Abstract
Background
The impact of SARS-CoV-2 variants of concern (VOCs) on disease severity is unclear. In this retrospective study, we compared the outcomes of patients infected with B.1.1.7, B.1.351, and B.1.617.2 with wild-type strains from early 2020.
Methods
National surveillance data from January to May 2021 were obtained and outcomes in relation to VOCs were explored. Detailed patient-level data from all patients with VOC infection admitted to our center between December 2020 and May 2021 were analyzed. Clinical outcomes were compared with a cohort of 846 patients admitted from January to April 2020.
Results
A total of 829 patients in Singapore in the study period were infected with these 3 VOCs. After adjusting for age and sex, B.1.617.2 was associated with higher odds of oxygen requirement, intensive care unit admission, or death (adjusted odds ratio [aOR], 4.90; 95% confidence interval [CI]: 1.43-30.78). Of these patients, 157 were admitted to our center. After adjusting for age, sex, comorbidities, and vaccination, the aOR for pneumonia with B.1.617.2 was 1.88 (95% CI: .95-3.76) compared with wild-type. These differences were not seen with B.1.1.7 and B.1.351. Vaccination status was associated with decreased severity. B.1.617.2 was associated with significantly lower polymerase chain reaction cycle threshold (Ct) values and longer duration of Ct value ≤30 (median duration 18 days for B.1.617.2, 13 days for wild-type).
Conclusions
B.1.617.2 was associated with increased severity of illness, and with lower Ct values and longer viral shedding. These findings provide impetus for the rapid implementation of vaccination programs.
Keywords: B.1.617.2, COVID-19, SARS-CoV-2, severity, variants of concern
In this retrospective cohort study we found an association between infection with B.1.617.2 (Delta) and increased disease severity. B.1.617.2 was also associated with higher viral loads and prolonged duration of viral shedding. Vaccination remained protective.
Since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease 2019 (COVID-19), numerous viral variants have been characterized. Most genetic variations arise as a result of drift and have no phenotypic effect. However, variants of concern (VOCs) have emerged that carry signature amino acid substitutions in key areas of the immunodominant spike protein, with evidence of altered virus characteristics [1]. For example, VOCs have been associated with increased transmissibility, evasion of immunity from infection and vaccination, and reduced susceptibility to monoclonal antibody therapies [2–4]. Several VOCs have temporally coincided with spikes in infection rates in countries where they first arose, such as B.1.1.7 (Alpha in the World Health Organization classification) [5], B.1.351 (Beta) [6], P.1 (Gamma) [7], and B.1.617.2 (Delta) [8].
While robust epidemiologic data have provided evidence for the increased transmissibility of these VOCs [4, 6, 9], their impact on clinical outcomes such as disease severity and mortality is less clear. Large population studies have found a significant trend toward increased mortality associated with B.1.1.7 [10, 11]; whereas for a cohort of hospitalized patients, no significant differences in disease severity and clinical outcomes were found compared with non-B.1.1.7–infected patients [12].
The emergence of B.1.617.2 had a catastrophic impact on infection and mortality rates in India. By May 2021, it had become the dominant strain identified in the United Kingdom and, in June 2021, the dominant strain identified worldwide [13]. Early data have shown an association between infections with B.1.617.2 and hospitalization [14]. However, there are limited data on the relationship between B.1.617.2 and clinical outcomes such as disease severity and mortality. National infection and mortality rates can be confounded by factors such as healthcare resource utilization, demographics, and sociobehavioral trends, and detailed cohort studies are required to distinguish the impact of viral vs nonpathogen factors [15].
Singapore has used a robust test-trace-isolate strategy in a largely successful effort to limit community transmission of SARS-CoV-2. This includes detailed contact-tracing efforts, rigorous screening of symptomatic and asymptomatic contacts, and quarantine of all positive cases and incoming travelers [16]. This system has been in place since the start of the outbreak in January 2020 with minor adjustments but without significant change to the overarching strategy. Active case finding provided a high rate of diagnosis and accumulation of clinical data across the entire spectrum of COVID-19 disease severity [17].
In this retrospective study of both a national cohort dataset of all confirmed COVID-19 cases and a smaller single-center cohort of hospitalized patients, we compared the outcomes of patients infected with B.1.1.7, B.1.351, and B.1.617.2 variants, with the aim of improving our understanding of the relationships between viral variants, disease severity, and viral shedding kinetics.
METHODS
Patient Recruitment
Individuals who met the following inclusion criteria were screened for VOC infection: confirmed to have SARS-CoV-2 infection by real-time reverse-transcriptase polymerase chain reaction (RT-PCR); admitted to the National Centre for Infectious Diseases (NCID), the national outbreak clinical management center in Singapore; and have a diagnosis between 20 December 2020 (when the first VOC infection, B.1.1.7, was detected) and 12 May 2021. As part of active genomic surveillance, whole-genome sequencing by the National Public Health Laboratory (NPHL) is performed for all individuals with SARS-CoV-2 detected by RT-PCR with a cycle threshold (Ct) value <30 (cutoff based on previous unpublished data from NPHL that showed sequencing was only successful for isolates with Ct value <30). Participants with B.1.1.7, B.1.351, or B.1.617.2 variants were included in the study. There were no exclusion criteria.
