Significance
Viruses rely on organelle remodeling for their replication and for the spread of infection. Alterations to mitochondrial functions and cellular metabolism are hallmarks of nearly all viral infections. However, how the widely spread HCMV remodels mitochondrial structure and function has presented a conundrum. It remained unclear how HCMV increases mitochondrial bioenergetics despite triggering mitochondrial fragmentation. Using a multidisciplinary approach, we address this question. We establish that the uncharacterized viral protein, pUL13, targets the mitochondria, interacts with the MICOS complex, and is sufficient for increasing oxidative phosphorylation during infection. Our findings provide evidence that viral pathogens have acquired mechanisms for manipulating cristae architecture and ETC function to achieve increased energy production and can have broad implications in understanding virus-driven pathologies.
Keywords: HCMV, pUL13, mitochondria, metabolism, proteomics
Abstract
Viruses modulate mitochondrial processes during infection to increase biosynthetic precursors and energy output, fueling virus replication. In a surprising fashion, although it triggers mitochondrial fragmentation, the prevalent pathogen human cytomegalovirus (HCMV) increases mitochondrial metabolism through a yet-unknown mechanism. Here, we integrate molecular virology, metabolic assays, quantitative proteomics, and superresolution confocal microscopy to define this mechanism. We establish that the previously uncharacterized viral protein pUL13 is required for productive HCMV replication, targets the mitochondria, and functions to increase oxidative phosphorylation during infection. We demonstrate that pUL13 forms temporally tuned interactions with the mitochondrial contact site and cristae organizing system (MICOS) complex, a critical regulator of cristae architecture and electron transport chain (ETC) function. Stimulated emission depletion superresolution microscopy shows that expression of pUL13 alters cristae architecture. Indeed, using live-cell Seahorse assays, we establish that pUL13 alone is sufficient to increase cellular respiration, not requiring the presence of other viral proteins. Our findings address the outstanding question of how HCMV targets mitochondria to increase bioenergetic output and expands the knowledge of the intricate connection between mitochondrial architecture and ETC function.
Viruses are obligate intracellular parasites that rely on host cells for metabolite and energy production to fuel their replication cycles. As mitochondria are the central metabolic hubs of cells, viruses are known to specifically target the mitochondria during infection to modulate mitochondrial processes that support virus replication (1, 2). Given the relationship between mitochondrial morphology and function, metabolic changes during infection are frequently accompanied by the dysregulation of mitochondrial structure (3). Some of the most striking changes in mitochondrial metabolism and structure are observed after infection with the widely spread β-herpesvirus human cytomegalovirus (HCMV) (4–7).
HCMV is estimated to infect over half of the world’s population (8). Although infection is typically asymptomatic, HCMV is a major concern for immunocompromised individuals and the leading infectious cause of congenital birth defects (9, 10). Mitochondrial structural and metabolic changes occur early during the HCMV replication cycle, which is completed over 120 h and is characterized by a temporal cascade of immediate-early (IE), delayed early (DE), and late (L) viral gene expression (11). By 24 h postinfection (hpi) with HCMV, the mitochondrial network becomes progressively fragmented (4). Although fragmentation is typically associated with mitochondrial dysfunction (12), mitochondrial metabolic processes are induced during HCMV infection (5–7, 13, 14).
It is now understood that HCMV globally alters cellular metabolism during its replication cycle, increasing glycolysis, glutaminolysis, and the tricarboxylic acid (TCA) cycle (5–7). The mitochondria play a central role in these metabolic processes, connecting glycolysis and glutaminolysis to the TCA cycle and oxidative phosphorylation (OXPHOS). In particular, an essential function for mitochondrial electron transport chain (ETC) activity and OXPHOS during HCMV infection has been demonstrated (5, 7, 13–15). The transcription and protein levels of ETC and ATP synthase proteins increase during infection (5, 13, 14). Additionally, the HCMV ncRNAβ2.7 has been shown to stabilize mitochondrial membrane potential, which is essential for ATP production (15). In a recent study, when ETC activity was disrupted, HCMV replication became significantly impaired (14). However, despite the demonstrated importance of OXPHOS during HCMV infection, little is understood regarding the mechanism underlying HCMV regulation of mitochondrial bioenergetics.
We previously showed that the viral protein pUL13 has a dynamic temporal localization to the mitochondria during HCMV infection (16), but its function remained unknown. Prior to our analysis, only one study had reported findings concerning pUL13, showing that pUL13 is an IE protein expressed as early as 24 hpi at the transcript level (17). Here, we demonstrate that pUL13 is necessary for productive virus replication and functions to increase mitochondrial bioenergetics during infection. We establish that pUL13 predominantly localizes to mitochondria by 48 hpi, coinciding with increased abundance of OXPHOS proteins. Using immunoaffinity-purification mass spectrometry (IP-MS) analyses throughout the HCMV replication cycle, we characterize the temporal pUL13 interactome. We identify and validate the interaction of pUL13 with cristae remodeling proteins and the mitochondrial contact site and cristae organizing system (MICOS) complex, a critical regulator of mitochondrial ultrastructure and ETC function (18). Indeed, using superresolution microscopy, we demonstrate that pUL13 alters cristae architecture. Furthermore, metabolic assays establish that pUL13 is sufficient to increase OXPHOS during infection.
Our results mechanistically define how HCMV targets the mitochondria to enhance bioenergetic output during infection. Furthermore, we provide evidence that intracellular pathogens have evolved mechanisms for manipulating cristae architecture to achieve increased energy production. These findings are broadly relevant for understanding mitochondria biology, contributing to the emerging knowledge of a functional relationship between cristae shaping complexes and ETC activity.
Results
pUL13 Localizes to the Mitochondria and Is Necessary for Productive HCMV Replication.
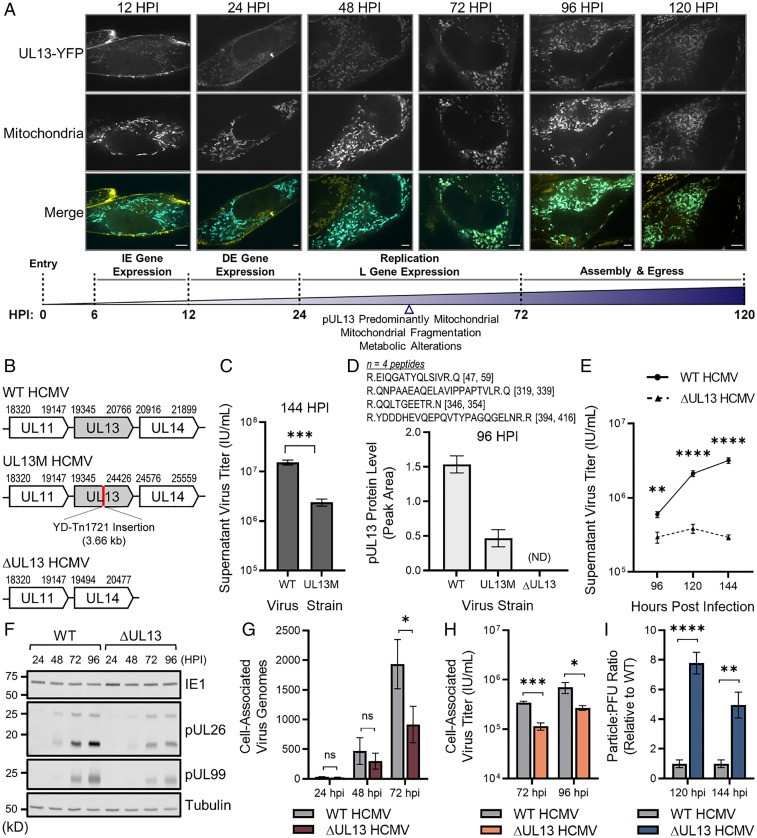
We previously demonstrated that the uncharacterized viral protein pUL13 has a temporal localization to the mitochondria during HCMV infection (16). To better define the temporality of pUL13 localization, we infected fibroblasts with a virus strain expressing pUL13 tagged with yellow fluorescent protein (YFP) and analyzed its localization using live-cell fluorescence microscopy at timepoints spanning the HCMV replication cycle (Fig. 1A). In agreement with our previous findings, pUL13 localized to the plasma membrane early in infection (12 hpi) (16). However, we also observed mitochondrial localization throughout HCMV replication, demonstrating that pUL13 localizes to the mitochondria earlier than anticipated. By 48 hpi, pUL13 gained a predominantly mitochondrial localization, which was retained for the remainder of the HCMV replication cycle. Mitochondrial morphological and metabolic changes have been reported to begin at 48 hpi with HCMV, raising the question of whether pUL13 is involved in modulating these processes during infection (4–7, 14).
