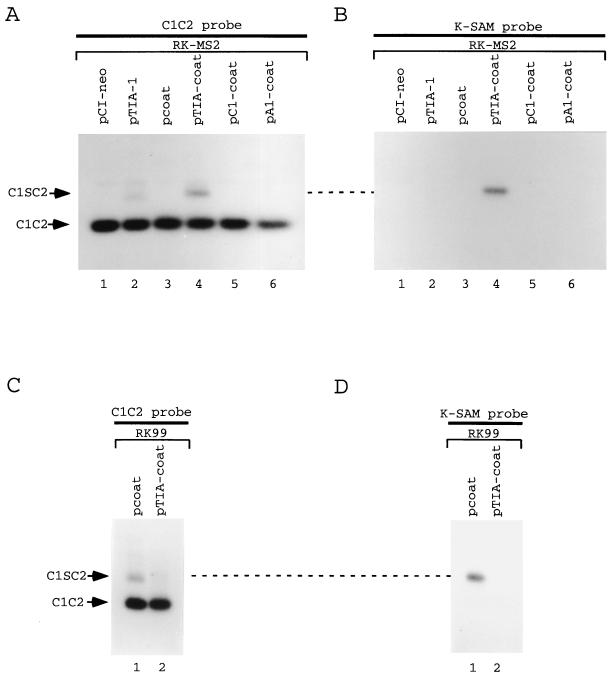
Our results suggest that IAS1 serves as a binding site to recruit TIA-1 close to the 5′ss. If so, artificial recruitment of TIA-1 downstream from the 5′ss might activate splicing of a K-SAM exon not linked to IAS1. In RK-MS2 (Fig. 7), which has the BEK exon deleted, nucleotides 15 to 505 (which include IAS1) of the intron downstream from the K-SAM exon have been replaced with a tandem copy of the bacteriophage MS2 coat protein operator. Proteins can thus be recruited to RK-MS2 pre-mRNA downstream from the K-SAM exon as fusions with coat protein. When RK-MS2 is transfected together with the empty expression vector pCI-neo or pcoat, the K-SAM exon is skipped. RT-PCR products detected with the C1C2 probe (Fig. 9A, lanes 1 and 3) reflect splicing of exon C1 to exon C2 (product C1C2). This result is expected, as IAS1 is required for efficient K-SAM exon splicing. Cotransfection of pTIA-1 with RK-MS2 does not detectably induce K-SAM exon inclusion: note that none of the RT-PCR products detected with the C1C2 probe (Fig. 9A, lane 2) hybridize with the K-SAM exon probe (Fig. 9B, lane 2). However, when RK-MS2 is cotransfected with an expression vector for a TIA-1–coat fusion protein, some inclusion of the K-SAM exon is induced: RT-PCR products which hybridize to the K-SAM probe and correspond to C1SC2 can be detected (Fig. 9A and B, lanes 4). No activation of K-SAM exon splicing was observed when RK-MS2 was cotransfected with expression vectors for hnRNP C1-coat fusions (Fig. 9A and B, lanes 5) or hnRNP A1-coat fusions (lanes 6).
FIG. 9.
TIA-1 activates splicing if recruited close to the 5′ss. Cells were cotransfected with minigenes and the empty expression vector pCI-neo or expression vectors for bacteriophage MS2 coat protein or TIA-1 or the following fusions with coat protein: TIA-1–coat fusion (TIA-coat), hnRNP C1-coat fusion (C1-coat), and hnRNP A1-coat fusion (A1-coat). RT-PCR was carried out on transfected cell RNA using primers P3 and P4 shown in Fig. 7, and products were subjected to Southern analysis. Hybridization was performed first to a probe corresponding to the K-SAM exon (B and D), followed by dehybridization and rehybridization to a probe made up of exons C1 and C2 (A and C). RT-PCR products are identified using names corresponding to structures shown in Fig. 8E.
