Abstract
Plasma membrane localization of Ras requires posttranslational addition of farnesyl and palmitoyl lipid moieties to a C-terminal CaaX motif (C is cysteine, a is any aliphatic residue, X is the carboxy terminal residue). To better understand the relationship between posttranslational processing and the subcellular localization of Ras, a yeast genetic screen was undertaken based on the loss of function of a palmitoylation-dependent RAS2 allele. Mutations were identified in an uncharacterized open reading frame (YLR246w) that we have designated ERF2 and a previously described suppressor of hyperactive Ras, SHR5. ERF2 encodes a 41-kDa protein with four predicted transmembrane (TM) segments and a motif consisting of the amino acids Asp-His-His-Cys (DHHC) within a cysteine-rich domain (CRD), called DHHC-CRD. Mutations within the DHHC-CRD abolish Erf2 function. Subcellular fractionation and immunolocalization experiments reveal that Erf2 tagged with a triply iterated hemagglutinin epitope is an integral membrane protein that colocalizes with the yeast endoplasmic reticulum marker Kar2. Strains lacking ERF2 are viable, but they have a synthetic growth defect in the absence of RAS2 and partially suppress the heat shock sensitivity resulting from expression of the hyperactive RAS2(V19) allele. Ras2 proteins expressed in an erf2Δ strain have a reduced level of palmitoylation and are partially mislocalized to the vacuole. Based on these observations, we propose that Erf2 is a component of a previously uncharacterized Ras subcellular localization pathway. Putative members of an Erf2 family of proteins have been uncovered in yeast, plant, worm, insect, and mammalian genome databases, suggesting that Erf2 plays a role in Ras localization in all eucaryotes.
Ras proteins are small membrane-associated GTP binding proteins that cycle between active (GTP-bound) and inactive (GDP-bound) states to regulate cell growth and differentiation. In Saccharomyces cerevisiae, two Ras proteins (Ras1 and Ras2) affect such diverse processes as vegetative growth, sporulation, carbon source utilization, stress response, and pseudohyphal growth (12, 25, 45, 61, 64). Ras-dependent growth requires plasma membrane localization, which in turn depends on a series of posttranslational modifications that occur on a carboxyl-terminal CaaX box (C is cysteine, a is any aliphatic residue, X is the carboxy-terminal residue) (15, 20, 21, 57). The first step is the farnesylation of the CaaX box cysteine by a soluble, heterodimer farnesyl protein transferase encoded by the RAM1 and RAM2 genes in yeast (34, 39, 53). The aaX sequence is removed by one of two endoplasmic reticulum (ER)-associated proteases encoded by RCE1 and AFC1/STE24 (9, 58). The newly exposed cysteinyl α-carboxyl is then methyl esterified by the product of STE14, an integral membrane protein which also colocalizes with the ER in yeast and mammalian cells (16, 18, 22, 54, 56).
The mature form of Ras is localized primarily on the cytoplasmic surface of the plasma membrane. Mutations in either the CaaX box or the genes encoding the posttranslational modification enzymes cause a reduction in Ras plasma membrane localization and a corresponding reduction in the ability of Ras to support growth (15, 57). However, the mechanism by which prenylation and subsequent posttranslational modifications direct Ras proteins to the plasma membrane is not known. It is clear that prenylation alone is not sufficient for efficient plasma membrane targeting of Ras. Yeast mutants that fail to carry out the palmitoylation step have reduced amounts of Ras protein at the plasma membrane and increased resistance to heat shock in the presence of activated Ras2(V19) alleles (6). Palmitoylation, unlike the CaaX processing steps, is reversible, making it a likely regulatory step. Interestingly, oncogenic forms of Ras are also rendered nontransforming by mutating the palmitoylation sites (69). Unfortunately, palmitoylation is the least well understood step in the Ras posttranslational modification pathway. A major obstacle has been the failure to identify a protein palmitoyltransferase, although reports of partial purification of palmitoyltransferase activities have appeared (3, 19, 37). The matter is further complicated by the fact that protein palmitoylation can occur nonenzymatically; however, the reaction rate is considerably lower than that observed in vivo (1, 23, 66). The issue is unlikely to be resolved until there is a better understanding of the requirements for and biological consequences of palmitoylation in vivo.
The multistep nature of Ras modification suggests that subcellular targeting may be an ordered process involving distinct intracellular membrane compartments. In support of this idea is the recent demonstration that the -aaX proteases (Afc1 and Rce1) and methyltransferase (Ste14) are associated with the ER in yeast and mammalian cells (9, 18, 54, 58). It would appear that prenylated Ras first associates with the ER membrane, although the mechanism by which it is targeted there is unknown. One possible mechanism is a receptor-mediated process that is prenylation dependent. Indeed, high-affinity association of prenylated peptides with microsomal membranes has been observed (62, 63). However, a receptor or anchor protein has not been identified. It is also not known at this time how Ras translocates from the ER to the plasma membrane. One possibility is that Ras associates with the cytoplasmic surface of the classical secretory pathway. However, studies on the prenylated yeast mating pheromone a-factor argue against this model. Similar to Ras, a-factor is posttranslationally processed by prenylation, -aaX proteolysis, and Ste14-dependent methylation. Export of a-factor was unaffected by temperature-sensitive blocks at several key steps of the yeast secretory pathway (36, 40). This has led to the proposal that Ras, a-factor, and possibly other prenylated proteins may utilize a nonclassical pathway to traffic from the ER to the plasma membrane. However, the components of such a pathway and the mechanism by which they act remain elusive.
To investigate in greater detail the relationship between posttranslational modification and Ras trafficking, we have employed a genetic screen using a palmitoylation-dependent Ras allele that we previously described (43). Due to the palmitate dependence of the Ras allele and its defect in localization, we predicted that mutations affecting palmitoylation or a component of the Ras localization pathway would be uncovered in this screen. In this report, we describe the isolation of two mutations designated erf2 and erf4 (effect on Ras function). ERF4 was found to encode a protein previously described by Jung et al. as a suppressor of a hyperactive Ras allele, SHR5 (31). ERF2 encodes a novel integral membrane protein harboring a recently described cysteine-rich domain (CRD) containing a DHHC motif (DHHC-CRD) (52). Characterization of Erf2 reveals that it is required for the proper subcellular localization and palmitoylation of Ras proteins and may represent a component of a previously uncharacterized Ras localization pathway.
MATERIALS AND METHODS
Strains, media, and yeast techniques.
The yeast strains used in this study are listed in Table 1. Solid and liquid media were prepared as described by Sherman et al. (59) and included synthetic complete (SC) medium lacking one or more specified amino acids and rich medium (YPD). Induction of expression of GAL1,10 promoters was achieved by adding 4% galactose to SC medium lacking uracil. Standard procedures were used for yeast manipulations (59). Yeast transformations were done by the lithium acetate procedure (30). Medium containing 5-fluoro-orotic acid (5-FOA; PCR, Inc.) was prepared as described elsewhere (7).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| RJY5 | MATa/α leu2-3,112/leu2-3,112 ura3-52/ura3-52 trp1-289/trp1-289 his3Δ1/his3Δ1 ade8Δ/ade8Δ RAS1/ras1::HIS3 RAS2/ras2::URA3 | Broach strain collection (Princeton University) |
| RJY248 | MATα ura3-52 his3Δ1 lys2-1 ras1::HIS3 ras2(ΔHpaI) [YCp50Ras1] | Broach strain collection |
| RJY266 | MATa/α leu2-3,112/leu2-3,112 ura3-52/ura3-52 his3Δ1/his3Δ1 trp1-289/trp1-289 | |
| RJY1081 | MATα leu2-3,112 ura3-52 trp1-289 ade2-101 ade8Δ lys2-1 ras1::HIS3 erf2-5 ras2(CSIIKLIKRK) [YCp52-Ras2] | |
| RJY1090 | MATα leu2-3,112 ura3-52 trp1-289 ade2-101 ade8Δ lys2-1 ras1::HIS3 shr5-5 ras2(CSIIKLIKRK) [YCp52-Ras2] | |
| RJY1106 | MATa leu2-3,112 ura3-52 trp1-289 ade2-101 ade8Δ ras1::HIS3 ras2(CSIIKLIKRK) [YCp52-Ras2] | |
| RJY1107 | MATα leu2-3,112 ura3-52 trp1-289 ade2-101 ade8Δ lys2-1 ras1::HIS3 ras2(CSIIKLIKRK) [YCp52-Ras2] | |
| RJY1270 | MATα leu2-3,112 ura3-52 trp1-289 ade2-101 ade8Δ lys2-1 ras1::HIS3 ras2(CSIIKLIKRK) [YCp52-Ras2] [pEG(KG)Ras2(CCIIS)] | |
| RJY1272 | MATα leu2-3,112 ura3-52 trp1-289 ade2-101 ade8Δ lys2-1 ras1::HIS3 erf2::TRP1 ras2(CSIIKLIKRK) [YCp52-Ras2] [pEG(KG)Ras2(CCIIS)] | |
| RJY1274 | MATα leu2-3,112 ura3-52 trp1-289 ade2-101 ade8Δ lys2-1 ras1::HIS3 shr5-5 ras2(CSIIKLIKRK) [YCp52-Ras2] [pEG(KG)Ras2(CCIIS)] | |
| RJY1277 | MATα leu2-3,112 ura3-52 trp1-289 ade2-101 ade8Δ lys2-1 ras1::HIS3 erf2::TRP1 ras2(CSIIKLIKRK) [YCp52-Ras2] | Obtained from RJY1107 by single-step gene replacement with a erf2::TRP1 fragment |
| RJY1280 | MATa/α leu2-3,112/leu2-3,112 ura3-52/ura3-52 his3Δ1/his3Δ1 trp1-289/trp1-289 ERF2/erf2::TRP1 | Obtained from RJY266 by single-step gene replacement with an erf2::TRP1 fragment |
| RJY1301 | MATa/α leu2-3,112/leu2-3,112 ura3-52/ura3-52 trp1-289/trp1-289 his3Δ1/his3Δ1 ade8Δ/ade8Δ ras2::URA3/ras2::URA3 ERF2/erf2::TRP1 | Obtained by sporulating RJY1302 and backcrossing resulting segregants |
| RJY1302 | MATa/α leu2-3,112/leu2-3,112 ura3-52/ura3-52 trp1-289/trp1-289 his3Δ1/his3Δ1 ade8Δ/ade8Δ RAS1/ras1::HIS3 RAS2/ras2::URA3 ERF2/erf2::TRP1 | Obtained from RJY5 by single-step gene replacement with an erf2::TRP1 fragment |
| RJY1318 | MATa/α leu2-3,112/leu2-3,112 ura3-52/ura3-52 his3Δ1/his3Δ1 erf2::TRP1/erf2::TRP1 | Obtained by backcrossing erf2Δ segregants arising from the sporulation of RJY1280 |
| LRB759 | MATα leu2 his3 ura3-52 | Panek et al. (50) |
| RJY1438 | MATα leu2 his3 ura3-52 erf2::TRP1 | Obtained from LRB79 by single-step gene replacement with an erf2::TRP1 fragment |
| RJY1412 | MATa/α leu2-3,112/leu2-3,112 ura3-52/ura3-52 trp1-289/trp1-289 his3Δ1/his3Δ1 ade8Δ/ade8Δ ras1::HIS3/ras1::HIS3 ERF2/erf2::TRP1 | Obtaining by sporulating RJY1302 and backcrossing resulting segregants |
Genetic screen for mutants that affect the subcellular localization of Ras.
