Abstract
Viral proteases are diverse in structure, oligomeric state, catalytic mechanism, and substrate specificity. This chapter focuses on proteases from viruses that are relevant to human health: human immunodeficiency virus subtype 1 (HIV-1), hepatitis C (HCV), human T-cell leukemia virus type 1 (HTLV-1), flaviviruses, enteroviruses, and coronaviruses. The proteases of HIV-1 and HCV have been successfully targeted for therapeutics, with picomolar FDA-approved drugs currently used in the clinic. The proteases of HTLV-1 and the other virus families remain emerging therapeutic targets at different stages of the drug development process. This chapter provides an overview of the current knowledge on viral protease structure, mechanism, substrate recognition, and inhibition. Particular focus is placed on recent advances in understanding the molecular basis of diverse substrate recognition and resistance, which is essential toward designing novel protease inhibitors as antivirals.
Keywords: HIV-1, HCV, HTLV-1, Flavivirus, Enterovirus, Coronavirus, Viral protease, Protease inhibitors, Substrate envelope, Drug resistance
1. Introduction
Many viruses encode one or more proteases as a common strategy to sustain replication with a compacted genome. The viral genome encodes a polyprotein with an embedded viral protease that cleaves the polyprotein at several specific sites to generate mature viral proteins. Viral proteases are therefore essential for replication, which makes them ideal therapeutic targets. The cleavage sites recognized by a given viral protease are generally diverse in amino acid sequence and processed at different rates, as polyprotein processing is an ordered and obligatory sequential process to produce infectious virus. This ability to specifically recognize and cleave diverse substrate sequences is conserved for proteases of different families of viruses. Although overall folds are shared between eukaryotic and viral proteases, diverse substrate recognition represents a unique feature of viral proteases. The unique substrate preference of viral proteases can be exploited in inhibitor design, toward developing selective and potent drug-like molecules. This has been achieved for HIV-1 and HCV where protease inhibitors have been part of antiviral combination therapies. However, drug resistance has emerged as a major problem and therefore needs to be considered as part of the drug-design process for evolving targets. The error-prone nature of replication allows viruses to rapidly select for resistance mutations under drug pressure. This chapter will focus on the viral protease structure, mechanism, substrate recognition, inhibition and resistance mutations for human immunodeficiency virus subtype 1 (HIV-1) and hepatitis C (HCV) virus where viral proteases have proven to be successful therapeutic targets. Furthermore, viral proteases from human T-cell leukemia virus type 1 (HTLV-1), flaviviruses, enteroviruses, and coronaviruses are discussed as emerging targets for drug development.
2. Viral proteases targeted with antiviral drugs
2.1. HIV-1 protease
HIV-1 protease is the most well-studied viral protease and was the first to be targeted for therapeutics. The success in targeting HIV-1 protease has paved the way for inhibiting other viral proteases in drug development. There are numerous reviews and book chapters highlighting the progress in HIV-1 protease research over the years [1], [2], [3]. Therefore, in this chapter, a general overview is provided with recent advances in understanding the molecular basis of drug-resistance mutations.
2.1.1. HIV-1 protease structure
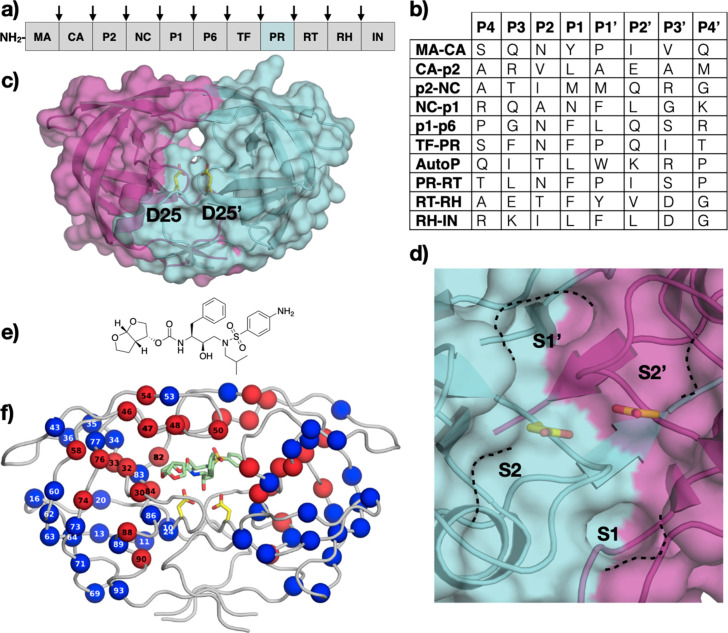
HIV-1 protease is expressed as part of a larger Gag-Pro-Pol polyprotein (Fig. 1A). The initial cleavage occurs in cis to release the protease itself, and the current model suggests that the polyprotein precursor must dimerize to activate the protease. Additionally, the protease cleaves nine other sites on the viral polyprotein (Fig. 1B). HIV-1 protease is composed of two identical chains of 99-amino acids each and functions as a pseudo-symmetric homodimer (Fig. 1C). The overall structure of HIV-1 protease is related to that of monomeric cellular aspartic proteases. However, HIV-1 protease contains two distinct flaps that cover the active site formed at the dimer interface. These flaps must open up to allow substrates to bind and then close to stably bind substrates to perform catalysis.
Fig. 1.
HIV-1 Protease. (A) Schematic representation of HIV-1 polyprotein (Gag-Pro-Pol), with viral protease (PR) cleavage sites between structural proteins (MA: matrix; CA: capsid; NC: nucleocapsid), enzymes (RT: reverse transcriptase; RH: RNase H; IN: integrase) and peptides (TF: trans-frame; spacer peptides P1, P2) indicated by arrows. (B) Amino acid sequence of polyprotein cleavage sites recognized by HIV-1 protease. The protease cleaves between P1-P1’. (C) The crystal structure of HIV-1 protease (PDB ID: 1T3R) with the two monomers colored cyan and magenta and the catalytic residue (D25) depicted in yellow sticks and labeled, and (D) a close-up view of the active site with S2-S2’ pockets annotated. (E) The chemical structure of the latest FDA-approved HIV-1 protease inhibitor, darunavir. (F) Cocrystal structure of darunavir (green sticks) bound to HIV-1 protease, with location of primary (red) and secondary (blue) resistance mutations indicated by spheres.
Panel F: Reprinted with permission from A.N. Matthew, F. Leidner, G.J. Lockbaum, M. Henes, J. Zephyr, S. Hou, et al., Drug design strategies to avoid resistance in direct-acting antivirals and beyond. Chem. Rev. 121(6) (2021) 3238–3270.
© 2021 American Chemical Society.
2.1.2. Substrate recognition and cleavage
HIV-1 protease cleaves the Gag-Pro-Pol polyprotein at a total of ten sites that are of diverse amino acid sequence which are processed at different rates (Fig. 1B). The active site cavity covers the S2-S2’ pockets with the S3/S3’ being partially solvent-exposed. The hydrophobic S2-S2’ pockets recognize mostly hydrophobic side chains, especially at P1/P1’ positions (Fig. 1B). Although the sequences differ, cleavage sites share a conserved shape upon binding to the protease active site. The model explaining diverse substrate recognition is termed the substrate envelope, which represents the consensus volume occupied by the substrates. Hence, HIV-1 protease recognizes peptide substrates based not on the sequence but the shape the peptide can adopt [4], [5], [6]. HIV-1 protease uses two catalytic aspartic acids (D25 /D25’), one from each monomer, to activate a water molecule for catalysis. Each monomer contributes one aspartic acid as part of the general aspartyl protease Asp-Thr-Gly catalytic motif located at the base of the active site. The hydroxyl ion then attacks the scissile carbonyl carbon to generate a tetrahedral intermediate that collapses to generate new N- and C-termini. The tetrahedral transition state has been exploited in HIV-1 protease inhibitor design, following the transition state theory which states that enzymes bind tighter to the transition state intermediates compared to substrate or product.
2.1.3. HIV-1 protease inhibitors as drugs
There are a total of ten FDA-approved drugs targeting HIV-1 protease [1]. The initial peptidomimetic HIV-1 protease inhibitors (saquinavir, ritonavir, indinavir, nelfinavir, and amprenavir) resulted in high metabolic clearance, poor oral bioavailability, and had a low barrier to resistance. Second-generation inhibitors, aimed at improving on these limitations, led to the development of darunavir (DRV), the latest and most potent FDA-approved HIV-1 protease inhibitor (Fig. 1E). DRV exhibits activity against a broad range of clinical isolates that demonstrated resistance against the earlier generation drugs.
2.1.4. Resistance mutations
HIV-1 readily develops resistance under drug pressure because of the error-prone replication and reverse transcription of the viral RNA. Resistance mutations occur throughout the HIV-1 protease and are broadly classified into two categories. Primary resistance mutations that directly alter contact with the inhibitor occur in the active site and are largely explained by being outside the substrate envelope. In contrast, secondary resistance mutations are located mostly distal from the active site and impact the dynamic ensemble of the protease altering the balance of substrate turnover versus inhibitor binding (Fig. 1F). [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19]. Recent studies combining parallel molecular dynamics simulations and machine learning have provided insights into how remote changes can confer resistance and which interactions are most indicative of these changes [7], [20]. Each of the FDA-approved HIV-1 protease inhibitors has specific signature mutations that cause resistance; detailed reviews can be found elsewhere [3], [19], [21], [22], [23], [24], [25].
2.1.5. Substrate envelope and mechanisms of resistance
HIV-1 protease substrate envelope, which explains the molecular mechanism of diverse substrate recognition, is also a useful tool to explain the mechanism of drug resistance and thus can guide the design of more robust protease inhibitors [26]. The substrate envelope explains how resistance mutations lead to a decrease in inhibitor binding but do not impair substrate processing. Comparing cocrystal structures of substrate and inhibitor-bound structures revealed that inhibitors protrude outside of the substrate envelope at specific locations that correspond closely with the site of drug resistance mutations. Moreover, inhibitors that fit within the envelope have a higher barrier for drug resistance. The HIV-1 protease substrate envelope has been used to guide the design of inhibitors that are more robust against resistance [27], [28], [29], [30].
The success in developing drugs against HIV-1 protease has demonstrated that viral proteases are viable therapeutic targets. Lessons learned from the drug design efforts targeting HIV-1 have been applied to other systems with varying levels of success, including the HCV NS3/4A protease.
2.2. HCV NS3/4A protease
The positive-sense single-stranded RNA genome of HCV encodes a viral polyprotein within which exists subsists the NS3/4A bifunctional protein (Fig. 2A). The 70 kDa NS3 protein contains two domains, a protease located at N-terminus and a helicase domain at the C-terminus [31]. Each enzymatic domain of NS3 can function independently of the other. The NS3 protease domain contains all the catalytic residues and the substrate binding pocket. However, the protease activity is significantly enhanced by the NS4A protein which serves as a cofactor. In total, four sites in the polyprotein are cleaved by the NS3/4A protease between non-structural (NS) viral proteins (Fig. 2B) [32], [33], [34], [35], [36], [37]. Cleavage at the NS2/NS3 junction is carried out by a second viral encoded protease, the NS2 autoprotease, that is responsible for cleaving only that site [38]. Similar to HIV-1 protease, there are multiple reviews on HCV NS3/4A protease as a therapeutic target. Here, we will provide a concise overview and highlight progress made over the past decade, specifically in direct-acting antivirals (DAAs) against the HCV NS3/4A protease, which represents a triumph in drug design.
Fig. 2.
