Abstract
The ability of distant cis-acting DNA elements to interact functionally has been proposed to be mediated by the interaction of proteins associated site specifically with those cis-acting elements. We have found that the DNA-linking regions of EBNA1 are essential for its contribution to both replication and transcription. The synthesis of plasmids containing the Epstein-Barr virus (EBV) origin of plasmid replication (oriP) can be mediated entirely by the cellular machinery; however, the replicated molecules are lost rapidly from proliferating cells. When EBNA1 is provided in trans, plasmids containing oriP (oriP plasmids) are synthesized during repeated S phases, and the newly formed daughter molecules are precisely segregated to the daughter cells. The contribution(s) of EBNA1 to the stable replication of oriP plasmids is therefore likely to be postsynthetic. In latently infected cells, EBNA1 also regulates the expression of multiple EBV promoters located as many as 10 kbp away. EBNA1 supports replication and transcription through binding to oriP; both the ability of EBNA1 to bind to DNA and the integrity of its binding sites in oriP are required. However, DNA binding by EBNA1 is not sufficient to support replication or transcription, indicating that an additional activity (or activities) is required. EBNA1 links DNAs to which it binds and can form a loop between the two subelements of oriP, the family of repeats and the region of dyad symmetry, each of which contains multiple binding sites for EBNA1. We have constructed a set of derivatives of EBNA1 which contain both, one, or neither of its linking regions in various contexts. Analyses of these derivatives demonstrate that the linking regions of EBNA1 are essential for its support of replication and transcription and that the ability of derivatives of EBNA1 to link DNAs correlates strongly with their support of these activities in cells. These findings indicate that protein-protein associations of the linking regions of EBNA1 underlie its long-range contributions to replication and transcription.
Upon infection of primary B lymphocytes, the Epstein-Barr virus (EBV) efficiently induces them to proliferate. The virus establishes a latent infection in these cells, and the viral plasmid replicates in a manner similar to that of a cellular chromosome (1). Specifically, viral DNA is replicated semiconservatively, once per cell cycle, during the S phase. The EBV origin of plasmid replication (oriP) is sufficient, when provided in cis, to confer upon exogenous plasmids the ability to replicate similarly (38, 50, 66, 68) and to be faithfully segregated to daughter cells (31, 60). EBNA1 is the only viral protein required in trans for the stable replication of plasmids containing oriP (oriP plasmids) (38, 50). In cells expressing EBNA1, oriP plasmids containing a selectable marker are maintained indefinitely as plasmids at a stable copy number under selective conditions (31). In the absence of selection, oriP plasmids are lost at a rate of 2 to 4% per cell per generation (31, 60), a rate of loss comparable to that of highly stable ARS-CEN plasmids in Saccharomyces cerevisiae (27, 36). Therefore, oriP is replicated once in each S phase and is efficiently and evenly partitioned to the nuclei of daughter cells.
oriP is an origin of DNA synthesis that can be recognized entirely by cellular enzymes (3). oriP plasmids support at least two rounds of DNA synthesis in the absence of EBNA1. In the first 48 h following transfection, replicated oriP plasmids accumulate to similar levels in the presence or absence of EBNA1. During subsequent cell cycles, oriP plasmids are maintained at similar numbers of copies per cell in the presence of EBNA1 but are lost rapidly in the absence of EBNA1. Thus, oriP is a cis-acting sequence sufficient to facilitate synthesis by the cellular replication apparatus, and EBNA1 acts postsynthetically to ensure continued replication of oriP.
oriP is composed of two clusters of EBNA1-binding sites (48, 50). The family of repeats (FR) consists of 20 degenerate copies of a 30-bp sequence to which EBNA1 binds with a high affinity; the region of dyad symmetry (DS) has four truncated copies of this repeat to which EBNA1 binds with a lower affinity (6, 30, 50). Individually, the FR with its flanking DNA and the DS both facilitate DNA synthesis in the absence of EBNA1 (3). Flanking the DS is an element called Rep*, which can partially substitute for the DS in the support of replication (33). Like the other elements of oriP, Rep* may also support the initiation of DNA synthesis. In cells expressing EBNA1, replication initiates predominantly at or near the DS (21). Like cellular origins (14), oriP contains multiple elements which can facilitate DNA synthesis and has one site at which synthesis predominantly initiates.
EBNA1 supports the stable replication of oriP plasmids through postsynthetic contributions which ensure their persistence in cells (3). oriP, or just the FR, is also a transcriptional enhancer through which EBNA1 supports the activity of at least two EBV promoters (22, 47, 49, 61). The ability of EBNA1 to bind to oriP is necessary for its support of both replication and transcription (46). The dimerization and DNA-binding domain of EBNA1 is insufficient to support these activities; in fact, this domain alone is a dominant negative inhibitor of these activities of wild-type EBNA1 (32). EBNA1 lacks the helicase and ATPase activities associated with origin-binding proteins of other viruses, an observation consistent with the fact that the synthesis of oriP is not dependent on EBNA1 (20, 44). Interaction assays with yeast have identified numerous molecules which associate with EBNA1, but the biological significance of these interactions remains unknown (3, 16, 63). Mutagenesis of EBNA1 outside the region which mediates dimerization and DNA binding has indicated that no single domain is required for the support by EBNA1 of replication and transcription (67). One interpretation of this result is that, in addition to its DNA-binding domain, EBNA1 contains multiple, redundant domains which contribute to its activities in cells. We wished to understand what, in addition to DNA binding, is necessary for the support by EBNA1 of stable replication and transcription.
We had previously shown that multiple domains of EBNA1 can independently mediate DNA linking (39). Therefore, the linking domains are candidates for those domains that contribute to the support of replication and transcription. EBNA1 links bound DNAs via direct protein-protein interactions between the linking domains (40). EBNA1 can link the FR and the DS of oriP and, in doing so, form a loop of the intervening DNA (19, 59). Numerous other viral and cellular proteins similarly form a DNA loop between sites to which they bind (34, 35, 37, 58). Members of this family of linking-looping proteins, which include EBNA1, E2 of bovine papillomavirus, and the cellular transcription factor Sp1, regulate replication and transcription; therefore, linking activity may contribute generally to the regulation of these processes in cells. We have tested the contribution of DNA linking to the support by EBNA1 of replication and transcription.
EBNA1 has two regions rich in basic residues and each containing domains sufficient to mediate DNA linking (39). These basic regions are termed linking region 1 (amino acids 40 to 89) and linking region 2 (amino acids 328 to 377). We have constructed a set of derivatives of EBNA1 designed to test the contributions of these regions to the activities of EBNA1. These derivatives lack one or both of the linking regions or have one of the linking regions replaced by the other. The ability of the derivatives to support replication and transcription was dependent on the linking regions. The efficiency with which the derivatives linked DNA was a function of the number of linking regions that they contained. Strong correlations between the efficiency of linking by derivatives of EBNA1 and their support of replication and transcription are consistent with linking contributing to the activities of EBNA1 in cells. Linking regions 1 and 2 were functionally redundant in their contributions to the support by EBNA1 of replication; either linking per se or another activity shared by the linking regions underlies these contributions. We propose several mechanisms by which linking may contribute to the support by EBNA1 of replication and transcription and infer how other viral and cellular proteins which can link DNA may function similarly.
MATERIALS AND METHODS
Plasmids.