Fig. 1.
pUL13 localizes to the mitochondria and is required for productive virus replication. (A) Confocal analysis of pUL13 localization in fibroblasts labeled with mito-BFP (mitochondria) and infected with UL13-YFP HCMV. (Scale bars, 5 μm.) Temporal stages of HCMV replication are shown. (B) Schematics of the UL13 genetic locus of WT, UL13M, and ∆UL13 HCMV. (C) Extracellular HCMV titers at 144 hpi during WT or UL13M infection. (D) pUL13 protein levels during WT, UL13M, and ∆UL13 infection at 96 hpi, measured by targeted MS. Peptide sequences and locations within the pUL13 sequence are shown. n = 4 peptides. (E) Extracellular HCMV titers during WT or ∆UL13 infection. (F) Representative Western blot showing IE (IE1), DE (pUL26), and L (pUL99) virus protein levels during WT or ∆UL13 infection. (G) Cell-associated virus genomes produced during ∆UL13 and WT infection, quantified using qPCR. (H) Cell-associated virus titers collected during WT or ∆UL13 infection. (I) Particle:PFU ratio of virus produced during WT or ∆UL13 infection. n = 3 biological replicates for C, E, G, H, and I. Significance determined by Student’s t test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001; ns, P > 0.05. Error bars indicate the SD.
To investigate the requirement of pUL13 for efficient HCMV replication, we used a mutant HCMV strain (UL13M) with disrupted pUL13 expression via a transposon insertion in the UL13 locus (Fig. 1B) (19). By measuring supernatant viral titer collected from fibroblasts infected with either wild-type HCMV (WT, AD169) or UL13M, we observed that UL13M resulted in a significant, 6.4-fold reduction in viral titer relative to WT (Fig. 1C). Given the potential of the transposon-mutagenized pUL13 to retain partial function, we also generated an HCMV strain with a full deletion of the UL13 locus (∆UL13) (Fig. 1B). We confirmed decreased pUL13 levels during infection with WT, UL13M, and ∆UL13 HCMV using targeted mass spectrometry (MS) to specifically monitor the levels of signature pUL13 peptides (Fig. 1D). Low-abundance pUL13 peptides were still detected during UL13M infection, whereas levels were completely ablated during ∆UL13 infection. Furthermore, infection with ∆UL13 resulted in a more significant, 10.9-fold decrease in supernatant viral titer relative to WT infection (Fig. 1E). Therefore, we utilized the ∆UL13 strain in subsequent experiments.
To pinpoint when the defect during ∆UL13 replication occurs, we analyzed temporal viral protein levels, replication of the viral genome, and generation of infectious viral progeny as parameters corresponding to the sequential stages of virus replication. We analyzed the abundance of the IE, DE, and L viral protein markers, IE1, pUL26, and pUL99, respectively, using Western blotting (Fig. 1F). While IE1 levels were similar at all time points during WT and ∆UL13 infections, pUL26 and pUL99 levels were decreased during ∆UL13 infection, beginning at 48 hpi. We measured cell-associated viral genome abundance during infection using qPCR and observed an over twofold decrease in viral genome abundance during ∆UL13 relative to WT infection by 72 hpi (Fig. 1G). To measure the production of new infectious viral progeny, we quantified cell-associated viral titers and observed a threefold decrease during ∆UL13 compared to WT infection, suggesting a defect in virus production (Fig. 1H). We additionally quantified the particle:plaque-forming unit (PFU) ratio of extracellular virus as a parameter of viral particle infectivity (i.e., the ability to form plaques) (Fig. 1I). ∆UL13 resulted in an ∼eightfold (120 hpi) and ∼fivefold (144 hpi) increase in the particle:PFU ratio, indicating that the virus produced during ∆UL13 infection is less infectious. As DE proteins regulate the replication of the viral genome, which is then required for the expression of L genes and viral assembly, it is likely that the observed defects in genome replication, virus production, and infectivity during ∆UL13 infection are tied to the decrease in DE protein levels. Together, these results demonstrate that pUL13 serves an important function during HCMV infection and that loss of pUL13 negatively impacts stages of the viral life cycle subsequent to the expression of IE genes.
The Temporal Profile of Viral Protein Levels Is Dysregulated during ∆UL13 Infection.
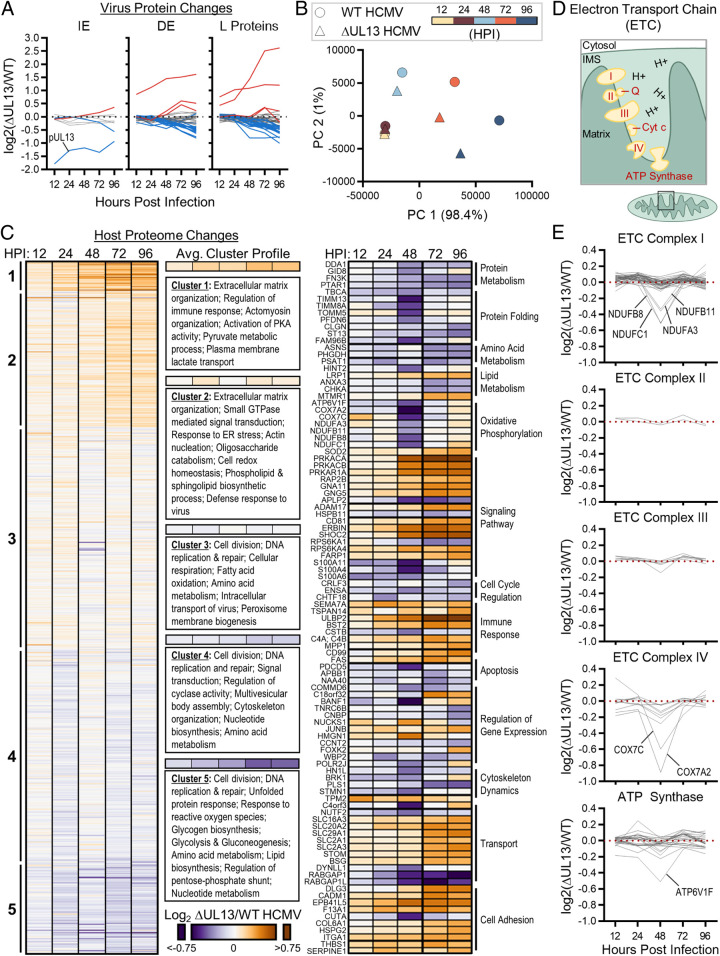
To further investigate the importance of pUL13 during HCMV replication, we explored the temporal changes in the virus and host proteomes during WT and ∆UL13 infections using tandem mass tag (TMT) labeling and MS quantification (SI Appendix, Fig. S1A). Infected fibroblasts were collected throughout the HCMV replication cycle (12, 24, 48, 72, and 96 hpi), thereby capturing time points at which the replication defect was observed during ∆UL13 infection. Prior to TMT labeling, a fraction of each sample from each replicate (30 samples total) was pooled to form an internal reference channel. This allowed for internal reference scaling and reduced variation between replicates (SI Appendix, Fig. S1 B–D, Dataset S1). A total of 9,250 proteins were identified, with 5,177 host proteins and 95 viral proteins passing our quality and quantification criteria.
As expected, pUL13 levels were robustly decreased at all time points during ∆UL13 relative to WT infection, further verifying the deletion of the UL13 locus (Fig. 2A and SI Appendix, Fig. S2A). Additionally, we observed a marked disruption in the temporal profile of viral protein abundances during ∆UL13 infection. Congruent with our Western blot results (Fig. 1F), DE and L viral proteins were more impacted by ∆UL13 than IE proteins (Fig. 2A and SI Appendix, Fig. S2A). By 48 hpi, the vast majority (81/95) of detected viral proteins were decreased in abundance during ∆UL13 relative to WT infection. The decrease in viral protein abundances became more pronounced at later time points of infection (i.e., after 48 hpi). In agreement, a principal component analysis (PCA) of viral protein abundances showed a high correlation between WT and ∆UL13 at 12 and 24 hpi (Fig. 2B). The virus strains diverged beginning at 48 hpi, becoming less clustered as infection progressed.
Fig. 2.
∆UL13 infection results in altered temporal viral protein levels and decreased abundance of cellular metabolic proteins. (A) The relative abundances of viral proteins detected by TMT-MS (n = 95 proteins), shown as fold change during ∆UL13 relative to WT infection. Proteins passing 1.25-fold change at any time point are highlighted in red (increased) and blue (decreased). (B) Principal component analysis of virus protein abundances at each analyzed time point during WT or ∆UL13 infection. (C) Relative abundances of host proteins during ∆UL13 relative to WT infection. The average k-means cluster profile and enriched gene ontology terms are shown. Proteins passing the 1.25-fold change and P value < 0.05 cutoffs at any time point are shown in the second heatmap (Center), organized by biological process. (D) Schematic of the ETC and ATP synthase within the mitochondrial inner membrane. IMS, intermembrane space. (E) The relative temporal abundances of ETC and ATP synthase protein subunits, shown organized by complex. Proteins passing the 1.25-fold change and P value < 0.05 cutoffs at any time point are labeled.