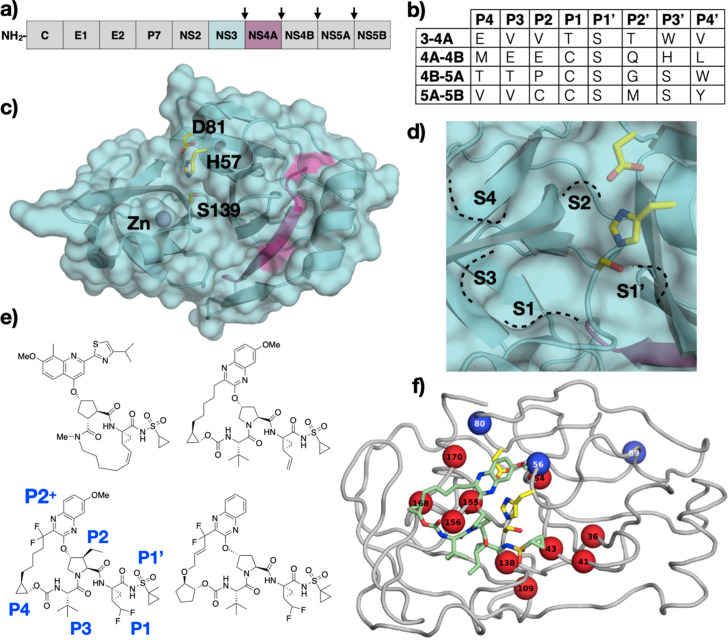
HCV NS3/4A Protease. (A) Schematic representation of HCV polyprotein, with viral protease cleavage sites between non-structural proteins (NS) indicated by arrows. (B) Amino acid sequence of polyprotein cleavage sites recognized by HCV NS3/4A protease. The protease cleaves between P1-P1’. (C) The crystal structure of HCV NS3/4A protease (PDB ID: 3M5O) with the catalytic residues depicted as yellow sticks and labeled, and (D) a close-up view of the active site with S4-S1’ pockets annotated. (E) The chemical structure of HCV NS3/4A protease inhibitors (top, left) simeprevir, (top, right) grazoprevir, (bottom, left) voxilaprevir, and (bottom, right) glecaprevir. (F) Cocrystal structure of grazoprevir (green sticks) bound to HCV NS3/4A protease, with primary (red) and secondary (blue) resistance-associated substitutions labeled.
Panel F: Reprinted with permission from A.N. Matthew, F. Leidner, G.J. Lockbaum, M. Henes, J. Zephyr, S. Hou, et al., Drug design strategies to avoid resistance in direct-acting antivirals and beyond. Chem. Rev. 121(6) (2021) 3238–3270.
© 2021 American Chemical Society.
2.2.1. HCV NS3/4A protease structure
The protease domain of NS3 is composed of 180 amino acids and has a chymotrypsin-like fold with a zinc binding site opposite to the active site that stabilizes the protease tertiary structure (Fig. 2C). A minimal domain at the C-terminus of the NS4A cofactor is sufficient to increase the activity of the NS3/4A protease. In complex with NS3, NS4A forms a β-strand that extends through the core of the NS3 protease domain, more than 10 Å away from the active site. The NS4A β-strand constitutes one of the eight β-strands that make up one of the two β-barrels of the NS3/4A protease. The Ser-His-Asp catalytic triad is located between the two β-barrels (Fig. 2D). Unlike cellular chymotrypsin-like serine proteases with similar overall folds, the HCV NS3/4A protease lacks extended surface loops leading to an active site that is relatively solvent exposed, except when the helicase domain covers the active site.
2.2.2. Substrate recognition
HCV NS3/4A protease cleaves the viral polyprotein at four sites and recognizes a ten amino acid sequence (P6-P4’), of which only three positions are conserved. All the substrates bind in a conserved β-sheet conformation in the active site with similar hydrogen bonding patterns. Furthermore, the P1 residue of the substrate, which is either a cysteine or threonine, forms van der Waals interactions with an aromatic residue (F154) of the S1 pocket. Despite only the P2 position of the NS4/NS5A cleavage site being a proline, the P2 residues at the other cleavage sites adopt a phi torsion angle to maintain a binding mode similar to that of the proline. The P6 residue of the substrate is either aspartate or glutamate and forms a salt bridge interaction with K165 in the S6 pocket or with R123 in the case of the NS4B/NS5A cleavage site [39]. The other sites in the substrate have no sequence conservation. The approach used in the HIV-1 protease to understand diverse substrate recognition was successfully applied to the HCV NS3/4A protease to determine the substrate envelope.
2.2.3. The HCV NS3/4A substrate envelope
The HCV NS3/4A protease also recognizes diverse substrate amino acid sequences in a conserved shape despite having a solvent exposed active site, further validating the substrate envelope hypothesis as a general approach to understanding diverse substrate recognition [39], [40]. Lack of cocrystal structures of the protease bound to peptide substrates spanning the non-prime and prime side limit our knowledge to only the non-prime side of the substrate. The prime side binds stronger to the protease active site where also all current inhibitors bind.
2.2.4. Enzyme catalysis
The HCV NS3/4A protease contains the canonical Ser-His-Asp catalytic triad and follows the general acyl transfer mechanism of serine proteases. Together, the catalytic histidine (H57) and aspartate (D81) increase the nucleophilicity of the serine (S139), thereby increasing the reaction rate with the scissile carbonyl carbon of the substrate [41]. The deprotonated S139 hydroxyl attacks the carbonyl carbon of the substrate, generating an acylated tetrahedral intermediate. The high-energy intermediate is stabilized by the oxyanion hole composed of backbone amide nitrogen atoms of residues 137–139 and the Nϵ of the catalytic histidine. The scissile amide nitrogen is then protonated by H57 generating a new N-terminus and leaving an ester acylated protease that is hydrolyzed by a water molecule to generate a new C-terminus and completing the catalytic cycle [42].
2.2.5. Inhibitors of HCV NS3/4A protease as drugs
The nucleophilic nature of the catalytic serine has been exploited in drug design leading to the development of the first-generation covalent HCV NS3/4A protease inhibitor drugs (telaprevir, boceprevir, and narlaprevir). The second-generation inhibitors departed from the ketoamide warhead and introduced bulky aromatic P2+ moieties that are attached via an ether linker to the P2 group (asunaprevir). Macrocyclization as a strategy to rigidify and pre-organize the inhibitors for binding gave rise to P1-P3 (simeprevir) and P2-P4 (grazoprevir) macrocyclic protease inhibitors (Fig. 2E). The latest generation inhibitors are P2-P4 macrocycles and addressed the need to treat genotype 3 (GT-3) infected patients [43]. These compounds (voxilaprevir and glecaprevir) are very similar to grazoprevir and incorporate fluorine atoms that are in part responsible for the pan-genotypic activity [44]. Only five of the twelve FDA-approved HCV NS3/4A inhibitors are currently used in the clinic primarily due to drug resistance [45].
2.2.5.1. P1-P3 macrocycles
P1-P3 macrocyclic inhibitors were designed to overcome the low barrier to resistance of the previous generation drugs [46]. The P1-P3 macrocyclic scaffold links the P1 and P3 groups that reside proximal to each other in the cocrystal structures. The P1-P3 macrocyclic protease inhibitors share the same cycloprolylsulfonamide P1’, P1, and five-membered cyclic P2 moieties found in the linear competitive inhibitors. However, the P3 tert-butyl group of the linear inhibitors is replaced with a alkyl linker that is connected to the terminal olefin of the P1 group. Simeprevir was the first macrocyclic inhibitor approved for the treatment of GT-1 and GT-4. The P1’ cyclopropyl sulfonamide group of simeprevir fits well into the S1’ pocket of the HCV NS3/4A protease. The P1-P3 macrocycle complements the groove formed by the S1 and S3 pockets. The P2+ moieties of these inhibitors adopt a binding mode similar to that of the linear competitive inhibitors, interacting with H57, D81, and R155. Simeprevir lacks a P4 group and therefore does not interact with the S4 pocket but still makes van der Waals interactions with A156 and A157 that form one side of the S4 pocket. Danoprevir, a P1-P3 macrocyclic inhibitor that contains a tert-butyl P4 group makes additional interactions with D168 in the S4 pocket.
2.2.5.2. P2-P4 macrocycles
Although the P1-P3 macrocyclic HCV NS3/4A inhibitors represented a significant improvement in potency and pharmacokinetic/pharmacodynamic properties, the lack of efficacy against the GT-3 variant and susceptibility to resistance drove the development of the P2-P4 macrocycles. The macrocycle of these inhibitors links the P2+ aromatic moiety and the P4 group. While the P2-P4 macrocycle in grazoprevir is an unsubstituted alkyl linker, that of voxilaprevir and glecaprevir contain two fluorine atoms at the benzylic position. Additionally, glecaprevir's macrocycle contains an alkene in the E conformation potentially to decrease flexibility. Voxilaprevir and glecaprevir contain a methyl substituent at the P1’ cyclosulfoaminde moiety that was present in previous generation drugs, which orients the P1’ group deeper into the S1’ pocket [47], [48]. Grazoprevir has the same terminal olefin unnatural amino acid at the P1 moiety found in the linear inhibitors. The terminal olefin of that P1 group is replaced with a difluoromethylene moiety in voxilaprevir and glecaprevir. All three P2-P4 macrocycles contain a P2 proline core bonded to a quinoxaline moiety. The quinoxaline substitution patterns differ, with grazoprevir and voxilaprevir containing a 7-OMe substituent while glecaprevir is unsubstituted at that position. All three drugs contain the identical P3 tert-butyl group found in the previous generation inhibitors, and the carbamate linkage to attach a cyclopropyl P4 group in the case of grazoprevir and voxilaprevir or cyclopentyl P4 group in the case of glecaprevir. While grazoprevir is approved against GT-1, voxilaprevir and glecaprevir are pan-genotypic inhibitors and are approved against all the genotypes (GT1–6), highlighting the biological significance of the subtle differences between these compounds [44].
These three latest inhibitors bind to the HCV NS3/4A protease in nearly identical binding poses (Fig. 2F). The P2+ quinoxaline adopts a binding pose distinct from the P2+ moieties of earlier generation inhibitors. The quinoxaline avoids contact with R155, a common resistance-associated substitution (RAS) site, and is oriented in such a way to interact primarily with invariant residues D75 and H57 of the catalytic triad (Fig. 2F). The cyclopropyl P4 group of grazoprevir and voxilaprevir binds but does not fully occupy the S4 pocket, while cyclopentyl P4 cap of glecaprevir can better fill in the S4 pocket [48]. In voxilaprevir and glecaprevir, the difluoromethylene moiety contributes fluorine-specific interactions, including two orthogonal multipolar interactions. Furthermore, the high electronegativity of fluorine compared to hydrogen promotes the hydrogen of the difluoromethylene moiety to form a “caged” fluorine-induced hydrogen bond with the backbone carbonyl oxygen of the protease and the carbonyl oxygen of the P1 group thereby pre-organizing the inhibitor for binding. These fluorine-specific interactions occur with the backbone atoms, thereby improving the potency against proteases from all genotypes, and decreasing the susceptibility to resistance [44]. The two fluorine atoms on the macrocycle of voxilaprevir and glecaprevir mainly contribute to improving metabolic stability.
2.2.6. Resistance associated substitutions (RAS) and genotypic differences
Although the chemical structure of the HCV NS3/4A inhibitors differs significantly, there are three major RASs that confer resistance to all, especially earlier generation, inhibitors (Fig. 2F). One of these RAS, R155K, leads to resistance by disrupting the electrostatic network spanning R123, D168, R155, and D81, which is important for constructing a flat binding surface for inhibitors [49], [50]. Inhibitors with bulky aromatic P2+ moieties that interact with R155 for potency are susceptible to this RAS because of the loss of a cation-π interaction with the guanidinium group [49]. The P2+ quinoxaline moiety of the P2-P4 macrocycles does not interact with R155 but instead interacts primarily with the catalytic triad. Therefore, the R155K RAS only causes subtle changes in the binding modes of grazoprevir and glecaprevir [43], [44], [51]. This unique binding mode of the quinoxaline P2+ moiety is independent of macrocyclization and underlies the improved potency of later generation inhibitors [52]. The RASs at A156 lead to resistance to all inhibitors by causing steric hindrance with either the P2, P4, or the P2-P4 macrocycle moiety. Interestingly, the capacity of the ketoamide warhead group of the first-generation inhibitors to covalently modify the protease is also compromised by the A156T RAS. The RAS and polymorphisms at D168 cause resistance by disrupting the salt bridge with R155 and thereby increasing the dynamics of the S4 pocket that accommodate the P4 group or the P2-P4 macrocycle and also impact genotypic differences [44], [51], [53], [54].