The EBNA1 expression plasmids p1160 and p1553, which contain the genes 1160 and 1553, respectively, have been described previously (2, 45). Standard PCR-based cloning strategies were used to generate the new derivatives. The expression of all the derivatives of EBNA1 is under the control of the cytomegalovirus (CMV) immediate-early (IE) promoter. The derivatives contain only amino acids from EBNA1, with the following exceptions: 1893 has a valine and a methionine following the Gly-Gly-Ala repeats and preceding amino acid 378, 1767 has a leucine and a methionine following the Gly-Gly-Ala repeats, 1763 has an alanine and a methionine following the Gly-Gly-Ala repeats, 1891 has a valine and a methionine preceding amino acid 378, and 1747 has a serine and a methionine following the Gly-Gly-Ala repeats. An ABI automated DNA sequencer (Applied Biosystems) was used to verify the sequence of each derivative. pCDNA3 (Invitrogen), which has the CMV IE promoter but expresses no protein, served as a negative control.
The replication reporter plasmids were 1591 (with oriP) and 1381 (without oriP). Plasmid 1381 has been described previously (32). Plasmid 1591 was constructed as follows. The AatII-to-BamHI fragment from 994 (31), which contains oriP, was introduced into the AatII and BamHI sites in pUC19. The resulting plasmid was then cut with StuI and HindIII (the HindIII site was blunted with the Klenow fragment), and a BsmBI-to-BamHI fragment (each site was blunted with the Klenow fragment), which contains the simian virus 40 (SV40) IE promoter driving the expression of the neomycin resistance gene, was introduced. The insert came from a derivative of pSV2Neo (56) which had previously been cut with NsiI, blunted with mung bean nuclease, and religated. This fragment, which contains only one of the 72-bp repeats from SV40, is oriented with the promoter from SV40 driving transcription away from the DS of oriP.
The transcription reporter plasmid FR-TK-Luciferase has been described previously (44). Briefly, this plasmid contains the FR of oriP upstream of the herpes simplex virus thymidine kinase promoter driving the expression of the luciferase gene. In the transcription assays, pEQ176 (52), which has the CMV IE promoter driving the expression of the β-galactosidase gene, was used as an internal control for transfection efficiency.
Cells and transfections.
143B (7) and 293 (23) cells were grown in Dulbecco modified Eagle medium supplemented with 10% bovine calf serum and fetal calf serum, respectively. Plasmids were transfected into both cell types as a calcium phosphate precipitate (51). The precipitate was placed on adherent 293 cells in a standard fashion. 143B cells were suspended by trypsinization, combined with the precipitate, and placed in a new tissue culture dish. For each cell type, the medium was refreshed 6 to 8 h after the addition of the precipitate.
Western blotting.
Between 1 and 6.4 μg of each EBNA1 expression plasmid was transfected into 293 cells. We adjusted the amount of each expression plasmid that was transfected in order to normalize the expression of the derivatives. The total amount of CMV IE promoter-containing plasmid used per transfection was kept constant by supplementation with pCDNA3. The cells were grown for 48 h prior to harvest. Western blotting was done by standard protocols (51). Briefly, extracts were separated on sodium dodecyl sulfate–10% polyacrylamide gels and transferred to nitrocellulose membranes. After blocking was done with 10% fat-free milk and 0.05% Tween 20, blots were exposed to a rabbit anti-EBNA1 polyclonal antiserum. This antiserum was raised against a fusion protein containing amino acids 7 to 37 and 420 to 617 of EBNA1 and primarily or entirely recognizes the DNA-binding domain (57). The secondary antibodies were either alkaline phosphatase-conjugated goat anti-rabbit antibodies (Boehringer Mannheim Biochemicals) (data not shown) or 35S-labeled donkey anti-rabbit antibodies (Amersham) (Fig. 1B). The 35S-labeled blot was visualized and quantified with a PhosphorImager (Molecular Dynamics).
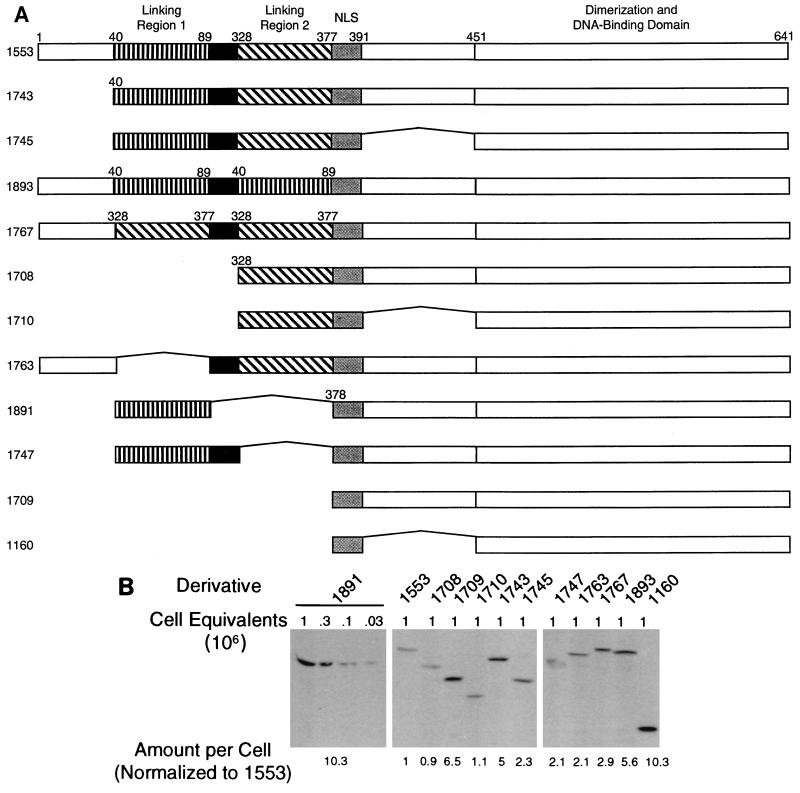
FIG. 1.
Structure and expression of derivatives of EBNA1 studied. (A) Linking region 1 (amino acids 40 to 89) and linking region 2 (amino acids 328 to 377) are regions rich in basic residues; each has been shown to link DNAs. All the derivatives of EBNA1 contain amino acids 451 to 641, which include a domain sufficient for dimerization and site-specific DNA binding. Amino acids 379 to 391 (gray box) contain a nuclear localization sequence (NLS) which is present in all the derivatives. The Gly-Gly-Ala repeats (black box) constitute a domain consisting solely of glycines and alanines. The Gly-Gly-Ala repeats in the derivatives contain only 15 amino acids of the 239 in EBNA1 from the B95-8 strain of EBV. However, the 1553 derivative of EBNA1 supports replication and transcription as well as does full-length EBNA1 and is considered wild type in this study. The amino acid numbers are from EBNA1 of the B95-8 strain. (B) Representative Western blot demonstrating that each derivative is expressed in cells in tissue cultures. Plasmids expressing the derivatives of EBNA1 were introduced into 293 cells as calcium phosphate precipitates. After 48 h, cells were harvested and counted, and the indicated numbers were run on sodium dodecyl sulfate-polyacrylamide gels. Following transfer to nitrocellulose, blots were probed with a rabbit polyclonal antiserum which detects amino acids 451 to 641 of EBNA1. A 35S-labeled secondary antibody (donkey anti-rabbit) was used to detect the bound anti-EBNA1 antibodies. Various numbers of cell equivalents expressing the 1891 derivative were used to generate a standard curve. The relative level of expression of each derivative (with 1553 set to 1) was interpolated from this curve and is shown below the blots. Independent qualitative experiments with an alkaline phosphatase-linked secondary antibody also indicated that the derivatives were expressed similarly (data not shown).
Replication assays.