The K-SAM exon inclusion induced by TIA-coat with RK-MS2 pre-mRNA is less than that induced by TIA-1 with pre-mRNAs containing IAS1 (compare Fig. 8 and 9), although approximately equal amounts of the two proteins are made following transfection of their expression vectors (data not shown). The TIA-coat fusion bound to the MS2 operator may be presented to the splicing apparatus suboptimally compared to TIA-1 in its natural position. Activation by the TIA-coat fusion is, as expected, position dependent. In RK99 (Fig. 7), the operator is placed 213 bp downstream from the 5′ss, and not 15 bp downstream as in RK-MS2. (Note that RK99 does not contain IAS1, which has been replaced with the random sequence of RK20.) When RK99 is transfected together with the TIA-coat expression vector, no activation of K-SAM exon splicing is observed (Fig. 9C and D). In fact, TIA-coat expression now represses K-SAM exon splicing (compare lanes 1 and 2).
TIA-1's effect on other exons in vivo.
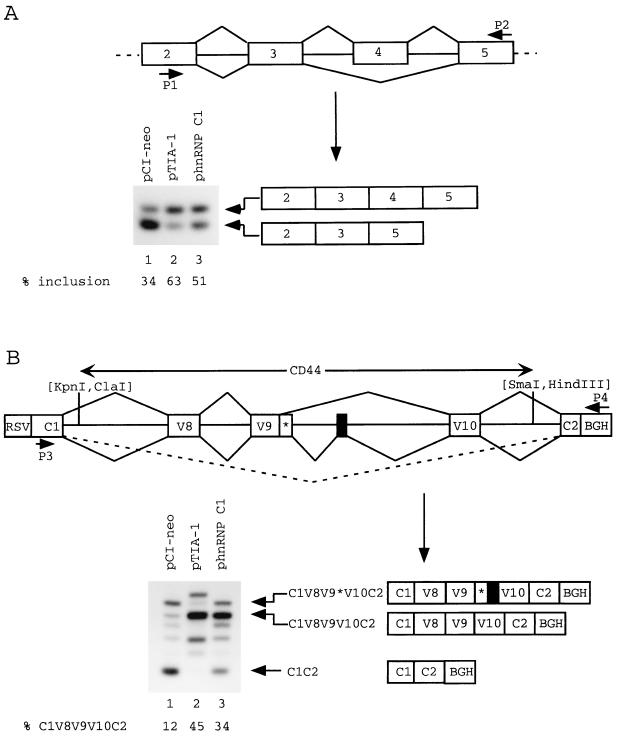
To test whether TIA-1 can influence alternative splicing of other exons in vivo, we used several minigenes reflecting well-documented cases of alternative exon splicing. Minigenes were cotransfected into cells with different expression vectors, and splicing patterns were investigated by RT-PCR analysis of transfected cell RNA using minigene-specific primers. Normally, splicing of preprotachykinin pre-mRNA involves preferential skipping of optional exon 4 (23, 37). In 293-EBNA cells cotransfected with a preprotachykinin minigene (containing exons 2 to 7) and the empty expression vector pCI-neo, inclusion of exon 4 is inefficient (34%) as judged by RT-PCR analysis (Fig. 10A, lane 1). When the minigene is cotransfected with the expression vector pTIA-1, exon 4 inclusion increases to 63% (lane 2). However, a similar increase also occurs (to 51%, lane 3) following cotransfection with the hnRNP C1 expression vector phnRNP C1.
FIG. 10.