To investigate the possibility that the decrease in viral protein levels observed during ∆UL13 infection stems from a temporal delay, we performed a distance matrix analysis (SI Appendix, Fig. S2B). Prior to 96 hpi, ∆UL13 was most similar to the corresponding WT time point. At 96 hpi, ∆UL13 became more similar to the 72 hpi WT time point. This demonstrates that the decreased protein levels seen during ∆UL13 infection, up to 96 hpi, are not due to a kinetic delay but rather a global decrease in protein levels.
Notably, the abundances of three viral proteins (pUL14, pUL15A, and pUL17) significantly increased during ∆UL13 infection relative to WT infection (SI Appendix, Fig. S2 A and C). These currently uncharacterized proteins are encoded directly downstream of the UL13 locus (SI Appendix, Fig. S2D). We validated increased pUL14 and pUL17 levels by targeted MS during both ∆UL13 and UL13M infections (SI Appendix, Fig. S2C). We also attempted to detect pUL15A but were not successful, likely due to the small mass of this protein (11 kDa) and the few tryptic peptides produced from its digestion. As there are no known regulatory regions in the UL13 locus, one possibility is that there is a compensatory function for these currently uncharacterized viral proteins during infection. Our previous finding that pUL15A localizes to the mitochondria during infection supports this possibility (16). Altogether, our TMT-MS analysis demonstrates that ∆UL13 significantly disrupts the temporal cascade of viral gene expression, supporting a critical role for pUL13 during HCMV replication.
Metabolic and OXPHOS Proteins Decrease in Abundance during ∆UL13 Infection.
Given our finding that pUL13 is important for productive HCMV replication, we next sought to understand its function. Using our TMT-MS proteome dataset, we monitored the levels of cellular proteins, assessing temporal protein profiles and enriched gene ontology (GO) terms in each cluster (Fig. 2C). Proteins functioning in the regulation of immune response and extracellular matrix organization were increased during ∆UL13 relative to WT infection, while proteins involved in cell division and DNA replication and repair were decreased (Fig. 2C). Notably, metabolic pathways were among the enriched GO terms for both temporally increased and decreased clusters, suggesting that pUL13 might function to regulate cellular metabolism during infection. Evident were the increase in proteins involved in the activation of protein kinase A (PKA) activity and oligosaccharide catabolism (clusters 1 and 2, respectively) and the decrease in glycogen and lipid biosynthetic proteins during ∆UL13 infection.
Functional network analysis of proteins that passed our fold-change and P-value significance cutoffs (1.25-fold change and P value < 0.05) pointed to a temporally regulated decrease in components of the TIM/TOM complex (TOMM5, TIMM8A, and TIMM13), which transports OXPHOS proteins across the mitochondrial membranes (Fig. 2 C, Right and SI Appendix, Fig. S2E). At the same time point (48 hpi), subunits of the ETC decreased in abundance (1.3- to 1.9-fold). We plotted the log2 abundances of all ETC protein components detected in our TMT-MS dataset (n = 81 proteins belonging to complexes I through IV of the ETC and ATP synthase) (Fig. 2 D and E). The decrease in proteins was complex specific: subunits of complex I, complex IV, and ATP synthase were decreased, while no significant change was observed for complex II or III subunits (Fig. 2E). Altogether, the differential alterations of the cellular proteome during infections with ∆UL13 and WT HCMV suggest a role for pUL13 in regulating cellular metabolism and OXPHOS.
pUL13 Is Necessary for Increased Oxidative Phosphorylation during HCMV Infection.
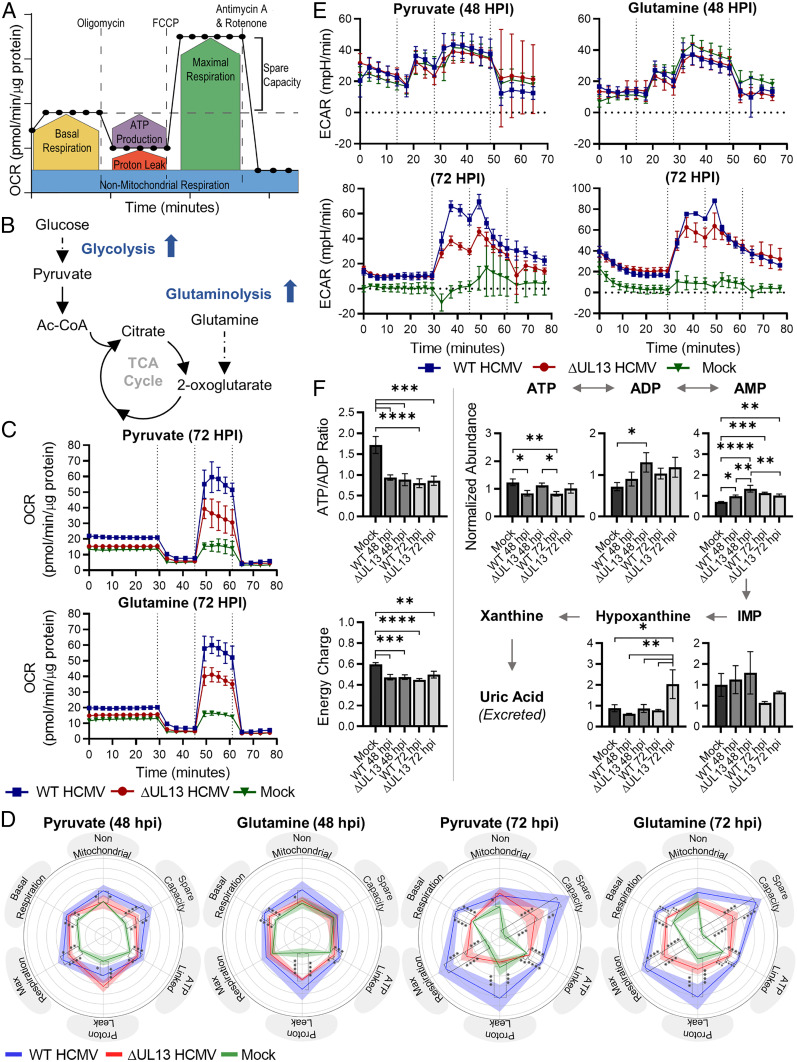
OXPHOS is known to increase during HCMV infection and to be required for virus replication (14). Given our finding that ETC proteins decrease in abundance during ∆UL13 infection, we hypothesized that pUL13 contributes to modulating OXPHOS during infection. Given that OXPHOS accounts for the majority of cellular oxygen consumption, we used a live-cell Seahorse assay to measure oxygen consumption rates (OCRs) in WT and ∆UL13-infected cells (Fig. 3A). Mock, WT, or ∆UL13-infected fibroblasts were provided with a respiratory substrate, and the OCR of the cells was measured using a series of respiratory modulators. We assessed several functional ETC parameters, including basal respiration, ATP-linked respiration, maximal respiration, proton leak, spare respiratory capacity, and nonmitochondrial respiration. Glycolysis and glutaminolysis, two major pathways for energy production, are increased during HCMV infection to promote viral replication (Fig. 3B) (5–7). Therefore, either glutamine or pyruvate were provided to cells as respiratory substrates during the Seahorse assay to determine if a particular metabolic pathway was preferentially utilized for respiration during infection.
Fig. 3.
pUL13 increases oxidative phosphorylation during HCMV infection. (A) Diagram illustrating the method of converting OCR into the six parameters of ETC function. Reproduced from ref. 51 with permission, © Agilent Technologies, Inc. (B) HCMV rewires the metabolism of infected cells, increasing metabolic flux through glycolysis and glutaminolysis (6, 7). Dashed lines indicate multistep pathways; solid lines indicate single-step pathways. (C) OCR of mock, WT, and ∆UL13-infected cells 72 hpi using either pyruvate or glutamine as the respiratory substrate. The carbon source was injected at 0 min; vertical lines indicate the injection time of oligomycin, FCCP, and Antimycin A + Rotenone. (D) Radar charts showing parameters of ETC function during mock, WT, and ∆UL13 infection at 48 and 72 hpi using either pyruvate or glutamine as the respiratory substrate. Solid lines indicate the mean, and shaded regions indicate the SD. (E) ECAR at 48 or 72 hpi during mock, WT, or ∆UL13 infection following the provision of pyruvate or glutamine as the respiratory substrate. The carbon source and ETC modulators were injected as in C. (F) LC-MS quantification of intracellular metabolites related to ATP metabolism. Metabolites were extracted from mock, WT, or ∆UL13 infected cells at 48 and 72 hpi. The energy charge was calculated from the abundance of ATP, ADP, and AMP as described in ref. 21. n = 3 biological replicates. For C–F: Significance was determined by one-way ANOVA using Tukey’s test for post hoc analysis. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001. Error bars indicate the SD. n ≥ 4 biological replicates, unless otherwise indicated.