Patients with VOC infection were compared with a cohort of 846 consecutive patients admitted to NCID from 22 January 2020 to 15 April 2020 (hereafter, referred to as “wild-type”). Viral shedding dynamics of the 3 VOCs were compared with wild-type virus using a database of RT-PCR Ct values in 63 patients admitted from January 2020 to April 2020 with available serial Ct value data, previously reported in a separate study [18].
Data Collection
Clinical information was extracted from the medical record using a standardized data-collection form adapted from the International Severe Acute Respiratory and Emerging Infection Consortium case record form [19]. Laboratory data including Ct values from RT-PCR and serologic tests were recorded. Collection of clinical data was censored at 28 days from the last onset of illness on 11 May 2021.
We also obtained anonymized national mandatory case report data submitted to the Ministry of Health (Singapore) from 1 January 2021 to 22 May 2021. The data submitted by all healthcare institutions are implemented and analyzed under the Infectious Diseases Act (Singapore). It included all known patients with COVID-19 infection in the country during this period. Available metadata were age, sex, VOC type, and clinical outcomes (oxygen requirement, intensive care unit [ICU] admission, and death). National-level data on whether patients developed pneumonia were not available.
Clinical Management
All patients were admitted to airborne-infection isolation rooms. Risk stratification was conducted at the discretion of managing physicians based on age, comorbidities, and radiographic and laboratory findings; low-risk patients were transferred to a separate designated community isolation facility. High-risk patients with pneumonia (presence of pulmonary opacities on chest radiograph) were treated with intravenous remdesivir for 5 days, while patients who required supplemental oxygen were treated with dexamethasone, per national guidelines [20]. Patients were de-isolated based on discharge criteria of 2 negative swabs for SARS-CoV-2 PCR, seroconversion with 1 negative swab for SARS-CoV-2 PCR, or 3 weeks from illness onset. Repeat PCR testing was conducted at intervals decided by individual managing physicians.
Viral RNA Sequencing and VOC Determination
SARS-CoV-2 PCR was performed with a range of commercially available assays used by the clinical laboratory. Sequencing was performed at a centralized laboratory (NPHL, the national infectious diseases reference laboratory). Pangolin COVID-19 Lineage Assigner and CoVsurver were used to assign lineage to each sequence [21, 22].
Outcomes
The primary end point for the retrospective NCID cohort study was development of pneumonia as defined by the presence of pulmonary opacities on chest radiograph. This was chosen as the primary end point due to the expected small numbers of patients with severe infection outcomes. For the national cohort, primary outcome was severe COVID-19, defined as a composite of any of the following outcomes: hypoxia requiring supplemental oxygen (both low-flow including nasal prongs or high-flow), ICU admission, and/or death. Secondary outcomes were comparison of viral PCR Ct values and duration of viral shedding between VOCs.
Statistical Analyses
Firth’s logistic regression was used to examine the association between variant type and development of pneumonia due to the potential for the presence of separation with zero cell frequency for a particular variant. The following covariates known to impact COVID-19 disease severity were selected for inclusion in the multivariable logistic regression model: age group, sex, comorbidities, COVID-19 vaccination history, and SARS-CoV-2 lineage. For the national cohort, the multivariable logistic regression model included only age group, sex, and lineage due to the lack of other available data.
We fitted a generalized additive mixed model (GAMM) to the serial Ct values with random intercept by patient ID in addition to the fixed factor of day of illness with smoothing terms, separately for cases infected with B.1.1.7, B.1.351, and wild-type. We included random slope of Ct values with respect to the day of illness (in addition to random intercept) for cases infected with B.1.617.2. We plotted Ct values with marginal effect of day of illness from GAMM for each of the infection groups: B.1.1.7, B.1.351, B.1.617.2, and wild-type.
To compare duration of infectivity, we used Kaplan-Meier curves to describe time from symptom onset to first Ct value >30 and the log-rank test for comparison between variants. An event was defined as the first Ct value >30 as a surrogate indicator for the end of the infective period, since Ct values above this cutoff have been shown to be associated with lack of positive viral culture [23, 24]. A patient was considered censored if he or she did not have a Ct value >30 as of the date of the last available Ct value.
Data analysis was done in the R statistical language (version 3.6.1). All statistical tests were 2-sided, and P values < .05 were considered statistically significant.
Informed consent for retrospective data collection was waived as approved by the institutional review board.