Effects of TIA-1 and hnRNP C1 on splicing in vivo. (A) The preprotachykinin minigene was cotransfected into SVK14 cells with pCI-neo (lane 1), pTIA-1 (lane 2), or phnRNP C1 (lane 3). RT-PCR was carried out on transfected cell RNA using primers P1 and P2, and products were subjected to Southern analysis with hybridization to a probe made up of exons 2 to 5. (B) A hybrid FGFR-2–CD44 minigene was cotransfected into 293-EBNA cells with pCI-neo (lane 1), pTIA-1 (lane 2), or phnRNP C1 (lane 3). RT-PCR was carried out on transfected cell RNA using primers P3 and P4 as marked, and products were subjected to Southern analysis with hybridization to a probe made up of exons C1 and C2 of the FGFR-2 gene. The 6.9-kb ClaI-SmaI fragment of the human CD44 gene used is identified by arrows and contains alternative exons v8, v9, and v10, as well as an additional alternative exon (50) represented by a black box. Note that exon v9 has two alternative 5′ss (50). On the minigene map, the three major splicing events seen in 293-EBNA cells transfected with the minigene and pCI-neo are illustrated. The corresponding RT-PCR products are illustrated below the map. RSV, Rous sarcoma virus long terminal repeat; BGH, bovine growth hormone polyadenylation signal. Radioactivities present in bands were determined by phosphorimager and used to calculate splicing percentages.
Exons v8, v9, and v10 of the CD44 gene's pre-mRNA are spliced to generate mRNA for the epithelial cell form of CD44 (9). A minigene was made in which these exons and their flanking introns were placed between two constitutively spliced exons, C1 and C2, of the human FGFR-2 gene (Fig. 10B). When this minigene was transfected into the epithelial cell line SVK14, exons v8, v9, and v10 were efficiently spliced to the flanking exons C1 and C2 (data not shown). However, when the minigene was transfected into 293-EBNA cells together with the control vector pCI-neo, RT-PCR analysis revealed a number of products (Fig. 10B, lane 1). The identity of some of these products was investigated by hybridization to a probe composed of v8, v9, and v10 sequences and by sequencing of subcloned fragments (data not shown). Unlike in SVK14 cells, product C1V8V9V10C2 is not the major product, representing only 12% of all products. More abundant products correspond either to skipping of all CD44 exons (product C1C2) or to splicing of exons v8, v9, and v10, with use of an alternative 5′ss for exon v9 and inclusion of an additional exon (represented by a black box) between v9 and v10 (product C1V8V9*V10C2). (This splicing possibility has already been described for the mouse CD44 gene [50]). The identity of other products was not established. When the CD44 minigene was transfected into 293-EBNA cells together with pTIA-1, the C1C2 product disappeared and the RT-PCR products (lane 2) shifted in favor of the C1V8V9V10C2 product, which now represents 45% of all products. A similar increase of this product (to 34% of all products, lane 3) also occurs following cotransfection with the hnRNP C1 expression vector.
In summary, for both the preprotachykinin and CD44 pre-mRNAs, TIA-1 overexpression and hnRNP C1 overexpression have qualitatively similar effects. The smaller effect of phnRNP C1 transfection could be explained if the transfection-induced increase in hnRNP C1 levels is lower than that in TIA-1 levels. On the other hand, our in vitro analysis has shown that TIA-1 can activate 5′ss not linked to IAS1, albeit less effectively than those so linked. This could also contribute to the increased effect of TIA-1 relative to that of hnRNP C1. TIA-1 overexpression does not lead to the activation of splicing of all exons, however, as TIA-1 had no detectable effect (data not shown) on splicing of another alternative exon, the poorly spliced EIIIb exon (24) of the rat fibronectin gene.
The similar effects on preprotachykinin and CD44 pre-mRNAs of TIA-1 and hnRNP C1 overexpression suggest that increasing the level of any protein which binds to U-rich sequences may suffice to perturb splicing in these cases. One possible mechanism could involve competition for pyrimidine-rich binding sites with a protein such as PTB. Insofar as PTB is known to repress splicing of a variety of exons (48a), limiting its access to the pre-mRNA could favor exon inclusion. It should be recalled that, in contrast to the preprotachykinin and CD44 exons, for the K-SAM exon, TIA-1 overexpression markedly stimulates splicing, while hnRNP C1 overexpression has no detectable effect. This suggests that TIA-1's stimulation of K-SAM exon splicing cannot be attributed to the same type of effect as that exerted on either preprotachykinin or CD44 exon splicing.