The screen was based on a sectoring assay in which the ability to lose an ADE8/URA3-marked RAS2-encoding plasmid (YCp52-Ras2) was monitored by the red/white sectoring of the colonies (see Fig. 1). Cultures of RJY1106 and RJY1107 (8 × 107 cells/ml) were plated on rich medium (YPD) plates (103 cells/plate) and mutagenized with UV light to obtain approximately an 80% kill rate. Plates were incubated for 5 days (30°C), and the sectoring patterns of 152,000 colonies were assessed. Nonsectored colonies (total of 1,346) were grown as patches on YPD plates and replicated to plates containing 5-FOA (1.0 g/liter). A recessive/dominant test of prospective mutants was performed by backcrossing to either RJY1106 or RJY1107. A complementation test based on restoration of sectoring and 5-FOA resistance was performed on all recessive mutants. A clone by complementation strategy was performed by using the acquisition of 5-FOA resistance as the primary screen to identify the wild-type gene corresponding to the erf mutations. For example, strain RJY1081 was transformed with a YSB32-based (ATCC 77162) genomic library, and transformants were plated onto SC medium plates lacking leucine. To select for cells capable of losing YCp52-Ras2, transformants were replicated to plates containing 5-FOA and plasmid DNA was isolated from the 5-FOA-resistant colonies. Bypass suppressors were eliminated by transforming plasmids into RJY248 (ras1Δ ras2Δ [YCp50Ras1]) and testing for 5-FOA sensitivity. The plasmid (B644) which complemented erf2 was mapped by a combination of deletion analysis and subcloning strategies which allowed the identification of open reading frame (ORF) YLR246w as the gene responsible for complementing the erf2 mutation.
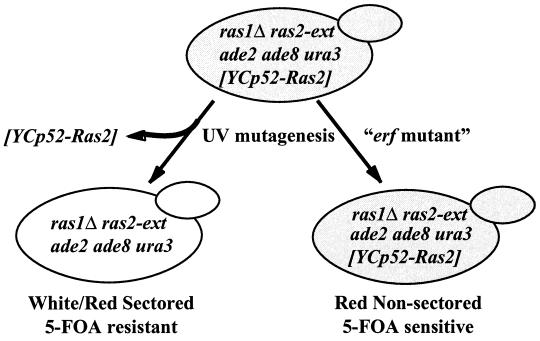
FIG. 1.
Genetic screen to identify mutants that have an effect on the function of a palmitoylation-dependent RAS2 allele (ras2-ext). RJY1106 (MATa) and RJY1107 (MATα) cells were mutagenized as described in Materials and Methods. Mutants are defined by the inability to lose YCp52-Ras2 and are detected by the failure to form a sectored colony and 5-FOA sensitivity.
Plasmid construction.
The ERF2 gene (YLR246w) was isolated by PCR amplification from the YSB32-based genomic library plasmid (B644) that complemented the 5-FOA sensitivity and red nonsectoring phenotypes of the erf2-5 mutant. Two oligonucleotide primers, OLI-222 (−165 to −150; 5′-CGCGAATTCTCTGTTTGGTTT-3′) and OLI-225 (+1061 to +1079; 5′ CGATGAGCTCTTAGCGGCCGCATATTTTCTGTATTTTT-3′), were designed to amplify the ERF2 ORF. The 1.2-kb PCR product was digested with EcoRI and SacI (sites incorporated into the PCR oligonucleotides) and ligated into EcoRI/SacI-digested pRS314 (60) to create B642. The OLI-225 primer was designed to include a NotI site into which a triply iterated version of the influenza hemagglutin epitope (HA3) was placed at the C terminus to create the plasmid B753. Erf2-HA3 protein expressed from B753 was shown to be expressed by immunoblotting and to be fully functional by complementation of the 5-FOA sensitivity and nonsector phenotype of RJY1277. Low-copy-number and multicopy plasmids expressing Erf2-HA3 were constructed by subcloning a XhoI/SacI fragment derived from pRS314-Erf2-HA3 (B753) into pRS315 (60) (B754) and YEp351 (27) (B745), respectively.
Construction of erf2::TRP1 (B777) and erf2::HIS3 (B778) plasmids utilized a PstI-BglII subclone of ERF2 in pLITMUS28 (New England Biolabs). pLIT28-ERF2 was digested with SalI and ApaI to remove the ERF2 ORF, and then either TRP1 or HIS3 was ligated to create pErf2::TRP (B777) and pErf2::HIS (B778), respectively. For yeast disruptions included in this study, a 3.3-kb erf2::TRP1 or 3.6-kb erf2::HIS3 fragment was generated by digesting B777 or B778 with SacI/SpeI.
Site-directed mutagenesis of ERF2 was done with a Stratagene QUICK CHANGE kit. Mutagenesis was done on pRS315-Erf2-HA3 (B754). For mutagenesis of Cys189 and Cys-192 to Ser-189 and Ser-192, OLI-350 (5′ CCACCTCGGTCTTCTCACTCTTCCACATCTAACGTCTGCG-3′) and OLI-351 (5′ CGCAGACGTTAGATGTGGAAGAGTGAGAAGACCGAGGTGG-3′) were used (underlining denotes nucleotide changes). For mutagenesis of Asp-200 to Ala, OLI-348 (5′GCGTAATGGTTCATGCCCACCATTGC-3′) and OLI-349 (5′-GCAATGGTGGGCATGAACCATTACGC3′) were used. After mutagenesis, plasmids were checked for expression and ability to complement the nonsectoring and 5-FOA sensitivity phenotypes of RJY1277.
The MET25-RAS1 expression plasmid was created by PCR amplification of a 380-bp region of the MET25 promoter, using OLI-452 (−382 to −368; 5′AGCTAGCTAAGCTTCGGATGCAAGGGTTCG3′) and OLI-457 (−25 to −8; 5′TCGATCGATCTAGAGATGGGGGTAATAGAATTG 3′). The MET25 PCR fragment was digested with HindIII and XbaI and ligated into pRS315 to create B803. RAS1 was amplified from the genome by PCR using OLI-455 (+1 to +19; 5′AGCTAGCTACTAGTATGCAGGGAAATAAATCAA 3′) and OLI-456 (+316 to +365; 5′CGTACGTAGAGCTCGCCGAGTTTATTGTTGCTAG 3′). The RAS1 PCR fragment was digested with SpeI and SacI and ligated into B803 cut with XbaI and SacI to create B804. As expected, the expression of RAS1 from the MET25 promoter construct was under the control of methionine concentrations in the medium (47) (data not shown).
Green fluorescent protein (GFP)-Ras2(SCIIS) (B763) was derived from GFP-Ras2 (9). The glutathione S- (GST)–Ras2 fusion plasmid pEG(KG)Ras2(CCIIS) has been described previously (44). pRS315-Ras2 (B250) was created by removing the 3.1-kb EcoRI-HindIII fragment from YCp50Ras2 (43) and placing into pRS315 (60). pRS315-Ras2 (V19) (B561) and pRS315-Ras2(V19,SCIIS) (B562) were created by site-directed mutagenesis.
Preparation of yeast extracts and immunoblot analysis.
Cultures were grown (25 ml) to a density of 1 × 107 to 2 × 107 cells/ml in synthetic medium and then collected by centrifugation, washed with water, and resuspended in sorbitol buffer (0.3 M sorbitol, 0.1 M NaCl, 5 mM MgCl2, 10 mM Tris-HCl [pH 7.4]) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 100 U of aprotinin per ml, 1 μM pepstatin, 100 μM leupeptin, and 1 μg of chymostatin per ml) and lysed as described previously (43). Soluble (S100) and membrane (P100) fractions were separated at 100,000 × g at 4°C, using a TLA100.2 rotor of a Beckman TLA100 ultracentrifuge. Protein loading buffer was added, extracts were denatured (95°C, 5 min), and proteins (50 μg) were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (12.5% gel). Proteins were transferred to nitrocellulose by semidry electrotransfer in transfer buffer (20 mM Tris base, 150 mM glycine, 0.1% SDS 20% [wt/vol] methanol [pH 8.0]) at 400 mA for 1 h. All subsequent manipulations were performed at room temperature. The filter was blocked for 1 h in buffer A (100 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.05% Tween 20) containing 5% nonfat dried milk. After one washing with buffer A, the membrane was incubated with the designated primary antibody (2 h), washed three times (15 min) in buffer A, and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (2 h). Following three buffer A washes (15 min), the blot was visualized by using a Pierce SUPER-SIGNAL kit.