143B cells were transfected with 1 to 6.4 μg of each EBNA1 expression plasmid, 10 μg of 1591, and 7.3 μg of 1381 (equimolar amounts of 1591 and 1381). After 96 h, the cells were harvested, and low-molecular-weight DNA was extracted by the method of Hirt (28) and digested exhaustively with DpnI. A modified version of a quantitative competitive PCR assay (32) was used to quantify the amounts of DpnI-resistant 1591 and 1381; the modified assay did not use radioactive primers. The PCR products of 1591, 1381, and a competitor were resolved in an agarose gel, visualized with ethidium bromide, and quantified with a charge-coupled device camera (IS-1000 digital imaging system; Alpha Innotech Corporation). The amounts of 1591 and 1381 were determined by identifying the amount of competitor DNA which yielded the same amount of product.
Transcription assays.
Cells were transfected with 0.1 to 0.64 μg of each EBNA1 expression plasmid (10% as much of each derivative as was used for Western blotting and replication assays), 100 ng of FR-TK-Luciferase, and 100 ng of pEQ176 (see above). Cells were grown for 48 h and harvested, and luciferase activity and β-galactosidase activity were measured (with kits from Bio-Rad and Tropix, respectively). The β-galactosidase activity was used to normalize the luciferase activity from each transfection.
DNA-linking assays.
293 cells were transfected with 1 to 6.4 μg of each EBNA1 expression plasmid, and whole-cell extracts were prepared. Cell pellets were resuspended at 108 cells/ml in a buffer consisting of 50 mM Tris (pH 7.5), 5 mM EDTA, 0.8 M NaCl, and 0.3% Nonidet P-40, sonicated, and incubated at room temperature for 15 min, and then an equal volume of 50 mM Tris (pH 7.5) was added. The extracts were microcentrifuged at 12,000 × g for 10 min at 4°C, and the supernatant was used. A similar extract was prepared from untransfected 293 cells. The total amount of extract in each reaction was kept constant. The probe DNA, which is 131 bp long and contains two binding sites for EBNA1, is the SalI-to-PstI fragment from p880 (45). This fragment was purified from an agarose gel, quantified, and labeled by filling in the ends with the Klenow fragment in the presence of [α-32P]dCTP (Amersham). Extracts, 4 fmol of probe, and 1.35 μg of poly(dl) · poly(dC) (Pharmacia) were combined in reaction mixtures with a final NaCl concentration of 0.3 M and incubated at room temperature for 30 min. Reactions were resolved by electrophoresis at 15 to 25 mA through 4% polyacrylamide gels in 0.5× TBE (45 mM Tris-borate, 1 mM EDTA). Gels were dried under vacuum at 80°C onto 3MM chromatography paper (Whatman), and a PhosphorImager was used to visualize and quantify the data.
RESULTS
Structures of derivatives of EBNA1.
We wished to determine whether DNA linking contributes to EBNA1 activities in cells. Because the linking regions are large and contain multiple domains which can independently mediate linking (39), subtle mutations which substantially affect linking are unlikely to be identified readily. Therefore, we constructed derivatives of EBNA1 with gross deletions or rearrangements of the linking regions (Fig. 1A). Derivatives of EBNA1 in one set each contain two linking regions. These derivatives contain either linking region 1 and linking region 2 in various contexts (1553, 1743, and 1745) or substitutions of linking region 1 for linking region 2 (1893) and of linking region 2 for linking region 1 (1767). Derivatives in a second set contain one linking region in various contexts (1708, 1710, 1763, 1891, and 1747). A final set consists of derivatives containing neither linking region (1709 and 1160). All of the derivatives contain the nuclear localization sequence within amino acids 379 to 391 (5). Each derivative also contains amino acids 451 to 641, which include the necessary residues for dimerization and site-specific DNA binding (5, 10, 12, 29). Several derivatives contain a truncated, 15-amino-acid version of the Gly-Gly-Ala repeats. Aside from the truncated Gly-Gly-Ala repeats, the 1553 derivative is identical in sequence to EBNA1 from the B95-8 strain of EBV (8). The 1553 derivative and EBNA1 from B95-8 indistinguishably support replication and transcription in tissue cultures (69); therefore, the 1553 derivative is considered wild type in this study.
Each derivative of EBNA1 was detectably expressed following its transfection into 293 cells (Fig. 1B). The amounts of DNA transfected were chosen to minimize both the range of expression of derivatives and the range in levels of DNA needed to provide this expression. From 1 to 6.4 μg of each DNA was transfected; these amounts provided levels of expression of each derivative within 10-fold of one another. The variation in both the amount of DNA transfected and the level of the derivatives expressed is independent of their activities (compare Fig. 1B, 2, and 3). The specific amount of each plasmid used in this calibration experiment was identical to the amounts used in the replication assays, and 1/10 as much of each plasmid was used in the transcription assays.
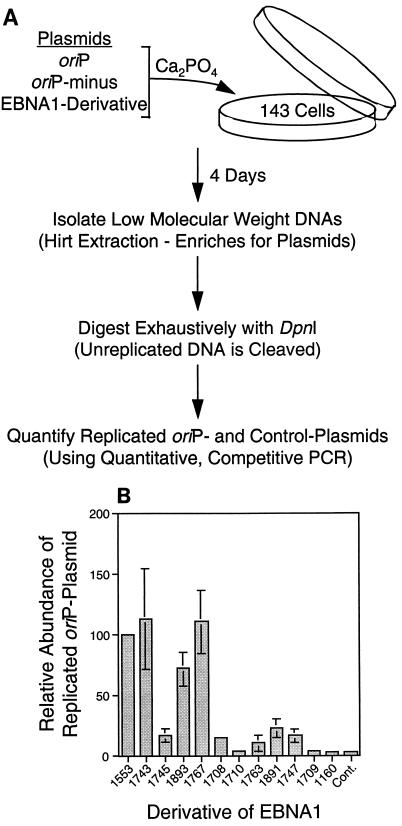
FIG. 2.
Derivatives of EBNA1 support the replication of oriP plasmids to different degrees. (A) Plasmids expressing derivatives of EBNA1 were introduced as calcium phosphate precipitates into 143B cells along with a replication reporter plasmid (containing oriP) and a replication control plasmid (lacking oriP). After 4 days, plasmid DNA was isolated by Hirt extraction, linearized with HindIII, and digested with DpnI, which cleaves input DNAs that have not undergone multiple rounds of DNA synthesis. A quantitative competitive PCR assay was used to measure the amounts of replicated plasmids containing and lacking oriP. (B) Average amount of replicated oriP plasmid measured in transfections with derivatives of EBNA1. The amount of replicated oriP plasmid in cells expressing 1553 was set to 100% for each experiment. The control plasmid (pCDNA3; Cont.) has only the CMV IE promoter and expresses no EBNA1. The amount of replicated plasmid lacking oriP, measured simultaneously for all points, was smaller than the amount of replicated plasmid containing oriP from cells transfected with pCDNA3 (data not shown). Shown are the averages and standard errors from four experiments (six data points for 1743 and 1893, five data points for 1767, and three data points for all other derivatives).
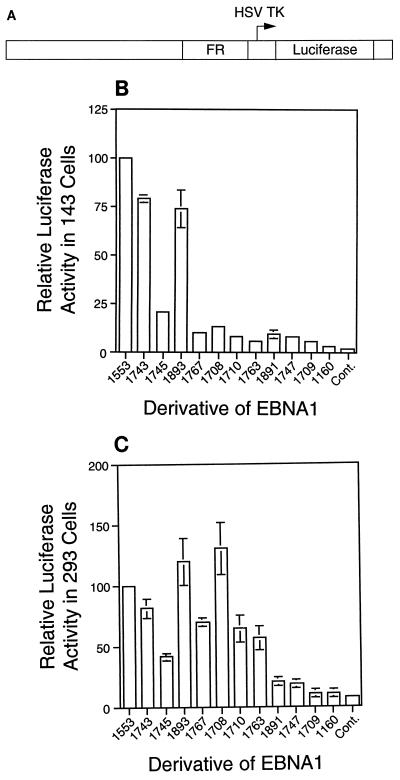
FIG. 3.