2.2.7. The HCV NS3/4A substrate envelope and resistance
The HCV NS3/4A substrate envelope explains the molecular basis of diverse substrate recognition and also serves as a tool to elucidate the susceptibility of inhibitors to resistance. As the consensus volume adopted by the four viral polyprotein substrates, the substrate envelope represents the essential recognition feature. Inhibitor-bound structures reveal areas where inhibitors protrude from the substrate envelope. The major RAS occur at A156, R155, and D168 where the inhibitors protrude beyond the envelope and contact the active site, rendering these residues more important for inhibitor binding compared to substrate recognition [21], [40], [45], [49], [52]. The P2+ aromatic group of the P1-P3 macrocyclic drugs interact outside the substrate envelope with R155 and in some cases Y56, which are residues not essential for substrate recognition [55]. Although the large aromatic P2+ quinoxaline moiety of the P2-P4 macrocyclic drugs protrude out of the substrate envelope, the interactions are limited to catalytic residues essential for enzymatic activity, D81 and H57, thereby decreasing the susceptibility to RAS at R155. The position of the macrocycle also plays a role in selecting for RASs. The P1-P3 macrocycle resides in the S1 and S3 pockets and fits within the substrate envelope. In contrast, the P2-P4 macrocycle protrudes out to contact A156, and selects for RAS primarily at this position and D168 (sometimes in combination with Y56) [55]. Inhibitors have been designed to decrease the susceptibility to resistance due to the P2-P4 macrocycle by relocating the macrocycle at the P1-P3 position while maintaining the unique binding mode of the P2+ quinoxaline moiety [43]. These P1-P3 macrocyclic inhibitors maintained potency and exhibited a flatter resistance profile compared to the P2-P4 macrocyclic analogs [48], [56].
3. Viral proteases as emerging therapeutic targets
The successful targeting of the HIV-1 protease established viral proteases as therapeutic targets and paved the way to the relatively rapid success in developing potent pan-genotypic inhibitors of the HCV NS3/4A protease. Other viruses of global health concern that are dependent on a viral protease for replication are also potentially tractable therapeutic targets. These emerging targets include proteases from the human T-cell leukemia virus type 1 (HTLV-1), flaviviruses, enteroviruses, and coronaviruses.
3.1. HTLV-1 protease
HTLV is a retrovirus, belonging to the Deltaretrovirus genus. HTLV-1 is associated with various diseases including adult T-cell leukemia lymphoma (ATL) and HTLV-1-associated myelopathy. HTLV was the first retrovirus to be identified and currently, an estimated 20 million people are infected worldwide. ATL caused by HTLV-1 has poor prognosis and is resistant to conventional and high-dose chemotherapy [57]. Unfortunately, there are no vaccines or direct-acting antivirals against HTLV-1. Similar to the replication of other retroviruses, the HTLV-1 virus genome encodes an aspartyl protease that is critical for polyprotein processing and viral maturation (Fig. 3A and B) [58]. Given the similarities in structure and function to the HIV-1 protease, the HTLV-1 protease can be a primary target for drug design. Certain aspects of the HTLV-1 protease have been previously reviewed [59], [60]; here we will provide an overview of the protease structure and substrate recognition, with recent advances in inhibitor development.
Fig. 3.
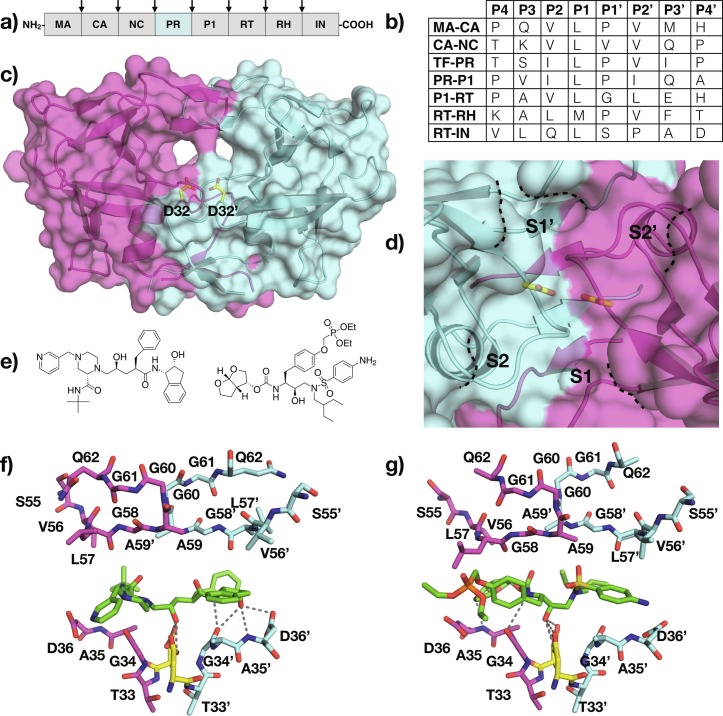
HTLV-1 Protease. (A) Schematic representation of HTLV-1 polyprotein, with viral protease cleavage sites indicated by arrows. (B) Amino acid sequence of polyprotein cleavage sites. The protease cleaves between P1-P1’. (C) The crystal structure of HTLV-1 protease (PDB ID: 6W6Q) with the catalytic residues depicted as yellow sticks and labeled, and (D) a close-up view of the active site with S2-S2’ pockets annotated. (E) (left) Indinavir and (right) PU6 (e), with their respective cocrystal structures bound to the HTLV-1 protease (PDB ID: 3WSJ and 6W6S) (F and G).
3.1.1. HTLV-1 protease structure
The HTLV-1 protease is a homodimeric aspartyl protease, similar to the HIV-1 protease (Fig. 3C). However, the HTLV-1 protease is larger with each monomer comprising of 125 amino acids instead of 99. The additional amino acids, relative to the HIV-1 protease, are located at the C-terminus and are a distinct feature of the HTLV-1 protease compared to other viral aspartyl proteases. The crystal structure of HTLV-1 protease revealed a very similar fold to that of the HIV-1 protease. Both proteases share a conserved core structure around the active site and at the dimerization interface, with structural divergences in the loops around the active site and the flaps [59]. A unique aspect of the HTLV-1 protease is an additional two residues in the flap regions relative to that of HIV-1 that increases the helical conformation of the flap regions [61].
The dimeric structure of HTLV-1 protease is held together by four-stranded antiparallel β sheets that involve the N and C termini of each monomer. The active site is located at the cleft region above the dimer interface where one aspartate from each monomer, positioned at the base of the cleft, forms the catalytic dyad. Each monomer contributes a β turn that together constitute the flaps and enclose the active site [59]. The additional residues at the flap region create a more extended substrate-binding site compared to that of the HIV-1 protease [58].
3.1.2. Substrate recognition
The substrates of HTLV-1 protease are highly hydrophobic and diverse in sequence, except for the P1 position where there is a preference for leucine (Fig. 3B). The side chain interactions confer specificity while backbone interactions properly orient the substrate in the active site [59]. The strongest interactions between the protease and substrates occurs within S3-S3’ pockets [59]. There are no substrate-bound structures of the HTLV-1 protease available to date. However, the cocrystal structure of the protease bound to a statine-containing peptide provides insights into substrate recognition. Each carbonyl oxygen and amide group of the peptidic inhibitor participates in direct hydrogen bond interaction with the enzyme in a manner identical to other viral aspartyl proteases [61]. The HTLV-1 protease can accommodate larger residues in S1/S1’ and S2/S2’ pockets relative to the HIV-1 protease (Fig. 3D). The flaps also play a crucial role in dictating substrate specificity although hydrogen bonding interactions with the interior of the protease are stronger compared to that of the flaps. The HTLV-1 S3/S3’ pockets differ significantly compared to those of HIV-1 protease. The S3 pocket of the HTLV-1 protease is partially solvent exposed; consequently, P3 of the substrates can either be hydrophobic or hydrophilic. The S4/S4’subsites of HTLV-1 are larger and more hydrophobic compared to those of the HIV-1 protease, consistent with the observed substrate preference for more hydrophobic P4 residues. Overall, substrate-binding pockets more distal from the catalytic site are more divergent between the two proteases.
Unlike HIV-1, which utilizes both viral and host molecular machinery to replicate, HTLV-1 is thought to replicate primarily in the DNA form and therefore has to force the host cell to divide [61], [62]. This aspect of the HTLV-1 life cycle does not require viral machinery for transcription, which tends to be error-prone; therefore, the HTLV-1 sequence is more conserved compared to that of HIV-1. This implies that HTLV-1 protease will be less likely to evolve under drug pressure to develop resistance.
3.1.3. Inhibitors of HTLV-1 protease
The current treatment options for ATL caused by HTLV-1, including chemotherapy and antibodies against the interleukin 2 receptor, offer limited success. Inhibitors targeting the HTLV-1 protease provide an attractive novel approach for developing therapeutics. The search for a drug targeting the HTLV-1 protease is in very early stages. The two main strategies being investigated are designing peptidomimetic inhibitors based on the substrate sequence, and repurposing HIV-1 protease inhibitors. Of the HTLV-1 protease peptidomimetic inhibitors reported, statine-containing peptides have reached nanomolar potencies [63], [64]. The similarity (28% overall and 45% in the active site) between HTLV-1 and HIV-1 proteases serves as the rationale to leverage FDA-approved HIV-1 protease inhibitors against the HTLV-1 protease. Three of these inhibitors, ritonavir, indinavir, and darunavir were found to inhibit HTLV-1 protease with varying effectiveness (Fig. 3E). These three peptidomimetic inhibitors contain hydrophobic P1/P1’ moieties and a secondary hydroxyl group as a transition state mimetic. Ritonavir, with a K i > 20 μM was found to induced apoptosis in HTLV-1 infected cells [57]. Indinavir weakly inhibits the HTLV-1 protease with a K i of 3.5 μM [65]. A cocrystal structure of indinavir bound to the HTLV-1 protease reveals that the central hydroxyl of indinavir contacts both catalytic aspartates and occupies S3-S3’ pocket (Fig. 3F) [66]. However, the larger HTLV-1 protease active site prevented optimal packing with the flap residues. DRV, a picomolar inhibitor of the HIV-1 protease, has a K i of about 1 μM against the HTLV-1 protease [67]. Cocrystal structures of DRV bound to HIV-1 and HTLV-1 proteases revealed a similar binding mode. Given the larger binding pockets of HTLV-1 protease, DRV analogues with bulkier substitutions were also investigated. A DRV analog, UM6, containing a bulky and hydrophobic P1’ moiety was 10-fold more potent against HTLV-1 protease compared to DRV. Moreover, substituting a diethylphosphonate group at the P1 phenyl group to generate inhibitor PU6, led to further improvement in the potency to a K i of 0.03 μM. The cocrystal structure of PU6 bound to the HTLV-1 protease revealed that the phosphonate moiety extends into the S1 pocket and increases van der Waals contacts with the protease (Fig. 3G). UM6 and PU6 were also effective at inhibiting Gag processing in biochemical assays and maturation of virus-like particles [67]. The cocrystal structures of DRV and analogs bound to HTLV-1 protease provide a promising path toward designing potent HTLV-1 protease inhibitors as therapeutics.
3.2. Flavivirus proteases
The flavivirus genus contains over 70 mosquito-borne viruses, including human pathogens such as Zika (ZIKV), dengue (DENV), and West Nile (WNV) virus [68]. Viruses within this genus are highly similar, with 56% sequence identity between ZIKV and dengue serotype 2 (DENV-2) [69]. The ZIKV outbreak in 2015 raised public concern due to sexual and maternal-fetal mode of transmission and infections causing neurological disorders, especially microcephaly in neonates [70], [71]. Meanwhile, the persistent DENV infections have been a global health concern since the 1950s [72]. Hemorrhagic fever is the most severe pathology of DENV-infected patients [73]. Similar to other positive-strand RNA viruses, flaviviruses encode a viral protease that processes the viral polyprotein at multiple sites. The flaviviral proteases share a high level of sequence identity (~ 30–80%) and are composed of two viral non-structural proteins, the NS2B cofactor and the NS3 protease (Fig. 4A) [74]. Flaviviral proteases require an Arg/Lys at the P1 and P2 positions of the substrate (Fig. 4B), which greatly influences chemical features of peptidomimetic inhibitors that are being developed (Fig. 4B and C). We will focus on the viral proteases from DENV and ZIKV, the two viruses that have caused the most health burden within the genus.
Fig. 4.