Subcellular fractionation of Erf2.
RJY1318 was transformed with an Erf2-HA3-expressing plasmid (B754), and the resulting strain was grown at 30°C to an A600 of 0.5 to 1.0 in SC medium lacking tryptophan (2% glucose). Subcellular fractionation was performed essentially as previously described (28). Briefly, cells were spheroplasted, lysed by Dounce homogenization, and subjected to centrifugation in a Beckman GS-15R to obtain 13,000 × g pellet (P13) and supernatant (S13) fractions. The P100 and S100 fractions were obtained and immunoblot analyses were performed as described above. Erf2-HA3 was detected by using mouse anti-HA antibody (BAbCo) followed by sheep anti-mouse–HRP conjugate (Amersham). Dpm1 was detected by using an anti-Dpm1 antibody (1:20) followed by sheep anti-mouse–HRP conjugate. Kar2 was detected by using an anti-Kar2 antibody (55) (1/20,000 dilution) followed by HRP-conjugated goat anti-rabbit (1:1,000) (Bio-Rad). Pma1 was detected with anti-Pma1 (1:2,000) antibody followed by HRP conjugated sheep anti-mouse–HRP.
In vivo labeling of GST-Ras2 with [3H]palmitate.
Metabolic labeling of GST-Ras was performed as described previously (43). The galactose-inducible GST-Ras2 expression plasmid, pEG(KG)Ras(CCIIS) (B529), was also described previously (43). RJY1270, RJY1272, and RJY1274 cells (100 ml) were grown to a density of 2 × 107 cells/ml in SC medium lacking uracil (4% raffinose), spun, and resuspended in 10 ml of the same medium containing 3 μg of cerulenin per ml, and the cells were incubated for 2 h. The expression of GST-Ras2 was induced by the addition of galactose (4%), and cells were labeled for 4 h with [3H]palmitic acid (1 mCi, 50 Ci/mmol; NEN). Cells were broken with glass beads, and the sample was subjected to low-speed centrifugation (100 × g, 2 min) to remove unbroken cells. The cleared lysate was resuspended in protein loading buffer without β-mercaptoethanol and heated (60°C, 5 min). After separation by SDS-PAGE, the gel was fixed and visualized as described elsewhere (43).
Immunofluorescence and fluorescence microscopy.
Immunolocalization of Erf2-HA3 was performed as described previously (4), with the following modifications. RJY1318 was transformed with YEp351-Erf2-HA3 (B745) and grown in SC medium lacking leucine (2% glucose) to 107 cells/ml. Erf2-HA3 was detected by using mouse anti-HA epitope antibody (1:400) (BAbCo) followed by Oregon Green 488-conjugated goat anti-mouse immunoglobulin G (IgG; 1:240 dilution; Molecular Probes Inc., Eugene, Oreg.). Kar2 was detected with anti-Kar2 antibody (55) (1/2,000 dilution) and lissamine rhodamine-conjugated goat anti-rabbit (1:240 dilution; Jackson ImmunoResearch Laboratories, West Grove, Pa.). To visualize GFP-Ras, cultures (5 ml) were grown to mid-exponential phase in synthetic medium under the same conditions as the palmitate labeling and concentrated by centrifugation. FM4-64 labeling of the vacuoles was done as described elsewhere (68). Images were obtained for Erf2-HA3 with a Zeiss Axioskop microscope (100× objective). GFP-Ras localization images were obtained with a laser scanning confocal system (MRC-1024; Bio-Rad) (60× objective).
Heat shock assay.
The heat shock assay was performed by growing LRB759 (ERF2) and RJY1438 (erf2::HIS3) harboring a pRS315 plasmid expressing RAS2 (B250), RAS2(V19) (B561), or RAS2(V19,SCIIS) (B562) in SC medium lacking leucine for 3 days 30°C. Equal numbers of cells (107) were aliquoted into wells of a 96-well titer plate, and a fivefold dilution series prepared. Heat shock was carried out by placing the microtiter plate, at 55°C for the indicated period of time (typically 10 to 30 min). An aliquot of cells was transferred to SC medium plates lacking leucine, using a CLONEMASTER (Immusine Laboratories), and allowed to grow for 2 days (30°C).
RESULTS
Isolation of mutants affecting Ras localization.
In a previous study, we described a group of RAS2 mutants in which the CaaX box (Cys-Cys-Ile-Ile-Ser) was replaced by the basic extension sequence Cys-Ser-Ile-Ile-Lys-Leu-Ile-Lys-Arg-Lys (Ras2-ext) (43). The membrane association of the Ras2-ext form of Ras2 requires the basic residues in the extension as well as palmitoylation but not prenylation (43). It was not possible in that study to determine how these nonprenylated Ras2-ext proteins functioned given the overall lack of information on Ras trafficking. In an attempt to shed light on this process, we performed a genetic screen that was designed to uncover mutants that affect Ras processing and/or subcellular trafficking. The screen was based on the assumption that the nonprenylated Ras2-ext alleles are partially defective in membrane binding and therefore more sensitive than wild-type Ras to defects in the processing or trafficking pathway.
Strains RJY1106 and RJY1107 were mutagenized as described in Materials and Methods and screened as illustrated in Fig. 1. Mutants exhibiting a synthetic lethality with the Ras2-ext allele were detected by a red/white sectoring assay and by monitoring the inability of the strain to lose an ade8/URA3 plasmid expressing RAS2 (YCp52-Ras2). The screen is a variation of a method described by Bender and Pringle to isolate synthetic lethal mutants (2), except that we used ade8 instead of ade3 to block the production of pigment accumulation of ade2 mutants. A total of 152,000 colonies were screened by the sectoring assay. To confirm that nonsectoring colonies represented cells unable to lose the (YCp52-Ras2) (URA3/ADE8) plasmid, patches (total of 1,346) were replicated to plates containing 5-FOA (1 g/liter). Approximately 0.9% of the colonies satisfied the criteria of being unable to sector and 5-FOA sensitive.
Three complementation groups, designated erf (effect on Ras function) mutants were identified. The erf1 complementation group consisted of two distinct alleles mapped to the Ras2-ext locus and was not pursued. A second complementation group, represented by six alleles of erf4, was determined to be allelic with SHR5, a previously described suppressor of the heat shock sensitivity of Ras2(V19) (31). Deletion of ERF4/SHR5 caused a decrease in palmitoylation and mislocalization of Ras (31). The final complementation group, erf2, consisted of six alleles and is the topic of this report.
Isolation of ERF2.
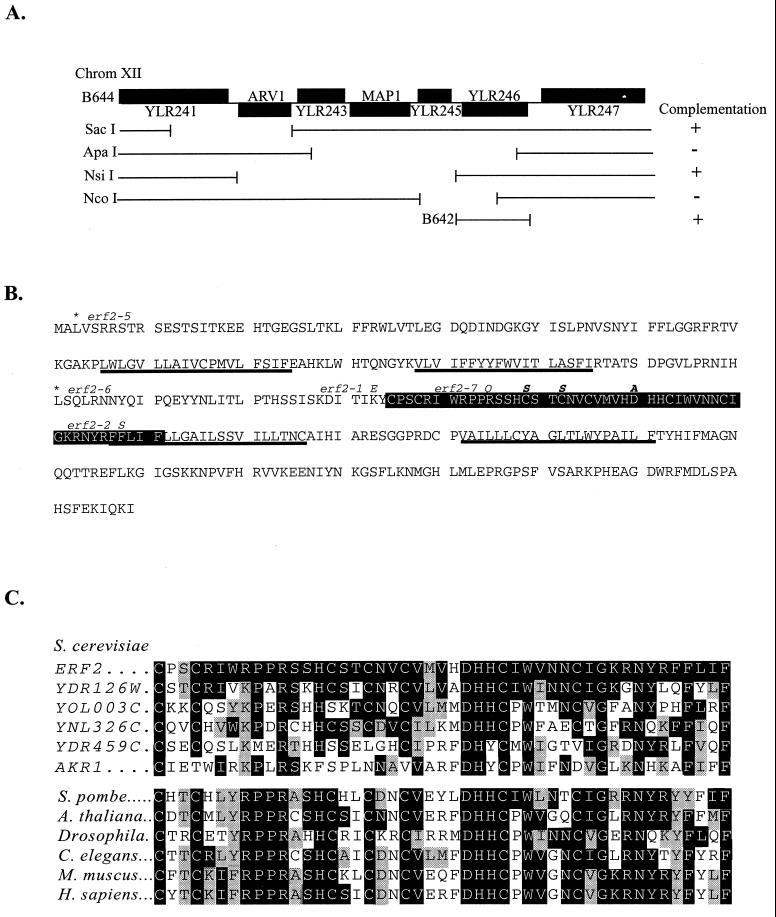
The ERF2 gene was isolated by complementation of the 5-FOA sensitivity and nonsectoring phenotype of RJY1081 (MATa ras1::HIS3 ras2-ext erf2-5 ura3 ade2 ade8 leu2 [YCp52-Ras2]), using a low-copy-number (YSB32) yeast genomic DNA library (ATCC 77162). Plasmid B644 was recovered and found by sequencing the plasmid/insert junctions to contain an insert encompassing 12.3 kb of chromosome XII. Two known genes, MAP1 and ARV1, and four uncharacterized ORFs are encoded in this interval (Fig. 2A). A series of deletion plasmids was constructed and tested for the ability to complement the 5-FOA sensitivity of an erf2 mutant. Deletions that removed YLR246w failed to complement erf2 (Fig. 2A), and a construct expressing only YLR246w was able to restore 5-FOA resistance (data not shown). In addition, a TRP1 gene disruption of YLR246w exhibits the nonsectoring phenotype of the original erf2 mutants. When a diploid arising from a cross between erf2-5 and a strain harboring the TRP1 disruption of YLR246w was sporulated, all of the resulting progeny were nonsectoring and 5-FOA sensitive. Finally, sequence analysis of all but one of the erf2 mutants uncovered either missense or nonsense mutations in YLR246w.