RESULTS
National Data
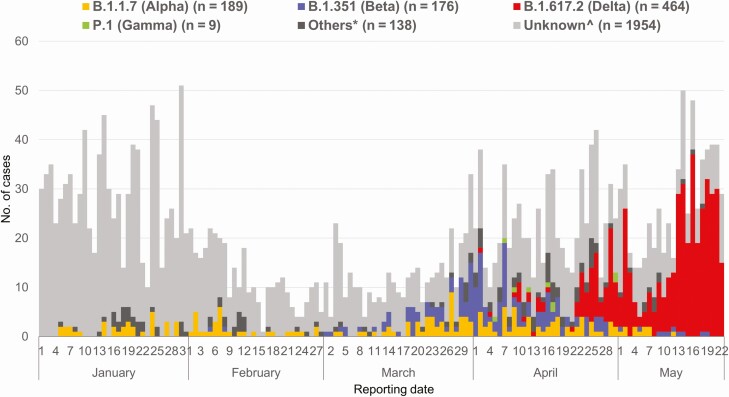
From 1 January 2021 to 22 May 2021, there were 2930 COVID-19 infections reported to the Ministry of Health. Of 976 (33%) with sequences available, 829 (85%) were by the VOCs B.1.1.7 (Alpha: 189, 20%), B.1.351 (Beta: 176, 18%), or B.1.617.2 (Delta: 464, 48%) (Figure 1) due to both importation and local transmission. Nine (1%) individuals with P.1 infection were excluded due to the small sample size.
Figure 1.
All reported coronavirus disease 2019 cases in Singapore, 1 January 2021 to 22 May 2021 (n = 2930). Of 829 infections with B.1.1.7 (Alpha), B.1.351 (Beta), or B.1.617.2 (Delta), 157 (19%) were admitted to the National Centre for Infectious Diseases. * refers to other non-variants of concern and wild-type. ^ indicates those that were unable to be sequenced and not sequenced.
The majority of infections by the 3 VOCs were nonsevere; 30 (4%) required supplemental oxygen and 10 (1%) were admitted to the ICU or died (Supplementary Table 1). After adjusting for age and sex in a multivariable logistic regression model, B.1.617.2 was associated with increased disease severity, as defined by a composite outcome of oxygen requirement, ICU admission, or death, compared with sequenced non-VOC cases (adjusted odds ratio [aOR], 4.90; 95% confidence interval [CI]: 1.43–30.78; Table 1). We could not adjust for comorbidities in this national analysis due to the lack of detailed patient data.
Table 1.
Odds Ratios of Candidate Predictors for Composite Outcome of Oxygen Requirement, Intensive Care Unit Admission, or Death in Cases With Sequences Available from 1 January 2021 to 22 May 2021 in Singapore
| Univariable Model | Multivariable Modela | |||
|---|---|---|---|---|
| Candidate Predictor | Crude OR (95% CI) | P Value | Adjusted OR (95% CI) | P Value |
| Variant | ||||
| Others | Ref | Ref | ||
| B.1.1.7 (Alpha) | 1.10 (.18–8.41) | .920 | 1.88 (.30–14.76) | .500 |
| B.1.351 (Beta) | 0.78 (.09–6.58) | .807 | 1.69 (.19–14.69) | .610 |
| B.1.617.2 (Delta) | 5.55 (1.66–34.44) | .020 | 4.90 (1.43–30.78) | .033 |
| Age group, years | ||||
| <45 | Ref | Ref | ||
| 45–64 | 7.91 (3.64–18.52) | <.001 | 6.62 (2.99–15.79) | <.001 |
| ≥65 | 19.73 (8.13–49.99) | <.001 | 13.84 (5.48–36.62) | <.001 |
| Female sex | 1.91 (1.03–3.58) | .041 | 1.42 (.74–2.75) | .291 |
P.1 excluded due to small sample size (n = 967).
Abbreviations: CI, confidence interval; OR, odds ratio.
aAdjusted for variant type, age group and sex.
NCID Cohort
For the cohort admitted to NCID, 157 patients infected with VOCs were identified (57 with B.1.1.7, 33 with B.1.351, and 67 with B.1.617.2). Four patients with P.1 were excluded from analysis due to the small number. Patients with the other 3 VOCs were compared with the cohort of 846 wild-type infections (lineage assignment provided in Supplementary Table 2). Patient demographics, symptoms, laboratory values, and clinical outcomes are summarized in Table 2. Patients with B.1.617.2 infection were older and with more comorbidities due to an outbreak among elderly in a healthcare facility. They also presented earlier in the illness course, partly due to aggressive contact tracing as part of this outbreak. Eighteen patients developed breakthrough infection despite prior vaccination with at least 1 dose of SARS-CoV-2 vaccine ≥14 days prior to infection (all Pfizer-BioNTech/Moderna mRNA vaccines except 1 with AstraZeneca vaccine).
Table 2.
