Abstract
ZEB is a zinc finger-homeodomain protein that represses transcription by binding to a subset of E-box sequences. ZEB inhibits muscle differentiation in mammalian systems, and its Drosophila orthologue, zfh-1, inhibits somatic and cardiac muscle differentiation during Drosophila embryogenesis. ZEB also binds to the promoter of pivotal hematopoietic genes (including those encoding interleukin-2, CD4, GATA-3, and α4-integrin), and mice in which ZEB has been genetically targeted show thymic atrophy, severe defects in lymphocyte differentiation, and increased expression of the α4-integrin and CD4. Here, we demonstrate that ZEB contains separate repressor domains which function in T lymphocytes and muscle, respectively. The most C-terminal domain inhibits muscle differentiation in mammalian cells by specifically blocking the transcriptional activity of the myogenic factor MEF2C. The more N-terminal domain blocks activity of hematopoietic transcription factors such as c-myb, members of the ets family, and TFE-III. Our results demonstrate that ZEB has evolved with two independent repressor domains which target distinct sets of transcription factors and function in different tissues.
Transcriptional repression is crucial in the regulation of developmental and differentiation processes from yeast to mammals (reviewed in references 21, 26, 30 and 46). Most transcriptional repressors and activators have an identifiable domain that regulates transcription and that is transferable to a heterologous DNA-binding domain (active repression or activation). The molecular mechanism of action of some of these transcriptional regulatory sequences is being uncovered, and these results may provide insight into relationships between regulatory sequences and how they selectively control tissue-specific processes. In most cases, there is a single regulatory domain. However, some activators and repressors contain more than one activator-repressor domain. For example, the T-cell factor NFAT-1 and the T-cell proto-oncogene RBTN-2 contain two independent transactivator domains (36, 39). Among repressors, Drosophila Krüppel contains two repressor domains that map to the N-terminal and the C-terminal regions of the protein (53). While the biological significance of multiple transcriptional regulatory domains remains unclear, it is likely that these domains are directed at different target genes or at different sets of transcription factors in a single promoter. Accordingly, there is some evidence that the repressor domains in Krüppel target a distinct but overlapping set of transcription factors in transfection assays (27).
We present evidence here that the zinc finger-homeodomain protein ZEB contains two independent repressor domains that target different sets of transcription factors and regulate distinct biologic processes. ZEB has been identified from Drosophila (where it was termed zfh-1) to vertebrates (where it received different names according to the species in which it was identified) (8, 16–20, 57). ZEB/zfh-1 is an active transcriptional repressor that binds to a subset of E boxes (with higher affinity for the CACCTG sequence) and E-box-like sequences in muscle and hematopoietic genes (20, 49–51, 56).
ZEB/zfh-1 regulates myogenic differentiation in both mammalian cells and Drosophila (49, 51, 56). In mammals, muscle differentiation is regulated by two families of positive factors: (i) the muscle regulatory factors (MRF; myoD, myf-5, myogenin, and MRF-4) which are basic helix-loop-helix proteins that induce muscle differentiation by binding E-box sequences in the promoter regions of muscle genes and activating their transcription (66); and (ii) the MEF-2 proteins, which synergize with MRF proteins to regulate muscle differentiation and activate transcription either by binding to specific DNA sequences or by interacting with the MRF proteins (44). Muscle differentiation is also under negative regulation by a number of factors such as ZEB, Id proteins, twist, and I-mfa (4, 11, 49, 57, 61); hence, myogenesis is the result of a fine balance between positive and negative factors. Binding sites for ZEB are present in the promoter regions of a number of muscle genes such as the MRF themselves (5, 15, 65), muscle creatine kinase (1), acethylcholine receptor δ (58), etc. ZEB/zfh-1 is an active transcriptional repressor, and regulation of muscle differentiation by ZEB/zfh-1 requires this repressor domain—the DNA binding domain alone does not block myogenesis (49, 51).
We also found that the Drosophila homologue of ZEB, zfh-1 (16, 32, 33), is also an active transcriptional repressor that negatively regulates the onset of somatic and cardiac muscle differentiation in Drosophila embryos (51). zfh-1 expression is downregulated before muscle differentiation proceeds, but maintenance of its expression (using a heat shock–zfh-1 construct) blocks somatic and cardiac myogenesis (reference 51 and unpublished results). We also found that vertebrate and Drosophila homologues are interchangeable, since zfh-1 also represses mammalian muscle differentiation and ZEB represses in Drosophila cells (51).
ZEB is also critical for T-cell differentiation and function in vertebrates, since mice with targeted deletion of the ZEB/zfh-1 gene show a drastic decrease in thymocyte numbers and defective T-cell differentiation at several stages (29). In fact, ZEB was originally discovered as a repressor of the IL2 gene and further studies have demonstrated that negative regulation of the IL2 gene correlates with ZEB activity (68, 73). ZEB also regulates the activity of the immunoglobulin (Ig) heavy-chain enhancer (20), GATA-3 (24), and the α4-integrin gene (29, 50). α4-integrin (as part of the heterodimer α4β1-integrin) is expressed on hematopoietic precursors and interacts with vascular cell adhesion molecule type 1 (VCAM-1) on stromal cells, and this interaction is crucial for hematopoietic differentiation (reviewed in reference 35). Binding to α4β1 can also transmit a costimulatory signal in T-cell activation (reviewed in reference 35). Additionally, α4β1 is critical in leukocyte trafficking to sites of inflammation (35). The α4 gene is dependent upon the combination of c-myb and the ets family of transcription factors (as are a number of other hematopoietic genes) (50). ZEB blocks the activity of c-myb and ets proteins individually, but together the proteins synergize to prevent repression by ZEB (50). This imposes the requirement for both c-myb and ets for expression of these hematopoietic genes. Both c-myb and ets are required for normal hematopoietic differentiation (2, 45, 55, 63), suggesting that alterations in ZEB expression may adversely affect hematopoietic differentiation. Indeed, as outlined above, mice lacking ZEB show defects in T-cell differentiation (29). Additionally, in these mice, two genes that are dependent upon the combination of c-myb, ets, and ZEB (α4-integrin and CD4) are upregulated (29).
Here we show that ZEB contains two independent repressor domains. One domain regulates muscle differentiation and specifically blocks the activity of the myogenic transcription factor MEF2C. The other domain functions in lymphocytes and regulates the activity of hematopoietic factors such as c-myb and ets family members. ZEB is thus, to the best of our knowledge, the first example of a transcriptional repressor that contains independent domains with distinct transcription factor specificities to regulate different biological processes.
MATERIALS AND METHODS
Cell culture.
The HT1080 fibrosarcoma (American Type Culture Collection [ATCC], Rockville, Md.) and C33A cervical carcinoma cell lines (ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies) containing 5% fetal calf serum and 5% calf serum (Life Technologies). C3H10T1/2 fibroblasts (ATCC) were grown in DMEM containing 13% fetal calf serum. The T-cell line Jurkat (ATCC) was grown in RPMI 1640 supplemented with 10% FCS. 293T cells were obtained from S. J. Korsmeyer (Dana Farber Cancer Institute, Boston, Mass.). CV1-MLP-CAT cells were obtained by stably cotransfecting CV1 cells (ATCC) with a chloramphenicol acetyltransferase (CAT) reporter driven by the major late promoter (MLP) and a neomycin-resistant vector.
Plasmid construction.