FIG. 2.
Isolation and sequence analysis of ERF2. (A) B644 was identified from a low-copy-number yeast genomic library (ATCC 77162) by complementation of the 5-FOA sensitivity of an erf2-5 strain (RJY1081). The insert of B644 encompasses a 12.3-kb region of chromosome (Chrom) XII starting at nucleotide 617299 and ending at 629649. B644 was digested with the indicated restriction enzymes and religated, and a complementation test was performed. YLR246w was subcloned in B642 and shown to complement erf2-5. (B) Deduced amino acid sequence of ERF2 (YLR246w). The conserved CRD is shown in the black box, and predicted TM regions are underlined. Mutations isolated from the ERF screen are shown in italic, and site-directed mutations are italicized and boldface. Nonsense mutations uncovered in the screen are indicated with asterisks. (C) Sequence alignment of the five putative S. cerevisiae ERF2 orthologs and a representative list of possible CRD homologs from other organisms: S. pombe (accession no. CAA20305), A. thaliana (CAA23000), Drosophila (AAD34351), C. elegans (CAA21738), M. muscus (AI322485), and H. sapiens (AA112746). Amino acid identities and similarities to Erf2 are shaded black and gray, respectively.
ERF2 encodes a novel protein containing a DHHC-CRD.
ERF2 is predicted to encode a 359-residue protein with a calculated molecular mass of 41 kDa and four predicted transmembrane (TM) segments (Fig. 2B). A possible metal binding site consisting of a DHHC-CRD is found between TM2 and TM3. The DHHC-CRD has been referred to as a NEW1 variant of the C2H2 zinc finger motif (8). The appearance of the DHHC motif within the CRD distinguishes this family from other cysteine-rich metal binding domains (42, 52). However, the function of this domain has not been established despite the presence of the sequence in genomic databases of S. cerevisiae, Schizosaccharomyces pombe, Arabidopsis thaliana, Drosophila melanogaster, Caenorhabditis elegans, and Homo sapiens (Fig. 2C). The region of highest sequence identity encompasses the DHHC-CRD and ranges from 65% (YDR126w) to 36% (YDR459c). Members of the Erf2 subgroup of this family share additional similarities. All are predicted to encode integral membrane proteins, often with four TM segments and a molecular mass of approximately 40 kDa. YDR459c and Akr1 (YDR264c) may be more distantly related to Erf2 because the DHHC motif has been replaced by DHYC, several cysteines within the CRD are not conserved, and the number of TM domains is less certain.
The CRD region of Erf2 appears to be critical for its function. Several of the mutant alleles isolated in the screen map to this region (Fig. 2B). For example, mutation of the conserved residue Arg-182 to Gln (erf2-7) and Lys-174 to Glu (erf2-1) abolishes Erf2 function without significantly affecting the expression level of the protein (data not shown). Site-directed mutagenesis was used to mutate two of the conserved Cys residues within the CRD to Ser (C189S and C192S). The erf2(C189S,C192S) mutant protein was expressed normally but failed to complement the nonsectoring and 5-FOA sensitivity of the erf2Δ mutant (RJY1277) (data not shown). The DHHC motif is also required for Erf2 function. Mutating Asp-200 to Ala inactivated Erf2, as measured by the failure to complement the nonsectoring and 5-FOA sensitivity in RJY1277 (data not shown). The screen also identified a F218S substitution (erf2-2) in the region predicted to be the start of TM3. The other alleles identified in the screen resulted in the insertion of stop codon at Leu-3 (erf2-5) or Leu-145 (erf2-6). In one case (erf2-4), we were unable to detect a change within the ORF and presume that the mutation affects the expression of the protein, but this was not confirmed.
No overlap in function was observed between Erf2 and the other yeast DHHC-CRD ORFs. First, deletion of YDR126, YOL003, YNL326, or YDR459 in RJY1107 had no effect on viability or 5-FOA sensitivity, and all strains sectored normally. Second, overexpression of YDR126, YOL003, YNL326, or YDR459 could not compensate for the loss of ERF2 in the sector or 5-FOA sensitivity assays in RJY1277 (data not shown). Finally, we deleted each of these ORFs alone or in combination with ERF2 and found that the double deletions did not alter the viability of the erf2Δ strain alone (data not shown). Therefore, the other Erf2-like proteins appear to function outside the Ras pathway.
Subcellular localization of Erf2-HA3.
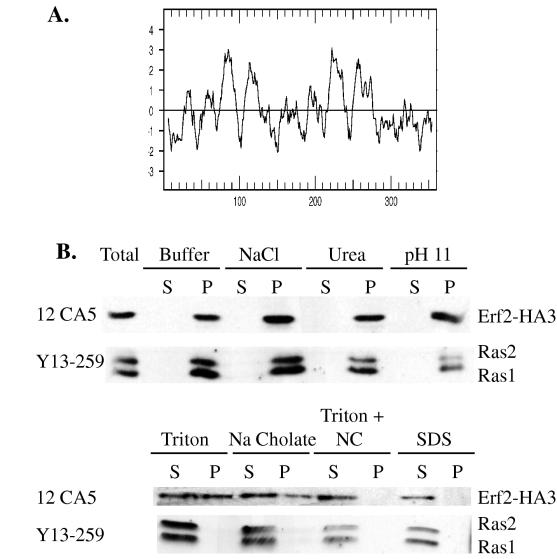
The subcellular localization of Erf2 was investigated next in an effort to understand its function. A hydropathy plot of the Erf2 sequence predicts that it is an integral membrane protein with four TM segments (Fig. 3A). To examine this directly, an HA3 tag was inserted at the C terminus of Erf2 in a low-copy-number Erf2-HA3 expression plasmid (B754). Expression of B754 in RJY1277 (erf2Δ) was able to complement the 5-FOA sensitivity and nonsectoring phenotypes (data not shown). Total cell lysates were fractionated by centrifugation to obtain soluble (S100) and high-speed crude membrane (P100) fractions, and Erf2-HA3 was found exclusively in the P100 fraction (Fig. 3B). Addition of 1.0 M NaCl, 1.6 M urea or 0.1 M sodium bicarbonate (pH 11) was unable to release Erf2-HA3 from the P100 fraction, consistent with it being an integral membrane protein. Ras1 and Ras2 proteins also fractionated with the P100 fraction under the same conditions. The nonionic detergent Triton X-100 (1%) extracted only a portion of the Erf2-HA3 from the membrane (Fig. 3B). Similar results were obtained with the ionic detergent sodium cholate (1%). In contrast, Ras proteins were effectively solubilized by Triton X-100 (1%) or sodium cholate (1%) (Fig. 3B). The combination of Triton X-100 and sodium cholate was able to complete the solubilization of Erf2-HA3.
FIG. 3.
Erf2 is an integral membrane protein. (A) Kyte-Doolittle analysis of hydrophobicity predicts the existence of four TM segments. (B) Membrane fractionation of Erf2-HA3. Total cell lysates were prepared from RJY1318 harboring B754 (Erf2-HA3) as described in Materials and Methods. The extracts were then treated with buffer, 0.6 M NaCl, 1.6 M urea, 0.1 M Na2CO3 (pH 11), 1% Triton X-100, 1% sodium cholate, 1% Triton X-100 and 1% sodium cholate (Triton + NC), or 1% SDS. Following incubation on ice for 30 min the extracts were separated into cytosolic (S100) and crude membrane (P100) fractions. Samples were resolved by SDS-PAGE and visualized by immunoblotting with anti-HA monoclonal antibody 12CA5 (BAbCo) or anti-Ras monoclonal antibody Y13-259. Immunocomplexes were visualized by using an HRP-conjugated secondary antibody and enhanced chemiluminescence (Pierce SUPER-SIGNAL).
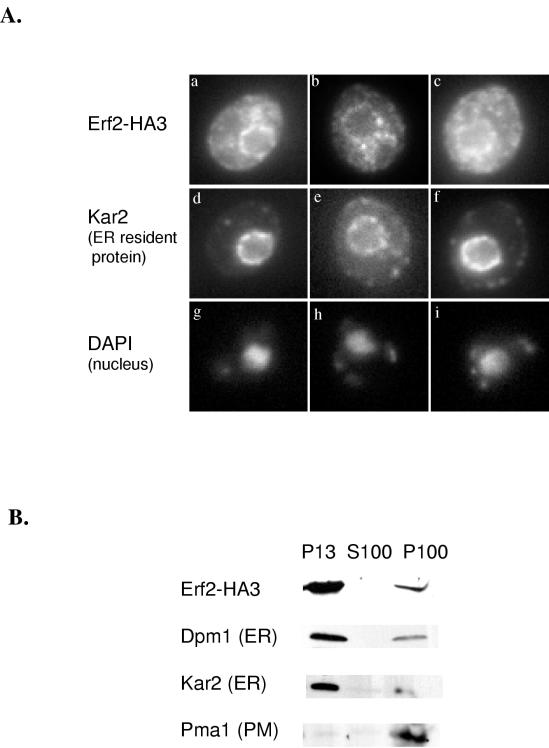
Indirect immunofluorescence of cells expressing Erf2-HA3 from a multicopy plasmid (B745) was performed to determine subcellular distribution of Erf2. As seen in Fig. 4A, Erf2-HA3 appears as a ring surrounding the nucleus which is stained by 4′,6-diamidino-2-phenylindole (DAPI). This perinuclear staining pattern is similar to that seen with the ER-resident protein Kar2 (Fig. 4A) (55). Because it was necessary to overexpress Erf2-HA3 in order to detect it by immunofluorescence, we also performed subcellular fractionation experiments using strains expressing Erf2-HA3 at physiological levels from a low-copy-number vector (B754). As seen in Fig. 4B, Erf2-HA3 is found primarily in the P13 fraction along with Dpm1 and Kar2, two ER-resident proteins. Very little Erf2-HA is seen in the P100 membrane fraction, where plasma membrane proteins such as Pma1 reside (Fig. 4B).