Derivatives of EBNA1 support transcription through the FR to different degrees. Plasmids expressing derivatives of EBNA1 or pCDNA3 (Cont.) and FR-TK-Luciferase (A) were introduced as calcium phosphate precipitates into 143B (B) and 293 (C) cells, and luciferase activity was measured 48 h later. A plasmid expressing β-galactosidase (pEQ176) was included with each transfection, and β-galactosidase activity was used to normalize each transfection. In each experiment, the normalized luciferase activity measured in cells expressing 1553 was set to 100%. Shown in panels B and C are the averages and standard errors for each derivative from four separate experiments in each cell type. HSV TK, herpes simplex virus thymidine kinase.
Ability of derivatives of EBNA1 to support replication.
We determined the ability of derivatives of EBNA1 to support replication in a short-term assay (Fig. 2A). Expression plasmids for each derivative of EBNA1 were cotransfected with a replication reporter plasmid (containing oriP) and a replication control plasmid (lacking oriP) into 143B cells. After 96 h, the cells were harvested and low-molecular-weight DNAs were isolated by Hirt extraction (28). The transfected DNA was prepared from dam+ Escherichia coli and thus was sensitive to cleavage by DpnI. DpnI cleaves DNA that is fully methylated or hemimethylated by dam methylase (3); therefore, only plasmids which have undergone two rounds of synthesis will yield DNA resistant to digestion with DpnI. A quantitative competitive PCR assay (32) was used to measure the extent of replication of the plasmids containing and lacking oriP. This assay has been validated previously by use of Southern blotting with related oriP plasmids in the same cells (32).
The ability of each derivative of EBNA1 to support the replication of an oriP plasmid in this short-term assay was measured in four independent experiments (Fig. 2B). Within each experiment, the amount of replicated oriP plasmid in cells expressing 1553 was set to 100%. In general, derivatives with two linking regions supported replication in a manner similar to that of 1553, derivatives with one linking region supported a reduced level of replication, and derivatives with neither linking region did not support replication above the background level observed with pCDNA3 (control plasmid expressing no EBNA1). Two exceptions to this generalization are 1745 and 1710, which are similar to 1743 and 1708, respectively, but lack amino acids 392 to 450 and are impaired in their support of replication. Thus, to support oriP-dependent replication fully, EBNA1 requires two linking regions and amino acids 392 to 450. In a parallel experiment, derivatives of EBNA1 supported the replication of oriP similarly in 293 cells and in 143 cells. Derivatives 1893 and 1767, each with two linking regions, had activities similar to that of 1553; derivatives 1891 and 1708, with only one linking region, had reduced activities; and derivative 1160, lacking linking regions, had activity close to background activity (data not shown).
Linking region 1 and linking region 2 are equivalent in their contributions to the support of replication by derivatives of EBNA1. Derivatives with either linking region alone, excluding 1710, which lacks amino acids 392 to 450, supported replication similarly (10 to 23% as efficiently as 1553). Furthermore, derivatives in which each linking region was substituted for the other (1893 and 1767) supported replication as well as did 1553. That linking region 1 and linking region 2 contribute equally to the support of replication by EBNA1 indicates that linking per se or some other activity shared between them contributes to the support of DNA replication by EBNA1.
Ability of derivatives of EBNA1 to support transcription.
Expression plasmids for each derivative of EBNA1 were cotransfected with FR-TK-Luciferase (Fig. 3A) into 143B (Fig. 3B) or 293 (Fig. 3C) cells. Cells were harvested 48 h after transfection, and luciferase activity was measured. Within each experiment, the luciferase activity in cells expressing 1553 was set to 100%. The average signals with the control plasmid, pCDNA3, were 1.3% in 143B cells and 7.9% in 293 cells.
The linking regions are not equivalent in their support of transcription. In 143B cells (Fig. 3B), the derivative containing two copies of linking region 2 (1767), which fully supported oriP-dependent replication, was significantly impaired in its support of transcription. Otherwise, the ability of the derivatives to support transcription mirrored their ability to support replication. Derivatives which contained only one linking region (1708, 1710, 1763, 1891, and 1747) were much less active than derivatives with both linking regions (1553 and 1743) or two copies of linking region 1 (1893) but were more active than derivatives with neither linking region. Application of the Wilcoxon rank sum test indicated that, in 143B cells, each derivative with one linking region supported transcription significantly better than 1709 and 1166 (P, <0.05), except for 1763 (P, 0.09). The ability of derivatives of EBNA1 to support transcription in 293 cells (Fig. 3C) differed from that in 143B cells. In 293 cells, derivatives containing one copy of linking region 2 (1708, 1710, and 1763) supported transcription in a manner comparable to that of 1553, and duplication of linking region 2 (1767) did not augment this activity. In 293 cells, as in 143B cells, derivatives containing one copy of linking region 1 (1891 and 1747) supported transcription slightly, although significantly better than 1709 and 1160 (P, <0.03), and the derivative with a duplication of linking region 1 (1893) supported transcription fully. The contribution of linking region 1 but not that of linking region 2 to the support of transcription by EBNA1 can be augmented by duplicating that region within EBNA1.
The deletion of amino acids 392 to 450 diminished the ability of EBNA1 to support transcription. In each cell type, 1745 and 1710 were less active than 1743 and 1708, respectively. However, this deletion decreased the support of transcription less markedly than it affected the support of replication.
Ability of derivatives of EBNA1 to link DNA.
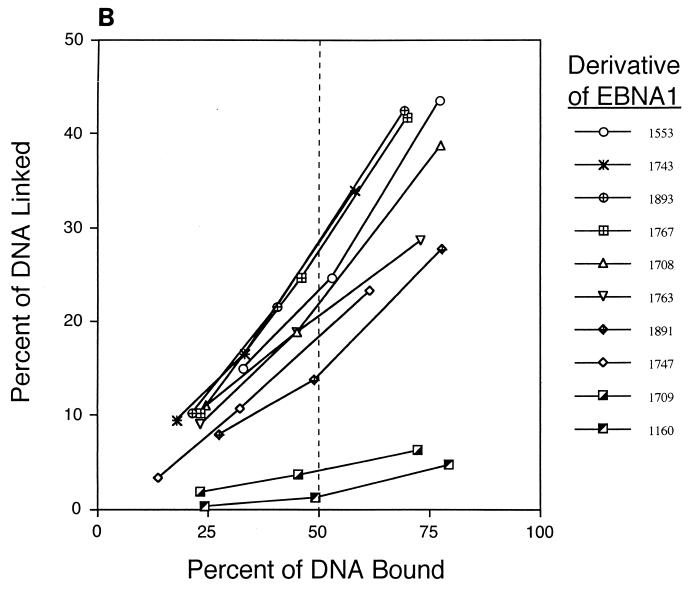
The ability of derivatives of EBNA1 to link DNA was determined with an electrophoretic mobility shift assay (39). In this assay, DNAs linked by EBNA1 are incorporated into large complexes which are unable to migrate appreciably into the gel. This assay has been validated previously by electron microscopic analyses of intramolecular linking (19, 59) and by enhancement of ligation of DNAs mediated by their linkage (39). A representative experiment for a subset of the derivatives is shown in Fig. 4A. Various amounts of whole-cell extract expressing derivatives of EBNA1 were incubated with a DNA containing two binding sites for EBNA1 in the presence of a constant, total amount of cell extract. The amounts of EBNA1-expressing extract were adjusted so that the percentage of bound DNA spanned 50% for each derivative. For each derivative tested, the percentage of DNA linked was plotted against the percentage bound (Fig. 4B). These plots were used to interpolate the percentage of DNA linked by each derivative when 50% of the DNA was bound, and this value was defined as the linking activity of that derivative.