DISCUSSION
The activating element IAS1 of the FGFR-2 gene participates with a variety of other elements in controlling splicing of the K-SAM exon (12). IAS1 lies immediately downstream from the K-SAM exon and indeed activates splicing only when so positioned. Our results demonstrate that (i) IAS1 can function independently of other FGFR-2 elements to activate heterologous 5′ss, (ii) TIA-1 binds to IAS1 in cell extracts if a 5′ss is adjacent to it, (iii) this binding is dependent on intact U1 snRNA, and (iv) TIA-1 activates use of 5′ss. The extent of activation observed depends on the sequence downstream from the 5′ss. 5′ss adjacent to IAS1 are preferentially activated, and increasing TIA-1 levels can provoke a switch to use of such sites. Replacing IAS1 downstream from the K-SAM exon with a binding site for the bacteriophage MS2 coat protein allows activation of this exon's splicing by a TIA-1–coat fusion. Activation occurs only if the fusion binds close to the 5′ss, indicating that TIA-1 needs to be close to the 5′ss to activate. Our results identify TIA-1 as a novel splicing activator and suggest that TIA-1 activates splicing by binding close to a 5′ss, for a direct or indirect interaction with U1 snRNP bound to the 5′ss. Does TIA-1 bind transiently to U1 snRNP before interaction of the resulting complex with the splice site? We have no evidence in favor of this. Though TIA-1 and U1 snRNP do coimmunoprecipitate weakly, we have not been able to rule out nonspecific RNA bridging as an explanation (our unpublished data). An alternative possibility is that TIA-1 and U1 snRNP associate in situ at the 5′ss region: their association, dependent on the 5′ss and the adjacent IAS1, would last only the time needed for splicing. Clearly, further experiments are required to define in detail the nature and the chronology of the interactions between TIA-1, U1 snRNP, and the 5′ss and flanking IAS1 sequence and to clarify whether TIA-1 enhances U1 snRNP binding to the 5′ss or acts at some later step. It is interesting to note, however, that incubation of the IAS1 up and RAN tropomyosin pre-mRNAs (Fig. 1) in nuclear extract under conditions allowing only formation of early (E) complexes (absence of ATP and Mg2+) specifically promotes U1 snRNP-dependent protection of the 5′ss linked to IAS1 (C. F. Bourgeois, L. Kister, and J. Stévenin, unpublished data). This suggests that IAS1 acts to facilitate the U1 snRNP-5′ss binding step.
We were led to investigate TIA-1 by recent work on yeast U1 snRNP (39, 53). Yeast U1 snRNP is more complex than mammalian U1 snRNP, containing in addition to the proteins found in mammalian U1 snRNP a number of specific proteins including Nam8p (21). When yeast U1 snRNP binds to a 5′ss, Nam8p is positioned so as to contact intron sequences downstream from the splice site (39, 53). Nam8p is required for splicing when the 5′ss is noncanonical, as for example in the MER2 pre-mRNA (36). Nam8p is also required in yeast strains lacking the nuclear CBC (16) and is presumably indispensable to facilitate 5′ss recognition in the absence of CBC activation. While Nam8p activity is maximal if sequences downstream of the 5′ss are U rich (39), this is not a requirement, as sequences immediately downstream of the MER2 5′ss, for example, are not particularly U rich. Nam8p, with its three RNA binding domains, can presumably interact with a variety of RNA sequences, albeit with different affinities, and activate splicing via them to different extents.