FIG. 4.
Immunolocalization of Erf2-HA3. (A) RJY1318 harboring B745 (Erf2-HA3) was grown and prepared for immunofluorescence as described in Materials and Methods. Erf2-HA3 was visualized with anti-HA monoclonal antibody 12CA5 (BAbCo) diluted 1/400 followed by Oregon Green 488-conjugated IgG (1:240) (a to c). Kar2 was visualized with an affinity-purified anti-Kar2 antibody (1:2,000) followed by lissamine rhodamine-conjugated anti-rabbit IgG (1:240) (d to f). DAPI staining of the nuclei is also shown (g to i). Cells were examined in a Zeiss Axioskop microscope. (B) P13 and P100 fractionation of RJY1318 harboring B754 (Erf2-HA3). Samples were resolved by SDS-PAGE and visualized by immunoblotting with an anti-HA (12CA5; BAbCo), anti-Dpm1, anti-Kar2, or anti-Pma1 antibody as described in Materials and Methods. Immunocomplexes were visualized as in Fig. 3. PM, plasma membrane.
Deletion of ERF2 affects the function of wild-type Ras.
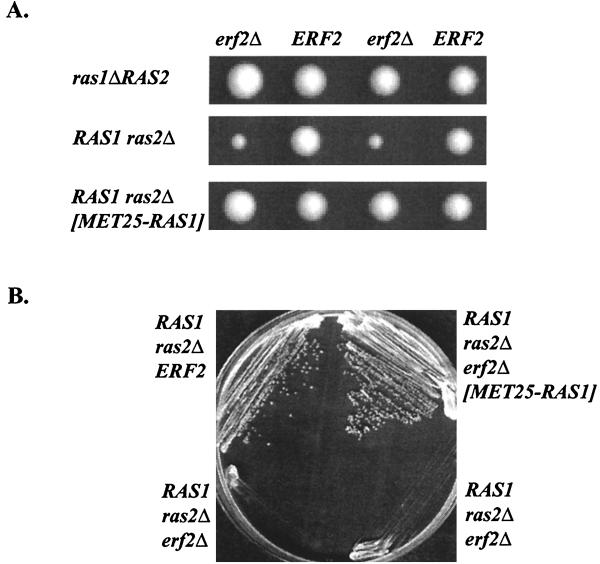
Deletion of ERF2 has no discernible effect on growth or viability of RAS1 RAS2 wild-type cells growing in rich or synthetic growth medium (Fig. 5 and data not shown). However, ERF2 is required when the CaaX mutant Ras2-ext is the only Ras expressed. This might indicate that Erf2 is essential only when Ras is not prenylated. Alternatively, there may be a differential requirement for Erf2 depending on the allele of RAS being expressed. We examined the requirement for ERF2 when either RAS1 or RAS2 is deleted. Tetrad analysis revealed that ras2Δ erf2Δ strains exhibited a severe growth defect, whereas a ras1Δ erf2Δ strain grew normally (Fig. 5A). This effect was not due to a sporulation or germination defects because the slow growth of ras2Δ erf2Δ strains was also observed when cells were streaked on plates (Fig. 5B) or growth curves were measured (data not shown). The apparent differential requirement of Erf2 depending on the RAS gene being expressed could arise from differences in the level of Ras activity in ras2Δ and ras1Δ cells or perhaps a functional difference between the two Ras proteins. RAS1 and RAS2 are differentially expressed depending on growth conditions and the position in the growth curve (10, 11). However, we did not observe a difference in the steady-state levels of Ras1 and Ras2 proteins in our strains (Fig. 3B). ras2Δ cells have lower cyclic AMP levels than isogenic ras1Δ cells, and purified recombinant Ras2 protein is a more effective activator of adenylyl cyclase than Ras1 (13, 29). We reasoned therefore that Erf2 is required in ras2Δ cells because the overall Ras activity in the cell is reduced. Consistent with this explanation, overexpression of RAS1 from the MET25 promoter rescues the growth defect of a ras2Δ erf2Δ strain (Fig. 5A). We cannot, however, rule out the possibility that Erf2 has a preference for Ras1 and that this preference can be overcome by overexpression of RAS1.
FIG. 5.
ras2Δ erf2Δ strains grow slowly. (A) Heterozygous diploids RJY1412 (top) and RJY1301 (middle) were sporulated, and a representative tetrad is shown. The bottom panel shows tetrads resulting from sporulation of RJY1301 harboring the MET25-RAS1 plasmid (B804). Colonies were grown on rich medium containing glucose (YEPD) for 3 days. (B) Indicated strains were streaked onto YEPD medium and shown to have a severe growth phenotype.
Overexpression of GPA2 fails to suppress the growth defect of a ras2-ext erf2Δ strain.
Our results demonstrate that Erf2 is required when the Ras CaaX box is mutated and under conditions of reduced Ras activity. Recently, the heterotrimeric G protein α subunit Gpa2 has been shown to act in parallel to Ras when Ras activity is compromised (17, 35, 70). Specifically, elevated expression of Gpa2 rescues the growth defect of a ras2ts strain at the nonpermissive temperature (49). A ras2Δ gpa2Δ strain grows very poorly, reminiscent of the growth defect of a ras2Δ erf2Δ strain (35, 38). This raises the possibility that the effect of Erf2 on the Ras pathway is actually mediated through modulation of the Gpa2 pathway. To test this hypothesis, we constructed a set of double and triple mutants and analyzed tetrads following sporulation. Strains harboring ras2Δ erf2Δ and ras2Δ gpa2Δ deletions were viable but grew slowly. In contrast, a ras2Δ erf2Δ gpa2Δ strain was inviable (data not shown). The additive effect of the erf2Δ and gpa2Δ mutations suggests that they operate in different pathways. Consistent with this interpretation, overexpression of GPA2 failed to rescue the inviability of a ras2-ext erf2Δ strain.
Deletion of ERF2 leads to a reduction in Ras palmitoylation.
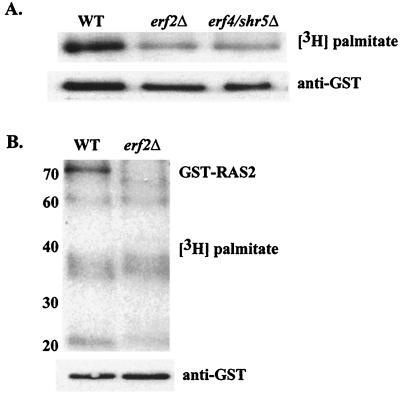
To examine the effect of Erf2 on Ras palmitoylation, wild-type and erf2Δ cells expressing GST-Ras2 were labeled with [3H]palmitate, and the fusion protein was analyzed by SDS-PAGE. As seen in Fig. 6A, deletion of either ERF2 or ERF4/SHR5 results in a decrease but not complete loss of steady-state Ras palmitoylation. The steady-state palmitoylation of other acylated proteins, however, was not affected in erf2Δ or erf4/shr5Δ strains (Fig. 6B). It is therefore unlikely that either ERF2 or ERF4/SHR5 encodes a component of a general palmitoyltransferase. It is possible that Erf2 is a Ras-specific palmitoylation system. Alternatively, Erf2 and Erf4/Shr5 could be involved in a step in Ras processing or trafficking that precedes palmitoylation.
FIG. 6.
In vivo labeling with [3H]palmitate. Strains RJY1270 wild type [WT]), RJY1272 (erf2Δ), and RJY1274 (erf4/shr5Δ) were labeled with [3H]palmitate as described in Materials and Methods. (A) GST-Ras2 was purified by glutathione affinity chromatography and resolved by SDS-PAGE, and the gel was subjected to fluorography (top) or anti-GST immunoblotting (bottom). (B) Total lysates from [3H]palmitate-labeled RJY1270 (WT) and RJY1272 (erf2Δ) were resolved by SDS-PAGE and subjected to fluorography (top) or anti-GST immunoblotting (bottom). The migration positions of prestained molecular weight markers (Gibco-BRL) are indicated on the left in kilodaltons.
Deletion of ERF2 causes mislocalization of Ras.
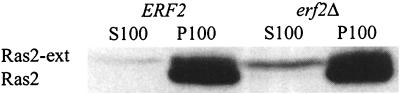
We performed subcellular fractionation and immunofluorescence experiments to examine the role of Erf2 in Ras membrane localization. Extracts from isogenic ERF2 and erf2::TRP1 deletion strains (RJY1107 and RJY1277) were separated into P100 and S100 fractions. As seen in Fig. 7, deletion of ERF2 did not significantly change the distribution of wild-type Ras proteins, but there was a redistribution of Ras2-ext protein from the membrane into the soluble fraction (Fig. 7). The prenyl group appears to be sufficient to retain wild-type Ras in the membrane, whereas the Ras2-ext protein requires palmitoylation for membrane association.
FIG. 7.
Crude membrane association of Ras2 and Ras2-ext in ERF2 and erf2Δ strains. Total cell lysates were prepared from RJY1107 (ERF2) or RJY1277 (erf2Δ) as described in Materials and Methods. The extracts were separated into S100 and P100 fractions. Ras2 and Ras2-ext were visualized by immunoblotting using a rat anti-ras monoclonal antibody (Y13-259) and anti-rat HRP-conjugated secondary antibody (Amersham). Immunocomplexes were visualized as in Fig. 3.