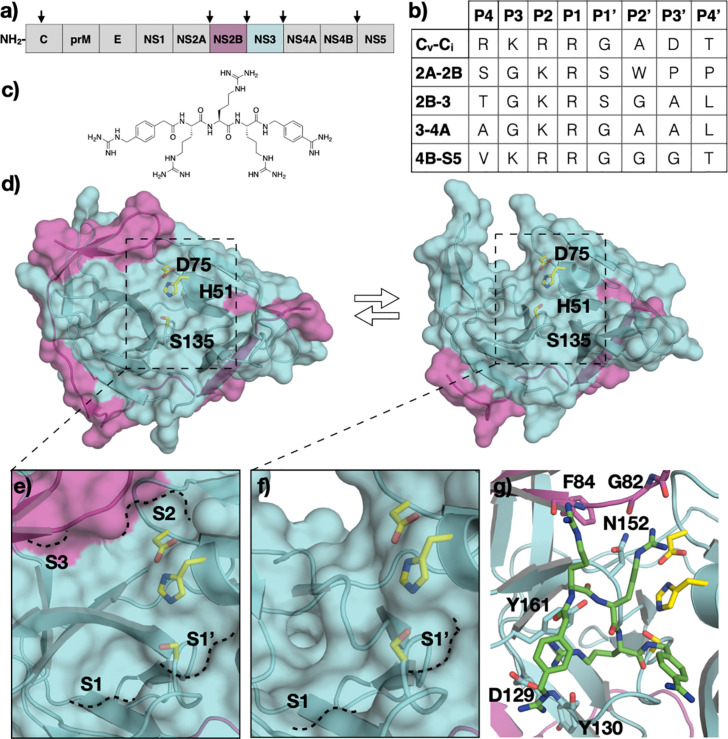
Flavivirus Proteases. (A) Schematic representation of flavivirus polyprotein, with viral protease cleavage sites indicated by arrows (B) Amino acid sequence of polyprotein cleavage sites. The protease cleaves between P1-P1’. (C) The chemical structure of flaviviral protease inhibitor, compound 1. (D) The crystal structure of Zika virus (ZIKV) NS2B/3 protease in the (left) closed and (right) open conformation (PDB ID: 5LC0 and 5GXJ), with the catalytic residues labeled. (E) The active site topology of the closed (E) and open (F) conformation with S3-S1’ pockets annotated. (G) The cocrystal structure of compound 1 bound to the ZIKV NS2B/3 protease in the closed conformation.
3.3. Structure of flavivirus NS2B/NS3 protease
The flavivirus NS2B/NS3 protease is a chymotrypsin-like viral protease, with a canonical Ser-His-Asp catalytic triad (Fig. 4D). Similar to HCV, the NS3 of flaviviruses contains the protease domain at the N-terminus and a helicase domain at the C-terminus. However, unlike HCV NS3 protease, the flaviviral NS3 protease is inactive without the NS2B cofactor, which stabilizes the NS3 protease structure and is essential for proper folding [75], [76], [77], [78]. Crystal structures of flaviviral proteases revealed that the NS2B is dynamic and can adapt an open or closed conformation. In the open conformation, only one β strand portion of the NS2B interacts with the NS3 protease, while the rest of the NS2B is disordered. In the closed conformation, the NS2B fully wraps around NS3 protease domain and forms an additional β turn that contributes residues to construct part of the active site S2 pocket and interact with P2 of the substrates. In the open conformation, the S2 pocket is not present; therefore, the protease cannot stably bind substrates (Fig. 4E and F). The active site of the flaviviral proteases are relatively flat and solvent exposed. Upon substrate binding, the reaction mechanism of flaviviral proteases follows that of the general chymotrypsin-like serine protease family, similar to that of the HCV NS3/4A protease.
3.3.1. Substrate recognition
The flaviviral polyprotein contains transmembrane domains that insert the polyprotein into the ER membrane as translation by the host ribosomes takes place. The NS2B/NS3 protease is responsible for processing all the cleavage sites on the cytosolic side of the polyprotein, including NS2A/NS2B, NS2B/NS3, NS3/NS4A, and NS4B/NS5 (5, 6). The binding mode of peptides corresponding to the non-prime side of the substrates has been revealed through a crystal structure of ZIKV protease caught in cis cleavage and other cocrystal structures with covalent peptidic inhibitors [79]. The C-terminus generated from scissile bond cleavage is stabilized in the oxyanion hole through backbone hydrogen bonding interactions. The P1 arginine extends into the S1 pocket and forms a salt bridge with Asp129 at the base of the pocket. The P1 guanidinium group also participates in hydrogen bonding interaction with the backbone carbonyl of Tyr130, and forms cation-π interaction with Tyr161. In the S2 pocket, the positive charge of P2 lysine is stabilized by side chain of Asp75 of the catalytic triad and Asp83 of the NS2B cofactor. In the cis cleavage structure, the P3 glycine residue adopts a rotamer that orients the P4 and P5 residues toward solvent. Therefore, this structure does not provide information on interactions at S3 and S4 pockets. The cocrystal structure with bound compound 1 revealed that the P3 arginine makes polar contacts with the carbonyl of F84 of NS3 and the sidechain of S85 of NS2B (Fig. 4C and G). The P4 group of compound 1 does not extend into the S4 pocket but instead is solvent exposed, with the compound binding in a cyclic conformation [80].
3.3.2. Inhibitors of flavivirus proteases
Given the overall high similarities between flaviviral proteases, inhibitors identified against one flaviviral protease should be generally effective against others, and support the ultimate aim of discovering pan-flaviviral protease inhibitors. Despite significant efforts, none of the flaviviral protease inhibitors have yet reached the clinical stage. The flat and highly negatively charged active site led to the design of inhibitors with poor cellular activity and pharmacokinetics [81], [82], [83]. The traditional approach of designing inhibitors against serine proteases, which involves the addition of electrophilic warhead groups to the non-prime side of the substrate, led to potent inhibitors with poor selectivity, cellular activity, and pharmacokinetics [84]. The initial peptidomimetic inhibitors led to the development of compound 1 (K i = 0.43 μM) which showcased potency could be achieved without a warhead group; however, with five guanidinium moieties compound 1 does not have desired drug-like properties [80]. The macrocyclization drug design strategy, which has proven successful in inhibiting the HCV NS3/4A protease, was applied to the ZIKV protease with limited success [85]. Non-peptidic and small aromatic active site inhibitors with low levels of potency have also been identified [86]. Other methods of identifying inhibitors such as high-throughput screening and fragment-based drug design have also been investigated with limited success [87], [88], [89]. There are several reviews available on the inhibition of flaviviral proteases [74], [81], [84], [90], [91]. The binding loop of aprotinin, a general protease inhibitor that is also active against flaviviral proteases, has been used as the basis for cyclic peptidic inhibitors, and to understand the contribution of prime side recognition to potency [92], [93]. Although there is significant structural information on the binding mode of the peptidomimetic inhibitors, the bias toward cationic scaffolds limits the progress of drug design to the clinic. To overcome this limitation, allosteric inhibitors have also been investigated, with reports of inhibitors aiming to interfere with the NS3 and NS2B binding interface [90], [94], [95], [96], [97], [98], [99], [100], [101]. However, there is no clear evidence to validate the binding mode of these inhibitors. Drug repurposing as an alternative approach has also been investigated. Specifically, efforts targeting the ZIKV protease have led to the identification of micromolar inhibitors, including bromocriptine, temoporfin, and hydroxychloroquine [89], [102], [103]. Similar to other allosteric inhibitors identified, detailed characterization of inhibitor binding mode is lacking, but is essential for structure-based drug design and to guide medicinal chemistry efforts.
3.4. Enterovirus proteases
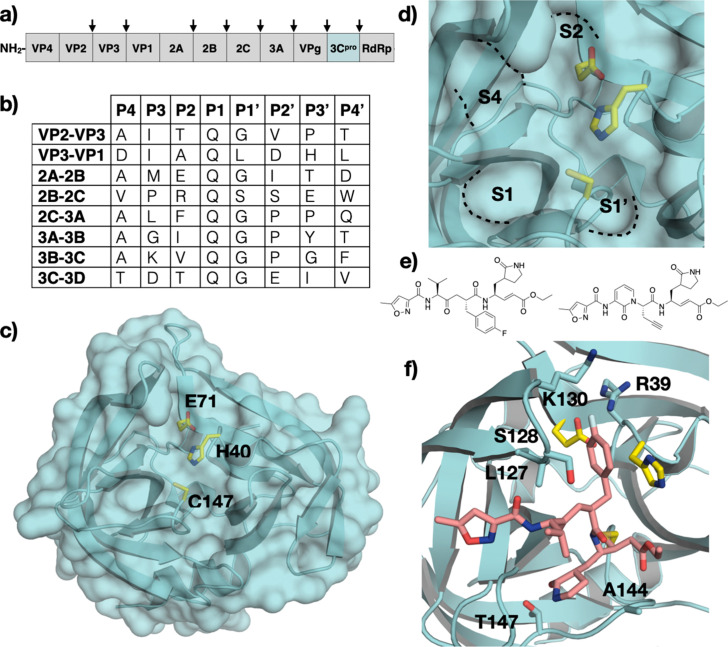
The genus Enterovirus (EV) of the Picornaviridae family includes poliovirus, numbered rhinoviruses and enteroviruses. Poliovirus is the best-known and one of the most studied enterovirus for causing the debilitating poliomyelitis disease [104]. Enteroviruses that do not cause poliomyelitis are classified as non-polio enteroviruses (NPEV), many of which are important human pathogens. EV-A71 and EV-D68 have recently emerged as serious public health threats. EV-A71 causes hand-foot-and-mouth disease (HFMD) with life-threatening neurological complications, such as brainstem encephalitis. EV-D68 can cause mild respiratory disorders as well as more severe symptoms such as acute flaccid paralysis in children [105], [106]. Human rhinoviruses are the predominant agents responsible for the common cold [107]. Two EV-A71 inactivated enteroviruses are approved in China as vaccines and represent the only preventive measure against NPEV. Enterovirus has a positive-strand RNA genome that is translated into a single large polyprotein to be proteolytically processed into ten structural and replication proteins by two virally encoded proteases, 2Apro and 3Cpro (Fig. 5A and B). Inhibition of these viral proteases represents a tractable approach to treat enterovirus infections, as both proteases play crucial roles in polyprotein processing: 2Apro cleaves only one site on the polyprotein while the 3Cpro cleaves all the other sites. The 3Cpro is a highly conserved cysteine protease among enteroviruses possessing a non-canonical Cys-His-Glu catalytic triad.
Fig. 5.
Enterovirus 3C Protease (3Cpro). (A) Schematic representation of the enterovirus polyprotein, with 3Cpro cleavage sites indicated by arrows. (B) Amino acid sequence of polyprotein cleavage sites. The protease cleaves between P1-P1’. (C) The crystal structure of the EV-A71 3Cpro (PDB ID: 7DNC) with the catalytic residues labeled, and (D) a close-up view of the active site with S4-S1’ pockets annotated. (E) Chemical structures of rupintrivir and AG7404. (F) Cocrystal structure of rupintrivir bound to EV-A71 3Cpro (PDB ID: 3SJO).
3.4.1. Structure of enterovirus 3Cpro proteases
Enterovirus 3Cpro proteases belong to the chymotrypsin-related endopeptidase protease family and are biologically active as monomers [108], [109]. The catalytic triad and the surrounding residues of 3Cpro are highly conserved among the Enterovirus genus. The EV-A71 3Cpro consists of two twisted β-ribbon folds packed in a perpendicular orientation (Fig. 5C). The substrate-binding pocket lies at the groove between the two β-ribbon folds. The catalytic cysteine in the active site acts as a nucleophile and attacks the carbonyl carbon of the scissile bond following a mechanism similar to that of the HCV NS3/4A serine protease.
3.4.2. Substrate recognition
Enterovirus 3Cpro cleaves the viral polyprotein at eight sites, each constituting a unique P4-P4’ sequence with a strong preference for glutamine at the P1 position and glycine at the P1’ position (Fig. 5B). The other positions within the P4 to P4’ substrate can accommodate diverse amino acids. The interactions at S3-S1’ pockets of the protease contribute most to substrate binding. There are no available crystal structure of enterovirus 3Cpro bound to a natural substrate peptide. However, cocrystal structures of EV-D68 3Cpro bound to peptidomimetic inhibitors provide insight into the molecular basis of substrate recognition [110], [111]. The S2 pocket is hydrophobic, hence the preference for hydrophobic P2 moieties. The EV-D68 3Cpro, similar to other homologous viral proteases, does not possess an S3 pocket which leads to a diverse set of amino acids at the P3 position of the substrates. The S4 pocket is very shallow and can accommodate only small residues (Fig. 5D).