Characteristics of 846 Wild-Type and 157 Variant of Concern Coronavirus Disease 2019 Cases
| B.1.1.7 | B.1.351 | B.1.617.2 | ||
|---|---|---|---|---|
| Variant: World Health Organization Label | Wild-type | (Alpha) | (Beta) | (Delta) |
| N | 846 | 57 | 33 | 67 |
| Demographics | ||||
| Age, median (IQR), years | 43 (28–55) | 38 (27–43) | 36 (31–41) | 48 (37–63) |
| Age ≥65 years, n (%) | 91 (11) | 0 (0) | 0 (0) | 14 (21) |
| Male sex, (n, %) | 466 (55) | 35 (61) | 24 (73) | 34 (51) |
| Charlson comorbidity index, median (IQR) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–1.5) |
| Diabetes mellitus, n (%) | 100 (12) | 4 (7) | 4 (12) | 16 (24) |
| Hypertension, n (%) | 144 (17) | 10 (18) | 2 (6) | 13 (19) |
| Hyperlipidemia, n (%) | 163 (19) | 9 (16) | 3 (9) | 19 (28) |
| Imported infection, n (%) | 456 (54) | 50 (88) | 31 (94) | 24 (36) |
| Reinfection, n (%) | 0 (0) | 0 (0) | 1 (3) | 0 (0) |
| Vaccination, n (%)a | 0 (0) | 0 (0) | 5 (15) | 18 (27) |
| Symptoms during illness, n (%) | ||||
| Symptom onset to admission, median (IQR), days | 4 (2–7) | 2 (1–4) | 2 (0.5–4) | 0 (1–2) |
| Asymptomatic | 5 (1) | 10 (18) | 10 (30) | 7 (10) |
| Fever | 581 (69) | 33 (58) | 14 (42) | 48 (72) |
| Cough | 541 (64) | 28 (49) | 10 (30) | 31 (46) |
| Dyspnea | 90 (11) | 3 (5) | 1 (3) | 13 (19) |
| Sore throat | 343 (41) | 15 (26) | 6 (18) | 23 (34) |
| Nasal congestion and/or rhinorrhea | 269 (32) | 11 (19) | 6 (18) | 11 (16) |
| Worst laboratory values, median (IQR) | ||||
| Neutrophil, 109/L | 2.7 (3.6–4.9) | 3.36 (2.7–4.4) | 4.25 (3.4–5.5) | 4.56 (3.7–6.3) |
| Lymphocyte, 109/L | 0.87 (1.2–1.7) | 1.43 (1.1–1.8) | 1.95 (1.5–2.7) | 0.81 (0.6–1.2) |
| Alanine aminotransferase, U/L | 26 (18–46) | 29 (17–40) | 29 (22–59) | 37 (19–67) |
| C-reactive protein, mg/L | 6.1 (1.4–28.3) | 5.6 (2.5–23.5) | 5.1 (2.2–12.1) | 28.7 (10–101) |
| Lactate dehydrogenase, U/L | 413 (347–567) | 371 (309–436) | 369 (314–441) | 488 (385–631) |
| Polymerase chain reaction cycle threshold value | 32.9 (29.3–35.6)b | 20.5 (16.4–26.5) | 17.8 (16.3–22.7) | 15.6 (13.6–19.1) |
| Treatment, n (%) | ||||
| Remdesivir | 7 (1) | 3 (5) | 1 (3) | 17 (25) |
| Corticosteroids | 0 (0) | 3 (5) | 1 (3) | 11 (16) |
| Outcome, n (%) | ||||
| Pneumonia | 320 (38) | 9 (16) | 3 (9) | 33 (49) |
| Supplemental oxygen | 96 (11) | 3 (5) | 1 (3) | 19 (28) |
| Intensive care unit admission | 50 (6) | 2 (4) | 0 (0) | 3 (4) |
| Death | 12 (1) | 0 (0) | 0 (0) | 1 (1) |
Abbreviation: IQR, interquartile range.
aOnset of symptoms or first positive polymerase chain reaction ≥14 days after first dose of coronavirus disease 2019 vaccine.
bn = 63.
Twenty-seven (17.2%) patients with VOC infection were asymptomatic. The spectrum and frequency of other presenting symptoms were similar to those of patients infected with wild-type SARS-CoV-2. More patients with B.1.617.2 had pneumonia (49%), supplemental oxygen requirement (28%), and ICU admission (4%); this greater disease severity is reflective of their older age and greater comorbidity burden in this cohort.
In a multivariable logistic regression model, after adjustment for age, sex, comorbidities, and vaccination status, B.1.617.2 was associated with higher odds of pneumonia, though this was not statistically significant (aOR, 1.88; 95% CI: .95–3.76; Table 3). B.1.1.7 and B.1.351, on the other hand, were significantly less likely to be associated with pneumonia. Despite small numbers, B.1.617.2 was associated with significantly higher odds of severe infection (aOR, 3.02; 95% CI: 1.41–6.32; Supplementary Table 3). Individuals with B.1.617.2 infection were also more likely to receive remdesivir and/or corticosteroid treatment.
Table 3.