Gal4 and LexA fusion proteins of different ZEB fragments were obtained by PCR of the corresponding regions and in-frame cloning of the product in the BamHI-XbaI site of PM1 and PBXL3, respectively. RD (repressor domain)-ZEB refers to the region between the two zinc finger clusters (from nucleotides 906 to 2706). Region 1 comprises the cDNA between nucleotides 906 and 1626; region 2 comprises the cDNA between nucleotides 1626 and 2280; and region 3 comprises the cDNA between nucleotides 2280 and 2706. Out-of-frame GAL4 and LexA constructs for RD-ZEB were obtained by out-of-frame insertion of RD-ZEB in PM2 and PBXL3 and used as controls (data not shown). Most Gal4 activators were either previously described (67) (Gal4-CTF, Gal4-VP16, Gal-Sp1, Gal4-MEF2C [amino acids 1 to 465], Gal4-NFκB-p65, Gal4-PU.1, Gal4–E2F-1 [amino acids 285 to 437], Gal4–c-fos [amino acids 21 to 380], Gal4–ITF-1 [amino acids 1 to 427], and Gal4–TFE-III [amino acids 2 to 216]) or obtained from the following investigators: Gal4–c-myb–CD (J. Lipsick, Stanford University, Stanford, Calif.), Gal4-myoD (amino acids 1 to 318) (G. Tomaselli, Johns Hopkins University, Baltimore, Md.), and Gal4–c-jun (C. Caelles, University of Barcelona, Barcelona, Spain).
pETS contains three ets sites from the α4 gene promoter as previously described (50). pETS-ZEB contains two ZEB binding sites (CACCTG) upstream of pETS; pETS-ZEB-MYB contains four myb binding sites upstream of pETS-ZEB. pETSmut-ZEB-MYB is as pETS-ZEB-MYB but with the ets sites mutated. Details on these constructs have been previously described (reference 50 and references therein). The transcriptional elements included in the deletional constructs 76α4-CAT, 400α4-CAT, and 2.0α4-CAT as well as details on their construction were previously reported (reference 50 and references therein). An expression vector for c-myb (pSV–c-myb) was obtained from B. Calabretta (Jefferson University, Philadelphia, Pa.).
Fusion proteins between the DNA binding domain of ZEB and the different regions in RD-ZEB were designed as follows. The C-terminal zinc fingers of ZEB (DB-ZEB) were cloned in the MluI-XbaI sites of pCI-neo (Promega Corp. Madison, Wis.). Annealed oligonucleotides of a Kozak sequence, an ATG codon, and a FLAG sequence (GCC ACC ATG GAC TAC AAG GAC GAC GAT GAC AAG) were cloned upstream of and in frame with DB-ZEB in the XhoI-MluI sites. PCR fragments of regions 1, 2, and 3 were then cloned in frame with DB-ZEB in the XbaI-NotI sites of pCI-neo. A simian virus 40 nuclear localization signal and an in-frame stop codon are immediately downstream of the cDNA sequence (CAA ATG GAA CCT AAG AAG AAG AGG AAG GTT TAA).
pGL contains six LexA sites 30 bp upstream of two (or five) Gal4 sites and was described previously (67). A new version of pGL (where the LexA sites are 1.7 kb upstream of the Gal4 sites) was constructed by cloning a BglII-HindIII 1.7-kb fragment corresponding to the Drosophila cDNA Nautilus (cloned into the EcoRI site of pSP73 [Promega Corp.] and obtained from A. Michelson, Brigham and Womens Hospital, Boston, Mass.) into the BglII-HindIII site of pGL.
CMVp300 was obtained from Dr. D. Livingston (Dana-Farber Cancer Institute, Boston, Mass.). Gal4 p300 1737–2414 was obtained from A. Giordano (Jefferson University). CMV-E1A-12S and E1A-12SΔ2–36 were obtained from E. Harlow (Massachusetts General Hospital, Charlestown, Mass.).
A CAT reporter driven by the MLP was obtained from D. E. Ayer (University of Utah, Salk Lake City, Utah).
An expression vector for mitogen-activated protein (MAP) p38 kinase (pSRα2-p38) was obtained from S. Chellappan (College of Physicians and Surgeons, Columbia University, New York, N.Y.).
Transient transfections and CAT assays.
HT1080 cells were transfected by the calcium phosphate method, and Jurkat cells were transfected by electroporation as previously described (49, 50). After 48 h, lysates were collected and the transfection efficiency was corrected as previously described by cotransfection of a thymidine kinase-driven luciferase vector (49, 50). CAT assays were performed as described previously (49). DNA was brought to a total of 6 μg for 60-mm dishes (or to 20 μg for 100-mm dishes) with pBluescript (Stratagene, La Jolla, Calif.).
For transfection of CV1-MLP-CAT cells, cells were plated into 150-mm dishes, cotransfected with 60 μg of Gal4-ZEB constructs and 5 μg of puro-BABE (a puromycin-resistant vector), and maintained in growth medium supplemented with 800 μg of G418 (Life Technologies) and 1.5 μg of puromycin for 4 days to both select transfected cells and assess newly synthesized CAT activity. In the last 48 h, 300 nM trichostatin A (TSA) (Wako Chemicals USA, Inc., Richmond, Va.) was added to the medium.
CAT results are means of duplicate assays and are all representative of at least five separate experiments with standard deviations below 15%.
Myogenic conversion assays and immunostaining.
For 10T1/2 cell myogenic conversion assays, 10T1/2 cells were transfected by the Lipofectamine method in Optimem medium (Life Technologies). After 12 h, medium was removed and replaced with differentiation medium (2% horse serum–DMEM). After 4 to 5 days, the cells were fixed in methanol and stained with anti-myosin heavy chain MF-20 (R. Kopan, Washington University, St. Louis, Mo.) monoclonal antibody as previously described (49). After being washed to remove unbound antibody, the cultures were incubated sequentially with horseradish peroxidase-conjugated anti-mouse IgG antibody (Jackson ImmunoResearch, West Grove, Pa.) and with diaminobenzidine substrate (Vector Laboratories, Burlingame, Calif.). Nuclei were counterstained with hematoxylin (Zymed, South San Francisco, Calif.).
Western blot analysis.
At 48 h after transfection with the appropriate plasmids, C33a cells were lysed in ELB (150 mM NaCl, 50 mM HEPES [pH 7.0], 5 mM EDTA, 0.1% Nonidet P-40), sonicated briefly, and centrifuged to remove nuclear debris as described previously (51). The lysates were then loaded onto a sodium dodecyl sulfate–4 to 15% polyacrylamide gradient gel (Bio-Rad, Hercules, Calif.) and transferred overnight to a polyvinylidene difluoride membrane (Immobilon P; Millipore Corp., Bedford, Mass.) in 10 mM CAPS [3(cyclohexylamino) 1-propanesulfonic acid] buffer (pH 11). The membrane was serially incubated with anti-Flag polyclonal antibody (Santa Cruz Biotechnologies, Santa Cruz, Calif.) and anti-rabbit IgG-horseradish peroxidase secondary antibody. After several washes, the membrane was then developed by the chemiluminescence method (Renaissance; NEN Life Sciences Products, Boston, Mass.) as specified by the manufacturer.
RESULTS
ZEB is a selective transcriptional repressor.
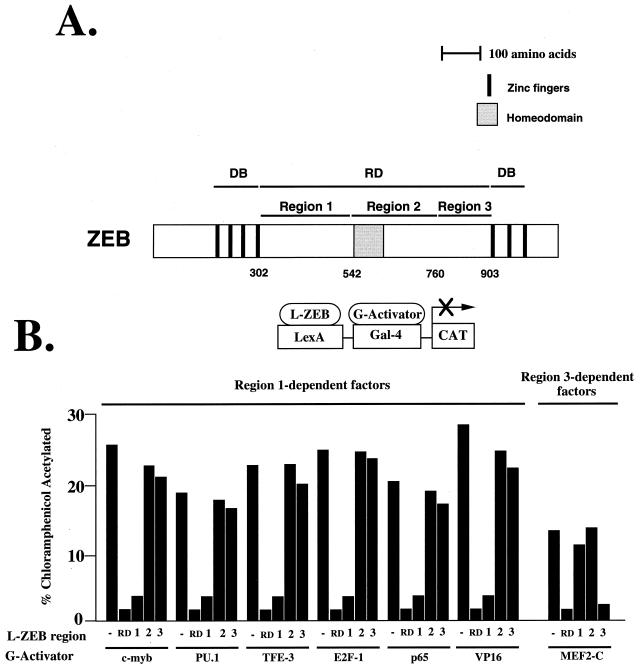
In previous studies, we found that the central region of ZEB, between the N-terminal and C-terminal zinc finger DNA binding domains, contains the repressor domain (RD) (schematized below in Fig. 2A) (49). We found that this RD could function independently to repress transcription when fused to the DNA binding domain from the yeast transcription factor Gal4 (49). In this study, we asked which transcription factors are repressed by ZEB. We found that ZEB blocked the activity of some transcription factors but not others (Fig. 1). It is of note that the set of transcription factors blocked by ZEB includes factors that are important for both myogenesis (MEF2C) and lymphoid differentiation (i.e., c-myb, ets family members, TFE-III, and the NF-κB protein p65).