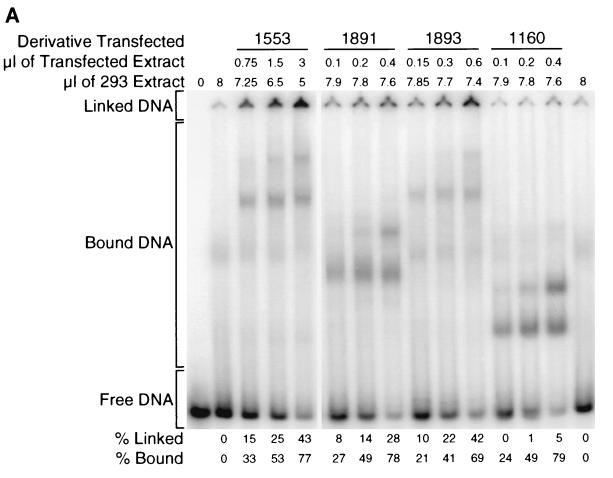
FIG. 4.
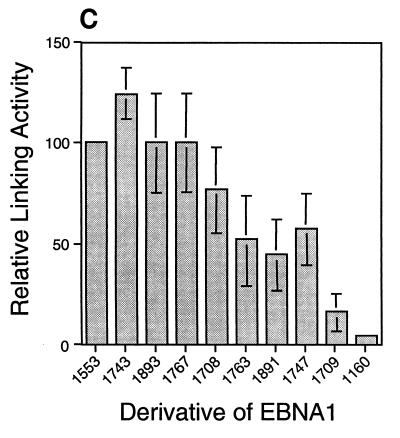
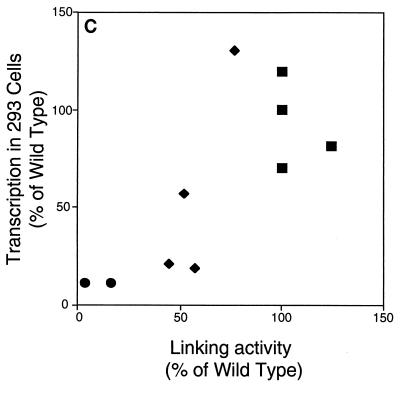
Derivatives of EBNA1 differ in linking activity measured in a gel shift assay. (A) Extracts of 293 cells expressing various derivatives of EBNA1 were incubated with a radiolabeled, 131-bp DNA containing two EBNA1-binding sites. When a derivative of EBNA1 only binds to this DNA (as with 1160), its mobility is reduced, but the protein-DNA complex still enters the gel. When a derivative of EBNA1 binds to and links this DNA (as with 1553), the linked complex does not migrate appreciably into the gel. The amounts of extracts expressing the derivatives of EBNA1 were varied so that the range of bound DNA spanned 50%. The total amount of 293 cell extract derived from transfected plus untransfected cells in each reaction was constant (8 μl). PhosphorImager analysis was used to quantify the amounts of DNA in the linked, bound, and free regions of each lane. The values for linked and bound DNAs in the lanes containing only 293 cell extract were set to zero. The percentage of linked DNA was determined by dividing the amount of linked DNA by the amount of total DNA in the lane. The percentage of bound DNA was determined by dividing the amounts of linked and bound DNAs by the amount of total DNA in the lane. (B) Graphic representation of all data from the experiment shown partially in panel A. The percentage of linked DNA versus that bound was plotted for the derivatives. This plot was used to determine the percentage of DNA linked when 50% of the DNA was bound. This value was defined as the linking activity for the derivative. (C) Graphic representation of the relative linking activities of derivatives of EBNA1. In each experiment, the linking activity of 1553 was set to 100%. Shown are the averages and standard errors from three separate experiments like the one shown in panel B.
The linking activity of derivatives of EBNA1 is a function of the number of linking regions that they contain. The linking assay was conducted three times (Fig. 4C), and in each experiment the linking activity of 1553 was set to 100%. In all three experiments, the derivatives with two linking regions (1553, 1743, 1893, and 1767) had the highest linking activities, the derivatives with one linking region (1708, 1763, 1891, and 1747) had intermediate linking activities, and the derivatives with neither linking region (1709 and 1160) had the lowest linking activities. Derivatives of EBNA1 with one linking region link DNAs more efficiently than those with neither linking region but less efficiently than those with two linking regions.
DNA linking correlates with biological activities.
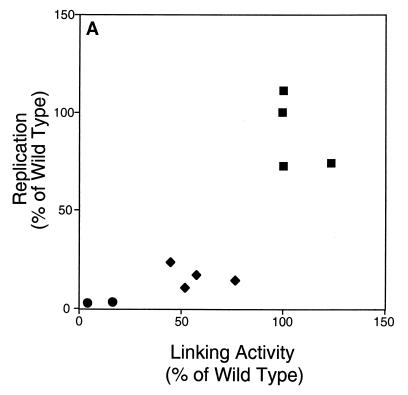
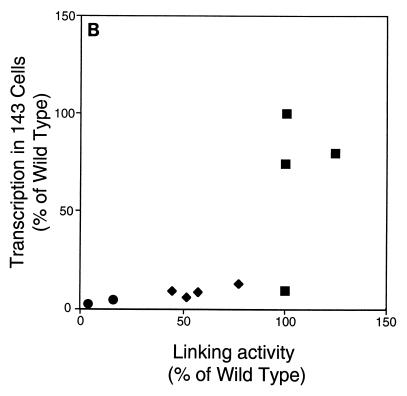
The linking activity of derivatives of EBNA1 measured in Fig. 4 was compared to their activities measured in cells in Fig. 2 and 3. Because amino acids 392 to 450 do not contribute to DNA linking (39) but do affect the support of replication (Fig. 2) and transcription (Fig. 3) by EBNA1, derivatives lacking them (1710 and 1745) are not included in these comparisons (Fig. 5). For the derivatives of EBNA1 tested in Fig. 4, their average linking activity was plotted against their support of oriP-dependent replication (Fig. 5A) and transcription in 143B (Fig. 5B) and 293 (Fig. 5C) cells. The application of the Kendall rank correlation test indicated that linking activity correlates with the support of replication (P, 0.003) and transcription in 143B (P, 0.003) and 293 (P, 0.02) cells. These correlations are consistent with DNA linking contributing to the support of replication and transcription by EBNA1.
FIG. 5.
Linking activity and support of replication and transcription correlate for derivatives of EBNA1. The linking activity of derivatives of EBNA1 is plotted against the ability of those derivatives to support replication (A) or transcription in 143B (B) and 293 (C) cells. Squares represent derivatives of EBNA1 with two linking regions, diamonds represent those with one linking region, and circles represent those with neither linking region. Application of the Kendall rank correlation test indicates that for this set of derivatives, linking activity correlates with the activation of replication (P, 0.003) and with the activation of transcription in 143B (P, 0.003) and 293 (P, 0.02) cells.