Odds Ratios of Candidate Predictors for Development of Coronavirus Disease 2019 Pneumonia from Firth’s Logistic Regression Analysis
| Univariable Model | Multivariable Modela | |||
|---|---|---|---|---|
| Candidate Predictor | Crude OR (95% CI) | P Value | Adjusted OR (95% CI) | P Value |
| Variant | ||||
| Wild-type | Ref | Ref | ||
| B.1.1.7 (Alpha) | 0.32 (.15–.63) | .00050 | 0.42 (.19–.86) | .016 |
| B.1.351 (Beta) | 0.19 (.051–.50) | .00034 | 0.33 (.086–.93) | .034 |
| B.1.617.2 (Delta) | 1.60 (.97–2.62) | .066 | 1.88 (.95–3.76) | .069 |
| Hypertension | 6.05 (4.23–8.78) | <.0001 | 2.04 (1.31–3.20) | .0016 |
| Diabetes mellitus | 6.23 (4.03–9.91) | <.0001 | 2.29 (1.37–3.90) | .0015 |
| Presence of other comorbidityb | 3.89 (2.64–5.79) | <.0001 | 1.50 (.92–2.43) | .10 |
| Age group, years | ||||
| <45 | Ref | Ref | ||
| 45–64 | 4.26 (3.17–5.76) | <.0001 | 2.92 (2.12–4.04) | <.0001 |
| ≥65 | 18.68 (11.2–32.7) | <.0001 | 7.06 (3.81–13.5) | <.0001 |
| Female sex | 0.88 (.68–1.14) | .32 | 0.80 (.59–1.07) | .14 |
| Coronavirus disease 2019 vaccinatedc | 0.29 (.077–.81) | .016 | 0.15 (.025–.62) | .0072 |
Abbreviations: CI, confidence interval; OR, odds ratio.
aAdjusted for lineage, hypertension, diabetes, presence of other comorbidities, age group, sex, and coronavirus disease 2019 (COVID-19) vaccination status.
bPresence of other comorbidity defined as Charlson score ≥1 after excluding diabetes mellitus from the calculation.
cOnset of symptoms or first positive polymerase chain reaction ≥14 days after first dose of COVID-19 vaccine.
Analysis of PCR Ct Values
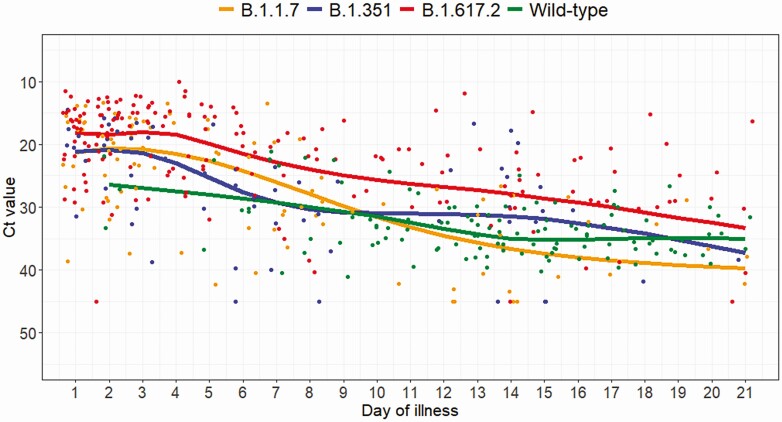
Comparing viral shedding patterns in the NCID cohort, patients with B.1.617.2 infection had significantly longer duration of positive PCR with Ct ≤30, a surrogate indicator of viable (therefore, infective) virus (Figure 2). When the infection group was included as a fixed factor with B.1.1.7 as the reference in the multivariable GAMM, B.1.617.2 was significantly associated with lower Ct values (P < .0005).
Figure 2.
Scatter plot of serial Ct values and marginal effect of day of illness from generalized additive mixed models for the 3 variants of concern and wild-type infections. B.1.17, n = 47; B.1.351, n = 21; B.1.617.2, n = 58; wild-type, n = 59. Abbreviation: Ct, cycle threshold. Negative PCR result is denoted by Ct value of 45.
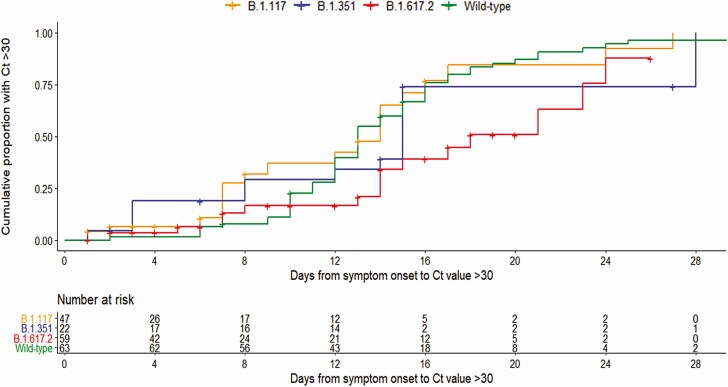
The Kaplan-Meier estimate of the median duration from symptom onset to first Ct value >30 was 13 days for wild-type, 14 days for B.1.1.7, 15 days for B.1.351, and 18 days for B.1.617.2. The estimated duration from symptom onset to first Ct value >30 was longer for B.1.617.2 compared with wild-type (P = .01) and B.1.1.7 (P = .029), and it was similar when compared with B.1.351 (P = .35; Figure 3). There was no significant difference in estimated duration from symptom onset to first Ct value >30 between B.1.1.7 and B.1.351 (P = .24). The estimated duration from symptom onset to first Ct value >30 for wild-type was not significantly different from that of B.1.1.7 (P = .54) and B.1.351 (P = .38).