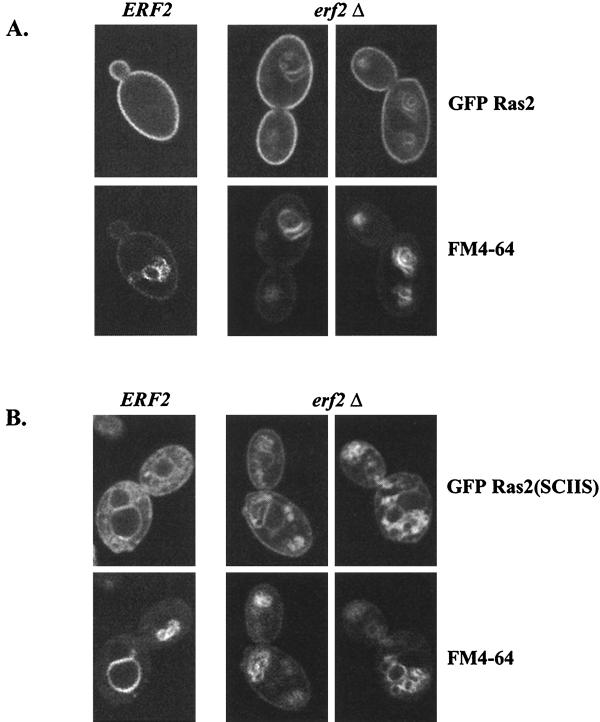
While the fractionation experiments described above suggest that the membrane association of wild-type Ras is not affected by deleting ERF2, palmitoylation of wild-type Ras is diminished in an erf2Δ strain (Fig. 6). We therefore examined the subcellular localization of GFP-Ras by fluorescence microscopy. As seen in Fig. 8A (upper left panel), GFP-Ras2 appears on the cell perimeter, consistent with its localization on the plasma membrane. In contrast, a shift of GFP-Ras to internal membranes occurs when ERF2 is deleted (Fig. 8A, top panels). The internal membrane compartment is stained with FM4-64, indicative of the vacuole (68). A more pronounced Ras mislocalization defect is observed with GFP-Ras2(SCIIS), a Ras mutant that cannot be palmitoylated (Fig. 8B). GFP-Ras2(SCIIS) is found primarily on internal membranes that include, but are not restricted to, the vacuole (Fig. 8B).
FIG. 8.
Subcellular localization of palmitoylated and nonpalmitoyled GFP-Ras proteins in ERF2 and erf2Δ strains. (A) RJY266 (ERF2) and RJY1318 (erf2Δ) were transformed with GFP-Ras2 (B701) (A) or GFP-Ras2(SCIIS) (B763) (B), and cells were grown in SC medium lacking uracil (4% galactose) to an optical density at 660 nm of 1.0. Vacuoles were visualized by staining with FM4-64. Cells were visualized directly by confocal microscopy (Bio-Rad MRC1024) using a 60× objective.
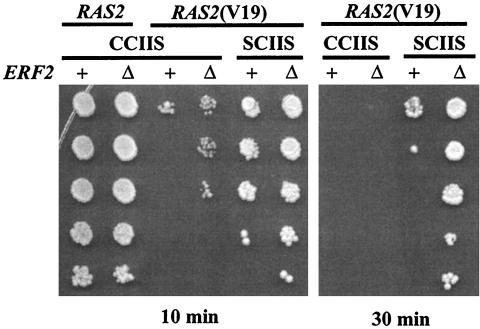
Deletion of ERF2 reduces the heat shock sensitivity caused by expression of Ras2(V19).
Yeast strains expressing Ras2(V19), a form that cannot hydrolyze GTP, are sensitive to heat shock. We examined whether Erf2 affected the heat shock sensitivity of strains expressing wild-type and Ras2(V19) proteins. Deletion of ERF2 had no measurable effect on the heat shock sensitivity of wild-type Ras2 but did have a protective effect on strains expressing Ras2(V19) (Fig. 9, 10-min heat shock treatment). It is important to note that the protection afforded by deleting ERF2 is less than the protection seen when palmitoylation is prevented by mutating Cys-318 to Ser [Ras2(V19,SCIIS)]. However, deletion of ERF2 provides further heat shock protection to the nonpalmitoylated Ras2(V19,SCIIS) allele. Since Erf2 affects the function of a nonpalmitoylated form of Ras2(SCIIS), it is not likely to be a component of the palmitoyltransferase; rather, Erf2 appears to play a role in Ras trafficking prior to palmitoylation.
FIG. 9.
Heat shock sensitivity of ERF2 and erf2Δ strains. ERF2 (+; LRB759) or erf2Δ (Δ; RJY1438) strains were transformed with pRS315 plasmids expressing Ras2(CCIIS) (B250), Ras2(V19,CCIIS) (B561), or Ras2(V19,SCIIS) (B562). The strains were subjected to heat shock for 10 min or 30 min and a serial dilution of cells was plated on SC medium plates lacking leucine as described in Materials and Methods.
DISCUSSION
It is now well established that subcellular targeting of Ras proteins requires a series of posttranslational modifications of a C-terminal CaaX box motif. The modifications include prenylation, -aaX proteolysis, carboxy methylation, and either palmitoylation or a patch of basic amino acid residues. It is also apparent that these modifications are not unique to Ras. In fact, it has been estimated that 1% of cellular proteins undergo prenylation and the associated CaaX modification events (24). It is therefore reasonable to expect to find additional factors that are required for correct subcellular targeting of Ras and other CaaX proteins. In this study, a genetic screen was carried out to identify additional components involved in the Ras subcellular localization pathway. A key feature of the screen was that it used a previously described Ras CaaX mutant (Ras2-ext) that is not farnesylated, aaX proteolyzed, or carboxyl methylated. Ras2-ext proteins are functional but exhibit lower affinity for membranes than prenylated Ras. Membrane association relies on a patch of basic amino acid residues and palmitoylation. We reasoned that strains expressing only Ras2-ext would have an increased sensitivity to mutations that affect Ras binding to the membrane, Ras trafficking, and/or palmitoylation. To date, we have identified mutants that fall into two complementation groups. ERF4/SHR5 was previously identified as a suppressor of the hyperactivity of a RAS2(V19) allele (31). Loss of Erf4/Shr5 causes a reduction in Ras palmitoylation, but this apparently results from a defect in membrane trafficking and not a defect in palmitoylation per se. Our analysis involving an additional seven mutant alleles of erf4/shr5 is consistent with that conclusion (data not shown). The second mutant identified in our screen, erf2, encodes a 41-kDa ER-localized protein. As for Erf4/Shr5, loss of Erf2 function results in a reduction in Ras palmitoylation and causes a similar mislocalization of Ras. erf4/shr5Δ and erf2Δ strains also suppress the heat shock phenotype of Ras2(V19). Interestingly, the double mutant (erf2Δ erf4/shr5Δ) is indistinguishable from either single mutant alone, suggesting that Erf2 and Erf4/Shr5 may function in the same pathway (data not shown).
DHHC motif proteins represent a distinct subgroup of zinc finger proteins (8, 42, 52). Although it is not known whether DHHC proteins bind zinc, it is clear from the mutations identified in ERF2 that the CRD region is important for function. ERF2 is the first member of this family found to have an effect on a known pathway. In the absence of Erf2, yeast Ras2 protein is mislocalized, leading to a growth defect under conditions of low Ras activity. The DHHC-CRD could form a protein-protein or protein-lipid binding domain. In this respect, the DHHC-CRD may be similar to the zinc clusters or zinc ring proteins Raf, Vav, Nore1, and rabphilin that interact with G proteins including metazoan Ras (33, 41, 65, 67). The structure of the Raf CRD has been solved and compared to the lipid binding domain of the protein kinase C cysteine-rich region (46). It would be interesting to determine the structure of the Erf2 DHHC-CRD and compare it to structures of these other domains.
Despite the high level of sequence similarity, the four other yeast DHHC-CRD proteins do not have functional overlap with Erf2. Gene disruptions of the other members (YDR126, YOL003, YNL326, and YDR459) of the group have no effect on Ras-dependent growth, and their overexpression does not suppress the Ras localization defect observed in an erf2Δ strain (data not shown). Thus, these other DHHC-CRD proteins likely function in pathways outside Ras. One member, YDR126w, has been uncovered in a synthetic lethal screen using a hypomorphic profilin gene and designated PSL10 (26a). Deletion of other members of the yeast Erf2 family have no discernible defects in growth on fermentable or nonfermentable carbon sources, mating, or bud polarity. Among the yeast DHHC-CRD proteins, only the most distantly related member, AKR1, has received attention. Akr1 physically associates with the cytoplasmic tail of the a-factor receptor (Ste3) and with the associated βγ (Ste4/Ste18) subunit of the heterotrimeric G protein activated during mating (26, 51). Genetic interactions have also been observed between AKR1 and the yeast Rho family G gene CDC42 (32). Finally, Akr1 may also play a role in trafficking due to its interaction with the ARF GTPase-activating protein Gcs1 (32). It is not known at this time whether the CRD of Akr1 contributes to these interactions.
The localization of Erf2 to the ER places it in the same subcellular membrane compartment as the -aaX proteases Rce1 and Afc1/Ste24 and the prenyl protein carboxymethyltransferase Ste14. It is tempting to speculate that Erf2, Rce1, and Ste14 may exist in a complex. Although there is no indication of a complex at this time, attempts to extract Erf2 with Triton X-100 were only partially successful, leading us to consider the possibility that Erf2 associates with a complex. Triton X-100 insolubility has been interpreted as evidence of interaction with the cytoskeleton, but sodium cholate, a detergent reported to extract cytoskeletal proteins, was only partially effective at extracting Erf2 from the P100 fraction. The combination of Triton X-100 and sodium cholate effectively solubilizes the bulk of Erf2. Perhaps there are two distinct pools of Erf2 within the cell that can be distinguished by detergent solubility. Perhaps Erf2 resides in two distinct microdomains of the ER; alternatively, Erf2 may cycle between the ER and another organelle such as the Golgi complex. Results of subcellular localization experiments were consistent with an ER localization, but we cannot rule out the existence of another pool of Erf2.