DISCUSSION
The ability of derivatives of EBNA1 to link DNA is a function of the number of linking regions that they contain (Fig. 4), as is their ability to support the replication of oriP plasmids (Fig. 2). The support of replication correlates strongly with DNA linking (Fig. 5A). The ability of derivatives of EBNA1 to support transcription in two cell types (Fig. 3) also correlates with DNA linking (Fig. 5B and C). The correlation of DNA linking by EBNA1 with its support of replication and transcription is consistent with linking contributing to its support of these activities in cells. Amino acids 392 to 450 of EBNA1 contribute to the support of replication and transcription (Fig. 2 and 3) but do not contribute to DNA linking by EBNA1 in vitro (39). Derivatives with a deletion of these residues, which may function in vivo as a structural element, are therefore excluded from comparisons of these activities.
Linking region 1 and linking region 2 contribute equally to the support of oriP-dependent replication (Fig. 2). Derivatives of EBNA1 which contain one copy of either linking region support similar levels of oriP-dependent replication (compare 1708, 1763, 1891, and 1747). Derivatives which contain two linking regions also support similar levels of replication (compare 1553, 1743, 1893, and 1767). The linking regions contribute combinatorially to the support of replication; derivatives with two linking regions are more active than those with one linking region. The linking regions are also functionally redundant. Derivatives in which one linking region is replaced by the other (1893 and 1767) support replication as well as derivatives containing both linking regions (1553 and 1743). Therefore, the contributions of the linking regions to the support of DNA replication by EBNA1 are equal, combinatorially active, and functionally redundant. The contributions of the linking regions to DNA linking mediated by EBNA1 are also equal, combinatorially active, and functionally redundant (Fig. 4), indicating that linking underlies the support of replication by EBNA1. However, the linking regions may share an activity other than linking which contributes to the support of replication by EBNA1. Aside from being rich in glycines and arginines, the linking regions show no significant sequence identity or similarity. EBNA1 has been shown to bind RNA nonspecifically, and three RGG motifs within EBNA1 are thought to contribute to this binding (55). Linking region 2 contains two of these RGG motifs, but linking region 1 contains only part of the third motif. The derivative of EBNA1 with two copies of linking region 1 (1893) has no intact RGG motifs and supports replication and transcription as well as wild-type EBNA1. Therefore, we find it most likely that linking per se underlies the support of replication by EBNA1.
The linking regions are not equivalent in their support of transcription. The activities of these regions, relative to one another, differ in 293 and 143B cells. In 293 cells, derivatives of EBNA1 containing only linking region 2 support transcription better than those with only linking region 1 (Fig. 3C, compare 1708, 1710, and 1763 with 1891 and 1747). In 143B cells, these derivatives all support similar, low levels of transcription (Fig. 3B). Linking region 1, but not linking region 2, is combinatorially active. A derivative of EBNA1 containing two copies of linking region 1 supports transcription better than derivatives with one copy of linking region 1 (Fig. 3, compare 1893 with 1891 and 1747). However, a derivative of EBNA1 with two copies of linking region 2 does not support transcription better than derivatives with one copy of linking region 2 (Fig. 3, compare 1767 with 1708, 1710, and 1763). The simplest interpretation of these results is that the contributions of the linking regions to the support of transcription by EBNA1 differ. The interaction of linking region 1 and linking region 2 with distinct cellular factors (discussed below) could account for this difference.
The linking activities of derivatives of EBNA1 with two linking regions, as assayed in vitro, do not differ dramatically from those of derivatives with only one linking region; however, differences in the support of replication and transcription by the two sets of derivatives in vivo are pronounced. In this study, we measured in vitro linking of DNAs containing only two binding sites for EBNA1. In cells, EBNA1 supports both replication and transcription through the FR, a cluster of 20 binding sites. We suggest that the differences observed in the assays in vitro are magnified in vivo as a result of the high concentration of EBNA1 bound to the FR.
The abilities of EBNA1 to support replication and transcription previously have not been genetically separated (46, 67). The derivative of EBNA1 with two copies of linking region 2 (1767) supports replication and transcription unequally. In 143B cells, 1767 supports replication slightly better than 1553 (Fig. 2B) but supports transcription less than 10% as well as 1553 (Fig. 3B). The levels of expression of 1553 and 1767 are unlikely to account for this difference because they are nearly identical (Fig. 1B). Therefore, a derivative of EBNA1 in which linking region 1 is replaced with linking region 2 differentially supports replication and transcription in a particular cell type.
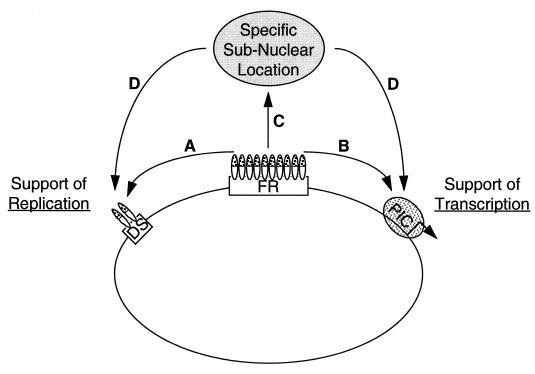
The linking regions of EBNA1 may mediate several types of molecular interactions. The linking regions of EBNA1 interact homotypically to mediate the linking of bound DNAs (40). The assays used previously and in this work measure this type of homotypic association of the linking regions. The linking regions of EBNA1 may also interact heterotypically with similar linking domains of other proteins. For example, the E2 protein of bovine papillomavirus and the cellular transcription factor Sp1 each homotypically link DNAs to which they bind (34, 58). E2 and Sp1 also heterotypically link to one another (37). EBNA1 could link heterotypically to proteins that bind to DNA directly, such as Sp1; to proteins that bind to DNA indirectly, such as components of the basal transcriptional machinery; or to proteins that are not associated with DNA, such as nuclear matrix components. Homotypic and heterotypic interactions of the linking regions of EBNA1 each may contribute to its support of replication and transcription (Fig. 6).
FIG. 6.
Model of modes by which the linking regions may contribute to the support of replication and transcription by EBNA1. The large circle represents a plasmid containing oriP and a promoter (arrow). The FR and the DS of oriP are bound by EBNA1 (small ovals), with the DNA-binding domain in white and the linking domains mottled. (A) EBNA1-mediated linking of the FR and the DS is likely to contribute to sustained replication of oriP. (B) The linking domains of EBNA1 may interact with basal transcription factors within the PIC, possibly stabilizing PIC binding at the promoter. (C) The linking domains of EBNA1 may interact with specific nuclear factors, targeting EBNA1 and DNAs bound by it to a specific subnuclear location. Linking may also occur between EBNA1 molecules bound to a plasmid and to cellular DNA, thereby localizing the plasmid to a particular chromatin domain. (D) Once localized to a specific site within the nucleus, origins of replication or promoters may more efficiently interact with cellular trans-acting factors.
How might homotypic linking between the FR and the DS contribute to the support of replication by EBNA1? Cellular factors alone can mediate the synthesis of plasmids containing the DS (3); however, EBNA1 must bind to sites in the DS to support the stable replication of oriP (24). EBNA1 binds to sites in the DS with a low affinity (relative to sites in the FR) (6, 30), perhaps because it needs to be displaced for the initiation of DNA synthesis to occur at the DS. We propose that the binding of EBNA1 to the DS prevents the formation of a chromatin structure which would preclude recognition of the DS by cellular replication factors. The placement of histones affects the efficiency with which DNA synthesis initiates at an ARS (53), and modulation of chromatin structure has been proposed as a general contribution of transcription factors at origins of DNA synthesis (62). Linking between the FR and the DS increases the apparent affinity of EBNA1 for sites in the DS (18, 59) and thus may facilitate its rebinding to the DS after the replication complex vacates it (Fig. 6A). This model parallels that for the cellular origin of DNA synthesis at the β-globin locus. It is thought that the locus control region (LCR) of the β-globin locus supports replication at the origin located 50 kbp away via its effect on chromatin structure throughout the locus (4, 17).