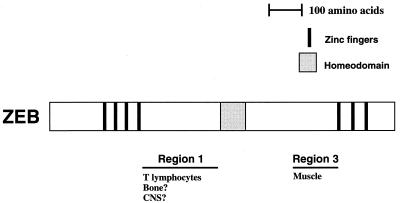
FIG. 2.
ZEB contains two independent repressor domains that target distinct sets of transcription factors. (A) Scheme of ZEB. The RD of ZEB lies between the zinc finger regions (the DNA binding domains [DB]) (49). Numbers indicate amino acids. (B) RD-ZEB contains two independent repressor regions. Regions 1, 2, and 3 of RD-ZEB (indicated by 1, 2, and 3) were fused to the DNA binding domain of LexA and tested for their ability to repress different transcriptional activators fused to the DNA binding domain of yeast GAL4 (G-activators as in Fig. 1). Eight-tenths microgram of pGL (as in Fig. 1) was cotransfected in HT1080 cells with 2 μg of L-ZEB constructs and 0.1 to 0.5 μg of different G-activators. After 36 to 48 h, cells were harvested and the CAT activity was determined. Equal molar amounts of the control LexA expression vector did not affect the activity of any of the different Gal4 activators (data not shown). CAT results are the mean of duplicate assays and are all representative of at least five separate experiments with standard deviations below 15%.
FIG. 1.
ZEB is a selective transcriptional repressor that blocks the activity of some transcription factors but not others. The repressor domain (RD-ZEB) was fused to the DNA binding domain of the bacterial protein LexA (L-ZEB) and tested for its activity to repress several transcriptional activators fused to the DNA binding domain of yeast Gal4 (G-activators). Eight-tenths microgram of the pGL construct containing LexA sites 30 bp upstream of Gal4 sites was cotransfected into HT1080 cells with 2 μg of L-ZEB and 0.1 to 0.5 μg of different G-activators. After 36 to 48 h, cells were harvested and the CAT activity was determined as described in Materials and Methods. Equal molar amounts of the control Lex A expression vector did not affect the activity of any of the different G-activators (data not shown). CAT results are the mean of duplicate assays and are all representative of at least five separate experiments with standard deviations below 15%.
ZEB contains two independent repressor domains.
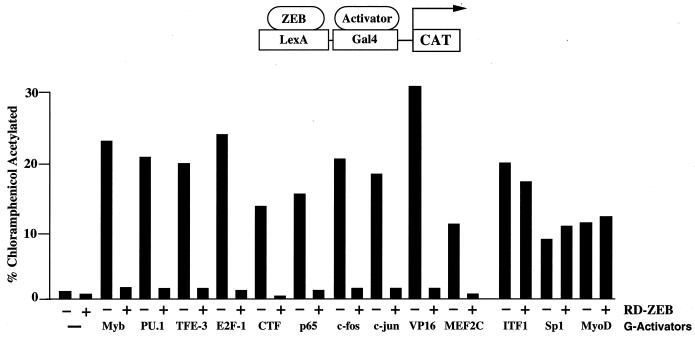
To further localize sequences important for repressor activity, we divided the RD into three parts (Fig. 2A). Region 1 includes sequences N-terminal of the central homeodomain; region 2 contains the homeodomain, and region 3 contains the C-terminal region of the RD. Region 2 did not show any repressor activity in these assays (Fig. 2B). We found that region 1 was sufficient to repress all of the transcription factors repressed by RD, as shown in Fig. 1, with the exception of the myogenic factor, MEF2C (Fig. 2). Factors repressed by region 1 include hematopoietic factors (e.g., c-myb, ets family members, and TFE-III) as well as several more general and ubiquitous factors (Fig. 2B and data not shown). Interestingly, region 3 blocked the activity of MEF2C but had no effect on the activity of any other transcription factor we tested (Fig. 2B and data not shown). These results indicate that ZEB has two independent repressor domains, which target different subsets of transcription factors.
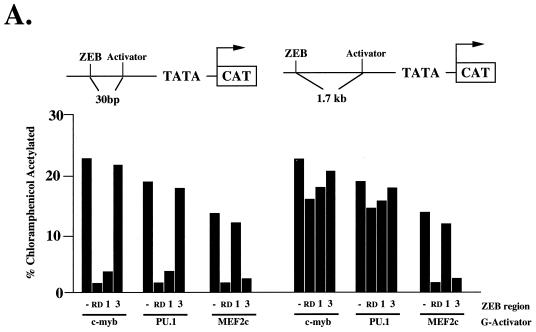
Recent reports indicate that in Drosophila, transcriptional repressors fall into two categories according to their ability to repress at short (<50 bp) or long (>300 bp) distances (22, 23). This distinction between long- and short-range repressors is thought to be important in establishing patterns of gene expression in Drosophila (21–23). Short-range repressors permit enhancer autonomy in modular promoters—repressors would affect the activity only of enhancers nearby. Long-range repressors function to block expression without regard to promoter organization. We tested the two repressor regions of RD-ZEB to determine whether they act as long- or short-range repressors (Fig. 3A). We constructed reporter plasmids in which the distance between the transcriptional activator binding site and the ZEB binding site was either 1.7 kb (long range) or 30 bp (short range). Region 1 repressed only at short range, whereas region 3 was equally active at both short and long ranges (Fig. 3A). These results provide additional evidence that regions 1 and 3 repress through distinct mechanisms.
FIG. 3.
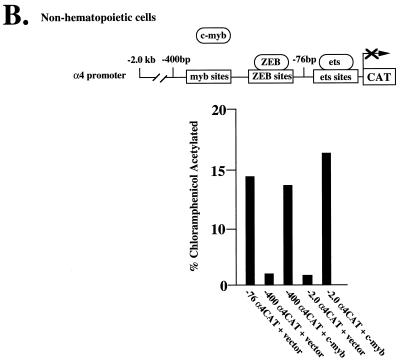
Effect of promoter position on repression by regions 1 and 3 of ZEB and regulation of c-myb and ets by domain 1. (A) Region 3 is a long-range repressor, whereas region 1 is a short-range repressor. Eight-tenths microgram of a reporter construct containing LexA sites either 30 or 1.7 kb upstream of Gal4 sites was cotransfected with 2 μg of expression vectors for fusion proteins between LexA and regions 1 and 3 along with 0.1 to 0.3 μg of G–c-myb, G-PU.1, and G-MEF2 expression vectors. LexA alone did not have any effect on the activity of the different Gal4 activators (data not shown). (B) ets and c-myb synergize in the α4-integrin promoter to overcome repression by ZEB. Three micrograms of a reporter containing 2.0 kb, 400 bp, or 76 bp of the α4 promoter region (52) was cotransfected in c-myb-negative HT1080 cells along with 3 μg of vector alone or with a vector expressing c-myb. (C) Region 1 regulates the activity of ets and c-myb sites in the α4-integrin promoter. Sixty micrograms of each of the reporters pETS, pETS-ZEB-MYB, pETS-ZEB plus pETSmut-ZEB-MYB, pETSmut-MYB, and pETS-MYB (50) was cotransfected into Jurkat cells by electroporation along with 30 μg of expression vectors encoding DB-ZEB or equal molar amounts of full-length ZEB (FL) or DB-ZEB fused to regions 1, 2, and 3. Note that domain 1 represses transcriptional activity mediated by c-myb and ets independently but not when the two factors are combined. CAT results are the mean of duplicate assays and are all representative of at least five separate experiments with standard deviations below 15%.