Mutations in ERF2 were isolated by using a Ras2-ext allele which terminates in a nonfunctional CaaX box. Several lines of evidence indicate that Erf2 is required for the function of wild-type RAS alleles as well. The growth of a RAS1 ras2Δ strain is significantly impaired by the deletion of ERF2. GFP-Ras2 is mislocalized upon deletion of ERF2. Finally, the heat shock sensitivity of the activated Ras2(V19) allele is partially suppressed in the absence of ERF2. Deletion of ERF2 also reduces the steady-state level of Ras palmitoylation (Fig. 6). Protein palmitoylation has received considerable attention as a potential regulatory mechanism in cell signaling (48, 66). Palmitoylation often occurs in conjunction with myristoylation or farnesylation, but unlike these other lipid modifications, palmitoylation is reversible and therefore a potential point of regulation. Unfortunately, little is known about the enzymology of protein palmitoylation. Protein palmitoyltransferase activities have been reported, but the purification of the enzyme or identification of the genes has not been accomplished (5, 19, 37). This has led some to suggest that protein palmitoylation may occur nonenzymatically (1, 23); however, the rates of the nonenzymatic reaction are probably too low to account for palmitoylation observed in vivo (1). The screen that we describe in this report was based on a palmitoylation-dependent Ras allele, and thus mutants derived from the screen might shed light on the palmitoylation debate. Are Erf2 and Erf4/Shr5 components of the elusive palmitoyltransferase? Several observations argue against this hypothesis. First, the sequences of Erf2 and Erf4/Shr5 bear no relationship to those of known acyltransferases. Second, palmitoylation of GST-Ras2 is not completely abolished in an erf2Δ strain, and when crude extracts were analyzed for [3H]palmitate incorporation, no major differences were apparent between erf2Δ and wild-type extracts (Fig. 6). Third, phenotypic differences exist between an erf2Δ strain and an ERF2 strain expressing a mutant Ras2 protein that cannot be palmitoylated [Ras2(SCIIS)]. Nonpalmitoylated Ras is virtually undetectable on the plasma membrane, whereas deletion of ERF2 causes a less severe localization defect. A difference is also observed when we analyze heat shock sensitivities in strains expressing a Ras2(V19) allele. Preventing palmitoylation by mutating the palmitoylated Cys of Ras to Ser affords a better heat shock protection than deleting ERF2. Furthermore, deleting ERF2 has an additive protective effect even in the case of a nonpalmitoylated Ras protein. We therefore conclude that the reduction of Ras palmitoylation in the absence of Erf2 and Erf4/Shr5 is due to a step prior to palmitoylation such as Ras trafficking.
There are still many gaps in our knowledge of how Ras assembles into an active signaling complex at the plasma membrane. Processing of the CaaX clearly plays a central role and has provided important insights into the trafficking pathway. Once prenylylation occurs, all subsequent Ras CaaX processing events occur on membranes. For example, the -aaX proteases (Rce1 and Afc1) and the carboxymethyltransferase (Ste14) are localized on the ER membrane, suggesting that translocation of Ras to the plasma membrane begins on the surface of the ER (9, 18, 54, 58). However the steps from ER to plasma membrane are not known. It has been hypothesized that a second signal in the form of palmitoylation or a patch of basic amino acids is required for plasma membrane localization (14). This stems from observations that nonpalmitoylated Ras mutant protein accumulates on intracellular membranes. But it is not known at this time whether palmitoylation serves as a signal that directs Ras to the plasma membrane or as an anchor once Ras arrives. A more fundamental question is whether Ras is a passenger on the surface of other secretory pathway organelles or if there a novel translocation pathway that delivers Ras from the ER to the plasma membrane. The further characterization of Erf2 and Erf4/Shr5 may provide some insight into these questions. The aberrant localization of Ras in an erf2Δ deletion strain suggests that Erf2 does indeed play a role in Ras trafficking.
An intriguing idea is that Erf2 may be part of a scaffold or receptor for Ras binding to the ER. A similar scaffolding function has been proposed for Akr1 with the heterotrimeric Gβγ proteins in the mating pheromone pathway (51). However, to date we have been unable to demonstrate a direct interaction between Ras and Erf2 by using two-hybrid analysis or immunoprecipitation. Perhaps other components such as Erf4/Shr5 are needed to form a functional scaffold. Future work will investigate whether Erf2 and Erf4/Shr5 comprise a membrane-organizing center for the Ras signaling complex.
ACKNOWLEDGMENTS
We thank Mark Rose (Princeton) and Tom Stevens (Oregon) for antibodies, Jasper Rine (Berkeley) for the GFP-Ras plasmid, and Lucy Robinson for strains. In addition, we thank Lois Weisman and Jan Fassler for many helpful discussions and for reading the manuscript. Immunolocalization studies were done with the assistance of Cherie Malone and of Tom Monniger, University of Iowa Central Microscopy Research Facility. FM4-64 labeling was done with the skillful assistance of Cecilia Bonangelino of the Weisman lab. We especially thank members of the lab (Cherie Malone, Hong Lin, Addison Ault, and Lihong Zhao) for their input throughout the study.
This work was supported by grant CA50211 from the National Cancer Institute.
REFERENCES
- 1.Bano M C, Jackson C S, Magee A I. Pseudo-enzymatic S-acylation of a myristoylated Yes protein tyrosine kinase peptide in vitro may reflect non-enzymatic S-acylation in vivo. Biochem J. 1998;330:723–731. doi: 10.1042/bj3300723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender A, Pringle J R. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger M, Schmidt M F G. Cell-free fatty acid acylation of Semliki forest viral polypeptides with microsomal membranes from eucaryotic cells. J Biol Chem. 1984;259:7245–7252. [PubMed] [Google Scholar]
- 4.Berkower C, Loayza D, Michaelis S. Metabolic instability and constitutive endocytosis of STE6, the a-factor transporter of Saccharomyces cerevisiae. Mol Cell Biol. 1994;5:1185–1198. doi: 10.1091/mbc.5.11.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthiaume L, Resh M D. Biochemical characterization of a palmitoyl acyltransferase activity that palmitoylates myristoylated proteins. J Biol Chem. 1995;270:22399–22405. doi: 10.1074/jbc.270.38.22399. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharya S, Chen L, Broach J R, Powers S. Ras membrane targeting is essential for glucose signaling but not for viability in yeast. Proc Natl Acad Sci USA. 1995;92:2984–2988. doi: 10.1073/pnas.92.7.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 8.Bohm S, Frishman D, Mewes H W. Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res. 1997;25:2464–2469. doi: 10.1093/nar/25.12.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyartchuk V L, Ashby M N, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- 10.Breviario D, Hinnebusch A, Cannon J, Tatchell K, Dhar R. Carbon source regulation of RAS1 expression in Saccharomyces cerevisiae and the phenotypes of ras2 cells. Proc Natl Acad Sci USA. 1986;83:4152–4156. doi: 10.1073/pnas.83.12.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breviario D, Hinnebusch A G, Dhar R. Multiple regulatory mechanisms control the expression of the RAS1 and RAS2 genes of Saccharomyces cerevisiae. EMBO J. 1988;7:1805–1813. doi: 10.1002/j.1460-2075.1988.tb03012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broach J R, Deschenes R J. The function of RAS genes in Saccharomyces cerevisiae. Adv Cancer Res. 1990;54:79–140. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- 13.Broek D, Samiy N, Fasano O, Fujiyama A, Tamanoi F, Northup J, Wigler M. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell. 1985;41:763–769. doi: 10.1016/s0092-8674(85)80057-x. [DOI] [PubMed] [Google Scholar]
- 14.Cadwallader K A, Paterson H, Macdonald S G, Hancock J F. N-terminally myristoylated Ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol Cell Biol. 1994;14:4722–4730. doi: 10.1128/mcb.14.7.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke S. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu Rev Biochem. 1992;61:355–386. doi: 10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- 16.Clarke S, Vogel J P, Deschenes R J, Stock J. Posttranslational modification of the Ha-ras oncogene protein: evidence for a third class of protein carboxyl methyltransferases. Proc Natl Acad Sci USA. 1988;85:4643–4647. doi: 10.1073/pnas.85.13.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colombo S, Ma P, Cauwenberg L, Winderickx J, Crauwels M, Teunissen A, Nauwelaers D, de Winde J H, Gorwa M F, Colavizza D, Thevelein J M. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signaling in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:3326–3341. doi: 10.1093/emboj/17.12.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai Q, Choy E, Chiu V, Romano J, Slivka S R, Steitz S A, Michaelis S, Philips M R. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J Biol Chem. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- 19.Das A K, Dasgupta B, Bhattacharya R, Basu J. Purification and biochemical characterization of a protein-palmitoyl acyltransferase from human erythrocytes. J Biol Chem. 1997;272:11021–11025. doi: 10.1074/jbc.272.17.11021. [DOI] [PubMed] [Google Scholar]
- 20.Der C J, Cox A D. Isoprenoid modification and plasma membrane association: critical factors for ras oncogenicity. Cancer Cells. 1991;3:331–340. [PubMed] [Google Scholar]
- 21.Deschenes R J, Resh M D, Broach J R. Acylation and prenylation of proteins. Curr Opin Cell Biol. 1990;2:1108–1113. doi: 10.1016/0955-0674(90)90164-a. [DOI] [PubMed] [Google Scholar]
- 22.Deschenes R J, Stimmel J B, Clarke S, Stock J, Broach J R. RAS2 protein of Saccharomyces cerevisiae is methyl-esterified at its carboxyl terminus. J Biol Chem. 1989;264:11865–11873. [PubMed] [Google Scholar]
- 23.Duncan J A, Gilman A G. Autoacylation of G protein alpha subunits. J Biol Chem. 1996;271:23594–23600. doi: 10.1074/jbc.271.38.23594. [DOI] [PubMed] [Google Scholar]
- 24.Epstein W W, Lever D, Leining L M, Bruenger E, Rilling H C. Quantitation of prenylcysteines by a selective cleavage reaction. Proc Natl Acad Sci USA. 1991;88:9668–9670. doi: 10.1073/pnas.88.21.9668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gimeno C J, Fink G R. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Givan S A, Sprague G F., Jr The ankyrin repeat-containing protein Akr1p is required for the endocytosis of yeast pheromone receptors. Mol Biol Cell. 1997;8:1317–1327. doi: 10.1091/mbc.8.7.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Haarer, B. Personal communication.