3.4.3. Inhibitors of enterovirus 3Cpro proteases
There are no FDA-approved inhibitors targeting the enterovirus 3Cpro. Rupintrivir, a peptidomimetic inhibitor of rhinovirus 3Cpro and an analogue (AG7404) have advanced to clinical trials, but failed due to limited efficacy (Fig. 5E) [112], [113]. Cocrystal structures of rupintrivir bound to the EV-A71 3Cpro reveal the structural basis for antiviral activity [114], [115]. The warhead group of rupintrivir forms a covalent bond with nucleophilic C147 and the rupintrivir scaffold binds via an extensive set of hydrogen bonds and hydrophobic interactions that span the S1’-S4 pockets (Fig. 5F). The majority of 3Cpro inhibitors are of similar peptidomimetic scaffolds, although non-peptidomimetic inhibitors have also been investigated [116].
3.5. Coronavirus proteases
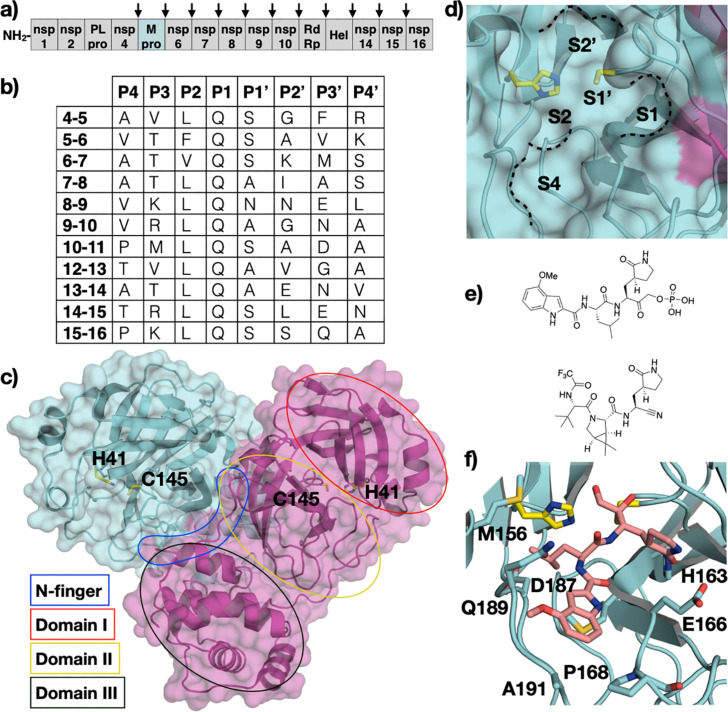
The coronavirus disease 2019 (COVID-19) pandemic is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [117]. Over 170 million confirmed infection cases have been reported worldwide as of mid-2021. The 2019 pandemic marks the third coronavirus outbreak in the 21st century [118], [119]. SARS-CoV, Middle East respiratory syndrome (MERS-CoV), and SARS-CoV-2 belong to the Coronaviridae family under the Betacoronavirus genus. Similar to the other positive-sense single-stranded RNA viruses discussed in this chapter, coronaviruses encode viral proteases, main protease (Mpro, also known as 3CLpro, or nsp5), and papain-like protease (PLpro, also known as nsp3), to cleave the viral polyprotein [120]. Mpro is seen as a more tractable therapeutic target because of its significant role in polyprotein processing (Fig. 6A and B) [121]. Coronaviruses share a high overall sequence identity. Consequently, Mpro from SARS-CoV-1 and SARS-CoV-2 have ~ 96% sequence identity [122], [123]. Here we will focus on the structure, mechanism, and inhibition of Mpro from SARS-CoV-2.
Fig. 6.
Coronavirus Main Protease (Mpro). (A) Schematic representation of the coronovirus polyprotein, with Mpro cleavage sites indicated by arrows. (B) Amino acid sequence of polyprotein cleavage sites. The protease cleaves between P1-P1’. (C) The crystal structure of SARS-CoV-2 Mpro homodimer (PDB ID: 7KHP) with the catalytic residues labeled and the N-finger, domains I, II, and III colored accordingly on one of the monomers, and (D) a close-up focused view of the active site with S4-S2’ pockets annotated. (E) Chemical structures of Mpro inhibitors PF-07304814 and PF-07321332. (F) Cocrystal structure of PF-00835231 bound to SARS-CoV-2 Mpro (PDB ID: 6XHM).
3.5.1. Structure of coronavirus main protease
Mpro is a 306 residue protease with a catalytic dyad consisting of cysteine and histidine. The biologically active state of Mpro is dimeric, where two monomers are arranged in an orthogonal fashion (Fig. 6C) [124]. Each monomer is made up of three domains; domain I and domain II together construct two anti β-parallel barrels with a fold similar to that of trypsin-like serine proteases, and domain III contains five ⍺-helices that are connected via a linker to domain II. Dimerization of the protomers occurs at the N-terminus (N-finger) located between domains II and III. The N-finger also contributes to the formation of the substrate-binding site between domains I and II [125], [126].
3.5.2. Substrate recognition
In addition to the catalytic dyad composed of H47 and C145, Mpro contains a buried water molecule in the active site that substitutes for the aspartate residue that usually accompany cysteine and histidine in the canonical catalytic triad of other proteases (Fig. 6D) [121], [127]. The catalytic process follows a multi-step mechanism, where H47 deprotonates the catalytic cysteine generating a nucleophilic thiolate that attacks the carbonyl carbon of the scissile bond and forms a tetrahedral intermediate. The tetrahedral intermediate collapses leaving an acylated intermediate. The histidine activates a water molecule that hydrolyzes the intermediate regenerating the thiol sidechain [128], [129]. Mpro cleaves the viral polyproteins (pp1a and pp1ab) at 11 sites (Fig. 6B). The amino acid sequences at the cleavage sites are diverse, except at P1 which is always glutamine, and P2 which has a preference for leucine. The P3 position can accommodate hydrophobic and hydrophilic residues, and P4 has a preference for amino acids with small sidechains. The prime side of the cleavage sites lacks sequence conservation except at P1’ where there is a preference for small amino acids. Mpro is expected to recognize substrates spanning P4-P1’ based on peptide substrate analysis [130].
An acyl-enzyme intermediate crystal structure of SARS-CoV-2 Mpro bound to the C-terminal autoproteolysis product (nsp5-nsp6) provides details of non-prime side substrate recognition [131]. Cocrystals structures of SARS-CoV-1 Mpro bound to an 11-mer peptide (TSAVLQSGFRK) representing the nsp4-nsp5 elucidated the interactions that drive recognition of the prime side [132]. Comparing apo and substrate-bound structures revealed an induced-fit binding mechanism, where the width of the binding groove increases to accommodate the side chains of the substrates to bind in an extended β-sheet conformation. The backbone nitrogen of C145 and G143 stabilize the acyl-enzyme tetrahedral intermediate. The P1 glutamine side chain is stabilized through hydrogen bonding with polar sidechains in the S1 pocket. The hydrophobic P2 group forms packing interactions with M165 in the deep S2 pocket. The P3 residue is partially solvent exposed, which may explain the lack of specificity at this position. The shallow S1’ pocket explains the preference for small amino acids at the P1’ position of the substrates. The S2’ pocket is long and deep, explaining the diverse amino acids seen at the P2’ position of the substrates.
3.5.3. Inhibitors of coronavirus main protease
Inhibitors of SARS-CoV-1 and -2 Mpro have been reviewed extensively [123], [128], [133], [134]. Peptidomimetics and small molecules that contain Michael acceptors such as aldehydes and ketones make a significant portion of the designed inhibitors. A common feature of these inhibitors is a P1 γ-lactam group as a glutamine mimetic. Two dipeptidyl inhibitors, PF-07304814 and PF-07321332, are entering phase I clinical trials (NCT04535167 and NCT04756531) (Fig. 6E) [135], [136]. PF-07304814, the prodrug form of PF-00835231, contains a phosphonate group for better solubility that gets cleaved by alkaline phosphatase enzymes in tissue. The orally available PF-07321332 was designed to overcome the intravenous method of delivery for PF-07304814. In addition, extensive high-throughput screening (HTS) of FDA-approved drugs and fragment-based libraries has been investigated against Mpro [133]. Notably, boceprevir, an FDA-approved HCV NS3/4A protease inhibitor, was found to inhibits SARS-CoV-2 Mpro with single digit IC50 value [133]. The rapid discovery of SARS-CoV-2 Mpro inhibitors as clinical candidates surpass that of other viral protease drug targets, and underlies the importance of understanding the structure and mechanism of viral proteases.
4. Conclusion
Viruses utilize encoded proteases as a general strategy to survive with a compact genome. The viral proteases are essential to replication and the viral life cycle. Proteases of various viral families are diverse in structure, catalytic mechanism, and substrate preference. The success in targeting the proteases of HIV-1 and HCV proved that viral proteases are tractable drug targets, and simultaneously revealed significance of drug resistance as one of the major limitations of current approaches in drug design against rapidly evolving targets. The substrate envelope model provides a tool to aid in overcoming this limitation. The strategies and lessons of the past will be valuable in developing protease inhibitors as drugs against emerging viral therapeutic targets.
Acknowledgments
Research in the Schiffer lab on viral proteases is supported by NIH grants R01-GM135919, R21-AI149716, and R01-AI085051. JZ is supported by F3-GM131635.