Figure 3.
Kaplan-Meier curves of cumulative events of Ct value >30 stratified by variants of concern and wild-type. Abbreviation: Ct, cycle threshold.
DISCUSSION
In this retrospective cohort study comparing 3 VOCs with wild-type SARS-CoV-2, we found a possible association between B.1.617.2 infection and increased severity of COVID-19. This finding was similar in a broad analysis of Singapore’s national-level data and a detailed cohort of patient-level data with outcomes of severe infection or pneumonia. It is uncertain the extent to which this association reflects an effect of the variant, a chance finding due to relatively small sample sizes, or differences in the groups of individuals who were not accounted for in the multivariable models.
As part of broad-based disease surveillance, all international arrivals to Singapore, close contacts of confirmed cases, and patients who present with respiratory symptoms are screened for SARS-CoV-2 infection via PCR. Additionally, individuals in high-risk frontline occupations are subjected to regular routine testing. All confirmed infections are admitted for clinical evaluation. As such, our cohort depicts the entire spectrum of disease caused by VOCs, as opposed to only severe infections in cohorts of hospitalized patients in other settings.
Due to the relatively small number of COVID-19 cases in Singapore and a large healthcare resource buffer capacity, local healthcare systems have not been overwhelmed by large case numbers, obviating the significant confounding factor of resource limitations on clinical outcomes and mortality. The high mortality rate reported from the outbreak of B.1.617.2 (and other variants) in India has raised concerns regarding increased pathogenicity, especially as novel VOCs continue to increase in frequency throughout the world. Our finding of an association between VOCs and clinical severity is concerning if replicated in other situations and has been corroborated in recent technical reports from Public Health England and Public Health Scotland [14, 25]. Both studies reported an increased risk of hospitalization within 14 days of a positive test when infection was by B.1.617.2 compared with B.1.1.7.
The association of VOCs with lower Ct values (a surrogate for higher viral loads) and a longer duration of viral shedding has important implications on transmissibility. B.1.1.7 has been shown in multiple studies to have increased respiratory shedding [12, 26, 27]. We demonstrate that this is seen in B.1.617.2 as well. Although patients with B.1.617.2 presented earlier in the illness course in our cohort, the observed higher viral load is sustained across the entire duration of illness up to day 21, which would not have been impacted by timing of presentation or diagnosis. These virologic data provide a mechanism to support the epidemiologic data that suggest greater transmissibility of these VOCs [28]. Infection control policy with regard to duration of isolation of infected patients and public health control policies to reduce population-level transmission may have to be reevaluated given higher viral loads and prolonged shedding. In addition, future studies should quantify the duration of infectivity by subjecting interval respiratory specimens to viral culture. Higher viral loads have been associated with COVID-19 morbidity and mortality in some studies, though inconsistently, and it is not clear whether this reflects cause or effect [29, 30].
While there were a number of vaccine breakthrough infections in our cohort (18 patients with B.1.617.2, 5 with B.1.351; all received 2 vaccine doses prior), these patients had only mild disease, with none developing pneumonia or requiring supplemental oxygen. This provides real-world evidence of the protective efficacy of vaccination against severe disease by emerging VOCs, underscoring the importance of rapid implementation of vaccination programs. Clinical trials and post-implementation surveillance studies have demonstrated vaccine efficacy against B.1.1.7, B.1.351, and B.1.617.2 [31–34]. A separate study of vaccine-breakthrough infections by B.1.617.2 has also shown reduction in clinical severity with faster decline in viral loads in vaccinated individuals [35]. Detailed epidemiologic investigation was beyond the scope of this study, and we could not perform calculations of vaccine effectiveness against VOCs in our cohort. The population vaccination program in Singapore started in January 2021, and coverage increased gradually to 26% at the end of the study period [36]. Additional studies should be conducted to evaluate vaccine effectiveness against emerging variants in terms of both transmission and severe disease.
There are several limitations to this study. First, due to the small sample size, there were few patients who required supplemental oxygen or ICU admission, and there were fewer deaths. Smaller but statistically significant differences in clinical presentation with VOCs could have been missed due to inadequate power. For example, large population studies in the United Kingdom have found a signal toward increased mortality associated with B.1.1.7, which was not observed in a smaller cohort of hospitalized patients [10, 11, 37]. Larger cohort studies to compare different VOCs should be conducted to corroborate our findings.
Second, as this was a retrospective study, testing was not protocolized and PCR testing was conducted on the basis of assessment by individual managing physicians. Initial respiratory samples were also not processed in a centralized laboratory; therefore, estimation of viral load and shedding using Ct value is only a surrogate measure. However, the SARS-CoV-2 testing protocol in Singapore is conducted in a standardized manner using only validated PCR assays, and any possible interassay variability would have been distributed equally across the entire cohort.