Region 1 of the ZEB RD regulates the activity of the hematopoietic transcription factors ets and c-myb.
c-myb is essential for normal hematopoietic differentiation (45). The ets family contains a number of members, some of which are hematopoiesis specific and some of which are more widely acting (2). Mutations in hematopoietic ets genes have demonstrated that these proteins, as with c-myb, are essential for hematopoietic differentiation (55, 63). A number of hematopoietic genes contain binding sites for both c-myb and ets, and it has been demonstrated that the two factors synergize to activate transcription (40). In fact, the E26 virus encodes a v-myb–v-ets fusion protein, and it has been shown that the synergy between these two factors is essential in the generation of erythroleukemias in animal models (41, 42).
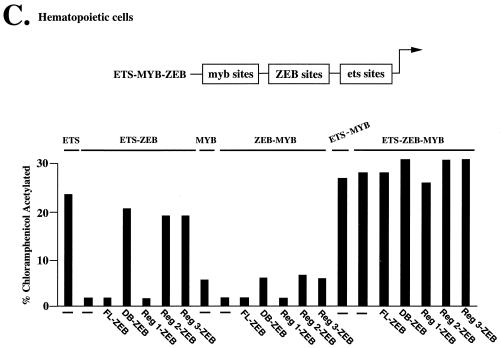
ZEB sites are found in a number of c-myb- and ets-dependent genes including those encoding α4-integrin, p56lck, and CD4, where they function as silencers (40, 50, 54). ZEB blocks the activity of c-myb or ets alone, but together these factors synergize to resist repression by ZEB (50). This arrangement may have two consequences: (i) it would ensure that some c-myb- and ets-dependent genes are not expressed until both factors appear during hematopoietic differentiation, and (ii) since ets factors contain a common binding domain and some are ubiquitous, ZEB would also ensure that c-myb- and ets-dependent genes are not ectopically expressed in nonhematopoietic cells. This is illustrated in Fig. 3B, where the activity of ets sites located in bp 1 to −76 of the α4-integrin gene promoter is repressed in nonhematopoietic cells (lacking c-myb expression) by the activity of ZEB sites located at bp −361 and −399 (50 and Figure 3B). Expression of c-myb is able to restore the activity of the α4 promoter. The c-myb sites in the α4 gene promoter are also sensitive to ZEB repression when the ets sites are not present or are mutated (see below and data not shown).
We wondered whether region 1 of the ZEB repressor domain is sufficient to recapitulate the activity of ZEB toward c-myb and ets. The results shown in Fig. 3C indicated that region 1 inhibits the activity of c-myb and ets products individually. To demonstrate this regulation in the context of a more normal promoter setting than that shown in Fig. 2B, individual regions of RD-ZEB were fused to the DNA binding region of ZEB (C-terminal zinc fingers, which have the highest DNA binding affinity [56]). Expression of these C-terminal zinc fingers (DB-ZEB) alone did not have any repressor activity and released repression by endogenous ZEB (50) (Fig. 3C). Neither, region 2 nor 3 (fused to DB-ZEB) had any effect on c-myb or ets activity. In contrast, region 1 efficiently blocked the activity of c-myb and ets sites individually. Also, as with full-length ZEB, region 1 was not able to repress the combination of c-myb and ets sites. The level of expression of these fusion proteins is shown below in Fig. 5C. These results suggest that region 1 is responsible for repression of α4-integrin (and probably other hematopoietic genes) by ZEB in lymphoid cells and thus mediates its role in lymphoid differentiation and function.
FIG. 5.
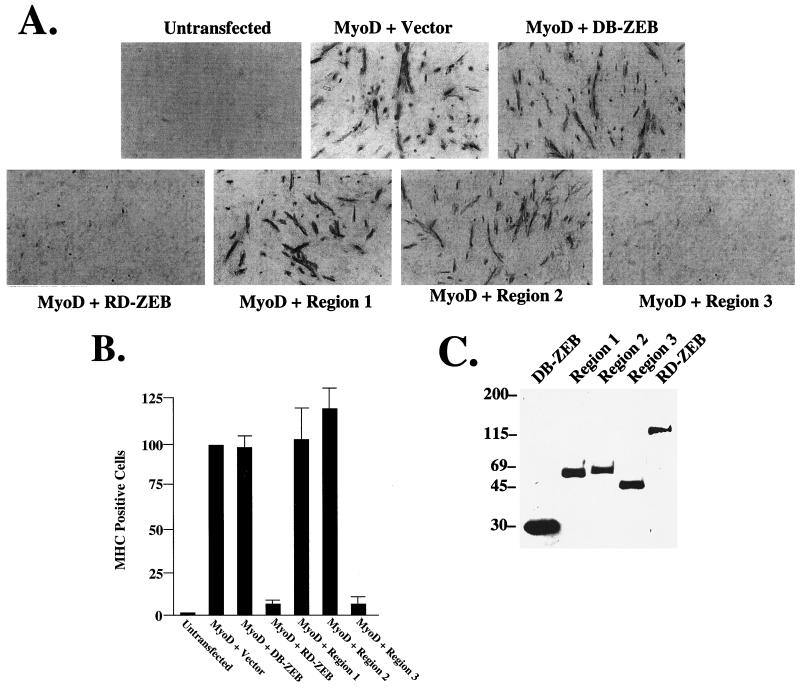
Region 3 of RD-ZEB selectively inhibits myogenic differentiation. (A) CH3-10T1/2 cells were transfected with 0.5 μg of a myoD expression vector and equal molar amounts of expression vectors for the DNA binding domain of ZEB (DB-ZEB) and DB-ZEB fused to RD-ZEB and regions 1, 2, and 3. The cells were switched to differentiation medium, and the formation of myotubes was assayed by immunostaining with myosin heavy chain as previously described (49). (B) Quantification of the results in panel A. Values represent the number of nuclei in myosin heavy-chain (MHC)-positive myotubes with the corresponding standard deviation. One hundred is an arbitrary value for MyoD + Vector. (C) Western blot analysis of the transfected proteins. Proteins were Flag tagged and detected with an anti-Flag antibody.
Region 1 of ZEB represses transcription by a mechanism involving histone deacetylase activity.
p300/CREB binding protein (CBP) interacts with a number of transcription factors and activates transcription by acetylation of histones and disruption of the nucleosome structure (reviewed in reference 38). While p300/CBP itself has histone acetyltransferase (HAT) activity on its own, the C-terminal region of the protein interacts with PCAF, which contains HAT activity essential for p300/CBP function (47). p300/CBP was initially discovered by its interaction with the adenovirus protein E1A (62). The amino-terminal region of E1A (amino acids 2 to 36) also binds to the C-terminal region of p300/CBP and displaces PCAF (62, 72). In addition, E1A can directly block p300/CBP HAT activity (9).