TIA-1 is a distant mammalian relative of Nam8p, and our results identify functional similarities between the two proteins. Both proteins can activate 5′ss use, and their activity is maximal if downstream intron sequences are U rich. There is, nevertheless, a clear difference between Nam8p and TIA-1: the latter is not an integral part of mammalian U1 snRNP. However, oligonucleotide-directed degradation of the 5′ extremity of U1 snRNA virtually abolishes TIA-1 binding to IAS1 in cell extracts. This implies that TIA-1 binding to IAS1 requires U1 snRNP binding to an adjacent 5′ss and suggests that there may not be a fundamental difference between TIA-1 and Nam8p to be found here. It has been proposed that splicing of a given vertebrate intron may require assembly at the corresponding exon's 5′ss of a complex containing core U1 snRNP and a particular subset of the relatives of the yeast U1 snRNP-specific proteins mentioned above (15). The subset would be chosen in view of the individual characteristics of the 5′ss in question. One way of activating a weak 5′ss would be to associate it with an intronic sequence recognized by TIA-1, to allow assembly of a complex containing core U1 snRNP and TIA-1. It should be possible to modulate the extent of such activation. Strong activation could be achieved by using an intronic sequence like IAS1 similar to the optimal U-rich sequence for TIA-1 binding. Weaker activation would ensue if the intronic sequence bound TIA-1 with less affinity (TIA-1 can probably bind, like Nam8p, to a variety of RNA sequences with different affinities). If so, it would be possible to use TIA-1 to modulate splice site choice in alternative splicing. The results of our in vitro analysis (Fig. 3 and 4) are in agreement with this model: TIA-1 can activate 5′ss not linked to IAS1, but when two 5′ss are in competition, one linked to IAS1 and the other not, TIA-1 markedly favors use of the former at the expense of the latter. Our data show that TIA-1 overexpression can have a profound effect on alternative splicing, both in vivo and in vitro. Although our results are based on overexpression data alone and it may be difficult to define the true physiological role of a protein with certainty from them, our additional observation that TIA-1 binding to IAS1 in cell extracts is U1 snRNP dependent does provide a further link to a defined part of the cellular splicing machinery. Taken together, our observations strongly suggest that one physiological role for TIA-1 is in regulation of splicing. Clearly, it will be important to identify exons which require TIA-1 for their splicing.
Which splice sites represent important targets for TIA-1 activation in vivo? It is interesting to recall that if Nam8p can apparently activate a variety of 5′ss (as implied by its requirement in strains lacking nuclear CBC [see above]), Nam8p activity is normally required at only a few, specific, noncanonical 5′ss. Indeed, Nam8p is not needed for vegetative growth. It is possible that in a similar fashion TIA-1 is normally required in vivo only for use of a subset of 5′ss. One candidate exon is the K-SAM exon. Splicing of this exon is repressed by an ESS. IAS1 is required for K-SAM exon splicing, but only to overcome the activity of this ESS (12). Our results show that TIA-1 has the characteristics required for a protein activating K-SAM exon splicing naturally: it binds to IAS1 and activates the K-SAM exon 5′ss in vitro and favors K-SAM inclusion in vivo. While this does not prove that TIA-1 is the actual protein which activates K-SAM exon splicing in vivo, it does make TIA-1 (or, if not TIA-1, a protein with very similar properties) a very good candidate.
We have detected no significant difference in levels of TIA-1 between SVK14 cells which splice the K-SAM exon normally and 293-EBNA cells which do not (F. Del Gatto-Konczak, unpublished data). While this excludes a simple model for tissue-specific K-SAM exon splicing based on tissue-specific expression of TIA-1, it is not in contradiction with the proposed role of TIA-1 in activating K-SAM exon splicing via IAS1. Thus, unlike K-SAM exon splicing, action of IAS1 is definitely not tissue specific per se: IAS1 will fully activate splicing of a heterologous fibronectin exon both in SVK14 cells and in HeLa cells, which normally do not splice the K-SAM exon (F. Del Gatto-Konczak, unpublished data). Thus, the protein which acts via IAS1 is not expected to have a marked tissue-specific distribution or activity.