Third, the proportion of successfully sequenced isolates differed throughout the study period, with a greater likelihood of successful sequencing in the later part of the study period. In Singapore, genomic surveillance has been conducted at NPHL for all isolates with a Ct value <30 throughout 2021. Changes in the proportion sequenced therefore reflects changes in the proportion of infections diagnosed with PCR Ct values <30. This may be due to a trend toward patients being diagnosed earlier during the illness course (consequently, with lower Ct values) or having a greater frequency of VOCs with higher viral loads (consequently, lower Ct values). There may be other unrecognized factors affecting the degree of success of genome sequencing; however, since we compared successfully sequenced VOCs with each other, this potential bias should have been mitigated.
Last, there are important differences between the control cohort from 2020 and the VOC cohort from 2021, which introduce bias to comparison between these cohorts. However, better management and treatment protocols and improved testing and contact tracing (with a resultant higher proportion of asymptomatic [17.2% in the VOC cohort vs 0.6% in the wild-type cohort] and early infections) in the VOC cohort are expected to have biased toward decreased severity compared with retrospective controls. Due to the limited ability to adjust for other unrecognized confounders, these findings, while of concern, should be interpreted with caution. There were also limited virologic and sequencing data available from wild-type infections in 2020, though this would not have impacted the difference in viral loads observed between B.1.617.2 and other 2 VOCs studied.
CONCLUSIONS
In this retrospective study, we found a possible association between infection by the B.1.617.2 variant and the odds of developing pneumonia or severe COVID-19. B.1.617.2 was associated with increased viral load and prolonged viral shedding in respiratory samples. This has implications in infection control and public health policy. Vaccinated patients appeared to have less severe illness. The increased transmissibility and possible greater severity of emerging variants provide a strong impetus for the rapid implementation of widespread vaccination programs.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank all clinical and nursing staff who provided care for the patients; Jeremy Nicholas Cutter at the National Public Health and Epidemiology Unit of the National Centre for Infectious Diseases who assisted with data management and analysis; and staff in the Infectious Disease Research and Training Office of the National Centre for Infectious Diseases who assisted with data collection.
Disclaimer. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, and approval of the manuscript; and decision to submit the manuscript for publication. The corresponding authors have full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Financial support. This study was funded by grants from the Singapore National Medical Research Council (COVID19RF-001, COVID19RF-008).
Conflict of interest disclosures. B. E. Y. reports personal fees from Roche and Sanofi outside the submitted work. All remaining authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Sean Wei Xiang Ong, National Centre for Infectious Diseases, Singapore, Singapore; Department of Infectious Diseases, Tan Tock Seng Hospital, Singapore, Singapore.
Calvin J Chiew, National Centre for Infectious Diseases, Singapore, Singapore; Ministry of Health, Singapore, Singapore.
Li Wei Ang, National Centre for Infectious Diseases, Singapore, Singapore.
Tze Minn Mak, National Centre for Infectious Diseases, Singapore, Singapore.
Lin Cui, National Centre for Infectious Diseases, Singapore, Singapore.
Matthias Paul H S Toh, National Centre for Infectious Diseases, Singapore, Singapore; Saw Swee Hock School of Public Health, National University of Singapore, Singapore, Singapore.
Yi Ding Lim, Ministry of Health, Singapore, Singapore.
Pei Hua Lee, National Centre for Infectious Diseases, Singapore, Singapore; Department of Infectious Diseases, Tan Tock Seng Hospital, Singapore, Singapore.
Tau Hong Lee, National Centre for Infectious Diseases, Singapore, Singapore; Department of Infectious Diseases, Tan Tock Seng Hospital, Singapore, Singapore; Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore; Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Po Ying Chia, National Centre for Infectious Diseases, Singapore, Singapore; Department of Infectious Diseases, Tan Tock Seng Hospital, Singapore, Singapore; Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore.
Sebastian Maurer-Stroh, Yong Loo Lin School of Medicine, National University of Singapore, Singapore; Bioinformatics Institute, Agency for Science, Technology and Research, Singapore, Singapore; A*STAR Infectious Diseases Labs (A*STAR ID Labs), Agency for Science, Technology and Research (A*STAR), Singapore; Department of Biological Sciences, National University of Singapore, Singapore.
Raymond T P Lin, National Centre for Infectious Diseases, Singapore, Singapore; Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Yee Sin Leo, National Centre for Infectious Diseases, Singapore, Singapore; Department of Infectious Diseases, Tan Tock Seng Hospital, Singapore, Singapore; Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore; Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Vernon J Lee, Ministry of Health, Singapore, Singapore; Saw Swee Hock School of Public Health, National University of Singapore, Singapore, Singapore.
David Chien Lye, National Centre for Infectious Diseases, Singapore, Singapore; Department of Infectious Diseases, Tan Tock Seng Hospital, Singapore, Singapore; Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore; Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Barnaby Edward Young, National Centre for Infectious Diseases, Singapore, Singapore; Department of Infectious Diseases, Tan Tock Seng Hospital, Singapore, Singapore; Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore.