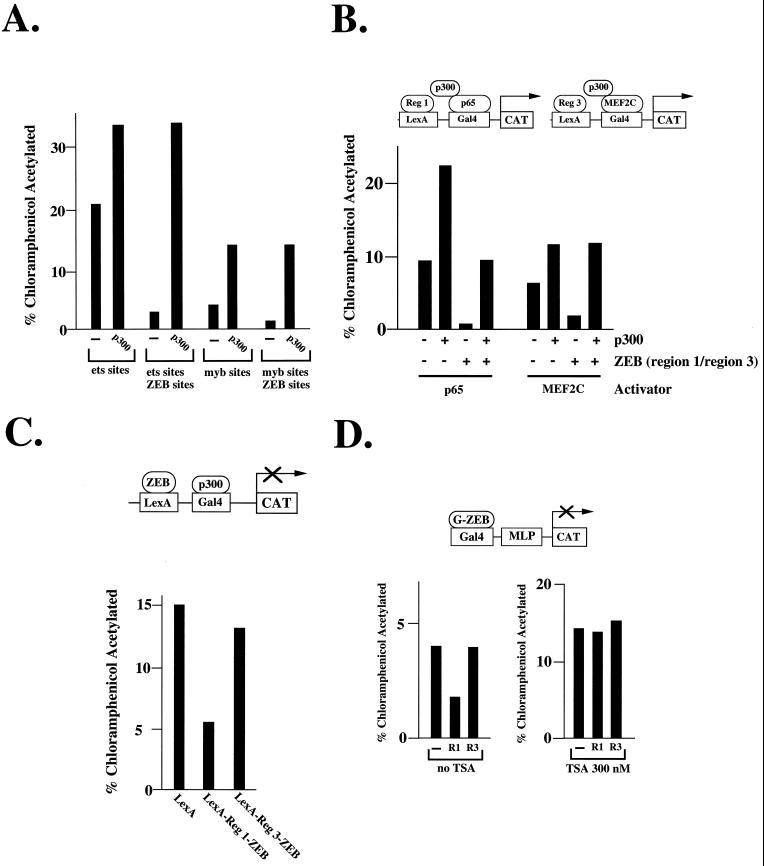
Factors repressed by ZEB (including c-myb, ets, and MEF2C) interact with p300/CBP and synergize with it in transcriptional activation (references 14, 70, and 71) and unpublished results. That prompted us to investigate whether ZEB may repress transcription by a mechanism involving p300/CBP. Inclusion of ZEB binding sites upstream of the ets or c-myb sites blocked the activity of both transcription factors (Fig. 3C) (50). We reasoned that if region 1 of ZEB were acting to block p300/CBP activity, overexpression of p300 should overcome repression by ZEB. Indeed, we found that overexpression of p300 completely reversed the repression of c-myb or ets sites by ZEB in hematopoietic cells (Fig. 4A), further suggesting that region 1 of ZEB could be targeting the p300 co-activator. Similar rescue of ZEB repression was obtained for NF-κB (Fig. 3B). We also found that repression of MEF2C by region 3 was fully rescued by overexpressing p300 (Fig. 3B).
FIG. 4.
Region 1 of ZEB blocks transcriptional activity of c-myb and ets by targeting the coactivator p300/CBP. (A) Repression of ets and c-myb activity by ZEB is overcome by overexpression of p300. Thirty micrograms of each of the reporter constructs containing ets sites (76α4CAT [40]) or c-myb sites (as described in the legend to Fig. 3C and reference 50) with or without ZEB sites upstream (50) was cotransfected by electroporation with 60 μg of CMV-p300. The empty cytomegalovirus vector did not have any effect on transcriptional activity (data not shown). After 36 to 48 h, cells were harvested and the CAT activity was determined as described in Materials and Methods. (B) Repression of MEF2C by region 3 is rescued by overexpression of p300. Eight-tenths microgram of the pGL reporter construct containing LexA sites 30 bp upstream of Gal4 sites was cotransfected in 293T cells with 0.2 μg of Gal4-p65 or 0.5 μg of Gal4-MEF2C and with or without 2 μg of LexA-region 1-ZEB or LexA-region 3-ZEB, respectively, and with or without 2.5 μg of CMV-p300. (C) Region 1 of ZEB represses transcriptional activity mediated by p300. Eight-tenths microgram of the reporter pGL construct containing LexA sites 30 bp upstream of Gal4 sites was cotransfected into C33a cells with 0.5 μg of Gal4-p300 (amino acids 1737 to 2414) (74). After 36 to 48 h, the cells were harvested and the CAT activity was determined as described in Materials and Methods. (D) Region 1 of ZEB represses the activity of the histone acetylase-dependent MLP. CV1-MLP-CAT cells were transfected as described in Materials and Methods. After 4 days in selection medium, 300 nM trichostatin A (TSA) was added to the medium and the cells were cultured for an additional 48 h. Six days after transfection, the cells were collected and CAT activity was assayed as described in Materials and Methods. The CAT results are the mean of duplicate assays and are all representative of at least five separate experiments with standard deviations below 15%.
Next, we wanted to determine whether ZEB domains could block p300/CBP transcriptional activity. Since p300/CBP does not bind directly to DNA, we used a fusion protein where the C-terminal region of p300, which binds PCAF, was linked to the DNA binding domain of Gal4. This fusion protein is a potent transcriptional activator (74) (Fig. 4C). When region 1 of ZEB was targeted to the promoter, transcriptional activation by p300/CBP was blocked (Fig. 4C). Region 3 of ZEB (or the retinoblastoma protein, which is also a transcriptional repressor [67]) did not have significant effect on p300 activity (Fig. 4C and data not shown). This experiment suggested that although p300/CBP was able to overcome ZEB repression by both regions 1 and 3, only region 1 is actively repressing p300 transcriptional activity.
Since we were not able to detect interaction between ZEB and p300/CBP (data not shown), we wondered whether region 1 of ZEB may offset p300 activity by recruitment of histone deacetylase activity. To investigate this, we studied the ability of regions 1 and 3 of ZEB to inhibit the activity of the major late promoter (MLP). It has been shown that repression of this promoter by the Mad and retinoblastoma repressors depends completely upon recruitment of histone deacetylase activity (28, 37, 38). As shown in Fig. 4D, region 1 but not region 3 repressed the activity of the MLP reporter. Moreover, the histone deacetylase inhibitor trichostatin A rescued repression by region 1, further suggesting that this region of ZEB offsets p300 activity by a histone deacetylase mechanism.
Region 3 of RD-ZEB blocks myogenesis.
Expression of myoD is sufficient to trigger the myogenic pathway in a number of cell types (66). MyoD activates a cascade of transcription factors, including members of the MEF2 family, which collaborate with myoD to amplify the muscle differentiation program (44). A dominant negative form of MEF2 blocks vertebrate myogenesis (48), and somatic and cardiac myogenic differentiation is disrupted in Drosophila embryos with mef2 null mutations (6, 34), indicating that MEF2 function is essential in myogenesis from Drosophila to mammals. Negative regulation of muscle by ZEB is not due to ZEB binding E boxes and displacing myogenic factors (49). ZEB binds only a small subset of E boxes (and it also binds a subset of non-E-box sequences), and thus it cannot efficiently displace MRF proteins. Indeed, DB-ZEB has no effect on myogenesis (49). Instead, the entire molecule was necessary for inhibition of muscle differentiation (49); thus, ZEB is functioning as an active repressor when bound to promoters.
We wondered which region of RD-ZEB is important for inhibition of myogenesis. For these studies, we fused RB-ZEB and each of the three regions of the RD to the DNA binding domain of ZEB (these are the same constructs used in the lymphocyte experiments in Fig. 3C). Expression vectors for these constructs were then tested in myogenesis assays, where myoD was used to trigger myogenesis in 10T1/2 cells. Myogenic conversion was followed by expression of the muscle-differentiation specific myosin heavy chain (Fig. 5A). We found that the RD is sufficient to block myogenesis. While neither region 1 nor region 2 had any effect on myogenic differentiation, region 3 blocked it efficiently (Fig. 5A and B). Western blots of these ZEB constructs are shown in Figure 5C. Even though DB-ZEB had no effect in these assays, it was expressed at higher levels than the other constructs were. Note that even though region 1 had no effect in these assays, the same construct efficiently repressed transcription in lymphoid cells (where the region 3 construct was inactive) (Fig. 3B). Together, these results suggest that region 1 of RD-ZEB blocks selectively the activity of several hematopoietic factors and that it is responsible for the regulation of hematopoietic genes and therefore for lymphoid differentiation and function. In contrast, region 3 functions in muscle cells, where it appears to block myogenesis and specifically inhibits the activity of MEF2.