References
- 1.Ghosh A.K., Osswald H.L., Prato G. Recent progress in the development of HIV-1 protease inhibitors for the treatment of HIV/AIDS. J. Med. Chem. 2016;59(11):5172–5208. doi: 10.1021/acs.jmedchem.5b01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brik A., Wong C.H. HIV-1 protease: mechanism and drug discovery. Org. Biomol. Chem. 2003;1(1):5–14. doi: 10.1039/b208248a. [DOI] [PubMed] [Google Scholar]
- 3.Anderson J., Schiffer C., Lee S.K., Swanstrom R. Viral protease inhibitors. Handb. Exp. Pharmacol. 2009;189:85–110. doi: 10.1007/978-3-540-79086-0_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prabu-Jeyabalan M., Nalivaika E., Schiffer C.A. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure. 2002;10(3):369–381. doi: 10.1016/s0969-2126(02)00720-7. [DOI] [PubMed] [Google Scholar]
- 5.Chellappan S., Kairys V., Fernandes M.X., Schiffer C., Gilson M.K. Evaluation of the substrate envelope hypothesis for inhibitors of HIV-1 protease. Proteins. 2007;68(2):561–567. doi: 10.1002/prot.21431. [DOI] [PubMed] [Google Scholar]
- 6.Ozen A., Haliloglu T., Schiffer C.A. Dynamics of preferential substrate recognition in HIV-1 protease: redefining the substrate envelope. J. Mol. Biol. 2011;410(4):726–744. doi: 10.1016/j.jmb.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leidner F., Kurt Yilmaz N., Schiffer C.A. Deciphering complex mechanisms of resistance and loss of potency through coupled molecular dynamics and machine learning. J. Chem. Theory Comput. 2021;17(4):2054–2064. doi: 10.1021/acs.jctc.0c01244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Özen A., Schiffer C.A. Structural Biology in Drug Discovery. 2020. Integrating evolution of drug resistance into drug discovery; pp. 521–543. [Google Scholar]
- 9.Henes M., Kosovrasti K., Lockbaum G.J., Leidner F., Nachum G.S., Nalivaika E.A., Bolon D.N.A., Kurt Yilmaz N., Schiffer C.A., Whitfield T.W. Molecular determinants of epistasis in HIV-1 protease: elucidating the interdependence of L89V and L90M mutations in resistance. Biochemistry. 2019;58(35):3711–3726. doi: 10.1021/acs.biochem.9b00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henes M., Lockbaum G.J., Kosovrasti K., Leidner F., Nachum G.S., Nalivaika E.A., Lee S.K., Spielvogel E., Zhou S., Swanstrom R., Bolon D.N.A., Kurt Yilmaz N., Schiffer C.A. Picomolar to micromolar: elucidating the role of distal mutations in HIV-1 protease in conferring drug resistance. ACS Chem. Biol. 2019;14(11):2441–2452. doi: 10.1021/acschembio.9b00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishima R., Kurt Yilmaz N., Schiffer C.A. NMR and MD studies combined to elucidate inhibitor and water interactions of HIV-1 protease and their modulations with resistance mutations. J. Biomol. NMR. 2019;73(6–7):365–374. doi: 10.1007/s10858-019-00260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leidner F., Kurt Yilmaz N., Paulsen J., Muller Y.A., Schiffer C.A. Hydration structure and dynamics of inhibitor-bound HIV-1 protease. J. Chem. Theory Comput. 2018;14(5):2784–2796. doi: 10.1021/acs.jctc.8b00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ragland D.A., Nalivaika E.A., Nalam M.N., Prachanronarong K.L., Cao H., Bandaranayake R.M., Cai Y., Kurt-Yilmaz N., Schiffer C.A. Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease. J. Am. Chem. Soc. 2014;136(34):11956–11963. doi: 10.1021/ja504096m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Y., Myint W., Paulsen J.L., Schiffer C.A., Ishima R., Kurt Yilmaz N. Drug resistance mutations Alter dynamics of inhibitor-bound HIV-1 protease. J Chem Theory Comput. 2014;10(8):3438–3448. doi: 10.1021/ct4010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foulkes-Murzycki J.E., Rosi C., Kurt Yilmaz N., Shafer R.W., Schiffer C.A. Cooperative effects of drug-resistance mutations in the flap region of HIV-1 protease. ACS Chem. Biol. 2013;8(3):513–518. doi: 10.1021/cb3006193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Y., Yilmaz N.K., Myint W., Ishima R., Schiffer C.A. Differential flap dynamics in wild-type and a drug resistant variant of HIV-1 protease revealed by molecular dynamics and NMR relaxation. J. Chem. Theory Comput. 2012;8(10):3452–3462. doi: 10.1021/ct300076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King N.M., Prabu-Jeyabalan M., Bandaranayake R.M., Nalam M.N., Nalivaika E.A., Ozen A., Haliloglu T., Yilmaz N.K., Schiffer C.A. Extreme entropy-enthalpy compensation in a drug-resistant variant of HIV-1 protease. ACS Chem. Biol. 2012;7(9):1536–1546. doi: 10.1021/cb300191k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai Y., Schiffer C. In: Computational Drug Discovery and Design. Baron R., editor. Springer New York; New York, NY: 2012. Decomposing the energetic impact of drug-resistant mutations: the example of HIV-1 protease–DRV binding; pp. 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ali A., Bandaranayake R.M., Cai Y., King N.M., Kolli M., Mittal S., Murzycki J.F., Nalam M.N., Nalivaika E.A., Ozen A., Prabu-Jeyabalan M.M., Thayer K., Schiffer C.A. Molecular basis for drug resistance in HIV-1 protease. Viruses. 2010;2(11):2509–2535. doi: 10.3390/v2112509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ragland D.A., Whitfield T.W., Lee S.K., Swanstrom R., Zeldovich K.B., Kurt-Yilmaz N., Schiffer C.A. Elucidating the interdependence of drug resistance from combinations of mutations. J. Chem. Theory Comput. 2017;13(11):5671–5682. doi: 10.1021/acs.jctc.7b00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King N.M., Prabu-Jeyabalan M., Nalivaika E.A., Schiffer C.A. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem. Biol. 2004;11(10):1333–1338. doi: 10.1016/j.chembiol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Matthew A.N., Leidner F., Lockbaum G.J., Henes M., Zephyr J., Hou S., Rao D.N., Timm J., Rusere L.N., Ragland D.A., Paulsen J.L., Prachanronarong K., Soumana D.I., Nalivaika E.A., Kurt Yilmaz N., Ali A., Schiffer C.A. Drug design strategies to avoid resistance in direct-acting antivirals and beyond. Chem. Rev. 2021;121(6):3238–3270. doi: 10.1021/acs.chemrev.0c00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yilmaz N.K., Schiffer C.A. In: Antimicrobial Drug Resistance: Mechanisms of Drug Resistance, Volume 1. Mayers D.L., et al., editors. Springer International Publishing; Cham: 2017. Drug resistance to HIV-1 protease inhibitors: molecular mechanisms and substrate coevolution; pp. 535–544. [Google Scholar]
- 24.Kurt Yilmaz N., Swanstrom R., Schiffer C.A. Improving viral protease inhibitors to counter drug resistance. Trends Microbiol. 2016;24(7):547–557. doi: 10.1016/j.tim.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Özen A., Schiffer C.A. In: Handbook of Antimicrobial Resistance. Berghuis A., et al., editors. 149-173. Springer New York; New York, NY: 2017. Substrate-envelope-guided design of drugs with a high barrier to the evolution of resistance. [Google Scholar]
- 26.Nalam M.N., Ali A., Altman M.D., Reddy G.S., Chellappan S., Kairys V., Ozen A., Cao H., Gilson M.K., Tidor B., Rana T.M., Schiffer C.A. Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance. J. Virol. 2010;84(10):5368–5378. doi: 10.1128/JVI.02531-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rusere L.N., Lockbaum G.J., Henes M., Lee S.K., Spielvogel E., Rao D.N., Kosovrasti K., Nalivaika E.A., Swanstrom R., Kurt Yilmaz N., Schiffer C.A., Ali A. Structural analysis of potent hybrid HIV-1 protease inhibitors containing Bis-tetrahydrofuran in a Pseudosymmetric dipeptide Isostere. J. Med. Chem. 2020;63(15):8296–8313. doi: 10.1021/acs.jmedchem.0c00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rusere L.N., Lockbaum G.J., Lee S.K., Henes M., Kosovrasti K., Spielvogel E., Nalivaika E.A., Swanstrom R., Yilmaz N.K., Schiffer C.A., Ali A. HIV-1 protease inhibitors incorporating Stereochemically defined P2' ligands to optimize hydrogen bonding in the substrate envelope. J. Med. Chem. 2019;62(17):8062–8079. doi: 10.1021/acs.jmedchem.9b00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lockbaum G.J., Leidner F., Rusere L.N., Henes M., Kosovrasti K., Nachum G.S., Nalivaika E.A., Ali A., Yilmaz N.K., Schiffer C.A. Structural adaptation of Darunavir analogues against primary mutations in HIV-1 protease. ACS Infect. Dis. 2019;5(2):316–325. doi: 10.1021/acsinfecdis.8b00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nalam M.N., Ali A., Reddy G.S., Cao H., Anjum S.G., Altman M.D., Yilmaz N.K., Tidor B., Rana T.M., Schiffer C.A. Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance. Chem. Biol. 2013;20(9):1116–1124. doi: 10.1016/j.chembiol.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindenbach B.D., Rice C.M. Molecular biology of flaviviruses. Adv. Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 32.Bartenschlager R., Ahlborn-Laake L., Mous J., Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 1993;67(7):3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eckart M.R., Selby M., Masiarz F., Lee C., Berger K., Crawford K., Kuo C., Kuo G., Houghton M., Choo Q.L. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem. Biophys. Res. Commun. 1993;192(2):399–406. doi: 10.1006/bbrc.1993.1429. [DOI] [PubMed] [Google Scholar]
- 34.Grakoui A., McCourt D.W., Wychowski C., Feinstone S.M., Rice C.M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 1993;67(5):2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grakoui A., Wychowski C., Lin C., Feinstone S.M., Rice C.M. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 1993;67(3):1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hijikata M., Mizushima H., Akagi T., Mori S., Kakiuchi N., Kato N., Tanaka T., Kimura K., Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 1993;67(8):4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hijikata M., Mizushima H., Tanji Y., Komoda Y., Hirowatari Y., Akagi T., Kato N., Kimura K., Shimotohno K. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 1993;90(22):10773–10777. doi: 10.1073/pnas.90.22.10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grakoui A., McCourt D.W., Wychowski C., Feinstone S.M., Rice C.M. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. U. S. A. 1993;90(22):10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romano K.P., Ali A., Royer W.E., Schiffer C.A. Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc. Natl. Acad. Sci. U. S. A. 2010;107(49):20986–20991. doi: 10.1073/pnas.1006370107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozen A., Sherman W., Schiffer C.A. Improving the resistance profile of hepatitis C NS3/4A inhibitors: dynamic substrate envelope guided design. J. Chem. Theory Comput. 2013;9(12):5693–5705. doi: 10.1021/ct400603p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedstrom L. Serine protease mechanism and specificity. Chem. Rev. 2002;102(12):4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 42.Copeland R.A. Wiley–VCH (Wiley–India); New Delhi, India: 2008. Enzymes : a Practical introduction to structure, mechanism, and data analysis. [Google Scholar]