Infection with WT or ∆UL13 increased OCRs over mock at 48 and 72 hpi for both glutamine and pyruvate (Fig. 3C and SI Appendix, Fig. S3A). This reveals that both pyruvate and glutamine are utilized as fuel sources for supporting increased cellular respiration during HCMV infection. Additionally, this finding uncovers that HCMV induces increased cellular respiration by 72 hpi relative to 48 hpi. However, ∆UL13 decreased OCR relative to WT infection at 48 and 72 hpi, supporting a function for pUL13 in increasing cellular respiration during HCMV infection. We converted the raw OCR traces into parameters of ETC function to investigate the impact of pUL13 on cellular respiration (Fig. 3D). We observed similar levels of nonmitochondrial respiration for the two virus strains, supporting the idea that pUL13 specifically alters mitochondrial OCR. In agreement with previous literature (14), HCMV infection resulted in increased basal, maximal, and ATP-linked respiration relative to uninfected cells. However, infection with ∆UL13 resulted in reduced basal, maximal, and ATP-linked respiration on both tested respiratory substrates. The dysregulation of cellular respiration during ∆UL13 infection was more pronounced at 72 compared to 48 hpi, suggesting a progressive defect.
The Seahorse assay media was unbuffered, allowing for quantification of extracellular acidification rate (ECAR), another measurement of metabolic activity. Two major contributors to ECAR are lactate produced from pyruvate and CO2 produced from TCA cycle reactions (20). Glutaminolysis contributes primarily to TCA cycle–derived acidification, as glutamine bypasses glycolytic fermentation and enters directly into the TCA cycle. Pyruvate contributes to both lactate and TCA cycle–derived acidification. We found no change in ECAR between infected and uninfected cells at 48 hpi (Fig. 3E). However, by 72 hpi, there is a substantial increase in ECAR of infected cells relative to uninfected cells. This indicates that flux through pyruvate fermentation and TCA-cycle pathways are up-regulated during infection, in agreement with previous findings (6, 7). ∆UL13 did not substantially alter ECAR relative to WT levels when glutamine was provided as the fuel source. However, ∆UL13 resulted in reduced ECAR following oligomycin treatment when pyruvate was provided. Oligomycin inhibits ATP synthase, thereby shifting cellular metabolism toward glycolytic ATP production and pyruvate fermentation. Therefore, this finding suggests that ∆UL13 results in decreased capacity for metabolic fuel switching between mitochondrial respiration and glycolysis.
To validate our findings regarding decreased cellular respiration and ATP production during ∆UL13 infection, we performed a liquid chromatography mass spectrometry (LC-MS)–based metabolite profiling experiment to monitor the levels of metabolites related to ATP metabolism in uninfected, WT, and ∆UL13-infected cells at 48 and 72 hpi (Fig. 3F). We used the quantified abundances of ATP, ADP, and AMP to calculate the ATP/ADP ratio and energy charge of uninfected and infected cells as indices of bioenergetic status (21). The ATP/ADP ratio was significantly decreased in infected cells relative to uninfected cells (Fig. 3F). Given that HCMV infection results in increased cellular respiration, this finding suggests that ATP consumption is also increased during infection. Similarly, the energy charge of mock-infected cells was 0.6, within the typical range observed for confluent fibroblasts (22). HCMV infection resulted in a significant decrease in energy charge to below 0.5, indicating a relative increase in AMP abundance during infection. This previously unrecognized decrease in energy charge during infection could reflect that, despite increased respiratory activity, cells still do not keep up with energy demand during HCMV infection. Interestingly, ∆UL13 did not impact the ATP/ADP ratio or energy charge of cells relative to WT infection, pointing to similar energetic status. We observed ATP levels to be slightly elevated during ∆UL13 infection relative to WT infection, suggesting decreased ATP consumption during ∆UL13 relative to WT infection. Additionally, we found an over twofold increase in hypoxanthine levels during ∆UL13 relative to WT infection at 72 hpi. When ATP synthesis is decreased, AMP is degraded to hypoxanthine to maintain the cellular energy charge (23). Altogether, these findings support that ATP metabolism is dysregulated, both in production and usage, during ∆UL13 relative to WT infection.
It has previously been shown that another HCMV protein, pUL37 × 1, contributes to increased cellular respiration during infection by increasing mitochondrial biogenesis (24). Therefore, we investigated the possibility that pUL13 functions similarly during infection. We quantified mitochondrial DNA (mtDNA) abundance during WT and ∆UL13 infection (SI Appendix, Fig. S3B). mtDNA levels increased during infection, in line with previous studies (24), and we found no difference in mtDNA levels between the two viral infections. We additionally analyzed mitochondrial morphology using confocal microscopy during WT and ∆UL13 infection (SI Appendix, Fig. S3C). We observed similar morphologies during infection with either virus. Together, these results demonstrate that pUL13 increases mitochondrial respiration, and unlike pUL37 × 1, this occurs without alteration of mitochondrial biogenesis.
pUL13 Interacts with Cristae-Shaping Proteins at 48 hpi.
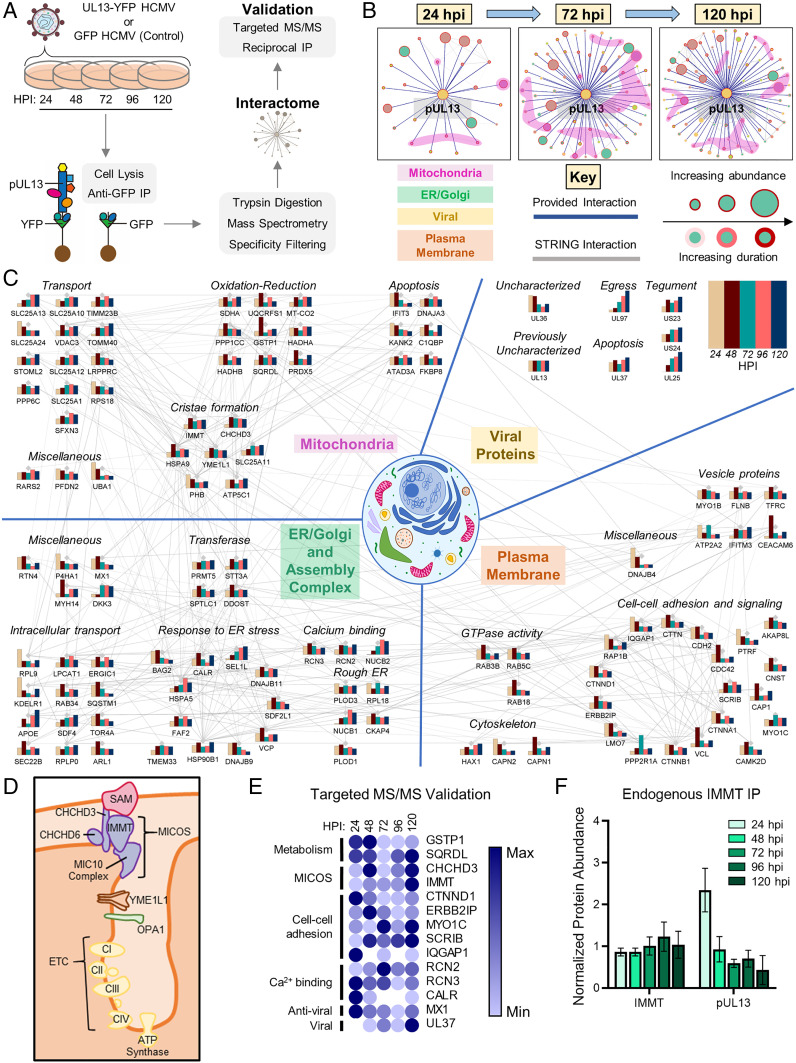
Having shown that pUL13 localizes to the mitochondria, impacts the abundance of ETC proteins, and increases OXPHOS without increasing mitochondrial biogenesis, we next investigated how pUL13 can accomplish this function. Given the temporally distinct localization of pUL13 during HCMV infection, we assessed its localization-dependent functions by analyzing its temporal interactions using IP-MS. The use of the UL13-YFP virus strain provided an affinity tag for isolating pUL13. By comparing protein markers of virus replication during UL13-YFP and WT infections, we observed similar levels of IE (IE1), DE (pUL26), and L (pUL99) virus proteins (SI Appendix, Fig. S4A). These observations support the idea that the tag does not affect the temporality of virus replication and therefore is suitable for interaction studies. We infected cells with UL13-YFP HCMV and immunopurified pUL13 at 24, 48, 72, 96, and 120 hpi (Fig. 4A). Using the Significance Analysis of INTnteractome (SAINT) algorithm (25, 26) for filtering specific protein interactions, 117 proteins were predicted to be pUL13 interactions. We visualized their temporal dynamics using our Inter-ViSTA computational platform (27) (Fig. 4B and Movie S1). As expected, pUL13 associated primarily with proteins localizing at the plasma membrane and mitochondria, with additional interactions observed with proteins localized at the endoplasmic reticulum (ER)/Golgi/assembly complex. K-means clustering highlighted four temporal trends of pUL13 interactions (SI Appendix, Fig. S4 B–C). In agreement with our microscopy analyses, many mitochondrial-localized proteins were found to interact with pUL13 beginning 48 hpi (cluster 1, SI Appendix, Fig. S4 B and C).