- 27.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 28.Horazdovsky B F, Emr S D. The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J Biol Chem. 1993;268:4953–4962. [PubMed] [Google Scholar]
- 29.Hurwitz N, Segal M, Marbach I, Levitzki A. Differential activation of yeast adenylyl cyclase by Ras1 and Ras2 depends on the conserved N terminus. Proc Natl Acad Sci USA. 1995;92:11009–11013. doi: 10.1073/pnas.92.24.11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung V, Chen L, Hofmann S L, Wigler M, Powers S. Mutations in the SHR5 gene of Saccharomyces cerevisiae suppress Ras function and block membrane attachment and palmitoylation of Ras proteins. Mol Cell Biol. 1995;15:1333–1342. doi: 10.1128/mcb.15.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao L R, Peterson J, Ji R, Bender L, Bender A. Interactions between the ankyrin repeat-containing protein Akr1p and the pheromone response pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:168–178. doi: 10.1128/mcb.16.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kazanietz M G, Bustelo X R, Barbacid M, Kolch W, Mischak H, Wong G, Pettit G R, Bruns J D, Blumberg P M. Zinc finger domains and phorbol ester pharmacophore. Analysis of binding to mutated form of protein kinase C zeta and the vav and c-raf proto-oncogene products. J Biol Chem. 1994;269:11590–11594. [PubMed] [Google Scholar]
- 34.Kohl N E, Diehl R E, Schaber M D, Rands E, Soderman D D, He B, Moores S L, Pompliano D L, Ferro-Novick S, Powers S, Thomas K A, Gibbs J B. Structural homology among mammalian and Saccharomyces cerevisiae isoprenyl-protein transferases. J Biol Chem. 1991;266:18884–18888. [PubMed] [Google Scholar]
- 35.Kubler E, Mosch H U, Rupp S, Lisanti M P. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- 36.Kuchler K, Thorner J. Secretion of peptides and proteins lacking hydrophobic signal sequences: the role of adenosine triphosphate driven membrane translocators. Endocrine Rev. 1992;13:499–514. doi: 10.1210/edrv-13-3-499. [DOI] [PubMed] [Google Scholar]
- 37.Liu L, Dudler T, Gelb M H. Purification of a protein palmitoyltransferase that acts on H-Ras protein and on a C-terminal N-Ras peptide. J Biol Chem. 1996;271:23269–23276. doi: 10.1074/jbc.271.38.23269. [DOI] [PubMed] [Google Scholar]
- 38.Lorenz M C, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manne V, Roberts D, Tobin A, O’Rourke E, De Virgilio M, Meyers C, Ahmed N, Kurz B, Resh M, Kung H-F, Barbacid M. Identification and preliminary characterization of protein-cysteine farnesyltransferase. Proc Natl Acad Sci USA. 1990;87:7541–7545. doi: 10.1073/pnas.87.19.7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGrath J P, Varshavsky A. The yeast STE6 gene encodes a homologue of the mammalian multidrug resistance P-glycoprotein. Nature. 1989;340:400–404. doi: 10.1038/340400a0. [DOI] [PubMed] [Google Scholar]
- 41.McKiernan C J, Stabila P F, Macara I G. Role of the Rab3A-binding domain in targeting of rabphilin-3A to vesicle membranes of PC12 cells. Mol Cell Biol. 1996;16:4985–4995. doi: 10.1128/mcb.16.9.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mesilaty-Gross S, Reich A, Motro B, Wides R. The Drosophila STAM gene homolog is in a tight gene cluster, and its expression correlates to that of the adjacent gene ial. Gene. 1999;231:173–186. doi: 10.1016/s0378-1119(99)00053-0. [DOI] [PubMed] [Google Scholar]
- 43.Mitchell D A, Farh L, Marshall T K, Deschenes R J. A polybasic domain allows nonprenylated Ras proteins to function in Saccharomyces cerevisiae. J Biol Chem. 1994;269:21540–21546. [PubMed] [Google Scholar]
- 44.Mitchell D A, Marshall T K, Deschenes R J. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 45.Morishita T, Mitsuzawa H, Nakafuku M, Nakamura S, Hattori S, Anraku Y. Requirement of Saccharomyces cerevisiae Ras for completion of mitosis. Science. 1995;270:1213–1215. doi: 10.1126/science.270.5239.1213. [DOI] [PubMed] [Google Scholar]
- 46.Mott H R, Carpenter J W, Zhong S, Ghosh S, Bell R M, Campbell S L. The solution structure of the Raf-1 cysteine-rich domain: a novel ras and phospholipid binding site. Proc Natl Acad Sci USA. 1996;93:8312–8317. doi: 10.1073/pnas.93.16.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mumby S M. Reversible palmitoylation of signaling proteins. Curr Opin Cell Biol. 1997;9:148–154. doi: 10.1016/s0955-0674(97)80056-7. [DOI] [PubMed] [Google Scholar]
- 49.Nakafuku M, Obara T, Kaibuchi K, Miyajima I, Miyajima A, Itoh H, Nakamura S, Arai K, Matsumoto K, Kaziro Y. Isolation of a second yeast Saccharomyces cerevisiae gene (GPA2) coding for guanine nucleotide-binding regulatory protein: studies on its structure and possible functions. Proc Natl Acad Sci USA. 1988;85:1374–1378. doi: 10.1073/pnas.85.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panek H R, Stepp J D, Engle H M, Marks K M, Tan P K, Lemmon S K, Robinson L C. Suppressors of YCK-encoded yeast casein kinase 1 deficiency define the four subunits of a novel clathrin AP-like complex. EMBO J. 1997;16:4194–4204. doi: 10.1093/emboj/16.14.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pryciak P M, Hartwell L H. AKR1 encodes a candidate effector of the G beta gamma complex in the Saccharomyces cerevisiae pheromone response pathway and contributes to control of both cell shape and signal transduction. Mol Cell Biol. 1996;16:2614–2626. doi: 10.1128/mcb.16.6.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Putilina T, Wong P, Gentleman S. The DHHC domain: a new highly conserved cysteine-rich motif. Mol Cell Biochem. 1999;195:219–226. doi: 10.1023/a:1006932522197. [DOI] [PubMed] [Google Scholar]
- 53.Reiss Y, Seabra M C, Armstrong S A, Slaughter C A, Goldstein J L, Brown M S. Nonidentical subunits of p21H-ras farnesyltransferase: peptide binding and farnesyl pyrophosphate carrier functions. J Biol Chem. 1991;266:10672–10677. [PubMed] [Google Scholar]
- 54.Romano J D, Schmidt W K, Michaelis S. The Saccharomyces cerevisiae prenylcysteine carboxyl methyltransferase Ste14p is in the endoplasmic reticulum membrane. Mol Cell Biol. 1998;9:2231–2247. doi: 10.1091/mbc.9.8.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rose M D, Misra L M, Vogel J P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 56.Sapperstein S, Berkower C, Michaelis S. Nucleotide sequence of the yeast STE14 gene, which encodes farnesylcysteine carboxyl methyltransferase, and demonstration of its essential role in a-factor export. Mol Cell Biol. 1994;14:1438–1449. doi: 10.1128/mcb.14.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schafer W R, Rine J. Protein prenylation: genes, enzymes, targets, and functions. Annu Rev Genet. 1992;26:209–237. doi: 10.1146/annurev.ge.26.120192.001233. [DOI] [PubMed] [Google Scholar]
- 58.Schmidt W K, Tam A, Fujimura-Kamada K, Michaelis S. Endoplasmic reticulum membrane localization of rce1p and ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc Natl Acad Sci USA. 1998;95:11175–11180. doi: 10.1073/pnas.95.19.11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sherman F, Fink G R, Hicks J B. Laboratory course manual: methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 60.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith A, Ward M P, Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thissen J A, Casey P J. Microsomal membranes contain a high affinity binding site for prenylated peptides. J Biol Chem. 1993;268:13780–13783. [PubMed] [Google Scholar]
- 63.Thissen J A, Gross J M, Subramanian K, Meyer T, Casey P J. Prenylation-dependent association of Ki-Ras with microtubules—evidence for a role in subcellular trafficking. J Biol Chem. 1997;272:30362–30370. doi: 10.1074/jbc.272.48.30362. [DOI] [PubMed] [Google Scholar]
- 64.Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- 65.Vavvas D, Li X, Avruch J, Zhang X F. Identification of Nore1 as a potential Ras effector. J Biol Chem. 1998;273:5439–5442. doi: 10.1074/jbc.273.10.5439. [DOI] [PubMed] [Google Scholar]
- 66.Veit M, Schmidt M F. Membrane targeting via protein palmitoylation. Methods Mol Biol. 1998;88:227–239. doi: 10.1385/0-89603-487-9:227. [DOI] [PubMed] [Google Scholar]
- 67.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y X, Zhao H, Harding T M, Gomes de Mesquita D S, Woldringh C L, Klionsky D J, Munn A L, Weisman L S. Multiple classes of yeast mutants are defective in vacuole partitioning yet target vacuole proteins correctly. Mol Biol Cell. 1996;7:1375–1389. doi: 10.1091/mbc.7.9.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willumsen B M, Cox A D, Solski P A, Der C J, Buss J E. Novel determinants of H-Ras plasma membrane localization and transformation. Oncogene. 1996;13:1901–1909. [PubMed] [Google Scholar]
- 70.Xue Y, Batlle M, Hirsch J P. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]