The linking regions of EBNA1 may directly activate transcription via heterotypic interactions with cellular factors (Fig. 6B). EBNA1 bound to oriP could interact with a component of a PIC via an interaction of the linking regions of EBNA1 with similar motifs present in other components of the PIC. Interactions of linking region 1 and linking region 2 with distinct components of the PIC may explain the nonequivalence of these regions in their support of transcription. EBNA1 can activate an EBV promoter located 10 kbp away from oriP (22). The linking of EBNA1 to a PIC at this promoter would juxtapose an activation domain with the promoter and could explain how the activation of such a distal element is mediated. Multiple examples are consistent with this suggestion. The activation domain of E2, which mediates DNA linking (34), also interacts with a component of the PIC (9, 65). Specific interactions (or links) between the activation domains of Sp1 and NTF-1 bound at an enhancer and specific components of the PIC underlie the activation of transcription in vitro (11). Linking between the β-globin LCR and distinct β-globin promoters likely accounts for the competition observed in vivo between promoters for the LCR (15).
The subnuclear localization of DNA is an alternative, nonexclusive mechanism by which EBNA1 may support both replication and transcription. EBNA1 may target DNAs bound to it to discrete sites within the nucleus (Fig. 6C). Homotypic linking between EBNA1 bound to cellular DNA and EBNA1 bound to an oriP plasmid would target the plasmid to a specific chromatin domain. Alternatively, EBNA1 could target an oriP plasmid to a specific site in the nucleus via heterotypic linking between EBNA1 and a nuclear protein. The linking domains of EBNA1 are sufficient to target green fluorescent protein to metaphase chromosomes (43). Subnuclear localization may facilitate stable replication of and transcription from oriP plasmids (Fig. 6D). Similarly, the linking of other viral genomes to specific subnuclear sites may facilitate their replication or transcription. For example, the binding of Sp1 to the SV40 core origin is not required for the replication of SV40 DNA in vitro (13) but does contribute to SV40 replication in vivo (26). In this model, Sp1 bound to the 21-bp repeats may link SV40 DNA to a nuclear site where its core origin is efficiently replicated. Also, the UL9 protein of herpes simplex virus type 1 (HSV-1) links DNA (35, 41) and is required for in vivo but not in vitro replication of HSV-1 (25, 54, 64). By linking to cellular components, UL9 may localize HSV-1 DNA to replication factories where it is synthesized (42). Cellular origins of DNA synthesis may similarly be localized to nuclear replication factories by linking of cellular replication factors to specific nuclear components.
The linking activity of EBNA1 is essential for its ability to support replication and transcription from distant binding sites. This activity provides a mechanism by which EBNA1 and other proteins act from distant cis-acting elements to regulate their targets.
ACKNOWLEDGMENTS
We thank Ashok Aiyar, Paul Lambert, Elizabeth Leight, and Dan Loeb for comments on the manuscript, Eric Seversen for excellent technical support, and Norman Drinkwater for assistance in statistical analysis.
This work was supported by grants from the NIH (CA 22443, CA 09075, and CA 07175).
REFERENCES
- 1.Adams A. Replication of latent Epstein-Barr virus genomes in Raji cells. J Virol. 1987;61:1743–1746. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar A, Sugden B. Fusions between EBNA-1 of EBV and the large T-antigen of SV40 replicate their cognate origins. J Biol Chem. 1998;273:33073–33081. doi: 10.1074/jbc.273.49.33073. [DOI] [PubMed] [Google Scholar]
- 3.Aiyar A, Tyree C, Sugden B. The Plasmid replicon of EBV consists of multiple elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aladjem M I, Groudine M, Brody L L, Dieken E S, Fournier R E, Wahl G M, Epner E M. Participation of the human beta-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- 5.Ambinder R F, Mullen M, Chang Y-N, Hayward G S, Hayward S D. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J Virol. 1991;65:1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambinder R F, Shah W A, Rawlins D R, Hayward G S, Hayward S D. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J Virol. 1990;64:2369–2379. doi: 10.1128/jvi.64.5.2369-2379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bacchetti S, Graham F L. Transfer of the gene for thymidine kinase to thymidine kinase-deficient human cells by purified herpes simplex viral DNA. Proc Natl Acad Sci USA. 1977;74:1590–1594. doi: 10.1073/pnas.74.4.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Séguin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 9.Benson J D, Lawande R, Howley P M. Conserved interaction of the papillomavirus E2 transcriptional activator proteins with human and yeast TFIIB proteins. J Virol. 1997;71:8041–8047. doi: 10.1128/jvi.71.10.8041-8047.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bochkarev A, Barwell J A, Pfuetzner R A, Bochkareva E, Frappier L, Edwards A M. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell. 1996;84:791–800. doi: 10.1016/s0092-8674(00)81056-9. [DOI] [PubMed] [Google Scholar]
- 11.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 12.Chen M-R, Middeldorp J M, Hayward S D. Separation of the complex DNA binding domain of EBNA-1 into DNA recognition and dimerization subdomains of novel structure. J Virol. 1993;67:4875–4885. doi: 10.1128/jvi.67.8.4875-4885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean F B, Borowiec J A, Ishimi Y, Deb S, Tegtmeyer P, Hurwitz J. Simian virus 40 large tumor antigen requires three core replication origin domains for DNA unwinding and replication in vitro. Proc Natl Acad Sci USA. 1987;84:8267–8271. doi: 10.1073/pnas.84.23.8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DePamphilis M L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- 15.Dillon N, Trimborn T, Strouboulis J, Fraser P, Grosveld F. The effect of distance on long-range chromatin interactions. Mol Cell. 1997;1:131–139. doi: 10.1016/s1097-2765(00)80014-3. [DOI] [PubMed] [Google Scholar]
- 16.Fischer N, Kremmer E, Lautscham G, Mueller L N, Grasser F A. Epstein-Barr virus nuclear antigen 1 forms a complex with the nuclear transporter karyopherin alpha2. J Biol Chem. 1997;272:3999–4005. doi: 10.1074/jbc.272.7.3999. [DOI] [PubMed] [Google Scholar]
- 17.Forrester W C, Epner E, Driscoll M C, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes Dev. 1990;1:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 18.Frappier L, Goldsmith K, Bendell L. Stabilization of the EBNA1 protein on the Epstein-Barr virus latent origin of DNA replication by a DNA looping mechanism. J Biol Chem. 1994;269:1057–1062. [PubMed] [Google Scholar]
- 19.Frappier L, O’Donnell M. Epstein-Barr nuclear antigen 1 mediates a DNA loop within the latent replication origin of Epstein-Barr virus. Proc Natl Acad Sci USA. 1991;88:10875–10879. doi: 10.1073/pnas.88.23.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frappier L, O’Donnell M. Overproduction, purification, and characterization of EBNA1, the origin binding protein of Epstein-Barr virus. J Biol Chem. 1991;266:7819–7826. [PubMed] [Google Scholar]