Fig. 4.
pUL13 interacts with components of the ETC and associated cristae-shaping proteins during infection. (A) IP-MS workflow for studying pUL13 interactions during infection. (B) InterViSTA (27) spatiotemporal analysis of pUL13 interactions. (C) Functional network of pUL13 interactions during infection, depicting only proteins that passed specificity criteria. Bars indicate the scaled protein abundance, normalized to bait (pUL13), at each time point. (D) Schematic of cristae-remodeling proteins and complexes identified in the pUL13 interactome. (E) Heatmap of pUL13 interactions, validated and quantified using PRM following IP-MS of pUL13 during UL13-YFP infection, scaled from minimum to maximum association with pUL13. White spaces indicate lack of detection. n = 2 to 3 peptides per protein. (F) PRM analysis of pUL13 abundance in endogenous anti-IMMT IP samples during WT HCMV infection. Protein abundances were quantified using two unique peptides per protein and normalized to IMMT abundance. Error bars denote the SD. n = 2 biological replicates.
Functional analysis of its entire temporal interactome implicated pUL13 in a range of cellular processes, including cell adhesion, cell polarity, protein folding, and calcium regulation (Fig. 4C). In agreement with our TMT-MS proteome results, many pUL13 interactions pointed to a likely function in the regulation of cellular metabolism during infection. We found pUL13 to interact with calcium/calmodulin-dependent protein kinase type II (CAMK2) and its downstream effector MYO1C, an unconventional myosin (Fig. 4 C and E). These proteins stimulate glucose uptake through the GLUT4 transporter, thereby implicating pUL13 in regulating glucose transport, which is known to increase during HCMV infection (28, 29). Also in line with our TMT-MS results, which implicated pUL13 in regulating PKA signaling, we found pUL13 to interact with the PKA anchoring protein AKAP8L, which is involved in PKA nuclear shuttling (30).
At 48 hpi, when pUL13 is predominantly localized in the mitochondria, we observed increased interaction with components of complexes II, III, and IV of the ETC (Fig. 4C). pUL13 also interacted with the prohibitin complex, an inner mitochondrial membrane (IMM) complex that associates with and is necessary for optimal ETC function, as well as with LRPPRC, which is necessary for translation of several ETC components (31, 32). Further suggesting a role in regulating OXPHOS during infection, pUL13 formed interactions with IMMT and CHCHD3, core components of the MICOS (33). The MICOS complex bridges the mitochondrial intermembrane space and regulates the architecture of cristae, which house ETC complexes (34, 35) (Fig. 4D). Dysregulation of cristae structure via perturbation of the MICOS complex is linked to impaired cellular respiration (36, 37).
To validate these temporal associations, we designed a targeted MS method for more accurate quantification of pUL13 interactions (Fig. 4E). A UL13-YFP IP was performed, and targeted MS was used to quantify the abundance of proteins of interest from our initial untargeted IP-MS assay, which included proteins involved in mitochondrial structure, metabolism, and calcium signaling (Fig. 4C). Targeted MS by parallel reaction monitoring (PRM) offered a more accurate quantification of the relative abundance of pUL13-interacting proteins at time points spanning the HCMV replication cycle. To further validate the pUL13–MICOS interaction, we performed reciprocal isolations of endogenous IMMT following infection with UL13-YFP (SI Appendix, Fig. S4D). Additionally, to ensure that the interaction does not occur through the YFP tag, endogenous IMMT IPs were performed during WT HCMV infection (Fig. 4F). Quantification of either pUL13 or pUL13-YFP abundances using targeted MS confirmed association with IMMT and further refined the temporal profile of the interaction, revealing that pUL13 most abundantly interacts with IMMT at 24 and 48 hpi.
To further validate our IP-MS findings, we investigated whether pUL13 is a soluble or integral membrane protein by performing sodium carbonate extraction analysis of mitochondria purified from UL13-YFP–infected cells at 48 hpi (SI Appendix, Fig. S4E). pUL13-YFP was detected almost exclusively in the membrane pellet fraction, indicating that it is an integral membrane protein. This submitochondrial localization supports the interaction of pUL13 with the membrane-spanning MICOS complex known to be embedded in the inner mitochondrial membrane (33). Altogether, the pUL13 interactome highlighted the likely multifunctional nature of this viral protein, in agreement with its diverse temporal localizations. Furthermore, it uncovered an interaction between pUL13 and the MICOS complex, which may underlie the ability of pUL13 to increase mitochondrial bioenergetics during infection.
pUL13 Is Sufficient to Alter Cristae Architecture and Increase Mitochondrial Bioenergetics.
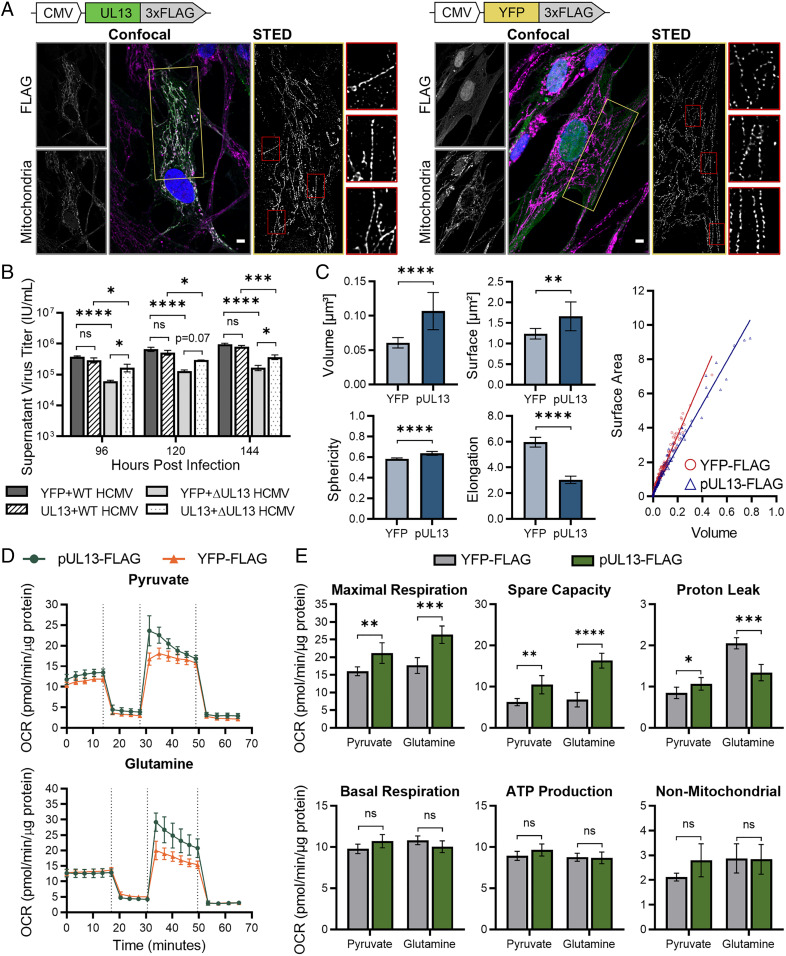
Our finding that pUL13 interacts with the MICOS complex suggested that pUL13 is positioned to modulate cristae architecture, which could facilitate pUL13-dependent increases in OXPHOS. However, we also observed and confirmed an interaction between pUL13 and pUL37 × 1, a viral protein known to modulate mitochondrial functions (4, 24, 38). Therefore, to investigate whether pUL13 alone is able to regulate mitochondrial structure and bioenergetics, we generated a fibroblast line stably expressing pUL13 tagged with 3xFLAG and a control YFP-3xFLAG cell line and verified construct expression using Western blotting (SI Appendix, Fig. S5A). Confocal microscopy showed that pUL13 localized to the mitochondria in the stable cell lines, illustrating that other viral factors are not required for its mitochondrial localization (Fig. 5A).