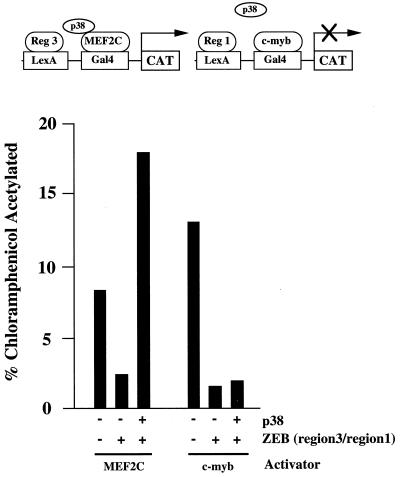
p300/CBP interacts and transcriptionally synergizes with MEF2C (52). The MAP kinase p38 is required for MEF2C activity, as shown by experiments examining mutants for phosphorylation sites on MEF2C or overexpression of a p38 dominant negative (25). Phosphorylation of cyclic AMP response element binding protein (CREB) by the calcium calmodulin CAMIV kinase is required for efficient recruitment of CBP/p300 by CREB (10, 66a). A similar mechanism could be postulated for MEF2C. We therefore investigated whether p38 kinase was able to rescue repression of MEF2C by region 3 of ZEB. Indeed, and as shown in Fig. 6, overexpression of p38 overcomes repression by region 3 of ZEB. We found that CAMIV kinase also overcomes repression by region 3 of ZEB (see Discussion) (data not shown). However, repression of c-myb by region 1 of ZEB remained unaffected by overexpression of p38 (or CAMIV kinase) (Fig. 6 and data not shown), further suggesting different mechanisms of repression for regions 1 and 3 of ZEB.
FIG. 6.
Repression of MEF2C activity by region 3 of ZEB is rescued by MAP p38 kinase. A 0.8-μg portion of the reporter pGL construct containing LexA sites 30 bp upstream of Gal4 sites was cotransfected into C33a cells with 0.5 μg of Gal4-MEF2C or 0.2 μg of Gal4–c-myb and 2 μg of either LexA-region 1 or LexA-region 3 of ZEB in the presence or absence of 1 μg of a plasmid encoding MAP p38 kinase (pSRα2-p38). Expression of the empty vector corresponding to p38 did not have any effect (data not shown). After 36 to 48 h, the cells were harvested and the CAT activity was determined as described in Materials and Methods. CAT results are the mean of results of duplicate assays and are all representative of at least five separate experiments with standard deviations below 15%.
DISCUSSION
ZEB/zfh-1 is a transcriptional repressor that functions in both muscle and T-cell differentiation (29, 49–51, 56). In this report we present evidence that distinct and independent repressor domains are responsible for these two biological activities (Fig. 7). To our knowledge, ZEB is the first example of a protein with independent repressor domains that target distinct sets of transcription factors.
FIG. 7.
ZEB contains independent repressor regions that target distinct sets of transcription factors and regulate different tissues. Region 1 of ZEB represses a number of hematopoietically restricted transcription factors including c-myb, ets family members and TFE-III, but it also represses other, more general transcription factors such as NFκB p65 protein, E2F-1, CTF, c-fos, and c-jun. These results suggest that in addition to its role in regulating T lymphocyte differentiation, region 1 may be involved in other reported ZEB/zfh-1 functions such as bone morphogenesis and central nervous system (CNS) differentiation (29, 64). Region 3 is very specific in its activity—it inhibits MEF2C activity to regulate muscle differentiation.
We have found that p300/CBP is able to overcome repression by regions 1 and 3 of ZEB. Most factors repressed by ZEB are known to bind p300 and are dependent on p300 for their activity. However, recent studies indicate that the requirements for the different components of the HAT coactivator complex vary among different transcription factors, with some factors requiring the HAT activity of p300, others requiring the HAT activity of pCAF, etc., (31). Therefore, it is likely that repression of different factors involves offsetting of different component of the HAT coactivator complex.
Although repression by both regions 1 and 3 is overcome by overexpression of p300/CBP, only region 1 represses p300 transcriptional activity. One mechanism by which region 1 is able to repress p300 activity could be by direct interaction with p300 and blocking of its ability to recruit other components of the histone acetylase complex. Another mechanism would be offsetting of p300 activity by recruitment of a deacetylase activity. Accordingly, we found that region 1 but not region 3 was able to repress the activity of the MLP and that the histone deacetylase inhibitor TSA is able to reverse that effect. Repression of this reporter by other repressors such as Mad or the retinoblastoma protein is fully dependent on the recruitment of histone deacetylase complexes (28, 37, 38). However, the precise mechanism of repression by region 1 remains to be determined, since we have been unable to detect interaction of RD-ZEB with either p300/CBP or histone deacetylase-1 (reference 37 and data not shown).
A number of hematopoietic gene promoters depend on the synergy between c-myb and ets for their activity. This is the case for p56lck and CD13, where mutation of either the c-myb or ets sites abolished their promoter activity (40). In other genes, such as the α4 gene, ets sites are active even in the absence of c-myb (the case in α4-negative cells), but both factors are necessary to resist repression by ZEB (as in α4-positive hematopoietic cells) (50). While the molecular basis for the transcriptional cooperation between ets and c-myb is not understood yet, it is clear that the factors together are still dependent on p300/CBP, since overexpression of E1A-12S (but not a 2–36 deletion mutant of E1A-12S) abolishes c-myb and ets activity (data not shown). Therefore, it is possible that the binding of both factors to p300/CBP renders the complex inaccessible to repression by ZEB. A similar mechanisms has been proposed for the cooperation between c-myb and C/EBP in the regulation of the myelomonocitic genes (43).
Results of experiments with mice where the ZEB gene has been knocked out indicate that this protein is essential for lymphoid differentiation, which correlates with the fact that ZEB is highly expressed during hematopoietic differentiation but it is downregulated as T cells differentiate and move into circulation (reference 29 and unpublished results). We have previously found that ZEB inhibits the activity of the factors c-myb and ets, which regulate the activity of several hematopoietic genes (including α4 integrin, CD4, and p56lck) (40, 49, 54, 59, 60). ZEB is able to block the activity of each of these factors separately, but together they synergize to resist repression (50). Although the mechanism of this synergy is not understood, the activity of both factors is essential for the expression of all of the above genes. Moreover, the E26 virus, which carries a fusion protein between v-myb and ets-1, is able to induce erythroleukemias where v-myb alone does not (41). This also points to a critical role for the synergy between c-myb and ets in the regulation of hematopoietic genes and indicates that the activity of these genes must be under strict control. Therefore, regulation of the activity of c-myb and ets is critical and to our knowledge ZEB is the only factor playing this regulatory role.
We found that region 3 of RD-ZEB regulates myogenic differentiation by specifically blocking the activity of the transcriptional factor MEF2C. MEF2C is the only target that we have found to date for this region. Although repression by region 3 is overcome by overexpression by p300, this region of ZEB failed to alter p300 transcriptional activity. Activation of MEF2C by p300 seems to be more complex than in the case of other factors. Transcriptional activity of MEF2C is completely dependent on its phosphorylation by MAP kinase p38, and mutations of the target residues for this kinase in MEF2C (or use of a dominant negative p38 kinase) render MEF2C inactive (25). It is tempting to speculate that phosphorylation by p38 may facilitate the formation of an active MEF2C-p300 complex. A similar mechanism regulates the activity of CREB, since phosphorylation by the calcium calmodulin-dependent CAMIV-kinase regulates its ability to interact with CBP/p300 (10, 66a). Calcium-dependent mechanisms have been recently linked to regulation of gene expression in muscle and to muscle differentiation and hypertrophy (12). We have found that overexpression of CAMIV kinase is also able to rescue repression of MEF2C by region 3 of ZEB (without affecting the repression of c-myb by region 1 of ZEB) (unpublished results), further suggesting a model where phosphorylation of MEF2C is critical for its transcriptional activity and where ZEB may work to offset the effects of this posttranslational regulation.