- 43.Soumana D.I., Kurt Yilmaz N., Prachanronarong K.L., Aydin C., Ali A., Schiffer C.A. Structural and thermodynamic effects of macrocyclization in HCV NS3/4A inhibitor MK-5172. ACS Chem. Biol. 2016;11(4):900–909. doi: 10.1021/acschembio.5b00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timm J., Kosovrasti K., Henes M., Leidner F., Hou S., Ali A., Kurt Yilmaz N., Schiffer C.A. Molecular and structural mechanism of Pan-genotypic HCV NS3/4A protease inhibition by Glecaprevir. ACS Chem. Biol. 2020;15(2):342–352. doi: 10.1021/acschembio.9b00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozen A., Prachanronarong K., Matthew A.N., Soumana D.I., Schiffer C.A. Resistance outside the substrate envelope: hepatitis C NS3/4A protease inhibitors. Crit. Rev. Biochem. Mol. Biol. 2019;54(1):11–26. doi: 10.1080/10409238.2019.1568962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X. Direct anti-HCV agents. Acta Pharm. Sin B. 2016;6(1):26–31. doi: 10.1016/j.apsb.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matthew A.N., Zephyr J., Hill C.J., Jahangir M., Newton A., Petropoulos C.J., Huang W., Kurt-Yilmaz N., Schiffer C.A., Ali A. Hepatitis C virus NS3/4A protease inhibitors incorporating flexible P2 Quinoxalines target drug resistant viral variants. J. Med. Chem. 2017;60(13):5699–5716. doi: 10.1021/acs.jmedchem.7b00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matthew A.N., Zephyr J., Nageswara Rao D., Henes M., Kamran W., Kosovrasti K., Hedger A.K., Lockbaum G.J., Timm J., Ali A., Kurt Yilmaz N., Schiffer C.A. Avoiding drug resistance by substrate envelope-guided design: toward potent and robust HCV NS3/4A protease inhibitors. mBio. 2020;11(2):e00172–20. doi: 10.1128/mBio.00172-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romano K.P., Ali A., Aydin C., Soumana D., Ozen A., Deveau L.M., Silver C., Cao H., Newton A., Petropoulos C.J., Huang W., Schiffer C.A. The molecular basis of drug resistance against hepatitis C virus NS3/4A protease inhibitors. PLoS Pathog. 2012;8(7) doi: 10.1371/journal.ppat.1002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soumana D.I., Ali A., Schiffer C.A. Structural analysis of asunaprevir resistance in HCV NS3/4A protease. ACS Chem. Biol. 2014;9(11):2485–2490. doi: 10.1021/cb5006118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soumana D.I., Kurt Yilmaz N., Ali A., Prachanronarong K.L., Schiffer C.A. Molecular and dynamic mechanism underlying drug resistance in genotype 3 hepatitis C NS3/4A protease. J. Am. Chem. Soc. 2016;138(36):11850–11859. doi: 10.1021/jacs.6b06454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ali A., Aydin C., Gildemeister R., Romano K.P., Cao H., Ozen A., Soumana D., Newton A., Petropoulos C.J., Huang W., Schiffer C.A. Evaluating the role of macrocycles in the susceptibility of hepatitis C virus NS3/4A protease inhibitors to drug resistance. ACS Chem. Biol. 2013;8(7):1469–1478. doi: 10.1021/cb400100g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonsall D., Black S., Howe A.Y., Chase R., Ingravallo P., Pak I., Brown A., Smith D.A., Bowden R., Barnes E. Characterization of hepatitis C virus resistance to grazoprevir reveals complex patterns of mutations following on-treatment breakthrough that are not observed at relapse. Infect Drug Resist. 2018;11:1119–1135. doi: 10.2147/IDR.S156581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeuzem S., Foster G.R., Wang S., Asatryan A., Gane E., Feld J.J., Asselah T., Bourliere M., Ruane P.J., Wedemeyer H., Pol S., Flisiak R., Poordad F., Chuang W.L., Stedman C.A., Flamm S., Kwo P., Dore G.J., Sepulveda-Arzola G., Roberts S.K., Soto-Malave R., Kaita K., Puoti M., Vierling J., Tam E., Vargas H.E., Bruck R., Fuster F., Paik S.W., Felizarta F., Kort J., Fu B., Liu R., Ng T.I., Pilot-Matias T., Lin C.W., Trinh R., Mensa F.J. Glecaprevir-pibrentasvir for 8 or 12 weeks in HCV genotype 1 or 3 infection. N Engl J Med. 2018;378(4):354–369. doi: 10.1056/NEJMoa1702417. [DOI] [PubMed] [Google Scholar]
- 55.Matthew A.N., Leidner F., Newton A., Petropoulos C.J., Huang W., Ali A., KurtYilmaz N., Schiffer C.A. Molecular mechanism of resistance in a clinically significant double-mutant variant of HCV NS3/4A protease. Structure. 2018;26(10):1360–1372. doi: 10.1016/j.str.2018.07.004. (e5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rusere L.N., Matthew A.N., Lockbaum G.J., Jahangir M., Newton A., Petropoulos C.J., Huang W., Kurt Yilmaz N., Schiffer C.A., Ali A. Quinoxaline-based linear HCV NS3/4A protease inhibitors exhibit potent activity against drug resistant variants. ACS Med. Chem. Lett. 2018;9(7):691–696. doi: 10.1021/acsmedchemlett.8b00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dewan M.Z., Uchihara J.N., Terashima K., Honda M., Sata T., Ito M., Fujii N., Uozumi K., Tsukasaki K., Tomonaga M., Kubuki Y., Okayama A., Toi M., Mori N., Yamamoto N. Efficient intervention of growth and infiltration of primary adult T-cell leukemia cells by an HIV protease inhibitor, ritonavir. Blood. 2006;107(2):716–724. doi: 10.1182/blood-2005-02-0735. [DOI] [PubMed] [Google Scholar]
- 58.Tozser J., Weber I.T. The protease of human T-cell leukemia virus type-1 is a potential therapeutic target. Curr. Pharm. Des. 2007;13(12):1285–1294. doi: 10.2174/138161207780618849. [DOI] [PubMed] [Google Scholar]
- 59.Shuker S.B., Mariani V.L., Herger B.E., Dennison K.J. Understanding HTLV-I protease. Chem. Biol. 2003;10(5):373–380. doi: 10.1016/s1074-5521(03)00104-2. [DOI] [PubMed] [Google Scholar]
- 60.Shibata D., Brynes R.K., Rabinowitz A., Hanson C.A., Slovak M.L., Spira T.J., Gill P. Human T-cell lymphotropic virus type I (HTLV-I)-associated adult T-cell leukemia-lymphoma in a patient infected with human immunodeficiency virus type 1 (HIV-1) Ann. Intern. Med. 1989;111(11):871–875. doi: 10.7326/0003-4819-111-11-871. [DOI] [PubMed] [Google Scholar]
- 61.Li M., Laco G.S., Jaskolski M., Rozycki J., Alexandratos J., Wlodawer A., Gustchina A. Crystal structure of human T cell leukemia virus protease, a novel target for anticancer drug design. Proc. Natl. Acad. Sci. U. S. A. 2005;102(51):18332–18337. doi: 10.1073/pnas.0509335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wattel E., Vartanian J.P., Pannetier C., Wain-Hobson S. Clonal expansion of human T-cell leukemia virus type I-infected cells in asymptomatic and symptomatic carriers without malignancy. J. Virol. 1995;69(5):2863–2868. doi: 10.1128/jvi.69.5.2863-2868.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Satoh T., Li M., Nguyen J.T., Kiso Y., Gustchina A., Wlodawer A. Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease. J. Mol. Biol. 2010;401(4):626–641. doi: 10.1016/j.jmb.2010.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang M., Nguyen J.T., Kumada H.O., Kimura T., Cheng M., Hayashi Y., Kiso Y. Locking the two ends of tetrapeptidic HTLV-I protease inhibitors inside the enzyme. Bioorg. Med. Chem. 2008;16(14):6880–6890. doi: 10.1016/j.bmc.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 65.Bagossi P., Kadas J., Miklossy G., Boross P., Weber I.T., Tozser J. Development of a microtiter plate fluorescent assay for inhibition studies on the HTLV-1 and HIV-1 proteinases. J. Virol. Methods. 2004;119(2):87–93. doi: 10.1016/j.jviromet.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 66.Kuhnert M., Steuber H., Diederich W.E. Structural basis for HTLV-1 protease inhibition by the HIV-1 protease inhibitor indinavir. J. Med. Chem. 2014;57(14):6266–6272. doi: 10.1021/jm500402c. [DOI] [PubMed] [Google Scholar]
- 67.Lockbaum G.J., Henes M., Talledge N., Rusere L.N., Kosovrasti K., Nalivaika E.A., Somasundaran M., Ali A., Mansky L.M., Kurt Yilmaz N., Schiffer C.A. Inhibiting HTLV-1 protease: a viable antiviral target. ACS Chem. Biol. 2021;16(3):529–538. doi: 10.1021/acschembio.0c00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferguson N.M., Cucunuba Z.M., Dorigatti I., Nedjati-Gilani G.L., Donnelly C.A., Basanez M.G., Nouvellet P., Lessler J. EPIDEMIOLOGY. Countering the Zika epidemic in Latin America. Science. 2016;353(6297):353–354. doi: 10.1126/science.aag0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chang H.H., Huber R.G., Bond P.J., Grad Y.H., Camerini D., Maurer-Stroh S., Lipsitch M. Systematic analysis of protein identity between Zika virus and other arthropod-borne viruses. Bull. World Health Organ. 2017;95(7):517–525I. doi: 10.2471/BLT.16.182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Russell P.K. The Zika pandemic—a perfect storm? PLoS Negl. Trop. Dis. 2016;10(3) doi: 10.1371/journal.pntd.0004589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang L., Valderramos S.G., Wu A., Ouyang S., Li C., Brasil P., Bonaldo M., Coates T., Nielsen-Saines K., Jiang T., Aliyari R., Cheng G. From mosquitos to humans: genetic evolution of Zika virus. Cell Host Microbe. 2016;19(5):561–565. doi: 10.1016/j.chom.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kyle J.L., Harris E. Global spread and persistence of dengue. Annu. Rev. Microbiol. 2008;62:71–92. doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 73.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O., Myers M.F., George D.B., Jaenisch T., Wint G.R., Simmons C.P., Scott T.W., Farrar J.J., Hay S.I. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kang C., Keller T.H., Luo D. Zika virus protease: an antiviral drug target. Trends Microbiol. 2017;25(10):797–808. doi: 10.1016/j.tim.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 75.Noble C.G., Seh C.C., Chao A.T., Shi P.Y. Ligand-bound structures of the dengue virus protease reveal the active conformation. J. Virol. 2012;86(1):438–446. doi: 10.1128/JVI.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robin G., Chappell K., Stoermer M.J., Hu S.H., Young P.R., Fairlie D.P., Martin J.L. Structure of West Nile virus NS3 protease: ligand stabilization of the catalytic conformation. J. Mol. Biol. 2009;385(5):1568–1577. doi: 10.1016/j.jmb.2008.11.026. [DOI] [PubMed] [Google Scholar]
- 77.Luo D., Xu T., Hunke C., Gruber G., Vasudevan S.G., Lescar J. Crystal structure of the NS3 protease-helicase from dengue virus. J. Virol. 2008;82(1):173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Erbel P., Schiering N., D'Arcy A., Renatus M., Kroemer M., Lim S.P., Yin Z., Keller T.H., Vasudevan S.G., Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006;13(4):372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 79.Phoo W.W., Li Y., Zhang Z., Lee M.Y., Loh Y.R., Tan Y.B., Ng E.Y., Lescar J., Kang C., Luo D. Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat. Commun. 2016;7:13410. doi: 10.1038/ncomms13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Phoo W.W., Zhang Z., Wirawan M., Chew E.J.C., Chew A.B.L., Kouretova J., Steinmetzer T., Luo D. Structures of Zika virus NS2B-NS3 protease in complex with peptidomimetic inhibitors. Antiviral Res. 2018;160:17–24. doi: 10.1016/j.antiviral.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 81.Nitsche C., Holloway S., Schirmeister T., Klein C.D. Biochemistry and medicinal chemistry of the dengue virus protease. Chem. Rev. 2014;114(22):11348–11381. doi: 10.1021/cr500233q. [DOI] [PubMed] [Google Scholar]
- 82.Yin Z., Patel S.J., Wang W.L., Chan W.L., Ranga Rao K.R., Wang G., Ngew X., Patel V., Beer D., Knox J.E., Ma N.L., Ehrhardt C., Lim S.P., Vasudevan S.G., Keller T.H. Peptide inhibitors of dengue virus NS3 protease. Part 2: SAR study of tetrapeptide aldehyde inhibitors. Bioorg. Med. Chem. Lett. 2006;16(1):40–43. doi: 10.1016/j.bmcl.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 83.Yin Z., Patel S.J., Wang W.L., Wang G., Chan W.L., Rao K.R., Alam J., Jeyaraj D.A., Ngew X., Patel V., Beer D., Lim S.P., Vasudevan S.G., Keller T.H. Peptide inhibitors of dengue virus NS3 protease. Part 1: warhead. Bioorg. Med. Chem. Lett. 2006;16(1):36–39. doi: 10.1016/j.bmcl.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 84.Poulsen A., Kang C., Keller T.H. Drug design for flavivirus proteases: what are we missing? Curr. Pharm. Des. 2014;20(21):3422–3427. doi: 10.2174/13816128113199990633. [DOI] [PubMed] [Google Scholar]
- 85.Nitsche C., Onagi H., Quek J.P., Otting G., Luo D., Huber T. Biocompatible macrocyclization between cysteine and 2-Cyanopyridine generates stable peptide inhibitors. Org. Lett. 2019;21(12):4709–4712. doi: 10.1021/acs.orglett.9b01545. [DOI] [PubMed] [Google Scholar]
- 86.Li Y., Zhang Z., Phoo W.W., Loh Y.R., Li R., Yang H.Y., Jansson A.E., Hill J., Keller T.H., Nacro K., Luo D., Kang C. Structural insights into the inhibition of Zika virus NS2B-NS3 protease by a small-molecule inhibitor. Structure. 2018;26(4):555–564. doi: 10.1016/j.str.2018.02.005. (e3) [DOI] [PubMed] [Google Scholar]
- 87.Lee H., Ren J., Nocadello S., Rice A.J., Ojeda I., Light S., Minasov G., Vargas J., Nagarathnam D., Anderson W.F., Johnson M.E. Identification of novel small molecule inhibitors against NS2B/NS3 serine protease from Zika virus. Antiviral Res. 2017;139:49–58. doi: 10.1016/j.antiviral.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Z., Li Y., Loh Y.R., Phoo W.W., Hung A.W., Kang C., Luo D. Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science. 2016;354(6319):1597–1600. doi: 10.1126/science.aai9309. [DOI] [PubMed] [Google Scholar]
- 89.Chan J.F., Chik K.K., Yuan S., Yip C.C., Zhu Z., Tee K.M., Tsang J.O., Chan C.C., Poon V.K., Lu G., Zhang A.J., Lai K.K., Chan K.H., Kao R.Y., Yuen K.Y. Novel antiviral activity and mechanism of bromocriptine as a Zika virus NS2B-NS3 protease inhibitor. Antiviral Res. 2017;141:29–37. doi: 10.1016/j.antiviral.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 90.Voss S., Nitsche C. Inhibitors of the Zika virus protease NS2B-NS3. Bioorg. Med. Chem. Lett. 2020;30(5):126965. doi: 10.1016/j.bmcl.2020.126965. [DOI] [PubMed] [Google Scholar]