Fig. 5.
pUL13 alters cristae architecture and is sufficient for increased OXPHOS. (A) Confocal and STED analysis of pUL13 localization and cristae architecture in pUL13-3xFLAG and YFP-3xFLAG stably expressing fibroblasts. Cristae were visualized using an antibody against COXIV, a component of ETC complex IV. Yellow boxes indicate the regions sampled for STED analysis. Mitochondrial sections (boxed in red) have been enlarged to highlight cristae ultrastructure. (Scale bars, 5 μm.) (B) Extracellular HCMV titers during WT or ∆UL13 infection of pUL13-3xFLAG and YFP-3xFLAG stably expressing fibroblasts. n = 3 biological replicates. Significance was determined by one-way ANOVA. (C) Quantification of cristae morphology in STED images acquired from pUL13-3xFLAG and YFP-3xFLAG stably expressing fibroblasts. Error bars indicate a 95% CI. (D) OCR of pUL13-3xFLAG and YFP-3xFLAG stably expressing cells using either pyruvate or glutamine as the respiratory substrate. The carbon source and ETC modulators were injected as in Fig 3C. (E) Parameters of ETC function in pUL13-3xFLAG and YFP-3xFLAG stably expressing cells. n = 5 biological replicates for D and E. Significance was determined by Student’s t test for C–E. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001; n.s., P > 0.05. Error bars indicate the SD unless otherwise stated.
To further confirm that the pUL13 construct is functional, we investigated whether pUL13 expression can rescue the defect in viral titer observed during ∆UL13 infection (Fig. 5B). In line with our previous results, infection of the YFP control cell line with ∆UL13 resulted in decreased viral titer relative to WT. However, infection of the pUL13 cell line with ∆UL13 partially rescued the defect in viral titer at all analyzed infection time points. This result supports the functionality of the pUL13-3xFLAG construct and the fact that the defect in replication of ∆UL13 stems from the absence of pUL13. The partial rescue of viral titer likely stems from the tightly regulated temporality of pUL13 localization during infection, which is lost in the pUL13-expressing stable lines. We next considered whether a rescue in the levels of pUL14 or pUL17, the viral proteins found to be increased during ∆UL13 infection, contributes to this increase in viral titer. Using targeted MS, we quantified pUL14 and pUL17 levels during ∆UL13 infection of the pUL13 or YFP cell lines. We found that, although pUL13 expression partially rescues virus titer during ∆UL13 infection (Fig. 5B), we continue to see similar levels of pUL14 and pUL17 during ∆UL13 infection in both YFP- and pUL13-expressing cells (SI Appendix, Fig. S5B). This suggests that the partial rescue in viral titer is not due to a return of pUL14 and pUL17 abundances to WT infection levels but is instead due to the partial recovery of pUL13 expression.
We next investigated mitochondrial ultrastructure in the stable cell lines using stimulated emission depletion (STED) microscopy (Fig. 5A). The cristae were visualized using an anti-COXIV antibody. This probe was chosen because COXIV, a component of ETC complex IV, preferentially localizes to cristae, and the presence or absence of pUL13 did not change COXIV abundance in our TMT-MS dataset. We observed that pUL13 expression resulted in more heterogeneity in cristae shape and spacing along the mitochondria. Quantification of cristae morphology supported this observation, revealing that cristae are larger and more spherical in pUL13-expressing cells relative to control (Fig. 5C). These data align with our IP-MS findings, supporting that pUL13 modulates mitochondrial ultrastructure.
The modulation of cristae architecture and partial rescue of viral titer resulting from pUL13 expression led us to hypothesize that pUL13 is sufficient for regulating OXPHOS. Using the live-cell Seahorse assay, we measured ECAR and OCR in the pUL13 stable cell line relative to control. By monitoring ECAR during the Seahorse assay, we observed that pUL13 expression results in increased ECAR when pyruvate is provided as the fuel source. This suggests that pUL13 is capable of increasing glycolytic flux (SI Appendix, Fig. S5C). pUL13 expression also increased OCR on glutamine and pyruvate respiratory substrates (Fig. 5D). pUL13 expression did not significantly alter nonmitochondrial respiration, basal respiration, or ATP-linked respiration on any respiratory substrate (Fig. 5E). Excitingly, pUL13 expression resulted in significantly increased maximal respiration and the ability of the ETC to respond to increased energy demand (spare respiratory capacity). These increases were most significant when cells were fed glutamine, the predominant respiratory substrate during HCMV infection (7). pUL13 expression also resulted in a significant decrease in proton leak when glutamine was utilized as the respiratory substrate. These observations demonstrate that pUL13 does not increase basal levels of respiration in cells, but instead functions to prime the ETC for increased energy demand. Energy demand is increased during HCMV infection (14), providing a biological context for this pUL13 function. Together, these results establish that pUL13 modulates cristae architecture and is sufficient to support increased bioenergetic output during HCMV infection.
Discussion
The impact of HCMV infection on mitochondrial structure and function has historically presented a paradox. Although HCMV induces mitochondrial fragmentation during its replication, which is typically associated with mitochondrial dysfunction (12), it succeeds to maintain metabolic and bioenergetic output through induction of ETC activity (13, 14). A recent study demonstrated that ETC activity is essential for productive HCMV replication; ETC inhibition resulted in reduced viral titers and decreased DE and L viral protein abundances, pointing to a defect in replication progression past IE gene expression (14). However, the mechanism underlying HCMV-induced ETC activity remained unknown.
Here, we establish that the uncharacterized HCMV viral protein pUL13 targets the mitochondria during infection to increase mitochondrial bioenergetics. Using a combination of proteomic and molecular virology techniques, we show that absence of pUL13 restricts virus replication beginning at 48 hpi. At this infection time point, pUL13 is localized to the mitochondria, where we found it to interact with cristae-shaping proteins including the MICOS complex. We further demonstrate that pUL13 modulates mitochondrial ultrastructure and is sufficient to increase bioenergetic potential during HCMV infection.
Prior to this analysis, only one other HCMV protein, pUL37 × 1, had been implicated in regulating cellular respiration during HCMV infection (24). Infection with a ∆UL37 HCMV strain resulted in a partial decrease in cellular respiration relative to WT infection (24). However, whether pUL37 × 1 is sufficient for increasing cellular respiration was not investigated. This previous finding was connected to a deficiency in cellular mtDNA content during ∆UL37 HCMV infection, likely due to the function of pUL37 × 1 in promoting mitochondrial biogenesis (4). pUL37 × 1 is known to localize to the outer mitochondrial membrane during infection, supporting the idea that its impact on cellular respiration is indirect. Conversely, we demonstrate that cellular mtDNA content did not change during ∆UL13 infection relative to WT infection (SI Appendix, Fig. S3B). It is possible that this maintenance of increased mtDNA content during ∆UL13 infection underlies the increased cellular respiration when pUL13 is absent relative to uninfected cells (Fig. 3 C and D). Additionally, the ETC components that we found to decrease during ∆UL13 infection are nuclear encoded (Fig. 2C) (39). This further supports the idea that pUL13 specifically targets the mitochondria to directly increase cellular respiration through a mechanism independent of mitochondrial biogenesis. It is possible that there are as-yet unrecognized additional virus gene products that contribute to increased cellular respiration during HCMV infection, especially given how important respiratory output is for HCMV, a virus that dynamically reorganizes cellular machinery over a lengthy replication cycle.
Our proteomic analysis uncovered that deletion of UL13 within the virus genome leads to increased abundances of a specific subset of viral proteins that are encoded near the UL13 locus (SI Appendix, Fig. S2D). In contrast to UL13, which has been reported to be an IE gene (17), these genes are expressed with DE (UL17) or L (UL14 and UL15A) kinetics. Although it is possible that the increase in these viral proteins is influencing viral titers and cellular respiration, our results established that stable expression of pUL13 alone is sufficient to induce increased cellular respiration. We previously reported that pUL15A has mitochondrial localization during HCMV infection (16), albeit at 96 hpi, which is later than the respiratory increase that we measured using Seahorse assays. Given that we observed pUL13 interaction with the MICOS complex at 24 to 48 hpi, the localization of pUL15A in the mitochondria is unlikely to influence the pUL13 interaction with the MICOS complex and their impact on cellular respiration at these earlier stages of infection. However, there is still the possibility that these viral proteins contribute to the regulation of cellular metabolism and virus replication. As these affected proteins are viral, their effects would likely be proviral and therefore suppress the defect that ∆UL13 causes. If these viral proteins are impacting metabolism in a similar manner to pUL13, this could be an uncharacterized compensatory mechanism during HCMV infection, representing layers of protection to ensure cellular respiration is increased. This would be intriguing, as only a limited number of HCMV gene products are known to directly interact with the ETC (e.g., ncRNAβ2.7) (15).
pUL13 Modulates Cristae Structure to Increase OXPHOS.