MEF2 activity is essential for muscle differentiation, since its mutation results in a block of embryonic muscle differentiation in Drosophila (6, 34) and a dominant negative form of MEF2 blocks myogenesis in mammalian muscle differentiation assays (48). Thus, blocking of MEF2 activity is sufficient to inhibit myogenic differentiation from Drosophila to mammals. The Drosophila homologue of ZEB, zfh-1, blocks somatic and cardiac muscle differentiation in embryos and targets MEF2 in vivo and in transfection assays (reference 51 and data not shown). These results provide compelling evidence that regulation of myogenesis by inhibiting MEF2 is a property of these orthologues that has been conserved during evolution. It is also important to note that ZEB/zfh-1 is the only active repressor of muscle differentiation that targets MEF2 activity. Whereas MRF proteins are essential myogenic triggers in mammals, this role in Drosophila is played by twist (3), which directly activates MEF2 transcription (13). Nevertheless, MEF2 is essential for myogenesis through this evolutionary period, and thus it is a logical target for inhibition of myogenesis by an evolutionarily conserved protein like ZEB/zfh-1.
In addition to muscle, Drosophila zfh-1 is important for differentiation of the central nervous system, heart, gonadal cells, and fat body (7, 32, 33). Thus, region 1 could be important in regulation of one or more of these other tissues. It is then of note that in addition to transcription factors specific for hematopoiesis, region 1 of the ZEB targets transcription factors that are expressed more generally. Moreover, ZEB also has functions beyond lymphoid and muscle differentiation in mammals, since mice lacking ZEB also have skeletal defects (64).
All members of the zfh family (as ZEB/zfh-1) contain zinc fingers and homeodomains. However, the family members differ dramatically in the number and location of these domains. The general structure of these proteins suggests that the zfh family is very modular and is the result of extensive genetic recombination events. Interestingly, the ZEB gene maps to chromosome 10p11 in a region of frequent translocations in leukemias (69), making it tempting to speculate that the independent repressor domains in ZEB arose from such a recombination event involving two repressor genes. Given the similarity in zinc finger and homeodomain modules in zfh family members, it would not be surprising if at least some other family members have functions analogous to ZEB/zfh-1.
ACKNOWLEDGMENTS
We acknowledge C. Caelles, B. Calabretta, S. Chellappan, T. Genetta, A. Giordano, E. Harlow, T. Kadesch, R. Kopan, S. J. Korsmeyer, T. Lipsick, D. Livingston, J. Molkentin, E. Olson, G. Tomaselli, and H. Weintraub for providing us with plasmids, cell lines, and antibodies.
A.A.P. was supported by the Leukemia Society of America. This work was funded by grants from the National Institutes of Health to D.C.D.
REFERENCES
- 1.Amacher S L, Buskin J N, Hauschka S D. Multiple regulatory elements contribute differentially to muscle creatine kinase enhancer activity in skeletal and cardiac muscle. Mol Cell Biol. 1993;13:2753–2764. doi: 10.1128/mcb.13.5.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassuk A G, Leiden J M. The role of Ets transcription factors in the development and function of the mammalian immune system. Adv Immunol. 1997;64:65–104. doi: 10.1016/s0065-2776(08)60887-1. [DOI] [PubMed] [Google Scholar]
- 3.Baylies M K, Bate M. Twist: a myogenic switch in Drosophila. Science. 1996;272:1481–1484. doi: 10.1126/science.272.5267.1481. [DOI] [PubMed] [Google Scholar]
- 4.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 5.Black B L, Martin J F, Olson E N. The mouse MRF4 promoter is trans-activated directly and indirectly by muscle-specific transcription factors. J Biol Chem. 1995;270:2889–2892. doi: 10.1074/jbc.270.7.2889. [DOI] [PubMed] [Google Scholar]
- 6.Bour B A, O’Brien M A, Lockwood W L, Goldstein E S, Bodmer R, Taghert P H, Abmayr S M, Nguyen H T. Drosophila Mef2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;15:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- 7.Broihier H, Moore L, Doren M, Newman S, Lehmann R. zfh-1 is required for germ cell migration and gonadal mesoderm development in Drosophila. Development. 1998;125:655–666. doi: 10.1242/dev.125.4.655. [DOI] [PubMed] [Google Scholar]
- 8.Cabanillas A M, Darling D S. Alternative splicing gives rise to two isoforms of Zfhep, a zinc finger/homeodomain protein that binds T3-response elements. DNA Cell Biol. 1996;15:643–651. doi: 10.1089/dna.1996.15.643. [DOI] [PubMed] [Google Scholar]
- 9.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;97:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 10.Chawla S, Hardingham G E, Quinn D R, Bading H. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- 11.Chen C M A, Kraut N, Groudine M, Weintraub H. I-mf, a novel myogenic repressor, interacts with members of the MyoD family. Cell. 1996;86:731–741. doi: 10.1016/s0092-8674(00)80148-8. [DOI] [PubMed] [Google Scholar]
- 12.Chin E R, Olson E N, Richardson J A, Yang Q, Humphries C, Shelton J M, Wu H, Zhu W, Bassel-Duby R, Williams R S. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cripps R M, Black B L, Zhao B, Lien C L, Schulz R A, Olson E N. The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev. 1998;12:422–434. doi: 10.1101/gad.12.3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai P, Akimaru H, Tanaka Y, Hou D X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 15.Edmondson D G, Cheng T C, Cserjevi P, Chakrabarty T, Olson E N. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol Cell Biol. 1992;12:3665–3677. doi: 10.1128/mcb.12.9.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fortini M E, Lai Z C, Rubin G M. The Drosophila zfh-1 and zfh-2 genes encode novel proteins containing both zinc-finger and homeodomain motifs. Mech Dev. 1991;34:113–122. doi: 10.1016/0925-4773(91)90048-b. [DOI] [PubMed] [Google Scholar]
- 17.Franklin A J, Jetton T L, Shelton K D, Magnuson M A. BZP, a novel serum-responsive zinc finger protein that inhibits gene transcription. Mol Cell Biol. 1994;14:6773–6788. doi: 10.1128/mcb.14.10.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funahashi J, Seikido R, Murai K, Kamachi Y, Kondoh H. Delta-crystallin enhancer binding protein delta EF1 is a zinc finger-homeodomain protein implicated in postgastrulation embryogenesis. Development. 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 19.Genetta T, Kadesch T. Cloning of a cDNA encoding a mouse transcriptional repressor displaying striking sequence conservation across vertebrates. Gene. 1996;169:289–290. doi: 10.1016/0378-1119(95)00824-1. [DOI] [PubMed] [Google Scholar]
- 20.Genetta T, Ruezinsky D, Kadesch T. Displacement of an E box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray S, Levine M. Transcriptional repression in development. Curr Opin Cell Biol. 1996;8:358–364. doi: 10.1016/s0955-0674(96)80010-x. [DOI] [PubMed] [Google Scholar]
- 22.Gray S, Szymanski P, Levine M. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev. 1994;8:1829–1838. doi: 10.1101/gad.8.15.1829. [DOI] [PubMed] [Google Scholar]
- 23.Gray S, Levine M. Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes Dev. 1996;10:700–710. doi: 10.1101/gad.10.6.700. [DOI] [PubMed] [Google Scholar]
- 24.Gregoire J M, Romeo P H. T-cell expression of the human GATA-3 gene is regulated by a non-lineage-specific silencer. J Biol Chem. 1999;274:6567–6578. doi: 10.1074/jbc.274.10.6567. [DOI] [PubMed] [Google Scholar]
- 25.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 26.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcriptional repressors. Trends Gen. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 27.Hanna-Rose W, Licht J D, Hansen U. Two evolutionary conserved repression domains in the Drosophila Kruppel protein differ in activator specificity. Mol Cell Biol. 1997;17:4820–4829. doi: 10.1128/mcb.17.8.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 29.Higashi Y, Moribe H, Takagi T, Sekido R, Kawakami K, Kikutani H, Kondoh H. Impairment of T cell development in deltaEF1 mutant mice. J Exp Med. 1997;185:1467–1479. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson A D. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 31.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T-M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 32.Lai Z, Fortini M E, Rubin G M. The embryonic expression patterns of zfh-1 and zfh-2, two Drosophila genes encoding novel zinc-finger homeodomain proteins. Mech Dev. 1991;34:123–134. doi: 10.1016/0925-4773(91)90049-c. [DOI] [PubMed] [Google Scholar]