How then could tissue-specific splicing of the K-SAM exon be achieved? Control of the K-SAM exon is complex. The exon contains an ESS, which functions by binding hnRNP A1 (13). Its 5′ss is weak, and splicing of the exon requires not only IAS1 but also two additional intron-activating sequences, IAS2 and IAS3 (6, 12). It may be that each of these elements shows a low degree of tissue specificity individually but that when their effects are added together the sum is enough to confer tissue specificity. A similar model invoking the necessary cooperation among a variety of regulatory elements, none of which is absolutely tissue specific, has been proposed previously for splicing of a neuron-specific exon (35). We favor, however, an alternative model in which TIA-1 bound to IAS1 and hnRNP A1 bound to the ESS exert opposing influences on the 5′ss and poise the K-SAM exon on the brink of splicing, without any tissue specificity. Then, additional tissue-specific activation, possibly via IAS2 and IAS3, would suffice to tip the balance in favor of K-SAM exon splicing. Such additional activation need perhaps be only mild, the competing BEK exon being itself repressed in cells which splice the K-SAM exon (6, 12, 19).
Other speculative targets for TIA-1 action are pre-mRNAs coding for proteins implicated in apoptosis. Expression of several key proteins in apoptosis is regulated by alternative splicing (for a review, see reference 26). TIA-1 itself has been linked to apoptosis. The serine-threonine kinase FAST is activated during Fas-mediated apoptosis in Jurkat cells and phosphorylates TIA-1 prior to the onset of DNA fragmentation (48). The TIA-1-related protein TIAR is very similar to TIA-1 and likely to exert the same type of activity as TIA-1. Indeed, overexpression of TIAR in cells by transfection leads to the same effects on splicing as does overexpression of TIA-1 (C. Le Guiner, unpublished data). TIAR is translocated from the nucleus to the cytoplasm during Fas-mediated apoptosis (45). In the mouse, TIAR is essential for primordial germ cell development, as it appears to be necessary for cell survival (2). It is tempting to speculate that proapoptotic stimuli modify the activities of TIA-1 and TIAR, leading to changes in the splicing patterns of key pre-mRNAs. It may prove possible to test this hypothesis by inactivation of the TIA-1 and TIAR genes in chicken cells, using an approach similar to that used recently to inactivate the gene coding for another splicing factor, ASF/SF2 (49).
ACKNOWLEDGMENTS
We thank G. Dreyfuss, P. Grabowski, R. Hynes, and J. Marie for kindly providing materials. We also thank G. Hildwein for excellent technical assistance and the staff of the IGBMC facilities for their assistance.
This work was supported by funds from the INSERM, the CNRS, the Hôpitaux Universitaires de Nantes et de Strasbourg, the Association pour la Recherche sur le Cancer, and the Ligue Nationale contre le Cancer, Comité Departemental de Loire-Atlantique. C.F.B. was supported by fellowships from the Association pour la Recherche sur le Cancer and the Ligue Nationale contre le Cancer.
F.D.G.-K. and C.F.B. contributed equally to the work.
REFERENCES
- 1.Balvay L, Libri D, Gallego M, Fiszman M Y. Intronic sequence with both negative and positive effects on the regulation of alternative transcripts of the chicken beta tropomyosin transcripts. Nucleic Acids Res. 1992;20:3987–3992. doi: 10.1093/nar/20.15.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck A, Miller I J, Anderson P, Streuli M. RNA-binding protein TIAR is essential for primordial germ cell development. Proc Natl Acad Sci USA. 1998;95:2331–2336. doi: 10.1073/pnas.95.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck A R, Medley Q G, O'Brien S, Anderson P, Streuli M. Structure, tissue distribution and genomic organization of the murine RRM-type RNA binding proteins TIA-1 and TIAR. Nucleic Acids Res. 1996;24:3829–3835. doi: 10.1093/nar/24.19.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black D L. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgeois C F, Popielarz M, Hildwein G, Stevenin J. Identification of a bidirectional splicing enhancer: differential involvement of SR proteins in 5′ or 3′ splice site activation. Mol Cell Biol. 1999;19:7347–7356. doi: 10.1128/mcb.19.11.