The cristae create a distinct bioenergetic microdomain within the mitochondria, where ETC complexes are localized and the proton gradient is concentrated (35, 40). Several proteins and complexes have been shown to modulate cristae architecture. Of these, the MICOS complex is the key factor involved in regulating cristae dynamics and junction formation (18, 34, 41–44). We observed that, at 48 hpi, pUL13 interacts with the core MICOS subunits, IMMT and CHCHD3 (33), which we validated using targeted MS and reciprocal IP. Cristae architecture, and therefore cristae-shaping complexes, are intricately linked to mitochondrial bioenergetics and ETC function. Knockdowns of CHCHD3 and IMMT (MIC60) have been shown to substantially reduce mitochondrial respiration (18, 41, 42). Several mechanisms contribute to these MICOS functions. By controlling cristae junction formation, MICOS concentrates the proton gradient within cristae, restricting proton diffusion away from ATP synthase (44, 45). Additionally, deletion or knockdown of MICOS components has been shown to decrease the abundance and assembly of inner mitochondrial membrane complexes (41, 43, 46). We observed significant decreases in the abundance of IMM proteins during ∆UL13 infection, including TIMM8A-13 complex components and ETC proteins (Fig. 2C). Interestingly, the complex IV subunit COX7A2L was the most substantially decreased ETC subunit during ∆UL13 infection (Fig. 2E). COX7A2L is known to regulate the formation of ETC supercomplexes, supramolecular assemblies that function to increase ETC efficiency (47).
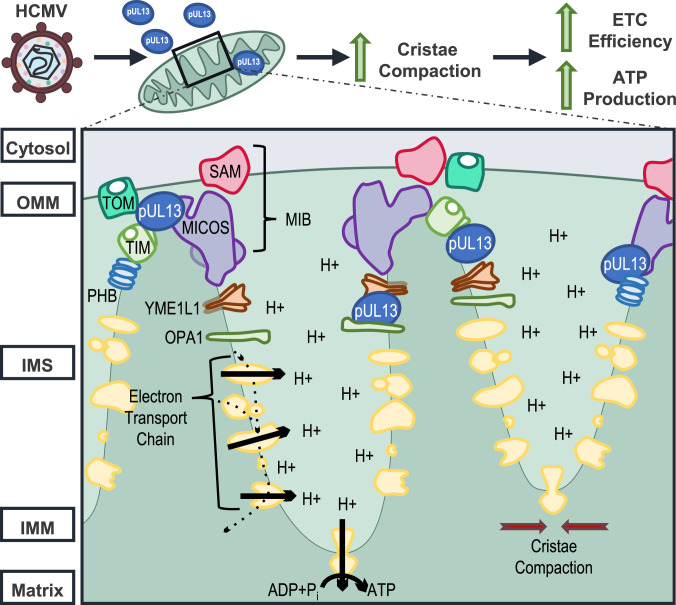
We propose that pUL13 stabilizes MICOS during infection, potentially by functioning as a chaperone (Fig. 6). Moreover, our superresolution microscopy suggests that this interaction functions to maintain cristae integrity and promote tight cristae-junction formation, thereby providing a means for increasing the proton gradient and ETC assembly to promote ETC efficiency and ATP production. This model is supported by several lines of evidence: 1) The ETC has already been demonstrated to be more efficient during HCMV infection (14, 24), suggesting that HCMV has evolved a mechanism to effectively couple electron transport and proton pumping to ATP production; 2) pUL13 does not increase mitochondrial biogenesis during infection, as evidenced by the similar mtDNA abundance during WT and ∆UL13 infection (SI Appendix, Fig. S3B), supporting the idea that pUL13-mediated induction of OXPHOS stems from modulation of ETC function rather than increased mitochondrial content; 3) pUL13 interacts with the core MICOS subunits, IMMT and CHCHD3, during infection; 4) The mitochondrial ultrastructure is visibly perturbed upon expression of pUL13 (Fig. 5A); and 5) Expression of pUL13 alone is sufficient for increasing the maximal respiration rate and spare respiratory capacity of the ETC (Fig. 5E). These findings establish that pUL13 serves to prime the mitochondria, promoting formation of a cristae architecture that is capable of supporting increased OXPHOS in response to the energy demand driven by HCMV replication.
Fig. 6.
Proposed model for the pUL13-induced increase in mitochondrial bioenergetics during HCMV infection. OMM, outer mitochondrial membrane; IMS, intermembrane space; IMM, inner mitochondrial membrane.
Our alignment of the pUL13 amino acid sequence across the laboratory-adapted strain AD169 and the clinical strains MERLIN and TB40/E shows a high level of homology (SI Appendix, Fig. S5D). The pUL13 sequence between TB40/E and AD169 shares 99.4% of its amino acid sequence homology, with only three amino acid substitutions and no insertions or deletions, and the MERLIN and AD169 share 97.9% of their homology. The overall similarity in sequences across these strains suggests that pUL13 functions may be conserved during infections with other HCMV strains.
Viral perturbation of ETC function has been typically studied in the context of viral modulation of reactive oxygen species generation, apoptosis, and innate immune sensing (48). Our findings contribute to an emerging narrative in which viruses, particularly those with long replication cycles, target the ETC to increase mitochondrial bioenergetics (49). By uncovering a function for pUL13 in increasing OXPHOS during HCMV infection, we establish a mechanism through which viruses can increase cellular respiration via modulation of MICOS function and IMM architecture.
Materials and Methods
Complete materials and methods are provided in SI Appendix.
Cell Culture and Viral Infection.
Human fibroblasts (HFs) were cultured under standard conditions in a complete growth medium. HFs stably expressing pUL13-3xFLAG or YFP-3xFLAG were generated via lentivirus transduction followed by puromycin selection.
Stocks of HCMV AD169, AD169-GFP, AD169 UL13-YFP, AD169 UL13M, and AD169 ∆UL13 were produced from bacterial artificial chromosomes in HFs and titered using tissue culture infectious dose (TCID50).
Infections were carried out at 37 °C for 1 h, followed by a phosphate-buffered saline (PBS) wash and addition of growth media. Infectious units were quantified by using cell-culture supernatant to infect a reporter plate of HFs. At 24 hpi, the reporter plate was stained with anti-IE1 antibody and DAPI. IE1-positive nuclei were quantified using Operetta imaging.
Immunofluorescence Microscopy.
Immunofluorescence microscopy was conducted using a Nikon Ti-E confocal microscope equipped with a spinning disk module. A Nikon A1R confocal microscope equipped with an STED detector was utilized for confocal and STED analysis of pUL13-3xFLAG and YFP-3xFLAG stably expressing HFs.
qPCR.
Viral, mitochondrial, and nuclear genomes were quantified by qPCR using the SYBR green PCR master mix and the ViiA7 Real-Time PCR System. Primers were specific to the virus IE1 gene, mtDNA 16s ribosomal RNA gene, or the nuclear β2-microglobulin gene. To quantify extracellular virus particles, the cell-culture supernatant was treated with DNase I to degrade nonpackaged viral DNA prior to qPCR sample preparation.
TMT-MS.
HFs infected with WT or ∆UL13 HCMV in biological triplicate were collected at 12, 24, 48, 72, and 96 hpi. Equal protein concentrations for each sample were reduced, alkylated, precipitated, and digested with trypsin. Prior to TMT labeling, equal volumes of all samples from each of the three replicates were pooled and then split into three aliquots to be used as internal reference channels. Samples were labeled using the TMT10plex and TMT11-131C Isobaric Label Reagents and analyzed via nano liquid chromatography–tandem mass spectrometry (LC-MS/MS).
IP-MS.
pUL13-YFP IPs, along with the matched GFP-virus control, were performed in biological duplicate at each time point (24, 48, 72, 96, and 120 hpi) for discovery and targeted experiments. For anti-GFP IPs, preconjugated GFP-Trap magnetic beads were used. For reciprocal endogenous IMMT IPs, anti-IMMT antibodies were conjugated to protein A/G magnetic beads. Bound proteins were eluted from the beads and reduced, alkylated, and digested with trypsin. Samples were analyzed via LC-MS/MS, and the total spectral count data were analyzed by SAINT (26) using the Resource for Evaluation of Protein Interaction Networks (REPRINT) (25) interface to determine interaction specificity.
Targeted MS.
Targeted MS samples were analyzed via LC-MS/MS with targeted MS2 scans controlled by a peptide inclusion list containing two to four unique peptides for each targeted protein. Data analysis was performed using the Skyline software (50). A summed area under the curve of three transitions per peptide was used for the quantitation.
Quantification of Cellular Respiration.
Cellular respiration was quantified using the Seahorse XF Cell Mito Stress Test Kit according to manufacturer specifications. The first injection contained the carbon source (either 5 mM pyruvate or 5 mM glutamine), and subsequent injections consisted of 2 μM oligomycin, 1 μM FCCP, and 0.5 μM rotenone/antimycin A. OCR results were normalized to the total protein abundance.