- 33.Lai Z, Rushton E, Bate M, Rubin G M. Loss of function of the Drosophila zfh-1 gene results in abnormal development of mesodermally derived tissues. Proc Natl Acad Sci USA. 1993;90:4122–4126. doi: 10.1073/pnas.90.9.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lilly B, Zhao B, Ranganayakulu G, Paterson B M, Schulz R A, Olson E N. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 35.Lobb R R, Hemler M E. The pathophysiologic role of alpha 4 integrins in vivo. J Clin Investig. 1994;94:1722–1728. doi: 10.1172/JCI117519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo C, Burgeon E, Rao A. Mechanisms of transactivation by nuclear factor of activated T cells-1. J Exp Med. 1996;184:141–147. doi: 10.1084/jem.184.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 38.Luo R X, Dean D C. Chromatin remodeling and transcriptional regulation. J Natl Cancer Inst. 1999;91:1288–1294. doi: 10.1093/jnci/91.15.1288. [DOI] [PubMed] [Google Scholar]
- 39.Mao S, Neale G A, Goorha R M. T-cell proto-oncogene rhombotin-2 is a complex transcription regulator containing multiple activation and repression domains. J Biol Chem. 1997;272:5594–5599. doi: 10.1074/jbc.272.9.5594. [DOI] [PubMed] [Google Scholar]
- 40.McCracken S, Leung S, Bosselut R, Ghysdael J, Miyamoto N G. Myb and Ets related transcription factors are required for activity of the human lck type I promoter. Oncogene. 1994;9:3609–3615. [PubMed] [Google Scholar]
- 41.Metz T, Graf T. Fusion of the nuclear oncoproteins v-Myb and v-Ets is required for the leukomogenicity of E26 virus. Cell. 1991;66:95–105. doi: 10.1016/0092-8674(91)90142-l. [DOI] [PubMed] [Google Scholar]
- 42.Metz T, Graf T. v-myb and v-ets transform chicken erythroid cells and cooperate both in trans and in cis to induce distinct differentiation phenotypes. Genes Dev. 1991;5:369–380. doi: 10.1101/gad.5.3.369. [DOI] [PubMed] [Google Scholar]
- 43.Mink S, Haenig B, Klempnauer K H. Interaction and functional collaboration of p300 and C/EBPbeta. Mol Cell Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 45.Mucenski M L, McLain K, Kier A B, Swerdlow S H, Schreiner C M, Miller T A, Pietryga D W, Scott W J, Jr, Potter S S. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 46.Ogbourne S, Antalis T M. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem J. 1998;331:1–14. doi: 10.1042/bj3310001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 48.Ornatsky O I, Andreucci J J, McDermott J C. A dominant-negative form of transcription factor MEF2 inhibit myogenesis. J Biol Chem. 1997;272:33271–33278. doi: 10.1074/jbc.272.52.33271. [DOI] [PubMed] [Google Scholar]
- 49.Postigo A A, Dean D C. ZEB, a vertebrate homologue of Drosophila zfh-1, is a negative regulator of muscle differentiation. EMBO J. 1997;16:3935–3943. doi: 10.1093/emboj/16.13.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Postigo A A, Sheppard A M, Mucenski M L, Dean D C. c-myb and ets factors synergize to overcome transcriptional repression by ZEB. EMBO J. 1997;16:3924–3934. doi: 10.1093/emboj/16.13.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Postigo A A, Ward E, Skeath J B, Dean D C. zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol Cell Biol. 1999;19:7255–7263. doi: 10.1128/mcb.19.10.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;117:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sauer F, Jackle H. Concentration-dependent transcriptional activation or repression by Kruppel from a single binding site. Nature. 1991;353:563–566. doi: 10.1038/353563a0. [DOI] [PubMed] [Google Scholar]
- 54.Sawada S, Scarborough J D, Killeen N, Littman D R. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 55.Scott E W, Simon M C, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 56.Sekido R, Murai K, Funahashi J, Kamachi Y, Fujisawa-Sehara A, Nabeshima K, Kondoh H. The delta-crystallin enhancer-binding protein delta EF1 is a repressor of E2-box-mediated gene activation. Mol Cell Biol. 1994;14:5692–5700. doi: 10.1128/mcb.14.9.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sekido R, Takagi T, Okanami M, Moribe H, Yamamura M, Higashi Y, Kondoh H. Organization of the gene encoding transcriptional repressor dEF1 and cross-species conservation of its domains. Gene. 1996;173:227–232. doi: 10.1016/0378-1119(96)00185-0. [DOI] [PubMed] [Google Scholar]
- 58.Simon A M, Burdens J. An E box mediates activation and repression of the acetylcholine receptor delta-subunit gene during myogenesis. Mol Cell Biol. 1993;13:5133–5140. doi: 10.1128/mcb.13.9.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siu G, Wurster A L, Lipsick J S, Hedrick S M. Expression of the CD4 gene requires a Myb transcription factor. Mol Cell Biol. 1992;12:1592–1604. doi: 10.1128/mcb.12.4.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siu G, Wurster A L, Duncan D D, Soliman T M, Hedrick S M. A transcriptional silencer controls the developmental expression of the CD4 gene. EMBO J. 1994;13:3570–3579. doi: 10.1002/j.1460-2075.1994.tb06664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spicer D B, Rhee J, Cheung W L, Lassar A B. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- 62.Stein R W, Corrigan M, Yaciuk P, Whelan J, Moran E. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J Virol. 1990;64:4421–4427. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Su G H, Chen H M, Muthusamy N, Garrett-Sinha L A, Baunoch D, Tenen D G, Simon M C. Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J. 1997;16:7118–7129. doi: 10.1093/emboj/16.23.7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takagi T, Moribe H, Kondoh H, Higashi Y. DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development. 1998;125:21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- 65.Tapscott S J, Lassar A B, Weintraub H. A novel myoblast enhancer element mediates MyoD transcription. Mol Cell Biol. 1992;12:4994–5003. doi: 10.1128/mcb.12.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thayer M J, Weintraub H. Activation and repression of myogenesis in somatic cell hybrids: evidence for trans-negative regulation of MyoD in primary fibroblasts. Cell. 1990;63:23–32. doi: 10.1016/0092-8674(90)90285-m. [DOI] [PubMed] [Google Scholar]
- 66a.Tokumitsu H, Enslen H, Soderling T R. Characterization of a Ca2+/calmodulin-dependent protein kinase cascade. Molecular cloning and expression of calcium/calmodulin-dependent protein kinase kinase. J Biol Chem. 1995;270:19320–19324. doi: 10.1074/jbc.270.33.19320. [DOI] [PubMed] [Google Scholar]
- 67.Weintraub S J, Chow K N, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 68.Williams T M, Moolten D, Burlein J, Romano J, Bhaerman R, Godillot A, Mellon M, Rauscher F J, Kant J A. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science. 1991;254:1791–1794. doi: 10.1126/science.1840704. [DOI] [PubMed] [Google Scholar]
- 69.Williams T M, Montoya G, Wu Y, Eddy R L, Byers M G, Shows T B. The TCF8 gene encoding a zinc finger protein (Nil-2-a) resides on human chromosome 10p11.2. Genomics. 1992;14:194–196. doi: 10.1016/s0888-7543(05)80307-6. [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto H, Kihara-Negishi F, Yamada T, Hashimoto Y, Oikawa T. Physical and functional interactions between the transcription factor PU.1 and the activator CBP. Oncogene. 1998;18:1495–1501. doi: 10.1038/sj.onc.1202427. [DOI] [PubMed] [Google Scholar]
- 71.Yang C, Shapiro L H, Rivera M, Kumar A, Brindle P K. A role for CREB binding protein and p300 transcriptional coactivators in ets-1 transactivation functions. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 73.Yasui D H, Genetta T, Kadesch T, Williams T M, Swain S L, Tsui L V, Huber B T. Transcriptional repression of the IL-2 gene in Th cells by ZEB. J Immunol. 1998;160:4433–4440. [PubMed] [Google Scholar]
- 74.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]