- 91.Behnam M.A., Nitsche C., Boldescu V., Klein C.D. The medicinal chemistry of dengue virus. J. Med. Chem. 2016;59(12):5622–5649. doi: 10.1021/acs.jmedchem.5b01653. [DOI] [PubMed] [Google Scholar]
- 92.Lin K.H., Nalivaika E.A., Prachanronarong K.L., Yilmaz N.K., Schiffer C.A. Dengue protease substrate recognition: binding of the prime side. ACS Infect. Dis. 2016;2(10):734–743. doi: 10.1021/acsinfecdis.6b00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin K.H., Ali A., Rusere L., Soumana D.I., Kurt Yilmaz N., Schiffer C.A. Dengue Virus NS2B/NS3 protease inhibitors exploiting the prime Side. J. Virol. 2017:91(10). doi: 10.1128/JVI.00045-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shiryaev S.A., Farhy C., Pinto A., Huang C.T., Simonetti N., Elong Ngono A., Dewing A., Shresta S., Pinkerton A.B., Cieplak P., Strongin A.Y., Terskikh A.V. Characterization of the Zika virus two-component NS2B-NS3 protease and structure-assisted identification of allosteric small-molecule antagonists. Antiviral Res. 2017;143:218–229. doi: 10.1016/j.antiviral.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brecher M., Li Z., Liu B., Zhang J., Koetzner C.A., Alifarag A., Jones S.A., Lin Q., Kramer L.D., Li H. A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B-NS3 protease. PLoS Pathog. 2017;13(5) doi: 10.1371/journal.ppat.1006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Millies B., von Hammerstein F., Gellert A., Hammerschmidt S., Barthels F., Goppel U., Immerheiser M., Elgner F., Jung N., Basic M., Kersten C., Kiefer W., Bodem J., Hildt E., Windbergs M., Hellmich U.A., Schirmeister T. Proline-based allosteric inhibitors of Zika and dengue virus NS2B/NS3 proteases. J. Med. Chem. 2019;62(24):11359–11382. doi: 10.1021/acs.jmedchem.9b01697. [DOI] [PubMed] [Google Scholar]
- 97.Lim L., Dang M., Roy A., Kang J., Song J. Curcumin allosterically inhibits the dengue NS2B-NS3 protease by disrupting its active conformation. ACS Omega. 2020;5(40):25677–25686. doi: 10.1021/acsomega.0c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nitsche C., Passioura T., Varava P., Mahawaththa M.C., Leuthold M.M., Klein C.D., Suga H., Otting G. De novo discovery of nonstandard macrocyclic peptides as noncompetitive inhibitors of the Zika virus NS2B-NS3 protease. ACS Med. Chem. Lett. 2019;10(2):168–174. doi: 10.1021/acsmedchemlett.8b00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yildiz M., Ghosh S., Bell J.A., Sherman W., Hardy J.A. Allosteric inhibition of the NS2B-NS3 protease from dengue virus. ACS Chem. Biol. 2013;8(12):2744–2752. doi: 10.1021/cb400612h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Raut R., Beesetti H., Tyagi P., Khanna I., Jain S.K., Jeankumar V.U., Yogeeswari P., Sriram D., Swaminathan S. A small molecule inhibitor of dengue virus type 2 protease inhibits the replication of all four dengue virus serotypes in cell culture. Virol. J. 2015;12:16. doi: 10.1186/s12985-015-0248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bodenreider C., Beer D., Keller T.H., Sonntag S., Wen D., Yap L., Yau Y.H., Shochat S.G., Huang D., Zhou T., Caflisch A., Su X.C., Ozawa K., Otting G., Vasudevan S.G., Lescar J., Lim S.P. A fluorescence quenching assay to discriminate between specific and nonspecific inhibitors of dengue virus protease. Anal. Biochem. 2009;395(2):195–204. doi: 10.1016/j.ab.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 102.Li Z., Brecher M., Deng Y.Q., Zhang J., Sakamuru S., Liu B., Huang R., Koetzner C.A., Allen C.A., Jones S.A., Chen H., Zhang N.N., Tian M., Gao F., Lin Q., Banavali N., Zhou J., Boles N., Xia M., Kramer L.D., Qin C.F., Li H. Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction. Cell Res. 2017;27(8):1046–1064. doi: 10.1038/cr.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kumar A., Liang B., Aarthy M., Singh S.K., Garg N., Mysorekar I.U., Giri R. Hydroxychloroquine inhibits Zika virus NS2B-NS3 protease. ACS Omega. 2018;3(12):18132–18141. doi: 10.1021/acsomega.8b01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Baggen J., Thibaut H.J., Strating J.R.P.M., van Kuppeveld F.J.M. The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 2018;16(6):368–381. doi: 10.1038/s41579-018-0005-4. [DOI] [PubMed] [Google Scholar]
- 105.Messacar K., Schreiner T.L., Maloney J.A., Wallace A., Ludke J., Oberste M.S., Nix W.A., Robinson C.C., Glode M.P., Abzug M.J., Dominguez S.R. A cluster of acute flaccid paralysis and cranial nerve dysfunction temporally associated with an outbreak of enterovirus D68 in children in Colorado, USA. Lancet. 2015;385(9978):1662–1671. doi: 10.1016/S0140-6736(14)62457-0. [DOI] [PubMed] [Google Scholar]
- 106.Holm-Hansen C.C., Midgley S.E., Fischer T.K. Global emergence of enterovirus D68: a systematic review. Lancet Infect. Dis. 2016;16(5):e64–e75. doi: 10.1016/S1473-3099(15)00543-5. [DOI] [PubMed] [Google Scholar]
- 107.Winther B. In: Rhinoviruses, in International Encyclopedia of Public Health. Heggenhougen H.K., editor. Academic Press; Oxford: 2008. pp. 577–581. [Google Scholar]
- 108.Seipelt J., Guarne A., Bergmann E., James M., Sommergruber W., Fita I., Skern T. The structures of picornaviral proteinases. Virus Res. 1999;62(2):159–168. doi: 10.1016/s0168-1702(99)00043-x. [DOI] [PubMed] [Google Scholar]
- 109.Costenaro L., Kaczmarska Z., Arnan C., Janowski R., Coutard B., Sola M., Gorbalenya A.E., Norder H., Canard B., Coll M. Structural basis for antiviral inhibition of the main protease, 3C, from human enterovirus 93. J. Virol. 2011;85(20):10764–10773. doi: 10.1128/JVI.05062-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tan J., George S., Kusov Y., Perbandt M., Anemuller S., Mesters J.R., Norder H., Coutard B., Lacroix C., Leyssen P., Neyts J., Hilgenfeld R. 3C protease of enterovirus 68: structure-based design of Michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J. Virol. 2013;87(8):4339–4351. doi: 10.1128/JVI.01123-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dai W., Jochmans D., Xie H., Yang H., Li J., Su H., Chang D., Wang J., Peng J., Zhu L., Nian Y., Hilgenfeld R., Jiang H., Chen K., Zhang L., Xu Y., Neyts J., Liu H. Design, synthesis, and biological evaluation of Peptidomimetic aldehydes as broad-Spectrum inhibitors against enterovirus and SARS-CoV-2. J. Med. Chem. 2021 doi: 10.1021/acs.jmedchem.0c02258. [DOI] [PubMed] [Google Scholar]
- 112.Hayden F.G., Turner R.B., Gwaltney J.M., Chi-Burris K., Gersten M., Hsyu P., Patick A.K., Smith G.J., 3rd, Zalman L.S. Phase II, randomized, double-blind, placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob. Agents Chemother. 2003;47(12):3907–3916. doi: 10.1128/AAC.47.12.3907-3916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Patick A.K., Brothers M.A., Maldonado F., Binford S., Maldonado O., Fuhrman S., Petersen A., Smith G.J., 3rd, Zalman L.S., Burns-Naas L.A., Tran J.Q. In vitro antiviral activity and single-dose pharmacokinetics in humans of a novel, orally bioavailable inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 2005;49(6):2267–2275. doi: 10.1128/AAC.49.6.2267-2275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lu G., Qi J., Chen Z., Xu X., Gao F., Lin D., Qian W., Liu H., Jiang H., Yan J., Gao G.F. Enterovirus 71 and coxsackievirus A16 3C proteases: binding to rupintrivir and their substrates and anti-hand, foot, and mouth disease virus drug design. J. Virol. 2011;85(19):10319–10331. doi: 10.1128/JVI.00787-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang J., Fan T., Yao X., Wu Z., Guo L., Lei X., Wang J., Wang M., Jin Q., Cui S. Crystal structures of enterovirus 71 3C protease complexed with rupintrivir reveal the roles of catalytically important residues. J. Virol. 2011;85(19):10021–10030. doi: 10.1128/JVI.05107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guo Y., Wang Y., Cao L., Wang P., Qing J., Zheng Q., Shang L., Yin Z., Sun Y. A conserved inhibitory mechanism of a Lycorine derivative against enterovirus and hepatitis C virus. Antimicrob. Agents Chemother. 2016;60(2):913–924. doi: 10.1128/AAC.02274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang Y., Peng F., Wang R., Yange M., Guan K., Jiang T., Xu G., Sun J., Chang C. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J. Autoimmun. 2020;109:102434. doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.da Costa V.G., Moreli M.L., Saivish M.V. The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Arch. Virol. 2020;165(7):1517–1526. doi: 10.1007/s00705-020-04628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ziebuhr J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 2004;7(4):412–419. doi: 10.1016/j.mib.2004.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ceraolo C., Giorgi F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020;92(5):522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020;30(17):127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Grum-Tokars V., Ratia K., Begaye A., Baker S.C., Mesecar A.D. Evaluating the 3C-like protease activity of SARS-coronavirus: recommendations for standardized assays for drug discovery. Virus Res. 2008;133(1):63–73. doi: 10.1016/j.virusres.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhong N., Zhang S., Zou P., Chen J., Kang X., Li Z., Liang C., Jin C., Xia B. Without its N-finger, the main protease of severe acute respiratory syndrome coronavirus can form a novel dimer through its C-terminal domain. J. Virol. 2008;82(9):4227–4234. doi: 10.1128/JVI.02612-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xia B., Kang X. Activation and maturation of SARS-CoV main protease. Protein Cell. 2011;2(4):282–290. doi: 10.1007/s13238-011-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ho B.L., Cheng S.C., Shi L., Wang T.Y., Ho K.I., Chou C.Y. Critical assessment of the important residues involved in the dimerization and catalysis of MERS coronavirus Main protease. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0144865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59(14):6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang C., Wei P., Fan K., Liu Y., Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43(15):4568–4574. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- 130.Muramatsu T., Takemoto C., Kim Y.T., Wang H., Nishii W., Terada T., Shirouzu M., Yokoyama S. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl. Acad. Sci. U. S. A. 2016;113(46):12997–13002. doi: 10.1073/pnas.1601327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee J., Worrall L.J., Vuckovic M., Rosell F.I., Gentile F., Ton A.T., Caveney N.A., Ban F., Cherkasov A., Paetzel M., Strynadka N.C.J. Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site. Nat. Commun. 2020;11(1):5877. doi: 10.1038/s41467-020-19662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X.C., Liao M., Bartlam M., Rao Z. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82(5):2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Amin S.A., Banerjee S., Ghosh K., Gayen S., Jha T. Protease targeted COVID-19 drug discovery and its challenges: insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorg. Med. Chem. 2021;29:115860. doi: 10.1016/j.bmc.2020.115860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liang R., Wang L., Zhang N., Deng X., Su M., Su Y., Hu L., He C., Ying T., Jiang S., Yu F. Development of small-molecule MERS-CoV inhibitors. Viruses. 2018;10(12):721. doi: 10.3390/v10120721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hoffman R.L., Kania R.S., Brothers M.A., Davies J.F., Ferre R.A., Gajiwala K.S., He M., Hogan R.J., Kozminski K., Li L.Y., Lockner J.W., Lou J., Marra M.T., Mitchell L.J., Jr., Murray B.W., Nieman J.A., Noell S., Planken S.P., Rowe T., Ryan K., Smith G.J., 3rd, Solowiej J.E., Steppan C.M., Taggart B. Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 2020;63(21):12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Boras B., Jones R.M., Anson B.J., Arenson D., Aschenbrenner L., Bakowski M.A., Beutler N., Binder J., Chen E., Eng H., Hammond J., Hoffman R., Kadar E.P., Kania R., Kimoto E., Kirkpatrick M.G., Lanyon L., Lendy E.K., Lillis J.R., Luthra S.A., Ma C., Noell S., Obach R.S., O’Brien M.N., O’Connor R., Ogilvie K., Owen D., Pettersson M., Reese M.R., Rogers T., Rossulek M.I., Sathish J.G., Steppan C., Ticehurst M., Updyke L.W., Zhu Y., Wang J., Chatterjee A.K., Mesecar A.D., Anderson A.S., Allerton C. Discovery of a novel inhibitor of coronavirus 3CL protease as a clinical candidate for the potential treatment of COVID-19. BioRxiv. 2020 [Google Scholar]