References
- 1. Harvey WT, Carabelli AM, Jackson B, et al. ; COVID-19 Genomics UK Consortium. . SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 2021; 19:409–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang P, Nair MS, Liu L, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021; 593:130–5. [DOI] [PubMed] [Google Scholar]
- 3. Gupta RK. Will SARS-CoV-2 variants of concern affect the promise of vaccines? Nat Rev Immunol 2021; 21:340–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Campbell F, Archer B, Laurenson-Schafer H, et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill 2021; 26:2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Volz E, Mishra S, Chand M, et al. ; COVID-19 Genomics UK Consortium. . Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 2021; 593:266–9. [DOI] [PubMed] [Google Scholar]
- 6. Tegally H, Wilkinson E, Giovanetti M, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021; 592:438–43. [DOI] [PubMed] [Google Scholar]
- 7. Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021; 372:815–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vaidyanathan G. Coronavirus variants are spreading in India—what scientists know so far. Nature 2021; 593:321–2. [DOI] [PubMed] [Google Scholar]
- 9. Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021; 372:eabg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davies NG, Jarvis CI, Edmunds WJ, Jewell NP, Diaz-Ordaz K, Keogh RH; CMMID COVID-19 Working Group. . Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature 2021; 593:270–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Challen R, Brooks-Pollock E, Read JM, Dyson L, Tsaneva-Atanasova K, Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ 2021; 372:n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frampton D, Rampling T, Cross A, et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect Dis 2021; 21:1246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tracking of variants. https://www.gisaid.org/hcov19-variants/. Accessed 13 August 2021.
- 14. Sheikh A, McMenamin J, Taylor B, Robertson C; Public Health Scotland and the EAVE II Collaborators. . SARS-CoV-2 delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet 2021; 397:2461–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ong SWX, Young BE, Lye DC. Lack of detail in population-level data impedes analysis of SARS-CoV-2 variants of concern and clinical outcomes. Lancet Infect Dis 2021; 21:1195–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee VJ, Chiew CJ, Khong WX. Interrupting transmission of COVID-19: lessons from containment efforts in Singapore. J Travel Med 2020; 27:taaa039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pung R, Cook AR, Chiew CJ, et al. Effectiveness of containment measures against COVID-19 in Singapore: implications for other national containment efforts. Epidemiology 2021; 32:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee PH, Tay WC, Sutjipto S, et al. Associations of viral ribonucleic acid (RNA) shedding patterns with clinical illness and immune responses in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Clin Transl Immunology 2020; 9:e1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. COVID-19 CRF - ISARIC. https://isaric.tghn.org/novel-coronavirus/. Accessed 1 September 2021.
- 20. Treatment guidelines for COVID-19. https://www.ncid.sg/Health-Professionals/Diseases-and-Conditions/Pages/COVID-19.aspx. Accessed 1 June 2021.
- 21. Rambaut A, Holmes EC, O’Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020; 5:1403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. CoVsurver: mutation analysis of hCoV-19. https://www.gisaid.org/epiflu-applications/covsurver-mutations-app/. Accessed 1 June 2021.
- 23. Young BE, Ong SWX, Ng LFP, et al. Viral dynamics and immune correlates of COVID-19 disease severity. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bullard J, Dust K, Funk D, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis 2020; 71:2663–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. SARS-CoV-2 variants of concern and variants under investigation in England: technical briefing 14. PHE publications gateway number GOV-8530. Available at: https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201/. Accessed 1 September 2021.
- 26. Kidd M, Richter A, Best A, et al. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral loads in samples tested by ThermoFisher TaqPath RT-qPCR. J Infect Dis 2021; 223:1666–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Calistri P, Amato L, Puglia I, et al. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. Int J Infect Dis 2021; 105:753–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Threat assessment brief: emergence of SARS-CoV-2 B.1.617 variants in India and situation in the EU/EEA. https://www.ecdc.europa.eu/en/publications-data/threat-assessment-emergence-sars-cov-2-b1617-variants. Accessed 21 May 2021.
- 29. Volz E, Hill V, McCrone JT, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 2021; 184:64–75.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fajnzylber J, Regan J, Coxen K, et al. ; Massachusetts Consortium for Pathogen Readiness. . SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun 2020; 11:5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abu-Raddad LJ, Chemaitelly H, Butt AA; National Study Group for COVID-19 Vaccination. . Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N Engl J Med 2021; 385:187–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shinde V, Bhikha S, Hoosain Z, et al. ; 2019nCoV-501 Study Group. . Efficacy of NVX-CoV2373 Covid-19 vaccine against the B.1.351 variant. N Engl J Med 2021; 384:1899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of the Pfizer-BioNTech and Oxford-AstraZeneca vaccines on covid-19 related symptoms, hospital admissions, and mortality in older adults in England: test negative case-control study. BMJ 2021; 373:n1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med 2021; 385:585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chia PY, Xiang Ong SW, Chiew CJ, et al. Virological and serological kinetics of SARS-CoV-2 Delta variant vaccine-breakthrough infections: a multi-center cohort study. medRxiv 2021. Doi: 10.1101/2021.07.28.21261295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Coronavirus (COVID-19) Vaccinations. https://ourworldindata.org/covid-vaccinations?country=SGP/. Accessed 1 September 2021.
- 37. Grint DJ, Wing K, Williamson E, et al. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England, 16 November to 5 February. Eurosurveillance 2021; 26:2100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.