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carstens R P, McKeehan W L, Garcia-Blanco M A. An intronic sequence element mediates both activation and repression of rat fibroblast growth factor receptor 2 pre-mRNA splicing. Mol Cell Biol. 1998;18:2205–2217. doi: 10.1128/mcb.18.4.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavaloc Y, Bourgeois C F, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavaloc Y, Popielarz M, Fuchs J P, Gattoni R, Stevenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper D L, Dougherty G, Harn H J, Jackson S, Baptist E W, Byers J, Datta A, Phillips G, Isola N R. The complex CD44 transcriptional unit; alternative splicing of three internal exons generates the epithelial form of CD44. Biochem Biophys Res Commun. 1992;182:569–578. doi: 10.1016/0006-291x(92)91770-q. [DOI] [PubMed] [Google Scholar]
- 10.Del Gatto F, Breathnach R. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1995;15:4825–4834. doi: 10.1128/mcb.15.9.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Gatto F, Gesnel M C, Breathnach R. The exon sequence TAGG can inhibit splicing. Nucleic Acids Res. 1996;24:2017–2021. doi: 10.1093/nar/24.11.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Gatto F, Plet A, Gesnel M C, Fort C, Breathnach R. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1997;17:5106–5116. doi: 10.1128/mcb.17.9.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Gatto-Konczak F, Olive M, Gesnel M C, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dember L M, Kim N D, Liu K Q, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 15.Fortes P, Bilbao-Cortes D, Fornerod M, Rigaut G, Raymond W, Seraphin B, Mattaj I W. Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev. 1999;13:2425–2438. doi: 10.1101/gad.13.18.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, Mattaj I W. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol Cell Biol. 1999;19:6543–6553. doi: 10.1128/mcb.19.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallego M E, Gattoni R, Stevenin J, Marie J, Expert-Bezancon A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 1997;16:1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia B M, Jamison S F, Sharp P A. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989;3:1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert E, Del Gatto F, Champion A P, Gesnel M C, Breathnach R. Control of BEK and K-SAM splice sites in alternative splicing of the fibroblast growth factor receptor 2 pre-mRNA. Mol Cell Biol. 1993;13:5461–5468. doi: 10.1128/mcb.13.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorlach M, Burd C G, Dreyfuss G. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J Biol Chem. 1994;269:23074–23078. [PubMed] [Google Scholar]
- 21.Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot H V, Mann M, Seraphin B, Rosbash M, Luhrmann R, Fabrizio P. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- 22.Gueydan C, Droogmans L, Chalon P, Huez G, Caput D, Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J Biol Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- 23.Hoffman B E, Grabowski P J. U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 1992;6:2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- 24.Huh G S, Hynes R O. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 1994;8:1561–1574. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- 25.Izaurralde E, Stepinski J, Darzynkiewicz E, Mattaj I W. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J Cell Biol. 1992;118:1287–1295. doi: 10.1083/jcb.118.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Z H, Wu J Y. Alternative splicing and programmed cell death. Proc Soc Exp Biol Med. 1999;220:64–72. doi: 10.1046/j.1525-1373.1999.d01-11.x. [DOI] [PubMed] [Google Scholar]
- 27.Kawakami A, Tian Q, Duan X, Streuli M, Schlossman S F, Anderson P. Identification and functional characterization of a TIA-1-related nucleolysin. Proc Natl Acad Sci USA. 1992;89:8681–8685. doi: 10.1073/pnas.89.18.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kedersha N L, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 30.Lambermon M H, Simpson G G, Kirk D A, Hemmings-Mieszczak M, Klahre U, Filipowicz W. UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J. 2000;19:1638–1649. doi: 10.1093/emboj/19.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis J D, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj I W. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 32.Lopez A J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 33.Lowin B, French L, Martinou J C, Tschopp J. Expression of the CTL-associated protein TIA-1 during murine embryogenesis. J Immunol. 1996;157:1448–1454. [PubMed] [Google Scholar]