- 21.Gahn T A, Schildkraut C L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 22.Gahn T A, Sugden B. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J Virol. 1995;69:2633–2636. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 24.Harrison S, Fisenne K, Hearing J. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1994;68:1913–1925. doi: 10.1128/jvi.68.3.1913-1925.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez T R, Dutch R E, Lehman I R, Gustafsson C, Elias P. Mutations in a herpes simplex virus type 1 origin that inhibit interaction with origin-binding protein also inhibit DNA replication. J Virol. 1991;65:1649–1652. doi: 10.1128/jvi.65.3.1649-1652.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hertz G Z, Mertz J E. Bidirectional promoter elements of simian virus 40 are required for efficient replication of the viral DNA. Mol Cell Biol. 1986;6:3513–3522. doi: 10.1128/mcb.6.10.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hieter P, Mann C, Snyder M, Davis R W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- 28.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 29.Inoue N, Harada S, Honma T, Kitamura T, Yanagi K. The domain of Epstein-Barr virus nuclear antigen 1 essential for binding to oriP region has a sequence fitted for the hypothetical basic-helix-loop-helix structure. Virology. 1991;182:84–93. doi: 10.1016/0042-6822(91)90651-q. [DOI] [PubMed] [Google Scholar]
- 30.Jones C H, Hayward D, Rawlins D R. Interaction of the lymphocyte-derived Epstein-Barr virus nuclear antigen EBNA-1 with its DNA-binding sites. J Virol. 1989;63:101–110. doi: 10.1128/jvi.63.1.101-110.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirchmaier A L, Sugden B. Plasmid maintenance of derivatives of oriP of Epstein-Barr virus. J Virol. 1995;69:1280–1283. doi: 10.1128/jvi.69.2.1280-1283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirchmaier A L, Sugden B. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J Virol. 1997;71:1766–1775. doi: 10.1128/jvi.71.3.1766-1775.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirchmaier A L, Sugden B. Rep*: a viral element that can partially replace the origin of plasmid DNA synthesis of Epstein-Barr virus. J Virol. 1997;72:4657–4666. doi: 10.1128/jvi.72.6.4657-4666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knight J D, Li R, Botchan M. The activation domain of the bovine papillomavirus E2 protein mediates association of DNA-bound dimers to form DNA loops. Proc Natl Acad Sci USA. 1991;88:3204–3208. doi: 10.1073/pnas.88.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koff A, Schwedes J F, Tegtmeyer P. Herpes simplex virus origin binding protein (UL9) loops and distorts the viral origin. J Virol. 1991;65:3284–3292. doi: 10.1128/jvi.65.6.3284-3292.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koshland D, Kent J C, Hartwell L H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985;40:393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- 37.Li R, Knight J D, Jackson S P, Tjian R, Botchan M R. Direct interaction between Sp1 and the BPV enhancer E2 protein mediates synergistic activation of transcription. Cell. 1991;65:493–505. doi: 10.1016/0092-8674(91)90467-d. [DOI] [PubMed] [Google Scholar]
- 38.Lupton S, Levine A J. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985;5:2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackey D, Middleton T, Sugden B. Multiple regions within EBNA1 can link DNAs. J Virol. 1995;69:6199–6208. doi: 10.1128/jvi.69.10.6199-6208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackey D, Sugden B. Studies on the mechanism of DNA linking by Epstein-Barr virus nuclear antigen 1. J Biol Chem. 1997;272:29873–29879. doi: 10.1074/jbc.272.47.29873. [DOI] [PubMed] [Google Scholar]
- 41.Makhov A M, Boehmer P E, Lehman I R, Griffith J D. The herpes simplex virus type 1 origin-binding protein carries out origin specific DNA unwinding and forms stem-loop structures. EMBO J. 1996;15:1742–1750. [PMC free article] [PubMed] [Google Scholar]
- 42.Malik A K, Shao L, Shanley J D, Weller S K. Intracellular localization of the herpes simplex virus type-1 origin binding protein, UL9. Virology. 1996;224:380–389. doi: 10.1006/viro.1996.0545. [DOI] [PubMed] [Google Scholar]
- 43.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J.-C. Nicholas. 1998. Personal communication. [DOI] [PMC free article] [PubMed]
- 44.Middleton T, Sugden B. EBNA1 can link the enhancer element to the initiator element of the Epstein-Barr virus plasmid origin of DNA replication. J Virol. 1992;66:489–495. doi: 10.1128/jvi.66.1.489-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Middleton T, Sugden B. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J Virol. 1994;68:4067–4071. doi: 10.1128/jvi.68.6.4067-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polvino-Bodnar M, Schaffer P A. DNA binding activity is required for EBNA 1-dependent transcriptional activation and DNA replication. Virology. 1992;187:591–603. doi: 10.1016/0042-6822(92)90461-w. [DOI] [PubMed] [Google Scholar]
- 47.Puglielli M T, Desai N, Speck S H. Regulation of EBNA gene transcription in lymphoblastoid cell lines: characterization of sequences downstream of BCR2 (Cp) J Virol. 1997;71:120–128. doi: 10.1128/jvi.71.1.120-128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 49.Reisman D, Sugden B. trans-Activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 52.Schleiss M R, Degnin C R, Geballe A P. Translational control of human cytomegalovirus gp48 expression. J Virol. 1991;65:6782–6789. doi: 10.1128/jvi.65.12.6782-6789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simpson R T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 54.Skaliter R, Lehman I R. Rolling circle DNA replication in vitro by a complex of herpes simplex virus type 1-encoded enzymes. Proc Natl Acad Sci USA. 1994;91:10665–10669. doi: 10.1073/pnas.91.22.10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snudden D K, Hearing J, Smith P R, Grasser F A, Griffin B E. EBNA-1, the major nuclear antigen of Epstein-Barr virus, resembles ‘RGG’ RNA binding proteins. EMBO J. 1994;13:4840–4847. doi: 10.1002/j.1460-2075.1994.tb06810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Southern P J, Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1:327–348. [PubMed] [Google Scholar]
- 57.Sternås L, Middleton T, Sugden B. The average number of molecules of Epstein-Barr nuclear antigen 1 per cell does not correlate with the average number of Epstein-Barr virus (EBV) DNA molecules per cell among different clones of EBV-immortalized cells. J Virol. 1990;64:2407–2410. doi: 10.1128/jvi.64.5.2407-2410.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su W, Jackson S, Tjian R, Echols H. DNA looping between sites for transcriptional activation: self-association of DNA-bound Sp1. Genes Dev. 1991;5:820–826. doi: 10.1101/gad.5.5.820. [DOI] [PubMed] [Google Scholar]
- 59.Su W, Middleton T, Sugden B, Echols H. DNA looping between the origin of replication of Epstein-Barr virus and its enhancer site: stabilization of an origin complex with Epstein-Barr nuclear antigen 1. Proc Natl Acad Sci USA. 1991;88:10870–10874. doi: 10.1073/pnas.88.23.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugden B, Warren N. Plasmid origin of replication of Epstein-Barr virus, oriP, does not limit replication in cis. Mol Biol Med. 1988;5:84–94. [PubMed] [Google Scholar]
- 61.Sugden B, Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Vliet P C. Roles of transcription factors in DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. pp. 87–1182. [Google Scholar]
- 63.Wang Y, Finan J E, Middeldorp J M, Hayward S D. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology. 1997;236:18–29. doi: 10.1006/viro.1997.8739. [DOI] [PubMed] [Google Scholar]
- 64.Wu C A, Nelson N J, McGeoch D J, Challberg M D. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao J M, Breiding D E, Androphy E J. Functional interaction of the bovine papillomavirus E2 transactivation domain with TFIIB. J Virol. 1998;72:1013–1019. doi: 10.1128/jvi.72.2.1013-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yates J L, Camiolo S M. Dissection of DNA replication and enhancer activation functions of Epstein-Barr virus nuclear antigen 1. Cancer Cells. 1988;6:197–205. [Google Scholar]
- 68.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]