Metabolite Profiling.
Cellular metabolites were extracted using ice-cold 80% MeOH containing 1 mM N-ethylmaleimide and analyzed by LC-MS. Data analysis was performed using EL-MAVEN software, and metabolite abundances were normalized to the total protein abundance. The cellular energy charge was calculated as (ATP+1/2ADP)/(ATP+ADP+AMP).
Supplementary Material
Acknowledgments
We thank Gary Laevsky (Confocal Imaging Facility, Princeton University) for instrument use and technical support, Todd Greco and Josiah Hutton III for MS support, Elizabeth Rowland for the ∆UL13 HCMV strain, and Thomas Shenk for HCMV strains and antibodies. We are grateful for funding from the NIH (grant GM114141) and the Edward Mallinckrodt Foundation to I.M.C., an NIH National Research Service Award (F31AI154796) to C.N.B., a Harold W. Dodds Fellowship to P.M.J.B., and a National Institute of General Medical Sciences training grant (T32GM007388) to W.A.H. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101675118/-/DCSupplemental.
Data Availability
MS proteomic raw files have been deposited in ProteomeXchange via the PRIDE repository (https://www.ebi.ac.uk/pride/) under database identifier PXD020009. All other study data are included in the article and/or supporting information.
References
- 1.Sanchez E. L., Lagunoff M., Viral activation of cellular metabolism. Virology 479–480, 609–618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisenreich W., Rudel T., Heesemann J., Goebel W., How viral and intracellular bacterial pathogens reprogram the metabolism of host cells to allow their intracellular replication. Front. Cell. Infect. Microbiol. 9, 42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan M., Syed G. H., Kim S. J., Siddiqui A., Mitochondrial dynamics and viral infections: A close nexus. Biochim. Biophys. Acta 1853 (10 Pt B), 2822–2833 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCormick A. L., Smith V. L., Chow D., Mocarski E. S., Disruption of mitochondrial networks by the human cytomegalovirus UL37 gene product viral mitochondrion-localized inhibitor of apoptosis. J. Virol. 77, 631–641 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munger J., Bajad S. U., Coller H. A., Shenk T., Rabinowitz J. D., Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2, e132 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munger J., et al., Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 26, 1179–1186 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers J. W., Maguire T. G., Alwine J. C., Glutamine metabolism is essential for human cytomegalovirus infection. J. Virol. 84, 1867–1873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannon M. J., Schmid D. S., Hyde T. B., Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol. 20, 202–213 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Burny W., Liesnard C., Donner C., Marchant A., Epidemiology, pathogenesis and prevention of congenital cytomegalovirus infection. Expert Rev. Anti Infect. Ther. 2, 881–894 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Britt W., Manifestations of human cytomegalovirus infection: Proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol. 325, 417–470 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Van Damme E., Van Loock M., Functional annotation of human cytomegalovirus gene products: An update. Front. Microbiol. 5, 218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westermann B., Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta 1817, 1833–1838(2012). [DOI] [PubMed] [Google Scholar]
- 13.Karniely S., et al., Human cytomegalovirus infection upregulates the mitochondrial transcription and translation machineries. MBio 7, e00029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Combs J. A., et al., Human cytomegalovirus alters host cell mitochondrial function during acute infection. J. Virol. 94, e01183–e01119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reeves M. B., Davies A. A., McSharry B. P., Wilkinson G. W., Sinclair J. H., Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 316, 1345–1348 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Jean Beltran P. M. M., Mathias R. A. A., Cristea I. M. M., A portrait of the human organelle proteome in space and time during cytomegalovirus infection. Cell Syst. 3, 361–373.e6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang N., et al., Transcription characteristics of the human cytomegalovirus UL13 gene. Arch. Virol. 158, 473–477 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Stephan T., et al., MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation. EMBO J. 39, e104105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu D., Silva M. C., Shenk T., Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U.S.A. 100, 12396–12401 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mookerjee S. A., Goncalves R. L. S., Gerencser A. A., Nicholls D. G., Brand M. D., The contributions of respiration and glycolysis to extracellular acid production. Biochim. Biophys. Acta 1847, 171–181 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Atkinson D. E., Walton G. M., Adenosine triphosphate conservation in metabolic regulation. Rat liver citrate cleavage enzyme. J. Biol. Chem. 242, 3239–3241 (1967). [PubMed] [Google Scholar]
- 22.Bereiter-Hahn J., Münnich A., Woiteneck P., Dependence of energy metabolism on the density of cells in culture. Cell Struct. Funct. 23, 85–93 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Johnson T. A., Jinnah H. A., Kamatani N., Shortage of cellular ATP as a cause of diseases and strategies to enhance ATP. Front. Pharmacol. 10, 98 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaarbø M., et al., Human cytomegalovirus infection increases mitochondrial biogenesis. Mitochondrion 11, 935–945 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Mellacheruvu D., et al., The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi H., et al., SAINT: Probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Federspiel J. D., et al., Mitochondria and peroxisome remodeling across cytomegalovirus infection time viewed through the lens of Inter-ViSTA. Cell Rep. 32, 107943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yip M. F., et al., CaMKII-mediated phosphorylation of the myosin motor Myo1c is required for insulin-stimulated GLUT4 translocation in adipocytes. Cell Metab. 8, 384–398 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Yu Y., Maguire T. G., Alwine J. C., Human cytomegalovirus activates glucose transporter 4 expression to increase glucose uptake during infection. J. Virol. 85, 1573–1580 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han I., et al., Protein kinase A associates with HA95 and affects transcriptional coactivation by Epstein-Barr virus nuclear proteins. Mol. Cell. Biol. 22, 2136–2146 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsutsumi T., et al., Proteomics analysis of mitochondrial proteins reveals overexpression of a mitochondrial protein chaperon, prohibitin, in cells expressing hepatitis C virus core protein. Hepatology 50, 378–386 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Xu F., Morin C., Mitchell G., Ackerley C., Robinson B. H., The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: Mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem. J. 382, 331–336 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott C., Dorsch E., Fraunholz M., Straub S., Kozjak-Pavlovic V., Detailed analysis of the human mitochondrial contact site complex indicate a hierarchy of subunits. PLoS One 10, e0120213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harner M., et al., The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30, 4356–4370 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilkerson R. W., Selker J. M. L., Capaldi R. A., The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 546, 355–358 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Anand R., Strecker V., Urbach J., Wittig I., Reichert A. S., Mic13 is essential for formation of crista junctions in mammalian cells. PLoS One 11, e0160258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guarani V., et al., QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology. eLife 4, 1–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xi Y., Harwood S., Wise L. M., Purdy J. G., Human cytomegalovirus pUL37x1 is important for remodeling of host lipid metabolism. J. Virol. 93, e00843-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taanman J. W., The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta 1410, 103–123 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Rieger B., Junge W., Busch K. B., Lateral pH gradient between OXPHOS complex IV and F(0)F(1) ATP-synthase in folded mitochondrial membranes. Nat. Commun. 5, 3103 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Darshi M., et al., ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J. Biol. Chem. 286, 2918–2932 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hessenberger M., et al., Regulated membrane remodeling by Mic60 controls formation of mitochondrial crista junctions. Nat. Commun. 8, 15258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman J. R., Mourier A., Yamada J., McCaffery J. M., Nunnari J., MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. eLife 4, 1–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding C., et al., Mitofilin and CHCHD6 physically interact with Sam50 to sustain cristae structure. Sci. Rep. 5, 16064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf D. M., et al., Individual cristae within the same mitochondrion display different membrane potentials and are functionally independent. EMBO J. 38, e101056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang R. F., et al., Suppression of Mic60 compromises mitochondrial transcription and oxidative phosphorylation. Sci. Rep. 5, 7990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lobo-Jarne T., et al., Human COX7A2L regulates complex III biogenesis and promotes supercomplex organization remodeling without affecting mitochondrial bioenergetics. Cell Rep. 25, 1786–1799.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Escoll P., Platon L., Buchrieser C., Roles of mitochondrial respiratory complexes during infection. Immunometabolism 1, e190011 (2019). [Google Scholar]
- 49.Claus C., et al., Activity increase in respiratory chain complexes by rubella virus with marginal induction of oxidative stress. J. Virol. 87, 8481–8492 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacLean B., et al., Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Technologies Agilent, Agilent Seahorse XF Cell Mito Stress Test Kit. https://www.agilent.com/cs/library/usermanuals/public/XF_Cell_Mito_Stress_Test_Kit_User_Guide.pdf (2019). Accessed 19 May 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MS proteomic raw files have been deposited in ProteomeXchange via the PRIDE repository (https://www.ebi.ac.uk/pride/) under database identifier PXD020009. All other study data are included in the article and/or supporting information.