- 34.Medley Q G, Kedersha N, O'Brien S, Tian Q, Schlossman S F, Streuli M, Anderson P. Characterization of GMP-17, a granule membrane protein that moves to the plasma membrane of natural killer cells following target cell recognition. Proc Natl Acad Sci USA. 1996;93:685–689. doi: 10.1073/pnas.93.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Modafferi E F, Black D L. Combinatorial control of a neuron-specific exon. RNA. 1999;5:687–706. doi: 10.1017/s1355838299990155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa T, Ogawa H. Involvement of the MRE2 gene of yeast in formation of meiosis-specific double-strand breaks and crossover recombination through RNA splicing. Genes Cells. 1997;2:65–79. doi: 10.1046/j.1365-2443.1997.d01-283.x. [DOI] [PubMed] [Google Scholar]
- 37.Nasim F H, Spears P A, Hoffmann H M, Kuo H C, Grabowski P J. A sequential splicing mechanism promotes selection of an optimal exon by repositioning a downstream 5′ splice site in preprotachykinin pre-mRNA. Genes Dev. 1990;4:1172–1184. doi: 10.1101/gad.4.7.1172. [DOI] [PubMed] [Google Scholar]
- 38.Ornitz D M, Xu J, Colvin J S, McEwen D G, MacArthur C A, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 39.Puig O, Gottschalk A, Fabrizio P, Seraphin B. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 1999;13:569–580. doi: 10.1101/gad.13.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt P, Gattoni R, Keohavong P, Stevenin J. Alternative splicing of E1A transcripts of adenovirus requires appropriate ionic conditions in vitro. Cell. 1987;50:31–39. doi: 10.1016/0092-8674(87)90659-3. [DOI] [PubMed] [Google Scholar]
- 41.Screaton G R, Bell M V, Jackson D G, Cornelis F B, Gerth U, Bell J I. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc Natl Acad Sci USA. 1992;89:12160–12164. doi: 10.1073/pnas.89.24.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharp P A. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 44.Swanson M S, Dreyfuss G. RNA binding specificity of hnRNP proteins: a subset bind to the 3′ end of introns. EMBO J. 1988;7:3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taupin J L, Tian Q, Kedersha N, Robertson M, Anderson P. The RNA-binding protein TIAR is translocated from the nucleus to the cytoplasm during Fas-mediated apoptotic cell death. Proc Natl Acad Sci USA. 1995;92:1629–1633. doi: 10.1073/pnas.92.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor P J, Purkis P, Lane E B, McKay I A, Chang S E. Effects of SV40 transformation on the cytoskeleton and behavioural properties of human keratinocytes. Cell Differ. 1982;11:169–180. doi: 10.1016/0045-6039(82)90008-2. [DOI] [PubMed] [Google Scholar]
- 47.Tian Q, Streuli M, Saito H, Schlossman S F, Anderson P. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell. 1991;67:629–639. doi: 10.1016/0092-8674(91)90536-8. [DOI] [PubMed] [Google Scholar]
- 48.Tian Q, Taupin J, Elledge S, Robertson M, Anderson P. Fas-activated serine/threonine kinase (FAST) phosphorylates TIA-1 during Fas-mediated apoptosis. J Exp Med. 1995;182:865–874. doi: 10.1084/jem.182.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48a.Valcarcel J, Gebauer F. Post-transcriptional regulation: the dawn of PTB. Curr Biol. 1997;7:R705–R708. doi: 10.1016/s0960-9822(06)00361-7. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Takagaki Y, Manley J L. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- 50.Yu Q, Toole B P. A new alternatively spliced exon between v9 and v10 provides a molecular basis for synthesis of soluble CD44. J Biol Chem. 1996;271:20603–20607. doi: 10.1074/jbc.271.34.20603. [DOI] [PubMed] [Google Scholar]
- 51.Zahler A M, Lane W S, Stolk J A, Roth M B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 52.Zamore P D, Green M R. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci USA. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang D, Rosbash M. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 1999;13:581–592. doi: 10.1101/gad.13.5.581. [DOI] [PMC free article] [PubMed] [Google Scholar]